Abstract
Contamination of toxic spore-forming bacteria is problematic since spores can survive a plethora of disinfection chemicals and it is hard to rapidly detect if the disinfection chemical has inactivated the spores. Thus, robust decontamination strategies and reliable detection methods to identify dead from viable spores are critical. In this work, we investigate the chemical changes of Bacillus thuringiensis spores treated with sporicidal agents such as chlorine dioxide, peracetic acid, and sodium hypochlorite using laser tweezers Raman spectroscopy. We also image treated spores using SEM and TEM to verify if we can correlate structural changes in the spores with changes to their Raman spectra. We found that over 30 min, chlorine dioxide did not change the Raman spectrum or the spore structure, peracetic acid showed a time-dependent decrease in the characteristic DNA/DPA peaks and ∼20% of the spores were degraded and collapsed, and spores treated with sodium hypochlorite showed an abrupt drop in DNA and DPA peaks within 20 min and some structural damage to the exosporium. Structural changes appeared in spores after 10 min, compared to the inactivation time of the spores, which is less than a minute. We conclude that vibrational spectroscopy provides powerful means to detect changes in spores but it might be problematic to identify if spores are live or dead after a decontamination procedure.
Introduction
A spore is an inactive seed-like form that some bacteria species can take to survive in a hostile environment. When faced with unfavorable conditions such as lack of food, these bacteria form spores to protect themselves in a process called sporulation.1 During sporulation, the vegetative cell undergoes an asymmetric division and engulfs the future spore (called the forespore). The mother cell then builds multiple protective layers around the forespore before finally bursting and releasing the completed spore into the environment.2 Spores are metabolically inactive but they contain the complete genome of the species as well as the cellular machinery and receptors needed to germinate back into vegetative cells again upon contact with favorable conditions. As long as the bacteria remain in spore form, they can survive circumstances that would kill a vegetative cell. For example, spores can survive temperatures below freezing and above 100 °C, exposure to strong acids (including stomach acid), antibiotics, ethanol, quaternary ammonium, and peroxide-based agents.3 Further, spores can survive in the environment for a very long time, easily into decades, such as with B. anthracis spores, unless decontaminated with strong chemical agents like formaldehyde or sodium hypochlorite.4
This extreme durability poses many problems for society as spores cause diseases in both humans and animals. For example, spores such as B. cereus and C. perfringens are common causes of food poisoning5 and C. difficile is a cause of colitis diarrhea. Canned food can become contaminated with C. botulinum spores producing dangerous botulin toxin. In cases of infection by these bacteria, their durability puts extra strain on society due to the harsh decontamination methods needed to deal with them.6 For example, hospital fabrics from C. difficile patients in hospitals cannot be washed with other fabrics as the spores will survive the high-temperature washes and contaminate all fabrics in the batch.7 Further, spores from the Bacillus genus such as those of B. anthracis present a potential biological warfare hazard since these spores are lethal and difficult to decontaminate.8
Several effective decontamination methods using chemicals exist. Chlorination is a popular approach; however, many strains of the Bacillus genus exhibit apparent resistance toward chlorination disinfection.9 Other proven effective decontamination chemicals for Bacillus strains are chlorine dioxide, sodium hypochlorite, and peracetic acid.10 Even though these are indeed effective, care must be taken since these compounds are unstable in regular conditions: chlorine dioxide and sodium hypochlorite decay and release chlorine (in itself a toxic gas), especially in the sunlight, while peracetic acid decays back to acetic acid and hydrogen peroxide (which, in turn, decays to water and oxygen).11 While these chemicals decay into nonlethal components, they are initially very toxic to a plethora of organisms as well as human skin cells.12 Thus, to assess if a decontamination procedure is successful without overusing the sporicidal chemical, it is important to detect if a bacterial spore is dead or alive.
To identify viable bacteria in samples, vibrational spectroscopy techniques, such as Raman spectroscopy, have been suggested and used.13−16 With Raman spectroscopy, it is also possible to identify key molecular components of bacterial spores not found in vegetative cells. For example, dipicolinic acid (DPA), a major protective component in the spore core in dormant spores,17 and amide peaks related to the spore protein content.18 Raman spectroscopy has also been used for species-specific spore detection and assessment of inactivation procedures.19 However, whether Raman methods can reliably differentiate between intact, damaged, and inactivated spores has not been investigated thoroughly. Therefore, in this work, we use laser tweezers Raman spectroscopy (LTRS) and electron microscopy (SEM/TEM) imaging to determine whether spore inactivation affects the Raman spectrum and the spore structure. LTRS allows us to isolate and move a single spore (“trap it”) and simultaneously measure its Raman spectrum to gain insight into its molecular changes during chemical exposure, and EM imaging allows us to observe exterior and interior cell structure changes.
Experimental Section
Laser Tweezers Raman Spectroscopy Setup
We used our optical trap and an LTRS instrument that is built around a modified inverted microscope (IX71, Olympus).20,21 We have shown an illustration of the system in Figure 1. To trap spores and acquire their corresponding Raman spectra, we used a Gaussian laser beam (TEM00, M2 < 1.1–1.3) from a continuous wave laser (CRL-DL808-120-S-US-0.5, CrystaLaser) operating at 808 nm. To measure the Raman spectrum, we collected the back-scattered light with the microscope objective. First, we passed the back-scattered light through a notch filter (NF808-34, Thorlabs) to remove the Rayleigh scattered light.22 Then, to maximize the spectral resolution, we expanded the beam to fill the spectrometer’s numerical aperture using a telescope. Further, to increase the signal to noise ratio, we placed a 150 μm diameter pinhole in the focal point of the telescope to avoid collecting unwanted light. Finally, we coupled the light into our spectrometer (Model 207, McPherson) through a 150 μm wide entrance slit where an 800 lp/mm holographic grating disperses the light, and the spectrum was imaged using a Peltier cooled CCD detector (Newton 920N-BR-DD XW-RECR, Andor) operated at −95 °C. Please see the Supporting Information for more details.
