Abstract
PURPOSE:
Imaging of glioblastoma patients after maximal safe resection and chemoradiation commonly demonstrates new enhancement concerning for true tumor progression (TP). However, in 30–50% of patients, this enhancement primarily represents treatment effect, or pseudo-progression (PsP). We hypothesize that quantitative machine learning (ML) analysis of clinically-acquired multi-parametric magnetic resonance imaging (mpMRI) can identify subvisual imaging characteristics to provide robust, non-invasive, imaging signatures that can distinguish TP and PsP.
METHODS:
We evaluated independent discovery (n=40) and replication (n=23) cohorts of glioblastoma patients who underwent second resection due to progressive radiographic changes suspicious for recurrence. Deep learning and conventional feature extraction methods were used to extract quantitative characteristics from the mpMRI scans. Multivariate analysis of these features revealed radio-phenotypic signatures distinguishing among TP, PsP, and mixed response that compared with similar categories blindly defined by board-certified neuropathologists. Additionally, inter-institutional validation was performed on 20 new patients.
RESULTS:
Patients categorized as TP on neuropathology are significantly different (p<0.0001) from those with PsP, showing imaging features reflecting higher angiogenesis, higher cellularity, and lower water concentration. The accuracy of the proposed signature in leave-one-out-cross-validation was 87% for predicting PsP (AUC=0.92) and 84% for predicting TP (AUC=0.83), whereas in the discovery/replication cohort, it was 87% for predicting PsP (AUC=0.84) and 78% for TP (AUC=0.80). The accuracy on the inter-institutional cohort was 75%(AUC=0.80).
CONCLUSION:
Quantitative mpMRI analysis via ML reveals distinctive non-invasive signatures of TP vs PsP after treatment of glioblastoma. Integration of the proposed method into clinical studies can be performed via the freely available CaPTk software.
Keywords: pseudo-progression, glioblastoma, true progression, radiographic biomarker, machine learning
Introduction
Glioblastoma is the most common malignant primary adult brain tumor 1. It is associated with a grim prognosis, with median overall survival ranging from 16–20 months despite maximal treatment 2–4. Initial treatment of glioblastoma involves a combined approach of maximal safe surgical resection and adjuvant chemoradiation 3. Surveillance of patients after completion of this initial treatment relies heavily on follow-up serial magnetic resonance imaging (MRI) to detect disease recurrence. As such, distinguishing treatment-induced MRI changes from true progression (TP) of tumor has critical implications to clinical decision-making.
Unfortunately, standardization of radiographic metrics for treatment response and for determining disease progression in glioblastoma has proven quite difficult. The widespread variance in definitions of “progressive disease” and “stable disease” led to the development of the Macdonald criteria in 1990, which rely upon crude radiographic measurement of areas of contrast enhancement on post-treatment MRI 5. With the addition of temozolomide to the standard of care, clinicians soon determined that these criteria were unable to accurately distinguish between true progression and treatment effects. The phenomenon of pseudo-progression (PsP) was identified, which is a subacute treatment-related effect, usually occurring within three months of completion of chemoradiation 6,7, with imaging characteristics mimicking TP, as defined by the Macdonald criteria 5.
The diagnosis of PsP is usually made on the basis of spontaneous improvement or stabilization of imaging findings over several months, i.e., in the setting of continuation of the chemotherapy for at least six months. Previous studies suggest that nearly half (30–50%) of glioblastoma patients with worsened radiographic findings after standard chemoradiation do not suffer from TP, but from PsP 7–14. Since traditional MRI cannot reliably distinguish TP from PsP, clinicians caring for patients with glioblastoma must frequently choose between declaring TP (and modifying the patient’s current therapy) versus proceeding with invasive brain surgery for diagnostic clarity. On histopathologic analysis, such surgeries may reveal recurrent glioblastoma tumor, therapy-related changes (PsP), or a mixed response consisting of a combination of the two.
The Response Assessment in Neuro-Oncology (RANO) Working Group developed new criteria 15 to address some of the limitations of the Macdonald criteria. As clinicians recognized the high prevalence of PsP in the months immediately following completion of chemoradiation, subtle changes in post-treatment imaging are no longer deemed to represent progressive disease, unless there is evidence of clinical deterioration or obvious new disease outside of the treatment field. This classification allows patients to continue maintenance therapy safely, with the goal of achieving some delayed improvement. Nonetheless, some of these patients do actually show TP, and identifying these patients without tissue sampling would allow for earlier change in therapy.
The goal of this study is to non-invasively evaluate radiographic changes in glioblastoma patients treated with chemoradiation, by multivariate analysis of pre-operative multi-parametric MRI (mpMRI), in order to identify a radio-phenotypic signature to distinguish between TP, mixed response, and PsP. Analysis of radiographic data via advanced computational analytics has been increasingly shown to provide rich and highly informative characterizations of glioblastoma and its surrounding brain tissue 11,16–21, extending the evaluation of tissue properties beyond the capabilities of human visual interpretation. We hypothesize that quantification of subtle, yet spatially complex, quantitative imaging phenomic (QIP) features extracted from mpMRI can facilitate non-invasive classification of TP vs PsP, with sufficient sensitivity and specificity to allow discrimination on an individual patient basis.
Materials and Methods
Study Patient Population
The study population was identified on the basis of retrospective review of the electronic medical record of patients diagnosed with glioblastoma at the Hospital of the University of Pennsylvania from 2011 to 2015. The criteria for inclusion comprised a) initial gross total resection of the tumor followed by standard radiation therapy and temozolomide chemotherapy, b) demonstration of new/increasing enhancement areas on follow-up MRI, within 6 months after completion of radiation therapy, c) second resection, for histopathological tissue evaluation, and d) acquisition of mpMRI (i.e., T1, T1-Gd, T2, T2-FLAIR, DTI, DSC) within 15 days prior to the second resection. We identified 63 patients (men/women=38/25; average age=57.28; age range=[32.79–81.60]) satisfying these inclusion criteria and we randomly divided them to independent discovery (n=40; 23 TP, 6 PsP, 11 mixed response) and replication (n=23; 12 TP, 4 PsP, 7 mixed response) cohorts. Isocitrate dehydrogenase 1 (IDH1) was wild-type, mutant, and not otherwise specified for 52, 2, and 9 patients, respectively (Suppl. Table 1). This study was approved by the Institutional Review Board of the University of Pennsylvania, and was compliant with the Health Insurance Portability and Accountability Act.
