Abstract
Neuroblastoma (NB) is considered a highly prevalent extracranial solid tumor in young children, and the upregulation of N-myc proto-oncogene (MYCN) is closely associated with the late stages of NB and poor prognostic outcomes. The current study was designed to evaluate the effects of exosomal microRNA (miRNA/miR)-17-5p from MYCN-amplified NB cells on the proliferative and migratory potential of non-MYCN amplified NB cells. miR-17-5p was found to activate the PI3K/Akt signaling cascade by targeting PTEN, and the overexpression of miR-17-5p was found to promote cellular migration and proliferation in vitro. Further experimentation revealed that the elevated expression of miR-17-5p in SK-N-BE(2) cell-derived exosomes significantly promoted the proliferative and migratory capacities of SH-SY5Y cells by inhibiting PTEN. Collectively, these findings demonstrated that miR-17-5p derived from MYCN-amplified NB cell exosomes promoted the migration and proliferation of non-MYCN amplified cells, highlighting an exosome-associated malignant role for miR-17-5p.
Keywords: cell proliferation, exosomes, microRNA-17-5p, N-myc proto-oncogene, neuroblastoma
Introduction
Neuroblastoma (NB) originates from undifferentiated neural crest cells and is the most common extracranial solid tumor in children, accounting for 15% of deaths from pediatric tumors (1). The clinical manifestations of NB are variable, ranging from a spontaneously developing mass to aggressive drug-resistant tumors, associated with high levels of metastasis and poor prognosis rates (2). In ~50% of patients with NB, metastasis occurs in the bone marrow, skin, liver and lymph nodes (3). Amplification of the N-myc proto-oncogene (MYCN) has been identified in 20–30% of patients, and is associated with advanced disease stage and poor prognosis (4). The transcription factor N-Myc (encoded by MYCN) contributes to the malignant phenotype by both up- and downregulating target genes (5). According to previous studies, N-myc directly interacts with the microRNA (miRNA/miR)-17–92 promoter site. Furthermore, there is a positive association between MYCN expression and members of the miR-17- 92 cluster in NB cell lines and primary tumors (6–8).
miRNAs belong to a group of single-stranded, non-coding RNAs of 18–25 nucleotides in length, which regulate gene expression at the post-transcriptional level (9). Abnormal regulation of miRNA expression has been linked to the development of various diseases, including NB (10–12). These molecules exert their regulatory effects by interacting with the 3′untranslated regions (3′UTRs) of mRNAs, and either inhibiting translation or facilitating mRNA degradation (13). Exosomes are membranous extracellular microvesicles with a diameter of 30–150 nm that contain various macromolecules, including miRNAs (14). These microvesicles originate from a variety of different cell types, and can act as key mediators between cancerous cells and facilitate stroma intercellular communication by delivering molecular cargo into the tumor microenvironment (15). Exosome-mediated miRNA transfer is considered an efficient strategy for miRNAs to exert their effects on different tumor progression processes (16). A number of recently reported studies have identified exosomal miRNAs as key biomarkers for the detection and prognosis of NB (17,18). Although exosomes from MYCN-amplified NB cells are able transmit aggressive cancer cell phenotypes (19), their regulatory mechanisms in NB cell functional variation and miRNA expression modification are not fully understood.
The present study aimed to investigate the transfer of aggressive cell phenotypes from MYCN-amplified to non-MYCN amplified NB cells via exosome-mediated miRNAs. It was hypothesized that anti-oncogenes in recipient cells are targeted by exosomal miRNAs, which are present in high levels in the exosomes of MYCN-amplified NB cells.
Materials and methods
Cell culture
Human SH-SY5Y NB cells were procured from American Type Culture Collection, and SK-N-BE(2) cells were purchased from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. The cells were cultured in DMEM (Hyclone; Cytiva) containing FBS (10%; Gibco; Thermo Fisher Scientific, Inc.), penicillin and streptomycin (100 U/ml and 100 µg/ml, respectively), and maintained at 37°C in an atmosphere of 5% CO2. Routine passage was conducted when the cells had reached 80–90% confluency.
Bioinformatics analysis
The miRNA expression profiles of MYCN-amplified and non-MYCN amplified NB tissues were obtained directly from the original text of three previous studies (6–8), with no additional processing. To identify the differential expression of miR-17-92a between these tissue types, miRNAs with significantly different expression levels were summarized from the results of these three studies, and then subjected to further analysis. The TargetScan (http://www.targetscan.org/vert_72/), DIANA (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index) and miRanda (http://mirdb.org/) databases were then used to predict the target genes of these miRNAs. The immunohistochemical profiles of PTEN (determined using a tissue microarray containing human NB sections) obtained from the results of a previously published article (20) were statistically analyzed to identify the differential expression of PTEN in MYC-amplified NB tissues (compared with their non-MYCN amplified counterparts).
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from cells using a TRIzol® RNA purification kit (Thermo Fisher Scientific, Inc.). Reverse transcription was performed with 1 µg total RNA using the miRNA 1st Strand cDNA Synthesis kit (Vazyme Biotech Co., Ltd.) for mature miRNA expression, and the TransScript® qPCR kit (TransGen Biotech Co., Ltd.) for mRNA expression. Reverse transcription was performed according to the manufacturer's protocols. qPCR were subsequently performed with the ChamQ Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.), according to the manufacturer's protocols. The thermocycling conditions were 94 C for 5 min, then 40 cycles of 93°C for 30 sec, 55°C for 30 sec and 72°C for 60 sec. Relative mRNA and miRNA expression levels were evaluated using the 2−∆∆Cq method (21), and normalized to the levels of GAPDH and U6, respectively. The primer sequences are presented in Table I.
Table I.
