Abstract
During embryonic cortical development, radial glial cells (RGCs) are the major source of neurons, and these also serve as a supportive scaffold to guide neuronal migration. Similar to Vimentin, glial fibrillary acidic protein (GFAP) is one of the major intermediate filament proteins present in glial cells. Previous studies confirmed that prenatal ethanol exposure (PEE) significantly affected the levels of GFAP and increased the disassembly of radial glial fibers. GFAPδ is a variant of GFAP that is specifically expressed in RGCs; however, to the best of our knowledge, there are no reports regarding how PEE influences its expression during cortical development. In the present study, the effects of PEE on the expression and distribution of GFAPδ during early cortical development were assessed. It was found that PEE significantly decreased the expression levels of GFAP and GFAPδ. Using double immunostaining, GFAPδ was identified to be specifically expressed in apical and basal RGCs, and was co-localized with other intermediate filament proteins, such as GFAP, Nestin and Vimentin. Additionally, PEE significantly affected the morphology of radial glial fibers and altered the behavior of RGCs. The loss of GFAPδ accelerated the transformation of RGCs into astrocytes. Using co-immunostaining with Ki67 or phospho-histone H3, GFAPδ+ cells were observed to be proliferative or mitotic cells, and ethanol treatment significantly decreased the proliferative or mitotic activities of GFAPδ+ RGCs. Taken together, the results suggested that PEE altered the expression patterns of GFAPδ and impaired the development of radial glial fibers and RGC behavior. The results of the present study provided evidence that GFAPδ may be a promising target to rescue the damage induced by PEE.
Keywords: prenatal ethanol exposure, glial fibrillary acidic protein δ, radial glial fiber, radial glial cells, embryonic cerebral cortex
Introduction
Prenatal ethanol exposure (PEE) is considered a significant risk factor underlying several functional and structural disorders, including stillbirth, spontaneous abortion, premature birth, intrauterine growth retardation and low birth weight, as well as developmental abnormalities of the central nervous system (CNS), which are collectively referred to as fetal alcohol spectrum disorders (1,2). These deficits may result in lifelong behavioral abnormalities. In the human fetus, PEE-induced abnormalities exhibit several characteristics, including facial abnormalities, growth deficits, lowered intelligence and microcephaly (3,4). In several animal models, including mouse (5), rat (6), macaque (7,8), zebrafish (9) and chick models (10), PEE causes similar birth defects and permanent neurodevelopmental disorders. Although alcohol usage is preventable and a well-established hazard to the fetus, the global consumption of alcohol during pregnancy is still prevalent (9.8%, 95% CI 8.9–11.1) (1,2,11). As previously reported, alcohol usage is an urgent public health issue in China (12). A population-based Chinese study reported that pregnant women in China had a high prevalence rate of alcohol consumption (6.2% in rural and 10.3% in urban areas), although women in China are discouraged from alcohol consumption (13). Therefore, it is important to investigate the mechanisms underlying the effects of prenatal ethanol or alcohol exposure on brain disorders.
Microcephaly is one of the major developmental brain defects associated with maternal ethanol exposure (14). PEE can result in a range of cortical dysfunctions, consisting of abnormal neuronal migration (15), reduced cell viability (16), changes to cortical thickness (17,18) and loss of neurons (8). Several factors are involved in the process of embryonic cortex development, such as morphogens, including fibroblast growth factor (19), bone morphogenetic protein 7 (20) and transforming growth factor β1 (21,22), as well as Notch and Wnt signaling (23,24), radial glial scaffolds for neuronal migration (25,26) and cell-cell communications between cortical neurons and cortical precursor cells (27,28). Among these, several studies have revealed that alterations of radial glial development contribute to the effects of alcohol on the development of the cortex (29–31). In vivo experimental results indicated that ethanol exposure decreases glial fibrillary acidic protein (GFAP) mRNA expression levels in the brains of pups from ethanol-fed Sprague Dawley or Wistar rat mothers and impairs the morphology of radial glial cells (RGCs) (29,31). In vitro studies also revealed that ethanol could affect the distribution and content of the radial glial cytoskeletal proteins GFAP, Vimentin and Nestin (32). Thus, investigation of the effects of prenatal alcohol exposure on radial glial fiber proteins, such as Vimentin, Nestin and GFAP, may shed light on the mechanisms involved in alcohol-induced cortical malformation.
Previous studies have reported that neurogenesis occurs in two primary sites in the mammalian brain where the neural stem cells (NSCs) reside: The subventricular zone (SVZ) of the lateral ventricle and the subgranular zone (SGZ) of the hippocampal dentate gyrus (33). RGCs derived from neuroepithelial cells are ubiquitously present in the developing brain from the start of fetal neurogenesis until their final transformation into astrocytes in certain regions of the brain (34,35). It has been confirmed that RGCs are specialized cells that can generate both neurons and astrocytes (34). In addition to the generation of neurons and intermediate progenitors, RGCs also provide radial glial scaffolding for neuronal migration via a long radial process (36,37). As suggested by their name, RGCs have two characteristics: A long radial process, which spans the entire thickness of the wall of the neural tube and an (astro) glial property, which is indicated by the expression of glial proteins, including GFAP (38). Previous studies observed that GFAP-expressing neural progenitor cells are the major source of continuous neurogenesis (39–41). RGCs are ultrastructurally similar to astrocytes with regards to the expression of the filament protein GFAP and glycogen accumulation (42). During early cortical neurogenesis, GFAP expression is specifically found in RGCs, where co-localization with Vimentin and Nestin is also observed. However, GFAP positive cells exhibit a more lineage-restricted phenotype (34). Hence, the investigation of GFAP and the dynamic changes of its major variants, such as GFAPα and GFAPδ, during the developmental process may elucidate the mechanisms underlying how GFAP regulates fetal cortical development.
In the present study, the developmental profile of GFAPδ was investigated in the cortex in the control and ethanol-exposed mice, with the aim of characterizing the roles of GFAPδ in alcohol-induced cortex maldevelopment.
Materials and methods
Animals
Adult C57BL/6J mice (age, 8–12 weeks; mean body weight, 24.0±1.8 g) were purchased from Hunan Silaike Jingda Laboratory Animal Co., Ltd. A single male mouse was housed with five female mice per standard polycarbonate cage. Mice were housed in a temperature- and humidity-controlled environment (22°C; 50% humidity) with a 12-h light/dark cycle. Before the initiation of the experiments, the mice were maintained with ad libitum access to standard laboratory chow and water. A total of 46 adults (male 6 and female 40) and 215 fetal mice were used in the present study. There were 14 dams and 102 fetuses in the ethanol (EtOH) group, and 14 dams and 113 fetuses in the Control group. There were 6 dams in the EtOH group and 6 dams in the Control group that were used specifically for a blood alcohol assessment. All animal maintenance and experimental procedures were approved by the Institutional Animal Care and Use Committee of Xi'an Medical University and animal experiments were performed in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications no. 80-23) (43).
Animal maintenance and ethanol acclimation
Before ethanol administration, the mice were acclimated for ≥1 week after arrival. For the generation of the prenatal alcohol exposure models, female mice were fed on a liquid diet containing ethanol. The rodent liquid diet (Lieber-DeCarli ′82, Bio-Serv 14-726-629 and 14-726-630) was purchased from Thermo Fisher Scientific, Inc. Control mice were maintained for the same period on the same diet without ethanol, but instead supplemented with maltodextrin to provide the appropriate amount of ethanol-derived calories. For alcohol exposure, the female mice were provided a Lieber-DeCarli liquid diet containing 2.4% (v/v) ethanol for 2 days, then the ethanol concentration in the diet was increased to 4.8% (v/v) and maintained for a further 14 days; 5% sucrose to Lieber-DeCarli liquid diet was added to increase palatability as described previously (44,45). During alcohol exposure, the liquid diet was changed every day with a freshly prepared diet. After the acclimation/acquisition period for 16 days, the liquid diet was changed into normal laboratory chow and water for an ethanol deprivation period, during which the mice could recover from any potential alcohol withdrawal symptoms. This ethanol deprivation period averaged 16 days. During this ethanol deprivation period, the mice were bred by placing two females in a cage with one male for 1 h during the dark cycle and then were assessed for the presence of a copulation plug. If the plug was present, the time at the initiation of the 1-h mating period was considered as the gestational day (GD) 0. On GD7.0 at the beginning of the dark cycle, the mice were provided a liquid diet containing ethanol 4.8% (v/v, ethanol group) or a control liquid diet. The control liquid diet, which contained an isocaloric amount of maltodextrin instead of ethanol to meet the calories derived from ethanol, served as the pair-fed control. Maternal bodies were weighed, and diet consumptions were recorded based on the previous 24 h change of liquid diet from 30-ml graduated screw-cap tubes. Hence, the control and ethanol groups received the same amount of diet (ml) based on the ml diet per g body weight (g). This matching of calorific content and diet feeding was to reduce the possible effects of any potential nutritional differences (44). Subsequently, the mice were maintained on this diet for 48 h through GDs 7 and 8 and returned to the standard laboratory chow and water until the fetuses were collected on GD14.
Maternal blood alcohol assessment
A set of dams was used to assess blood alcohol concentration (BAC). A total of 30 µl tail blood was collected into heparinized capillary tubes from mice with sperm plugs on the 8th day of pregnancy (at 21:00 and 23:00 h). BACs were measured using an Analox alcohol analyzer (Model AM1; Analox Instruments USA, Inc.). For each time point, ≥3 mice were sampled and analyzed, the total samples of each mouse were controlled for no more than three times. None of the fetuses from these animals were used for histological analysis or western blot analysis.
