Key Points
Question
Is a novel 3-dimensionally printed nasopharyngeal swab (3DP swab) accurate in detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)?
Findings
In this diagnostic study for coronavirus disease 2019 (COVID-19) across 2 institutions of 79 patients with COVID-19 and 10 controls, the overall agreement and positive percentage agreement of the 3DP swab was 91.1% and 93.5%, respectively, compared with the traditional FLOQSwab (COPAN Diagnostics) and Dacron swab (Deltalab). The positive percentage agreement was 100% for COVID-19 cases tested within the first week of illness, and reverse-transcriptase polymerase chain reaction cycle threshold values for the ORF1ab and E-gene targets showed a strong correlation between the 3DP and traditional swab on independent testing at each institution despite differences in sample processing.
Meaning
The 3DP swab performed accurately and consistently across health care institutions and may help mitigate strained resources in the escalating COVID-19 pandemic.
Abstract
Importance
Three-dimensionally printed nasopharyngeal swabs (3DP swabs) have been used to mitigate swab shortages during the coronavirus disease 2019 (COVID-19) pandemic. Clinical validation for diagnostic accuracy and consistency, as well as patient acceptability, is crucial to evaluate the swab’s performance.
Objective
To determine the accuracy and acceptability of the 3DP swab for identifying severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Design, Setting, and Participants
A diagnostic study was conducted from May to July 2020 at 2 tertiary care centers in Singapore with different reference swabs (FLOQSwab [COPAN Diagnostics] or Dacron swab [Deltalab]) and swab processing techniques (wet or dry) to evaluate the performance of the 3DP swab compared with traditional, standard-of-care nasopharyngeal swabs used in health care institutions. The participants were patients with COVID-19 in the first 2 weeks of illness and controls with acute respiratory illness with negative test results for SARS-CoV-2. Paired nasopharyngeal swabs were obtained from the same nostril and tested for SARS-CoV-2 by reverse-transcriptase polymerase chain reaction. The sequence of swabs was randomized based on odd and even participant numbers.
Main Outcomes and Measures
Primary outcome measures were overall agreement (OA), positive percentage agreement (PPA), and negative percentage agreement of the 3DP swab compared with reference swabs. Secondary outcome measures were the correlation of cycle threshold (Ct) values of both swabs.
Results
The mean (SD) age of participants was 45.4 (13.1) years, and most participants were men (87 of 89 [97.8%]), in keeping with the epidemiology of the COVID-19 pandemic in Singapore. A total of 79 patients with COVID-19 and 10 controls were recruited. Among the patients with COVID-19, the overall agreement and PPA of the 3DP swab was 91.1% and 93.5%, respectively, compared with reference swabs. The PPA was 100% for patients with COVID-19 who were tested within the first week of illness. All controls tested negative. The reverse-transcriptase polymerase chain reaction Ct values for the ORF1ab and E-gene targets showed a strong correlation (intraclass correlations coefficient, 0.869-0.920) between the 3DP and reference swab on independent testing at each institution despite differences in sample processing. Discordant results for both gene targets were observed only at high Ct values.
Conclusions and Relevance
In this diagnostic study of 79 patients with COVID-19 and 10 controls, the 3DP swab performed accurately and consistently across health care institutions and could help mitigate strained resources in the escalating COVID-19 pandemic.
This diagnostic study examines the accuracy and acceptability of a 3-dimensionally printed swab for identifying SARS-CoV-2.
Introduction
Since December 2019, the coronavirus disease 2019 (COVID-19) pandemic has spread to 191 countries, with 76 million cases worldwide and more than 1.6 million fatalities.1 One of the World Health Organization’s strategic objectives is to control clusters and prevent community transmission by rapidly identifying all cases.2 The identification of cases is typically performed using material collected from a nasopharyngeal swab, which is tested for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using reverse-transcriptase polymerase chain reaction (RT-PCR). Even as the pandemic continues to escalate, many countries that initially achieved good control of the pandemic have experienced resurgences and will continue to have high testing requirements to reestablish control.3,4,5 The gradual opening of borders between countries and the testing of travelers will also continue to place pressure on testing capacities.
Testing for COVID-19 has thus been limited by worldwide shortages of critical components, including RNA extraction kits, PCR reagents, transport media, and nasopharyngeal swabs.6,7,8 While 3-dimensionally printed nasopharyngeal swabs (3DP swabs) have been reported to be used during COVID-19, to our knowledge, clinical validation has been limited to single-institution experiences in suspected COVID-19 cases9 and, in some instances, using human gene targets, such as RNase-P, as a surrogate for effectiveness.10 In this article, we report our experience in designing a novel 3DP nasopharyngeal swab (the Python swab) and its clinical validation across 2 health care institutions for the identification of SARS-CoV-2.