Primer sequences used for reverse transcription- quantitative PCR.
| Gene | Primer sequences (5′→3′) |
|---|---|
| miR-17-5p | F: GCGCAAAGTGCTTACAGTGC |
| R: AGTGCAGGGTCCGAGGTATT | |
| miR-18a | F: CGCGTAAGGTGCATCTAGTGC |
| R: AGTGCAGGGTCCGAGGTATT | |
| miR-19a | F: GCGTGTGCAAATCTATGCAA |
| R: AGTGCAGGGTCCGAGGTATT | |
| miR-19b | F: CGTGTGCAAATCCATGCAA |
| R: AGTGCAGGGTCCGAGGTATT | |
| miR-20a | F: GCGCGTAAAGTGCTTATAGTGC |
| R: AGTGCAGGGTCCGAGGTATT | |
| miR-92a | F: GCGTATTGCACTTGTCCCG |
| R: AGTGCAGGGTCCGAGGTATT | |
| U6 | F: AGAGAAGATTAGCATGGCCCCTG |
| R: ATCCAGTGCAGGGTCCGAGG | |
| PTEN | F: GACCAGAGACAAAAAGGGAGTA |
| R: ACAAACTGAGGATTGCAAGTTC | |
| GAPDH | F: TCCAGAGTGCAAGGCTTCAG |
| R: GACAGCACGCAGTAGCAGTAG |
F, forward; R, reverse; miR, microRNA.
Isolation of exosomes by ultracentrifugation
Exosomes were isolated from the cell culture supernatants based on a previously reported procedure (22). Briefly, when the cells had reached 60–80% confluency, the media were replaced with DMEM containing 10% exosome-depleted FBS (Systems Biosciences, LLC). After incubation for 48 h, the supernatants were removed and centrifuged at 2,000 × g (10 min) and 10,000 × g (30 min) for the removal of the cells and cellular debris, respectively. The resulting supernatant was centrifuged again for 70 min (110,000 × g) to pellet the exosomal fraction. The pellets were washed with PBS, followed by ultracentrifugation for a further 70 min at 110,000 × g, and then resuspended in PBS. The pellets were stored −80°C and each centrifugation step was carried out at 4°C. For subsequent experimentation, 50 µg SK-N-BE(2) cell exosomes were incubated with 50,000 SH-SY5Y cells for 48 h at 37°C. The untreated cells were used as a control group.
Exosome characterization
Transmission electron microscopy (TEM)
Isolated exosomes (10 µl) were placed in copper grids and allowed to settle for 2 min at room temperature. The samples were blotted to remove excess fluid, followed by staining with phosphotungstic acid (3%, v/v) in ddH2O for 1 min at room temperature. The samples were carefully blotted with soft paper before being left in a dry environment for 10–15 min at room temperature. Finally, the samples were evaluated using the JEM1200-EX transmission electron microscope (JEOL, Ltd.) at a voltage of 80 kV. In total, five fields of view were randomly selected for each sample (magnification, ×200,000).
Nanoparticle tracking analysis (NTA)
To obtain an estimated number of vesicles (≤1×107), exosomal dilutions were prepared with PBS at a factor of 500. Exosome concentration and size were determined using the ZetaView® PMX 110 and the corresponding software (ZetaView® 8.04.02 SP2; both Analytik Jena AG).
Western blot analysis
Total cellular proteins were extracted using RIPA lysis buffer (Wuhan Servicebio Technology Co., Ltd.) 96 h after transfection. The native protein lysate was collected, and the protein concentration was measured using a BCA protein concentration assay kit (Wuhan Servicebio Technology Co., Ltd.), according to the manufacturer's instructions. Proteins (20–40 µg/per well) were separated via SDS-PAGE on a 10% gel. The separated proteins were transferred to PVDF membranes (EMD Millipore), which were then blocked for 2 h in non-fat milk (5%) in PBS with 0.1% Tween-20 (PBST) at room temperature. The membranes were treated with the following primary antibodies: Anti-β-actin (1:1,000; cat. no. GB12001; Wuhan Servicebio Technology Co., Ltd.), anti-CD63 (1:1,000; cat. no. NBP2-67425; Novus Biologicals, LLC), anti-phosphorylated (p)-Akt Ser473 (1:1,000; cat. no. 4060; Cell Signaling Technology, Inc.) anti-tumor susceptibility gene 101 protein (TSG101; 1:1,000; cat. no. ab125011; Abcam), anti-PTEN (1:1,000; cat. no. 9188; Cell Signaling Technology, Inc.) and anti-Akt (1:1,000; cat. no. 4691; Cell Signaling Technology, Inc.). After incubation at 4°C for 24 h, the membranes were washed three times with TBS with 0.1% Tween-20, followed by a 1-h incubation at ~25°C with a HRP-conjugated goat anti-rabbit IgG secondary antibody (1:1,000; cat. no. GB23303; Wuhan Servicebio Technology Co., Ltd.). An ECL immunoblotting kit (Dalian Meilun Biology Technology Co., Ltd.) was used to detect protein expression levels relative to the β-actin loading control. The proteins were visualized using chemiluminescent film and analyzed using ImageJ software (version 1.52; National Institutes of Health). Each experiment was independently conducted in triplicate.
Transfection
The miR-17-5p mimics (5′-CAAAGUGCUUACAGUGCAGGUAG-3′), inhibitor (5′-CUACCUGCACUGUAAGCACUUUG-3′) and negative control [mimics NC (5′-UUCUCCGAACGUGUCACGUTT-3′) and inhibitor NC (5′-CAGUACUUUUGUGUAGUACAA-3′)] were acquired from Shanghai GenePharma Co., Ltd. For subsequent experimentation, SH-SY5Y cells were seeded on a 6-well plate at a density of 1×106 cells/ml and cultured overnight at 37°C. The next day, cells were transfected with these molecular products (100 nM mimic, inhibitor or NC) using TransIntro™ EL Transfection Reagent (Beijing Transgen Biotech Co., Ltd.), following the manufacturer's protocols. The media were replaced with fresh growth medium and transfection was carried out for 6 h. Cells were cultured at 37°C with 5% CO2, and the medium was replaced after 24 h. Then, 48 h after changing the medium, the cells were harvested for subsequent experiments. Transfection efficiency was confirmed by RT-qPCR.
