Abstract
We aimed to isolate circulating tumor cells (CTCs) using a microfluidic technique with a novel lateral magnetophoretic microseparator. Prostate cancer–specific gene expressions were evaluated using mRNA from the isolated CTCs. A CTC‐based multigene model was then developed for identifying advanced prostate cancer. Peripheral blood samples were obtained from five healthy donors and patients with localized prostate cancer (26 cases), metastatic hormone‐sensitive prostate cancer (mHSPC, 10 cases), and metastatic castration‐resistant prostate cancer (mCRPC, 28 cases). CTC recovery rate and purity (enriched CTCs/total cells) were evaluated according to cancer stage. The areas under the curves of the six gene expressions were used to evaluate whether multigene models could identify mHSPC or mCRPC. The number of CTCs and their purity increased at more advanced cancer stages. In mHSPC/mCRPC cases, the specimens had an average of 27.5 CTCs/mL blood, which was 4.2 × higher than the isolation rate for localized disease. The CTC purity increased from 2.1% for localized disease to 3.8% for mHSPC and 6.7% for mCRPC, with increased CTC expression of the genes encoding prostate‐specific antigen (PSA), prostate‐specific membrane antigen (PSMA), and cytokeratin 19 (KRT19). All disease stages exhibited expression of the genes encoding androgen receptor (AR) and epithelial cell adhesion molecule (EpCAM), although expression of the AR‐V7 variant was relatively rare. Relative to each gene alone, the multigene model had better accuracy for predicting advanced prostate cancer. Our lateral magnetophoretic microseparator can be used for identifying prostate cancer biomarkers. In addition, CTC‐based genetic signatures may guide the early diagnosis of advanced prostate cancer.
Keywords: circulating tumor cells, genetic signature, magnetophoresis, prognostication, prostate cancer
CTCs have useful biological information for understanding the prostate cancer aggressiveness. CTC‐based gene signatures have potential for early diagnosis of metastatic prostate cancer. Multigene profile (AR, AR‐V7, PSA, PSMA, EpCAM, and KRT19) may guide the diagnosis and treatment of metastatic prostate cancer.
Abbreviations
- AR
androgen receptor
- AR‐V7
androgen receptor variant 7
- CTCs
circulating tumor cells
- ddPCR
droplet digital PCR
- EpCAM
epithelial cell adhesion molecule
- KRT19
cytokeratin 19
- mCRPC
metastatic castration‐resistant prostate cancer
- mHSPC
metastatic hormone‐sensitive prostate cancer
- MS
multigene score
- PSA
prostate‐specific antigen
- PSMA
prostate‐specific membrane antigen
- WBCs
white blood cells
1. INTRODUCTION
Prostate cancer is the second most common cancer among men, and is the most frequently diagnosed cancer in many countries, with 1.3 million new cases and a mortality rate of 3.8% in 2018. 1 , 2 Although prostate‐specific antigen (PSA) testing can help guide the diagnosis and management of prostate cancer, it remains difficult to achieve an early and accurate diagnosis of aggressive prostate cancer. 1 , 3 Furthermore, it remains unclear how localized prostate cancer progresses to metastatic hormone‐sensitive prostate cancer (mHSPC) or metastatic castration‐resistant prostate cancer (mCRPC). In addition, patients with mCRPC no longer respond to medications that target androgen receptor (AR) and experience continued deterioration. Therefore, it is important to clarify the mechanisms underlying prostate cancer progression, which may guide the development of better treatment strategies for patients with metastatic prostate cancer.
There is also a need for better biomarkers to identify prostate cancer progression, which can help guide treatment decisions. 4 The discovery that tumors shed circulating tumor cells (CTCs) has raised the possibility of minimally invasive cancer monitoring, which may help determine the tumor burden and molecular or genetic markers that are linked to the risk of cancer progression. Many studies have confirmed that CTCs and their numbers are associated with survival among patients with prostate cancer. 5 , 6 , 7 , 8 , 9 Genomic analysis of CTCs has also emerged as a promising tool since the discovery that resistance to AR‐targeting agents in patients with mCRPC was associated with CTC‐based expression of the gene for AR splice variant 7 (AR‐V7). 10 , 11 , 12 , 13 However, despite the development of next‐generation sequencing and droplet digital PCR (ddPCR), it remains difficult to obtain accurate genomic information regarding CTCs, based on their extreme rarity in the blood. Therefore, advanced techniques are needed to help isolate CTCs and identify clinically useful genomic biomarkers.
Most clinical studies have used commercially available CTC isolation methods, including the CellSearch system (Menarini Silicon Biosystems), the EPIC platform (Epic Sciences), and the AdnaTest (QIAGEN). 11 , 14 , 15 , 16 , 17 , 18 , 19 Nevertheless, many microfluidic techniques are superior to these commercial macroscale methods for isolating CTCs. While most studies have only described the recovery rate of CTCs, some studies have also described the proportion of CTCs and coisolated white blood cells (WBCs). 20 , 21 The results indicate that approximately 90% of CTCs can be recovered, although the WBC depletion rate remains in the range of 1.29‐3.25 log, and the purity of CTCs in the sample remains only 0.01%‐1%. 22 , 23 , 24
This study aimed to isolate CTCs from prostate cancer patients using a microfluidic technique with a disposable lateral magnetophoretic microseparator. Furthermore, we aimed to evaluate the expressions of various prostate cancer–specific genes using mRNA from the isolated CTCs and the ddPCR method. The ultimate aim of this approach is to achieve early detection of metastatic prostate cancer, which can help guide prognostication and clinical decision‐making. Unfortunately, conventional imaging has insufficient sensitivity when staging patients with advanced prostate cancer, and we hypothesized that a CTC‐based gene expression profile could help better identify mHSPC and mCRPC. Thus, we evaluated the CTC expression profiles of six genes encoding AR, AR‐V7, PSA, prostate‐specific membrane antigen (PSMA), epithelial cell adhesion molecule (EpCAM), and cytokeratin 19 (KRT19). That information was used to develop a CTC‐based multigene model for identifying metastatic prostate cancer.
