Abstract
Objectives:
In this study we describe the tumor microenvironment, the signaling pathways and genetic alterations associated with the presence or absence of CD8+ T-cell infiltration in primary squamous cell carcinoma of the head and neck (SCCHN) tumors.
Materials and Methods:
Two SCCHN multi-analyte cohorts were utilized, the Cancer Genome Atlas (TCGA) and the Chicago Head and Neck Genomics (CHGC) cohort. A well-established chemokine signature classified SCCHN tumors into high and low CD8+ T-cell inflamed phenotypes (TCIP-H, TCIP-L respectively). Gene set enrichment and iPANDA analyses were conducted to dissect differences in signaling pathways, somatic mutations and copy number aberrations for TCIP-H versus TCIP-L tumors, stratified by HPV status.
Results:
TCIP-H SCCHN tumors were enriched in multiple immune checkpoints irrespective of HPV-status. HPV-positive tumors were enriched in markers of T-regulatory cells (Tregs) and HPV-negative tumors in protumorigenic M2 macrophages. TCIP-L SCCHN tumors were enriched for the β-catenin/WNT and Hedgehog signaling pathways, had frequent mutations in NSD1 and amplifications in EGFR, NSD3 and FGFR1. TCIP-H SCCHN tumors were associated with the MAPK/ERK, JAK/STAT and mTOR/AKT signaling pathways, and were enriched in CASP8, EP300, HRAS mutations, and CD274, PDCD1LG2 and JAK2 amplifications.
Conclusions:
Our findings support that combinatorial immune checkpoint blockade and depletion strategies targeting Tregs in HPV-positive and M2 macrophages in HPV-negative tumors may lead to improved antitumor immune responses in patients with TCIP-H SCCHN. We highlight novel pathways and genetic events that may serve as candidate biomarkers and novel targeted therapies to enhance the efficacy of immunotherapy in SCCHN patients.
Introduction
Squamous cell carcinoma of the head and neck (SCCHN) is the sixth most common cancer worldwide [1]. It is pathogenetically subdivided in the tobacco-associated and the human papilloma virus (HPV)-positive SCCHN. Despite advances in chemoradiotherapy treatment standards, the 5-year overall survival of patients with locoregionally advanced SCCHN is approximately 50% [2], while patients with recurrent/metastatic disease have a median survival of 10 months. New treatment options are thus urgently awaited. Recently, immunotherapy through inhibition of immune checkpoints, specifically programmed-cell-death-1 (PD-1), has shown meaningful activity with objective response rates of 18% and durable responses for more than 6 months in 80% of the responders with platinum-refractory recurrent/metastatic SCCHN [3]. Furthermore, a randomized phase III trial of nivolumab versus investigator’s choice of standard systemic chemotherapy showed a significantly higher 1-year overall survival rate and a more favorable toxicity profile in patients treated with nivolumab [4]. This led to the expedited approval of pembrolizumab and nivolumab in patients with recurrent/metastatic SCCHN.
This activity of anti-PD-1 treatment supports that, in at least a fraction of patients, SCCHN is sufficiently immunogenic for checkpoint inhibition to trigger an effective anti-tumor immune response. However, only a minority of patients respond and both constitutive and acquired resistance are common, with patients progressing immediately or after limited treatment benefit. This underlines the presence of additional immune escape mechanisms in patients with SCCHN. Biomarkers of response to PD-1/PD-L1 axis blockade are currently not used in SCCHN, however it has been shown that PD-L1 expression measured by immunohistochemistry enriches for patients with a higher chance of benefit.
The induction of antigen-specific antitumor CD8+ T-cells at an amplitude sufficient to confer a significant antitumor response relies on an interplay of a multitude of factors which include the following: (1) the immunogenicity of tumor-associated antigens, (2) the functional robustness of the antigen presentation machinery, (3) the ability of T-cells to migrate into the tumor microenvironment (“T-cell trafficking”), (4) the expression of immune inhibitory or agonist checkpoint receptors and their respective ligands on T-cells and antigen-presenting cells or tumor cells, and (5) the relative ratios of specific immune cell subsets, such as cytotoxic CD8+ T-cells, T-regulatory cells (Tregs), myeloid-derived suppressor cells (MDSCs), antigen-presenting cells and CD4+ T-helper cells. This interplay is unique, in that different immune checkpoint pathways and immune cell subsets may dominate in different cancer types and even within a tumor entity [5]. Furthermore, the above suggests that strategies to improve response to immunotherapy should take all the aforementioned factors into consideration.
Based on the above, detailed dissection of the immune checkpoint landscape and tumor microenvironment would be necessary for a biologically rational administration of immunotherapy in order to increase its efficacy, as well as to introduce it in the treatment of earlier stages of SCCHN. In this context, no studies have systematically examined the relationship of the PDL-1/PD-1 axis with other checkpoints and specific immune cell subtypes in primary SCCHN. Since CD8+ T-cell infiltration is a sine qua non condition for immunotherapy to have an antitumor effect, we reasoned that an established gene signature that differentiates inflamed from non-inflamed tumors could be used to select SCCHN patients for immunotherapy. Harlin et al [6] had previously described a chemokine gene signature consisting of 10 chemokines (CCL2, CCL3, CCL4, CCL5, CCL19, CCL21, CXCL9, CXCL10, CXCL11 and CXCL13) which were found to induce CD8+ T-cell infiltration in melanoma metastases. In a subsequent study, Messina et al [7] described a similar signature consisting of 12 chemokines (CCL2, CCL3, CCL4, CCL5, CCL8, CCL18, CCL19, CCL21, CXCL9, CXCL10, CXCL11 and CXCL13) which was also associated with better overall survival in patients with metastatic melanoma.
In this study, using two genomic databases of patients with primary SCCHN, we implemented the Messina 12-chemokine gene signature to identify subgroups of patients with high or low CD8+ T-cell infiltration, phenotypes which we called “high T-cell-inflamed phenotype” (TCIP-H) or “low T-cell-inflamed phenotype” (TCIP-L), and to describe the immune checkpoints and immune cell subtypes that characterize each subgroup. Signaling pathways and genetic alterations associated with the T-cell inflamed phenotypes were interrogated. Results from this analysis provide insight into immune escape mechanisms in locoregionally advanced SCCHN tumors and suggest signaling pathways and genetic events that may underlie these mechanisms. This study could also provide the basis for rational combinatorial immune checkpoint blockade and/or immune cell subtype-specific therapies in the curative intent setting for SCCHN.
Materials and Methods
Definition of T-cell-inflamed phenotype (TCIP)
To identify SCCHN tumors with CD8+ T-cell enrichment, a 12-gene chemokine gene expression signature consisting of chemokines CCL2, CCL3, CCL4, CCL5, CCL8, CCL18, CCL19, CCL21, CXCL9, CXCL10, CXCL11 and CXCL13 was implemented. This signature was previously described by Messina et al [7], showing that expression of these chemokines by melanoma cells correlated with immune cell infiltration and improved overall survival in patients with metastatic melanoma. Furthermore, Harlin et al [6] showed that the expression of 10 of these chemokines by metastatic melanoma cells was associated with the migration of CD8+ T-cells within melanoma metastases, while lack of expression of this signature correlated with lack of CD8+ T-cell infiltration. SCCHN tumors with CD8+ T-cell enrichment based on the Messina signature were defined as having a high T-cell-inflamed phenotype (TCIP-H), whereas tumors with no CD8+ T-cell enrichment were defined as having a low T-cell-inflamed phenotype (TCIP-L).
Patient samples and RNA extraction
Two large SCCHN multi-analyte cohorts of patients with locoregionally advanced disease were utilized, the Cancer Genome Atlas [8] and the Chicago HNC (head and neck cancer) Genomics Cohort (CHGC) [9]. Clinical characteristics of each cohort are described in Table 1, whereby the CHGC is more enriched in HPV-positive (43% compared to 13% in the TCGA cohort) and stage IVA/B locoregionally advanced SCCHN tumors (96% compared to 58% in the TCGA cohort). The HPV status for the TCGA dataset was obtained from the Firehouse - Broad Institute. For the CHGC, OCT frozen tissue blocks of 171 SCCHN tumor samples of patients with locoregionally advanced disease prior to treatment were obtained from the University of Chicago Head and Neck Cancer tissue bank (Institutional Review Board-approved protocols IRB#8980/16-1269), as previously described [9]. Tumor content was confirmed to be more than 60%. HPV-status was determined using HPV-PCR (data available in 134 samples).
Table 1.