Cellular proliferation assay
In vitro cellular proliferative capacity was measured using the Cell Counting Kit-8 (CCK-8; Dalian Meilun Biology Technology Co., Ltd.), according to the manufacturer's protocols. Transfected cells were seeded into 96-well plates to a final volume of 100 µl complete medium (3×103 cells/well; five wells per concentration) and cultured for 4 days; 10 µl CCK-8 reagent was added to each well at the indicated time intervals (0, 1, 2, 3 and 4 days), after which the cultured plates were incubated for a further 2 h at 37°C. The OD of each well was recorded at 450 nm using a microplate reader (Synergy H1; BioTek Instruments, Inc.).
Colony formation assay
Transfected cells (500 cells/well) were seeded into 6-well plates in DMEM enriched with 10% FBS, followed by incubation at 37°C (5% CO2). After culturing for 2 weeks, the cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min at 37°C and stained with 1% crystal violet for 25 min at room temperature. Colonies were defined as >50 cells and counted under a fluorescence microscope (magnification, ×400) in five randomly selected fields.
Cellular migration assay
The migratory capacity of SH-SY5Y cells was observed using Transwell chambers with a pore size of 8 µm. After 24 h, transfected cells were isolated and resuspended in serum-free DMEM. The resuspended cells (~2×104 cells/well) were added into the upper chambers, and the lower chambers were filled with 500 µl DMEM containing 20% FBS. The plates were cultured for 12–20 h at 37°C with 5% CO2. The migrated cells were fixed with 4% paraformaldehyde for 25 min, followed by staining with 1% crystal violet for 20 min. Both the fixing and staining assays were conducted at room temperature. Migratory cells were counted in five randomly selected fields, and images were captured with a light microscope (magnification, ×200; Carl Zeiss AG).
Dual-luciferase reporter assay
miR-17-5p was predicted to interact with the 3′-UTR of PTEN. The wild-type (wt) and mutant (mut) 3′-UTR of PTEN mRNA were purchased from Shanghai GenePharma Co., Ltd., and were introduced between the NotI and XhoI restriction sites of the psiCHECK-2 vector (Promega Corporation), and the resulting vectors were termed psiCHECK-2-PTEN-wt and psiCHECK-2-PTEN-mut (the binging site ‘GCA CUU U’ was replaced by ‘CGU GAA A’), accordingly. The following primers were used for the amplification of particular fragments: PTEN-wt forward, 5′-CACAACTCGAGGCCCTGTACCATCCCAAGTC-3′ and reverse, 5′-AAGGAAAAAAGCGGCCGCACTGGCAGGTAGAAGGCAAC-3′; and PTEN-mut forward, 5′-CTAGAAATTTTCACGTTAATGTTCATAACGATGGCTGT-3′ and reverse, 5′-ACATTAACGTGAAAATTTCTAGAACTAAACATTAAAC-3′. 293T cells (1×105 cells/well; American Type Culture Collection) were co-transfected with 0.1 mg psiCHECK-2-PTEN-wt or 0.1 mg psiCHECK-2-PTEN-mut and 10 nm miR-17-5p mimics or the corresponding NC mimic using LipoFiter™ (Hanbio Biotechnology Co., Ltd.); 48 h post-transfection, the cells were collected and evaluated using the Dual-Luciferase reporter assay (Promega Corporation), according to the manufacturer's protocols. Firefly luciferase activity was normalized to that of Renilla luciferase.
Statistical analysis
The obtained data were statistically analyzed using SPSS version 21.0 (IBM Corp.) and graphically represented using GraphPad Prism 7 (GraphPad Software, Inc.). Each experiment was conducted in triplicate and the data are presented as the mean ± SD. An unpaired t-test was used to evaluate comparisons between two independent groups. One-way ANOVA was used to assess variations among multiple groups, followed by Tukey's multiple comparison tests. The P-values were two-sided, and P<0.05 was considered to indicate a statistically significant difference.
Results
PTEN and miR-17–92 are involved in NB
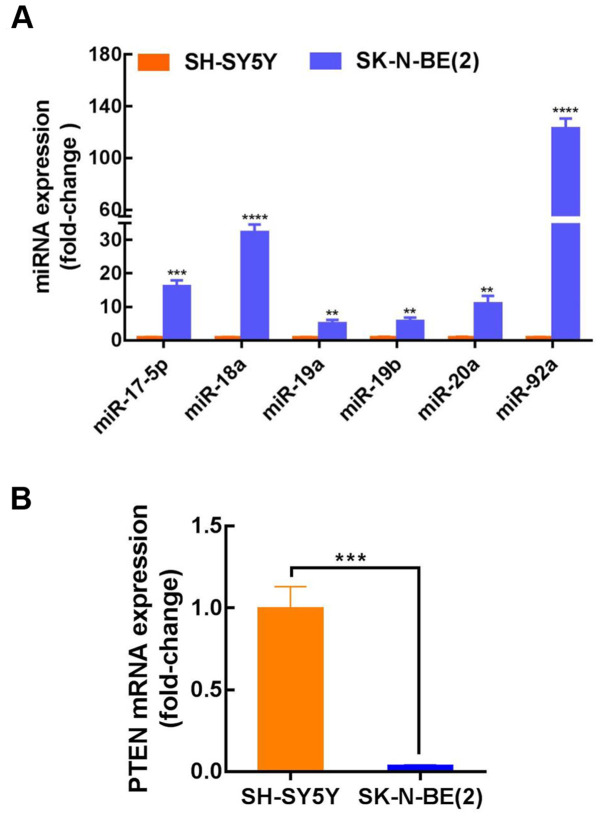
The published results from three independent studies were collected and analyzed (Table II), and the expression of the miR-17–92 cluster in MYCN-amplified NB tissues was found to be considerably higher than that in non-MYCN amplified NB. For further clarification of the miR-17–92 cluster mechanism in NB, three prediction databases (TargetScan, DIANA and miRanda) were used to identify potential binding targets. The target gene prediction results indicated a conserved binding site for miR-17-5p, miR-92a, miR-20a, miR-19a and miR-19b within the PTEN 3′-UTR (Table III). However, no PTEN binding sites were identified for miR-18a. SK-N-BE(2) and SH-SY5Y cells were then used to further investigate the role of the miR-17–92 cluster in NB progression. SK-N-BE(2) cells exhibit MYCN amplification and are known to be more aggressive than non-MYCN-amplified SH-SY5Y cells. The results indicated that the expression level of miR-17-92 in SK-N-BE(2) cells (MYCN-amplified) was considerably higher than that in SH-SY5Y cells (non-MYCN amplified); however, the expression of PTEN was considerably decreased (Fig. 1A and B). These findings demonstrated that the expression of PTEN was lower, while miR-17-92 expression was elevated in MYCN-amplified NB, and that miR-17-5p, miR-92a, miR-20a, miR-19a and miR-19b may target PTEN. To confirm these findings, miR-17-5p was selected for further investigation. The expression level of miR-17-5p in SK-N-BE(2) cells was ~16-fold higher than that in SH-SY5Y cells, though there are almost no published reports on the role of miR-17-5p in NB.