Figure 1.
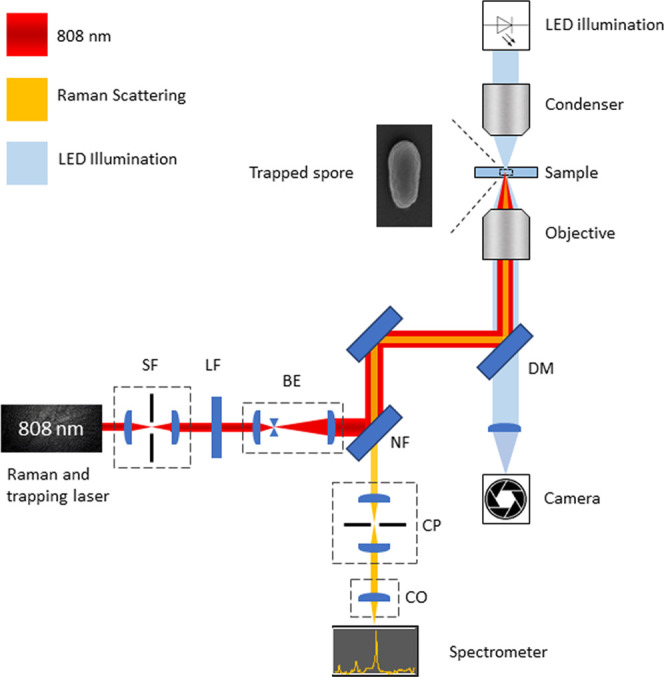
Illustration of the LTRS setup used to acquire Raman spectra of individual spores. The optical system consists of a spatial filter (SF), beam expander (BE), 808 nm line filter (LF), 808 nm notch filter (NF), dichroic 650 shortpass mirror (DM), confocal pinhole (CP), and coupling optics for a spectrometer (CO). To illuminate a sample, we used an LED and acquired images using a CMOS camera.
Verifying Viability of Treated Spores
Spores were incubated with the sporicidal compound for 1, 10, or 30 min. Treated spore samples were centrifuged twice and the supernatant was discarded to remove the leftover sporicidal agent and resuspended in a 5% sodium thiosulphate neutralizing solution for at least 10 min. The untreated control was centrifuged in a similar manner for consistency. Neutralized samples were then serially diluted in deionized water to 10 m–7 concentration and 10 μL drops were plated onto TSA plates and grown at 30 °C overnight. Colonies were counted and compared with the untreated control.
SEM Imaging
For SEM imaging of samples, we first prepared a glass coverslip by adding a 20 μL drop of a 0.1% poly-l-lysine solution (Sigma-Aldrich) to the coverslip and allowed the drop to evaporate. We marked the location of the drop on the opposite side to make it easier to find the spores. Excessive lysine was removed by gently pouring 2 mL of water to flow over the slide. A 3 μL drop of the spore suspension was then added on top on the poly-l-lysine drop. When imaging with sporicidal chemicals, we added the chemical on top of the spores and incubated for 30 min. Then, the sample was cleaned by again allowing 2 mL of water to flow over the sample to remove the sporicidal chemical and the sample was left to dry completely. We then coated the sample with a 5 nm layer of platinum, using a Quorum Q150T-ES sputter coater. The samples were then imaged using a Carl Zeiss Merlin FESEM electron microscope to see the spores using InLens and SE-2 imaging modes at a magnification of ×50,000. To ensure that the observed spores are representative of the sample, 20 fields of view were imaged for each sample.
TEM Imaging
Samples for TEM were prepared as liquid suspensions of spores after 30 min of treatment with peracetic acid, sodium hypochlorite, and chlorine dioxide as before, as well as an untreated sample suspended in water. After the incubation, samples with sporicidal chemicals were centrifuged and resuspended in MQ water twice to wash off the chemical. Spores were fixed with 2.5% glutaraldehyde (TAAB Laboratories, Aldermaston, England) in 0.1 M PHEM buffer and further postfixed in 1% aqueous osmium tetroxide. They were further dehydrated in ethanol and acetone and finally embedded in Spurr’s resin (TAAB Laboratories, Aldermaston, England). Ultrathin sections (70 nm) were then post contrasted in uranyl acetate and Reynolds lead citrate. Samples were examined using a Talos L120C (FEI, Eindhoven, The Netherlands) operating at 120 kV. Micrographs were acquired with a Ceta 16M CCD camera (FEI, Eindhoven, The Netherlands) using TEM Image and Analysis software ver. 4.17 (FEI, Eindhoven, The Netherlands).