Demographic and clinical characteristics of the CHGC and the TCGA patient cohorts.
| CHGC Patients N=134 | ||
|---|---|---|
| Age | Median(yrs) | 57 |
| HPV status | Positive | 57 (43%) |
| Negative | 77 (57%) | |
| Tumor Stage | I-II | 2 (1.5%) |
| III | 3 (2.5%) | |
| IV | 129 (96%) | |
| Anatomic Site | Larynx | 31 (23%) |
| Oral Cavity | 25 (19%) | |
| Oropharynx | 76 (57%) | |
| Others | 2 (1%) | |
| TCGA Patients N=424 | ||
| Age | Median(yrs) | 59 |
| HPV status | Positive | 55 (13%) |
| Negative | 369 (87%) | |
| Tumor Stage | I-II | 83 (24%) |
| III | 61 (18%) | |
| IV | 200 (58%) | |
| Anatomic Site | Larynx | 97 (24%) |
| Oral Cavity | 242 (61%) | |
| Oropharynx | 58 (15%) | |
Identification of TCIP phenotypes and correlation with SCCHN gene expression subtypes, signaling pathways and genomic alterations
To identify the TCIP phenotypes and correlate them with SCCHN gene expression subtypes and signaling pathways, previously generated gene expression data (Agilent, n=134) from the CHGC were used [9]. Validation was performed using the TCGA SCCHN dataset. For the genomic alterations in the TCGA samples, mutation data and copy number data (categorized as putative copy-number calls determined using GISTIC 2.0, where −2 = homozygous deletion; −1 = hemizygous deletion; 0 = neutral / no change; 1 = gain; 2 = high level amplification) were downloaded via the cBioPortal and analyzed using Spearman correlation.
Gene set enrichment analysis
Java GSEA package was obtained from the GSEA official website (www.broadinstitute.org/gsea/index.jsp) and all the input data files were prepared according to the GSEA User guide available at http://software.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html. Only expression levels for differentially expressed genes (group t-test P value <0.05) were used as the input. The KEGG pathway database was converted to the GSEA file format using the same package. The KEGG pathways included in the analysis are listed in the Supplementary Table 1. The parameter ‘Number of permutations’ was set to 1,000, ‘Collapse data set to gene symbols’ was set to ‘false’, ‘Permutation type’ was set to ‘gene_set’, ‘Enrichment statistics’ was set to ‘weighted’, ‘Scoring scheme’ was set to ‘weighted’, ‘Metric for ranking genes’ was set to ‘Signal2Noise’, ‘Gene list sorting mode’ was set to ‘real’, ‘Gene list ordering mode’ was set to ‘descending’, ‘Collapsing mode for probe sets’ was set to ‘max_probe’, ‘Normalization mode’ was set to ‘meandiv’, ‘Randomization mode’ was set to ‘no balance’. Normalized enrichment score values were extracted from GSEA report for further analysis.
Expression data processing for iPANDA pathway analysis
Raw RNA-Seq data were retrieved from the publicly available TCGA database. RNA-Seq or internally generated microarray data preprocessing and normalization steps were performed in R version 3.1.0 using DEseq package from Bioconductor. The resulting matrix contained mRNA expression information for over 20,000 genes across all analyzed samples. Normalized gene expression data were loaded into iPANDA [10]. The software combines precalculated gene coexpression data with gene importance factors based on the degree of differential gene expression and pathway topology decomposition for obtaining pathway activation scores (PAS) for each pathway analyzed, a value which serves as a quantitative measure of differential pathway activation. A collection of 374 intracellular signaling pathways (which cover a total of 2,294 unique genes) strongly implicated with various solid malignancies (64 main signaling pathways and 310 branched axes radiating from the main pathways) was obtained from the SABiosciences, and used for the computational algorithm as described previously [10-13].
RNA-Seq deconvolution
Pre-processed gene expression data was loaded into the version of iPANDA algorithm [10] with disabled gene grouping and topological weights. The ‘off’ state of topology coefficients means that they are equal to 1 for all genes during the calculation. The ‘off’ state for the gene grouping means that all the genes are treated as individual genes. Deconvolution was performed using a collection of differentially expressed genes that facilitate annotation of the four T-cell subtypes as naive-like, regulatory, cytotoxic, and exhausted [14], and the estimated proportion of each subtype was calculated for each sample [15]. Activation signs for all the genes was obtained from Puram et al [14] and iPANDA algorithm was used for the dimension reduction in gene expression data prior to the clustering and data visualization.
Immunohistochemistry
SCCHN tumour sections were stained for PD-L1 and CD8. PD-L1 staining was performed using the SP142 antibody (Ventana) and its expression was evaluated on tumor cells (TC), as well as tumor-infiltrating immune cells (IC) as previously described [3, 16]. PD-L1 staining was defined as positive in the presence of >=5% of PD-L1-expressing TC or IC, a cut-off used by the Ventana SP142 immunohistochemistry method which has already been tested in several studies of head and neck cancer patients [17, 18]. CD8 staining of SCCHN tumours was performed by the University of Chicago Pathology Department. Specifically, tissue sections from 73 SCCHN tumor blocks were deparaffinized and rehydrated through xylenes and serial dilutions of EtOH to deionized water. The sections were then incubated in antigen retrieval buffer (DAKO, S1699) and heated in steamer at 97°C for 20 minutes. Anti-CD8 antibody (1:100, Spring Bio, clone Sp16, #M3162) was applied on tissue slides for 1 hour at room temperature. The antigen-antibody binding was detected using Envision+ system (DAKO, K4003) and DAB+ chromogen (DAKO, K3468). Tissue sections were briefly immersed in hematoxylin for counterstaining and were covered with cover glasses. For the CD8 scoring, a head and neck pathologist blinded to the T-cell inflamed phenotype performed semiquantitative analysis assigning the following scores: 0: no infiltration, 1: low (<25%), 2: moderate (25-50%), 3: high (>50%) CD8+ T-cell infiltration, either within the tumor, at the tumor periphery or both.
Statistical analysis
All statistics tests were performed using R. Fisher’s exact test was used to determine the relationship of PD-L1 IHC score to TCIP status. One-sided t-test and Mann-Whitney test were used to determine the significance of the difference of gene mRNA expression and IHC score between TCIP-H and TCIP-L groups. Associations between the TCIP-H and clinicopathological characteristics were assessed in logistic regression models, where TCIP-H status was used as dependent factor and the clinicopathological characteristics were used as independent factors in both the TCGA and the CHGC datasets.
Results
Identification and validation of T-cell inflamed phenotypes in SCCHN.
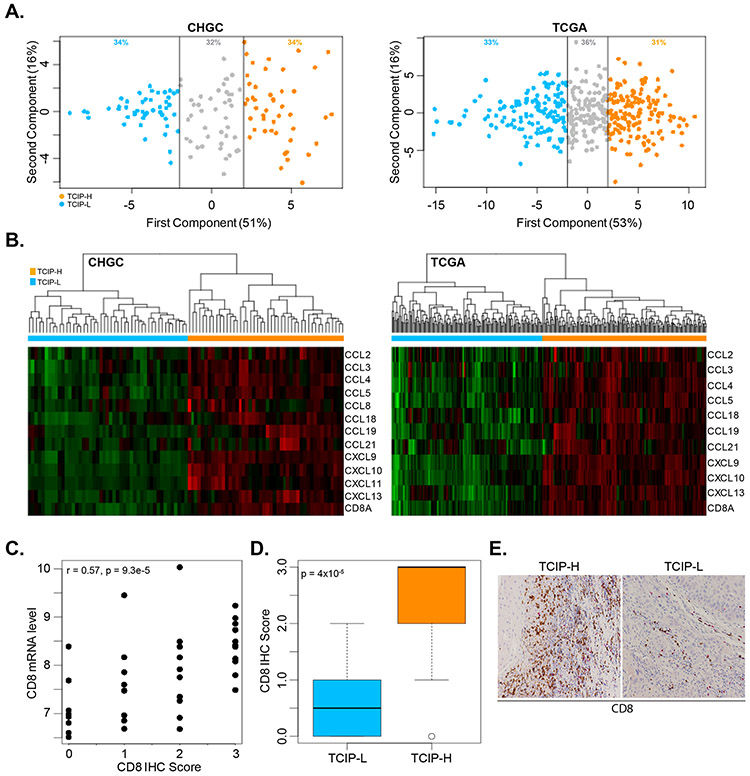
To examine whether SCCHN tumors can be categorized based on their level of CD8+ T-cell infiltration, the 12-chemokine gene expression signature (CCL2, CCL3, CCL4, CCL5, CCL8, CCL18, CCL19, CCL21, CXCL9, CXCL10, CXCL11 and CXCL13) described by Messina et al [7] was initially interrogated across the gene expression profiles of 134 locoregionally advanced SCCHN tumors of the CHGC. Using this chemokine signature, principal component analysis (PCA) was performed in the CHGC expression dataset and a PCA score was calculated using the first principal component (PC1). The PC1 approximated an average Z-score based on the log2 intensities for each of the 12 genes in the chemokine gene expression signature [7]. Using this approach, the SCCHN tumors were segregated into three main subgroups: the “high” T-cell inflamed phenotype (TCIP-H), the “low” TCIP (TCIP-L) and the “intermediate” TCIP (Figure 1A). 34% of samples (46 out of 134) were defined as TCIP-H, while another 34% of specimens were defined as TCIP-L (Supplementary Table 2). The remaining 42 samples (32%) showed intermediate expression of the 12 chemokine genes. SCCHN tumors with TCIP-H had increased expression of almost all chemokine genes, and the inverse was observed in TCIP-L tumors. These results were validated in the TCGA cohort of 464 patients with locoregionally advanced SCCHN tumors with similar percentages for the three TCIP phenotypes (Supplementary Table 3). As anticipated, the TCIP-H group was enriched in CD8 expression, while the TCIP-L group showed a very low signal for CD8. Thus, this signature could reproducibly identify a subgroup of SCCHN tumors with CD8+ T-cell infiltration (Figure 1B). To assess if this result was also reproducible at the protein level, we performed immunohistochemical analysis of CD8 expression in 73 patients from the CHGC with available tissue blocks. Overall, CD8 mRNA levels correlated well with CD8 protein levels (Figure 1C, Pearson’s correlation coefficient r=0.57, p=9.3×10−8). Patients with the TCIP-H phenotype had significantly higher CD8 protein levels compared to patients with the TCIP-L phenotype (Figure 1D, t-test, p=0.00004). Figure 1E shows an example of immunohistochemistry for CD8 in a CHGC patient with TCIP-H phenotype and rich CD8+ T-cell infiltration, and a patient with TCIP-L where CD8+ T-cell infiltration was lower.