Table II.
Differentially expressed miRNAs in human NB tumor.
| miRNA | Levels | Tissue | Refs. |
|---|---|---|---|
| miR-17-5p | Up | MYCN-amplified NB | (6,7) |
| miR-18a | Up | MYCN-amplified NB | (6–8) |
| miR-19a | Up | MYCN-amplified NB | (7,8) |
| miR-19b | Up | MYCN-amplified NB | (7) |
| miR-20a | Up | MYCN-amplified NB | (7,8) |
| miR-92a | Up | MYCN-amplified NB | (6–8) |
miR/miRNA, microRNA; MYCN, N-myc proto-oncogene; NB, neuroblastoma.
Table III.
Putative binding sites for miRNAs in the 3′UTR of PTEN.
| Position in the 3′UTR of PTEN | Predicted consequential pairing of target region and miRNA |
|---|---|
| Position 272–278 | |
| PTEN 3′UTR | 5′-GGAUUAAUAAAGAUGGCACUUUC-3′ |
| hsa-miR-17-5p | 3′-GAUGGACGUGACUUCGUGAAAC-5′ |
| Position 272–278 | |
| PTEN 3′UTR | 5′-GGAUUAAUAAAGAUGGCACUUUC-3′ |
| hsa-miR-20a-5p | 3′-GAUGGACGUGAUAUUCGUGAAAU-5′ |
| Position 1221–1228 | |
| PTEN 3′UTR | 5′-AAUGAAUUUUGCAGUUUUGCACA-3′ |
| hsa-miR-19a-3p | 3′-AGUCAAAACGUAUCUAAACGUGU-5′ |
| Position 1221–1228 | |
| PTEN 3′UTR | 5′-AAUGAAUUUUGCAGUUUUGCACA-3′ |
| hsa-miR-19b-3p | 3′-AGUCAAAACGUACCUAAACGUGU-5′ |
| Position 4003–4010 | |
| PTEN 3′UTR | 5′-AGUAAAUGAAAAAAUGUGCAAUA-3′ |
| hsa-miR-92a-3p | 3′-UGUCCGGCCCUGUUCACGUUAU-5′ |
Binding regions are highlighted in bold. UTR, untranslated region; miRNA/miR, microRNA.
Figure 1.
miR-17-92 is upregulated in N-myc proto-oncogene-amplified neuroblastoma cells and predicted to target PTEN. (A) Relative expression levels of miR-17-92 in SH-SY5Y and SK-N-BE(2) cells. Relative fold-levels (SK-N-BE(2)/SH-SY5Y): miR-17-5p, ~16-fold; miR-18a, ~32-fold; miR-19a, ~5-fold; miR-19b, ~5-fold; miR-20a, ~11-fold; and miR-92a, ~123-fold. (B) Relative expression levels of PTEN in SH-SY5Y and SK-N-BE (2) cells. PTEN expression was ~0.04-fold lower in SK-N-BE(2) cells. **P<0.01, ***P<0.001 and ****P<0.0001 vs. SH-SY5Y cells. Data are presented as the mean ± SD. The unpaired t-test was used for the comparison of mean values. miR, microRNA.
SK-N-BE(2) cells deliver miR-17-5p to SH-SY5Y cells via exosomes
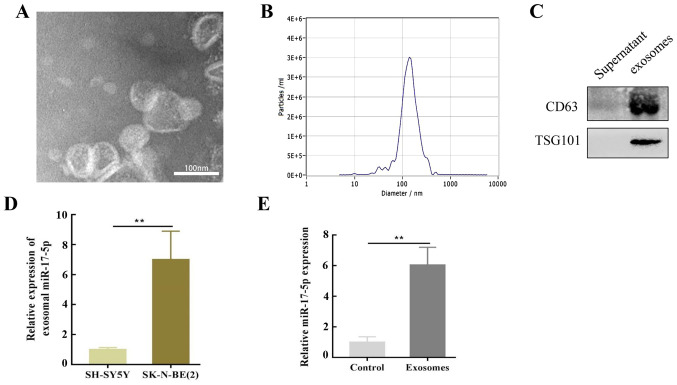
It was hypothesized that SK-N-BE(2) cells may influence SH-SY5Y cells via exosomes secreted into the culture medium. To verify this assumption, exosomes were isolated from SK-N-BE(2) cells using differential ultracentrifugation and characterized by TEM. The TEM results revealed that the purified exosomes were primarily cup-shaped in structure, with a diameter of 50–150 nm. An abnormal lipid bilayer was also observed (Fig. 2A), and NTA revealed that the number of purified exosomes peaked at a mean diameter of 50–150 nm (Fig. 2B). Moreover, these vesicles were enriched with exosomal markers, including CD63 and TSG101 (Fig. 2C).