Histopathological Tissue Evaluation
Following resection, the surgically extracted tissue specimens were entirely fixed in 10% buffered formalin, routinely processed, and embedded in paraffin. Five-micron thick sections of each specimen were cut onto glass slides, stained with hematoxylin and eosin (H&E), and assessed by two board-certified neuropathologists (M.M-L, M.P.N.) (blinded to our imaging assessment and the other rater) for presence of apparent tumor features and reactive treatment-related changes 11. The presence or absence of pseudopalisading necrosis and microvascular proliferation, which are features of recurrent glioblastoma; the presence or absence of dystrophic calcification and vascular hyalinization, and the percentage of geographic necrosis, representative of treatment-related changes, were quantified (Figure 1). Proliferative activity was determined by quantification of the number of mitotic figures in 10 high-power fields and semi-quantitative assessment of Ki-67 proliferative index by immunostaining (mouse monoclonal, MIB-1, IR62661; Dako, Carpinteria, California). Based on the combined assessment of these features, the entire resected specimen was scored from 1 to 6. Score 1 for <10% malignant features, score 2 for 10–25% malignant features, score 3 for 25–50% malignant features, score 4 for 50–75% malignant features, score 5 for 75–90% malignant features, and score 6 for >90% malignant features. A score of 1–2 was defined as pseudo-progression (PsP), 3–4 as a mixture of true progression (TP) and PsP, and 5–6 as TP. This combination was performed for clinical applicability. PsP (score 1–2) will continue the treatment as it was before. The mixture of TP and PsP (score 3–4) will change the treatment or continue the current treatment based on clinical status of the patient. The TP patients (score 5–6) will be recommended for repeat resection. We used linear weighted Cohen’s kappa to calculate the inter-rater agreement.
Figure 1.
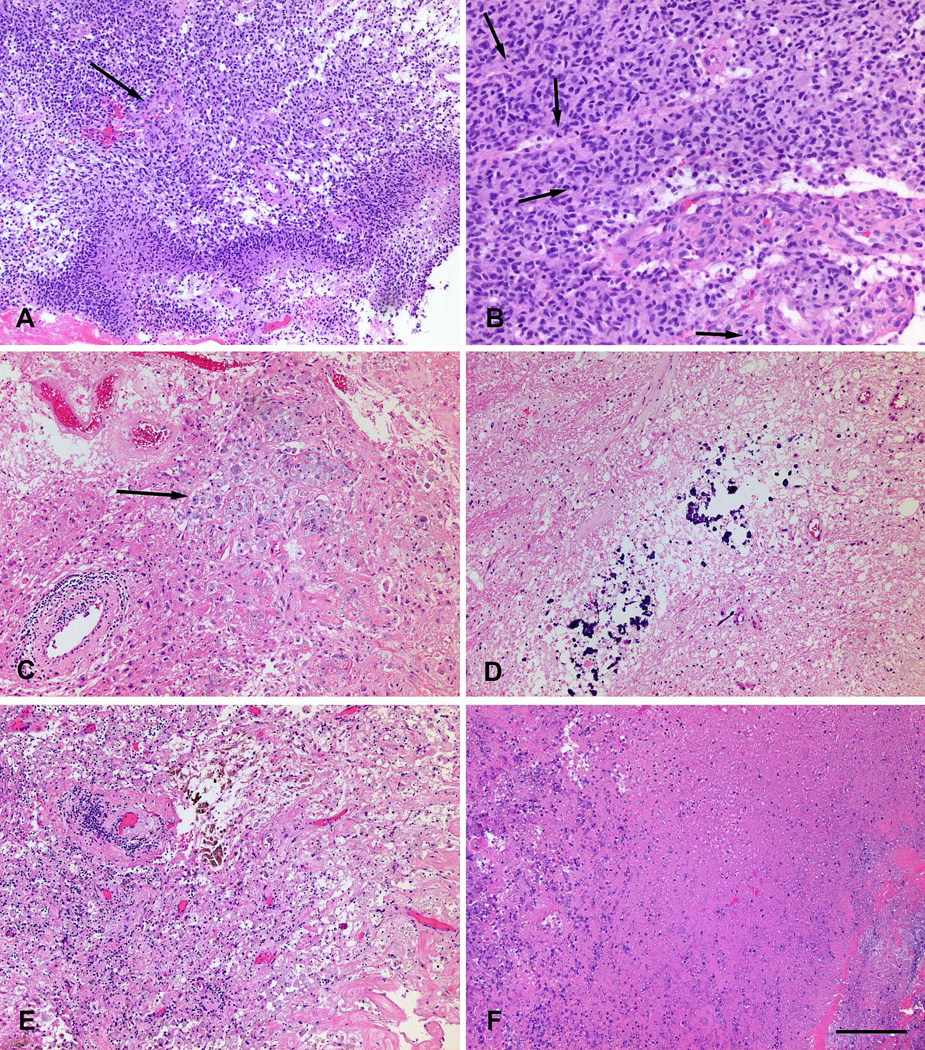
Histological features of true progression (malignant tumor, A and B) and pseudo-progression (treatment-related changes, C - F). A. Highly cellular tumor with pseudopalisading necrosis and microvascular proliferation (arrow). B. Mitotically active (arrows: mitoses) cellular tumor with microvascular proliferation. C. Thickened hyalinized vessel walls with focal lymphocytic inflammation, and macrophages (arrow). D. Dystrophic calcification. E. Hyalinized vessels, lymphocytes, macrophages and hemosiderin-laden macrophages. E. Geographic necrosis. Scale bar, lower right, indicates 200 microns in A, C - F; 100 microns in B.
MRI Acquisition Protocol
All MRI scans were performed on a Magnetom Tim Trio 3 Tesla scanner (Siemens, Erlangen, Germany) by using a 12-channel phased array head coil. Routine sequences included axial T1-weighted (T1): matrix 192×256×192, resolution 0.98×0.98×1.00 mm3, repetition time (TR in ms): 1760, echo time (TE in ms): 3.1; T1-weighted contrast enhanced with gadolinium (T1-Gd): matrix 192×256×192, resolution 0.98×0.98×1.00, TR: 1760, TE: 3.1; T2-weighted (T2): matrix 208×256×64, resolution 0.94×0.94×3.00, TR: 4680, TE: 85; T2 fluid-attenuated inversion recovery (T2-FLAIR): matrix 192×256×60, resolution 0.94×0.94×3.00, TR: 9420, TE: 141; and diffusion tensor imaging (DTI): matrix 128×128×40, resolution 1.72×1.72×3.00, 30 gradient directions, from which fractional anisotropy (DTI-FA), radial diffusivity (DTI-RAD), axial diffusivity (DTI-AX), and trace (DTI-TR) maps were calculated. DSC-MRI: FOV 22cm, 128×128×20, resolution 1.72×1.72×3 mm3, TR: 2000, TE: 45. The DSC-MRI sequences were acquired as follows: After an initial loading dose of 3mL of MultiHance (gadobenate dimeglumine) was administered to reduce the effect of contrast agent leakage, another bolus injection was given after five minutes with the remaining dose (for a total of 0.3mL/kg or 1.5 times single dose) during image acquisition (15 patients). With evolution of clinical protocols, dynamic contrast enhanced (DCE, also known as permeability) acquisitions have been routinely obtained on more recent studies. In these instances, DCE is obtained first with half of the total contrast and serves as the loading dose to reduce the effect of contrast agent leakage, followed by an additional bolus after a similar delay with the second half of the total contrast volume for the DSC acquisition (total 0.3 mL/kg, 48 patients).