Methods
Swab Design, Ex Vivo Testing, and Clinical Study Design
Details of the swab design, 3D printing and sterilization, mechanical testing, fluid absorption, and release testing, as well as ex-vivo testing with murine coronavirus, can be found in the eMethods in the Supplement. We performed a diagnostic study to compare the accuracy of the 3DP swab with standard-of-care nasopharyngeal swabs for detecting SARS-CoV-2. We chose to perform the study in patients with COVID-19 who were within the first 2 weeks of illness, rather than patients with newly diagnosed or suspected COVID-19 because we wanted to test the swab’s accuracy across a spectrum of viral loads, including those at the limit of detection as the viral burden declines. Two tertiary care centers in Singapore with different reference nasopharyngeal swabs were selected. At the National University Hospital, the COPAN Flexible Minitip FLOQSwab (Becton, Dickinson, and Company) was the reference swab. At the Singapore General Hospital, the polyester Dacron swab (Deltalab) was the reference swab.
Primary outcome measures were the overall agreement (OA), positive percentage agreement (PPA), and negative percentage agreement (NPA) of the 3DP swab compared with reference swabs. Secondary outcome measures were the correlation of the cycle threshold (Ct) values of both swabs.
Sample size calculations were performed in R (R Foundation) using the packages Basic Functions for Power Analysis (pwr), version 1.3-0, and Sample Size Estimation Functions for Studies of Interobserver Agreement (kappaSize), version 1.2.11 To identify a modest correlation between the results of the 3DP and reference swab at r = 0.5, 28, COVID-19–positive participants would be required (power, 0.8; 2-sided significance, P = .05). Assuming that 75% of patients with COVID-19 remain positive for SARS-CoV-2 during the first 2 weeks of illness, a minimum of 38 COVID-19–positive cases would have to be recruited for each site. Similarly, to identify a substantial intertest agreement between the results of the 3DP and reference swab at κ = 0.7, 36 patients with COVID-19 would have to be recruited for each site (power, 0.8; significance, P = .05; null hypothesis, κ = 0.2), assuming a 75% positivity rate.
Participants
Between May 11 and June 28, 2020, we recruited 80 adults with a laboratory-confirmed diagnosis of COVID-19 who were admitted to the National University Hospital and the Singapore General Hospital. Forty patients with COVID-19 were recruited from each health care institution. Only adults age 21 to 90 years with a nasopharyngeal swab test result that was positive for SARS-CoV-2 on RT-PCR were eligible. Patients with known bleeding diathesis were excluded. In addition, 10 adults who were admitted to the National University Hospital for acute respiratory illness who were tested to be negative for SARS-CoV-2 were recruited as controls.
All participants were recruited with written informed consent, and the study was approved by the institutional review board of the National Healthcare Group, Singapore. All parts of the study were performed in compliance with the Human Biomedical Research Act, Singapore.
Demographic information, such as age, sex, and race, as well as day of illness, was recorded. Participants received 2 consecutive nasopharyngeal swabs in the same nostril using the respective institution’s reference swab and the 3DP swab. All patients with COVID-19 were swabbed within the first 14 days of onset of symptoms. Participants who were assigned an even case number received the reference swab first, followed by the 3DP swab, while the reverse was true for participants who were assigned with an odd case number. Swabs were performed in full personal protective equipment by trained infectious disease or otolaryngology physicians who routinely administer nasopharyngeal swab testing in both institutions during the COVID-19 pandemic. For each participant, both swabs were administered by the same physician. The number of turns for each swab was standardized at 5 to 10 360° rotations, with consistency in the number of rotations between both swabs emphasized during the study initiation briefing.
Adverse Event Reporting and Acceptability Questionnaire
Immediately after the swabs were administered, the pain score, as well as any adverse events, including epistaxis and nausea, were recorded for each swab. An acceptability questionnaire was then administered that recorded the participant’s perception of the comfort and discomfort of the swabs, as well as acceptability for future testing.
