Abstract
Background
Due to the lack of a sensitive, specific and rapid detection method, aetiological diagnosis of pneumonia caused by Mycoplasma pneumoniae (M. pneumoniae, MP) is a constantly challenging issue. This retrospective study aimed to compare the diagnostic methods for Mycoplasma pneumoniae in children and evaluate their values.
Methods
From November 2018 to June 2019, 830 children with community-acquired pneumonia were selected from the Department of Respiratory Medicine, Shanghai Children’s Medical Center. On the first day of hospitalization, sputum, throat swab and venous blood samples were collected to analyse MP-IgM (particle agglutination, PA), MP-IgM (immune colloidal gold technique, GICT), MP-DNA, MP-RNA (simultaneous amplification and testing, SAT) and MP-DNA (real-time polymerase chain reaction, RT-PCR).
Results
Among these 830 children, RT-PCR showed that the positive rate was 36.6% (304/830), in which the positive rate of macrolide resistance (A2063G mutation) accounted for 86.2% of cases (262/304). Using RT-PCR as the standard, MP-RNA (SAT) had the highest specificity (97.5%), and MP-IgM (PA) had the highest sensitivity (74.0%) and Youden index (53.7%). If MP-RNA (SAT) was combined with MP-IgM (PA), its Kappa value (0.602), sensitivity (84.2%), specificity (78.7%) and Youden index (62.9%) were higher than those of single M. pneumoniae detection.
Conclusions
Our research indicated that a combination of MP-RNA (SAT) plus MP-IgM (PA) might lead to reliable results as an early diagnostic method for children with clinical manifestations of Mycoplasma pneumoniae pneumonia.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12887-021-02523-4.
Keywords: Detection methods, Mycoplasma pneumoniae, Children, Community-acquired pneumonia
Background
Mycoplasma pneumoniae (M. pneumoniae, MP) is known as a common cause of community-acquired pneumonia (CAP) in children throughout the world. M. pneumoniae is the smallest prokaryotic microorganism, lacks a cell wall and has a high affinity for respiratory epithelial cells. A total of 40% or more cases of CAP in children are caused by M. pneumoniae, and approximately 18% of cases require hospitalization [1]. At present, it is believed that the pathogenic mechanism of M. pneumoniae is to cause direct injury through community-acquired respiratory distress syndrome (CARDS) toxin, which induces eosinophilia, increases cytokine production and induces the hyperreactivity of the airway, such as asthma, in animal models [2–4]. In addition, the immunological response resulting from infection by M. pneumoniae causes pulmonary and extrapulmonary symptoms [5].
A worldwide increase in the prevalence of macrolide-resistant mycoplasma pneumoniae (MRMP) strains has been witnessed since 2000 [6]. In some regions of Asia, the resistance rates have been reported to be over 90% [7], whereas in North America and Europe, the rates have reached 25% [8, 9]. Compared to other patients with CAP, children with M. pneumoniae infection may be older and have prolonged symptoms of fever and cough [10]. The period of signs and symptoms will be shortened if accurate antimicrobial treatment is started early in the course of diseases. However, Mycoplasma pneumoniae pneumonia (MPP) in children cannot be diagnosed exactly in terms of clinical manifestations [11].
Mycoplasma pneumoniae pneumonia has the characteristics of insidious onset, mild pulmonary signs and low specificity of clinical manifestations and imaging. Therefore, effective and sensitive laboratory diagnostic methods are the main basis for the diagnosis of M. pneumoniae infection. At present, there are many clinical methods for detecting M. pneumoniae, such as culture, serology and molecular-based methods. However, as the gold standard, culture is limited in clinical utility because it is time-consuming and insensitive [12]. Both serology and molecular assays have their own advantages and disadvantages in clinical practice. In this paper, we aimed to evaluate the clinical application value of different methods to detect M. pneumoniae infection by comparing and analysing them to improve diagnostic efficiency.
Methods
Study population
We conducted a retrospective study that included children aged 1 month to 15 years with radiologically confirmed community-acquired pneumonia. Venous blood, throat swab and sputum specimens were obtained from these patients on the day of hospitalization at the Department of Respiratory Medicine, Shanghai Children’s Medical Center (SCMC) from November 1, 2018, to June 30, 2019. These specimens were tested for MP-IgM (particle agglutination, PA), MP-IgM (immune colloidal gold technique, GICT), MP-RNA (simultaneous amplification and testing, SAT), 7 respiratory tract RNAs (7 RNAs) and MP-DNA (real-time polymerase chain reaction, RT-PCR).