MRI Pre-processing
All MRI volume scans of each individual patient were affinely co-registered intra-patient using the Functional MRI of the Brain Software Library (FSL) 22. Subsequently all scans were smoothed to remove any high frequency intensity variations (i.e., noise) 23, corrected for magnetic field inhomogeneities 24 and skull-stripped using FSL BET 25 followed by manual revision when needed. We extracted commonly used measurements 26 from the acquired DTI volumes, i.e., DTI-TR, DTI-AX, DTI-RAD, DTI-FA. The DSC-MRI volumes were used to computationally extract parametric brain maps of the relative cerebral blood volume (rCBV), peak height (PH) and percentage signal recovery (PSR) after considering leakage correction 27,28. Also, all DSC curves were aligned/normalized for inter-patient for baseline and maximum drop. Principal component analysis (PCA) was also employed to extract a summarized signal of the complete temporal perfusion dynamics encapsulated in the DSC-MRI modality, instead of using just isolated measurements such as the rCBV alone 29. Finally, the ML approach we adapted considers all four structural MRI images (T1, T1-Gd, T2, T2-FLAIR), the subtraction of T1 from T1-Gd and T2-FLAIR from T2 (following intensity normalization), four DTI-derived measurements, perfusion derived PCA volumes, and isolated perfusion derivative parametric brain maps. In our study we collectively refer to all these image volumes as mpMRI.
Defining target tissue
To define the target region of interest (ROI) we firstly registered the pre-operative and post-operative images of the timepoint suspicious for TP by using the Deformable Registration via Attribute Matching and Mutual-Saliency Weighting (DRAMMS) software 30. We then delineate the ROI describing the resected tissue. The regions in pre-surgery image that corresponds to the resected tissue in post-surgery image was defined as ROI. Therefore, the imaging properties and the pathology features correspond to the same resected region. Following the definition of these ROIs, all mpMRI sequences were analyzed to extract relevant comprehensive QIP features from the corresponding ROIs, in order to create our predictive model.
Feature Extraction
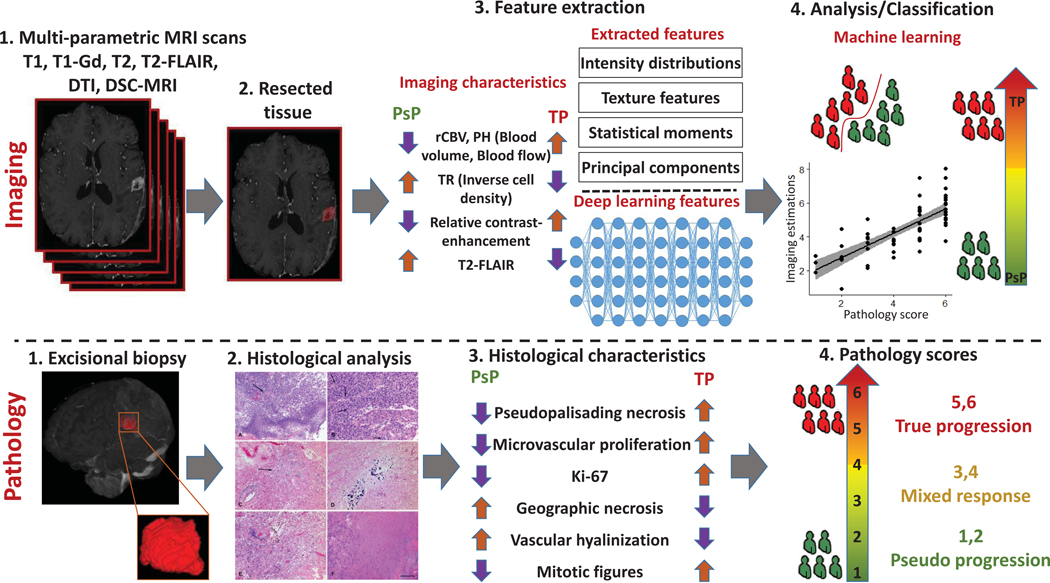
Considering the complexity of the problem our model is trying to address, we utilized two distinct approaches to ensure the comprehensiveness of the extracted QIP features from the mpMRI volumes, and hence that they describe all aspects of the radiographic appearance. The two approaches were distinctively based on deep learning and a priori selected (APS) feature extraction. We used and evaluated these two approaches both in combination and in comparison (Figure 2).
Figure 2.
Processing pipeline of the proposed radiomic analysis. Top row (imaging): 1) T1-w pre- and post-contrast, T2-w, T2-flair, DTI, DSC-MRI images are acquired. 2) Defining the resected enhancing tissues after registration of the pre-operative with post-operative images using DRAMMS. 3) Features are extracted from each region, quantifying intensity, shape, principal component analysis, statistics and texture. 4) Features analysis and classification between PsP versus non-PsP and TR versus non-TR. Bottom row (pathology): 1) Excisional biopsy 2) Histological analysis 3) Histological characteristics 4) Pathology scores.
Deep Learning Features
For obtaining the deep learning features, we used a pre-trained neural network 31,32 and adapted a convolutional neural network (CNN) model pre-trained on 1.2 million 3-channel images of the ImageNet LSVRC-2010, for classifying real-world images into different classes 31. The exact CNN model we utilized (imagenet_vgg_f 31) was provided by the VLfeat library 32 as part of their MatLab toolbox (MatConvNet) for computer vision applications. This CNN is a type of deep feed forward neural network that utilizes multilayer perceptrons with hidden layers. The hidden layers of CNN comprise convolutional (i.e., cross-correlation) layers, pooling layers, fully connected layers, and normalization layers 33,34. The convolution layer, which makes CNNs different from other types of deep neural networks, is a main layer of CNN and consists of several adaptive filters (as kernels) with small receptive fields. To apply this pre-trained model in our data, we created seven artificial 3-channel images, each channel of which describe an individual sequence from all the mpMRI considered (Suppl. Table 2).
APS radiomic features
We extracted 1040 APS radiomic features using the Cancer Imaging Phenomics Toolkit (CaPTk, www.cbica.upenn.edu/captk) 35. Specifically, the features extracted describe the first-order statistical distribution of voxel intensities within each ROI (comprising mean, median, maximum, minimum, skewness, and standard deviation) in all mpMRI sequences, the PCA summarized signal of the intensity distribution histogram of each mpMRI sequence, and texture features (second-order statistics) based on gray-level co-occurrence matrix (GLCM) 36 and gray-level run length matrix (GLRLM) 37. To obtain these texture features in 3 dimensions, all mpMRI volumes were first quantized to 16 gray levels within the ROI. GLCM and GLRLM were then populated by taking into account 13 main directions. A neighborhood of 3×3×3 was considered for GLCM. These features were first computed for each direction independently, and then averaged to find their final value. All features were rescaled via z-score normalization before machine learning (ML) analysis. To identify which of these 1040 extracted features had actual predictive value, we applied a sequential feature selection in the training data until convergence, based on a threshold in the accuracy improvement.