Collection of the embryos
On the morning of GD 14.0, the embryos of pregnant dams were collected after sacrifice using CO2 inhalation according to the NIEHS euthanasia methods for rodent fetuses and neonates (44). For CO2 asphyxiation, a clear acrylic chamber with a gasket lid (VWR International, LLC) was used. The total chamber volume was 2 l (0.5 gal). The gas flow was set to 400 ml/min to achieve a 20% per min volume displacement rate. The death of the pregnant female mice was confirmed by the cessation of blood circulation (absence of heartbeat) and the absence of a breathing and pedal reflex. Fetal death was confirmed using ultrasound determination of the absence of a heartbeat. Subsequently, embryos were isolated from the uterus and extra-embryonic membranes under a dissecting microscope. The embryos were immersed in ice-cold 0.1 M 1X PBS. Tissues were fixed in 4% PFA in 1X Dulbecco's (D)PBS overnight at 4°C with gentle shaking for immunohistochemistry. The dissected cortical tissues were collected and pooled from each dam for western blot analysis.
Protein extraction and western blot analysis
For protein extraction, the cortical tissues from mice embryos (n=5–10 samples per litter) were pooled and dissected, meninges were removed under a stereomicroscope and dissolved in 1X lysis buffer (Cell Signaling Technology, Inc.; cat. no. 9803) supplemented with 1 mM PMSF. Subsequently, the tissues were mechanically homogenized with a tissue homogenizer and incubated on ice for 15 min, followed by centrifugation at 12,000 × g for 15 min at 4°C to obtain the cleared supernatants. The supernatants were collected, and protein concentrations were measured using a Pierce™ BCA Protein assay kit (Thermo Fisher Scientific, Inc.; cat. no. 23225). The samples were then adjusted and dissolved in 2X Laemmli Sample buffer (Bio-Rad Laboratories, Inc.; cat. no. 1610737). Subsequently, the samples were boiled at 95°C for 5 min, loaded on a 12% SDS-gel and resolved using SDS-PAGE; a BLUEstain™ Protein ladder (Goldbio; cat. no. P007-1500) was used to determine the molecular weight. The gels were blotted on PVDF membranes (Immobilon; EMD Millipore) at 200 mA for 1.5 h at 4°C. A total of 15 µg protein lysate was loaded into each well of the 12% SDS gel. After transferring and washing three times with PBS with 0.1% Tween® detergent (PBST), 5 min per wash, the membranes were blocked using 3% bovine serum albumin (BSA; cat. no. ab64009, Abcam) in 1X PBST for 1 h at room temperature. Primary antibodies were diluted in 3% BSA-PBST buffer and incubated at 4°C on a shaker. After three washes (10 min each), the secondary horseradish peroxidase-conjugated anti-rabbit (1:2,000; cat. no. ab6721; Abcam) or anti-mouse antibodies (1:2,000; cat. no. ab6789; Abcam) were incubated with the membrane at room temperature for 1 h. After three washes (10 min each), the membranes were developed using SuperSignal™ West Femto Maximum Sensitivity substrate (Thermo Fisher Scientific, Inc.; cat. no. 34096) and imaged using Image Lab™ Software Version 6.0.1 (Bio-Rad Laboratories, Inc.). For normalization, β-actin was used as the loading control. The following antibodies were used: Mouse anti-GFAP antibody (Sigma-Aldrich; Merck KGaA; cat. no. G3893; 1:2,000), rabbit anti-GFAPδ antibody (Abcam; cat. no. ab93251; 1:2,000) or β-actin antibody (Abcam; cat. no. ab8227; 1:5,000). Densitometry analysis was performed using ImageJ version 1.6.0 (National Institutes of Health).
Embryo collections, fixation, sectioning and immunohistochemistry
On E14.5, control and ethanol-exposed mice were euthanized by CO2 inhalation, and cervical dissociation was performed. The maternal abdomen was opened and the fetuses were removed from the uterus. The number of fetuses was recorded and imaged for each litter using a digital camera (Micropublisher 5.0; Qimaging Corp.), which was mounted on an Olympus binocular dissecting microscope (Olympus America, Inc.).
For the histological analysis, the fetuses were collected from individual extraembryonic membranes and fixed in 4% paraformaldehyde (PFA, cat. no. 158127, Sigma-Aldrich; Merck KGaA) in 1XDPBS overnight at 4°C. After immersion in 30% sucrose in 1X DPBS, 20-µm-thick coronal sections were cut on a cryostat microtome (Leica CM3050 S; Leica Microsystems GmbH). The sections were washed with PBS three times (each for 10 min), then blocked using 10% donkey or goat serum (Abcam) in DPBS supplemented with 0.25% Triton X-100 (Sigma-Aldrich; Merck KGaA) for 2 h at room temperature. Sections were subsequently incubated with primary antibodies at 4°C overnight. The following antibodies were used: Rabbit anti-SOX2 (Cell Signalling Technology, Inc.; cat. no. 23064; 1:200), mouse anti-Glial Fibrillary Acidic Protein (GFAP) antibody (Sigma; cat. no. G3893, 1:200), Mouse Anti-SOX2 antibody [9-9-3] (1:200; cat. no. ab79351; Abcam), rabbit anti-GFAPδ antibody (1:200; cat. no. ab93251; Abcam), mouse anti-Hop antibody (E-1) (HOPX; Santa Cruz Biotechnology, Inc.; cat. no. sc-30216; 1:200), rabbit anti-Nestin antibody (1:200 cat. no. ab92391; Abcam), rabbit anti-Vimentin antibody (1:200; cat. no. ab92547; Abcam), mouse anti-S100β (β-Subunit; Sigma-Aldrich; Merck KGaA; cat. no. S2532; 1:200), mouse anti-Ki67 antibody (Sigma-Aldrich; Merck KGaA; cat. no. MAB4190; 1:200) mouse anti-phospho-histone H3 (Ser10; 6G3) (pHH3; Cell Signaling Technology, Inc.; cat. no. 9706; 1:200), Rat anti-Ctip2 (1:200; cat. no. ab18465; Abcam). After washing three times with 1X DPBS, the fluorophore-conjugated secondary antibodies were used at a dilution of 1:200 and incubated for 2 h at room temperature. The secondary antibodies used were all purchased from Thermo Fisher Scientific, Inc. and were: Alexa Fluor 488 goat anti-rabbit IgG (H+L) Cross-Adsorbed Secondary antibody (cat. no. A11008) and Alexa Fluor 594 donkey anti-mouse IgG (H+L) Highly Cross-Adsorbed Secondary antibody (cat. no. A21203). Cell nuclei were counterstained at room temperature for 10 min with DAPI (Sigma-Aldrich; Merck KGaA; cat. no. D9542; 1:2,000). Subsequently, the slides were sealed and images were captured using a Nikon A1R confocal microscope (Nikon Corporation) with ×200 magnification with the appropriate laser channels. Images were processed using ImageJ 1.50i (National Institutes of Health). The ventricular zone (VZ), SVZ, intermediate zone (IZ), subcortical plate (SP) and cortical plate (CP) layers were delineated using DAPI staining.
Quantification of the proliferative or mitotic GFAPδ+ RGCs
The number of Ki67+ GFAPδ+ or pHH3+ GFAPδ+ double-labeled cells, as well as total GFAPδ+ RGCs, were quantified using ImageJ 1.50i with the co-localization finder plugin (46). For each brain region, six representative coronal sections were acquired using the Nikon A1R confocal microscope with ×200 magnification. Of these images, three images were captured at the level of the ventricular surface, including the VZ and the inner SVZ. The other three images were captured above the initial images, including outer the SVZ, IZ and CP. Hence, cells were counted in ~6 images per brain region (6 images per region × 2 brain regions per section × 4 sections × 10 embryonic brains). Furthermore, the ratio of Ki67+ GFAPδ+ or pHH3+ GFAPδ+ cells to total GFAPδ+ cells was calculated.
Statistical analysis
For quantification of immunostaining, the images were processed in parallel and were captured using the same settings and laser power for confocal microscopy. The cell numbers were manually counted using the ImageJ 1.50i cell counter plugin (National Institutes of Health). The fluorescence intensity was measured using ImageJ 1.51i and normalized to the size of the aggregates. All quantitative results are presented as the mean ± SEM. To determine the statistical difference, an unpaired Student's t-test was performed. P<0.05 was considered to indicate a statistically significant difference. Experiments were repeated at least three times.
Results
Ethanol intake and blood concentration measurements
The aforementioned ethanol exposure protocol resulted in an average ethanol intake on GD8 of 25.1 g/kg/day. This mean amount of ethanol intake resulted in BACs of 168–216 mg/dl (mean; 178.4 mg/dl).
Western blot analysis of GFAP and GFAPδ expression levels in the cortex
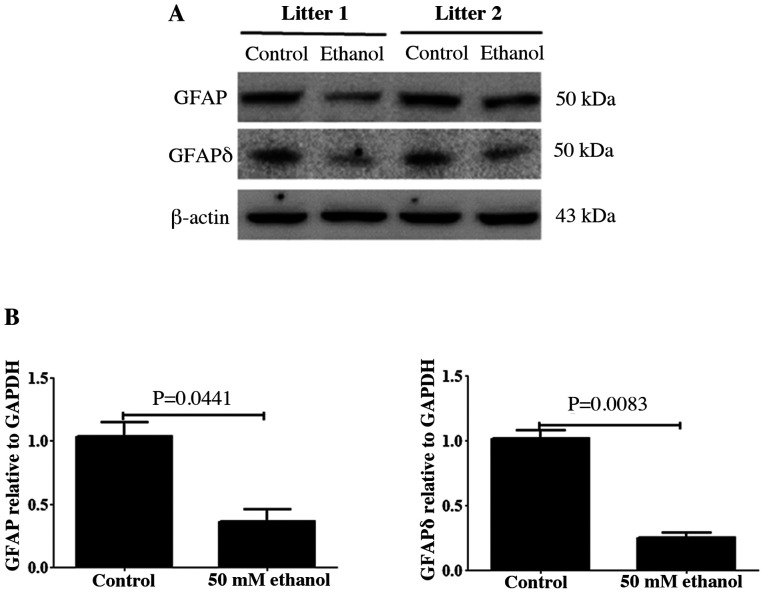
GFAP and its variant GFAPδ are two major components of radial glial fibers, and both proteins are expressed in RGCs (41). The relative expression levels of GFAP and GFAPδ in the control and PEE brain on E14.5 were analyzed using western blotting. As presented in Fig. 1A, immunoblots of GFAP and GFAPδ showed a single band of ~50 kDa, which corresponded to the molecular mass of GFAP and GFAPδ. After PEE, the relative densities of GFAP and GFAPδ were significantly decreased compared with those of the controls (Fig. 1B).