MP-IgM (particle agglutination, PA)
Particle agglutination (PA) antibody titres for M. pneumonia were assayed using SERODIA MYCO-II (Fuji Rebio Ltd., Tokyo, Japanese), which was performed using artificial gelatine particles sensitized with cell membrane components of M. pneumoniae. The result was considered positive if the titre was 1:160 or more (≥1:160).
MP-IgM (immune colloidal gold technique, GICT)
According to the manufacturer’s (Alere Diagnostics, Shanghai, China) instructions, colloidal gold was used as a tracer marker to detect M. pneumoniae antibodies using the principle of immunoreaction of specific antigen antibodies. The results of this method showed only negative, weak positive or positive titres.
MP-RNA (simultaneous amplification and testing, SAT)
Throat swab specimens of the children were collected and tested using M. pneumoniae nucleic acid detection kits (Rendu Biotechnology, Shanghai, China), including nucleic acid extraction and isothermal amplification detection.
Seven respiratory tract RNAs (7 RNAs)
Based on the double-amplification method of RNA isothermal amplification and multiple biotin signals, throat swab samples were collected to identify seven common respiratory pathogen RNAs (Zhongzhi Biotechnologies, Wuhan, China), namely influenza A, influenza B, respiratory syncytial virus (RSV), parainfluenza virus, adenovirus, Mycoplasma pneumoniae and Chlamydia pneumoniae, in a short period of time.
MP-DNA (real-time polymerase chain reaction, RT-PCR)
According to the specific point mutations in domain V of MP 23S rRNA (at positions 2063 and 2064), the primers were designed as follows: forward primer: 5′-AACTATAACGGTCCTAAGGTAGCG-3′; reverse primer: 5′-GCTCCTACCTATTCTCTACATGAT-3′. Sputum aspirated from the patients was collected for sample processing, and DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). PCR was performed, and the PCR product was verified using gel electrophoresis. Finally, the positive specimen was sequenced and then analysed using SnapGene software (from Insightful Science; available at snapgene.com). The measured sequence was compared to the standard sequence of the M129 strain in the NCBI database. If the sequence was not consistent, the A2063G transition was considered to be involved in macrolide resistance. Unfortunately, we did not find a mutation at position 2064.
Statistical analysis
SPSS software package v21.0 was used for all statistical analyses. Categorical variables are expressed as frequencies and percentages. Four-table diagnostic tests were used to calculate the Kappa value, sensitivity, specificity and Youden index. In a previous report [13], the value of the Kappa statistic was interpreted as follows: < 0.01, poor; 0 to 0.20, slight; 0.21 to 0.4, fair; 0.41 to 0.6, moderate; 0.61 to 0.80, substantial; and 0.81 to 1, almost perfect. The other indices were calculated using the following formulas: sensitivity = true positive cases / (true positive cases + false negative cases) × 100%; specificity = true negative cases / (true negative cases + false positive cases) × 100%; and Youden index = sensitivity + specificity − 1. The Youden index was used to evaluate the effectiveness and authenticity of experiments.
Results
Clinical characteristics of patients
A total of 830 patients diagnosed with community-acquired pneumonia, aged 1 month to 15 years, were enrolled in the present study between November 1, 2018, and June 30, 2019. Congenital heart disease, congenital biliary atresia, epilepsy, hydrocephalus and cerebral palsy were the most frequently observed underlying diseases in these patients (Table 1).
Table 1.
General characteristics of the patients
| Characteristic | Value for patients |
|---|---|
| Total | 830 |
| Age, No. (%) | |
| < 3 years | 421 (50.7) |
| 3–5 years | 215 (25.9) |
| 6–15 years | 194 (23.4) |
| Sex, No. (%) | |
| Male | 447 (53.9) |
| Female | 383 (46.1) |
| Underlying diseases, No. (%) | |
| None | 680 (81.9) |
| Congenital heart disease | 101 (12.2) |
| Congenital biliary atresia | 7 (0.8) |
| Brain lesions | 15 (1.8) |
| Other diseases | 27 (3.3) |
| Need of oxygen | |
| Yes | 61 (7.3) |
| No | 769 (92.7) |
| Prognosis after treat, No. (%) | |
| Alive | 825 (99.4) |
| Deatha | 5 (0.6) |
aFour children died from congenital heart disease, and one died from primary immunodeficiency disease
Comparison of positive rates of M. pneumoniae detection methods in different age groups
All the patients were grouped by age as follows: infants (age: < 3 years), preschoolers (age: 3–5 years) and school-aged children (age: 6–15 years). The highest specimen positivity rate was in school-aged children. There were significant differences in the positivity rate of the different age groups by each detection method, except for the 7 RNAs method (Table 2). In addition, the rate of macrolide resistance was highest in school-aged children (87.9%, 123/140) and lowest in infants (82.3%, 65/79), but it was not statistically significant (X2 = 1.396, P < 0.001).