Results and Discussion
Mapping Vibration Peaks of the Spores Using LTRS
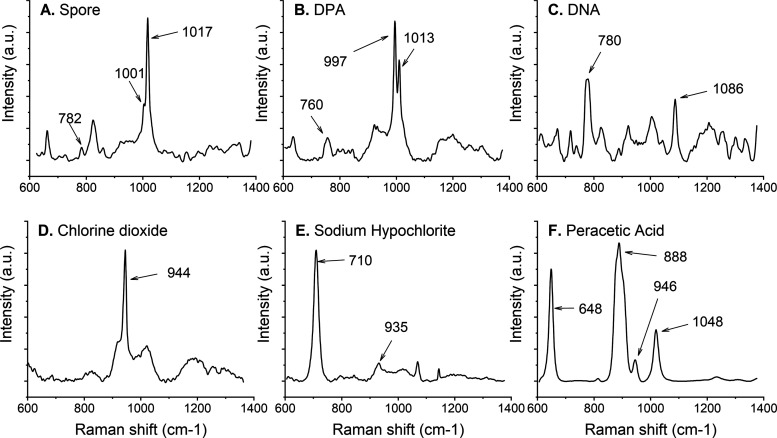
To measure the impact of the sporicidal chemicals on the spores’ Raman spectra, we first assessed the vibrational peaks in the absence of chemicals on purified B. thuringiensis spores using LTRS. One of the main constituents and the most common biomarker for these spores is CaDPA, which accounts for approximately 20% of the spore core weight.23 CaDPA is a protective component located in the core and is essential for a spore’s full resistance to wet heat. Figure 2A top panel shows the average Raman spectrum of (A) three dormant spores sequentially trapped in the LTRS, a spectrum of pure DPA (B), and DNA (C), and the spectra of purified sporicidal chemicals used in this study (D–F). In previous studies, it has been reported that there are no significant changes in the amide bands >1400 cm–1 in the presence of chemicals.19 To verify this, we investigated the 1580 cm–1 amide band for changes in the Raman intensity (Figure S1). We saw no change for chlorine dioxide or peracetic acid. For sodium hypochlorite, the changes in amide I band intensity followed the changes in the DPA peak intensity. Therefore, to allow for a fast acquisition rate of Raman signals, we limited the spectral measurement range of our system to 600–1400 cm–1, which is where CaDPA and DNA peaks are to be found.17
Figure 2.
Raman spectra of spores, their major components, and the sporicidal chemicals used. We marked the major peaks with arrows. Each panel is an average of three spectra. (A) B. thuringiensis spores, (B) dipicolinic acid (DPA), (C) double-stranded DNA, (D) chlorine dioxide, (E) sodium hypochlorite, and (F) peracetic acid.
Raman spectra of purified DPA and DNA are seen in Figure 2B,2C, respectively. We marked the major CaDPA and DNA peaks at 1017 and 782 cm–1 in the top panel. These peaks are slightly shifted in the purified DPA and DNA spectra as in the purified solutions, the bond length may be slightly different than when located in the spore. In particular, this is true with regards to pH and interactions with other spore components. For DNA, the 782 cm–1 peak is related to the O–P–O backbone or the cytosine ring breathing mode. Another visible peak in the pure DNA Raman spectrum is at 1086 cm–1, related to the phosphodiester stretching peak. This peak, however, is difficult to observe in the whole spore spectrum.24−26 We also marked the phenylalanine peak, a major structural component of the spore, with the Raman peak at 1001 cm–1. Overall, the peaks observed using our LTRS are consistent with what is found in the literature using similar approaches. We have also added SEM and TEM images of untreated spores in Figures 4A, 5A, and S2.
Figure 4.
SEM of B. thuringiensis spores treated with different sporicidal chemicals for 30 min, imaged at 50,000 magnification. The loose exosporium surrounding the spore (white arrows) can be seen for spores in (A) purified and deionized water, (B) chlorine dioxide, and (D) peracetic acid. No exosporium in spores treated with (C) sodium hypochlorite is seen. A partially degraded spore is also seen in the image of (D) a peracetic acid-treated spore (yellow arrow). Scale bars are 1 μm.
Figure 5.
TEM of B. thuringiensis spores treated with different sporicidal chemicals for 30 min, imaged at 73,000 magnification and 27,000 magnification (inset). We labeled the structural layers of the spore as follows: green arrows: exosporium; black arrows: spore coat; blue arrows: cortex; and yellow arrows: core. (A) Untreated spore in water. (B) Spores treated with chlorine dioxide appear no different than control. (C) Spores treated with sodium hypochlorite are degraded, and the core is no longer electron-dense, indicating DPA release. (D) Spores treated with peracetic acid have a fragmented spore coat.
Chlorine Dioxide Treatment Does Not Affect the Raman Signal and the Spore Structure
Chlorine dioxide is a microbicidal and sporicidal chemical, and it has been proven effective during decontamination of spores without being very harmful to human beings. However, the reported mechanism of action is not consistent in the literature, especially for DNA. For example, Zhu et al. reported that chlorine dioxide at concentrations higher than 100 ppm damages DNA in Saccharomyces cerevisiae.27 At the same time, other publications suggest that chlorine dioxide does not damage DNA directly.28,29 Another proposed effect is protein denaturation by oxidation of tryptophan and tyrosine.30 The concentrations used in these studies vary significantly from only a few ppm to several hundred ppm. To investigate the impact of chlorine dioxide on the Raman peaks and the spore structure, we treated spores with chlorine dioxide at concentrations of 200, 400, and 750 ppm, with the latter being the upper end of the concentrations used in the literature and carefully recorded the time of exposure.