Figure 2.
SK-N-BE(2) cells deliver miR-17-5p to SH-SY5Y cells through exosomes. (A) Transmission electron micrograph of SK-N-BE(2) cell-derived exosomes. Scale bar, 100 nm. (B) Nanoparticle tracking analysis was employed for analyzing exosomes. (C) Markers of exosomes such as CD63 and TSG101 proteins, were detected by western blot assay in SK-N-BE(2) cell-derived exosomes and cell culture supernatants. (D) miR-17-5p expression in SK-N-BE(2) and SH-SY5Y cell-derived exosomes. miR-17-5p expression in SK-N-BE(2) exosomes was 7.0±1.9-fold higher compared with SH-SY5Y cells. (E) miR-17-5p expression in SH-SY5Y co-cultured with exosomes derived from SK-N-BE(2) cells via reverse transcription-quantitative PCR. **P<0.01. miR, microRNA; TSG101, tumor susceptibility gene 101 protein.
To determine the molecular mechanism of SK-N-BE(2) cell-induced regulation, SH-SY5Y cells were co-cultured with exosomes isolated from SK-N-BE (2) cells, and RT-qPCR was performed to evaluate the levels of miR-17-5p expression. The results revealed that miR-17-5p was upregulated in SK-N-BE(2)-associated exosomes compared with those from SH-SY5Y cells (Fig. 2D). This upregulation in MYCN-amplified NB cell exosomes may reveal that oncomiRs and the cancerous phenotype can be transferred to other cells. Notably, a high expression level of miR-17-5p was observed in SH-SY5Y cells co-cultured with SK-N-BE(2)-derived exosomes (Fig. 2E).
miR-17-5p upregulation enhances the proliferative and migratory potential of SH-SY5Y cells
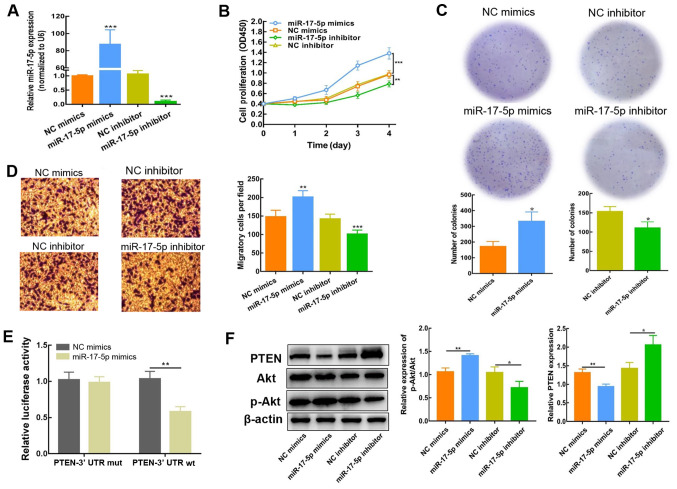
To determine whether miR-17-5p impacts the biological activity of NB cells, SH-SY5Y cells were transfected with miR-17-5p mimics or inhibitor; miR-17-5p expression was detected using RT-qPCR 2 days post-transfection. The expression of miR-17-5p was significantly increased following transfection with miR-17-5p mimics, but reduced as a result of miR-17-5p inhibitor transfection (Fig. 3A). CCK-8, colony formation and Transwell assays were subsequently conducted to evaluate the influence of miR-17-5p expression on the proliferation and migration of SH-SY5Y cells. As depicted in Fig. 3B-D, high miR-17-5p expression enhanced SH-SY5Y cell proliferation and migration. By contrast, inhibiting miR-17-5p expression resulted in a decreased level of proliferation and migration. These findings demonstrated that miR-17-5p significantly influenced the regulation of NB cellular motility, including proliferation and metastatic capacity.
Figure 3.
Upregulation miR-17-5p enhances the proliferative and migratory capability of SH-SY5Y. (A) miR-17-5p expression was identified via reverse transcription-quantitative PCR in SH-SY5Y cells after transfection of miR-17-5p mimic or inhibitor. (B) Cell proliferation was determined using a Cell Counting Kit-8 assay. (C) Colony formation assay was employed for the determination of cell proliferation in SH-SY5Y cells. (D) Migratory ability of transfected cells was determined using a Transwell assay and the obtained results are presented as the number of migrated cells per field (magnification, ×400). (E) Detection of luciferase activity. (F) PTEN, Akt and p-Akt protein expression in SH-SY5Y cells (transfected with miR-17-5p mimics or inhibitor) were measured using western blotting. The results are presented as the average of three experiments run individually (mean ± SD). *P<0.05, **P<0.01 and ***P<0.001 vs. NC mimic or NC inhibitor. NC, negative control; miR, microRNA; UTR, untranslated region; mut, mutant; wt, wild-type; p-, phosphorylated.
miR-17-5p stimulates the PI3K/Akt signaling cascade in NB cells by downregulating PTEN
For further evaluation of the regulatory effect of miR-17-5p on PTEN, the predicted interaction sites between miR-17-5p and PTEN were identified using TargetScan. As indicated in Table III, the PTEN 3′UTR contains a number of miR-17-5p interaction sites. Moreover, a dual-luciferase reporter assay was conducted using the PTEN-Wt 3′-UTR sequence, which was co-transfected into NB cells with miR-17-5p mimics or NC. Compared with the NC mimics, the luciferase activity of PTEN-Wt was blocked in the presence of miR-17-5p mimics (Fig. 3E), which indicated that miR-17-5p interacted with PTEN. Consistently, the results of western blot analysis showed that the expression of PTEN protein in SH-SY5Y cells was decreased, and that the p-Akt/Akt ratio was significantly elevated after transfection with miR-17-5p mimics (Fig. 3F). By contrast, PTEN protein expression levels were elevated, whereas the p-Akt/Akt ratio was reduced, following miR-17-5p inhibition, demonstrating that PTEN expression was attenuated via miR-17-5p. Collectively, these results indicated that the PI3K/Akt signaling cascade was activated by miR-17-5p via the downregulation of PTEN.