2. MATERIALS AND METHODS
2.1. Patients and sample preparation
Between May 2018 and May 2019, peripheral blood samples were obtained from five healthy donors and 64 patients who had localized prostate cancer (26 patients), mHSPC (10 patients), or mCRPC (28 patients; Table 1). All subjects provided written informed consent, and the study protocol was approved by our institutional review board.
Table 1.
Demographic characteristics and clinical features of the patients with prostate cancer
| Localized stage | Metastatic stage | ||||
|---|---|---|---|---|---|
| T2 | T3 | T4 | mHSPC | mCRPC | |
| (N*=11, n†=11) | (N = 9, n = 9) | (N = 6, n = 6) | (N = 10, n = 10) | (N = 23, n = 28) | |
| Age, median, IQR, years | 70 (55‐84) | 71 (62‐76) | 72 (59‐80) | 75 (64‐80) | 75 (57‐84) |
| PSA, median, IQR, ng/ml | 8.9 (4.2‐11.6) | 12.5 (8.3‐16.9) | 23.3 (14.5‐36.8) | 126.2 (85.2‐155.2) | 98.4 (30.6‐150.8) |
| Gleason score (%) | |||||
| 6 | 7 (63.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| 7 | 2 (18.2) | 4 (44.5) | 1 (16.7) | 0 (0.0) | 2 (8.7) |
| 8 | 2 (18.2) | 1 (11.1) | 2 (33.3) | 5 (50.0) | 6 (26.1) |
| 9 | 0 (0.0) | 4 (44.4) | 3 (50) | 3 (30.0) | 11 (47.8) |
| 10 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (20.0) | 4 (17.4) |
| Node metastasis (%) | |||||
| Yes | 0 (0.0) | 0 (0.0) | 1 (16.7) | 6 (55.6) | 11 (4.3) |
| No | 11(100.0) | 9 (100.0) | 5 (83.3) | 4 (44.4) | 12 (4.3) |
| Visceral or bone metastasis (%) | |||||
| Yes | 0 (0.0) | 0 (0.0) | 0 (0.0) | 10 (100.0) | 23 (100.0) |
| No | 11 (100.0) | 9 (100.0) | 6 (100.0) | 0 (0.0) | 0 (0.0) |
| Prior therapy (%) | |||||
| None | 11 (100.0) | 9 (100.0) | 6 (100.0) | 2 (20.0) | 0 (0.0) |
| Radiation | — | — | — | 1 (10.0) | 11 (39.2) |
| Taxane | — | — | — | 0 (0.0) | 5 (17.9) |
| Estramustine Phosphate | — | — | — | 0 (0.0) | 1 (3.5) |
| GnRH | — | — | — | 7 (70.0) | 4 (14.3)) |
| Antiandrogen | — | — | — | 0 (0.0) | 11 (39.2) |
| Abiraterone | — | — | — | 0 (0.0) | 2 (7.1) |
| Enzalutamide | — | — | — | 0 (0.0) | 5 (17.9) |
Abbreviations: GnRH, gonadotropin‐releasing hormone; mCRPC, metastatic castration‐resistant prostate cancer; mHSPC, metastatic hormone‐sensitive prostate cancer; N*, The number of patients; n†, The number of blood samples; PSA, prostate‐specific antigen.
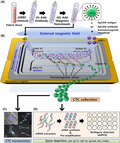
The blood samples were drawn into 10‐mL Vacutainer tubes (367525, BD Vacutainer) that contained EDTA (K2EDTA, 18.0 mg) and were stored at 4°C until processing, which was performed within 4 hours. Red blood cells (RBCs) were removed via density gradient centrifugation (700 g, 30 minutes) using a 1.119 g/mL Ficoll solution (Histopaque‐1119, Sigma‐Aldrich). The buffy coat layer was then transferred into 10 mL of ice‐cold PBS with 0.2% bovine serum albumin in a 50‐mL conical tube. After washing the nucleated cells, they were suspended in 200 μL of ice‐cold PBS with 0.2% bovine serum albumin in a 1.5‐mL microcentrifuge tube. Antibodies targeting EpCAM and magnetic nanobeads (STEMCELL Technologies) were sequentially added to the 200‐μL sample, with incubations on ice for 60 and 90 minutes, respectively, according to the manufacturer's instructions (Figure 1A). The final sample was prepared by dilution using 800 μL of ice‐cold PBS with 0.2% bovine serum albumin.
Figure 1.