Swab Processing
At the National University Hospital, a wet swab processing protocol was used. Immediately after each swab was administered, it was placed into 3 millimeters of BD Universal Viral Transport media (Becton, Dickinson and Company) and transported to the hospital’s clinical laboratory. At the Singapore General Hospital, a dry swab processing protocol was used. After each swab was administered, it was placed in a standardized sheath and transported without media to the hospital’s clinical laboratory. One thousand microliters of lysis buffer was then added to the specimen sheath, which was followed by vortexing and washing of the swab.
Clinical Testing for SARS-CoV-2
All swabs were tested at their respective hospital laboratories using the cobas SARS-CoV-2 test, which is a dual-target qualitative real-time RT-PCR assay for use on the high-throughput automated cobas 6800 System (Roche Molecular Systems, Inc). Briefly, 600 μL of the test media was transferred to a secondary tube for automated sample preparation (nucleic acid extraction and PCR amplification). Positive and negative controls were included in each run. The assay targets the ORF1ab (nonstructural protein) sequence, which is specific to SARS-CoV-2, and the E-gene (envelope protein) sequence, which is highly conserved among sarbecoviruses. Following the manufacturer guidelines for interpretation and the protocol for clinical samples from both institutions, samples that were positive for either gene target were considered to have a positive result. Samples that were positive for the E-gene only were considered to be presumptive positive.
Statistical Analysis
Statistical analysis for the mechanical testing and fluid experiments were performed in GraphPad Prism, version 8.4.2 (GraphPad Software). Results for tensile strength, flexural strength, and fluid absorption/release volumes are reported as means with 95% CIs.
Statistical calculations for the clinical study were performed in R, version 4.0.0 (R Foundation for Statistical Computing). Positive percentage agreement and NPA were calculated for the 3DP swab based on 2 × 2 contingency tables using the categorical results from the reference swab as the reference standard. Overall accuracy was determined by the fraction of true positives and true negatives. Cohen κ was used to measure the agreement between the categorical outcomes from both swabs.
Intraclass correlation coefficient (ICC) was used to evaluate the correlation of RT-PCR Ct values between both swabs when paired positive results were available. The ICC estimates at a 95% level of confidence were calculated using the R package Various Coefficients of Interrater Reliability and Agreement (irr), version 0.84.1, based on a single-rating, absolute-agreement, 2-way random effects model.12 The discordant samples (positive on only 1 swab) were assigned a Ct value of 2 or greater than the highest values for the ORF1ab and E-gene observed at each institution in this study. The Pearson correlation coefficient was used to evaluate the correlation of Ct values with the time course of illness.
Differences in Ct values and pain score for both swabs were reported together with 95% CIs and effect size (Cohen d). Bland-Altman plots were prepared using the paired.plotBA function from the R package Paired Data Analysis (PairedData), version 1.1.1.
Results
We first considered the clinical requirements of the swab regarding its mechanical, material, and biological properties. In particular, while some friction is required to enhance the collection of material from the nasopharynx, sharp edges had to be avoided to avoid trauma and epistaxis. The absorbent portion of the swab was thus designed as a reinforced helix, comprising outer blades and inward reverse blades to channel collected material into an internal reservoir, which ensured sufficient cellular and extracellular matrix could be collected from the nasopharynx (Figure 1, A and B). We chose medical-grade resin polymer as the material and allowed flexibility up to 180° to permit maneuverability within the narrow nasopharyngeal space. Poststerilization, the mechanical properties of the 3DP swab were tested and benchmarked against the FLOQSwab. The 3DP swab had increased mean (95%) tensile strength (39.5 [31.9-47.0] N vs 18.6 [15.1-22.0] N) and mean (95%) flexural strength (1.15 [1.08-1.22] N vs 0.17 [0.17-0.17] N) compared with the FLOQSwab, and was able to tolerate torsional twisting of more than 800° on itself (Figure 1C). While its mean (95 CI) capacity to absorb fluids was inferior to the FLOQSwab (36.7 [30.4-43.0] μL vs 89.1 [86.9-91.4] μL), the 3DP swab was able to release a similar amount of fluid on testing with a roll plate approach (22.0 [18.8-25.2] μL vs 20.9 [17.4-24.5] μL; Figure 1D; eFigure 1 in the Supplement).13
Figure 1. Design and Ex Vivo Testing of the 3-Dimensionally Printed (3DP) Swab.