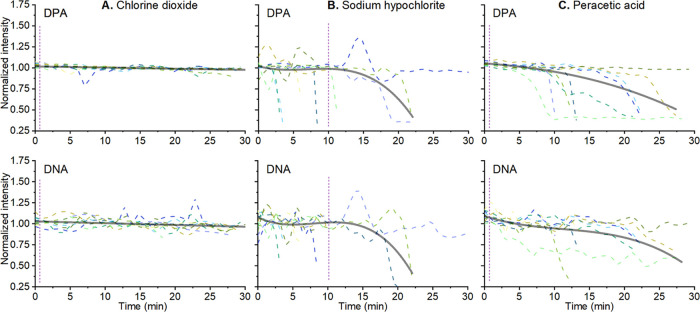
Spores were trapped using our LTRS and the Raman spectrum of individual spores was acquired. We found that the Raman spectrum of spores was not affected by incubation with chlorine dioxide. That is, we saw no detectable spore protein degradation or DNA disruption even at the highest concentration of 750 ppm (Figures 3A and S5A). Note that this is 75 times higher than the 10 ppm reported lethal to spores.28 Indeed, the spores were confirmed inactivated using a viability study with a 3 log reduction in CFU after 1 min and a minimum 5 log reduction compared to the control after 10 min, (Figures S6 and S7). We noted a gradual minor decrease in the signal intensity of 4% for DPA and 6% for DNA over the 30 min measurement time. Due to the smaller absolute intensity of the DNA peak (Figure 3A), the changes in the normalized intensity had more noise than in the DPA peak. Since these observed drifts are small and occur over a 30 min timescale, they do not influence the results.
Figure 3.
Change in the normalized intensity of the Raman (smoothed using Loess smoothing) peak associated with DPA (1017 cm–1) and DNA (cm–1) of single B. thuringiensis spores (n = 10 for each panel). (A) Spores were treated with 0.075% chlorine dioxide (B), 0.5% sodium hypochlorite (C), and 1% peracetic acid. The solid gray lines are averages of all data. Vertical purple lines indicate the time for a minimum 3 log reduction in viable spores.
Chlorine dioxide has been reported to react with and damage proteins, targeting the sulfur bonds in cysteine and methionine in particular.31−33 However, we did not see a decrease in Raman peaks associated with proteins, such as the amide I and amide II band. We used SEM and TEM imaging to look for surface and internal structural differences between untreated and treated spores that may not be seen with Raman alone. Both SEM (n = 112) and TEM (n = 107) images of spores treated with chlorine dioxide (Figures 4B, 5B, and S8) do not show any visible damage to the exosporium, spore coat, or internal structure, confirming the data obtained using Raman spectroscopy.
It is notable that although the spore coat is rich in cysteine,34,35 there appears to be no significant sign of spore protein degradation in Raman spectra and EM images. This may be due to the way the chlorine dioxide reacts with cysteine. After attacking the sulfur bond and forming a free radical, the likely reaction product when not at low pH is dimerization into cystine.36 While this works for free-floating amino acid solutions, this will be far slower with a tightly packed spore coat protein in a suspension in water. The degradation of tryptophan would be prevented for the same reasons and any localized degradation of tryptophan would be difficult to see in a spore Raman spectrum, as the main peak of tryptophan is at 1005 cm–1 and would be masked by the phenylalanine and DPA peaks. Overall, our data show that chlorine dioxide inactivates spores but not by general protein degradation.
Note that the specific Raman peak of chlorine dioxide at 944 cm–1 in Figure 2 is consistent with previously published data.37 This peak decreased in intensity during the measurement, being completely gone after approximately 15 min. This disappearance indicates that chlorine dioxide left the solution, either by chemical decomposition into chlorine or by diffusion out of the water. In summary, we conclude that spores inactivated by treatment with chlorine dioxide did not show any major changes in their Raman spectra as measured by LTRS, or exosporium and spore coat as visualized using SEM, or internally as visualized using TEM.
Sodium Hypochlorite Treatment Causes Rapid DPA Loss after a Lag Time and Spore Decomposition
Next, we analyzed spores treated with a 0.5% solution sodium hypochlorite (bleach) with a pH of 11.55. This concentration was previously reported to be sporicidal.38 Sodium hypochlorite is a cheap and prevalent decontamination agent that works by degrading organic material in several reactions: saponification of fatty acids and neutralization and chloramination of amino acids.39 As such, hypochlorite causes oxidative damage to lipids, protein, and DNA. We found that sodium hypochlorite causes the largest change in the Raman spectrum of a spore and strongly affects the DPA peak (Figures 3B and S5B). We observe that only one spore shows no DPA peak reduction over the entire measurement time. Though initially unaffected, the rest of the spores completely lose their major DPA peak at 1017 cm–1. However, there is a lag time before the decrease in the Raman-associated DPA peaks begins. The lag time varies from 4 to 22 min, but when the DPA starts to decrease, it does so rapidly. In general, the DPA signal can go from a full peak to a complete loss in less than a minute (2 min for the slowest spore). Finally, we note that the Raman peak of DNA correlates with the DPA peak in both the lag time and the rate of decrease. The loss of DPA in hypochlorite-treated spores is interesting. It was previously reported that spores treated with hypochlorite do not lose their DPA from treatment, but they do germinate slowly and then release their DPA but cannot grow further.40,41 When observed using SEM, the spores treated with sodium hypochlorite appear to have significant changes to the exosporium layer (Figure 4C), whereas the spore coat is still intact. A total of 33 of the 189 (17%) spores imaged had a visible exosporium, while it was missing in the rest. Exosporium degradation is expected since the exosporium is a thin protein layer that is permeable to small molecules like hypochlorite, and will lose integrity more easily than the spore coat.39
Our TEM observations confirm that the spores lose their DPA (Figures 5C, S3, and S9) and the core appears very discolored in 24 of the 27 spores imaged, (88%) compared to untreated spores Figure 5A. There is also visible degradation of the cortex, spore coat, and exosporium. The level of disruption of the spores varied from still having recognizable spore structural features to very pale outlines with completely unstained internal content. The TEM samples of hypochlorite-treated spores had very few visible spores in general, despite starting from the same high concentration stock. This is why we only imaged 27 spores.
