Abstract
Neuroblastoma, a type of cancer that is common in children, is composed of two genetically clonal but epigenetically distinct cell types: mesenchymal (MES) and adrenergic (ADRN) types, controlled by super‐enhancer‐associated lineage‐specific transcription factor networks. Mesenchymal‐type cells are more migratory, resistant to chemotherapy, and prevalent in relapse tumors. Importantly, both cell types spontaneously transdifferentiate into one another, and this interconversion can be induced by genetic manipulations. However, the mechanisms of their spontaneous transdifferentiation and extracellular factors inducing this phenomenon have not yet been elucidated. Using a unique approach involving gene set enrichment analysis, we selected six ADRN and 10 MES candidate factors, possibly inducing ADRN and MES phenotypes, respectively. Treatment with a combination of 10 MES factors clearly induced the MES gene expression profile in ADRN‐type SH‐SY5Y neuroblastoma cells. Considering the effects on gene expression profile, migration ability, and chemoresistance, a combination of tumor necrosis factor alpha (TNF‐α) and epidermal growth factor (EGF) was sufficient to synergistically induce the ADRN‐to‐MES transdifferentiation in SH‐SY5Y cells. In addition, human neuroblastoma cohort analysis revealed that the expression of TNF and EGF receptors was strongly associated with MES gene expression signatures, supporting their important roles in transdifferentiation in vivo. Collectively, we propose a mechanism of neuroblastoma transdifferentiation induced by extracellular growth factors, which can be controlled in clinical situations, providing a new therapeutic possibility.
Keywords: cell transdifferentiation, epidermal growth factor, intratumor heterogeneity, neuroblastoma, tumor necrosis factor‐alpha
Neuroblastoma is composed of two epigenetically controlled cell types, mesenchymal (MES) and adrenergic (ADRN) types, which spontaneously transdifferentiate into one another. We found a synergistic combination of tumor necrosis factor and epidermal growth factor that induced the transdifferentiation from ADRN to MES state in ADRN‐type SH‐SY5Y neuroblastoma cells.
1. INTRODUCTION
Intertumor and intratumor genetic heterogeneity is prevalent in many types of tumors, including neuroblastoma, posing a difficult challenge to personalized medicine. 1 Neuroblastoma is a type of tumor that is common in children. It arises in the adrenal gland or sympathetic ganglia and is derived from the sympathoadrenal lineage of neural crest stem cells. 2 Neuroblastoma shows inter‐ and intratumor genetic heterogeneity characterized by abnormal telomere maintenance mechanisms, MYCN amplification, and mutations in RAS and/or p53 pathways, thus leading to poor prognosis. 3 , 4 In addition, there are two genetically clonal but phenotypically distinct cell types in neuroblastoma cell lines: parental SK‐N‐SH cells and its sublines SH‐EP (substrate‐adherent [S‐type]) and SH‐SY5Y (neuroblastic [N‐type]) cells. 5 More recent studies have reported that this intratumor heterogeneity is epigenetically controlled by super‐enhancer‐associated transcription factor networks and is composed of two cell types: undifferentiated mesenchymal (MES; previously known as S‐type) and committed adrenergic (ADRN; previously known as N‐type). 6 , 7 Compared with lineage‐committed ADRN‐type cells, MES‐type cells expressing the stem cell marker CD133 showed gene expression patterns similar to that of neural crest cells, were highly migratory, more resistant to chemotherapy, and more prevalent in tumors that relapse after chemotherapy. 6 Importantly, these MES‐type and ADRN‐type cells can spontaneously transdifferentiate into one another, 8 and the interconversion can be controlled by lineage‐specific genes. For example, the expression of a MES transcription factor, paired related homeobox 1 (PRRX1), 6 and notch receptor 3 (NOTCH3) intracellular domain, 9 or depletion of a chromatin remodeling component AT‐rich interacting domain‐containing 1A (ARID1A) 10 reprogrammed the ADRN cells toward a MES state. Although MES‐type and ADRN‐type cells are well characterized and experimentally controllable, the mechanisms of their spontaneous transdifferentiation driven by external or environmental stimuli have not yet been elucidated.
In this study, we aimed to identify the extracellular factors present in our body that can induce/promote transdifferentiation between MES‐ and ADRN‐type cells and understand the molecular mechanisms of the epigenetically controlled intratumor heterogeneity of neuroblastoma. We found that a combination of tumor necrosis factor‐alpha (TNF‐α) and epidermal growth factor (EGF) induced a MES phenotype in ADRN‐type cells, suggesting the bona fide mechanism of transdifferentiation in neuroblastoma.