Figure 1. Interrogation of a 12-chemokine gene expression signature in the CHGC and TCGA cohorts.
(A) Principal component analysis (PCA) was performed in the CHGC and TCGA expression datasets and a PCA score was calculated using the first principal component (PC1) with 51% and 53% of variance in the CHGC and TCGA cohorts respectively. The PC1 approximated an average Z-score based on the log2 intensities for each of the 12 genes in the chemokine gene expression signature. A low, intermediate and high T-cell inflamed phenotype was identified (blue dots: TCIP-L, grey dots: TCIP-intermediate, orange dots: TCIL-H). Data shown for the CHGC (left) and TCGA (right). (B) Heatmap of the chemokine gene expression signature across the CHGC and the TCGA. TCIP phenotypes correlate well with CD8+ T-cell enrichment. Data shown for the CHGC and the TCGA. (C) CD8 mRNA levels correlate positively with CD8 protein levels as assessed by immunohistochemistry (IHC) in 73 patients with SCCHN. CD8 IHC scoring was performed using a semi-quantitative scale of 0 (no CD8 T-cells), 1 (1-25% of CD8 T-cells), 2 (26-50% of CD8 T-cells) and 3 (>50% CD8 T-cells). r : Pearson’s correlation coefficient. (D) Boxplot showing the average CD8 IHC score in TCIP-L versus TCIP-H patients. TCIP-L patients had significantly lower CD8 IHC score compared to TCIP-H patients (t-test, p=0.0004). (E) Example of CD8 immunohistochemistry (IHC) in a TCIP-H versus a TCIP-L patient.
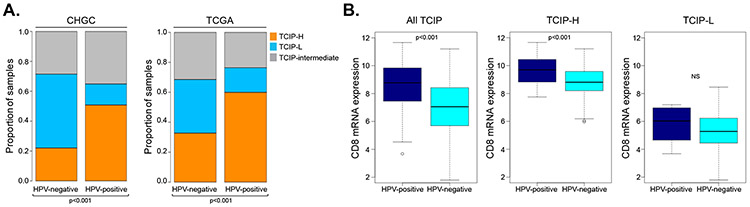
We then assessed if the TCIP segregation changes based on HPV-status. In the CHGC, 51% of HPV-positive tumors had a TCIP-H phenotype, compared to 21% of HPV-negative tumors. This result supports that HPV-positive tumors are more frequently infiltrated by CD8+ T-cells compared to HPV-negative tumors (Figure 2A). This observation was further validated in the TCGA cohort (Figure 2A). CD8 gene expression analysis further showed that HPV-positive SCCHN tumors not only had more frequent CD8+ T-cell infiltration, but also a significantly higher degree of infiltration compared to HPV-negative SCCHN tumors. This difference was observed across all TCIP types together (Mann-Whitney test, p=2×10−8) and the TCIP-H tumors (Mann-Whitney test, p=0.0001), but dissipated in the TCIP-L subset (Mann-Whitney test, p=0.39) (Figure 2B).
Figure 2. Interrogation of the 12-chemokine gene expression signature in SCCHN tumors by HPV-status.
(A) Histogram of distribution of TCIP-L (blue), TCIP-intermediate (gray) and TCIP-H (orange) phenotypes in HPV-negative versus HPV-positive SCCHN tumors across the CHGC and TCGA cohorts. (B) CD8 gene expression box-plot analysis in the CHGC SCCHN tumors in all TCIPs, TCIP-H only and TCIP-L only tumors based on HPV-status. The degree of CD8+ T-cell infiltration was significantly higher in HPV-positive compared to HPV-negative tumors in the categories of all TCIP tumors (Mann-Whitney test, p=2×10−8) and in TCIP-H tumors (Mann-Whitney, p=0.0001), but was insignificant for TCIP-L tumors (Mann-Whitney, p=0.39).
TCIP-H SCCHN tumors are enriched in markers of T-cell exhaustion and suppressive immune cell subtypes.
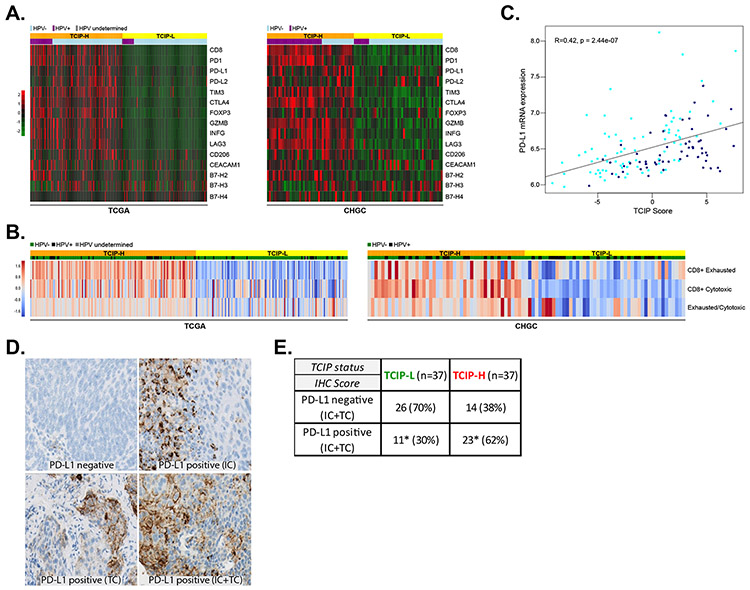
To explore the relationship between various immune checkpoints and immune cell markers with the TCIP phenotypes, a supervised analysis of the expression of immune checkpoints and various immune cell markers was performed using the 12-chemokine gene expression signature in the CHGC and TCGA datasets. TCIP-H tumors were significantly enriched for PD-L1, PD-1, CTLA4, TIM3, CEACAM1, LAG3, macrophage mannose receptor 1 (CD206) and FOXP3, compared to TCIP-L SCCHN tumors, suggesting that these checkpoints, as well as certain suppressive immune cell subtypes, such as M2 macrophages (CD206) and T-regulatory cells (FOXP3), may be contributing to immune escape in CD8+ T-cell inflamed SCCHN tumors. Granzyme B (GRZB) and interferon gamma (IFNγ) levels were also significantly higher in TCIP-H compared to TCIP-L SCCHN tumors, confirming the presence of T-cell infiltration in the TCIP-H group. No significant differences were observed for B7-H2, B7-H3 and B7-H4 between TCIP-H and TCIP-L tumors (Figure 3A).
Figure 3. Correlation of immune checkpoints and immune cell markers with T-cell-inflamed phenotypes.
(A) The 12-chemokine gene expression signature was interrogated across the TCGA and CHGC cohorts and immune checkpoints PD-L1, PD-L2, B7-H2, B7-H3, B7-H4, PD-1, CTLA4, TIM3, CEACAM1 and LAG3 are presented in a supervised analysis in a heatmap. Immune cell markers CD8, CD206 and FOXP3, as well as GRZB and INFG are also presented. Orange: TCIP-H tumors, yellow: TCIP-L tumors, light blue: HPV-negative, violet: HPV-positive. (B) Relative abundance of the CD8+ T-cell subtypes (cytotoxic and exhausted) was estimated from bulk RNA-Seq data by mathematical deconvolution using iPANDA algorithm (units on the color bar represent iPANDA score for corresponding gene set). The exhausted/cytotoxic CD8+ T-cell ratios were calculated to serve as numerical surrogates of the relative infiltration of tumors with different subsets of CD8+ T-cells. (C) Correlation between PD-L1 mRNA expression and TCIP score in SCCHN tumors (CHGC) (Pearson’s correlation coefficient R=0.42, p=2.44×10−7). (D) Immunohistochemistry for PD-L1 in SCCHN tumors. Examples of PD-L1 negative SCCHN tumor, a tumor with PD-L1 positive immune cells (IC), a tumor with PD-L1 positive tumor cells (TC) and a tumor with PD-L1 positive immune and tumor cells (IC and TC). (E) PD-L1 IHC (IC+TC) and correlation with TCIP phenotypes. TCIP-H tumors were more frequently PD-L1 positive (62%), though 38% of them were PD-L1 negative. 30% of TCIP-L tumors were PD-L1 positive (Fisher’s exact test, * signifies compared percentages, p=0.009).