Loss of DPA from spores exposed to chemicals has been reported with spores releasing DPA rapidly after an initial lag phase.28,40 A similar behavior was also seen in spores germinating in nutrient broth and germinants.42 However, in these studies, the DPA release from the spore took several minutes from the end of the lag phase to a complete loss of the DPA signal. By contrast, in our experiments, the DPA release takes only 1 min, typically the time between two individual measurements. It is unlikely for this difference to be due to temperature or germinants (the spores were suspended in germinant-free MQ water at 25 °C). It is also unlikely that heating from the Raman laser beam catalyzed the DPA release since the laser power was only 5 mW, which is comparable to 3 mW used for B. subtilis in studies such as by Peng et al.41
We attribute the observed differences in similar studies to the different species used. We used B. thuringiensis, whereas B. subtilis was used in the decontamination studies.28,40 A key difference is that unlike several other Bacillus species, B. subtilis spores lack an exosporium, which means that their germination mechanics is independent of exosporium damage.1 This means that sodium hypochlorite acts on the spore coat and cortex, starting its breakdown.43,44 In exosporium-producing spores, the exosporium has been reported to be involved in regulating germination.1,45 Therefore, its degradation can lead to DPA release in B. thuringiensis. This effect also needs to be thoroughly studied for other Bacillus species. Thus, we conclude from our experiments with 0.5% sodium hypochlorite that it affects the Raman spectra of the spores by significantly reducing the DPA and DNA peaks after a lag time of a few minutes up to about 22 min. After the lag time, the DPA release is initiated, and within 1 min, all DPA is released. SEM and TEM images together show significant changes to the exosporium layer and moderate degradation of the spore coat, cortex, and core. We verified that spores were inactivated by growing the treated spore suspension and noted a 1 log reduction after 1 min and a 5 log reduction after 10 min (Figures S6 and S7).
Peracetic Acid Treatment Causes Rapid DPA Loss after a Lag Time and Spore Coat Fragmentation
Peracetic acid is an oxidizing disinfectant agent efficient in inactivating microorganisms. It inactivates via denaturation of proteins, enzymes, and metabolites by oxidation of sulfhydryl and sulfur bonds.46 Peracetic acid has been shown to work against spores and it is effective in solution.47 We first measured the Raman spectrum of peracetic acid itself (Figure 2F) and confirmed that it is consistent with previous studies and that it does not decrease over the measurement time.48 We treated and investigated spores incubated with 1% peracetic acid. This concentration was chosen as the upper end of the reported sporicidal concentrations of peracetic acid.49 As with sodium hypochlorite, there was a variation in the lag time before DPA loss, ranging from 5 to 18 min. The speed with which the spores lost the DPA also varied. Out of 10 spores, only 2 lost their DPA in a minute, similar to the spores treated with sodium hypochlorite, while 7 lost the DPA peak more slowly, taking from 2 to 10 min (Figures 3C and S5C). One spore did not lose its DPA over the measurement time. There is a similar downward trend in the peak intensity of DNA. This trend continues over the measurement time (30 min), which is significantly slower than the reported inactivation time of the spores; at the concentration of 1% peracetic acid, spores are expected to be inactivated in less than a minute.10 As with the other experiments, we verified that the treated spores were inactivated by growing the treated spore suspension and noted a 5 log reduction in CFU after 1 min (Figure S7).
When observed using SEM, some spores appeared broken down and degraded, while others were still intact (Figure 4D). The panel shows both a degraded and intact spore in the same field of view. Out of the 148 spores imaged, 25 were degraded (17%). The degraded spores may correlate with the ones that lost their Raman DPA and DNA signal rapidly in the LTRS experiments. This variation observed in the SEM is plausible since spores are heterogeneous.50
When observed under TEM, spores treated with a peracetic acid showed a clear difference from the untreated spores. In the untreated spores, the spore coat can be seen as several dark layers (Figure 5A), consistent with its dense multilayer structure.1 The spore coat in 63 out of 64 (99%) peracetic acid-treated spores (Figures 5D, S4, and S10) appeared fragmented, separating and breaking into small pieces. The core, cortex, and exosporium appeared intact. This is consistent with the SEM observations, as spores with a damaged spore coat can lose their structural integrity. The exosporium and the core did not change visually.