2. MATERIALS AND METHODS
2.1. Cell culture
SH‐EP cells were provided by Professor M. Schwab from the German Cancer Research Center. The SH‐SY5Y cells were purchased from ATCC (CRL‐2266). The cells were maintained in RPMI‐1640 medium supplemented with 10% heat‐inactivated FBS in a humidified 5% CO2 incubator at 37°C. The following organic compounds and growth factors were used: estradiol (HY‐B0141; MedChemExpress) dissolved in DMSO, fibroblast growth factor‐basic (100‐18B; PeproTech), brain‐derived neurotrophic factor (450‐02; PeproTech), galectin‐1 (450‐39; PeproTech), granulocyte‐macrophage colony‐stimulating factor (300‐03; PeproTech), interleukin‐4 (200‐04; PeproTech), 5α‐androstan‐17β‐ol‐3‐one (dihydrotestosterone) (A8308; Merck) dissolved in ethanol, leukotriene D4 (20310; Cayman Chemical) dissolved in ethanol, interferon (IFN)‐α 2A (HZ‐1066; Proteintech), IFN‐α 2B (HZ‐1072; Proteintech), IFN‐β (300‐02BC; PeproTech), IFN‐γ (300‐02; PeproTech), transforming growth factor‐β1 (100‐21; PeproTech), TNF‐α (300‐01A; PeproTech), VEGF165 (100‐20; PeproTech), and EGF (AF‐100‐15; PeproTech). All growth factors were dissolved in 0.1% BSA/Dulbecco’s PBS. The final volumes of DMSO or ethanol and 0.1% BSA/Dulbecco’s PBS in media were 0.05% and 0.1%, respectively. The final concentrations of these factors are summarized in Table 1.
TABLE 1.
Six adrenergic (ADRN) and 10 mesenchymal (MES) candidate factors and their final concentrations (conc.)
| ADRN factors | Final conc. | 10 MES factors | Final conc. |
|---|---|---|---|
| Estradiol | 10 nmol/L | Dihydrotestosterone (DHT) | 10 nmol/L |
| FGF‐basic | 40 ng/mL | Leukotriene D4 (LTD4) | 100 nmol/L |
| BDNF | 50 ng/mL | IFN‐α 2A | 10 ng/mL |
| Galectin‐1 | 100 ng/mL | IFN‐α 2B | 10 ng/mL |
| GM‐CSF | 10 ng/mL | IFN‐β | 10 ng/mL |
| IL‐4 | 10 ng/mL | IFN‐γ | 10 ng/mL |
| TGF‐β1 | 10 ng/mL | ||
| TNF‐α | 10 ng/mL | ||
| VEGF165 | 15 ng/mL | ||
| EGF | 20 ng/mL |
BDNF, brain‐derived neurotrophic factor; EGF, epidermal growth factor; FGF, fibroblast growth factor; GM‐CSF, granulocyte‐macrophage colony‐stimulating factor; IFN, interferon; IL, interleukin; TGF, transforming growth factor; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
2.2. Western blot analysis
Cultured cells were lysed with RIPA buffer (25 mmol/L Tris‐Cl pH 7.6, 150 mmol/L NaCl, 1% NP‐40, 1% sodium deoxycholate, and 0.1% SDS) containing protease inhibitor cocktail (04080‐24; Nacalai Tesque). Protein concentrations were determined using the Pierce BCA Protein Assay Kit (23225; Thermo Fisher Scientific). Whole cell extracts were mixed with 2× sample buffer (125 mmol/L Tris‐Cl pH 6.8, 4% SDS, 20% glycerol, 3.1% DTT, and 0.02% bromophenol blue) and heated for 5 minutes at 95°C. Whole cell extracts containing equal amounts of protein were separated by SDS‐PAGE and transferred to PVDF membranes (10600023; Cytiva). The following Abs were used: monoclonal anti‐ß‐actin clone C‐15 (A5441; Merck), YAP (D8H1X) XP rabbit mAb (#14074; Cell Signaling Technology), GATA‐3 (D13C9) XP Rabbit mAb (#5852; Cell Signaling Technology), Phox2a (37K‐2) (sc‐81978; Santa Cruz Biotechnology), Phox2b (B‐11) (sc‐376997; Santa Cruz Biotechnology), and peroxidase‐conjugated rabbit anti‐mouse IgG or goat anti‐rabbit IgG (315‐035‐048 or 111‐035‐144; Jackson ImmunoResearch). Chemiluminescence was developed using the Immobilon Classico or Forte substrate (WBLUC0100 or WBLUF0100; Merck) and detected on the Amersham Imager 680 (Cytiva).
2.3. Quantitative RT‐PCR
Total RNA was isolated using the RNeasy Plus Mini Kit and QIAshredder (74134 and 79656; Qiagen). cDNA was synthesized using ReverTra Ace qPCR RT Master Mix with gDNA remover (FSQ‐301; Toyobo). Quantitative PCR (qPCR) was undertaken using Thunderbird SYBR qPCR Mix (QPS‐201; Toyobo) in the Mx3000P or Mx3005P qPCR system (Agilent). The relative abundances of mRNA transcripts were calculated following the 2−ΔΔCT method and normalized to the housekeeping gene ACTB. The primers used are summarized in Table S1. The qPCR experiments were repeated twice.
2.4. Scratch wound cell migration assay
IncuCyte ZOOM 96‐well Scratch Wound Cell Migration Assay (Essen BioScience) was carried out according to the manufacturer’s instructions. Briefly, the IncuCyte ImageLock 96‐well plates (4379; Essen BioScience) were coated with 100 μg/mL Matrigel Matrix Basement Membrane (354234; Corning) overnight. SH‐EP (30 000 cells/well) and SH‐SY5Y (50 000 cells/well) cells were seeded on the plate. The following day, the cell layers were scratched using the Wound Maker (Essen BioScience). After washing the wells twice with culture media, culture media with or without test factors were added. Phase‐contrast images were captured every 1 hour for 18 hours and analyzed using the IncuCyte ZOOM system with a 4× objective lens (Essen Bioscience). For each scratch, the wound confluence (%) was calculated based on the percentage of initial wound area occupied by cells.