Recently, single-cell RNA sequencing was performed on ~6,000 cells from 18 HNSCC patients, and immune cell content in tumor tissue was estimated by using a collection of differentially expressed genes that facilitate annotation of four T-cell subtypes as cytotoxic, exhausted, naive-like and regulatory [14]. To assess whether the expression of the aforementioned checkpoint axes and suppressive immune cell subtype markers correlated with the induction of an exhausted CD8+ T-cell phenotype, we used these T-cell-type-specific reference gene expression profiles (RGEPs) from the single-cell RNA sequencing [14] to estimate the relative abundance of the exhausted versus cytotoxic CD8+ T-cells from bulk gene expression data derived from the SCCHN TCGA or CHGC cohorts by mathematical deconvolution using the iPANDA algorithm. These analyses predicted that in both cohorts, TCIP-H tumors were enriched for both cytotoxic and exhausted CD8+ T-cells and the exhausted/cytotoxic CD8+ T-cell ratio was higher in these specimens, compared to the TCIP-L counterparts (Figure 3B). The enrichment of TCIP-H tumors with exhausted CD8+ T-cells further supports that the expression of co-inhibitory axes, such as PD-1/PD-L1 and TIM-3/CEACAM-1, which are key determinants of CD8+ T-cell exhaustion [19,20], and/or suppressive immune cell subtypes, such as Tregs and M2 macrophages, may play significant functional roles in inducing CD8+ T-cell exhaustion in this subset of SCCHN patients. Supporting this suggestion, the Treg/Th1 CD4+ T-cell ratio was also higher in tumors with TCIP-H phenotype (Supplementary Figure 1).
Correlation between T-cell inflamed phenotypes and PD-L1 expression in SCCHN tumors.
To investigate whether TCIP-H tumors were associated with increased PD-L1 expression, the relationship between PD-L1 mRNA levels and the TCIP score was explored. Using the CHGC, PD-L1 mRNA expression correlated significantly with the TCIP score, with higher PD-L1 mRNA levels correlating positively with higher TCIP scores (Pearson’s correlation coefficient, R=0.42, p=2.44×10−7) (Figure 3C). This was confirmed in the TCGA and reproduced both in HPV-positive and HPV-negative tumors (data not shown).
To further validate this finding at the protein level, immunohistochemistry (IHC) for PD-L1 was performed in tissue sections from all 134 SCCHN tumors of the CHGC dataset (Figure 3D). Of these sections, 37 TCIP-H and 37 TCIP-L tumors were evaluable and included in this analysis. The rest of the tumors were either of the intermediate TCIP or available sections were not of adequate amount and integrity. Scoring was done separately on the tumor and immune stroma cells and a positive result was defined as the presence of equal or more than 5% positive TC or IC cells in a given section. Based on these scoring criteria, 62% of TCIP-H tumors were PD-L1 positive compared to 30% of TCIP-L tumors, and this difference was statistically significant (Fisher’s exact test, p=0.009), supporting that PD-L1 is more frequently expressed in TCIP-H compared to TCIP-L tumors. However, despite a low level of CD8+ T-cell infiltration in TCIP-L tumors, 30% of them were PD-L1 positive. Inversely, 38% of TCIP-H tumors did not express PD-L1, implying that the presence of CD8+ T-cell infiltration is not necessarily accompanied by increase in PD-L1 expression (Figure 3E). These data underline that the presence of PD-L1 expression is not necessarily associated with increased CD8+ T-cell infiltration and provides insight into the observation that PD-L1 positive patients do not necessarily respond to anti-PD1/PD-L1 axis blockade. Detailed results of the IHC scoring at both ≥5% and ≥1% cut-offs are shown in Supplementary Table 4.
HPV-positive SCCHN tumors are enriched in markers of T-regs, whereas HPV-negative tumors are enriched in markers of M2 macrophages.
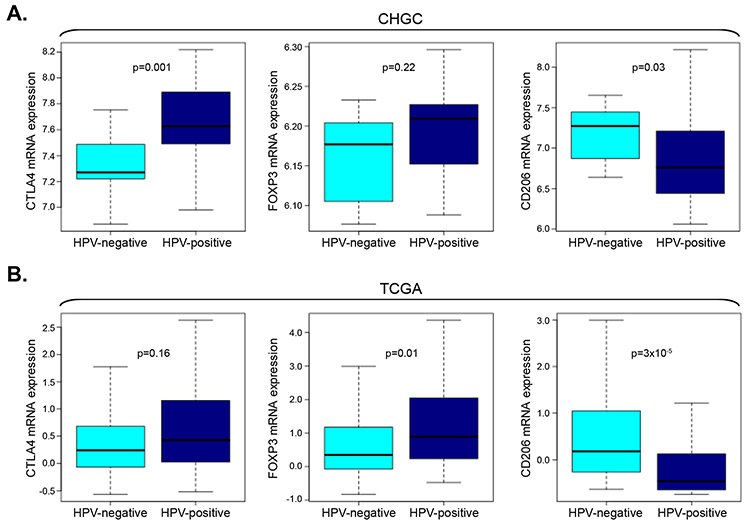
The immune microenvironment between HPV-positive and HPV-negative TCIP-H SCCHN tumors was also compared. Specifically, CTLA4 was significantly higher (Mann-Whitney, p=0.001, CHGC) in HPV-positive compared to HPV-negative tumors, supporting that this checkpoint may be more important in HPV-positive SCCHN tumors (Figure 4A). A relevant trend was observed in the TCGA cohort (Mann-Whitney, p=0.16) (Figure 4B). Accordingly, FOXP3, a marker of T-regulatory cells (T-regs), was higher in HPV-positive compared to HPV-negative tumors (Figure 4A). Though this difference did not reach statistical significance in the CHGC (Mann-Whitney, p=0.22), it was significant in the TCGA cohort (Mann-Whitney, p=0.01) (Figure 4B). A significant difference between HPV-positive and HPV-negative TCIP-H SCCHN tumors in both the CHGC and TCGA cohorts was also observed for CD206, a marker of M2 protumorigenic macrophages, which was higher in HPV-negative compared to HPV-positive SCCHN tumors (Mann-Whitney, p=0.03, CHGC; p=3×10−5, TCGA), signifying a role for M2 macrophages in HPV-negative SCCHN tumors (Figure 4A, B).
Figure 4. Differences in the tumor microenvironment in HPV-positive and HPV-negative SCCHN tumors.
(A) Box plot analysis of CTLA4, FOXP3 and CD206 mRNA expression in HPV-positive versus HPV-negative TCIP-H SCCHN tumors of the CHGC. CTLA4 mRNA is significantly higher in HPV-positive TCIP-H SCCHN tumors compared to HPV-negative tumors (Mann-Whitney, p=0.001). FOXP3 mRNA is higher in HPV-positive tumors but did not reach significance in this cohort (Mann-Whitney, p=0.22). CD206 mRNA is significantly higher in HPV-negative TCIP-H SCCHN tumors compared to HPV-positive tumors (Mann-Whitney, p=0.03). (B) Box plot analysis of CTLA4, FOXP3 and CD206 mRNA expression in HPV-positive versus HPV-negative TCIP-H SCCHN tumors of the TCGA. CTLA mRNA is higher HPV-positive tumors but did not reach statistical significance in this cohort (Mann-Whitney, p=0.16). FOXP3 mRNA is significantly higher in HPV-positive TCIP-H SCCHN tumors compared to HPV-negative tumors (Mann-Whitney, p=0.03). CD206 mRNA is significantly higher in HPV-negative TCIP-H SCCHN tumors compared to HPV-positive tumors (Mann-Whitney, p=3×10−5).
Correlation of T-cell inflamed phenotypes with SCCHN expression subtypes and signaling pathways.