Figure 1.
GFAP and GFAPδ protein expression levels in control and prenatal ethanol exposure embryonic cortex. Western blot analysis was used to detect the expression levels of GFAP and GFAPδ. Each line in the immunoblotting image represents one litter of mice. β-actin was used as a control. Values were normalized to those of the control group. Data are presented as the mean ± SEM of offspring brains from six litters/group. (A) Expression levels of both GFAP and GFAPδ. (B) Semi-quantitative densitometries for GFAP and GFAPδ were normalized to β-actin levels. The data indicated that ethanol significantly decreased both GFAP and GFAPδ expression levels. The P-value was obtained from an unpaired Student's t-test. GFAP, glial fibrillary acidic protein.
Effects of PEE on the expression of GFAP in RGCs
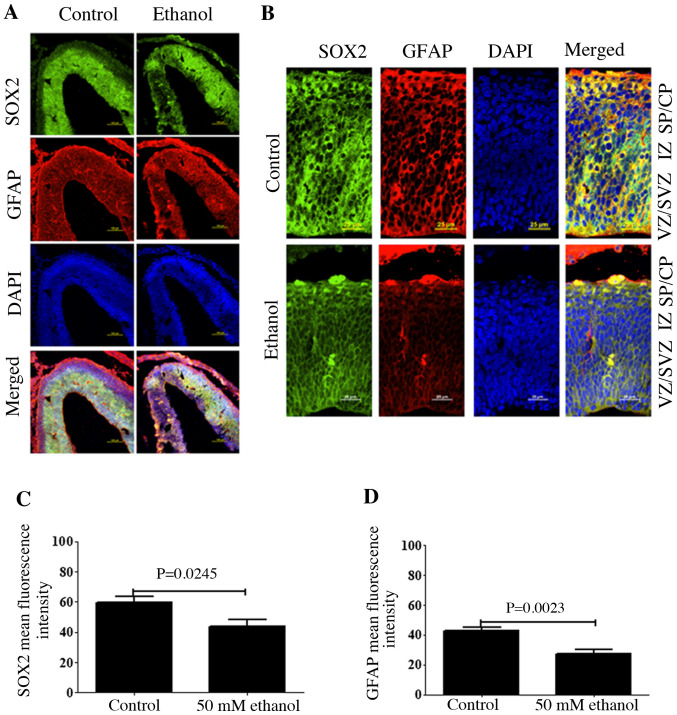
To assess the effects of ethanol exposure on the distribution of GFAP, cortical sections were fixed and co-immunostained for SOX2 and GFAP using the corresponding antibodies. The results demonstrated that SOX2+ cells were predominantly present in the VZ and SVZ, where almost all cells were SOX2+, and only a few SOX2+ cells in the IZ were present (Fig. 2). Moreover, few cells were SOX2+ in the SP/CP. SOX2+ cells also co-express GFAP. At E14.5, GFAP+ cells were observed in all layers. Some ventricular GFAP+ cells showed the characteristic morphologies of RGCs with an endfoot contacting the ventricular surface, a cell body within the ventricular zone and a long basal process extending toward the pia. Following ethanol treatment, the fluorescence intensities of both SOX2 and GFAP were significantly decreased (Fig. 2B and C), suggesting that ethanol exposure decreased the stem cell potential of RGCs.
Figure 2.
Effects of PEE on the expression level of GFAP in radial glial cells. (A) Coronal sections of control and PEE treated brain was stained with DAPI (blue), SOX2 (green), and GFAP (red). Immunostaining revealed the distributions of SOX2-positive neural progenitors and GFAP-positive cells in control and PEE-treated cortices. (B) The distribution of SOX2 and GFAP in cortical layers. Serial sections of embryonic cortices were double immunostained with the antibodies against SOX2 (green) and GFAP (red). Images indicated that GFAP was expressed in the whole embryonic cortex layers and colocalized with SOX2 in control group. PEE decreased the colocalization of SOX2 and GFAP. Quantification of mean fluorescence intensity of (C) SOX2 and (D) GFAP. PEE significantly decreased the numbers of SOX2+GFAP+ radial glial cells. Scale bars, 100 µm (A) and 25 µm (B). The P-value was obtained from an unpaired Student's t-test. PEE, prenatal ethanol exposure; GFAP, glial fibrillary acidic protein; SP, subcortical plate; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone.
Effect of PEE on the expression of GFAPδ in RGCs
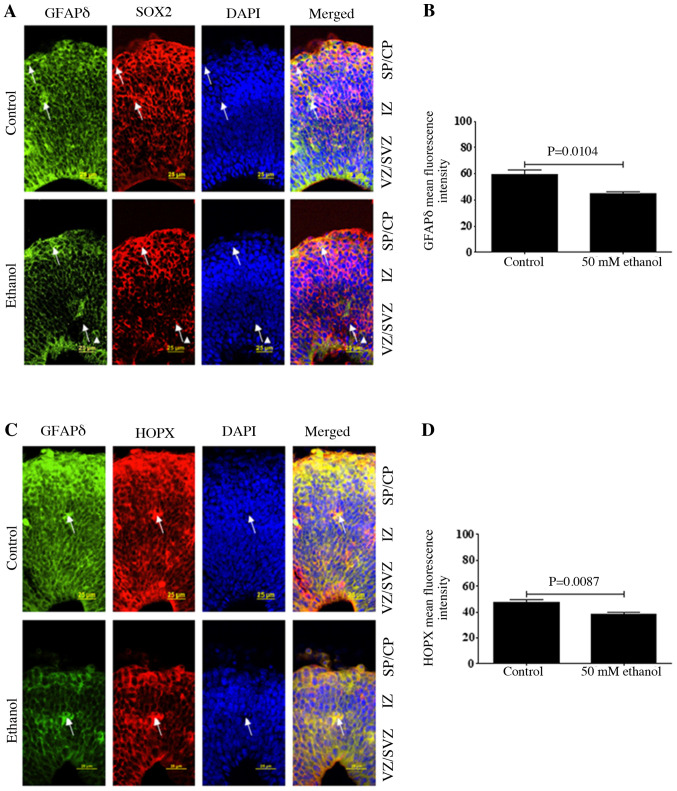
As GFAPδ expression was specifically limited to RGCs in the neurogenic regions, whether GFAPδ was co-expressed with the well-defined neural stem cell marker SOX2 and outer RGC (oRG) marker, HOPX expression was next determined. To confirmed the specificity of GFAPδ, GFAPδ and CTIP2 were co-immunostained and it was found that GFAPδ was not expressed in CTIP2-positive neurons (Fig. S1). The results demonstrated that GFAPδ had similar expression patterns with SOX2 and HOPX (Fig. 3). As a marker of oRG, HOPX was predominantly expressed in the VZ, SVZ and CP as indicated by the arrow in Fig. 3C. Double staining identified that GFAPδ expression strongly overlapped with both SOX2 and HOPX. Furthermore, the quantitative data indicated that ethanol treatment could significantly decrease the fluorescence intensity of GFAPδ and HOPX (Fig. 3B and D), suggesting that PEE could disrupt the assembly of oRG fibers.
Figure 3.
Effects of PEE on the expression level of GFAPδ in radial glial cells. Serial sections of embryonic cortices were subjected to the antibodies against GFAPδ and (A) radial glial marker SOX2 or (C) outer radial glial marker HOPX. At embryonic day 14.5, cells with overlapping expression levels of GFAPδ and SOX2 were shown in yellow. The nuclear stain was in blue. The data indicated that GFAPδ+ cells were also co-labeling with SOX2. Arrows pointed to representative SOX2+ cells with long process labeling with GFAPδ. In control mice, double labeling of GFAPδ with HOPX suggested that GFAPδ can also be expressed in outer radial glial cells. PEE decreases the intensity of GFAPδ in (B) SOX2 and (D) HOPX positive cells as shown in the right panels, indicating that PEE decreases the stem cell potential of radial glial cells. Scale bars, 25 µm. The P-value was obtained from an unpaired Student's t-test. PEE, prenatal ethanol exposure; GFAP, glial fibrillary acidic protein; SP, subcortical plate; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone; HOPX, Hop antibody (E-1).