2.5. Cell viability assay
SH‐SY5Y cells (6000 cells/well) were cultured in 96‐well plates in the absence or presence of TNF‐α and EGF for 48 hours. Then the cells were treated with the following chemotherapeutic reagents in a series of 10‐point, 2‐fold serial dilutions for 4 days: 10 μmol/L to 20 nmol/L for etoposide (E1383; Merck) and 1.5 μmol/L to 3 nmol/L for doxorubicin (D1515; Merck). Cell viability was measured using the CellTiter‐Blue Cell Viability Assay (G8080; Promega) according to the manufacturer’s instructions. The cell growth IC50 was calculated by undertaking a four‐parameter logistic curve fitting of the normalized dose‐response curves using Prism 8 (GraphPad software).
2.6. Statistical analysis
Ordinary one‐way ANOVA followed by Dunnett’s multiple comparisons with a single pooled variance was applied for statistical tests using Prism 8 (GraphPad software), and adjusted P values were used to determine the statistical significance.
2.6.1. Data analysis of human neuroblastoma cohort
We used the R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl http://r2platform.com) to investigate the expression of genes and their association with signatures, and to create graphs. The human neuroblastoma cohort including 649 samples (dataset; Kocak – 649 – custom – ag44kcwolf) was used. On the platform, MES and ADRN signature scores were defined as the average z‐score of a z‐score transformed dataset according to the previously identified 485 MES and 369 ADRN genes. 6 The analysis tool “Correlate Gene with track” was used to calculate the correlation coefficients between signature scores and gene expressions. The analysis tool “Relate 2 tracks” was used to draw graphs that show gene expressions in different colors.
3. RESULTS
3.1. Selection of possible candidate factors inducing either MES or ADRN genes using gene set enrichment analysis
To select candidate factors that induce either MES or ADRN genes, we utilized gene set enrichment analysis (GSEA), a computational method that is used to statistically compare a gene expression profile and a database of well‐annotated gene sets called the Molecular Signature Database (MSigDB). 11 For the expression dataset, we used the publicly available gene expression matrix comparing the four pairs of MES‐type and ADRN‐type cells from the Gene Expression Omnibus (accession number: GSE90803). 6 For gene sets, we selected C2‐CGP v6.2 (curated gene sets: chemical and genetic perturbations) from MSigDB, which is a collection of more than 3000 gene sets including upregulated or downregulated genes after certain experimental manipulations, such as gene expression/knockdown and treatment of cells with growth factors. After filtering (min = 15, max = 500 genes), 134 and 1338 of the 2675 gene sets were significantly enriched with a false‐discovery rate of <25% in ADRN and MES phenotypes, respectively. From the top 100 ADRN and 200 MES phenotype‐enriched gene sets, we manually selected the gene sets in which certain cells were treated with organic compounds or growth factors, not with inhibitors or gene manipulations (overexpression or knockdown). Therefore, we assumed that a single factor, or combinations of these factors, might induce gene expression profiles of ADRN or MES‐type cells. For example, we found that the gene sets “SANA_TNF_SIGNALING_UP” and “NAGASHIMA_EGF_SIGNALING_UP” were significantly enriched in MES‐type cells, suggesting that TNF and EGF possibly upregulate MES genes (Figure S1). The complete GSEA results are summarized in Table S2. After careful consideration, we selected six ADRN and 10 MES factors, including organic compounds and growth factors, that possibly induce either ADRN or MES genes (Table 1).
3.2. Ten candidate MES factors that induced MES gene expression profile in ADRN‐type SH‐SY5Y cells
To investigate the effects of ADRN or MES factors on the induction of ADRN or MES gene expression profiles, we selected a pair of neuroblastoma cell lines, SH‐EP (MES‐type) and SH‐SY5Y (ADRN‐type) cells, that were subcloned from parental SK‐N‐SH cells. 8 Initially, we determined whether a combination of six ADRN or 10 MES candidate factors could differentially regulate MES or ADRN genes. SH‐EP cells are larger and flatter, resembling epithelial cells, and highly substate adherent, whereas SH‐SY5Y cells are smaller, rounded, and loosely adherent, and have neurite‐like structures. When SH‐EP cells were treated with a combination of ADRN factors for 48 hours, significant changes in cell shape were observed, becoming smaller in size and showing neurite structures. The expressions of both MES and ADRN marker genes were evaluated by western blot analysis. Normally, the MES marker gene YAP is highly expressed, and the ADRN genes PHOX2A, PHOX2B, and GATA3 are absent in SH‐EP cells (Figure 1A). However, the expression of both MES and ADRN genes was not altered in SH‐EP cells treated with a combination of six ADRN factors, suggesting that candidate ADRN factors could not induce the ADRN gene profile (Figure 1A). In contrast, the structure of SH‐SY5Y cells treated with a combination of 10 MES factors for 48 hours resembled that of MES‐type cells, showing flattened cell shape with reduced neurite structures (Figure 1B). Importantly, a combination of 10 MES factors modestly upregulated YAP and strongly downregulated PHOX2A, PHOX2B, and GATA3 in SH‐SY5Y cells, indicating an ADRN to MES transition (Figure 1B).
FIGURE 1.