Conclusions
Rapid detection, whether a spore disinfection procedure was successful or not, is of significance in many areas. We treated B. thuringiensis spores with common disinfection chemicals, chlorine dioxide, peracetic acid, and sodium hypochlorite, and measured changes in the spore structure and Raman spectra. Chlorine dioxide does not change the Raman spectrum or the spore structure. Peracetic acid shows a time-dependent decrease in the characteristic DNA/DPA peaks; however, it happens much later than the spore inactivation itself. Approximately, 17% of the spore structure is degraded and collapsed, and TEM imaging shows the degradation of the spore coat. Sodium hypochlorite-treated spores show an abrupt drop in DNA and DPA Raman peaks within 20 min. The spore structure was overall intact, though internal structural degradation was observed using TEM and the exosporium layer was reduced in size or removed. In all of these experiments, structural changes appeared over several minutes, compared to the inactivation time of the spores, which is multiple logs in a minute for chlorine dioxide and peracetic acid and 1 log in a minute for sodium hypochlorite. We conclude that vibrational spectroscopy provides powerful means to detect changes in spores. However, it might be problematic to use Raman methods to identify if spores are live or dead directly after a decontamination procedure; no changes in the Raman spectrum occur for chlorine dioxide and changes for the other two chemicals occur significantly slower than the inactivation process itself.
Acknowledgments
This work was supported by the Swedish Research Council (2019-04016) and the Kempestiftelserna (JCK-1916.2). The authors thank Anna-Lena Johansson at FOI for preparing spores in this project. The authors acknowledge the facilities and technical assistance of the Umeå Core Facility for Electron Microscopy (UCEM) at the Chemical Biological Centre (KBC), Umeå University, a part of the National Microscopy Infrastructure NMI (VR-RFI 2016-00968).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c04519.
The laser tweezers Raman Spectroscopy (LTRS) setup; sample preparation and LTRS measurements; changes of the 1400 cm–1 amide peak; TEM of untreated spores; TEM of peracetic acid-treated spores; the change in the Raman spectrum of a single spore over time; serial dilution drop counts of samples treated with sporicidal chemicals for 1 min; and additional fields of view of B. thuringiensis treated with 1% peracetic acid for 30 min (24 SEM and 24 TEM fields of view) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Stewart G. C. The exosporium layer of bacterial spores: a connection to the environment and the infected host. Microbiol. Mol. Biol. Rev. 2015, 79, 437–457. 10.1128/MMBR.00050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D.; Dworkin J. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol. Rev. 2012, 36, 131–148. 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawley W. N.; Underwood S.; Freeman J.; Baines S. D.; Saxton K.; Stephenson K.; Owens R. C.; Wilcox M. H. Efficacy of Hospital Cleaning Agents and Germicides Against Epidemic Clostridium difficile Strains. Infect. Control Hosp. Epidemiol. 2007, 28, 920–925. 10.1086/519201. [DOI] [PubMed] [Google Scholar]
- Manchee R. J.; Broster M. G.; Anderson I. S.; Henstridge R. M.; Melling J. Decontamination of Bacillus anthracis on Gruinard Island?. Nature 1983, 303, 239–240. 10.1038/303239a0. [DOI] [PubMed] [Google Scholar]
- Andersson A.; Ronner U.; Granum P. E. What problems does the food industry have with the spore-forming pathogens Bacillus cereus and Clostridium perfringens?. Int. J. Food Microbiol. 1995, 28, 145–155. 10.1016/0168-1605(95)00053-4. [DOI] [PubMed] [Google Scholar]
- Wiegand P. N.; Nathwani D.; Wilcox M.; Stephens J.; Shelbaya A.; Haider S. Clinical and economic burden of Clostridium difficile infection in Europe: a systematic review of healthcare-facility-acquired infection. J. Hosp. Infect. 2012, 81, 1–14. 10.1016/j.jhin.2012.02.004. [DOI] [PubMed] [Google Scholar]
- Dyer C.; Hutt L. P.; Burky R.; Joshi L. T. Biocide resistance and transmission of Clostridium difficile spores spiked onto clinical surfaces from an American health care facility. Appl. Environ. Microbiol. 2019, 85, e01090-19 10.1128/AEM.01090-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A. K. Anthrax: A disease of biowarfare and public health importance. World. J. Clin. Cases 2015, 3, 20. 10.12998/wjcc.v3.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes F. A.; Hissa D. C.; Angelim A. L.; Pinto N. W.; Grangeiro T. B.; Melo V. M. Diversity of a Chlorine-Resistant Bacillus Population Isolated from a Wastewater Treatment Station. Water Environ. Res. 2012, 84, 274–281. 10.2175/106143012X13280358613462. [DOI] [PubMed] [Google Scholar]