Exosomal miR-17-5p enhances the proliferative and migratory abilities of SH-SY5Y cells
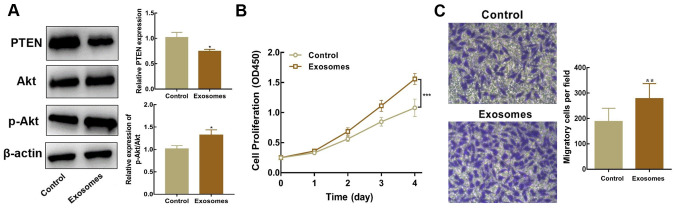
Exosomes isolated from SK-N-BE(2) cells were co-cultured with SH-SY5Y cells to identify whether exosomal miR-17-5p facilitates the activation of the PI3K/Akt signaling cascade (PTEN-mediated), and subsequently regulates the proliferation and migration of SH-SY5Y cells. Western blot analysis revealed that PTEN expression was lower, whereas the p-Akt/Akt ratio was higher, in SH-SY5Y cells co-cultured with exosomes than in untreated cells (Fig. 4A). Moreover, SH-SY5Y cells co-cultured with exosomes overexpressing miR-17-5p exhibited higher proliferative and migratory abilities (Fig. 4B and C). In summary, these results indicated that inhibiting PTEN expression and stimulating the PI3K/Akt signaling cascade via exosomal miR-17-5p enhanced the proliferative and migratory potential of SH-SY5Y cells.
Figure 4.
Exosomal microRNA-17-5p promotes the proliferation and migration of SH-SY5Y cells via inhibition of PTEN and activation of the PI3K/Akt signaling pathway. (A) PTEN, Akt and p-Akt protein expression in SH-SY5Y cells co-cultured with exosomes were measured using western blotting. (B) Cell proliferation was determined using a Cell Counting Kit-8 assay. (C) Cell migration was evaluated via a Transwell assay. *P<0.05, **P<0.01 and ***P<0.001 vs. untreated SH-SY5Y cells. p-, phosphorylated.
Discussion
MYCN amplification is the most effective prognostic marker of adverse disease outcome in NB (23). MYCN expression has been found to positively correlate with metastatic behavior, epithelial-mesenchymal transition, cell cycle progression and impaired immune surveillance in NB (4). Due to its secondary structure and no obvious sites for the binding of small molecules, the identification of therapeutics targeting MYCN has been challenging. To date, the direct targeting of myc family proteins has been unsuccessful, hence the potential for indirect targeting is currently under investigation (2,4). Exosomes are cell-secreted nanoscale vesicles that are derived from NB cells (MYCN-amplified), which mediate intercellular communication and impart aggressive NB phenotypes (24). Exosomes secreted from cancerous cells may serve to mediate the transfer of these phenotypes to sensitive recipient cells via miRNAs (25,26). However, there is still ambiguity regarding the ability of NB cells to transfer phenotypes in this manner. The present study revealed that exosomes derived from MYCN-amplified NB cells, with elevated miR-17-5p expression, inhibited PTEN and activated the PI3K/Akt signaling cascade, thus enhancing the proliferative and migratory capacities of non-MYCN amplified cells.
There is evidence to suggest that N-myc directly interacts with the promoter region of the miR-17-92 cluster, which results in its upregulation (27). The precursor transcript of the miR-17-92 gene comprises six tandem stem-loop hairpins that produce six mature miRNAs, including miR-92a, miR-20a, miR-19a, miR-19b, miR-18a and miR-17-5p (28). Consistently, the present study demonstrated that miR-17-92 cluster expression was upregulated in MYCN-amplified SK-N-BE(2) cells. Moreover, SK-N-BE(2)-derived exosomes were found to transfer miR-17-5p from the oncogenic miR-17-92 cluster, promoting the elevation of miR-17-5p expression in SH-SY5Y cells (non-MYCN amplified). Notably, the transfer of miR-17-5p via SK-N-BE(2)-derived exosomes enhanced the suppression of PTEN expression in SH-SY5Y cells.
Haug et al (24) analyzed the miRNA profiles of exosomes isolated from MYCN-amplified NB cells. Functional enrichment analysis revealed several well-characterized pathways, including the PTEN/PI3K/Akt signaling cascade. In previous years, several reports have also established the importance of PTEN/PI3K/Akt signaling in NB survival, angiogenesis, proliferation and invasion, including its association with MYCN (29,30). In the present study, the results of the dual-luciferase reporter assay demonstrated that PTEN was targeted, and its expression was downregulated, by miR-17-5p. Furthermore, Paul et al (20) immunohistochemically assessed the expression of PTEN using a tissue microarray containing human NB sections, revealing that PTEN was significantly downregulated in MYCN-amplified NB tissues. In the current study, miR-17-5p-upregulated exosomes derived from MYCN-amplified NB cells increased the proliferative and migratory potential of non-MYCN amplified NB cells in vitro. Previous studies have revealed that GDNF family receptor α2 plays a key role in the regulation of NB development by suppressing PTEN via the activation of the PI3K/Akt signaling cascade (31–33). In NB cells, the upregulation of miR-17-5p tends to elevate the levels of Akt phosphorylation, which indicates that in NB, the PI3K/Akt signaling cascade is activated via miR-17-5p.