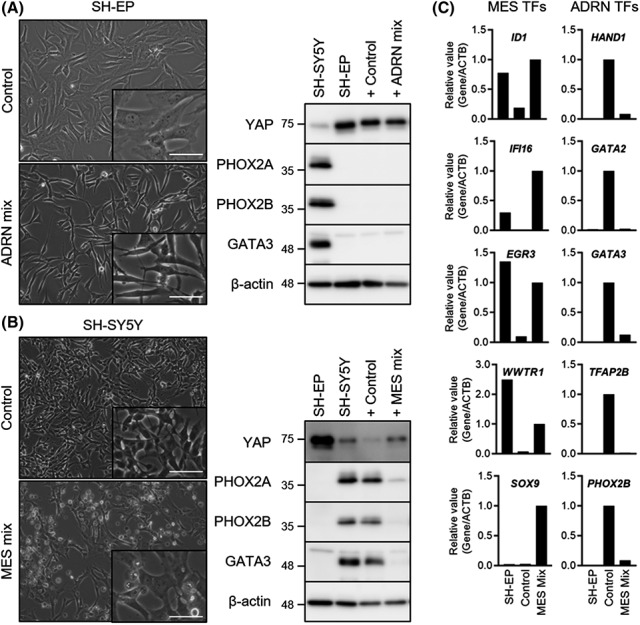
Treatment with a combination of 10 mesenchymal (MES) factors induced the MES gene expression profile in SH‐SY5Y neuroblastoma cells. A, Phase contrast images and western blotting of SH‐EP cells treated with control or a combination of six adrenergic (ADRN) factors for 48 h. B, Phase contrast images and western blotting of SH‐SY5Y cells treated with control or a combination of 10 MES factors for 48 h. C, Relative expression levels of five MES and five ADRN transcription factor (TF) genes normalized to ACTB in SH‐EP cells and SH‐SY5Y cells treated with control or a combination of 10 MES factors for 48 h. ACTB, β‐actin. Scale bar = 50 μm
Tim van Groningen et al 6 reported that both MES‐type and ADRN‐type cells showed different super‐enhancer landscapes and identified super‐enhancer‐associated lineage transcription factor (TF) networks, composed of either 20 MES or 18 ADRN TF genes. We wondered whether a combination of 10 MES factors could control these super‐enhancer‐associated lineage TFs. For qPCR experiments, five MES TF genes, including ID1, IFI16, EGR3, WWTR1, and SOX9, and five ADRN TF genes, including HAND1, GATA2, GATA3, TFAP2B, and PHOX2B, were selected; all of these genes were differentially expressed in SH‐EP and SH‐SY5Y cells (Figures 1C and S2A). Importantly, all five MES and five ADRN TF genes were upregulated and downregulated, respectively, in SH‐SY5Y cells treated with a combination of 10 MES factors (Figures 1C and S2A). These results indicate that a single component or a combination of these 10 MES factors can potentially induce a MES gene expression profile in ADRN‐type SH‐SY5Y neuroblastoma cells.
3.3. Narrowing down the candidate factors
To determine which of the 10 MES factors were essential, we examined the effect of withdrawal of individual factors from a combination of 10 MES factors on the expression of five MES and five ADRN TF genes in SH‐SY5Y cells. The removal of individual factors from the combination of 10 MES factors resulted in an altered expression pattern. Consistent with Figure 1C, all five MES or five ADRN TF genes were greatly upregulated or downregulated in SH‐SY5Y cells treated with a combination of 10 MES factors, respectively (Figures 2A and S2B). Importantly, this upregulation or downregulation of MES or ADRN TF genes was attenuated by the removal of TNF‐α or EGF, prominently observed in three MES TF genes (EGR3, WWTR1, and SOX9) and all five ADRN TF genes (Figures 2A and S2B). In contrast, the removal of IFN‐γ showed an effect similar to that of TNF‐α or EGF but with an upregulation of SOX9 (Figures 2A and S2B). This result suggests that three (IFN‐γ, TNF‐α, and EGF) of the 10 MES factors are important components that induce the MES gene expression profile. Accordingly, withdrawal of these three factors resulted in a more obvious attenuation of MES or ADRN TF gene upregulation or downregulation (Figures 2B and S2C).
FIGURE 2.