Table 2.
Positive rates of methods in different age groups
| Methods | < 3 years | 3-5 years | 6-15 years | X2 | p |
|---|---|---|---|---|---|
| n = 421(%) | n = 215(%) | n = 194(%) | |||
| MP-IgM (PA) | 91 (21.6) | 102 (47.4) | 139 (71.6) | 145.222 | < 0.001 |
| MP-IgM (GICT) | 66 (15.7) | 67 (31.2) | 89 (45.9) | 64.704 | < 0.001 |
| MP-RNA (SAT) | 36 (8.6) | 40 (18.6) | 70 (36.1) | 69.646 | < 0.001 |
| 7RNAs | 71 (16.9) | 29 (13.5) | 38 (19.6) | 2.772 | 0.25 |
| RT-PCR | 79 (18.8) | 85 (39.5) | 140 (72.2) | 164.208 | < 0.001 |
| Resistance | 65 (15.4) | 74 (34.4) | 123 (63.4) | 142.516 | < 0.001 |
SAT simultaneous amplification and testing
PA particle agglutination
GICT immune colloidal gold technique
RT-PCR real-time polymerase chain reaction
Positive rates of different detection methods for M. pneumoniae
In these 830 children, the positive rates of MP-IgM (PA), MP-IgM (GICT), MP-RNA (SAT) and 7 RNAs were 40% (332/830), 26.7% (222/830), 17.6% (146/830) and 16.6% (138/830), respectively. MP-DNA (RT-PCR) results showed that the positive rate was 36.6% (304/830), of which macrolide resistance accounted for 86.2% of cases (262/304) (Fig. 1). Through 7 RNAs, we found that the coinfection combination containing MP and RSV had the highest detection rate of 23.9% (33/138) followed by MP and parainfluenza virus (21.7%, 30/138).
Fig. 1.
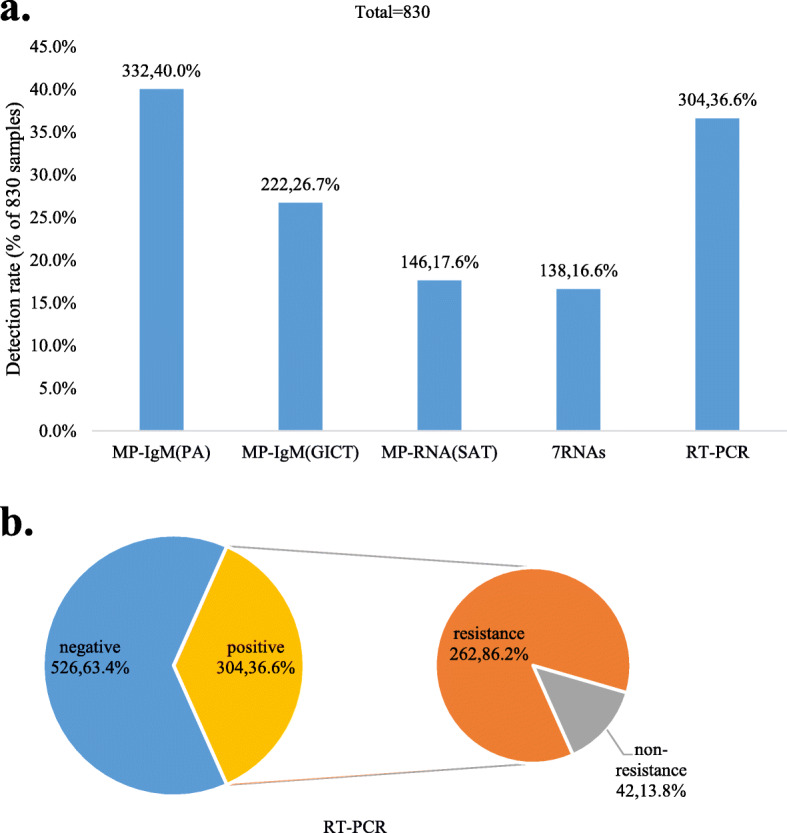
Positive rates of different detection methods for M. pneumoniae. A. The positive rate of MP-IgM (PA) was higher than that of the other methods. B. The positive rate of RT-PCR for M. pneumoniae was 36.6%, and the rate of macrolide resistance accounted for 86.2% of cases by sequencing analysis
Diagnostic value of different methods with a real-time polymerase chain reaction as the standard
We compared the diagnostic values of MP-IgM (PA), MP-IgM (GICT), MP-RNA (SAT) and 7 RNAs, considering MP-DNA (RT-PCR) as the standard. As shown in Table 3, MP-IgM (PA) had a good consistency (Kappa = 0.526), sensitivity (74.0%) and Youden index (53.7%). The specificity of MP-RNA (SAT) was the highest (97.5%) followed by 7 RNAs (89.2%). Moreover, comparing every two detection methods together, we found that the combination of MP-IgM (PA) and MP-RNA (SAT) was the best choice to detect M. pneumoniae because its substantial Kappa value (Kappa = 0.602) and its highest sensitivity (84.2%), specificity (78.7%) and Youden index (62.9%) were greater than those of the single method (Table 4).