A, The sample preparation procedure involved (i) removing red blood cells (RBCs) using a Ficoll gradient, (ii) adding antibodies that specifically bind to membrane epithelial cell adhesion molecule (EpCAM) on circulating tumor cells (CTCs), and (iii) adding immunomagnetic nanobeads (OD: 50 nm) to bind the anti‐EpCAM antibodies. B, An illustration presenting the working principle for using the lateral magnetophoretic microseparator to isolate the CTCs from prepared blood sample. C, A sample is used to determine the number of isolated CTCs and white blood cells (WBCs) using immunofluorescent image analysis. D, The other sample is used for genomic analysis using real‐time droplet digital PCR (RT‐ddPCR). AR, androgen receptor; AR‐V7, androgen receptor variant 7; KRT‐19, cytokeratin 19; PSA, prostate‐specific antigen; PSMA, prostate‐specific membrane antigen
2.2. Cell line spiking test
Prostate cancer cells were prepared using the LNCaP cell line to evaluate the performance of the lateral magnetophoretic microseparator based on the CTC recovery rate, the WBC depletion rate, and the purity of the CTCs. The LNCaP cells were initially stained using a green fluorescent nucleic acid dye (SYTO 13, Invitrogen). After washing, approximately 100 stained LNCaP cells were added into 5 mL of blood from the healthy donors, which resulted in spiked blood samples. The LNCaP cells were then isolated from the spiked blood samples according to the experimental methods described above.
2.3. Working principle and CTC isolation
The process for fabricating the disposable lateral magnetophoretic microseparator has been previously reported 25 and is briefly described with a schematic diagram in the Appendix S1 methods (Figure S1). The microseparator consists of a disposable superstrate, which contains a microchannel network, two inlets, two outlets, and a vacuum trench, as well as a reusable substrate that includes inlaid ferromagnetic wires (Figure 1B).
The reusable substrate is placed on two stacked neodymium‐iron‐boron permanent magnets, which applies a uniform external lateral magnetic field to the ferromagnetic wires. This creates a high‐gradient magnetic field around the wires and applies a magnetic force through the entire microchannel area. The disposable superstrate can be applied to the reusable substrate using a vacuum pressure of −50 kPa. Given the extremely close contact between the superstrate and substrate, the gradient magnetic field remains effective even through the micrometer‐thick PET film that forms the bottom of the microchannel in the superstrate. The thickness of the PET film is 12 µm, evaluated as optimal thickness for high recovery rate by previous study. 25 This fabricated device is used for cell line spiking and clinical study.
The CTC isolation process involves injecting the prepared blood sample through the sample inlet and PBS with 0.2% bovine serum albumin through the buffer inlet of the microchannel. The magnetically labeled CTCs are pulled laterally along the slanted ferromagnetic wires to the CTC outlet and ultimately collected into a 1.5‐mL sample tube, while the normal blood cells flow straight through the waste outlet (Figure 1B). After the CTC isolation is completed, the superstrate is removed by releasing the vacuum pressure and is replaced for the next experiment.
2.4. Detection and counting of CTCs
The CTC counting procedure involved fixing the isolated cells, which included CTCs and WBCs, using 100 µL of 4% paraformaldehyde for 10 minutes. The cells were then incubated at 4ºC for 30 minutes with a nucleic acid fluorescent dye (DAPI, Invitrogen) to identify nuclei and with Alexa Fluor 647 antibodies targeting CD45 (Biolegend) to identify WBCs. The cells were subsequently permeabilized for 10 minutes using 100 µL of 0.2% Triton X‐100 (AMRESCO) and incubated at 4ºC for 30 minutes with Alexa Fluor 488 antibodies targeting pan‐cytokeratin (eBioscience) to identify CTCs. The fluorescently stained cells were then classified as either CTCs or WBCs using confocal microscopy images (LSM800, Carl Zeiss; Figure 1C).
2.5. Gene expression analysis using RT‐ddPCR and the clinical specimens
The CTC expression profiles were analyzed using RT‐ddPCR (Figure 1D) and transcripts from six representative genes that reflect reactivity to androgen hormones (AR and AR‐V7), prostate cancer progression (PSA and PSMA), and epithelial phenotype (EpCAM and KRT19). The protocols for mRNA extraction and cDNA synthesis from the isolated cells are described in the Appendix S2 methods. The sensitivity of the genetic analyses was enhanced by preamplification of the six synthesized cDNAs using multiplex PCR before the ddPCR, as described in the Appendix S3 methods. The preamplification primers are listed in Table S1.
The validity of the preamplification process was evaluated by detecting AR‐V7 mRNA, which is the rarest among the six selected genes (0.4 copies/cell based on an average of 2,000 LNCaP cells). 26 The preamplified cDNAs were serially diluted using diethylpyrocarbonate (DEPC), and the expression levels of the six genes according to the dilution rate were quantified as copy numbers per microliter using ddPCR (QX200, Bio‐Rad), as explained in the Appendix S4 methods and Figure S2. The primer sequences and product sizes for the six genes selected for ddPCR are listed in Table S2. To ensure accurate genomic analyses, the detection thresholds for each gene were determined based on the maximum values that were measured in five no‐template controls (Figure S3).
The optimal dilution rate of the preamplified cDNA from the clinical samples was evaluated using a serial dilution test with blood samples from two patients with localized disease, two patients with mHSPC, and three patients with mCRPC (Appendix S5 methods, Figure S4, and Figure S5). The results indicated a 1/5 dilution ratio was optimal, and that dilution ratio was subsequently used for all blood samples.