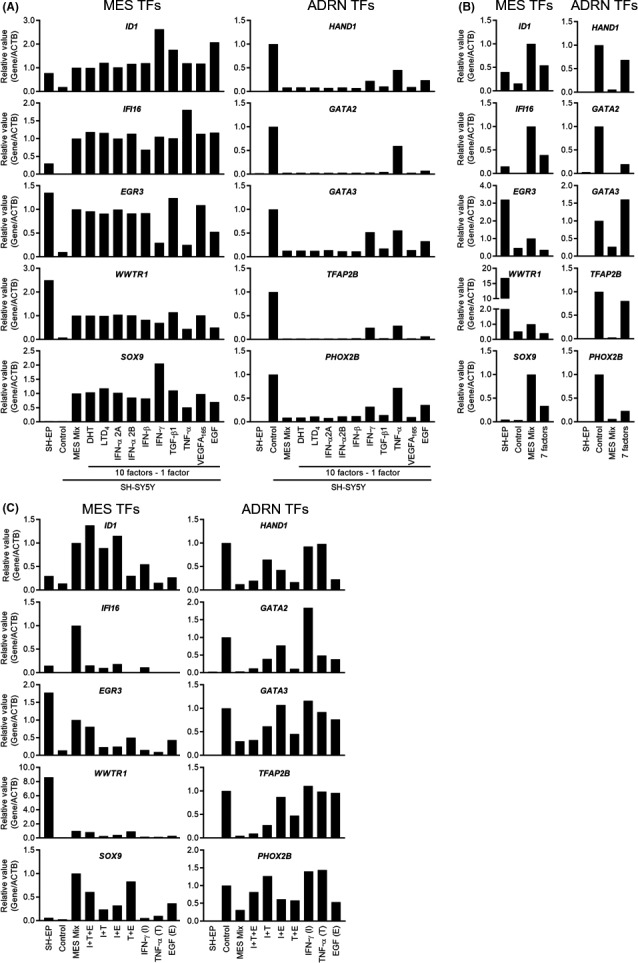
Narrowing down the candidate factors. A–C, Relative expression levels of five mesenchymal (MES) and five adrenergic (ADRN) transcription factor (TF) genes normalized to ACTB in SH‐SY5Y neuroblastoma cells treated with different conditions for 48 h. A, Each of the 10 MES factors were individually removed. B, Three (interferon‐γ [IFN‐γ], tumor necrosis factor‐α [TNF‐α], and epidermal growth factor [EGF]) of the 10 MES factors were individually removed. C, Single or combinations of three factors (IFN‐γ, TNF‐α, and EGF) were added. ACTB, β‐actin; DHT, dihydrotestosterone; LTD4, leukotriene D4; TGF‐β1, transforming growth factor‐β1; VEGFA, vascular endothelial growth factor A
To identify the significant factors that induce the MES gene expression profile, we determined whether treatment with a single or combination of three factors (IFN‐γ, TNF‐α, and EGF) could produce the effects comparable with those of a combination of 10 MES factors on the expression of MES and ADRN TF genes. As shown in Figures 2C and S2D, treatment with IFN‐γ or TNF‐α alone did not result in the upregulation or downregulation of MES or ADRN TF genes, while treatment with EGF alone resulted in mild changes. Importantly, treatment with a combination of TNF‐α and EGF did not completely but comparably upregulated or downregulated the MES or ADRN TF genes compared with treatment with a combination of all MES factors, which were prominently observed in three MES (EGR3, WWTR1, and SOX9) and four ADRN (HAND1, GATA2, GATA3, and PHOX2B) TF genes. Treatment with a combination of IFN‐γ with TNF‐α or EGF also showed comparable effects on some genes, but the overall effect was not stronger compared with that of a combination of TNF‐α and EGF. Combinations of all three factors almost completely mirrored the effect of a combination of 10 MES factors except that of IFI16 and PHOX2B. Therefore, treatment with all three components or a combination of TNF‐α and EGF among the 10 MES factors synergistically induced the MES gene expression profile.
3.4. Treatment with TNF‐α and EGF promotes migration ability and drug resistance in SH‐SY5Y cells
As shown in previous studies, one of the characteristics of MES‐type neuroblastoma cells is their higher migration ability. 6 Importantly, the migratory ability of this phenotype can be induced by genetic manipulations in ADRN‐type neuroblastoma cells, including overexpression of MES TF genes PRRX1 6 and NOTCH receptors, 9 , 12 and depletion of the chromatin remodeler ARID1A. 10 Hence, we investigated whether treatment with a single or a combination of three factors (IFN‐γ, TNF‐α, and EGF) promoted the migration ability of SH‐SY5Y cells. The scratch wound cell migration assay revealed that SH‐SY5Y cells migrated less than SH‐EP cells, confirming that MES‐type cells have higher migratory ability (Figure 3A). Under these conditions, a migration assay and the treatment of SH‐SY5Y cells with a single or combination of three factors were carried out simultaneously. As shown in Figure 3B,C, SH‐SY5Y cells treated with EGF alone or a combination of three factors showed significant migration compared with SH‐SY5Y nontreated cells. Importantly, the combination of TNF‐α and EGF significantly promoted the migration ability of SH‐SY5Y cells, as early as 10 hours after treatment (Figure 3A‐C). To determine the effect of cell proliferation on cell migration, the relative cell growth levels were measured. SH‐SY5Y cells treated with EGF alone or with IFN‐γ showed a slight increase in cell growth level, but treatment with TNF‐α and EGF did not affect the cell proliferation rate within 24 hours (Figure 3D). In contrast, treatment with a combination of 10 MES factors and a combination of three factors (IFN‐γ, TNF‐α, and EGF) reduced the cell proliferation rate (Figure 3D); cell death was also observed at later time points (data not shown). Based on these findings and the results of previous studies (Figure 2), we concluded that TNF‐α and EGF synergistically induced a MES phenotype in SH‐SY5Y cells without affecting their viability.
FIGURE 3.