Table 3.
Diagnostic values of different methods with a real-time polymerase chain reaction as the standard
| RT-PCR | Kappa | Sensitivity | Specificity | Accuracy | Youden Index | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| MP-IgM (PA) | |||||||
| Positive | 225 | 107 | 0.526 | 74.0% | 79.7% | 77.6% | 53.7% |
| Negative | 79 | 419 | |||||
| MP-IgM (GICT) | |||||||
| Positive | 151 | 71 | 0.384 | 49.7% | 86.5% | 73.0% | 36.2% |
| Negative | 153 | 455 | |||||
| MP-RNA (SAT) | |||||||
| Positive | 133 | 13 | 0.464 | 43.8% | 97.5% | 77.8% | 41.2% |
| Negative | 171 | 513 | |||||
| 7 RNAs | |||||||
| Positive | 81 | 57 | 0.179 | 26.6% | 89.2% | 66.3% | 15.8% |
| Negative | 223 | 469 | |||||
SAT simultaneous amplification and testing
PA particle agglutination
GICT immune colloidal gold technique
Table 4.
Diagnostic value of combining different methods with a real-time polymerase chain reaction as the standard
| RT-PCR | Kappa | Sensitivity | Specificity | Accuracy | Youden Index | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | ||||||
| MP-RNA (SAT) + MP-IgM (PA) | |||||||
| Positive | 256 | 112 | 0.602 | 84.2% | 78.7% | 80.7% | 62.9% |
| Negative | 48 | 414 | |||||
| MP-RNA (SAT) + MP-IgM (GICT) | |||||||
| Positive | 223 | 76 | 0.591 | 73.4% | 85.6% | 81.1% | 59.0% |
| Negative | 81 | 450 | |||||
| MP-IgM (GICT) + MP-IgM (PA) | |||||||
| Positive | 237 | 123 | 0.525 | 78.0% | 76.6% | 77.2% | 54.6% |
| Negative | 67 | 403 | |||||
| 7 RNAs + MP-IgM (PA) | |||||||
| Positive | 250 | 154 | 0.419 | 82.2% | 70.7% | 74.9% | 52.9% |
| Negative | 54 | 372 | |||||
| 7 RNAs + MP-IgM (GICT) | |||||||
| Positive | 198 | 120 | 0.419 | 65.1% | 77.2% | 72.8% | 42.3% |
| Negative | 106 | 406 | |||||
SAT simultaneous amplification and testing
PA particle agglutination
GICT immune colloidal gold technique
Discussion
In this study, we assessed the detection rate of different methods for M. pneumoniae pneumonia in different age groups and compared the value of these diagnostic methods.
M. pneumoniae is a major cause of infectious diseases worldwide. M. pneumoniae affects different tissues and organs, especially the respiratory tracts, of children in all age groups, and it has become the second leading pathogen after Streptococcus pneumoniae in children with community-acquired pneumonia [14]. The occurrence of CAP caused by M. pneumoniae varies with age. A previous study has reported that M. pneumoniae pneumonia is uncommon in children under 5 years of age but has a higher incidence among school-age children [1]. However, M. pneumoniae infections may occur in people from infancy through old age [15, 16]. From 2010 to 2012, 2638 children with pneumonia requiring hospitalization in three hospitals in the United States were enrolled in a study. Respiratory specimens were systematically collected and detected using real-time PCR, and M. pneumoniae was found to be more common in children ≥5 years (19% vs. 3%) [17]. In accordance with our data, the detection rates of M. pneumoniae by different methods in the three age groups were different, but the distributions were the same. School-aged children and adolescents were the most common ages affected, and there was rarely a significant infection before 3 years old, which was consistent with the epidemiological characteristics of M. pneumoniae.