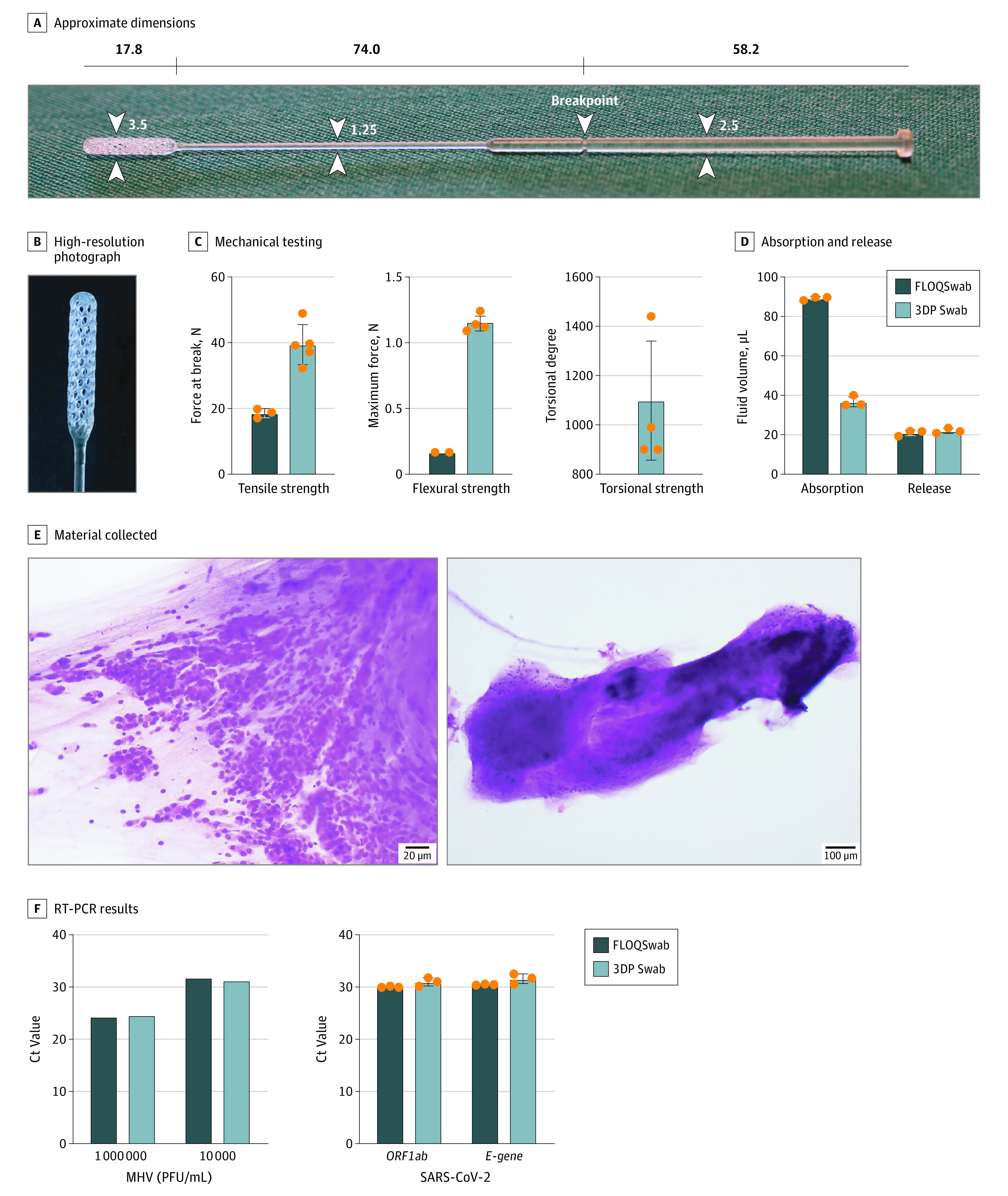
A, Overview and approximate dimensions (mm) The white numbers and arrowheads indicate the width of the swab (mm). B, High-resolution photograph of the swab tip. C, Mechanical testing of the swab demonstrating its tensile, flexural, and torsional strength. D, Viscous fluid absorption and release by roll plate approach. E, Hematoxylin-eosin stain of material collected from the swab smeared onto a glass slide, showing sheets of epithelial cells (left) and mucus (right). F, Reverse-transcriptase polymerase chain reaction (RT-PCR) results of swabs dipped in murine coronavirus (mouse hepatitis virus [MHV]) and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)–positive viral transport media. The orange dots represent individual data points for the respective tests. Error bars represent 1 standard deviation. Ct indicates cycle threshold; PFU, plaque-forming unit.