2.6. Determining the predictive potential of a multigene profile
The gene detection rates and expression levels were compared for each gene according to prostate cancer stage. To visualize these relationships, we created a heat map after the log‐transformed gene expression data had been converted into z‐scores. Composite scores were then created using the six genes in an attempt to identify metastatic prostate cancer (mCRPC or mHSPC). All patients were assigned scores based on their expression of each gene, and the data were grouped into tertiles (scores of 0 [lowest expression], 1 [medium expression], and 2 [highest expression]). The multigene score (MS) was defined as the sum of the individual gene scores and ranged from 0 to 12. For example, a patient with very high expression of all six genes would have a total score of 12 points (2 points per gene). The areas under receiver operating characteristic curves were evaluated to determine the relative values of the single‐gene and MSs for predicting mHSPC or mCRPC.
2.7. Statistical analysis
Continuous variables were compared using the Mann‐Whitney test, and categorical variables were compared using the chi‐squared test. A logistic regression classification model was constructed to predict mHSPC and/or mCRPC using the gene scores. The predictive power of each marker was evaluated using the C‐index, 27 which is a nonparametric measure of a predictive model's differential power (a C‐index of 0.5 is considered equivalent to random chance). The cutoff values for each gene's expression level were tested to assess their ability to identify patients with mHSPC and/or mCRPC (mCRPC or mHSPC [n = 38] vs. HD,T2,T3,T4 [n = 31]). All statistical analyses were performed using R software (version 3.6.2) and P‐values of < .05 were considered statistically significant.
3. RESULTS
3.1. Analyses of spiked blood samples from healthy donors
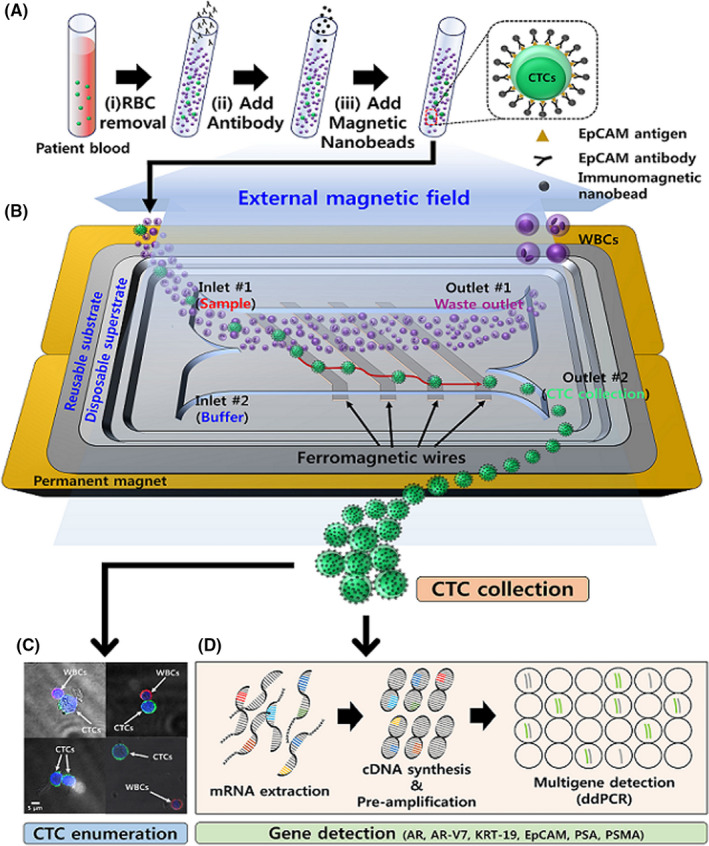
The performance of the lateral magnetophoretic microseparator was evaluated using 5‐mL blood samples from healthy donors that had been spiked with approximately 100 LNCaP cells (Figure 2A). The isolation performance (recovery rate) was measured in triplicate for flow rates of 1, 2, and 4 mL/h, with the spiked LNCaP cells collected and counted after they had passed through the microchannel. Counting was performed using cells that were spread on a glass slide and a fluorescence microscope, which confirmed the recovery rate of the LNCaP cells and the number of coisolated WBCs (Figure 2B). The number of coisolated WBCs was used to determine the WBC depletion rate and the purity of the retrieved LNCaP cells. The recovery rates were 95.1 ± 1.2% for a flow rate of 1 mL/h, 93.6 ± 0.7% for a flow rate of 2 mL/h, and 84.0 ± 7.0% for a flow rate of 4 mL/h (Figure 2C).
Figure 2.