At present, there are many methods for the laboratory diagnosis of M. pneumoniae infection, including culture, serological detection and molecular assays. Culture is often used for antimicrobial susceptibility testing or typing due to its high specificity. However, due to the complicated procedure, prolonged turnaround time and low sensitivity, culture is not recommended for routine testing [18, 19]. Serological tests are currently the most widely used detection methods of M. pneumoniae in clinical practice and are more sensitive than culture. Diagnostic sensitivity for serological tests of M. pneumoniae is determined by both the collection time of specimens and the performance characteristics of the methods. In our study, the positive rate of MP-IgM (PA) was 40%, which was higher than that of the others, which may have been attributed to the average time being 10 days before admission to our department. Although MP-IgM is the earliest antibody produced after M. pneumoniae infection, it still takes a certain period of time before it can be detected. In general, MP-IgM can be detected within approximately 1 week after infection, reaching a peak after 3 to 4 weeks, resulting in possible false negatives in the early stage [20]. It has been reported that the most accurate diagnosis of serology is obtained when paired sera collected at least 2 weeks apart are tested for both IgM and IgG at the same time, resulting in a 4-fold increase in titre [1]. However, in paediatrics, it is simply impossible to perform repeated blood sampling in a short time. MP-IgM (GICT), a rapid test kit based on immunochromatography, is a relatively new serological test. One study from Wei Li found that the specificity and sensitivity of MP-IgM (GICT) were 100 and 97.4%, respectively, compared with real-time PCR [21], which was not in line with our results. It might be that the dilution of the sample to only 100 μl caused false negatives. Due to the easy and rapid procedure (15 min), MP-IgM (GICT) is suitable for the identification of M. pneumoniae infections in paediatric outpatient departments. However, MP-IgM (PA) is still recommended for inpatients to reduce the false-negative rate.
With the development of molecular assays, nucleic acid amplification technology (NAAT) has gradually become an important method for the early rapid diagnosis of M. pneumoniae infection, which may help with early appropriate antibiotic therapy. Owing to the advantages of quick turnaround times, lower likelihood of contamination, higher sensitivity and higher specificity, as well as not being limited by time and immune function, NAAT is important in the early diagnosis of M. pneumoniae. In addition, NAAT has various detection formats, which can provide quantitative data, detect antimicrobial resistance genes and analyse the genetic relatedness of organisms [12]. However, there are a several limitations, such as contamination, which may result in false positives; difficulty in obtaining high-quality samples; sampling time point affecting results; and the possibility of PCR inhibitors leading to false negatives [22]. These limitations may have contributed to the lower positive rate of NAAT in this study.
We found that the rate of macrolide resistance accounted for 86.2% of cases. MRMP strains have been increasing in many countries since the first case was reported in Japan in 2000 [23] with the long-term widespread use of macrolides. Macrolides act on the ribosomal 50S subunit to inhibit protein synthesis. Mutations in the V region of the MP 23S rRNA domain can cause a decrease in affinity between the drug and the ribosome, leading to drug resistance [24]. Common mutations, at both position 2063 and position 2064, lead to high-level resistance, whereas positions 2067 and 2617 are associated with low-level resistance [25, 26]. In these 262 specimens, we found a mutation only at position 2063. MRMP in the Beijing population reached a high rate of more than 90% from 2008 to 2012 [27], which was similar to our results.
MP-DNA detection by RT-PCR technology amplifies gene fragments to diagnose pathogens, which has advantages in operation and sensitivity of detection [28]. We compared the diagnostic values of different methods for M. pneumoniae with RT-PCR as the standard. We found that MP-IgM (PA) had a high sensitivity of 74.0% and the highest Youden index and Kappa value, indicating that it was conducive to screening for MP infection. SAT is a recently developed method based on isothermal amplification of RNA [29, 30], which can be completed in approximately 3 h. In our study, the outcomes of SAT for M. pneumoniae, both MP-DNA and 7 RNAs, showed higher specificity but lower sensitivity. Specific RNA exists only in the proliferation stage of M. pneumoniae, indicating that increases in RNA levels may reflect bacterial multiplication [31]. Previous data have shown that the SAT positivity rates are significantly higher in untreated cases with MPP than in macrolide-treated MPP cases [32]. Therefore, one explanation for the “false-negative” SAT results is that long-term treatment might decrease the M. pneumoniae load. Our data showed that 78.3% of the patients received macrolide treatment before hospitalization. Another explanation might relate to the possible poor sampling procedural skills and the quality of the swab samples, leading some samples to be below the assay’s detection limit [33, 34]. In addition, multiplex PCR assays can be used to detect M. pneumoniae and other respiratory pathogens, but monoplex assays have higher sensitivity and specificity than multiplex assays [35], which was confirmed by our results.