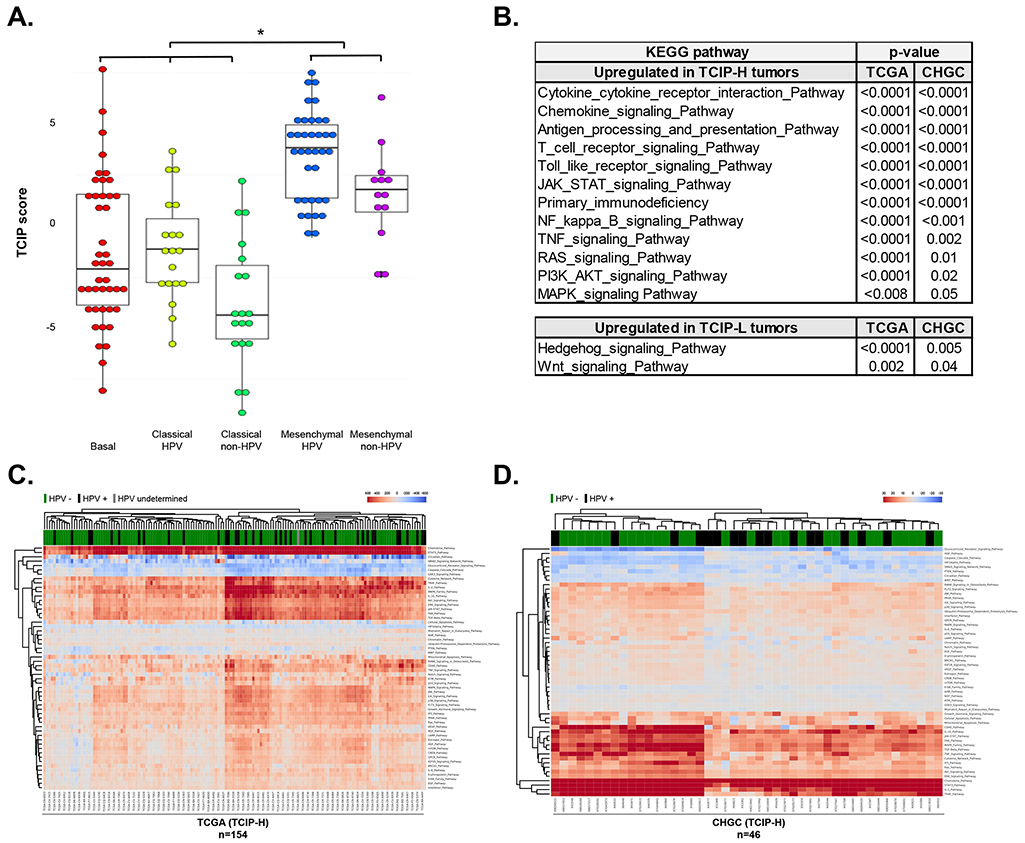
Previous work from our group and others [9, 21] has shown clustering of specific pathways within five SCCHN expression subtypes: the basal group, which is enriched in hypoxia and HER signaling pathways, the classical group, which is enriched in cell cycle and xenobiotic metabolism pathways and the mesenchymal group, both the latter being subclassified in HPV and non-HPV types. Notably, the mesenchymal subtype is highly enriched with immune-related genes irrespective of HPV status [9, 21]. To assess whether the TCIP groups correlated with any of these subtypes, the 12-chemokine gene expression signature (TCIP score) was interrogated across these SCCHN subtypes and results showed that the mesenchymal subtypes had significantly higher TCIP scores compared to the basal and classical tumors, irrespective of HPV status in both the CHGC and the TCGA cohorts (Figure 5A, Supplementary Figure 2).
Figure 5. Correlation of T-cell inflamed phenotypes with SCCHN expression subtypes and signaling pathways.
(A) TCIP-H tumors correlated significantly with the mesenchymal subtype, while the TCIP-L tumors correlated with the basal and classical subtypes, irrespective of HPV status (CHGC). (B) Table summarizing the results of gene set enrichment analysis for pathways upregulated in TCIP-H and TCIP-L SCCHN tumors. (C-D) RNA-Seq data (for TCGA cohort, C) or gene expression microarray data (for CHGC cohort, D) was processed and analyzed using the iPANDA software suite. The hierarchically clustered heatmap depicts main signaling pathways dysregulated in TCIP-H samples (transcriptomic data derived from TCIP-L tumors was used as a reference for analysis). Downregulated iPANDA values for each sample/pathway are indicated in blue, while upregulated values are shaded in red.
To examine whether specific signaling pathways were associated with the T-cell inflamed phenotypes, gene set enrichment analysis (GSEA) was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. TCIP-H tumors were enriched in multiple immune-related pathways, such as the cytokine-cytokine receptor interaction, chemokine signaling, antigen processing and presentation, and T-cell receptor signaling, as well as pathways associated with cell proliferation and survival, such as JAK-STAT, NFκB, TNF, RAS, PI3K/AKT and MAPK signaling pathways, similar to the mesenchymal subtype. TCIP-L tumors were significantly enriched for the Hedgehog and WNT signaling pathways. These correlations were independent of HPV status (Figure 5B).
Since pathway-based algorithms such as GSEA and its extensions rely solely on gene enrichment statistics treating pathways as unstructured sets of genes, the in silico pathway activation analysis (iPANDA) [10] algorithm was pursued to predict differential activation of cancer related pathways in the TCIP-H subsets from both the TCGA and CHCG datasets (tumors with TCIP-L status were used as a reference). Notably, 40 main pathways were commonly upregulated, and 5 main pathways were commonly downregulated among the TCIP-H tumors in both datasets analyzed (Supplementary Table 5). Focusing on the major pathways involved in cancer development, an hierarchically clustered heatmap of main pathways differentially activated in TCIP-H tumors was generated (Figure 5C, D). In both cohorts, pathways involved in chemokine and cytokine signaling, T cell survival (IL-2 and IL-10 pathways) and inflammation (IL-6, STAT3 and TNF pathways), antigen presentation (CD40) as well as cancer progression and maintenance, including MAPK/ERK, TGFβ, JAK/STAT, AKT/mTOR pathways, were significantly upregulated in the TCIP-H compared to the TCIP-L subset, whereas PTEN, HIF1-alpha and the SMAD signaling pathways were significantly downregulated in the TCIP-H tumors (Figure 5C, D). Similarly to GSEA analysis, these correlations were independent of HPV-status.
Although the direct comparison of the data generated by different methods should be addressed with caution, pathways involved in chemokine and cytokine signaling, inflammation and survival were among the most significantly upregulated axes identified in TCIP-H subsets of the TCGA and the CHGC cohorts by both bioinformatics approaches.
Correlation of T-cell inflamed phenotypes with genetic alterations and tumor mutational burden.
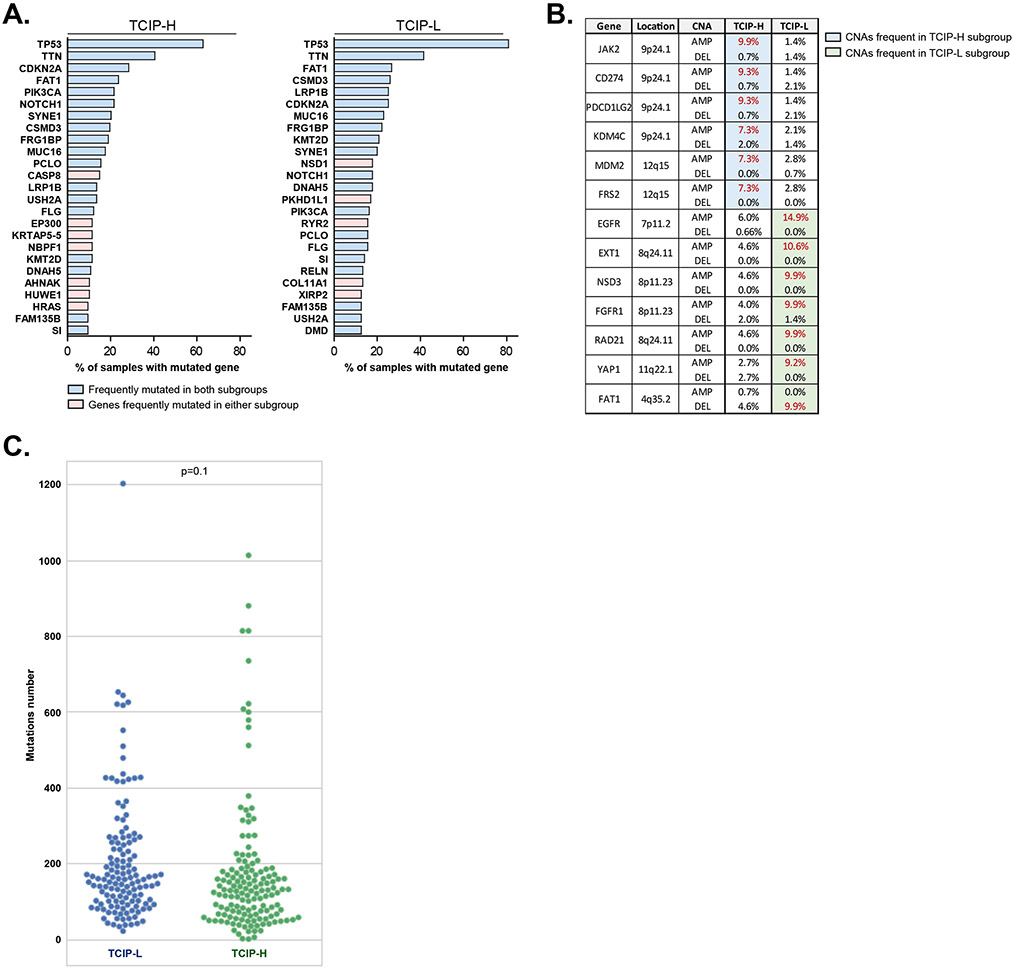
We then tried to identify mutations or copy number alterations (CNAs) that are associated with the T-cell inflamed phenotypes using the TCGA database. Mutations in caspase 8 (CASP8), the histone acetyltransferase EP300 and HRAS genes were substantially more frequent among the TCIP-H tumors, whereas the protein methyltransferase NSD1 gene was frequently mutated in TCIP-L tumors (Figure 6A, Supplementary Table 6). Furthermore, the TCIP-H samples were enriched for genetic amplification of the CD274 (gene encoding PD-L1), PDCD1LG2 (gene encoding PD-L2), JAK2 and KDM4C genes (all located on the chr9p24.1 locus termed the PDJ amplicon), whereas the TCIP-L subset had significantly higher frequency of EGFR, the protein methyltransferase NSD3 and FGFR1 amplifications (Figure 6B, Supplementary Table 7). As whole exome sequencing analysis was not available for the CHGC cohort, we could not use this database to perform validation of results obtained from the TCGA.