- Leggett M. J.; Spencer Schwarz J.; Burke P. A.; McDonnell G.; Denyer S. P.; Maillard J. Y. Mechanism of sporicidal activity for the synergistic combination of peracetic acid and hydrogen peroxide. Appl. Environ. Microbiol. 2016, 82, 1035–1039. 10.1128/AEM.03010-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper W. J.; Jones A. C.; Whitehead R. F.; Zika R. G. Sunlight-induced photochemical decay of oxidants in natural waters: implications in ballast water treatment. Environ. Sci. Technol. 2007, 41, 3728–3733. 10.1021/es062975a. [DOI] [PubMed] [Google Scholar]
- Stratilo C. W.; Crichton M. K.; Sawyer T. W. Decontamination efficacy and skin toxicity of two decontaminants against Bacillus anthracis. PLoS One 2015, 10, e0138491 10.1371/journal.pone.0138491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquharson S.; Gift A. D.; Maksymiuk P.; Inscore F. E. Rapid dipicolinic acid extraction from Bacillus spores detected by surface-enhanced Raman spectroscopy. Appl. Spectrosc. 2004, 58, 351–354. 10.1366/000370204322886735. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Young M. A.; Lyandres O.; Van Duyne R. P. Rapid detection of an anthrax biomarker by surface-enhanced Raman spectroscopy. J. Am. Chem. Soc. 2005, 127, 4484–4489. 10.1021/ja043623b. [DOI] [PubMed] [Google Scholar]
- Boardman A. K.; Wong W. S.; Premasiri W. R.; Ziegler L. D.; Lee J. C.; Miljkovic M.; Klapperich C. M.; Sharon A.; Sauer-Budge A. F. Rapid detection of bacteria from blood with surface-enhanced Raman spectroscopy. Anal. Chem. 2016, 88, 8026–8035. 10.1021/acs.analchem.6b01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse L. F.; Safir F.; Ho C.-S.; Hasbach X.; Khuri-Yakub B.; Jeffrey S. S.; Saleh A. A.; Dionne J. Toward rapid infectious disease diagnosis with advances in surface-enhanced Raman spectroscopy. J. Chem. Phys. 2020, 152, 240902 10.1063/1.5142767. [DOI] [PubMed] [Google Scholar]
- Huang S. S.; Chen D.; Pelczar P. L.; Vepachedu V. R.; Setlow P.; Li Y. Q. Levels of Ca2+-dipicolinic acid in individual Bacillus spores determined using microfluidic Raman tweezers. J. Bacteriol. 2007, 189, 4681–4687. 10.1128/JB.00282-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito A. P.; Talley C. E.; Huser T.; Hollars C. W.; Schaldach C. M.; Lane S. M. Analysis of single bacterial spores by micro-Raman spectroscopy. Appl. Spectrosc. 2003, 57, 868–871. 10.1366/000370203322102979. [DOI] [PubMed] [Google Scholar]
- Stöckel S.; Schumacher W.; Meisel S.; Elschner M.; Rösch P.; Popp J. Raman spectroscopy-compatible inactivation method for pathogenic endospores. Appl. Environ. Microbiol. 2010, 76, 2895–2907. 10.1128/AEM.02481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrisson J.; Singh B.; Svenmarker P.; Wiklund K.; Zhang H.; Hakobyan S.; Ramstedt M.; Andersson M. Detecting Bacterial Surface Organelles on Single Cells Using Optical Tweezers. Langmuir 2016, 32, 4521–4529. 10.1021/acs.langmuir.5b03845. [DOI] [PubMed] [Google Scholar]
- Stangner T.; Dahlberg T.; Svenmarker P.; Zakrisson J.; Wiklund K.; Oddershede L. B.; Andersson M. Cooke–Triplet tweezers: more compact, robust, and efficient optical tweezers. Opt. Lett. 2018, 43, 1990. 10.1364/OL.43.001990. [DOI] [PubMed] [Google Scholar]
- Dahlberg T.; Malyshev D.; Andersson P. O.; Andersson M.. Biophysical Fingerprinting of Single Bacterial Spores Using Laser Raman Optical Tweezers. In Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XXI; International Society for Optics and Photonics, 2020; p 28. [Google Scholar]
- Setlow B.; Atluri S.; Kitchel R.; Koziol-Dube K.; Setlow P. Role of dipicolinic acid in resistance and stability of spores of Bacillus subtilis with or without DNA-protective α/β-type small acid-soluble proteins. J. Bacteriol. 2006, 188, 3740–3747. 10.1128/JB.00212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasalvia M.; Scrima R.; Perna G.; Piccoli C.; Capitanio N.; Biagi P.; Schiavulli L.; Ligonzo T.; Centra M.; Casamassima G.; et al. Exposure to 1.8 GHz electromagnetic fields affects morphology, DNA-related Raman spectra and mitochondrial functions in human lympho-monocytes. PLoS One 2018, 13, e0192894 10.1371/journal.pone.0192894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draux F.; Gobinet C.; Sulé-Suso J.; Trussardi A.; Manfait M.; Jeannesson P.; Sockalingum G. D. Raman spectral imaging of single cancer cells: probing the impact of sample fixation methods. Anal. Bioanal. Chem. 2010, 397, 2727–2737. 10.1007/s00216-010-3759-8. [DOI] [PubMed] [Google Scholar]
- Jo S.; Son J.; Lee B. H.; Dugasani S. R.; Park S. H.; Kim M. K. Vibrational characteristics of DNA nanostructures obtained through a mass-weighted chemical elastic network model. RSC Adv. 2017, 7, 47190–47195. 10.1039/C7RA06125K. [DOI] [Google Scholar]
- Zhu C.; Chen Z.; Yu G. Fungicidal mechanism of chlorine dioxide on Saccharomyces cerevisiae. Ann. Microbiol. 2013, 63, 495–502. 10.1007/s13213-012-0494-8. [DOI] [Google Scholar]
- Young S.; Setlow P. Mechanisms of killing of Bacillus subtilis spores by hypochlorite and chlorine dioxide. J. Appl. Microbiol. 2003, 95, 54–67. 10.1046/j.1365-2672.2003.01960.x. [DOI] [PubMed] [Google Scholar]