Each detection method of M. pneumoniae has its own advantages and disadvantages. Although molecular assays are superior in detecting M. pneumoniae due to their rapid, sensitive and specific characteristics, they cannot take the place of serology [36]. A study in China analysed data from children hospitalized with MPP using IgM (PA) and RT-PCR [37]. The concordance was close to 90% for the two detection methods. However, 173 (7%) children with a positive PCR had a negative serological test, and there were only 72 (3%) IgM-positive children who were PCR negative. Different detection methods can be used not only to improve diagnostic specificity and sensitivity but also to reduce the false-negative rate and false-positive rate. No single test available can be reliable for the identification of M. pneumoniae infection, and a combination of various methods is the most reliable approach [38]. A positive PCR without serological evidence of infection may indicate that the specimen was collected too early in the course of M. pneumoniae infection for antibodies to develop. PCR results may become negative after a period of antibiotic treatment, while serological results would remain positive for a long time. Some reviews have concluded that no single test could reliably detect M. pneumoniae infection, but a combination of serological tests and PCR might be the most sensitive approach for early diagnosis in children [12, 39]. In our research, two different M. pneumoniae detection methods were combined and evaluated by RT-PCR, and the results showed that MP-IgM (PA) in combination with MP-RNA (SAT) can be used as a good screening method.
Our study had some limitations. First, this study was performed in a single centre and included no data on clinical features because there were no accurate ways to determine the specificity of these characteristics. Second, the lack of a gold standard diagnostic assay, such as microbiological culture and a 4-fold increase in IgM and IgG in paired serum, made it difficult to draw conclusions to confirm the most reliable and accurate method.
Conclusions
In conclusion, during the acute phases of M. pneumoniae pneumonia in children, the detection of MP-IgM (PA) together with MP-RNA (SAT) proved to be a reliable and precise diagnosis. Paediatricians need to select appropriate detection methods based on the age of children and the onset of the disease, and they need to consider various results to diagnose M. pneumoniae infection.
Supplementary Information
Acknowledgements
We would like to thank the patients and their parents for their support and cooperation in this work.
Abbreviations
- MP
Mycoplasma pneumoniae
- CAP
community-acquired pneumonia
- PA
particle agglutination
- GICT
immune colloidal gold technique
- SAT
simultaneous amplification and testing
- RT-PCR
real-time polymerase chain reaction
- MRMP
macrolide-resistant mycoplasma pneumoniae
- RSV
respiratory syncytial virus
Authors’ contributions
YY and QC initiated the study. MY-T and DW wrote the manuscript and analysed the data. XT and YF-W analysed the data and prepared figures. JZ and LZ conducted statistical analyses and provided technical support. QC provided technical support. YY provided critical feedback. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
The datasets generated and/or analysed during the current study are available in the Supplementary Material.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board and the Ethics Committee of Shanghai Children’s Medical Center (SCMCIRB-K2019060-1), and written informed consent was obtained from the parents of each patient. Yong Yin was granted permission to access raw data by Shanghai Children’s Medical Center.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Mingyu Tang and Dong Wang contributed equally to this work.
Contributor Information
Yong Yin, Email: yinyong@scmc.com.cn.
Qing Cao, Email: caoqing@scmc.com.cn.