Effect of PEE on co-localization of GFAPδ with Nestin or Vimentin in RGCs
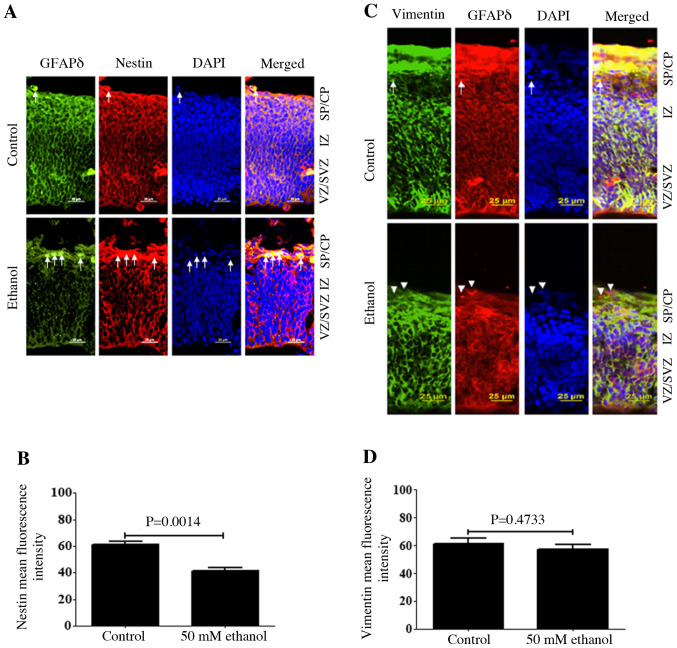
To determine whether PEE could impair other intermediate filament proteins, the expression distribution of Nestin and Vimentin was also assessed. The results demonstrated that Nestin was expressed on both the apical and basal surface processes of the RGCs, and its expression was co-localized with GFAPδ and surrounded the nuclei, similar to GFAPδ. Ethanol treatment significantly decreased both Nestin and GFAPδ expression (Fig. 4A and B). In the SVZ and IZ layers, Nestin expression, based on fluorescence intensity, was significantly decreased following ethanol exposure (Fig. 4B). Similar to Nestin, Vimentin expression was also co-localized with GFAPδ in the apical and basal surfaces of the process from the RGCs (Fig. 4C). Several RGCs in VZ showed strong Vimentin-positive apical processes. The majority of Vimentin-immunostaining encircled the nuclei of the cells and labeled the fibers of RGCs. At E14.5, Vimentin, similar to GFAPδ, was observed across almost the entire cortical wall. Compared with the staining of GFAPδ around the cell nucleus, Vimentin was more concentrated in the process of the RGCs. Ethanol exposure decreased the expression of Vimentin, as indicated by the presence of GFAPδ+ Vimentin− cells (Fig. 4C). Although ethanol exposure resulted in a slight decrease in the fluorescence intensity of Vimentin, there was no significant difference compared with control (Fig. 4C).
Figure 4.
Effects of PEE on the co-immunostaining of Nestin or Vimentin in GFAPδ+ cells. Serial sections of embryonic cortices were subjected to the antibodies against GFAPδ and intermediate filament protein (A) Nestin or (C) Vimentin. At embryonic day 14.5, cells with overlapping expression of GFAPδ and Nestin or Vimentin were shown in yellow. The nuclear stain was blue. The present data indicated that Nestin immunostaining had similar distribution as GFAPδ, which was surrounding the nuclear. In control mice, double immunostainings of GFAPδ and Nestin or Vimentin indicated colocalization of two immunoreactivities in most of the cells, as indicated by arrows. Quantification of mean fluorescence intensity of (B) Nestin and (D) Vimentin. PEE promoted the early appearance of GFAPδ+ astrocytes in the SP, as shown by single GFAPδ staining indicated by white arrowheads. It was indicated that PEE impaired the intermediate filamentous components of radial glial fibers and promoted the terminal differentiation of radial glial cells. Scale bars, 25 µm. The P-value was obtained from an unpaired Student's t-test. PEE, prenatal ethanol exposure; GFAPδ, glial fibrillary acidic protein δ; SP, subcortical plate; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone.
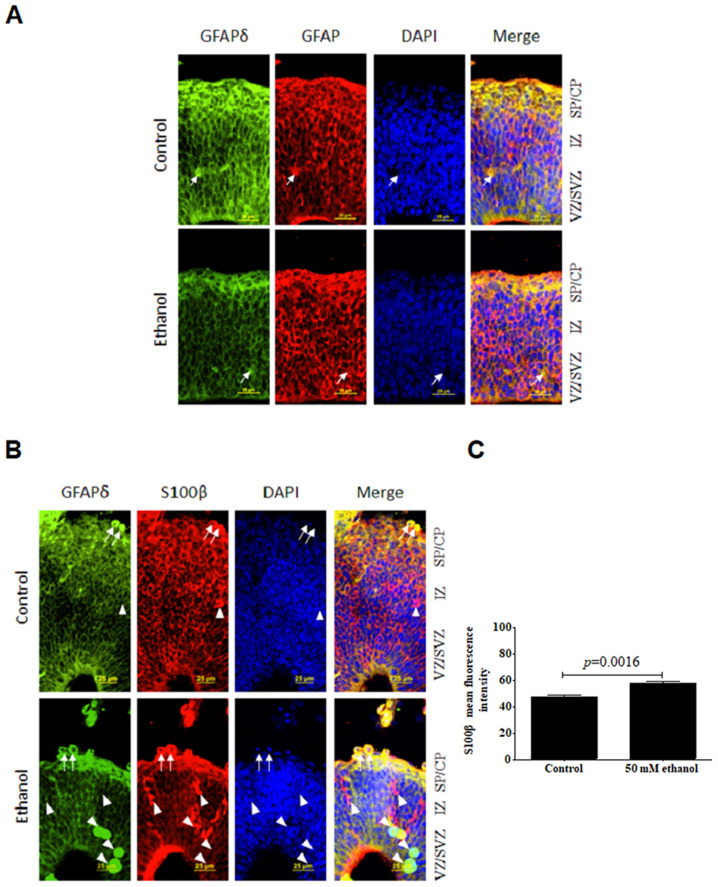
PEE decreases GFAP and GFAPδ protein expression levels in RGCs and promotes their maturation
According to previous reports, GFAP is a defined marker protein of astrocytes (40,47). To determine whether GFAPδ expression was similar to the GFAP expression in the developing cortex, double staining of GFAPδ together with GFAP was performed. The results demonstrated that both GFAP and GFAPδ were detected in the cortex at E14.5. These exhibited similar expression patterns in the developing VZ and SVZ, as well as the superficial layers, IZ and CP. GFAPδ-positive cells had a bipolar morphology with the soma in the VZ, short processes toward the ventricles and longer processes extending toward the CP (Fig. 5). Moreover, GFAPδ filaments were tangentially oriented, exhibiting unipolar or bipolar morphologies. From the staining, it was identified that GFAPδ filaments localized to the cell body and peri-nuclear regions. Similarly, GFAP was also observed both in the somata and processes of RGCs, which overlapped with the pattern of GFAPδ expression. Both GFAP and GFAPδ could be detected in the superficial layers at E14.5. Some GFAP+ RGCs exhibited only the basal processes, resembling basal RGCs of the neocortex (arrow; Fig. 5A). After ethanol treatment, the expression levels of both GFAP and GFAPδ were significantly decreased. In the SVZ region after ethanol exposure, there were some GFAP+ GFAPδ+ cells without apical and basal processes, and these would have eventually become astrocytes. Apical radial glia in the VZ had notable GFAP+GFAPδ+ staining compared with that of the control, which suggested ethanol treatment significantly influenced the formation of radial glial fibers. Thus, altering the expression of GFAP and GFAPδ may be how ethanol consumption during fetal cortical development exerts its detrimental effects.
Figure 5.
Effects of PEE on the co-immunostaining of GFAPδ and GFAP or S100β in radial glial cells. Serial sections of embryonic cortices were double immunostained with the antibodies against GFAPδ and GFAP or S100β. At embryonic day 14.5, cells with overlapping expression of GFAPδ and (A) GFAP or (B) S100β are shown in yellow. The nuclear stain is blue. The results suggested that GFAP+ cells exhibited long processes in control group. In control mice, double labelling of GFAPδ and GFAP or S100β revealed the colocalization of the two immunoreactivities. PEE notably decreased the expression of GFAPδ in GFAP-positive cells and increased the expression of S100β in GFAPδ-positive cells. Arrows indicated cells that have double staining of GFAPδ and GFAP or S100β in both the control and PEE cortex. Arrowheads showed cells had only express S100β or GFAPδ. It was indicated that PEE enhanced the maturation of GFAPδ+ radial glial cells. (C) Quantification of mean fluorescence intensity of S100β in control and ethanol groups. Scale bar, 25 µm. The P-value was obtained from an unpaired Student's t-test. PEE, prenatal ethanol exposure; GFAP, glial fibrillary acidic protein; SP, subcortical plate; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone.
It was hypothesized that loss of GFAP and GFAPδ may promote terminal differentiation of RGCs. S100β can be considered as a marker of the progression of neural progenitors. To evaluate whether GFAPδ could colocalize with S100β, double staining of GFAPδ and S100β was performed. Interestingly, S100β exhibited similar distribution patterns through all the layers of cortex with that of GFAPδ (Fig. 5B), and its expression was localized to the perinuclear area. The ethanol-exposed brain showed a significantly increased intensity of cytoplasmic staining compared with the control (Fig. 5C). However, there were more concentrated particles in RGCs in the ethanol group. Unlike GFAP immunostaining, S100β is a protein that can reliably define the maturation stage of GFAP+ cells (48). Increased expression of S100β in the developmental RGCs indicated the loss of stem cell differentiation potential by ethanol exposure. Quantification analysis demonstrated that ethanol resulted in a significant upregulation of S100β in the VZ and CP. Thus, it was suggested that GFAPδ+ RGCs gradually lost their stem cell potential and became more restricted following ethanol treatment.