As with other cytopathic viruses, SARS-CoV-2 invades and multiplies intracellularly; thus, the ability of a swab to pick up cellular material is imperative. Hematoxylin-eosin smears obtained from the 3DP swab demonstrated that the swab was able to collect sheets of epithelial cells (Figure 1E). We confirmed the ability of the 3DP swab to retain and release viral samples by dipping swabs in spiked samples with standard inocula of murine coronavirus (mouse hepatitis virus) and SARS-CoV-2, resuspending in fresh viral transport medium, followed by viral RNA extraction and RT-PCR (Figure 1F). On ex vivo testing, the mean ORF1ab Ct value was 31.0 (95% CI, 29.1-33.0) for the 3DP swab and 30.1 (95% CI, 29.8-30.4) for the FLOQSwab. For the E-gene, the mean Ct value was 31.6 (95% CI, 29.3-34.0) and 30.5 (95% CI, 30.3-30.8), respectively.
We proceeded to determine the clinical performance of the 3DP swab compared with standard-of-care swabs (FLOQSwab or Dacron swab) used in 2 health care institutions. Eighty patients with COVID-19 were swabbed at a median of the eighth day of illness (range, 2- 14 days). Paired nasopharyngeal swabs were obtained via the same nostril by trained infectious disease and otolaryngology physicians. The sequence of swabs was randomized based on the sample number assigned to the participant, and the swab technique was standardized. One case was excluded because of an error on sample labeling.
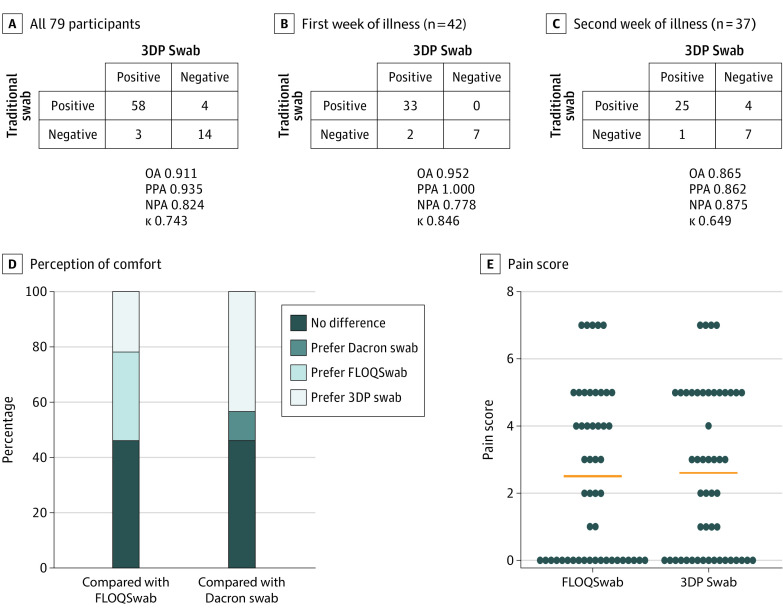
Among the 79 patients with COVID-19, the 3DP swab showed an excellent OA and PPA of 91.1% (95% CI, 82.6%-96.4%) and 93.5% (95% CI, 84.3%-95.5%), respectively, compared with reference swabs. The PPA was 100% (95% CI, 89.4%-100%) for 42 patients with COVID-19 (53.2%) who were tested within the first week of illness. In addition, there were 2 COVID-19 cases that were identified by the 3DP swab but not by the reference swab during the first week of illness (Figure 2, A-C). None of the swabs from the 10 control participants tested positive.
Figure 2. Clinical Performance of the 3-Dimensionally Printed (3DP) Swab Compared With Traditional Nasopharyngeal Swabs (FLOQSwab or Dacron Swab).
A-C, Overall categorical results of coronavirus disease 2019 (COVID-19) testing for all COVID-19 cases (A), COVID-19 cases in the first week of illness only (B), and COVID-19 cases in the second week of illness only (C). D, Participants’ perception of comfort on paired swab testing in the same nostril. E, Pain score comparing the FLOQSwab (COPAN) and 3DP swab (Deltalab). NPA indicates negative percentage agreement; OA, overall agreement; PPA, positive percentage agreement.
The OA, PPA, and NPA of the 3DP swab with each reference swab (FLOQSwab or Dacron swab) is presented in eFigure 2 in the Supplement. In general, the E-gene target identified more positive cases compared with the ORF1ab target.