References
- 1.Ken BW, Deborah FT. Mycoplasma pneumoniae and its role as a human pathogen. Clin Microbiol Rev. 2004;17:697–728. doi: 10.1128/CMR.17.4.697-728.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy RD, Coalson JJ, Peters J, et al. Analysis of pulmonary inflammation and function in the mouse and baboon after exposure to mycoplasma pneumoniae CARDS toxin. PLoS One. 2009;4:e7562. doi: 10.1371/journal.pone.0007562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Medina JL, Coalson JJ, Brooks EG, et al. Mycoplasma pneumoniae CARDS toxin induces pulmonary eosinophilic and lymphocytic inflammation. Am J Respir Cell Mol Biol. 2012;46:815–822. doi: 10.1165/rcmb.2011-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medina JL, Coalson JJ, Brooks EG, et al. Mycoplasma pneumoniae CARDS toxin exacerbates ovalbumin-induced asthma-like inflammation in BALB/c mice. PLoS One. 2014;9:e102613. doi: 10.1371/journal.pone.0102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer Sauteur PM, van Rossum AM, Vink C. Mycoplasma pneumoniae in children: carriage, pathogenesis, and antibiotic resistance. Curr Opin Infect Dis. 2014;27:220–227. doi: 10.1097/QCO.0000000000000063. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Zhou Y, Li S, et al. The clinical characteristics and predictors of refractory mycoplasma pneumoniae pneumonia in children. PLoS One. 2016;11:e0156465. doi: 10.1371/journal.pone.0156465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou Z, Li X, Chen X, et al. Macrolide-resistant mycoplasma pneumonia in adults in Zhejiang, China. Antimicrob Agents Chemother. 2015;59:1048–1051. doi: 10.1128/AAC.04308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Principi N, Esposito S. Macrolide-resistant mycoplasma pneumoniae: its role in respiratory infection. J Antimicrob Chemother. 2013;68:506–511. doi: 10.1093/jac/dks457. [DOI] [PubMed] [Google Scholar]
- 9.Bebear C, Pereyre S, Peuchant O. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol. 2011;6:423–431. doi: 10.2217/fmb.11.18. [DOI] [PubMed] [Google Scholar]
- 10.Fischer JE, Steiner F, Zucol F, et al. Use of simple heuristics to target macrolide prescription in children with community-acquired pneumonia. Arch Pediatr Adolesc Med. 2002;156:1005–1008. doi: 10.1001/archpedi.156.10.1005. [DOI] [PubMed] [Google Scholar]
- 11.Wang K, Gill P, Perera R, et al. Clinical symptoms and signs for the diagnosis of mycoplasma pneumoniae in children and adolescents with community-acquired pneumonia. Cochrane Database Syst Rev. 2012;10:CD009175. doi: 10.1002/14651858.CD009175.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waites KB, Li X, Yang L, et al. Mycoplasma pneumoniae from the Respiratory Tract and Beyond. Clin Microbiol Rev. 2017;30:747–809. doi: 10.1128/CMR.00114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 14.Miyashita N, Kawai Y, Tanaka T, et al. Diagnostic sensitivity of a rapid antigen test for the detection of mycoplasma pneumoniae: comparison with real-time PCR. J Infect Chemother. 2015;21:473–475. doi: 10.1016/j.jiac.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 15.Maheshwari M, Kumar S, Sethi GR, et al. Detection of mycoplasma pneumoniae in children with lower respiratory tract infections. Trop Dr. 2011;41:40–42. doi: 10.1258/td.2010.100149. [DOI] [PubMed] [Google Scholar]
- 16.Gadsby NJ, Reynolds AJ, McMenamin J, et al. Increased reports of Mycoplasma pneumoniae from laboratories in Scotland in 2010 and 2011-Impact of the epidemic in infants. Euro Surveill. 2012;17:20110. [PubMed]
- 17.Seema J, Derek JW, Sandra RA, et al. Community-acquired pneumonia requiring hospitalization among U.S. children. N Engl J Med. 2015;372:835–845. doi: 10.1056/NEJMoa1405870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song M, Zhang Y, Li S, et al. A sensitive and rapid immunoassay for mycoplasma pneumoniae in children with pneumonia based on single-walled carbon nanotubes. SciRep. 2017;7:16442. doi: 10.1038/s41598-017-16652-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrim M, Wolter N, Benitez AJ, et al. Epidemiology and molecular identification and characterization of mycoplasma pneumoniae, South Africa, 2012-2015. Emerg Infect Dis. 2018;24:506–513. doi: 10.3201/eid2403.162052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer Sauteur PM, Unger WWJ, Nadal D, et al. Infection with and carriage of mycoplasma pneumonia in children. Front Microbiol. 2016;7:329. doi: 10.3389/fmicb.2016.