Figure 6.
(A) Genes frequently mutated in TCIP-H (left) and TCIP-L (right) subgroups of SCCHN derived from TCGA. Genes frequently mutated in both subgroups are labeled in light blue, while genes frequently mutated in either TCIP-H or TCIP-L specimens are indicated in pink. (B) Frequent CNAs in TCIP-H or TCIP-L subgroups of SCCHN derived from TCGA. (C) Nonsynonymous mutational burden in the TCIP-L (blue circles) and TCIP-H (green circles) tumors.
It has been reported that higher tumor mutational burden is associated with higher neoantigen load and durable clinical benefit in response to PD-1/PD-L1 checkpoint blockade, implying neoantigen-specific cytotoxic CD8+ T-cell infiltration in tumors with highly mutated genomes [22-24]. Based on this, we reasoned that TCIP-H SCCHN tumors would be more likely to harbor a higher mutation number compared to TCIP-L tumors. To examine this hypothesis, we compared the tumor mutational burden in the TCIP-H and TCIP-L tumors of the TCGA cohort. Interestingly, we found no significant difference in the nonsynonymous mutational burden between the TCIP-H and TCIP-L tumors (Figure 6C). This is consistent with the previously reported lack of correlation between levels of immune infiltration and mutational load in SCCHN [25].
Association of TCIP status with overall survival and other clinicopathological factors.
To assess whether the TCIP status is associated with survival outcomes, Cox regression analysis was conducted using the TCGA database. Consistent with the observation that elevated CD8+ T-cell infiltration correlates with superior survival in SCCHN [25], TCIP-H status showed a trend for association with better overall survival, as shown by Kaplan–Meier survival curves (p=0.069) (Supplementary Figure 3A). However, multivariate analysis for HPV-status and stage diminished this trend (Supplementary Figure 3B), indicating that the survival benefit may be driven by these factors rather than the TCIP status. No trend for a survival benefit was observed in the TCIP-H status in the CHGC cohort (data not shown).
Cell type origin of chemokine signature in primary SCCHN tissues
We next examined the relevant contribution of each cell type within primary SCCHN tissues in the production of the Messina signature chemokines using the publicly available single-cell RNA sequencing data from 18 patients [14]. Although the relative contribution varies among patients due to the different number of cells isolated and sequenced for each tumor, our results indicate that in general, the Messina signature chemokines are produced predominantly by T-cells, macrophages and fibroblasts, with T-cells exhibiting the highest chemokine signature score (Supplementary Figure 4).
Discussion
With the advent of immune checkpoint inhibition for the treatment of SCCHN, the need for more robust characterization of the tumor microenvironment in SCCHN is becoming more prominent, especially as the tumor microenvironment varies and may require differing, rational approaches from patient to patient. Since the antitumor effect of immune checkpoint inhibition is mediated by CD8+ T-cell cytotoxicity, a pre-existing CD8+ T-cell infiltrate or successful migration of CD8+ T-cells within a tumor is theoretically a minimum prerequisite. This premise is supported by the observation that most clinical responders to anti-PD1/PD-L1 axis or CTLA4 blockade have a pre-existing T-cell infiltrate [26]. This report supports that the HPV status and potentially targetable pathways and genetic events may be important biologic modifiers for the immune microenvironment of SCCHN.
Harlin et al described [6] 10 chemokine genes (CCL2, CCL3, CCL4, CCL5, CCL19, CCL21, CXCL9, CXCL10, CXCL11 and CXCL13) which were found to be expressed by melanoma cells, with some of their corresponding receptors expressed on tumor infiltrating CD8+ T-cells. This chemokine signature could differentiate melanoma metastases that contained or lacked a CD8+ T-cell infiltrate, and a subset of these chemokines was shown to be necessary for the migration of CD8+ T-cells within the tumors. A similar signature was further validated by Messina et al in more than 30 tumor types, using gene expression analysis in 14,492 primary or metastatic tumor samples [7], whereby significant enrichment for CD8 mRNA expression and other immune-related genes was noted in the samples overexpressing this signature.
In this study, we used the Messina signature [7] to assess whether SCCHN tumors clustered based on CD8 mRNA expression. We identified a subset of SCCHN tumors with marked CD8+ T-cell enrichment, which we defined as high T-cell inflamed phenotype (TCIP-H). This group of SCCHN tumors correlated with the previously described mesenchymal intrinsic expression subtypes, both for HPV-positive and HPV-negative tumors [27]. Approximately one third of patients in the CHGC and the TCGA cohorts had TCIP-H tumors and another third exhibited a low T-cell inflamed phenotype (TCIP-L). Importantly, HPV-positive tumors had a significantly higher fraction of the TCIP-H phenotype compared to HPV-negative tumors (51% versus 21%, respectively in the CHGC), consistent with the observation that HPV-positive tumors exhibit more frequent CD8+ T-cell infiltration. The TCIP-H tumors were enriched for CD8 expression and demonstrated increased levels of multiple immune checkpoints, particularly PD-L1, PD-L2, PD-1, TIM3, CEACAM1, LAG3 and CTLA4. These markers may be upregulated as a secondary response to an intense and active inflammatory antitumor reaction, e.g. from interferon-gamma [28] or they may constitute primary mechanisms of immune escape orchestrated by the tumor cells, as suggested by the frequent PD-L1 and PD-L2 amplifications. Notably, the increase in exhausted CD8+ T-cells in TCIP-H patients, as demonstrated by the deconvolution analysis, suggests that elevation of these key immune checkpoints creates a dysfunctional tumor immune microenvironment which hinders CD8+ T-cell cytotoxicity and function [19, 20, 29, 30]. These results support that primary SCCHN tumors probably utilize multiple immune escape mechanisms and thus underline the importance of multitargeted immunotherapy. TIM3, LAG3 and CTLA4 may thus be rational targets for combination immunotherapy with PD-1/PD-L1 axis blockade for patients with TCIP-H SCCHN tumors.
Furthermore, the Messina signature provided insights into specific immune cell subtypes that could potentially be hindering the development of a robust immune response. HPV-positive TCIP-H SCCHN tumors were enriched in CTLA4 and FOXP3 markers, signifying that strategies to deplete Tregs may be more important in HPV-positive tumors. CD206, an M2 macrophage marker, was found to be more enriched in HPV-negative TCIP-H tumors, alluding to a more potent role of M2 macrophages in inducing immune escape in HPV-negative tumors. This suggests that targeting M2 macrophages could enhance the efficacy of PD-1/PD-L1 axis blockade in HPV-negative tumors. The above supports the importance of targeting specific immune cell subtypes to enhance the efficacy of immunotherapy in SCCHN.
Tumor cell expression of PD-L1 is induced by the RAS/MAPK/ERK, PI3K/AKT and JAK/STAT signaling pathways, which are frequently activated in SCCHN [31-33]. Concordantly, our findings indicate that TCIP-H tumors were enriched for the RAS/MAPK/ERK, PI3K/AKT and JAK/STAT pathways, as well as for the PD-L1/PD-1 axes. Furthermore, the TGFβ signaling pathway which skews the differentiation of CD4+ T-cells towards immunosuppressive Tregs [34] was also predicted to be elevated in TCIP-H tumors, in accordance with the enrichment of these tumors for FOXP3 and CTLA4. Additionally, frequent mutations in CASP8, EP300 and HRAS, as well as genomic amplifications of JAK2, KDM4C, which encodes a protein lysine demethylase, and MDM2, a negative regulator of the p53 tumor suppressor, were observed among the TCIP-H patients and have been shown to enhance tumor growth [35-39]. Coamplification of JAK2, CD274 (PD-L1) and PDCD1LG2 (PD-L2) was seen in approximately 9% of TCIP-H tumors. While the observation that the TCIP-H SCCHN tumors seem to be enriched in cell proliferation and survival signaling is interesting and each of the reported pathways have been associated with modulation of a few of the Messina signature chemokines, the specific molecular determinants that underlie the observed coordinated expression of these chemokines by the TCIP-H SCCHN tumors are undefined and require further investigation.