A, The lateral magnetophoretic microseparator and its experimental set‐up, including two stacked neodymium‐iron‐boron permanent magnets and a vacuum tube applying −50 kPa of pressure to assemble the disposable polymeric superstrate and the reusable substrate. Magnified views show the three spots in the microchannel where the circulating tumor cells (CTCs) are separated from the blood by the lateral magnetic force and flow into the CTC outlet. (i) A magnified view of the sample and buffer injection channel, with the blood sample entering the upper channel because of laminar flow. (ii) The middle region of the microchannel allows for lateral isolation of the CTCs by the ferromagnetic wires. (iii) The CTC isolation is completed via the CTC outlet, while most white blood cells (WBCs) continue to flow through the waste outlet. B, Immunofluorescent images of the retrieved LNCaP cells and coisolated WBCs, which were stained using SYTO 13 dye (green) for LNCaP cells and SYTO 64 dye (red) for nucleated cells (LNCaP cells and WBCs). C, The recovery rates for LNCaP cells from spiked healthy blood samples using the lateral magnetophoretic microseparator and various flow rates. D, The numbers of contaminated WBCs and the WBC depletion rates. E, The purity rates for LNCaP cells. Approximately 100 LNCaP cells were spiked into 5 mL of peripheral blood from a healthy donor and tagged using anti‐EpCAM*‐based magnetic nanobeads, with retrieval performed at sample and buffer flow rates of 1 mL/h, 2 mL/h, and 4 mL/h (external magnetic flux: 0.2 T). Error bars represent the standard deviation for three measured datasets
The average numbers of contaminating WBCs were 660.0 cells for a flow rate of 1 mL/h, 667.0 cells for a flow rate of 2 mL/h, and 374.3 cells for a flow rate of 4 mL/h (Figure 2D), with corresponding increases in the purity of the retrieved LNCaP cells (1 mL/h: 13.0%, 2 mL/h: 13.9%, and 4 mL/h: 19.7%) (Figure 2E). The mean contamination rate was 113.4 WBCs/mL (238‐786 WBCs per 5‐mL sample), which corresponded to an average WBC depletion rate of 44 083.1‐fold (4.64 log), based on assumption that there were 5 × 106 WBCs/mL in blood. 28 Higher flow rates had greater hydrodynamic drag forces acting on the cells passing through the microchannel, which leads to a lower recovery rate, less WBC contamination, and greater purity of the retrieved cells. For example, a flow rate of 1 mL/h provided the highest recovery rate (95.1%) but lower purity and throughput. Thus, the flow rate of 2 mL/h was considered optimal and used for all future tests, based on a recovery rate of 93.6%, purity of 13.9%, and a WBC depletion rate of 4.57 log.
3.2. CTC counting using patients’ blood
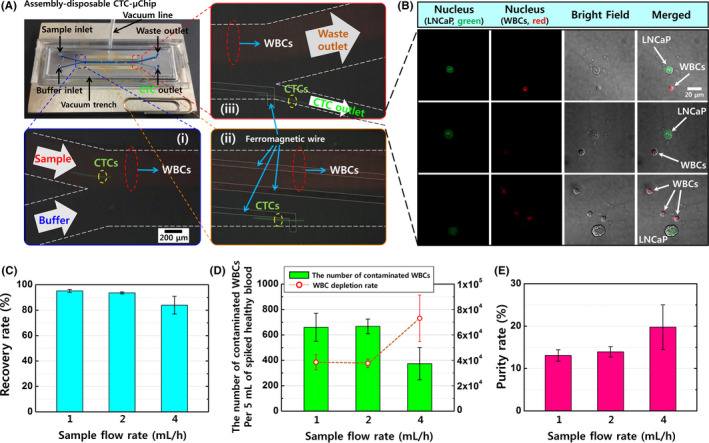
The clinical evaluations were performed using 3‐5‐mL peripheral blood samples from the healthy donors and patients with localized and metastatic prostate cancer. The collected cells were stained using immunofluorescent dyes to identify the CTCs and WBCs based on immunofluorescence and morphological analyses (Figure 3A and Figure S6). The lateral magnetophoretic microseparator isolated CTCs for 63 of 64 patients with prostate cancer (98.4%), despite many recent reports of similar technologies describing limited CTC detection rates (Table S3). Furthermore, the number of CTCs increased with increasing serum PSA levels, which indicated more advanced disease.
Figure 3.
A, Confocal microscopy of the circulating tumor cells (CTCs) and white blood cells (WBCs) isolated from blood samples obtained from patients with localized prostate cancer, metastatic hormone‐sensitive prostate cancer (mHSPC), and metastatic castration‐resistant prostate cancer (mCRPC). Positivity for pan‐cytokeratin (green) was used to identify CTCs, and positivity for CD45 (red) was used to identify WBCs. B, The numbers of isolated CTCs and WBCs per milliliter of blood and the serum PSA levels. C, The WBC depletion rate and (D) the purity of CTCs at each stage of prostate cancer
The average isolation results for localized disease were 6.5 CTCs/mL (range: 0.4‐30.5 CTCs/mL) and 514.4 WBCs/mL (range: 67.0‐1530.0 WBCs/mL). The average isolation results for mHSPC were 16.7 CTCs/mL (range: 8.0‐40.7 CTCs/mL) and 717.6 WBCs/mL (range: 98.6‐1192.5 WBCs/mL). The average isolation results for mCRPC were 31.0 CTCs/mL (range: 2.8‐160.4 CTCs/mL) and 627.6 WBCs/mL (range: 41.0‐2693.2 WBCs/mL; Figure 3B). Patients with mHSPC and mCRPC had an average of 27.5 CTCs/mL, which was 4.1 × higher than the isolate rate for localized disease. The average WBC contamination for all prostate cancer cases was 595.0 WBCs/mL (range: 41.0‐2693.2 WBCs/mL), which corresponded to a WBC depletion rate of 3.92 log (Figure 3C). The healthy donors had average values of 0.4 CTCs/mL (range: 0‐0.8 CTCs/mL) and 602.2 WBCs/mL (range: 297.4‐831.7 WBCs/mL).