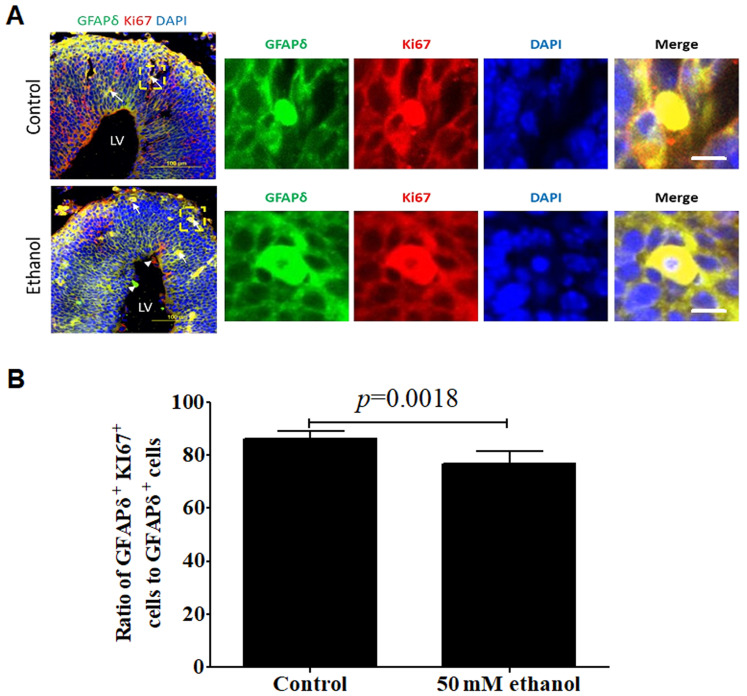
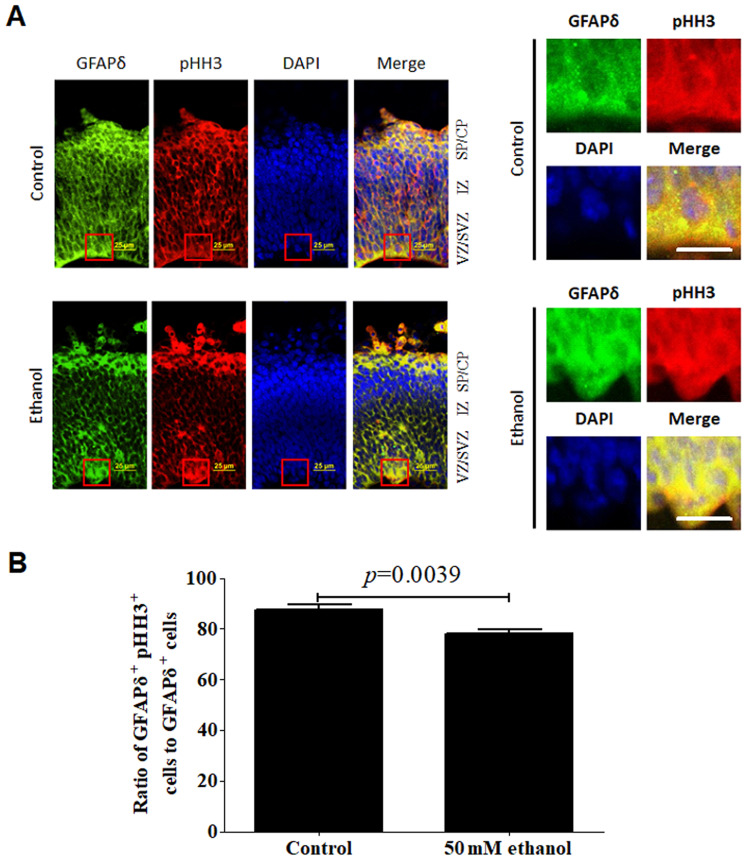
Effects of PEE on the proliferative or mitotic abilities of GFAPδ-positive cells
To determine whether GFAPδ positive cells were proliferative cells or dividing cells, the colocalization of GFAPδ with Ki67 (Fig. 6A and B) or pHH3 (Fig. 7A and B) was detected. The results demonstrated that GFAPδ-expressing cells expressed the proliferation marker Ki67 or mitotic marker pHH3 in both the control and ethanol groups. PEE remarkably inhibited the proliferative and mitotic activities of GFAPδ-positive cells compared with that of the control group. Furthermore, GFAPδ+ proliferative cells were distributed in all the layers. The proliferative cells had both basal and apical processes in the VZ region, suggesting these cells were apical RGCs. In the SVZ, the Ki67+ proliferative cells had their basal processes extending to the CP, which is one of the characteristic morphologies of oRG cells (Fig. 6A). Hence, the GFAPδ+Ki67+ cells were considered active RGCs.
Figure 6.
Effect of PEE on the proliferative activities in GFAPδ+ radial glial cells. (A) In embryonic day 14.5 cortices, co-labeling for GFAPδ and Ki67 demonstrated that GFAPδ+ cells were proliferating RGCs in the VZ, SVZ, IZ or CP. PEE significantly influenced the distribution of Ki67+ cells. In the ethanol cortex, radial glial cells were less abundant for GFAPδ+Ki67+ cells. The arrows indicate examples of GFAPδ+Ki67+ proliferative cells located in the control and ethanol-treated cortex. Arrowheads show the single labeling cells by GFAPδ or Ki67 in the ethanol-treated embryonic cortex. Immunostainings for representative GFAPδ+Ki67+ proliferative cells in control and ethanol groups are showed in the right magnification images. (B) Quantification of the ratio of GFAPδ+Ki67+ positive cells to total GFAPδ+ cells in the cortical layers. Scale bar, 100 µm (Left image) and 10 µm (right magnification images). The P-value was obtained from an unpaired Student's t-test. PEE, prenatal ethanol exposure; GFAP, glial fibrillary acidic protein; SP, subcortical plate; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone; LV, lateral ventricle.
Figure 7.
Effect of PEE on the mitotic activities in GFAPδ+ radial glial cells. (A) In embryonic day 14.5 cortices, co-labeling for GFAPδ and pHH3 indicated that GFAPδ+ cells were mitotic RGCs in the VZ, SVZ, IZ or CP. (B) PEE significantly influenced the distribution of GFAPδ+ pHH3+ cells. In the ethanol cortex, radial glial cells were less abundant for GFAPδ+pHH3+ cells, especially in SVZ and IZ layers. The representative mitotic cells by co-immunostaining of GFAPδ and pHH3 in the control and ethanol-treated cortex are showed in the right magnified images. Scale bar, 25 µm (left image) and 20 µm (right magnification images). The P-value was obtained from an unpaired Student's t-test. PEE, prenatal ethanol exposure; GFAP, glial fibrillary acidic protein; SP, subcortical plate; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subventricular zone; LV, lateral ventricle; pHH3, phospho-histone H3.
Regarding the co-expression of GFAPδ and pHH3, most of the GFAPδ+ cells exhibited active mitotic activities. pHH3+ mitotic cells were distributed in almost all the layers including the VZ, SVZ and CP. Ethanol treatment significantly affected the colocalization of GFAPδ and pHH3, particularly in the SVZ region (Fig. 7A). Following alcohol treatment, the ratio of co-localization of GFAPδ+Ki67+ or GFAPδ+pHH3+ to total GFAPδ+ cells was significantly decreased (Figs. 6B and 7B).
Discussion
During the developmental process of the dorsal telencephalon, RGCs and intermediate progenitor cells are the primary sources of excitatory neurons (49). SOX2 is a Sry-related HMG-box-type transcription factor, which is important for the maintenance of stem cell differentiation potential (50). SOX2 is a well-known and key regulator of cell fate decisions during neurogenesis, and it is important for the maintenance of the proliferative potential and the supervision of the generation of subpopulations of cells with varying phenotypes in the developing neocortex (51). A high expression level of SOX2 autonomously maintains stem cell potential, inhibits neuronal differentiation and maintains progenitor characteristics (49). Moreover, inhibition of SOX2 results in a loss of expression of progenitor markers and enhances early neuronal differentiation (50). Using conditional deletion, it has been revealed that SOX2 deletion could significantly influence the embryonic development of the ventral telencephalon (52). Thus, SOX2 is a key regulator between the stem cell and the dividing progenitors (53,54). It was previously confirmed that SOX2 is specifically expressed in neural epithelial cells and RGCs, but not in intermediate progenitor cells (49), thus it is considered a radial glial marker protein. Following the process of differentiation, SOX2 expression is decreased during the final fate decision of stem cells. During cortical development, SOX2 can be expressed together with GFAP in RGCs within the neurogenic regions SVZ and SGZ (39). In the present study, the findings demonstrated that prenatal ethanol exposure resulted in a significant decrease in both the number of SOX2+ cells and the intensity of GFAPδ and GFAP expression in SOX2+ cells.
Radial glial fibers are highly dynamic and are cell specific, providing the scaffold for neuroblast migration during normal neurodevelopment (25,26). Radial glial fibers promote the postnatal ventricular-subventricular zone (v-svz)-derived neuroblast to migrate toward to the lesion sites (36). GFAP is one of the major intermediate filament proteins in RGCs and astrocytes, which can colocalize with several other intermediate filaments, such as Vimentin and Nestin (41,55). Previous studies have confirmed that increased expression of intermediate filament proteins is an important step in the formation of reactive astrocytes (47,56,57). Dividing GFAP-expressing cells with bipolar or unipolar morphologies are considered RGCs in the human forebrain (58). In rodent models, GFAP expression is only detected in the astrocytic lineage after neurogenesis has ended and Vimentin expression is no longer detectable (58). GFAP is a member of class III intermediate filament proteins. There are 10 different transcripts of the GFAP gene and these can generate 10 differential splicing variants: GFAPα, GFAPβ, GFAPγ, GFAPδ, GFAPκ, GFAPζ, GFAPD135, GFAPD164, GFAPDexon6 and GFAPDexon7 (56,59). All these variants are similar, and the differences primarily reside in the C-terminal tail region. GFAPα is the most widely expressed protein. Moreover, GFAPδ expression is limited to specialized astrocyte populations and is more abundant in neurogenic regions, such as the SVZ of the lateral ventricle, the rostral migratory stream, the SGZ of the hippocampus and the subpial astrocytes (39,41,55,56). GFAPδ positive cells are specifically restricted to these neurogenic areas, where expression of Nestin and Vimentin is also observed, suggesting that GFAPδ may be a RGC-specific filament protein that is required for their substrate functions and may be a determining factor associated with neurogenesis (55,60). The ratio of GFAPα and GFAPδ is an important factor in the formation of the intermediate filament network, which is closely associated with cellular functions (such as neurogenesis) or cellular morphology (such as gliosis) (56). The changes in the expression levels of the GFAP splice variants could significantly influence the intermediate filament network, which is closely associated with cell morphology and cell motility (59).