Classification and Correlation Approach
A multivariate pattern classification method, known as Support Vector Machines (SVM), was used to construct two classifiers, to predict TP/PsP; one classifier to distinguish between PsP (scores 1–2) vs. everything else (scores 3–6), and another classifier to distinguish between TP (scores 5–6) vs. everything else (scores 1–4). We conducted this multivariate analysis through linear configuration of SVM. The parameter for the soft margin cost function (C) was optimized on the training data, based on a 5-fold cross-validated grid search; C=2α, where αϵ[−5,5]. This parameter controls the influence of each individual support vector that involves trading error penalty for stability. These classifiers were trained separately for each classification task and each time one of the two types of features was used, i.e., deep learning and APS features. The classifiers were trained on the discovery cohort (n=40) and tested on the replication cohort (n=23). To confirm the robustness, accuracy, and generalizability of the proposed method in a larger cohort, while avoiding optimistically biased estimates of performance, we have also evaluated the classifiers in all 63 patients using a leave-one-out cross-validation (LOOCV) schema, where in every iteration of cross-validation, the features were selected using data of n-1 patients and tested on the left-out nth patient. All steps including feature selection and model development were performed through cross-validation.
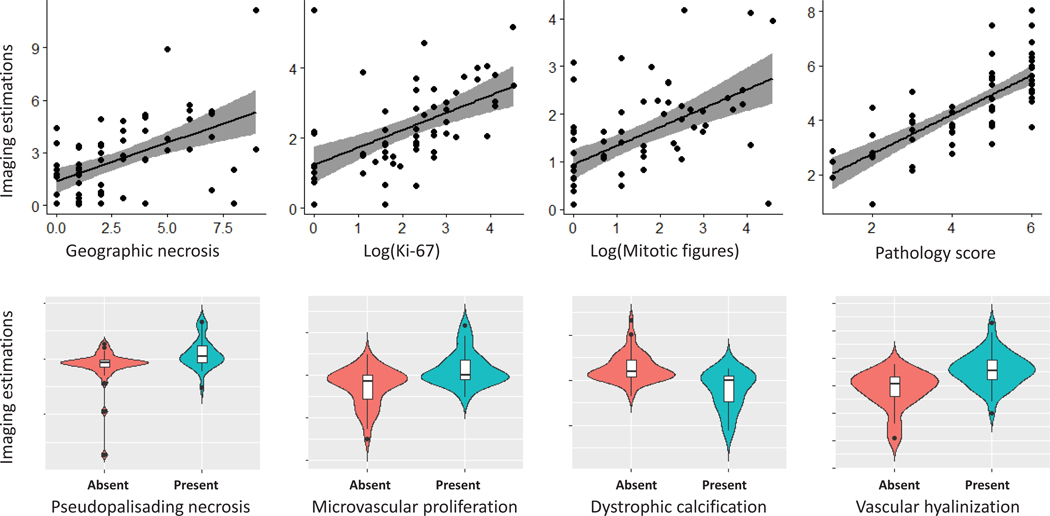
In addition to identifying a non-invasive signature to distinguish between TP and PsP, we also tried to find the correlations between the APS features and the histopathologic characteristics of the resected tissue specimen. This approach should identify complementary information of the extracted features by their correlations with pathological evaluations and quantitative scores. To achieve this, we used all 63 patients and trained separate support vector regression (SVR) models in a LOOCV configuration for the histopathologic characteristics with continuous values (i.e., mitotic figures, Ki-67, geographic necrosis, and the overall histopathology score), and SVM classification models for the histopathologic characteristics with discrete/binary values (i.e., pseudopalisading necrosis, microvascular proliferation, dystrophic calcification, and vascular hyalinization) (Figure 4).
Figure 4.
Histopathologic characteristics vs. machine learning estimations. The gray band represents the 95% confidence interval for the regression line.
Inter-institutional validation
We evaluated our method on an independent testing cohort from a different institute and with different acquisition protocols (20 patients, 10 TP, 10 PsP). In particular, we trained the model on the dataset acquired from University of Pennsylvania and applied the model on an independent cohort acquired from Thomas Jefferson University. Due to the lack of diffusion tensor imaging in Thomas Jefferson University dataset, we created a model using structural, DSC perfusion, and apparent diffusion coefficient imaging sequences from University of Pennsylvania and tested on Thomas Jefferson University patients. Due to lack of pathology, the follow-up serial MR imaging examinations were used to confirm prediction of PsP and TP by a board certified neuro-radiologist (M.B.). We selected time points that are distinct from therapy changes (systemic therapy and radiation therapy), to reduce the probability of treatment-related changes being measured on the scans. All MRI scans were performed on a 1.5 Tesla GE Signa HDx scanner (General Electric, Milwaukee, WI, USA), using an 8-channel phased array head coil. Routine sequences include T1: matrix 256(4) or 512(16)×256(4) or 512(16)×15–30, resolution 0.39–0.86×0.39–0.86×6–10mm3,TR: 12.9–583.3,TE: 4.1–12; T1-Gd: matrix 512×512×22–130, resolution 0.43–0.57×0.43–0.57×1.5(18) or 7.5(2),TR: 516.7–600,TE: 7.9–12; T2: matrix 512×512×20–30, resolution 0.39–0.49×0.39–0.49×5, TR: 2466.7–5952, TE: 90.7–102.1; T2-FLAIR: matrix 512×512×20–30, resolution 0.39–0.47×0.39–0.47×6(14) or 6.5(4) or 7.5(2), TR: 10000–10015, TE: 126–148.5; ADC: matrix 256×256×30–37, resolution 0.93(1) or 1.02(19)×0.93(1) or 1.02(19)×5, TR: 8000–10000, TE: 76.8–101; DSC-MRI: FOV 22cm(17) or 24cm(3), 128×128×15–27, resolution 1.7(16) or 1.9(3)×1.7(16) or 1.9(3)×6(2) or 8(14) or 10(3), TR: 9–22.4, TE: 400–2000.
Results
The imaging information captured via both the methods was multivariately integrated via SVM to build two classification models: i) TP vs non-TP (mixed response + PsP), and ii) PsP vs non-PsP (mixed response + TP). Table 1 provides a summary of these results. We have used linear weighted Cohen’s kappa to calculate inter-rater agreement. The observed agreement (po) for pathological scores of 1–6 was 0.9103 with random agreement (pe) of 0.6562, Cohen’s kappa, 0.7392, and kappa error, 0.1091. The observed agreement (po) for PsP/Mix/TP was 0.9138 with random agreement (pe) of 0.6231, Cohen’s kappa, 0.7713, and kappa error, 0.0978.