The CTC purity values were calculated based on the ratio of cytokeratin‐positive cells to CD45‐positive cells (CTCs/WBCs). Because the CTC count increased for more advanced prostate cancer, the average CTC purity values were 2.1% for localized disease, 3.7% for mHSPC, and 6.7% for mCRPC (Figure 3D). The average number of contaminated WBCs in these patients’ samples (595.0 WBCs/mL) was approximately 5.2 × higher than in the spiked healthy blood samples, as patients with prostate cancer are more likely to be elderly and infirm, thereby leading to greater coagulation. Thus, the average purity of CTCs was 3.5 × lower than for LNCaP cells that were retrieved from the spiked healthy blood samples.
3.3. Genetic analyses using isolated CTCs
The sample clustering of the six genes was well correlated with cancer stage for all patients (five healthy donors, 26 patients with localized disease, 10 patients with mHSPC, and 28 patients with mCRPC) based on the RT‐ddPCR findings. The gene detection rates according to cancer stage were evaluated as copies/µL above the detection threshold for each gene. For example, AR mRNA was detected in 80% of healthy cases, 76.9% of localized disease cases, 90.0% of mHSPC cases, and 100% of mCRPC cases. Relative to the other genes, the expression of the gene for AR was the highest and fairly stable for all prostate cancer stages (Figure 4A,B). The gene for AR‐V7 was detected in 20.0% of mHSPC cases and 35.7% of mCRPC cases (Figure 4C), with the expression level being 17.5 × higher for mCRPC cases than for mHSPC cases (Figure 4D).
Figure 4.
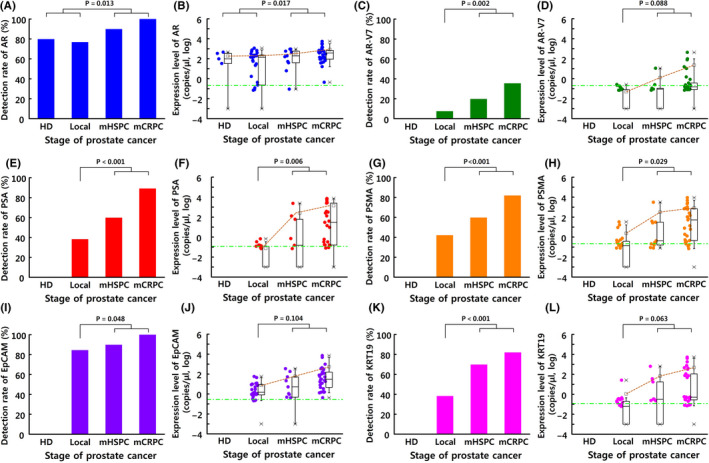
The detection rates and expression levels (copies/µL) of the target genes in circulating tumor cells (CTCs) according to prostate cancer stage: A, B, androgen receptor (AR); C, D, androgen receptor variant 7 (AR‐V7); E, F, prostate‐specific antigen (PSA); G, H, prostate‐specific membrane antigen (PSMA); I, J, epithelial cell adhesion molecule (EpCAM); K, L, cytokeratin 19 (KRT19). Blood samples were obtained from healthy donors (HD, n = 5) and patients with localized prostate cancer (n = 26), metastatic hormone‐sensitive prostate cancer (mHSPC) (n = 10), or metastatic castration‐resistant prostate cancer (mCRPC) (n = 28)
Interestingly, prostate cancer progression led to a greater increase in CTC expression of the gene for PSA, relative to serum PSA levels (Figure 3B and Figure 4F), which is a common biomarker for monitoring prostate cancer progression. The greatest increase in the PSA gene expression was observed between the localized disease and mCRPC cases (approximately 20 000×) (Figure 4E,F). Similarly, the expression of the gene for PSMA was significantly higher in cases of metastatic prostate cancer than in cases of localized disease (639.9 copies/µL vs. 2.4 copies/µL, P = .029) (Figure 4G,H).
Expression of the gene for EpCAM was approximately 8.7 × higher for mCRPC than for mHSPC, was significantly correlated with the increased number of isolated CTCs, and was detected in 100% of the CTCs isolated from patients with mCRPC (Figure 4I,J). The detection rate of the gene for KRT19 increased with prostate cancer stage (range: 38.46%‐82.14%) and was approximately 336.3 × higher in mHSPC/mCRPC cases than in localized disease cases (Figure 4K,L). This result is much higher than previously reported results, which is likely related to the microseparator's good ability to isolate CTCs and the high sensitivity of ddPCR. 25 , 29 , 30
The relationships between the numbers of isolated CTCs and the gene expressions were plotted (Figure 5A), and the six gene signatures were matched to individual patients using a heatmap (Figure 5B). These results revealed that most of the six genes were highly overexpressed in cases involving metastatic prostate cancer. Furthermore, progression of prostate cancer was associated with higher CTC counts, increased gene detection rates, and increased expression levels for the genes encoding AR, AR‐V7, PSA, PSMA, EpCAM, and KRT19 in the CTCs.
Figure 5.