- Roller S.; Olivieri V.; Kawata K. Mode of bacterial inactivation by chlorine dioxide. Water Res. 1980, 14, 635–641. 10.1016/0043-1354(80)90121-9. [DOI] [Google Scholar]
- Ogata N. Denaturation of protein by chlorine dioxide: oxidative modification of tryptophan and tyrosine residues. Biochemistry 2007, 46, 4898–4911. 10.1021/bi061827u. [DOI] [PubMed] [Google Scholar]
- Navalon S.; Alvaro M.; Garcia H. Chlorine dioxide reaction with selected amino acids in water. J. Hazard. Mater. 2009, 164, 1089–1097. 10.1016/j.jhazmat.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Noszticzius Z.; Wittmann M.; Kály-Kullai K.; Beregvári Z.; Kiss I.; Rosivall L.; Szegedi J. Chlorine dioxide is a size-selective antimicrobial agent. PLoS One 2013, 8, e79157 10.1371/journal.pone.0079157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V. K.; Sohn M. Reactivity of chlorine dioxide with amino acids, peptides, and proteins. Environ. Chem. Lett. 2012, 10, 255–264. 10.1007/s10311-012-0355-5. [DOI] [Google Scholar]
- Goldman R. C.; Tipper D. J. Bacillus subtilis spore coats: complexity and purification of a unique polypeptide component. J. Bacteriol. 1978, 135, 1091–1106. 10.1128/JB.135.3.1091-1106.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Romero P.; Castro-Córdova P.; Reyes-Ramírez R.; Milano-Céspedes M.; Guerrero-Araya E.; Pizarro-Guajardo M.; Olguín-Araneda V.; Gil F.; Paredes-Sabja D. Clostridium difficile exosporium cysteine-rich proteins are essential for the morphogenesis of the exosporium layer, spore resistance, and affect C. difficile pathogenesis. PLoS Pathog. 2018, 14, e1007199 10.1371/journal.ppat.1007199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison A.; Odeh I. N.; Margerum D. W. Kinetics and mechanisms of chlorine dioxide and chlorite oxidations of cysteine and glutathione. Inorg. Chem. 2006, 45, 8768–8775. 10.1021/ic0609554. [DOI] [PubMed] [Google Scholar]
- Esposito A. P.; Stedl T.; Jonsson H.; Reid P. J.; Peterson K. A. Absorption and Resonance Raman Study of the 2B1 (X)- 2A2 (A) Transition of Chlorine Dioxide in the Gas Phase. J. Phys. Chem. A 1999, 103, 1748–1757. 10.1021/jp984368i. [DOI] [Google Scholar]
- Omidbakhsh N. Evaluation of sporicidal activities of selected environmental surface disinfectants: carrier tests with the spores of Clostridium difficile and its surrogates. Am. J. Infect. Control 2010, 38, 718–722. 10.1016/j.ajic.2010.02.009. [DOI] [PubMed] [Google Scholar]
- Estrela C.; Estrela C. R.; Barbin E. L.; Spanó J. C. E.; Marchesan M. A.; Pécora J. D. Mechanism of action of sodium hypochlorite. Braz. Dent. J. 2002, 13, 113–117. 10.1590/S0103-64402002000200007. [DOI] [PubMed] [Google Scholar]
- Setlow B.; Yu J.; Li Y.-Q.; Setlow P. Analysis of the germination kinetics of individual Bacillus subtilis spores treated with hydrogen peroxide or sodium hypochlorite. Lett. Appl. Microbiol. 2013, 57, 259–265. 10.1111/lam.12113. [DOI] [PubMed] [Google Scholar]
- Peng L.; Chen D.; Setlow P.; Li Y.-q. Elastic and inelastic light scattering from single bacterial spores in an optical trap allows the monitoring of spore germination dynamics. Anal. Chem. 2009, 81, 4035–4042. 10.1021/ac900250x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.; Setlow P.; Li Y.-q. Slow leakage of Ca-dipicolinic acid from individual Bacillus spores during initiation of spore germination. J. Bacteriol. 2015, 197, 1095–1103. 10.1128/JB.02490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman S.; Scott E.; Hutchinson E. Hypochlorite effects on spores and spore forms of Bacillus subtilis and on a spore lytic enzyme. J. Appl. Bacteriol. 1984, 56, 295–303. 10.1111/j.1365-2672.1984.tb01350.x. [DOI] [PubMed] [Google Scholar]
- Russell A. Bacterial spores and chemical sporicidal agents. Clin. Microbiol. Rev. 1990, 3, 99–119. 10.1128/CMR.3.2.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 2014, 196, 1297–1305. 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonnell G.; Russell A. D. Antiseptics and Disinfectants: Activity, Action, and Resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. 10.1128/CMR.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaper S. Synergistic killing of spores of Bacillus subtilis by peracetic acid and alcohol. Int. J. Food Sci. Technol. 1984, 19, 355–360. 10.1111/j.1365-2621.1984.tb00359.x. [DOI] [Google Scholar]
- Popov E.; Eloranta J.; Hietapelto V.; Vuorenpalo V.-M.; Aksela R.; Jäkärä J. Mechanism of decomposition of peracetic acid by manganese ions and diethylenetriaminepentaacetic acid (DTPA). Holzforschung 2005, 59, 507–513. 10.1515/HF.2005.084. [DOI] [Google Scholar]
- Rutala W. A.; Weber D. J.. Guideline for Disinfection and Sterilization in Healthcare Facilities, 2008 (Updated May 2019); Centers for Disease Control and Prevention, 2019. [Google Scholar]
- Chen D.; Huang S. S.; Li Y. Q. Real-time detection of kinetic germination and heterogeneity of single Bacillus spores by laser tweezers Raman spectroscopy. Anal. Chem. 2006, 78, 6936–6941. 10.1021/ac061090e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.