GFAPδ and the GFAP expression are first observed at around gestational week 13 in the human fetal brain (41). In 2010, Middeldorp et al (41) first confirmed that GFAPδ was specifically found in radial glia and SVZ neural progenitors during human brain development. The GFAP transcript is first detectable at E9.5 in mice embryonic cortex, earlier than the corresponding time for a human (55). During the developmental process, GFAP expression continuously increases until a plateau arrives in the adult brain. In the second trimester, differentiated astrocytes from radial glia are observed in the germinal layers, corpus callosum and cavum septum pellucidum (61). However, it remains unknown what roles GFAPδ serves in the transition of RGCs into astrocytes. The present study found that PEE could markedly decrease the intensities of GFAP and GFAPδ, indicating that ethanol might impair the development of radial glial fibres and influence the supportive role for the migrating neuroblasts.
Nestin is a class VI intermediate filament protein that is expressed in the majority of mitotically active neural progenitors, including RGCs, which produce both neurons and astrocytes in the developing CNS (62). After differentiation, Nestin expression is downregulated. Using immunostaining, the present study identified that the majority of Nestin labeled RGCs were confined to the VZ and SVZ. Hence, Nestin was considered a marker of NSCs. Using Nestin–/– mice, Wilhelmsson et al (63) confirmed that Nestin negatively regulated neuronal differentiation and survival through its role in Notch signaling. Further experiments demonstrated that Nestin was co-expressed with GFAP during the transitional differentiation process of the neural progenitors into astrocytes (64) or neurons (65). The increased colocalization of GFAP and Nestin is a feature of reactive astrocytes (64). NSCs expressing both GFAP and Nestin were termed stem astrocytes (63). The Nestin+GFAP+ double labeling of neural progenitors may assist in determining the status of neural progenitors and their differentiation direction. In the present study, ethanol treatment significantly decreased both Nestin and GFAPδ expression levels in the SVZ. The co-expression of Nestin and GFAPδ expression was also decreased by ethanol treatment. These results indicated that ethanol treatment influenced the stem cell potential of oRG and their morphologies.
Vimentin (also known as nanofilament) is another intermediate filament protein expressed in neural stem/progenitor cells and astrocytes (41,55). Its phosphorylation is an important step required for the organization of the intermediate filament network and the subcellular distribution of its proteins (66–68). Mutations of Vimentin not only alter the cell morphology, proliferative abilities and motility but are also closely associated with alterations in neuronal differentiation of neural progenitor cells and neurogenesis (67). In astrocytes and RGCs, Vimentin forms an intermediate filament complex with GFAP and Nestin. The absence of GFAP and Vimentin could decrease Notch signaling and further increase neuronal differentiation (67,69). A recent report by Cunningham et al (70) revealed that all mitotic cells at the surface of the ventricle in the human and non-human primate dorsal telencephalon express GFAP and Vimentin. Following differentiation into astrocytes, RGCs stop expressing Vimentin and continually upregulate GFAP expression, which suggests that intermediate filament proteins can be transformed during the process of differentiation (67). In the present study, ethanol treatment accelerated the downregulation of Vimentin and promoted the appearance of GFAPβ+ cells.
HOPX is the smallest Homeodomain-only protein, which was identified as a temporally and spatially restricted gene in the neurogenic regions in the developing medial cortex and cortical hemisphere at a certain period during the process of neurogenesis (71). Using in situ hybridization, the HOPX gene was first detected at E12.5 forebrains in the most medial part of the cortex. From E14.5 to E16.5, HOPX expression in the forebrain increased from a low level at E14.5 to its peak at E16.5 (71). Thus, HOPX expression exhibits an expression gradient at a high level in the cortical VZ and a low level in the lateral caudal forebrain. In the dorsal forebrain, HOPX expression begins at E12.5 and reaches a peak at ~E16.5 with a rostromedial to the caudolateral gradient (71). At the same timepoint, RGCs are gradually produced. Several molecular markers including GFAP, SOX2 and Nestin are expressed in quiescent NSCs of this sub-region (71). Moreover, HOPX can be specifically expressed in slow-cycling or quiescent NSCs together with GFAP, Nestin and SOX2, but is not co-expressed with transit-amplifying cell progenitor markers, such as T-box brain protein 2 or the neuroblast marker Doublecortin (72). The aforementioned studies suggested that HOPX may modulate neurogenesis and self-renewal of neural stem cells. Furthermore, the present study demonstrated that ethanol treatment reduced the intensity of HOPX staining in GFAPδ+ cells.
The S100β protein is an acidic calcium-binding protein that can be expressed both in differentiated and undifferentiated proliferating glial cells (73). Additionally, S100β is regarded as a classical astrocyte marker, which suggests that S100β expression characterizes a terminal maturation stage (48). During embryonic neurodevelopment, the dynamic expression of S100β in NSCs and RGCs is associated with the proliferative potential and the migration of neural progenitors (73). A previous study reported that the SVZ microenvironment can repress S100β expression (48). Using transgenic S100β-EGFP cells, Raponi et al (48) confirmed that S100β expression defined a late neurodevelopmental stage coinciding with the loss of their stem cell potency. RGCs, as a type of neural stem cell, share certain similar characteristics with immature and mature astrocytes and one of these characteristics is the expression of the intermediate filament GFAP (38). Hence, RGCs have both neurogenic and gliogenic abilities during the development of the embryonic forebrain (74). The onset of S100β expression can be regarded as the acquisition of a more mature developmental stage-related to spatiotemporal maturation and the loss of neural stem cell potential (48). Amongst its multiple functions, S100β has been identified as a promoter of cytoskeletal stabilization, which serves an important role in normal corticogenesis (75). Decreased GFAP expression may impair the formation of filament complexes together with Nestin and further disrupt the supportive roles in the process of neuronal migration. Co-expression of Nestin and S100β or Nestin and GFAP may indicate that S100β+ or GFAP+ RGCs exhibit more restricted developmental potential, and are capable of differentiating into glial cells directed by factors in the local environment (65). In the present study, GFAPδ+ cells exhibited a strong intensity of S100β expression following ethanol treatment, which indicated that ethanol promoted the transformation of RGCs into astrocytes.
Ki67 is a nuclear protein, which is used as a marker for identifying dividing cells. It can label cells in the late G1, S, G2 and M phases, but not the G0 phase of the cell cycle (76,77). pHH3 is a proliferation marker that is closely associated with mitotic chromatin condensation in the late G2 and M phases of the cell cycle and HH3 is not phosphorylated during apoptosis (78). Compared with Ki67, PHH3 is a more reproducible proliferation and mitosis-specific marker (77). The results of the present study demonstrated that ethanol treatment notably inhibited the proliferative and mitotic activities of GFAPδ+ cells, which contributed to the decreased production of neurons or intermediate progenitor cells and impaired cortical thickness.
In summary, to the best of our knowledge, the present study was the first to identify the effects of prenatal alcohol exposure on the expression and distribution of GFAPδ during early neurogenesis. The impairment in GFAPδ+ RGCs contributed to prenatal alcohol exposure-induced abnormal cortical development. Although the mechanisms have not fully been elucidated, the results suggested that the regulation of GFAPδ may highlight a novel direction for the treatment and management of prenatal alcohol exposure associated with abnormal cortical development.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- GFAP
glial fibrillary acidic protein
- RGCs
radial glial cells
- oRGs
outer radial glial cells
- NSCs
neural stem cells
- PEE
prenatal ethanol exposure
- BACs
blood alcohol concentrations
- VZ
ventricular zone
- SVZ
subventricular zone
- IZ
intermediate zone
- SP
subcortical plate
- CP
cortical plate
- CNS
central nervous system
Funding Statement
This work was supported by grants from the Key Research and Development Foundation of Shaanxi Province, China (grant no. 2018SF-029), National Natural Science Foundation of China (grant no. 81760441; Principal Investigator, XLD) and Xi'an Science and Technology Plan Project [grant no. 20YXYJ0009(11)].
Funding
This work was supported by grants from the Key Research and Development Foundation of Shaanxi Province, China (grant no. 2018SF-029), National Natural Science Foundation of China (grant no. 81760441; Principal Investigator, XLD) and Xi'an Science and Technology Plan Project [grant no. 20YXYJ0009(11)].
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL, FYX and HZ designed the experimental plan. YL, LNZ, LC and YL performed the experiments. YL analysed the data and wrote the manuscript. FYX, HZ and XLD was the principal investigator and revised the manuscript. All authors read and approved the final version of the manuscript and agree to take responsibility for the published article.