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Liu Y, Yu Z, et al. Rapid diagnosis of Mycoplasma pneumoniae in children with pneumonia by an immuno-chromatographic antigen assay. Sci Rep. 2015;5:15539. doi: 10.1038/srep15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chong H, Peiting H, Jieying Y, et al. Pooled pooled analysis of nuclear acid sequence-based amplification for rapid diagnosis of mycoplasma Pneumoniae infection. J Clin Lab Anal. 2019;33:e22879. doi: 10.1002/jcla.22879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka M, Narita M, Okazaki N, et al. Characterization and molecular analysis of macrolide- resistant mycoplasma pneumoniae clinical isolates obtained in Japan. Antimicrob Agents Chemother. 2004;48:4624–4630. doi: 10.1128/AAC.48.12.4624-4630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang HJ, Song DJ, Shim JY, et al. Mechanism of resistance acquisition and treatment of macrolide-resistant mycoplasma pneumoniae pneumonia in children. Korean J Pediatr. 2017;60:167–174. doi: 10.3345/kjp.2017.60.6.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu X, Jiang Y, Chen X, et al. Drug resistance mechanisms of mycoplasma pneumoniae to macrolide antibiotics. Biomed Res Int. 2014;2014:320801. doi: 10.1155/2014/320801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zibo Z, Xiangzhi L, Xiaojian C, et al. Macrolide-resistant mycoplasma pneumoniae in adults in Zhejiang, China. Antimicrob Agents Chemother. 2015;59:1048–1051. doi: 10.1128/AAC.04308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao F, Liu G, Wu J, et al. Surveillance of macrolide-resistant mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob Agents Chemother. 2013;57:1521–1523. doi: 10.1128/AAC.02060-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amy ER, Lynn BD, Ken BW. Comparison of the illumigene mycoplasma DNA amplification assay and culture for detection of mycoplasma pneumonia. J Clin Microbiol. 2014;52:1060–1063. doi: 10.1128/JCM.02913-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei L, You-Hong F, Hong-Qiang S, et al. Evaluation of a real-time method of simultaneous amplification and testing in diagnosis of mycoplasma pneumoniae infection in children with pneumonia. PLoS One. 2017;12:e0177842. doi: 10.1371/journal.pone.0177842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Feng Z, Zhao M, et al. A comparison study between GeXp-based multiplex-PCR and serology assay for mycoplasma pneumoniae detection on children with community acquired pneumonia. BMC Infect Dis. 2017;17:518. doi: 10.1186/s12879-017-2614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao C, Zhao Y, Wang L. Establishment and validation of RNA-based predictive models for understanding survival of vibrio parahaemolyticus in oysters stored at low temperatures. Appl Environ Microbiol. 2017;83:e02765–e02716. doi: 10.1128/AEM.02765-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jieqiong L, Lin S, Xirong W, et al. Early diagnosis of mycoplasma pneumoniae in children: simultaneous amplification and testing (SAT) is the key. Front Pediatr. 2019;7:441. doi: 10.3389/fped.2019.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raty R, Ronkko E, Kleemola M. Sample type is crucial to the diagnosis of mycoplasma pneumoniae pneumonia by PCR. J Med Microbiol. 2005;54:287–291. doi: 10.1099/jmm.0.45888-0. [DOI] [PubMed] [Google Scholar]
- 34.Qu J, Gu L, Wu J, et al. Accuracy of IgM antibody testing, FQ-PCR and culture in laboratory diagnosis of acute infection by Mycoplasma pneumoniae in adults and adolescents with community-acquired pneumonia. BMC Infect Dis. 2013;13:172. doi: 10.1186/1471-2334-13-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surinder K. Mycoplasma pneumoniae: a significant but underrated pathogen in paediatric community-acquired lower respiratory tract infections. Indian J Med Res. 2018;147:23–31. doi: 10.4103/ijmr.IJMR_1582_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei Z, Zhi-Yong Z, Yan-Bin L, et al. PCR versus serology for diagnosing mycoplasma pneumoniae infection: a systematic review & meta-analysis. Indian J Med Res. 2011;134:270–280. [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao-yan H, Xing-bin W, Rong Z, et al. Investigation of mycoplasma pneumoniae infection in pediatric population from 12,025 cases with respiratory infection. Diagn Microbiol Infect Dis. 2013;75:22–27. doi: 10.1016/j.diagmicrobio.2012.08.027. [DOI] [PubMed] [Google Scholar]
- 38.Thurman KA, Walter ND, Schwartz SB, et al. Comparison of laboratory diagnostic procedures for detection of mycoplasma pneumoniae in community outbreaks. Clin Infect Dis. 2009;48:1244–1249. doi: 10.1086/597775. [DOI] [PubMed] [Google Scholar]
- 39.Loens K, Ieven M. Mycoplasma pneumoniae: current knowledge on nucleic acid amplification techniques and serological diagnostics. Front Microbiol. 2016;7:448. doi: 10.3389/fmicb.2016.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available in the Supplementary Material.