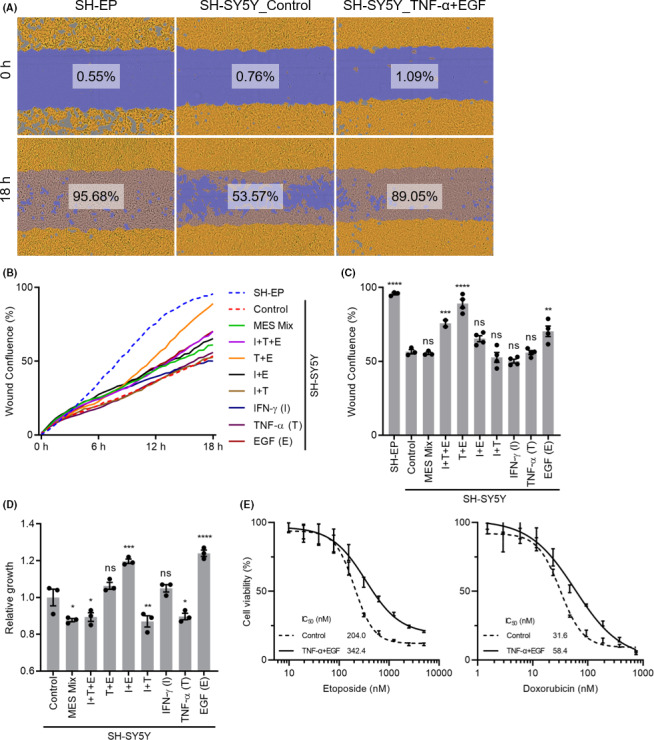
Treatment with a combination of tumor necrosis factor‐α (TNF‐α) and epidermal growth factor (EGF) promoted the migration ability and drug resistance of SH‐SY5Y neuroblastoma cells. A, Cell migration assay was carried out using IncuCyte ZOOM. Representative phase contrast images of SH‐EP and SH‐SY5Y cells treated with control or a combination of TNF‐α and EGF. Phase contrast images were taken by IncuCyte ZOOM every 1 h for 18 h, and the cell area (yellow) and initial wound area (blue) were measured. Percent wound confluences were calculated for an initial wound area covered with migrated cell area. B, Percent wound confluences for all conditions from 0 to 18 h. C, Quantification of percent wound confluences for all conditions at 18 h. D, Quantification of relative cell growth at 24 h after the treatment of SH‐SY5Y cells. E, Dose‐response curves to etoposide and doxorubicin at 4 days after 48 h of pretreatment with control or a combination of TNF‐α and EGF. Cell growth IC50 values were calculated using a four‐parameter logistic curve fitting. Statistical significance was reported against SH‐SY5Y cells treated with control. *P < .05; **P < .01; ***P < .001; ****P < .0001. MES, mesenchymal; ns, not significant
Another important characteristic of MES‐type neuroblastoma cells is chemoresistance. For example, cisplatin‐resistant neuroblastoma sublines achieved epithelial‐to‐mesenchymal transition, 13 SH‐EP cells were more resistant to standard anticancer drugs cisplatin, doxorubicin, and etoposide compared to SH‐SY5Y cells, 6 , 7 and the loss of ADRD1A induced an MES phenotype and resistance to cisplatin in ADRN‐type NGP cells. 10 Therefore, we investigated whether a combination of TNF‐α and EGF affects chemoresistance in SH‐SY5Y cells. The growth IC50 values were examined 4 days after the addition of etoposide and doxorubicin 48 hours following the pretreatment of SH‐SY5Y cells with either control or a combination of TNF‐α and EGF (Figure 3E). Interestingly, SH‐SY5Y cells treated with a combination of TNF‐α and EGF were more resistant to these drugs by two fold (Figure 3E). Collectively, a combination of TNF‐α and EGF induced MES‐type neuroblastoma phenotypes in SH‐SY5Y cells.
3.5. Positive correlation between expressions of TNF and EGF receptors and MES signatures in human neuroblastoma
Finally, we investigated whether our findings are relevant to human neuroblastoma. Using the R2 genomics platform, a publicly available web‐based analysis platform, we evaluated the expression of TNF, EGF, and its receptors, and their association with MES/ADRN signatures in a cohort including 649 human neuroblastomas. We calculated the MES and ADRN signature scores based on the 485 MES and 369 ADRN genes defined previously. 6 Then we calculated the correlation coefficients between the signature scores and gene expressions. The expression of NOTCH receptors such as NOTCH1 and NOTCH2 has previously been reported to be highly correlated with MES signature scores, 9 confirming the validity of this analysis (Figure S3). Unexpectedly, TNF and EGF were broadly expressed across tumors and their expression was not associated with either MES or ADRN signature scores (Figure 4A). In contrast, the expression of TNF receptors (TNFRSF1A and TNFRSF1B) and EGF receptor (EGFR) were strongly positively and negatively correlated with the MES and ADRN signature scores, with correlation coefficients of more than 0.5 and less than −0.5, respectively (Figure 4B). These results imply that human neuroblastoma with high MES signatures are more likely to receive extracellular TNF‐α and EGF proteins, which might be the result of the transdifferentiation of ADRN‐type cells to MES‐type cells.
FIGURE 4.