Ethics approval and consent to participate
All animal maintenance and experimental procedures were approved by the Institutional Animal Care and Use Committee of Xi'an Medical University and animal experiments were performed in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80-23).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Popova S, Lange S, Probst C, Gmel G, Rehm J. Estimation of national, regional, and global prevalence of alcohol use during pregnancy and fetal alcohol syndrome: A systematic review and meta-analysis. Lancet Global health. 2017;5:e290–e299. doi: 10.1016/S2214-109X(17)30021-9. [DOI] [PubMed] [Google Scholar]
- 2.Popova S, Lange S, Probst C, Parunashvili N, Rehm J. Prevalence of alcohol consumption during pregnancy and Fetal Alcohol Spectrum Disorders among the general and Aboriginal populations in Canada and the United States. Eur J Med Genet. 2017;60:32–48. doi: 10.1016/j.ejmg.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Riley EP, Infante MA, Warren KR. Fetal alcohol spectrum disorders: An overview. Neuropsychol Rev. 2011;21:73–80. doi: 10.1007/s11065-011-9166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhoit LF, Scott DA, Simecka BA. Fetal alcohol spectrum disorders: Characteristics, complications, and treatment. Community Ment Health J. 2017;53:711–718. doi: 10.1007/s10597-017-0104-0. [DOI] [PubMed] [Google Scholar]
- 5.Petrelli B, Weinberg J, Hicks GG. Effects of prenatal alcohol exposure (PAE): Insights into FASD using mouse models of PAE. Biochem Cell Biol. 2018;96:131–147. doi: 10.1139/bcb-2017-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis-Anderson KL, Wesseling H, Siebert LM, Lunde-Young ER, Naik VD, Steen H, Ramadoss J. Fetal regional brain protein signature in FASD rat model. Reprod Toxicol. 2018;76:84–92. doi: 10.1016/j.reprotox.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Creeley CE, Dikranian KT, Johnson SA, Farber NB, Olney JW. Alcohol-induced apoptosis of oligodendrocytes in the fetal macaque brain. Acta Neuropathol Commun. 2013;1:23. doi: 10.1186/2051-5960-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farber NB, Creeley CE, Olney JW. Alcohol-induced neuroapoptosis in the fetal macaque brain. Neurobiol Dis. 2010;40:200–206. doi: 10.1016/j.nbd.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerlai R. Embryonic alcohol exposure: Towards the development of a zebrafish model of fetal alcohol spectrum disorders. Dev Psychobiol. 2015;57:787–798. doi: 10.1002/dev.21318. [DOI] [PubMed] [Google Scholar]
- 10.Flentke GR, Smith SM. The avian embryo as a model for fetal alcohol spectrum disorder. Biochem Cell Biol. 2018;96:98–106. doi: 10.1139/bcb-2017-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popova S, Lange S, Probst C, Gmel G, Rehm J. Global prevalence of alcohol use and binge drinking during pregnancy, and fetal alcohol spectrum disorder. Biochem Cell Biol. 2018;96:237–240. doi: 10.1139/bcb-2017-0077. [DOI] [PubMed] [Google Scholar]
- 12.Cheng HG, Deng F, Xiong W, Phillips MR. Prevalence of alcohol use disorders in mainland China: A systematic review. Addiction. 2015;110:761–774. doi: 10.1111/add.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YY, D'Amato RC. Understanding fetal alcohol spectrum disorders in China. J Pediatr Neuropsychol. 2017;3:53–60. doi: 10.1007/s40817-017-0029-0. [DOI] [Google Scholar]
- 14.Clarren SK, Alvord EC, Jr, Sumi SM, Streissguth AP, Smith DW. Brain malformations related to prenatal exposure to ethanol. J Pediatr. 1978;92:64–67. doi: 10.1016/S0022-3476(78)80072-9. [DOI] [PubMed] [Google Scholar]
- 15.Miller MW. Migration of cortical neurons is altered by gestational exposure to ethanol. Alcohol Clin Exp Res. 1993;17:304–314. doi: 10.1111/j.1530-0277.1993.tb00768.x. [DOI] [PubMed] [Google Scholar]
- 16.Lotfullina N, Khazipov R. Ethanol and the developing brain: Inhibition of neuronal activity and neuroapoptosis. Neuroscientist. 2018;24:130–141. doi: 10.1177/1073858417712667. [DOI] [PubMed] [Google Scholar]
- 17.Sowell ER, Mattson SN, Kan E, Thompson PM, Riley EP, Toga AW. Abnormal cortical thickness and brain-behavior correlation patterns in individuals with heavy prenatal alcohol exposure. Cereb Cortex. 2008;18:136–144. doi: 10.1093/cercor/bhm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou D, Lebel C, Lepage C, Rasmussen C, Evans A, Wyper K, Pei J, Andrew G, Massey A, Massey D, Beaulieu C. Developmental cortical thinning in fetal alcohol spectrum disorders. NeuroImage. 2011;58:16–25. doi: 10.1016/j.neuroimage.2011.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Kang W, Wong LC, Shi SH, Hébert JM. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J Neurosci. 2009;29:14571–14580. doi: 10.1523/JNEUROSCI.3844-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segklia A, Seuntjens E, Elkouris M, Tsalavos S, Stappers E, Mitsiadis TA, Huylebroeck D, Remboutsika E, Graf D. Bmp7 regulates the survival, proliferation, and neurogenic properties of neural progenitor cells during corticogenesis in the mouse. PLoS One. 2012;7:e34088. doi: 10.1371/journal.pone.0034088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choe Y, Huynh T, Pleasure SJ. Migration of oligodendrocyte progenitor cells is controlled by transforming growth factor β family proteins during corticogenesis. J Neurosci. 2014;34:14973–14983. doi: 10.1523/JNEUROSCI.1156-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tiberi L, Vanderhaeghen P, van den Ameele J. Cortical neurogenesis and morphogens: Diversity of cues, sources and functions. Curr Opin Cell Biol. 2012;24:269–276. doi: 10.1016/j.ceb.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Bengoa-Vergniory N, Kypta RM. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell Mol Life Sci. 2015;72:4157–4172. doi: 10.1007/s00018-015-2028-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inestrosa NC, Varela-Nallar L. Wnt signalling in neuronal differentiation and development. Cell Tissue Res. 2015;359:215–223. doi: 10.1007/s00441-014-1996-4. [DOI] [PubMed] [Google Scholar]
- 25.Razavi MJ, Zhang T, Chen H, Li Y, Platt S, Zhao Y, Guo L, Hu X, Wang X, Liu T. Radial structure scaffolds convolution patterns of developing cerebral cortex. Front Comput Neurosci. 2017;11:76. doi: 10.3389/fncom.2017.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto H, Mandai K, Konno D, Maruo T, Matsuzaki F, Takai Y. Impairment of radial glial scaffold-dependent neuronal migration and formation of double cortex by genetic ablation of afadin. Brain Res. 2015;1620:139–152. doi: 10.1016/j.brainres.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Falk S, Götz M. Glial control of neurogenesis. Curr Opin Neurobiol. 2017;47:188–195. doi: 10.1016/j.conb.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 28.Yuzwa SA, Miller FD. Deciphering cell-cell communication in the developing mammalian brain. Neurogenesis (Austin) 2017;4:e1286425. doi: 10.1080/23262133.2017.1286425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller MW, Robertson S. Prenatal exposure to ethanol alters the postnatal development and transformation of radial glia to astrocytes in the cortex. J Comp Neurol. 1993;337:253–266. doi: 10.1002/cne.903370206. [DOI] [PubMed] [Google Scholar]
- 30.Rubert G, Miñana R, Pascual M, Guerri C. Ethanol exposure during embryogenesis decreases the radial glial progenitorpool and affects the generation of neurons and astrocytes. J Neurosci Res. 2006;84:483–496. doi: 10.1002/jnr.20963. [DOI] [PubMed] [Google Scholar]
- 31.Vallés S, Pitarch J, Renau-Piqueras J, Guerri C. Ethanol exposure affects glial fibrillary acidic protein gene expression and transcription during rat brain development. J Neurochem. 1997;69:2484–2493. doi: 10.1046/j.1471-4159.1997.69062484.x. [DOI] [PubMed] [Google Scholar]
- 32.Nash R, Krishnamoorthy M, Jenkins A, Csete M. Human embryonic stem cell model of ethanol-mediated early developmental toxicity. Exp Neurol. 2012;234:127–135. doi: 10.1016/j.expneurol.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Johansson PA, Cappello S, Götz M. Stem cells niches during development-lessons from the cerebral cortex. Curr Opin Neurobiol. 2010;20:400–407. doi: 10.1016/j.conb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Götz M, Hartfuss E, Malatesta P. Radial glial cells as neuronal precursors: A new perspective on the correlation of morphology and lineage restriction in the developing cerebral cortex of mice. Brain Res Bull. 2002;57:777–788. doi: 10.1016/S0361-9230(01)00777-8. [DOI] [PubMed] [Google Scholar]
- 35.Malatesta P, Appolloni I, Calzolari F. Radial glia and neural stem cells. Cell Tissue Res. 2008;331:165–178. doi: 10.1007/s00441-007-0481-8. [DOI] [PubMed] [Google Scholar]
- 36.Jinnou H, Sawada M, Kawase K, Kaneko N, Herranz-Pérez V, Miyamoto T, Kawaue T, Miyata T, Tabata Y, Akaike T, et al. Radial glial fibers promote neuronal migration and functional recovery after neonatal brain injury. Cell Stem Cell. 2018;22:128–137.e9. doi: 10.1016/j.stem.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron. 2016;91:1219–1227. doi: 10.1016/j.neuron.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barry DS, Pakan JM, McDermott KW. Radial glial cells: Key organisers in CNS development. Int J Biochem Cell Biol. 2014;46:76–79. doi: 10.1016/j.biocel.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 39.Garcia AD, Doan NB, Imura T, Bush TG, Sofroniew MV. GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci. 2004;7:1233–1241. doi: 10.1038/nn1340. [DOI] [PubMed] [Google Scholar]