Table 1.
Quantitative evaluation results
| PsP vs non-PsP | TP vs non-TP | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (%) | AUC | Sensitivity(%) | FNR (%) | Specificity(%) | FPR (%) | Accuracy (%) | AUC | Sensitivity(%) | FNR (%) | Specificity(%) | FPR (%) | |
| Deep Learning Features (Hold-out set) | 87.50 | 0.811 | 60.00 | 40 | 94.74 | 5.26 | 78.26 | 0.867 | 83.33 | 16.67 | 72.73 | 27.27 |
| APS Features (Hold-out set) | 86.96 | 0.842 | 75.00 | 25 | 89.47 | 10.53 | 78.26 | 0.803 | 83.33 | 16.67 | 72.73 | 27.27 |
| APS Features (LOOCV) | 87.30 | 0.919 | 80.00 | 20 | 88.68 | 11.32 | 84.13 | 0.835 | 80.00 | 20 | 89.29 | 10.71 |
| Combined Features (Hold-out set) | 69.57 | 0.776 | 75.00 | 25 | 68.42 | 31.58 | 78.26 | 0.712 | 83.33 | 16.67 | 72.73 | 27.27 |
Using Deep Learning Features
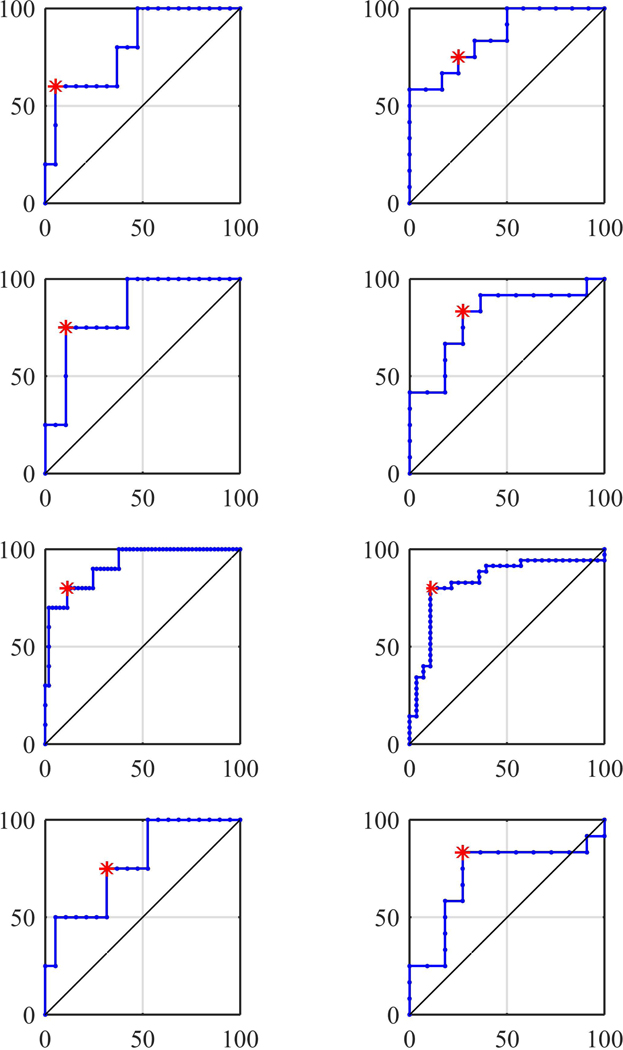
Similar performance was observed when using the deep learning features, which concluded in extracting >28000 features. Specifically, the classification performance was evaluated on the independent discovery and replication cohorts, and the accuracy for the ‘PsP vs non-PsP’ model was 87.50% (sensitivity=60.00%, specificity=94.74%, area under the curve (AUC)=0.8105, and for the ‘TP vs non-TP’ was equal to 78.26% (sensitivity=83.33%, specificity=72.73%, AUC=0.8636 (Table 1, Figure 3).
Figure 3.
ROC curves of different experiments: Left column shows PsP versus non-PsP, right column shows TP versus non-TP. Top to bottom rows show deep learning (hold-out set), APS features (hold-out set), APS features (LOOCV), and combined features (hold-out set), respectively. The vertical axis represents true positive rate and the horizontal axis represents false positive rate. The asterisk represents the shortest distance from the top left point.
Using APS Features
The PsP and TP models developed on the discovery cohort when applied to the replication cohort returned an accuracy of 86.96% (sensitivity=75.00%, specificity=89.47%) and 78.26% (sensitivity=83.33%, specificity=72.73%), respectively. A receiver operating characteristic (ROC) analysis also resulted in an AUC of 0.84 and 0.80 for ‘PsP vs non-PsP’ and ‘TP vs non-TP’ (Figure 3) classification models, respectively. While using only the APS radiomic features, the accuracy of our model using LOOCV in the pooled cohort was equal to 87.30% (sensitivity=80.00%, specificity=88.68%, AUC=0.9189) for ‘PsP vs non-PsP’, and 84.13% (sensitivity=80.00%, specificity=89.29%, AUC=0.8347) for TP vs non-TP, confirming its generalizability (Table 1, Figure 3). The most distinctive features for these classifiers can be found in Suppl. Table 3.
Integrating Deep Learning and APS Features
In addition to comparing the performance of each type of features, we also evaluated the performance of their integration (Table 1, Figure 3). Specifically, when combining these features, the accuracy for the ‘PsP vs non-PsP’ model was equal to 69.57% (sensitivity=75.00%, specificity=68.42%, AUC=0.7763) and for the ‘TP vs non-TP’ was 78.26% (sensitivity=83.33%, specificity=72.73%, AUC=0.7121).
Histopathologic Characteristics vs Machine Learning Estimates
The Pearson correlation coefficients between the SVR scores and logarithm of mitotic figures, logarithm of Ki-67, and geographic necrosis were estimated to be 0.54, 0.53, and 0.51, respectively. The Pearson correlation coefficients between the SVR scores and pathology scores was 0.76 (Figure 4). We also evaluated the trained SVM models for the presence or absence of pseudopalisading necrosis (AUC=0.7434), microvascular proliferation (AUC=0.7406), dystrophic calcification (AUC=0.8292), and vascular hyalinization (AUC=0.7939) (Figure 4).