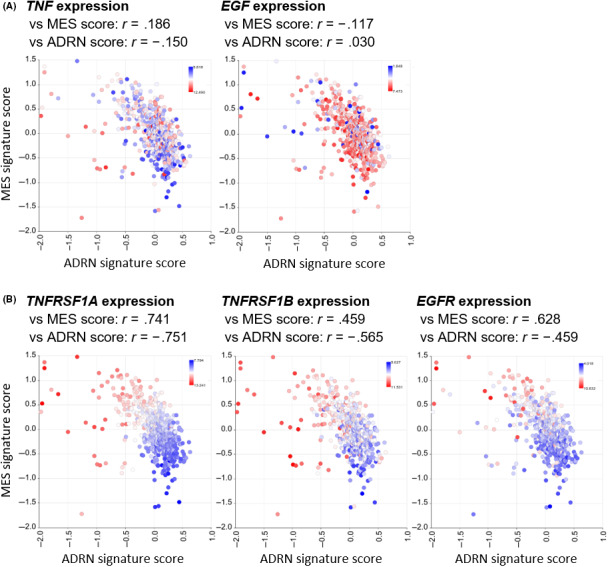
Comparison of mesenchymal (MES) and adrenergic (ADRN) signature scores based on expression levels of tumor necrosis factor (TNF), epidermal growth factor (EGF), and their receptors in human neuroblastoma. A, Expression levels of TNF and EGF in 649 human neuroblastoma cohort. X‐axis shows the ADRN signature scores, and Y‐axis shows the MES signature scores. Color indicates the log2‐transformed gene expression levels. Correlation coefficients between gene expressions and signature scores are indicated above the graphs. B, Same plots for TNF (TNFRSF1A and TNFRSF1B) and EGF (EGFR) receptors
4. DISCUSSION
Recent large‐scale genetic analysis of 702 neuroblastoma samples revealed that 40% of them contained at least one of the recurrent driver gene abnormalities, including MYCN, ATRX, and TERT alterations. 14 However, the remaining 60% of neuroblastomas do not show such gene‐level alterations, implying that neuroblastomas are epigenetically driven tumors, and thus clinicians and patients are facing more difficult situations to cure the disease. For better personalized medicine, the epigenetically regulated inter‐ and intratumor heterogeneities of neuroblastomas must be intensively studied. In the current study, a unique approach was used to understand the transdifferentiation between ADRN‐type and MES‐type neuroblastomas. MSigDB C2‐CGP in GSEA is a collection of well‐curated gene sets generated by undertaking a gene expression analysis after carrying out experimental interventions, including treatment of cells with growth factors. Therefore, if we have our own gene expression matrix comparing two conditions, we can find gene sets that were significantly enriched in one of the conditions. Because these gene sets were created by experimental manipulations, we can assume that the same manipulation would provide the gene expression profile we observed.
Using this unique approach, we selected six ADRN and 10 MES candidate factors that can possibly induce ADRN and MES expression profiles, respectively. Unfortunately, treatment with a combination of six ADRN factors did not induce the ADRN gene expression profile in SH‐EP cells (MES type), probably because the number of enriched gene sets after GSEA was not sufficient to select the candidate factors; 134 and 1338 gene sets were significantly enriched in ADRN and MES phenotypes (Table S2). In contrast, treatment with a combination of 10 MES factors clearly induced the MES gene expression profile in SH‐SY5Y cells (ADRN type). Based on the expression of super‐enhancer‐associated lineage TFs, migration ability, and chemoresistance, we narrowed down and identified that the synergistic combination of TNF‐α and EGF induced the MES state. Importantly, the expression of both TNF and EGF receptors was strongly associated with MES signatures in human neuroblastoma, suggesting that intracellular signaling mediated by these receptors might control the interconversion between ADRN and MES types or the identity of MES‐type neuroblastoma cells.
Interestingly, a previous study showed that the acquisition of cisplatin resistance (a characteristic of MES‐type cells) accompanied increased EGFR expression in neuroblastoma cells, making them highly sensitive to EGFR‐targeted therapy. 15 Epidermal growth factor promotes the proliferation of neuroblastoma cells, 16 as observed in this study (Figure 3D). Tumor necrosis factor‐α can also increase the cell proliferation rate in two neuroblastoma cell lines (SK‐N‐BE and SK‐N‐FI), and Abs targeting TNF or its receptors inhibited cell growth 17 ; however, we observed an opposite effect in SH‐SY5Y cells (Figure 3D). A combination of TNF‐α and IFN‐γ also suppressed cell growth in our study, probably due to the induction of neuroblastoma cell differentiation, as shown previously. 18 In addition, ionizing radiation induces a TNF‐α‐nuclear factor‐κB positive feedback loop, which is advantageous for the survival of neuroblastoma cells. 19 Because MES‐type neuroblastoma cells are more prevalent in relapsed neuroblastoma, 6 resistance to chemotherapy or radiation might be regulated partly by TNF signaling. Although TNF‐α and EGF alone or in combination with other factors have shown various effects in neuroblastoma, our finding that a combination of these factors induced the transdifferentiation of ADRN‐type cells to MES‐type cells confirms the previously unknown synergistic effects of TNF‐α and EGF.
We tested the combination of TNF‐α and EGF on other ADRN‐type neuroblastoma cell lines (Kelly, NB69, SK‐N‐BE2, and SK‐N‐FI); however, none of them showed obvious changes in the expression of ADRN or MES TF genes or migration abilities (data not shown), probably due to the context‐dependent phenomenon limited to SH‐SY5Y cells. Thus, our in vitro findings should be validated in vivo, preferably at the single‐cell level in humans. Nevertheless, our unique approach and findings showed that neuroblastoma transdifferentiation is induced by external/extracellular growth factors, which can be controlled in the clinical setting and thus can be used as a basis for establishing a new therapeutic option.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
Figure S1‐S3
Table S1
Table S2
ACKNOWLEDGMENTS
This study was supported by JSPS KAKENHI Grant Numbers JP18K15208 (S. Tsubota), JP20K16355 (S. Tsubota), JP18H02690 (Y. Takahashi), and JST CREST Grant Number JPMJCR1502 (K. Kadomatsu). Y. Huang would like to thank the Otsuka Toshimi Scholarship Foundation for its financial support (19‐S89, 20‐S37). The authors would like to thank Editage for English language editing.