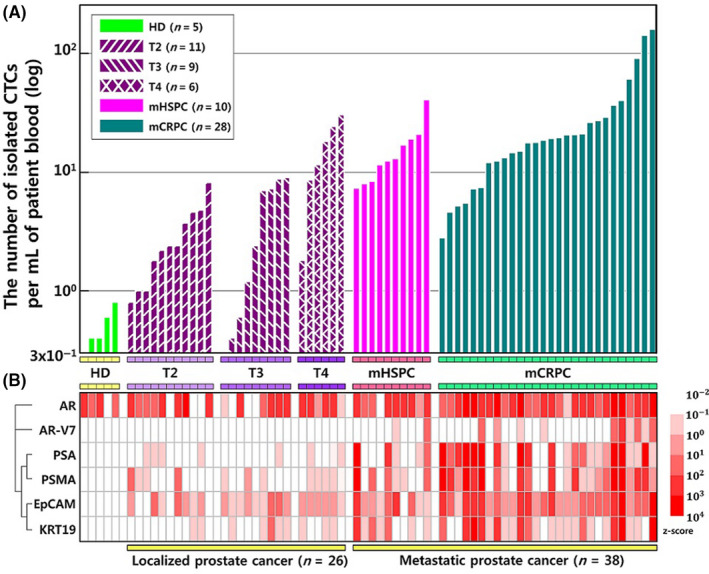
A, The numbers of isolated circulating tumor cells (CTCs) from healthy donors and prostate cancer patients. B, A two‐dimensional heat map with each column representing a different patient and each row representing gene expression levels matched to the number of isolated CTCs. The log‐transformed data for gene expression were converted to z‐scores [−2, 4]. The heatmap color spectrum represents relative overexpression (red) to underexpression (white). AR, androgen receptor; AR‐V7, androgen receptor variant 7; EpCAM, epithelial cell adhesion molecule; KRT‐19, cytokeratin 19; mCRPC, metastatic castration‐resistant prostate cancer; mHSPC, metastatic hormone‐sensitive prostate cancer; PSA, prostate‐specific antigen; PSMA, prostate‐specific membrane antigen
3.4. Development of a CTC‐based multigene model
Logistic regression was used to evaluate single‐gene models and multigene models for predicting metastatic prostate cancer (mCRPC or mHSPC). Their predictive values were evaluated based on the areas under the curves (AUCs) (Figure 6).
Figure 6.
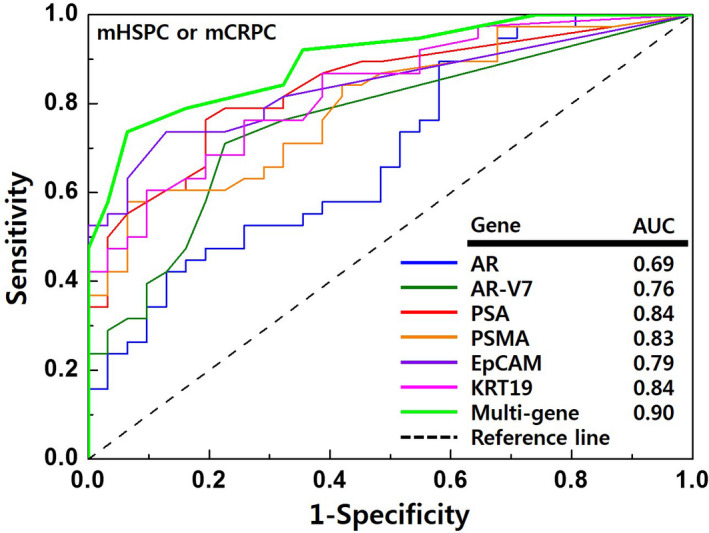
The accuracies of the models for predicting advanced prostate cancer. The receiver operating characteristic curves were used to compare the accuracies of androgen receptor (AR), androgen receptor variant 7 (AR‐V7), cytokeratin 19 (KRT19), epithelial cell adhesion molecule (EpCAM), prostate‐specific antigen (PSA), prostate‐specific membrane antigen (PSMA), and the multigene model for identifying metastatic hormone‐sensitive prostate cancer (mHSPC) or metastatic castration‐resistant prostate cancer (mCRPC). AUC, area under the curves
The multigene model for identifying mCRPC or mHSPC was:
with a = –3.28, b = 0.65, and MS = the sum of the six genes’ scores.
Figure 6 showed that this model had good performance (AUC: 0.90). Furthermore, relative to the individual gene models, the AUC values for identifying metastatic prostate cancer were significantly better for the multigene model (multigene model: 0.90 vs. AR: 0.69, AR‐V7: 0.76, PSA: 0.84, PSMA: 0.83, EpCAM: 0.79, KRT19: 0.84) (P < .001).
4. DISCUSSION
We have previously reported the isolation of CTCs from the blood of patients with breast cancer using a lateral magnetophoretic microseparator with an assembly‐disposable microfluidic device. 25 Although there are many techniques for isolating CTCs to identify biomarkers, most have focused on the rates of CTC isolation and detection, without clear data regarding the WBC depletion rate (ie, CTC purity). Thus, it remains unclear whether their findings regarding genomic markers were related to expression in CTCs and/or WBCs, and it is important to consider the purity of CTCs in this setting. A purity rate of > 1% is generally required for PCR analysis, and a rate of > 5% is generally required for sequencing. 31 , 32 The present study evaluated 5‐mL blood samples from healthy donors that were spiked with approximately 100 LNCaP cells, which revealed a high LNCaP cell recovery rate (93.60%) and good WBC depletion (4.57 log) at a flow rate of 2 mL/h, which resulted in high CTC purity (13.93%). Furthermore, we observed that WBC contamination remained relatively constant, while the number of CTCs increased at higher cancer stages, which was related to increasing CTC purity (2.1% for localized disease, 3.7% for mHSPC, and 6.7% for mCRPC). Therefore, it appears that we collected relatively accurate CTC‐based genomic information regarding advanced prostate cancer. Moreover, it appears that the CTCs isolated using our lateral magnetophoretic microseparator are suitable for use in advanced genomic analysis techniques, such as ddPCR or next‐generation sequencing.