Biologically Interpreted Features
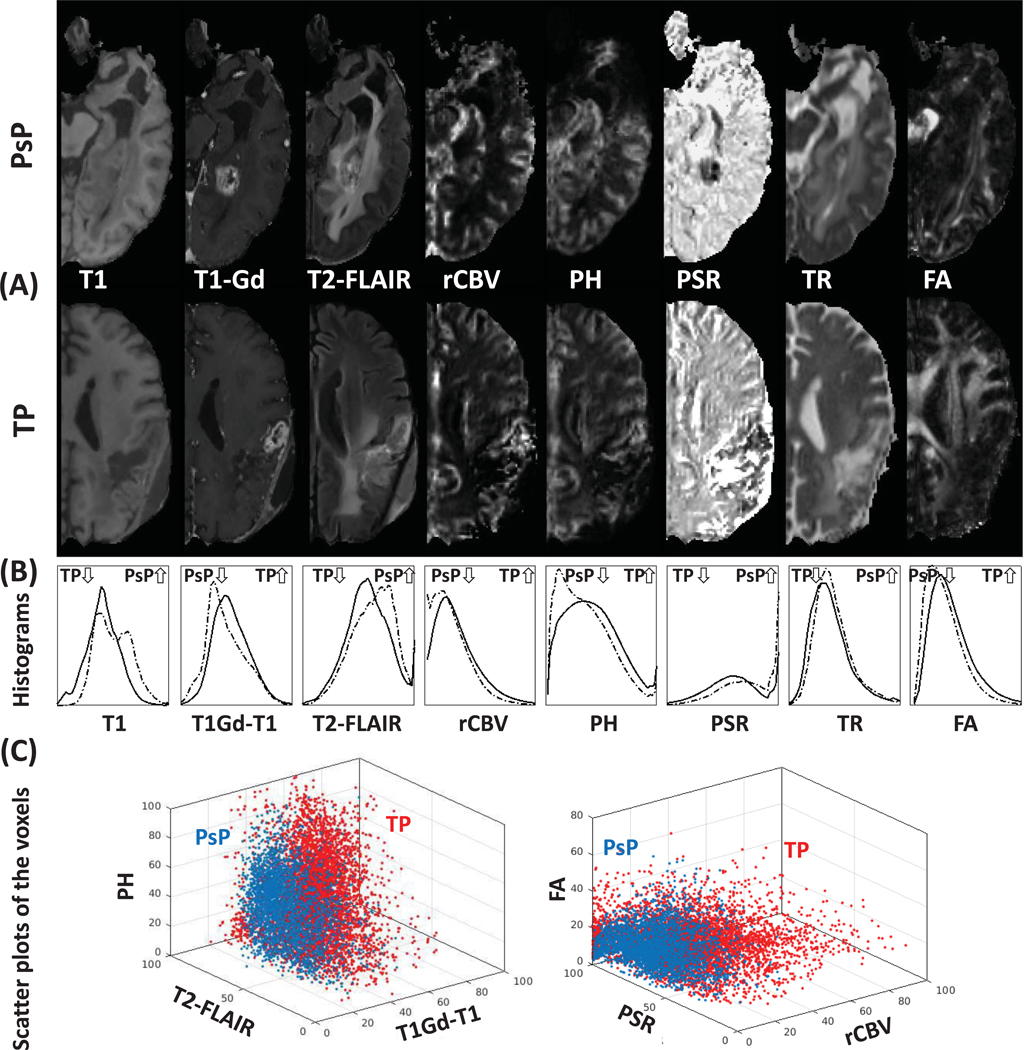
In our attempt to understand the underlying biological processes that are likely to give rise to the imaging signature of TP, we performed a detailed histogram analysis of all the imaging features. The Cohen’s d effect sizes are T1, 0.6477; normalized T1-Gd, 0.3103; FLAIR, 0.2599; rCBV, 0.2829; PH, 0.2907; PSR, 0.5020; TR, 0.1442; FA, 0.3319 for difference between TP and PsP tumors. Figure 5 shows the imaging characteristics of PsP (dashed lines) and TP (solid lines) in each modality across all patients. The main results of comparing the QIP features of TP with those of PsP patients, identify TP tumors as having:
Regions of higher blood volume and flow, which points towards hypervascularity, hyper-perfusion, and increased angiogenesis, based on the combination of rCBV and PH;
Regions of higher cellularity, suggestive of increased proliferation, as well as different tissue microarchitecture, based on the combination of measures extracted from DTI, namely DTI-TR and DTI-FA.
Regions of more compromised blood brain barrier (BBB), based on the combination of measures extracted from T1-Gd and T1-Gd subtracted T1, also consistent with infiltrating tumor characteristics;
Regions of lower water concentration, based on the combination of T2-FLAIR and DTI-TR, consistent with dense and non-necrotic tissue.
Figure 5.
(A) represented two examples of the imaging modalities in PsP and TP patients. (B) illustrated histograms of the most distinctive modalities, according to the progression status. The histograms were created using information from all PsP (dashed lines) and TP (solid lines) patients. (C) illustrated scatter plots of the voxels of patients with highest ratio of PsP (blue) and highest ratio of TP (red).
Inter-institutional validation
We evaluated the trained model on the dataset acquired from University of Pennsylvania on an independent testing cohort from Thomas Jefferson University with different acquisition protocols. 7 out of 10 TP patients and 8 out of 10 PsP patients correctly diagnosed by the model which reveals an overall accuracy of 75% (AUC=0.80). The training performance was 79% (AUC=0.80).
Discussion
In this study of glioblastoma patients treated with standard chemoradiation, we utilized advanced feature extraction and ML techniques to comprehensively capture the radiographic characteristics of a given ROI using structural MRI, the temporal dynamics of DSC-MRI, and DTI-derived modalities. Notably, our approach identified selected radiomic features within the given ROI in post-chemoradiation MRI that are significantly and robustly correlated with the histopathology of resected tissue, thereby offering non-invasive means of discriminating between TP and PsP in post-treatment glioblastoma. Critically, we have made these methods and models freely available through the CaPTk (www.cbica.upenn.edu/captk), a publicly available open-source software platform (Suppl. Figure 1), in order to facilitate clinical use and further validation of these results in other studies. This software has been designed for research purposes only and has neither been reviewed nor approved for clinical use by the Food and Drug Administration (FDA) or by any other federal/state agency and should not be used as the primary source of information for making clinical decisions.
Using advanced computational methodologies, our proposed non-invasive signature can quantify subtle imaging characteristics within an ROI that confer an estimate of the likelihood of TP vs PsP. It is important to emphasize that no single imaging feature was sufficiently discriminative by itself in our modeling. Rather, several QIP features were integrated by our multivariate approach to generate a discriminative score able to capture differences between TP and PsP. These results emphasize the importance of comprehensive multivariate analysis as opposed to imaging threshold based on isolated features, and the use of computational methods complementing traditional human interpretation. One of the important strengths of this signature is that it is generated from images acquired in the standard-of-care surveillance of glioblastoma patients. Thus, additional testing (invasive or otherwise) is not required, which is an advantage when considering clinical generalizability. Importantly, the QIP features are strongly associated with histopathologic scoring (Pearson correlation coefficient=0.76, p-value=5.5×10−13), and the predictive models have been validated in an independent replication cohort, unseen during their training, as well as via LOOCV.