It is commonly known that within a single tumor, the presence of different cell cancer subsets with unique genotypes results in intra-tumor heterogeneity (34,35). Clonal subsets of tumor cells with different mutant states may also exist in a single NB tumor. Exosomes may facilitate the interaction between different subsets of tumor cell clones, as well as with non-cancerous cells in the tumor microenvironment (19). The present study was also conducted in the context of intra-tumor heterogeneity. However, as the research is still at the preclinical stage, the internalization of PKH67-labeled exosomes by SH-SY5Y cells was not observed. Although several articles have reported that miR-17-5p can be regulated by N-myc (36–38), this was not confirmed using SH-SY5Y cells in the present study, and the mechanisms by which exosomal miRNAs influence NB are yet to be elucidated. Therefore, additional experimental approaches, such as Transwell invasion assays, apoptosis analysis and animal studies, are required to reveal the intrinsic pathways of exosomal miR-17-5p in NB, both in vitro and in vivo.
In conclusion, the results of the present study suggested that exosomes derived from MYCN-amplified SK-N-BE(2) cells transfer miR-17-5p into non-MYCN amplified SH-SY5Y cells, promoting the proliferation and migration of non-MYCN amplified NB cells. Therefore, exosomes from SK-N-BE(2) cells with upregulated miR-17-5p expression may be a key candidate target for the treatment of NB.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by Qingdao Minsheng Science and Technology Plan Project Task Book (grant no. 18-6-1-71-nsh) and Qingdao Outstanding Health Professional Development.
Funding
This work was supported by Qingdao Minsheng Science and Technology Plan Project Task Book (grant no. 18-6-1-71-nsh) and Qingdao Outstanding Health Professional Development.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LH and HL conceived the project and supervised the experiments. WC, BY, YZ and YH performed the experiments. JY, XH and JZ provided technical support. WC, LY and LS performed statistical analysis. WC and XH wrote the manuscript with help from all of the authors. WC, XH and BY confirm the authenticity of all the raw data. HL and LH drafted the article and critically revised it for important intellectual content. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Pastor ER, Mousa SA. Current management of neuroblastoma and future direction. Crit Rev Oncol Hematol. 2019;138:38–43. doi: 10.1016/j.critrevonc.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Johnsen JI, Dyberg C, Wickstrom M. Neuroblastoma-A neural crest derived embryonal malignancy. Front Mol Neurosci. 2019;12:9. doi: 10.3389/fnmol.2019.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3:a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buechner J, Einvik C. N-myc and noncoding RNAs in neuroblastoma. Mol Cancer Res. 2012;10:1243–1253. doi: 10.1158/1541-7786.MCR-12-0244. [DOI] [PubMed] [Google Scholar]
- 6.Schulte JH, Horn S, Otto T, Samans B, Heukamp LC, Eilers UC, Krause M, Astrahantseff K, Klein-Hitpass L, Buettner R, et al. MYCN regulates oncogenic MicroRNAs in neuroblastoma. Int J Cancer. 2008;122:699–704. doi: 10.1002/ijc.23153. [DOI] [PubMed] [Google Scholar]
- 7.Bray I, Bryan K, Prenter S, Buckley PG, Foley NH, Murphy DM, Alcock L, Mestdagh P, Vandesompele J, Speleman F, et al. Widespread dysregulation of miRNAs by MYCN amplification and chromosomal imbalances in neuroblastoma: Association of miRNA expression with survival. PLoS One. 2009;4:e7850. doi: 10.1371/journal.pone.0007850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mestdagh P, Fredlund E, Pattyn F, Schulte JH, Muth D, Vermeulen J, Kumps C, Schlierf S, De Preter K, Van Roy N, et al. MYCN/c-MYC-induced microRNAs repress coding gene networks associated with poor outcome in MYCN/c-MYC-activated tumors. Oncogene. 2010;29:1394–1404. doi: 10.1038/onc.2009.429. [DOI] [PubMed] [Google Scholar]
- 9.Babaei K, Shams S, Keymoradzadeh A, Vahidi S, Hamami P, Khaksar R, Norollahi SE, Samadani AA. An insight of microRNAs performance in carcinogenesis and tumorigenesis; an overview of cancer therapy. Life Sci. 2020;240:117077. doi: 10.1016/j.lfs.2019.117077. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X, Lu H, Li F, Hao X, Han L, Dong Q, Chen X. MicroRNA-429 inhibits neuroblastoma cell proliferation, migration and invasion via the NF-κB pathway. Cell Mol Biol Lett. 2020;25:5. doi: 10.1186/s11658-020-0202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X, Gu H, Wang L, Zhang P, Du J, Shen L, Jiang D, Wang J, Li X, Zhang S, et al. MicroRNA-23a-5p mediates the proliferation and differentiation of C2C12 myoblasts. Mol Med Rep. 2020;22:3705–3714. doi: 10.3892/mmr.2020.11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang D, Gao W, Yang J, Liu J, Zhao J, Ge J, Chen Q, Liu B. miR181d promotes cell proliferation via the IGF1/PI3K/AKT axis in glioma. Mol Med Rep. 2020;22:3804–3812. doi: 10.3892/mmr.2020.11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galardi A, Colletti M, Businaro P, Quintarelli C, Locatelli F, Di Giannatale A. MicroRNAs in neuroblastoma: Biomarkers with therapeutic potential. Curr Med Chem. 2018;25:584–600. doi: 10.2174/0929867324666171003120335. [DOI] [PubMed] [Google Scholar]