Overall, the 3DP swab was acceptable in terms of comfort and preference for most participants. Compared with the FLOQSwab, 23 participants (25.8%) had no preference between the swabs, 16 (18.0%) preferred the FLOQSwab, and 11 (12.4%) preferred the 3DP swab. The mean difference in pain score between the 3DP swab and FLOQSwab was 0.1 (95% CI, −0.45 to 0.65; Cohen d = 0.041). Compared with the Dacron swab, 18 participants (20.2%) had no preference between the swabs, 4 (4.5%) preferred the Dacron swab, and 17 (19.1%) preferred the 3DP swab (Figure 2, D and E). No participant experienced frank epistaxis or swab breakage in this study. There was 1 episode of mild giddiness noted after administration of the 3DP swab.
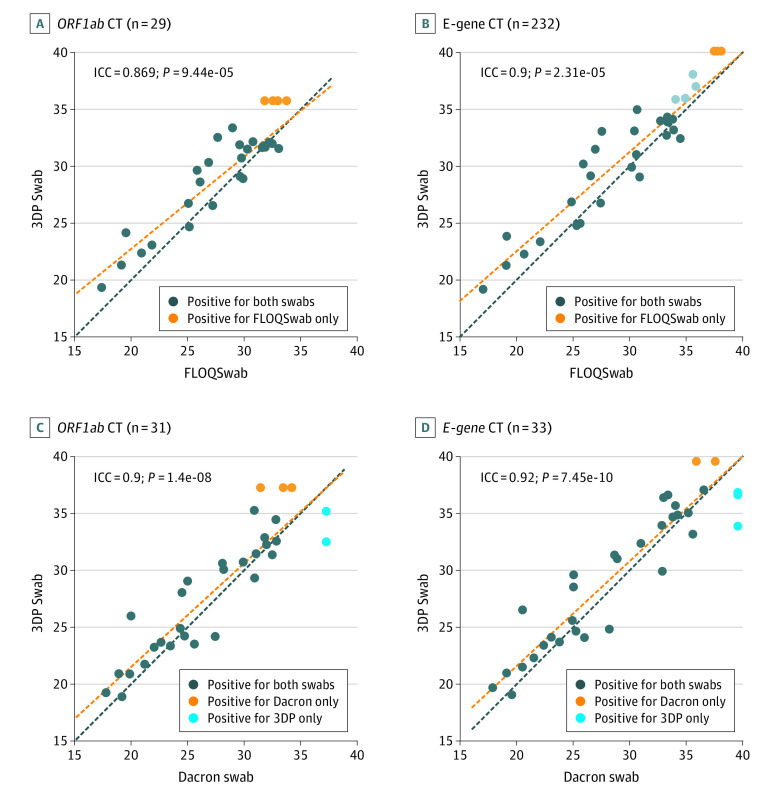
We observed a strong correlation of Ct values for the 3DP swab compared with the FLOQSwab and Dacron swab. The ICC ranged from 0.869 to 0.920 for the ORF1ab and E-gene targets for all paired comparisons (Figure 3). Discordant results, for which only the reference swab was positive, were observed only at high Ct thresholds (Ct value >31 for ORF1ab and Ct value >35 for E-gene). The 3DP swab also successfully identified cases that would have otherwise been missed by the Dacron swab (Figure 3, C and D). Four patients who tested positive for the ORF1ab target for the FLOQSwab only were successfully identified by the 3DP swab on E-gene testing, reflecting the importance of a dual-target testing strategy (Figure 3, A and B). Finally, we also observed that Ct values correlated positively with the day of illness, which was consistent with a reduction in viral load as a patient recovers (eFigure 3 in the Supplement).
Figure 3. Correlation of Cycle Threshold (Ct) Values Between the 3-Dimensionally Printed (3DP) Swab and Traditional Nasopharyngeal Swabs.
A-B, Correlation of Ct values between the 3DP swab and the FLOQSwab (COPAN) for the ORF1ab and E-gene targets on reverse-transcriptase polymerase chain reaction. Four samples that were ORF1ab negative for the 3DP swab were all positive for the E-gene target, which is represented as light gray dots in B. C-D, Correlation of Ct values between the 3DP swab and the Dacron swab for the ORF1ab and E-gene targets. Yellow and blue circles represent test-positive cases that were identified on only 1 of the swabs. In such cases, the negative sample was assigned a Ct value corresponding to the limit of detection for the purpose of illustration only. Correlation test (intraclass correlation coefficient [ICC]) includes only paired-positive cases. The red line represents line of best fit in a linear model.