We quantitatively evaluated the performance of our approach to distinguish between TP and PsP using models trained both independently on, and in combination with, two distinct feature extraction approaches, i.e., one based on deep learning and another using APS radiomic features. The comparison between the deep learning based model and the APS radiomic features based model yielded similar results, based on their evaluation on a relatively small independent replication cohort (Table 1). Interestingly, when combining the two feature types to create an integrated model we noted a drop (>17%) in the classifier distinguishing PsP from the rest, while the accuracy of the classifier distinguishing TP from the rest remained stable. It is worth noting that the evaluation of the APS features based model using a LOOCV scheme revealed a better performing model. A larger cohort of patients could possibly allow for further conclusions, and for developing a deep learning classifier instead of utilizing deep learning to create features for training an SVM model. However, considering the current results we tend to be in favor of the model trained using APS radiomic features due to the benefit of interpretability. Larger datasets might allow deep learning achieve more reproducible and accurate results in future studies.
The feature engineering approach we consider for estimating TP can offer potential insights into biological mechanisms via each MRI sequence that may uniquely reflect radiographic phenotypes of TP vs PsP. Specifically, regions of TP in our data showed increased contrast uptake on the T1-Gd sequences, which, consistent with existing literature, may be indicative of regional angiogenesis and associated with compromise of the BBB in areas of tumor infiltration 38. T2 and T2-FLAIR sequences provide information relevant in assessment of areas of non-enhancing and necrotic tumor, as well as the extent of the peritumoral edematous/invaded tissue 39. Our results identified regions of TP demonstrating lower signal intensity on T2 and T2-FLAIR, which indicate relatively lower water content and may thus reflect higher levels of tumor infiltration. This finding is consistent with the hypothesis that the TP regions harbor a higher ratio of malignant cells to water content. DTI maps the diffusion process of water in the brain, affected in part by tumor cellularity 40 and by integrity of white matter structures, as well as the underlying microstructure of tissue, e.g., via the DTI-FA measurements. Here, regions of TP showed lower DTI-TR and increased DTI-FA that may be expected in areas of higher cellular concentration (i.e., tumor cell proliferation). DTI-AX and DTI-RAD were also consistent with overall diffusivity captured by the DTI-TR volume. DSC imaging reflects various aspects of perfusion in the brain 29, which provide quantitative measures of regional microvasculature, perfusion hemodynamics, and permeability of blood vessels 38,41. Specifically, when brain tumors exceed a critical volume, the resultant ischemia triggers the secretion of angiogenic factors that promote vascular proliferation, leading to the formation and maintenance of tumor vessels 42–48. These new, immature vessels tend to be tortuous and leaky 49. In our analysis, the second principal component of the DSC signal (PC2) is inversely related to the magnitude of the signal drop, in relation to the baseline. Our results indicate a relatively lower PC2 in TP regions, which may be indicative of a higher degree of BBB compromise and leaky neovasculature. We also observed PC3, which reflects the steepness of the complete perfusion signal drop and its recovery rate. We found TP regions to show relatively higher values of PC3 that may suggest a relative time delay in the contrast agent reaching the TP tissue, possibly due to higher flow resistance, tortuosity and other characteristics of tumor vasculature 29,38,43,50. While these proposed biological associations are limited by the macroscopic nature of MRI, it must be pointed out that one does not require understanding of the mechanism to develop an effective signature – it merely requires rigorous validation to have potential clinical utility.
The limitations of our study include the fact that it was conducted in data from a single institution, and could further benefit from validation in multi-institutional data. Furthermore, the discovered signature relies upon features extracted from advanced mpMRI, which may not be routinely acquired in all clinical departments. Sample size is also a potential limitation, as the strength of deep learning methods is often improved as the number of subjects increases. The limited number of patients relative to the number of features utilized in deep learning methods, in particular, may increase the risk of overfitting. We addressed this potential pitfall by cross-validation of all steps when using APS features, i.e. feature selection, SVM and SVR parameters selections, and training and testing on different patients. Multi-institutional, prospective validation of our signature is necessary to establish inter-institutional reproducibility.
In summary, advanced computational analyses are increasingly used in the clinical evaluation of human gliomas and their response to treatment. The present study extracts subtle but informative QIP features from the temporal dynamics of DSC-MRI, DTI, and structural MRI modalities, and integrates them via multivariate ML, to develop an imaging signature that may non-invasively distinguish TP from PsP. Accurate stratification of the entities of TP and PsP may facilitate appropriate triage of patients to continuing maintenance, therapy, or evaluating them for new intervention, which carries great importance in the era of increasing personalization of therapy.
Supplementary Material
Two concise sentences that state the significant conclusions.
Artificial intelligence methods can accurately predict pseudo-progression in GBM.
Histopathologic characteristics of GBM progression correlate with radiomic features.
Acknowledgements
Funding: National Institutes of Health (NIH) R01 grant on “Predicting brain tumor progression via multiparametric image analysis and modeling” (R01-NS042645), and the National Institutes of Health (NIH) U24 grant of “Cancer imaging phenomics software suite: application to brain and breast cancer” (U24-CA189523), and a grant by the State of Pennsylvania to the University of Pennsylvania’s Abramson Cancer Center.
Footnotes
Conflict of Interest: Nothing to disclose.