- 14.Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 15.McAndrews KM, Kalluri R. Mechanisms associated with biogenesis of exosomes in cancer. Mol Cancer. 2019;18:52. doi: 10.1186/s12943-019-0963-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: 10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morini M, Cangelosi D, Segalerba D, Marimpietri D, Raggi F, Castellano A, Fruci D, de Mora JF, Canete A, Yanez Y, et al. Exosomal microRNAs from longitudinal liquid biopsies for the prediction of response to induction chemotherapy in high-risk neuroblastoma patients: A proof of concept SIOPEN Study. Cancers (Basel) 2019;11:1476. doi: 10.3390/cancers11101476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Xu M, Yin M, Hong J, Chen H, Gao Y, Xie C, Shen N, Gu S, Mo X. Exosomal hsa-miR199a-3p promotes proliferation and migration in neuroblastoma. Front Oncol. 2019;9:459. doi: 10.3389/fonc.2019.00459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fonseka P, Liem M, Ozcitti C, Adda CG, Ang CS, Mathivanan S. Exosomes from N-Myc amplified neuroblastoma cells induce migration and confer chemoresistance to non-N-Myc amplified cells: Implications of intra-tumour heterogeneity. J Extracell Vesicles. 2019;8:1597614. doi: 10.1080/20013078.2019.1597614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul P, Qiao J, Kim KW, Romain C, Lee S, Volny N, Mobley B, Correa H, Chung DH. Targeting gastrin-releasing peptide suppresses neuroblastoma progression via upregulation of PTEN signaling. PLoS One. 2013;8:e72570. doi: 10.1371/journal.pone.0072570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Purushothaman A. Exosomes from cell Culture-Conditioned medium: Isolation by ultracentrifugation and characterization. Methods Mol Biol. 2019;1952:233–244. doi: 10.1007/978-1-4939-9133-4_19. [DOI] [PubMed] [Google Scholar]
- 23.Zafar A, Wang W, Liu G, Wang X, Xian W, McKeon F, Foster J, Zhou J, Zhang R. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev. 2020 Nov 6; doi: 10.1002/med.21750. (Epub ahead of print). doi: 10.1002/med.21750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haug BH, Hald ØH, Utnes P, Roth SA, LØkke C, FlÆgstad T, Einvik C. Exosome-like extracellular vesicles from MYCN-amplified neuroblastoma cells contain oncogenic miRNAs. Anticancer Res. 2015;35:2521–2530. [PubMed] [Google Scholar]
- 25.Su T, Xiao Y, Xiao Y, Guo Q, Li C, Huang Y, Deng Q, Wen J, Zhou F, Luo XH. Bone marrow mesenchymal stem Cells-Derived Exosomal miR-29b-3p Regulates Aging-Associated insulin resistance. ACS Nano. 2019;13:2450–2462. doi: 10.1021/acsnano.8b09375. [DOI] [PubMed] [Google Scholar]
- 26.Babuta M, Furi I, Bala S, Bukong TN, Lowe P, Catalano D, Calenda C, Kodys K, Szabo G. Dysregulated autophagy and lysosome function are linked to exosome production by Micro-RNA 155 in alcoholic liver disease. Hepatology. 2019;70:2123–2141. doi: 10.1002/hep.30766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loven J, Zinin N, Wahlstrom T, Muller I, Brodin P, Fredlund E, Ribacke U, Pivarcsi A, Pahlman S, Henriksson M. MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc Natl Acad Sci USA. 2010;107:1553–1558. doi: 10.1073/pnas.0913517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khuu C, Utheim TP, Sehic A. The three paralogous MicroRNA clusters in development and disease, miR-17-92, miR-106a-363, and miR-106b-25. Scientifica (Cairo) 2016;2016:1379643. doi: 10.1155/2016/1379643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Opel D, Poremba C, Simon T, Debatin KM, Fulda S. Activation of Akt predicts poor outcome in neuroblastoma. Cancer Res. 2007;67:735–745. doi: 10.1158/0008-5472.CAN-06-2201. [DOI] [PubMed] [Google Scholar]
- 30.Hogarty MD, Maris JM. PI3King on MYCN to improve neuroblastoma therapeutics. Cancer Cell. 2012;21:145–147. doi: 10.1016/j.ccr.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 31.Li Z, Xie J, Fei Y, Gao P, Xie Q, Gao W, Xu Z. GDNF family receptor alpha 2 promotes neuroblastoma cell proliferation by interacting with PTEN. Biochem Biophys Res Commun. 2019;510:339–344. doi: 10.1016/j.bbrc.2018.12.169. [DOI] [PubMed] [Google Scholar]
- 32.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: New modes and prospects. Nat Rev Mol Cell Biol. 2018;19:547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 33.Wu Q, Shang Y, Shen T, Liu F, Xu Y, Wang H. Neuroprotection of miR-214 against isoflurane-induced neurotoxicity involves the PTEN/PI3K/Akt pathway in human neuroblastoma cell line SH-SY5Y. Arch Biochem Biophys. 2019;678:108181. doi: 10.1016/j.abb.2019.108181. [DOI] [PubMed] [Google Scholar]
- 34.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barranha R, Costa JL, Carneiro F, Machado JC. Genetic heterogeneity in colorectal cancer and its clinical implications. Acta Med Port. 2015;28:370–375. doi: 10.20344/amp.5398. [DOI] [PubMed] [Google Scholar]
- 36.Lu R, Zhao G, Yang Y, Jiang Z, Cai J, Zhang Z, Hu H. Long noncoding RNA HOTAIRM1 inhibits cell progression by regulating miR-17-5p/ PTEN axis in gastric cancer. J Cell Biochem. 2019;120:4952–4965. doi: 10.1002/jcb.27770. [DOI] [PubMed] [Google Scholar]
- 37.Liang W, Yue Z. Lycium barbarum polysaccharides promote osteoblasts viability by regulating microRNA-17/PTEN. Life Sci. 2019;225:72–78. doi: 10.1016/j.lfs.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Li Z, Bai J, Song W, Zhang F. miR-17-5p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting phosphatase and tensin homolog. Mol Med Rep. 2019;19:3132–3138. doi: 10.3892/mmr.2019.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.