Considering cases with paired positive results, the mean difference in Ct values between the 3DP swab and FLOQswab was 1.33 (95% CI, 0.56-2.09; Cohen d = 0.308) for ORF1ab and 1.36 (95% CI, 0.49-2.24; Cohen d = 0.274) for the E-gene (eFigure 4 in the Supplement). Compared with the Dacron swab, the mean difference in Ct values for the 3DP swab was 0.968 (95% CI, 0.144-1.79; Cohen d = 0.197) for ORF1ab and 0.998 (95% CI, 0.169-1.83; Cohen d = 0.172) for the E-gene.
Discussion
Given the escalation of the COVID-19 pandemic, the development of alternative technologies is important to mitigate the continued strain on testing resources. Here we observed that the novel 3DP swab had a strong performance compared with standard-of-care swabs used in routine clinical testing.
The accuracy of COVID-19 identification using nasopharyngeal swabs depends on several factors, including a patient’s viral burden in the nasopharynx, swabbing technique, and the laboratory assays used for testing. This is reflected in a meta-analysis that studied the accuracy of RT-PCR testing in COVID-19 that reported a pooled sensitivity of RT-PCR testing at 89% but with a wide range of sensitivities from 50% to 100% between studies.14 Deviations of RT-PCR results of up to 2 cycles (ie, 2 Ct values) have also been attributed to differences in RT-PCR machines (as indicated in the manufacturer’s manual)15 as well as interoperator differences in laboratory technique.16 Although the 3DP swab had slightly higher Ct values, it appears that this marginal difference did not have a significant association with the overall accuracy of RT-PCR testing, which is highly sensitive for small amounts of material.
Strengths and Limitations
A strength of our study is the multi-institutional design that allowed the 3DP swab to be compared with different reference swabs and processing techniques. The study design included COVID-19 cases up to 14 days of illness and thus allowed us to robustly evaluate the swab by sampling COVID-19 cases across various viral loads, including those at the limits of detection. Earlier studies that evaluated 3D-printed polymer swab designs were performed by using human RNase-P as a surrogate marker for testing,10 or by sampling participants presenting for COVID-19 diagnostic testing, with positive cases typically early in the course of illness when the viral burden is high. In such a setting, 3D-printed swab prototypes have shown a high level of OA (95.8%),17 which is similar to the agreement observed among patients with COVID-19 in the first week of illness in our study (95.2%). However, there have also been studies in which false-negative results have been observed even at low Ct values of less than 25, highlighting the need for robust testing of each 3D-printed swab prototype.9
The false-negative cases in our study had high Ct values and were identified only in the second week of disease; these may represent patients in recovery with low or no infectivity. A recent study demonstrated that the virus could not be isolated in cell cultures from patients who had E-gene Ct values of 34 or greater, suggesting a lack of infectivity.18 Another study observed cell culture infectivity only at Ct values of less than 24 and fewer than 8 days of illness.19 Similarly, in a multicenter cohort of 73 patients with COVID-19 in Singapore, no viable virus could be isolated in cell cultures when the Ct value was 30 or higher.20
The shortage of nasopharyngeal swabs during the pandemic has resulted in increasing prices as a result of robust demand. Given that the material cost of a 3DP swab is around $0.25,17,21 and that the price of commercially available flocked swabs is approximately $1 per piece, there is certainly room for competitive pricing of 3DP swabs. In addition, the presence of 3DP swab alternatives in the market plays an important role in keeping the price of traditional commercial swabs in check, especially because 3DP technologies are available worldwide. Based on our findings here, 3DP swabs that have been validated may be recommended for standard use and not just in the event of a shortage of existing options.
Conclusions
In this article, we show that with careful design and testing, 3DP polymer swabs can be highly accurate. The 3DP swab is safe, acceptable for patient use, and performs consistently across institutions. It is an excellent alternative to traditional swabs and will help mitigate strained resources in the escalating COVID-19 pandemic.
eMethods.