References
- 1.Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-oncology. 2017;19(suppl_5):v1–v88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stupp R, Taillibert S, Kanner A, et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. Jama. 2017;318(23):2306–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The lancet oncology. 2009;10(5):459–466. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert MR, Dignam JJ, Armstrong TS, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. New England Journal of Medicine. 2014;370(8):699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Macdonald DR, Cascino TL, Schold SC Jr, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J Clin Oncol. 1990;8(7):1277–1280. [DOI] [PubMed] [Google Scholar]
- 6.Taal W, Brandsma D, de Bruin HG, et al. Incidence of early pseudo‐progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer. 2008;113(2):405–410. [DOI] [PubMed] [Google Scholar]
- 7.O’Brien BJ, Colen RR. Post-treatment imaging changes in primary brain tumors. Current oncology reports. 2014;16(8):397. [DOI] [PubMed] [Google Scholar]
- 8.Chamberlain MC, Glantz MJ, Chalmers L, Van Horn A, Sloan AE. Early necrosis following concurrent Temodar and radiotherapy in patients with glioblastoma. Journal of neuro-oncology. 2007;82(1):81–83. [DOI] [PubMed] [Google Scholar]
- 9.Rowe LS, Butman JA, Mackey M, et al. Differentiating pseudoprogression from true progression: analysis of radiographic, biologic, and clinical clues in GBM. Journal of neuro-oncology. 2018:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young R, Gupta A, Shah A, et al. Potential utility of conventional MRI signs in diagnosing pseudoprogression in glioblastoma. Neurology. 2011;76(22):1918–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Martinez-Lage M, Sakai Y, et al. Differentiating tumor progression from pseudoprogression in patients with glioblastomas using diffusion tensor imaging and dynamic susceptibility contrast MRI. American Journal of Neuroradiology. 2016;37(1):28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verma N, Cowperthwaite MC, Burnett MG, Markey MK. Differentiating tumor recurrence from treatment necrosis: a review of neuro-oncologic imaging strategies. Neuro-oncology. 2013;15(5):515–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ismail M, Hill V, Statsevych V, et al. Shape Features of the Lesion Habitat to Differentiate Brain Tumor Progression from Pseudoprogression on Routine Multiparametric MRI: A Multisite Study. American Journal of Neuroradiology. 2018;39(12):2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prasanna P, Rogers L, Lam T, et al. Disorder in Pixel-Level Edge Directions on T1WI Is Associated with the Degree of Radiation Necrosis in Primary and Metastatic Brain Tumors: Preliminary Findings. American Journal of Neuroradiology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. Journal of clinical oncology. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 16.Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. The Lancet Oncology. 2019;20(5):728–740. [DOI] [PubMed] [Google Scholar]
- 17.Galbán CJ, Chenevert TL, Meyer CR, et al. Prospective Analysis of Parametric Response Map–Derived MRI Biomarkers: Identification of Early and Distinct Glioma Response Patterns Not Predicted by Standard Radiographic Assessment. Clinical Cancer Research. 2011;17(14):4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gevaert O, Mitchell LA, Achrol AS, et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273(1):168–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellingson BM. Radiogenomics and imaging phenotypes in glioblastoma: Novel observations and correlation with molecular characteristics. Current neurology and neuroscience reports. 2015;15(1):1–12. [DOI] [PubMed] [Google Scholar]
- 20.Zhang B, Chang K, Ramkissoon S, et al. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro-Oncology. 2016:now121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akbari H, Bakas S, Pisapia JM, et al. In vivo evaluation of EGFRvIII mutation in primary glioblastoma patients via complex multiparametric MRI signature. Neuro-Oncology. 2018;20(8):1068–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. NeuroImage. 2012;62(2):782–790. [DOI] [PubMed] [Google Scholar]
- 23.Smith SM, Brady JM. SUSAN—A new approach to low level image processing. International journal of computer vision. 1997;23(1):45–78. [Google Scholar]
- 24.Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. Ieee Transactions on Medical Imaging. 1998;17(1):87–97. [DOI] [PubMed] [Google Scholar]
- 25.Smith SM. Fast robust automated brain extraction. Human brain mapping. 2002;17(3):143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soares J, Marques P, Alves V, Sousa N. A hitchhiker’s guide to diffusion tensor imaging. Frontiers in neuroscience. 2013;7:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boxerman J, Schmainda K, Weisskoff R. Relative cerebral blood volume maps corrected for contrast agent extravasation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. American Journal of Neuroradiology. 2006;27(4):859–867. [PMC free article] [PubMed] [Google Scholar]
- 28.Cha S, Lupo J, Chen M-H, et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. American Journal of Neuroradiology. 2007;28(6):1078–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akbari H, Macyszyn L, Da X, et al. Pattern Analysis of Dynamic Susceptibility Contrast-enhanced MR Imaging Demonstrates Peritumoral Tissue Heterogeneity. Radiology. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ou Y, Akbari H, Bilello M, Da X, Davatzikos C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE transactions on medical imaging. 2014;33(10):2039–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatfield K, Simonyan K, Vedaldi A, Zisserman A. Return of the devil in the details: Delving deep into convolutional nets. arXiv preprint arXiv:14053531. 2014. [Google Scholar]
- 32.Vedaldi A, Fulkerson B. VLFeat: An open and portable library of computer vision algorithms. Paper presented at: Proceedings of the 18th ACM international conference on Multimedia 2010. [Google Scholar]
- 33.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:14091556. 2014. [Google Scholar]
- 34.Yousefi B, Kalhor D, Usamentiaga R, Lei L, Castanedo CI, Maldague XP. Application of Deep Learning in Infrared Non-Destructive Testing. [Google Scholar]
- 35.Davatzikos C, Rathore S, Bakas S, et al. Cancer imaging phenomics toolkit: quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. Journal of Medical Imaging 2018;5(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Transactions on systems, man, and cybernetics. 1973(6):610–621. [Google Scholar]
- 37.Galloway MM. Texture analysis using grey level run lengths. NASA STI/Recon Technical Report N. 1974;75. [Google Scholar]
- 38.Akbari H, Macyszyn L, Da X, et al. Imaging Surrogates of Infiltration Obtained Via Multiparametric Imaging Pattern Analysis Predict Subsequent Location of Recurrence of Glioblastoma. Neurosurgery. 2016;78(4):572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurki T, Lundbom N, Valtonen S. Tissue characterisation of intracranial tumours: the value of magnetisation transfer and conventional MRI. Neuroradiology. 1995;37(7):515–521. [DOI] [PubMed] [Google Scholar]
- 40.Lu S, Ahn D, Johnson G, Cha S. Peritumoral diffusion tensor imaging of high-grade gliomas and metastatic brain tumors. American Journal of Neuroradiology. 2003;24(5):937–941. [PMC free article] [PubMed] [Google Scholar]
- 41.Wintermark M, Sesay M, Barbier E, et al. Comparative overview of brain perfusion imaging techniques. Journal of neuroradiology. 2005;32(5):294–314. [DOI] [PubMed] [Google Scholar]
- 42.Lev MH, Hochberg F. Perfusion magnetic resonance imaging to assess brain tumor responses to new therapies. Cancer Control. 1998;5:115–123. [DOI] [PubMed] [Google Scholar]
- 43.Bullitt E, Zeng D, Gerig G, et al. Vessel tortuosity and brain tumor malignancy: a blinded study1. Academic radiology. 2005;12(10):1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. Journal of Clinical Oncology. 2005;23(5):1011–1027. [DOI] [PubMed] [Google Scholar]
- 45.Essock-Burns E, Lupo JM, Cha S, et al. Assessment of perfusion MRI-derived parameters in evaluating and predicting response to antiangiogenic therapy in patients with newly diagnosed glioblastoma. Neuro-oncology. 2011;13(1):119–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen RL, Mumert ML, Gillespie DL, Kinney AY, Schabel MC, Salzman KL. Preoperative dynamic contrast-enhanced MRI correlates with molecular markers of hypoxia and vascularity in specific areas of intratumoral microenvironment and is predictive of patient outcome. Neuro-oncology. 2013:not148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonald DM, Choyke PL. Imaging of angiogenesis: from microscope to clinic. Nature medicine. 2003;9(6):713–725. [DOI] [PubMed] [Google Scholar]
- 48.Swami M Cancer: Enhancing EGFR targeting. Nature medicine. 2013;19(6):682–682. [Google Scholar]
- 49.Thompson G, Mills S, Coope D, O’connor J, Jackson A. Imaging biomarkers of angiogenesis and the microvascular environment in cerebral tumours. British Journal of Radiology. 2011;84(Special Issue 2):S127–S144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jain RK, Di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nature Reviews Neuroscience. 2007;8(8):610–622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.