- 40.Johnson K, Barragan J, Bashiruddin S, Smith CJ, Tyrrell C, Parsons MJ, Doris R, Kucenas S, Downes GB, Velez CM, et al. Gfap-positive radial glial cells are an essential progenitor population for later-born neurons and glia in the zebrafish spinal cord. Glia. 2016;64:1170–1189. doi: 10.1002/glia.22990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Middeldorp J, Boer K, Sluijs JA, De Filippis L, Encha-Razavi F, Vescovi AL, Swaab DF, Aronica E, Hol EM. GFAPdelta in radial glia and subventricular zone progenitors in the developing human cortex. Development. 2010;137:313–321. doi: 10.1242/dev.041632. [DOI] [PubMed] [Google Scholar]
- 42.Gotoh H, Nomura T, Ono K. Glycogen serves as an energy source that maintains astrocyte cell proliferation in the neonatal telencephalon. J Cereb Blood Flow Metab. 2017;37:2294–2307. doi: 10.1177/0271678X16665380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.National Research Council, corp-author. 8th edition. The National Academies Press; Washington, DC: 2011. Guide for the Care and Use of Laboratory Animals. [Google Scholar]
- 44.Parnell SE, Chen SY, Charness ME, Hodge CW, Dehart DB, Sulik KK. Concurrent dietary administration of D-SAL and ethanol diminishes ethanol's teratogenesis. Alcohol Clin Exp Res. 2007;31:2059–2064. doi: 10.1111/j.1530-0277.2007.00524.x. [DOI] [PubMed] [Google Scholar]
- 45.Parnell SE, Dehart DB, Wills TA, Chen SY, Hodge CW, Besheer J, Waage-Baudet HG, Charness ME, Sulik KK. Maternal oral intake mouse model for fetal alcohol spectrum disorders: Ocular defects as a measure of effect. Alcohol Clin Exp Res. 2006;30:1791–1798. doi: 10.1111/j.1530-0277.2006.00212.x. [DOI] [PubMed] [Google Scholar]
- 46.Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pekny M, Pekna M. Astrocyte intermediate filaments in CNS pathologies and regeneration. J Pathol. 2004;204:428–437. doi: 10.1002/path.1645. [DOI] [PubMed] [Google Scholar]
- 48.Raponi E, Agenes F, Delphin C, Assard N, Baudier J, Legraverend C, Deloulme JC. S100B expression defines a state in which GFAP-expressing cells lose their neural stem cell potential and acquire a more mature developmental stage. Glia. 2007;55:165–177. doi: 10.1002/glia.20445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutton SR, Pevny LH. SOX2 expression levels distinguish between neural progenitor populations of the developing dorsal telencephalon. Dev Biol. 2011;352:40–47. doi: 10.1016/j.ydbio.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 50.Kamachi Y, Kondoh H. Sox proteins: Regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 51.Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, Sikorska M. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Ferri A, Favaro R, Beccari L, Bertolini J, Mercurio S, Nieto-Lopez F, Verzeroli C, La Regina F, De Pietri Tonelli D, Ottolenghi S, et al. Sox2 is required for embryonic development of the ventral telencephalon through the activation of the ventral determinants Nkx2.1 and Shh. Development. 2013;140:1250–1261. doi: 10.1242/dev.073411. [DOI] [PubMed] [Google Scholar]
- 53.Hagey DW, Muhr J. Sox2 acts in a dose-dependent fashion to regulate proliferation of cortical progenitors. Cell Rep. 2014;9:1908–1920. doi: 10.1016/j.celrep.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 54.Wang S, Chandler-Militello D, Lu G, Roy NS, Zielke A, Auvergne R, Stanwood N, Geschwind D, Coppola G, Nicolis SK, et al. Prospective identification, isolation, and profiling of a telomerase-expressing subpopulation of human neural stem cells, using sox2 enhancer-directed fluorescence-activated cell sorting. J Neurosci. 2010;30:14635–14648. doi: 10.1523/JNEUROSCI.1729-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mamber C, Kamphuis W, Haring NL, Peprah N, Middeldorp J, Hol EM. GFAPδ expression in glia of the developmental and adolescent mouse brain. PLoS One. 2012;7:e52659. doi: 10.1371/journal.pone.0052659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamphuis W, Mamber C, Moeton M, Kooijman L, Sluijs JA, Jansen AH, Verveer M, de Groot LR, Smith VD, Rangarajan S, et al. GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS One. 2012;7:e42823. doi: 10.1371/journal.pone.0042823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang Z, Wang KK. Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 2015;38:364–374. doi: 10.1016/j.tins.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Howard BM, Zhicheng M, Filipovic R, Moore AR, Antic SD, Zecevic N. Radial glia cells in the developing human brain. Neuroscientist. 2008;14:459–473. doi: 10.1177/1073858407313512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sullivan SM. GFAP variants in health and disease: Stars of the brain… and gut. J Neurochem. 2014;130:729–732. doi: 10.1111/jnc.12754. [DOI] [PubMed] [Google Scholar]
- 60.Zalfa C, Grasselli C, Santilli G, Ferrari D, Lamorte G, Vescovi AL, De Filippis L. GFAP delta as divergent marker of human glial progenitors. J Stem Cell Res Ther. 2018;8:9. doi: 10.4172/2157-7633.1000437. [DOI] [Google Scholar]
- 61.Rezaie P, Ulfig N, Male D. Distribution and morphology of GFAP-positive astrocytes in the human fetal brain at second trimester. Neuroembryology. 2003;2:50–63. doi: 10.1159/000071021. [DOI] [Google Scholar]
- 62.Sunabori T, Tokunaga A, Nagai T, Sawamoto K, Okabe M, Miyawaki A, Matsuzaki Y, Miyata T, Okano H. Cell-cycle-specific nestin expression coordinates with morphological changes in embryonic cortical neural progenitors. J Cell Sci. 2008;121:1204–1212. doi: 10.1242/jcs.025064. [DOI] [PubMed] [Google Scholar]
- 63.Wilhelmsson U, Lebkuechner I, Leke R, Marasek P, Yang X, Antfolk D, Chen M, Mohseni P, Lasič E, Bobnar ST, et al. Nestin regulates neurogenesis in mice through notch signaling from astrocytes to neural stem cells. Cereb Cortex. 2019;29:4050–4066. doi: 10.1093/cercor/bhy284. [DOI] [PubMed] [Google Scholar]
- 64.Messam CA, Hou J, Major EO. Coexpression of nestin in neural and glial cells in the developing human CNS defined by a human-specific anti-nestin antibody. Exp Neurol. 2000;161:585–596. doi: 10.1006/exnr.1999.7319. [DOI] [PubMed] [Google Scholar]
- 65.Vukojevic K, Petrovic D, Saraga-Babic M. Nestin expression in glial and neuronal progenitors of the developing human spinal ganglia. Gene Expr Patterns. 2010;10:144–151. doi: 10.1016/j.gep.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Eriksson JE, He T, Trejo-Skalli AV, Härmälä-Braskén AS, Hellman J, Chou YH, Goldman RD. Specific in vivo phosphorylation sites determine the assembly dynamics of vimentin intermediate filaments. J Cell Sci. 2004;117:919–932. doi: 10.1242/jcs.00906. [DOI] [PubMed] [Google Scholar]
- 67.Chen M, Puschmann TB, Marasek P, Inagaki M, Pekna M, Wilhelmsson U, Pekny M. Increased neuronal differentiation of neural progenitor cells derived from phosphovimentin-deficient mice. Mol Neurobiol. 2018;55:5478–5489. doi: 10.1007/s12035-017-0759-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chou YH, Khuon S, Herrmann H, Goldman RD. Nestin promotes the phosphorylation-dependent disassembly of vimentin intermediate filaments during mitosis. Mol Biol Cell. 2003;14:1468–1478. doi: 10.1091/mbc.e02-08-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Widestrand A, Faijerson J, Wilhelmsson U, Smith PL, Li L, Sihlbom C, Eriksson PS, Pekny M. Increased neurogenesis and astrogenesis from neural progenitor cells grafted in the hippocampus of GFAP−/− Vim−/− mice. Stem Cells. 2007;25:2619–2627. doi: 10.1634/stemcells.2007-0122. [DOI] [PubMed] [Google Scholar]
- 70.Cunningham CL, Martinez-Cerdeno V, Noctor SC. Diversity of neural precursor cell types in the prenatal macaque cerebral cortex exists largely within the astroglial cell lineage. PLoS One. 2013;8:e63848. doi: 10.1371/journal.pone.0063848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li D, Takeda N, Jain R, Manderfield LJ, Liu F, Li L, Anderson SA, Epstein JA. Hopx distinguishes hippocampal from lateral ventricle neural stem cells. Stem Cell Res. 2015;15:522–529. doi: 10.1016/j.scr.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Braun SM, Jessberger S. Adult neurogenesis: Mechanisms and functional significance. Development. 2014;141:1983–1986. doi: 10.1242/dev.104596. [DOI] [PubMed] [Google Scholar]
- 73.Patro N, Naik A, Patro IK. Differential temporal expression of S100β in developing rat brain. Front Cell Neurosci. 2015;9:87. doi: 10.3389/fncel.2015.00087. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 74.Docampo-Seara A, Santos-Durán GN, Candal E, Rodríguez Díaz MÁ. Expression of radial glial markers (GFAP, BLBP and GS) during telencephalic development in the catshark (Scyliorhinus canicula) Brain Struct Funct. 2019;224:33–56. doi: 10.1007/s00429-018-1758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aronne MP, Guadagnoli T, Fontanet P, Evrard SG, Brusco A. Effects of prenatal ethanol exposure on rat brain radial glia and neuroblast migration. Exp Neurol. 2011;229:364–371. doi: 10.1016/j.expneurol.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Malik S, Vinukonda G, Vose LR, Diamond D, Bhimavarapu BB, Hu F, Zia MT, Hevner R, Zecevic N, Ballabh P. Neurogenesis continues in the third trimester of pregnancy and is suppressed by premature birth. J Neurosci. 2013;33:411–423. doi: 10.1523/JNEUROSCI.4445-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, Jiao J. Molecular biomarkers for embryonic and adult neural stem cell and neurogenesis. Biomed Res Int. 2015;2015:727542. doi: 10.1155/2015/727542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van den Berge SA, van Strien ME, Korecka JA, Dijkstra AA, Sluijs JA, Kooijman L, Eggers R, De Filippis L, Vescovi AL, Verhaagen J, et al. The proliferative capacity of the subventricular zone is maintained in the parkinsonian brain. Brain. 2011;134:3249–3263. doi: 10.1093/brain/awr256. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.