eFigure 1. Fluid release and absorption
eFigure 2. Categorical results comparing the swabs
eFigure 3. Ct values and day of illness
eFigure 4. Bland-Altman plots for the comparison of Ct values between paired positive swabs
References
- 1.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533-534. doi: 10.1016/S1473-3099(20)30120-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . 2019 Novel coronavirus: strategic preparedness and response plan. Accessed November 26, 2020. https://www.who.int/publications/i/item/strategic-preparedness-and-response-plan-for-the-new-coronavirus
- 3.Bagdasarian N, Fisher D. Heterogenous COVID-19 transmission dynamics within Singapore: a clearer picture of future national responses. BMC Med. 2020;18(1):164. doi: 10.1186/s12916-020-01625-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Team C-NIRS . COVID-19, Australia: epidemiology report 20 (fortnightly reporting period ending 5 July 2020). Commun Dis Intell. 2020;44. doi: 10.33321/cdi.2020.44.75 [DOI] [PubMed] [Google Scholar]
- 5.Devi S. COVID-19 resurgence in Iran. Lancet. 2020;395(10241):1896. doi: 10.1016/S0140-6736(20)31407-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yelin I, Aharony N, Tamar ES, et al. Evaluation of COVID-19 RT-qPCR test in multi-sample pools. Clin Infect Dis. 2020;71(16):2073-2078. doi: 10.1093/cid/ciaa531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith KP, Cheng A, Chopelas A, et al. Large-scale, in-house production of viral transport media to support SARS-CoV-2 PCR testing in a multihospital healthcare network during the COVID-19 pandemic. J Clin Microbiol. 2020;58(8):e00913-e00920. doi: 10.1128/JCM.00913-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vermeiren C, Marchand-Senécal X, Sheldrake E, et al. Comparison of Copan Eswab and FLOQswab for COVID-19 PCR diagnosis: working around a supply shortage. J Clin Microbiol. 2020;58(6):e00669-e20. doi: 10.1128/JCM.00669-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callahan CJ, Lee R, Zulauf KE, et al. Open development and clinical validation of multiple 3D-printed nasopharyngeal collection swabs: rapid resolution of a critical COVID-19 testing bottleneck. J Clin Microbiol. 2020;58(8):e00876-e20. doi: 10.1128/JCM.00876-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox JL, Koepsell SA. 3D-printing to address COVID-19 testing supply shortages. Lab Med. 2020;51(4):e45-e46. doi: 10.1093/labmed/lmaa031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donner A, Rotondi MA. Sample size requirements for interval estimation of the kappa statistic for interobserver agreement studies with a binary outcome and multiple raters. Int J Biostat. 2010;6(1):31. doi: 10.2202/1557-4679.1275 [DOI] [PubMed] [Google Scholar]
- 12.Koo TK, Li MY. A guideline of selecting and reporting intraclass Correlation Coefficients for Reliability Research. J Chiropr Med. 2016;15(2):155-163. doi: 10.1016/j.jcm.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osterblad M, Järvinen H, Lönnqvist K, et al. Evaluation of a new cellulose sponge-tipped swab for microbiological sampling: a laboratory and clinical investigation. J Clin Microbiol. 2003;41(5):1894-1900. doi: 10.1128/JCM.41.5.1894-1900.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H, Hong H, Yoon SH. Diagnostic performance of CT and reverse transcriptase polymerase chain reaction for coronavirus disease 2019: a meta-analysis. Radiology. 2020;296(3):E145-E155. doi: 10.1148/radiol.2020201343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization . Molecular assays to diagnose COVID-19: Summary table of available protocols. Accessed August 14, 2020. https://www.who.int/publications/m/item/molecular-assays-to-diagnose-covid-19-summary-table-of-available-protocols
- 16.Chang MC, Hur J, Park D. Interpreting the COVID-19 test results: a guide for physiatrists. Am J Phys Med Rehabil. 2020;99(7):583-585. doi: 10.1097/PHM.0000000000001471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Decker SJ, Goldstein TA, Ford JM, et al. 3D printed alternative to the standard synthetic flocked nasopharyngeal swabs used for COVID-19 testing. Clin Infect Dis. 2020;ciaa1366. doi: 10.1093/cid/ciaa1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Scola B, Le Bideau M, Andreani J, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059-1061. doi: 10.1007/s10096-020-03913-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullard J, Dust K, Funk D, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020;ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.National Centre for Infectious Diseases SaCoIDP, Academy of Medicine, Singapore . Period of infectivity to inform strategies for de-isolation for COVID-19 patients. Accessed May 24, 2020. https://www.ams.edu.sg/policy-advocacy/covid-19-resource-page
- 21.Rybicki FJ. 3D printing in medicine: COVID-19 testing with 3D printed nasopharyngeal swabs. Clin Infect Dis. 2020;ciaa1437. doi: 10.1093/cid/ciaa1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods.
eFigure 1. Fluid release and absorption
eFigure 2. Categorical results comparing the swabs
eFigure 3. Ct values and day of illness
eFigure 4. Bland-Altman plots for the comparison of Ct values between paired positive swabs