Huang Y, Tsubota S, Nishio N, Takahashi Y, Kadomatsu K. Combination of tumor necrosis factor‐α and epidermal growth factor induces the adrenergic‐to‐mesenchymal transdifferentiation in SH‐SY5Y neuroblastoma cells. Cancer Sci. 2021;112:715–724. 10.1111/cas.14760
Funding information
Japan Society for the Promotion of Science KAKENHI, Grant/Award Numbers: JP18K15208, JP20K16355, JP18H02690; JST CREST, Grant/Award Number: JPMJCR1502; Otsuka Toshimi Scholarship Foundation, Grant/Award Number: 19‐S89, 20‐S37
Contributor Information
Shoma Tsubota, Email: tsubota@med.nagoya-u.ac.jp.
Kenji Kadomatsu, Email: kkadoma@med.nagoya-u.ac.jp.
REFERENCES
- 1. Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338‐345. [DOI] [PubMed] [Google Scholar]
- 2. Matthay KK, Maris JM, Schleiermacher G, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2:16078. [DOI] [PubMed] [Google Scholar]
- 3. Ackermann S, Cartolano M, Hero B, et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362:1165‐1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high‐risk neuroblastoma. Nat Genet. 2013;45:279‐284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ciccarone V, Spengler BA, Meyers MB, Biedler JL, Ross RA. Phenotypic diversification in human neuroblastoma cells: expression of distinct neural crest lineages. Cancer Res. 1989;49:219‐225. [PubMed] [Google Scholar]
- 6. van Groningen T, Koster J, Valentijn LJ, et al. Neuroblastoma is composed of two super‐enhancer‐associated differentiation states. Nat Genet. 2017;49:1261‐1266. [DOI] [PubMed] [Google Scholar]
- 7. Boeva V, Louis‐Brennetot C, Peltier A, et al. Heterogeneity of neuroblastoma cell identity defined by transcriptional circuitries. Nat Genet. 2017. [DOI] [PubMed] [Google Scholar]
- 8. Ross RA, Spengler BA, Biedler JL. Coordinate morphological and biochemical interconversion of human neuroblastoma cells. J Natl Cancer Inst. 1983;71:741‐747. [PubMed] [Google Scholar]
- 9. van Groningen T, Akogul N, Westerhout EM, et al. A NOTCH feed‐forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat Commun. 2019;10:1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shi H, Tao T, Abraham BJ, et al. ARID1A loss in neuroblastoma promotes the adrenergic‐to‐mesenchymal transition by regulating enhancer‐mediated gene expression. Sci Adv. 2020;6(29):eaaz3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge‐based approach for interpreting genome‐wide expression profiles. P Natl Acad Sci Usa. 2005;102:15545‐15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. van Nes J, Chan A, van Groningen T, van Sluis P, Koster J, Versteeg R. A NOTCH3 transcriptional module induces cell motility in neuroblastoma. Clin Cancer Res. 2013;19:3485‐3494. [DOI] [PubMed] [Google Scholar]
- 13. Piskareva O, Harvey H, Nolan J, et al. The development of cisplatin resistance in neuroblastoma is accompanied by epithelial to mesenchymal transition in vitro. Cancer Lett. 2015;364:142‐155. [DOI] [PubMed] [Google Scholar]
- 14. Brady SW, Liu Y, Ma X, et al. Pan‐neuroblastoma analysis reveals age‐ and signature‐associated driver alterations. Nat Commun. 2020;11:5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Michaelis M, Bliss J, Arnold SC, et al. Cisplatin‐resistant neuroblastoma cells express enhanced levels of epidermal growth factor receptor (EGFR) and are sensitive to treatment with EGFR‐specific toxins. Clin Cancer Res. 2008;14:6531‐6537. [DOI] [PubMed] [Google Scholar]
- 16. Ho R, Minturn JE, Hishiki T, et al. Proliferation of human neuroblastomas mediated by the epidermal growth factor receptor. Cancer Res. 2005;65:9868‐9875. [DOI] [PubMed] [Google Scholar]
- 17. Goillot E, Combaret V, Ladenstein R, et al. Tumor necrosis factor as an autocrine growth factor for neuroblastoma. Cancer Res. 1992;52:3194‐3200. [PubMed] [Google Scholar]
- 18. Ponzoni M, Casalaro A, Lanciotti M, Montaldo PG, Cornaglia‐Ferraris P. The combination of gamma‐interferon and tumor necrosis factor causes a rapid and extensive differentiation of human neuroblastoma cells. Cancer Res. 1992;52:931‐939. [PubMed] [Google Scholar]
- 19. Veeraraghavan J, Natarajan M, Aravindan S, Herman TS, Aravindan N. Radiation‐triggered Tumor Necrosis Factor (TNF) α‐NFκB cross‐signaling favors survival advantage in human neuroblastoma cells. J Biol Chem. 2011;286:21588‐21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1‐S3
Table S1
Table S2