TCIP-L SCCHN tumors were enriched for the β-catenin/WNT and Hedgehog signaling pathways, NSD1 mutations, as well as EGFR, NSD3 and FGFR1 amplifications. Recent reports indicate that the β-catenin/WNT signaling pathway strongly associates with a non-inflamed phenotype in multiple tumor types [40, 41] and efforts to target this pathway in combination trials are ongoing, β-catenin activation has been described to account for the lack of T-cell infiltration in 48% of melanoma tumors, and preliminary data have indicated a similar finding for urothelial bladder carcinomas [42]. Mechanistically, β-catenin induces the expression of the transcriptional repressor ATF3 in melanoma cells, which binds to the promoter of CCL4, a chemokine that recruits BATF3-lineage dendritic cells necessary for T-cell activation, inducing its silencing and thus inhibition of T-cell activation and trafficking [43]. The association of NSD1 inactivation and disruption of the Hedgehog signaling pathway with decreased immune cell infiltration of the tumor microenvironment are less well studied. Inactivating mutations of NSD1 define a SCCHN subtype with pronounced DNA hypomethylation and an “immune cold” phenotype [44]. While the therapeutic implications of these findings remain to be elucidated, identification of specific signaling pathways deregulated by NSD1 inactivation may facilitate novel targeted therapies for patients with TCIP-L SCCHN tumors. Furthermore, previous studies have highlighted that inhibition of the Hedgehog signaling pathway promotes CD8+ T-cell infiltration and induces profound alteration of the local chemokine/cytokine network [45]. While the relevance of EGFR amplification in [25]this context is unclear, it may be associated with hypoxia and a metabolically unfavorable tumor microenvironment leading to “cold” tumors [9]. Furthermore, NSD3 and FGFR1 (both present in the chromosomal locus 8p11.23) are coamplified and have been reported to have oncogenic functions in SCCHN [46-48], but their association with immune cell exclusion has not been studied yet. The mechanisms through which these pathways and genetic events may affect CD8+ T-cell trafficking merit further investigation.
The observation that the tumor mutational burden did not differ significantly between the TCIP-H and TCIP-L SCCHN phenotypes is interesting and suggests that the presence of neoantigens may be a necessary but not a sufficient factor for effective anti-tumor immunity to be mounted in head and neck cancer patients. This is in accordance with previous reports supporting that tumor mutational burden does not clearly correlate with immune infiltration [25]. Furthermore, while the decreased expression of the Messina signature chemokines by TCIP-L tumors may be one major determinant of immune cell exclusion in SCCHN, another possible mechanism may be a prohibitive tumor endothelial barrier, whereby endothelial cells of the tumor vasculature may be induced to relinquish their adhesive properties for circulating immune cells, thus leading to decreased influx of T-cells within the tumors. It can thus be speculated that combinatorial approaches that would upregulate the expression of T-cell attracting chemokines and enhance the adhesive properties of the vascular endothelium within tumors may increase intratumoral T-cell infiltration [49].
Although CD8 mRNA could have been implemented for the same analysis, the use of a multigene signature, such as the Messina signature, could be considered a more robust biomarker given that, if CD8 mRNA expression is not accurate or available in all samples due to technical reasons, a multigene signature could compensate for this. A major limitation of this study is its observational and correlative nature, and preclinical experiments will be necessary to validate the causative nature of any of the reported correlations. Additionally, the survival associations observed with the TCIP phenotypes were weak, though the relatively small sample sizes might have contributed to this. Finally, correlation of the TCIP phenotypes with response to PD-1/PD-L1 and other immune checkpoint inhibitors, as well as to suppressive immune cell depletion strategies would be necessary to validate the Messina chemokine signature as a biomarker of response to these immunotherapies.
In summary, TCIP-H primary SCCHN tumors may represent a promising candidate group for immunotherapy with checkpoint blockade, while combinatorial immune checkpoint approaches and depletion strategies for Tregs in HPV-positive and M2 macrophages in HPV-negative SCCHN tumors may be necessary for an effective antitumor response to be mounted. The WNT/β-catenin and Hedgehog signaling pathways, as well as pathways affected by NSD1 truncating mutations may be rational targets to induce CD8+ T-cell infiltration of TCIP-L SCCHN tumors. Preclinical experimentation would be important to validate these findings and to ultimately translate them to clinical trials aiming to broaden the number of SCCHN patients that could benefit from immunotherapeutic approaches. Finally, as the investigation of immunotherapy is transitioning in the earlier stages of SCCHN [50], this study provides rationale to personalize combination immunotherapy based on the underlying tumor microenvironment of SCCHN patients.
Conclusion
In this study we show that SCCHN tumors with high T-cell inflamed phenotype (TCIP-H) are enriched in multiple immune checkpoints, have frequent mutations in CASP8, EP300 and HRAS, and frequent coamplification of JAK2 and CD274. HPV-positive tumors were enriched in markers of Tregs and HPV-negative tumors in M2 macrophages. SCCHN tumors with low T-cell inflamed phenotype (TCIP-L) are enriched in the WNT/β-catenin and Hedgehog signaling pathways, have frequent NSD1 mutations and EGFR, NSD3 and FGFR1 amplifications. These results suggest that personalized combinatorial immune checkpoint blockade and targeting Tregs in HPV-positive and M2 macrophages in HPV-negative TCIP-H SCCHN tumors may improve antitumor immune responses. Targeting the WNT/β-catenin pathway may induce CD8+ T-cell infiltration in TCIP-L SCCHN tumors, while the Hedgehog signaling pathway, NSD1 mutations, NSD3 and FGFR1 amplifications merit further investigation as mediators of immune cell exclusion. These findings may inform rational clinical trial design in the neoadjuvant setting for SCCHN.
Supplementary Material
Supplementary Figure 1. Relative abundance of the CD4+ T cell subtypes (naive-like and regulatory) was estimated from bulk RNA-Seq data by mathematical deconvolution using iPANDA algorithm and Treg/Th1 ratios were calculated to serve as numerical surrogates of the relative infiltration of tumors with different subsets of CD4+ TILs.
Supplementary Figure 2. TCIP-H tumors correlated significantly with the mesenchymal subtype, while the TCIP-L tumors correlated with the basal and classical subtypes, irrespective of HPV status (TCGA) (* p<0.05, t-test).
Supplementary Figure 3. (A) Kaplan Meier estimates of overall survival based on TCIP status in the TCGA SCCHN patient cohort (TCIP-H - orange line, TCIP-L - blue line). (B) Multivariate analysis using Cox regression for TCIP status, stage and HPV status in the TCGA SCCHN patient cohort.
Supplementary Figure 4. The 12-chemokine gene expression signature was interrogated across different tumor cell components using the publicly available single-cell RNA sequencing data from 18 SCCHN patients.
Supplementary Table 1. List of KEGG pathways included for the gene set enrichment analysis.
Supplementary Table 2. Demographic and clinical characteristics of the patients from the CHGC cohort segregated by their TCIP status.
Supplementary Table 3. Demographic and clinical characteristics of the patients from the TCGA dataset segregated by their TCIP status.
Supplementary Table 4. Immunohistochemistry scoring for PD-L1 in immune and tumor cells or both from 134 SCCHN tumors of the CHGC dataset. Two different cut-offs were implemented.
Supplementary Table 5. List of iPANDA signaling pathways concurrently dysregulated (upregulated or downregulated) in TCIP-H patients from TCGA and CHGC cohorts. All shared pathways (left) and main shared pathways (right) are shown.
Supplementary Table 6. List of genes frequently mutated in TCIP-H and TCIP-L tumors derived from TCGA dataset.
Supplementary Table 7. List of frequent CNAs in TCIP-H or TCIP-L subgroups derived from TCGA dataset.
Footnotes
Conflicts of interest statement: TY Seiwert: Consultant: Loxo Oncology; Honoraria: Aduro, Astra Zeneca, Bayer, BMS, Celgene, Innate, Merck, Nanobiotix; Research Funding: BMS, Jounce, Merck. M Kowanetz, H Koeppen: employees of Genentech/Roche and owning stock in Roche. Other authors: no disclosures related to this project.
References
- 1.Chaturvedi AK, et al. , Worldwide trends In incidence rates for oral cavity and oropharyngeal cancers. J Clin Oncol, 2013. 31(36): p. 4550–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ang KK and Sturgis EM, Human papillomavirus as a marker of the natural history and response to therapy of head and neck squamous cell carcinoma. Semin Radiat Oncol, 2012. 22(2): p. 128–42. [DOI] [PubMed] [Google Scholar]
- 3.Seiwert TY, et al. , Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol, 2016. 17(7): p. 956–965. [DOI] [PubMed] [Google Scholar]
- 4.Ferris RL, et al. , Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med, 2016. 375(19): p. 1856–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pardoll DM, The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer, 2012. 12(4): p. 252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harlin H, et al. , Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res, 2009. 69(7): p. 3077–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messina JL, et al. , 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep, 2012. 2: p. 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas, N., Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature, 2015. 517(7536): p. 576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keck MK, et al. , Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res, 2015. 21(4): p. 870–81. [DOI] [PubMed] [Google Scholar]
- 10.Ozerov IV, et al. , In silico Pathway Activation Network Decomposition Analysis (iPANDA) as a method for biomarker development. Nat Commun, 2016. 7: p. 13427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozawa H, et al. , SMAD4 Loss Is Associated with Cetuximab Resistance and Induction of MAPK/JNK Activation in Head and Neck Cancer Cells. Clin Cancer Res, 2017. 23(17): p. 5162–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makarev E, et al. , In silico analysis of pathways activation landscape in oral squamous cell carcinoma and oral leukoplakia. Cell Death Discov, 2017. 3: p. 17022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maldonado L, et al. , Integrated transcriptomic and epigenomic analysis of ovarian cancer reveals epigenetically silenced GULP1. Cancer Lett, 2018. 433: p. 242–251. [DOI] [PubMed] [Google Scholar]
- 14.Puram SV, et al. , Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell, 2017. 171(7): p. 1611–1624 e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schelker M, et al. , Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun, 2017. 8(1): p. 2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehrenbacher L, et al. , Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet, 2016. 387(10030): p. 1837–46. [DOI] [PubMed] [Google Scholar]
- 17.Ionescu DN, et al. , Harmonization of PD-L1 testing in oncology: a Canadian pathology perspective. Curr Oncol, 2018. 25(3): p. e209–e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HR, et al. , PD-L1 expression on immune cells, but not on tumor cells, is a favorable prognostic factor for head and neck cancer patients. Sci Rep, 2016. 6: p. 36956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fourcade J, et al. , Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J Exp Med, 2010. 207(10): p. 2175–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wherry EJ and Kurachi M, Molecular and cellular insights into T cell exhaustion. Nat Rev Immunol, 2015. 15(8): p. 486–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung CH, et al. , Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell, 2004. 5(5): p. 489–500. [DOI] [PubMed] [Google Scholar]
- 22.Snyder A, et al. , Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med, 2014. 371(23): p. 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rizvi NA, et al. , Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science, 2015. 348(6230): p. 124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Allen EM, et al. , Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science, 2015. 350(6257): p. 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandal R, et al. , The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight, 2016. 1(17): p. e89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swanson MS and Sinha UK, Rationale for combined blockade of PD-1 and CTLA-4 in advanced head and neck squamous cell cancer-review of current data. Oral Oncol, 2015. 51(1): p. 12–5. [DOI] [PubMed] [Google Scholar]
- 27.Seiwert TY, et al. , Integrative and comparative genomic analysis of HPV-positive and HPV-negative head and neck squamous cell carcinomas. Clin Cancer Res, 2015. 21(3): p. 632–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spranger S, et al. , Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med, 2013. 5(200): p. 200ra116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao X, et al. , TIM-3 expression characterizes regulatory T cells in tumor tissues and is associated with lung cancer progression. PLoS One, 2012. 7(2): p. e30676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wing K, et al. , CTLA-4 control over Foxp3+ regulatory T cell function. Science, 2008. 322(5899): p. 271–5. [DOI] [PubMed] [Google Scholar]
- 31.Ritprajak P and Azuma M, Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol, 2015. 51(3): p. 221–8. [DOI] [PubMed] [Google Scholar]
- 32.Ferris RL, Immunology and Immunotherapy of Head and Neck Cancer. J Clin Oncol, 2015. 33(29): p. 3293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.D'Incecco A, et al. , PD-1 and PD-L1 expression in molecularly selected non-small-cell lung cancer patients. Br J Cancer, 2015. 112(1): p. 95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massague J, TGFbeta in Cancer. Cell, 2008. 134(2): p. 215–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, et al. , Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer. Oncogene, 2009. 28(50): p. 4491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, et al. , Caspase-8 mutations in head and neck cancer confer resistance to death receptor-mediated apoptosis and enhance migration, invasion, and tumor growth. Mol Oncol, 2014. 8(7): p. 1220–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riaz N, et al. , Unraveling the molecular genetics of head and neck cancer through genome-wide approaches. Genes Dis, 2014. 1(1): p. 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia J and Lizcano F, KDM4C Activity Modulates Cell Proliferation and Chromosome Segregation in Triple-Negative Breast Cancer. Breast Cancer (Auckl), 2016. 10: p. 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shangary S and Wang S, Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res, 2008. 14(17): p. 5318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spranger S and Gajewski TF, A new paradigm for tumor immune escape: beta-catenin-driven immune exclusion. J Immunother Cancer, 2015. 3: p. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spranger S, Bao R, and Gajewski TF, Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature, 2015. 523(7559): p. 231–5. [DOI] [PubMed] [Google Scholar]
- 42.Sweis RF, et al. , Molecular Drivers of the Non-T-cell-Inflamed Tumor Microenvironment in Urothelial Bladder Cancer. Cancer Immunol Res, 2016. 4(7): p. 563–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spranger S and Gajewski TF, Tumor-intrinsic oncogene pathways mediating immune avoidance. Oncoimmunology, 2016. 5(3): p. e1086862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brennan K, et al. , NSD1 inactivation defines an immune cold, DNA hypomethylated subtype in squamous cell carcinoma. Sci Rep, 2017. 7(1): p. 17064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otsuka A, et al. , Hedgehog pathway inhibitors promote adaptive immune responses in basal cell carcinoma. Clin Cancer Res, 2015. 21(6): p. 1289–97. [DOI] [PubMed] [Google Scholar]
- 46.Koole K, et al. , FGFR1 Is a Potential Prognostic Biomarker and Therapeutic Target in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res, 2016. 22(15): p. 3884–93. [DOI] [PubMed] [Google Scholar]
- 47.Saloura V, et al. , WHSC1L1-mediated EGFR mono-methylation enhances the cytoplasmic and nuclear oncogenic activity of EGFR in head and neck cancer. Sci Rep, 2017. 7: p. 40664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saloura V, et al. , WHSC1L1 drives cell cycle progression through transcriptional regulation of CDC6 and CDK2 in squamous cell carcinoma of the head and neck. Oncotarget, 2016. 7(27): p. 42527–42538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melero I, et al. , T-cell and NK-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov, 2014. 4(5): p. 522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uppaluri R, et al. , Neoadjuvant pembrolizumab in surgically resectable, locally advanced HPV negative head and neck squamous cell carcinoma (HNSCC). Journal of Clinical Oncology, 2017. 35(15_suppl): p. 6012–6012. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Relative abundance of the CD4+ T cell subtypes (naive-like and regulatory) was estimated from bulk RNA-Seq data by mathematical deconvolution using iPANDA algorithm and Treg/Th1 ratios were calculated to serve as numerical surrogates of the relative infiltration of tumors with different subsets of CD4+ TILs.
Supplementary Figure 2. TCIP-H tumors correlated significantly with the mesenchymal subtype, while the TCIP-L tumors correlated with the basal and classical subtypes, irrespective of HPV status (TCGA) (* p<0.05, t-test).
Supplementary Figure 3. (A) Kaplan Meier estimates of overall survival based on TCIP status in the TCGA SCCHN patient cohort (TCIP-H - orange line, TCIP-L - blue line). (B) Multivariate analysis using Cox regression for TCIP status, stage and HPV status in the TCGA SCCHN patient cohort.
Supplementary Figure 4. The 12-chemokine gene expression signature was interrogated across different tumor cell components using the publicly available single-cell RNA sequencing data from 18 SCCHN patients.
Supplementary Table 1. List of KEGG pathways included for the gene set enrichment analysis.
Supplementary Table 2. Demographic and clinical characteristics of the patients from the CHGC cohort segregated by their TCIP status.
Supplementary Table 3. Demographic and clinical characteristics of the patients from the TCGA dataset segregated by their TCIP status.
Supplementary Table 4. Immunohistochemistry scoring for PD-L1 in immune and tumor cells or both from 134 SCCHN tumors of the CHGC dataset. Two different cut-offs were implemented.
Supplementary Table 5. List of iPANDA signaling pathways concurrently dysregulated (upregulated or downregulated) in TCIP-H patients from TCGA and CHGC cohorts. All shared pathways (left) and main shared pathways (right) are shown.
Supplementary Table 6. List of genes frequently mutated in TCIP-H and TCIP-L tumors derived from TCGA dataset.