This study revealed that the number of CTCs increased at higher prostate cancer stages and proportionally to the serum PSA level. Several studies have already indicated that the CTC burden is correlated with the prognosis of patients with mCRPC. 8 , 33 However, the number of CTCs alone may not accurately reflect the patient's condition, as treatment decreases the number of CTCs. In addition, it is possible that the number of detected CTCs may change according to the timing of blood sampling during treatment. Moreover, the CTC number alone does not reflect the degree of malignancy or cancer cell characteristics. Therefore, CTC genomic characteristics, not just an absolute count, will likely be more useful for monitoring the patient's treatment response or predicting their prognosis. For example, mutations in cancer cells are likely to proceed even if the CTC count decreases during treatment.
It remains unclear what mutations occur in prostate cancer cells during the progression from localized disease to metastasis. The mechanism for progression from mHSPC to mCRPC is also unknown. To address these issues, the molecular characteristics of cancer cells will need to be analyzed for patients with disease ranging from localized to metastatic prostate cancer. However, repeated sampling of cancer tissues from patients is challenging, and the characteristics of cancer cells may vary according to the metastasis site. Moreover, sampling of metastatic tumors is highly invasive and posing a serious risk to the patient. In contrast, blood specimens for CTC detection can be obtained repeatedly and with minimal invasiveness, while still being representative of the patients’ cancer cell characteristics.
The main finding of the present study is that the CTC‐based expression signatures involving six genes were associated with metastatic prostate cancer, and that a multigene model could be used to predict metastatic prostate cancer. Aberrant expression of the genes encoding AR and AR‐V7 are associated with resistance to systemic treatment using AR‐targeting agents, with elevated AR expression being linked to increased generation of active truncated AR variants and greater resistance to AR‐targeting agents. 35 , 36 , 37 One of these variants (AR‐V7) is a marker than can accurately predict the response to AR‐targeting agents in patients with mCRPC. 11 , 12 , 13 Epithelial markers (EpCAM and KRT19) are detected in all stages of prostate cancer, as the CTCs are essentially derived from epithelial cells and only a few CTCs lose their epithelial phenotype via the epithelial‐mesenchymal transition. However, the present study confirmed that KRT19 and EpCAM expression was elevated at later disease stages, which suggests that these might be effective biomarkers for diagnosing advanced prostate cancer, especially as they were not expressed at all in the cells that were isolated from the healthy donors. The AR signaling pathway also influences PSA transcripts, which are associated with survival duration and response to AR‐targeting agents in patients with mCRPC. 35 , 38 , 39 The expression of PSMA (a transmembrane glycoprotein) on CTCs is another promising marker for measuring disease progression or response to therapy, 40 as PSMA expression is limited in benign prostatic hyperplasia and upregulated dramatically in prostate cancer and especially metastatic cases. 41 Moreover, PSMA‐targeted radioligands may be a potential treatment option for patients with mCRPC. 42 , 43 Finally, there are emerging data that PSMA‐based PET‐CT is more accurate than conventional imaging for detecting metastasis in cases involving high‐risk localized prostate cancer. 44 , 45 , 46 , 47
Although several studies have been reported on CTC‐based multigene models that predict the prognosis of prostate cancer, the aim of this study is to achieve early detection of metastatic prostate cancer, which can help clinical decision‐making. 48 , 49 The clinical significance of the six genes that we identified remains unclear for metastatic prostate cancer. Nevertheless, the diagnostic applications of CTC‐based multigene profiling continue to evolve, and this strategy may complement other biomarker measurements for evaluating disease progression and treatment response. This diagnostic method may also decrease the possibility of false‐negative findings for metastatic lesions during radiographic imaging. This is important because conventional imaging using computed tomography and bone scans has limited sensitivity for staging high‐risk localized prostate cancer. Thus, genomic information from CTCs might guide better metastasis identification and treatment selection, which could help improve survival outcomes because timely treatment at a low tumor burden is associated with better efficacy. 50
This study has a few limitations. First, our CTC assay may be confounded by the biased selection of cells, and the prognosis of patients with metastatic breast cancer is reportedly associated with the presence of mesenchymal and stem cell markers in CTCs. However, we used standard epithelial antigen–based technology to detect EpCAM on the CTCs. Second, given the broad and variable relationship between the epithelial‐mesenchymal transition and patient prognosis, the mechanism for prostate cancer progression may not be completely explained by the biomarkers that we evaluated (AR, AR‐V7, PSA, PSMA, EpCAM, KRT19). Third, the present study involved a relatively small sample of patients and controls, and larger prospective studies are needed to validate our findings.
5. CONCLUSION
This study revealed that our lateral magnetophoretic microseparator was effective for isolating CTCs from the blood of patients with prostate cancer, and the resulting CTCs could be used for accurate analyses of genetic information via RT‐ddPCR. The results indicated that the CTC count increased at later stages of prostate cancer and in proportion to the serum PSA levels. Moreover, the CTC‐based genetic information suggests that prostate cancer progression was related to the expression of prostate‐ and epithelial‐specific genes that encode AR, AR‐V7, PSA, PSMA, EpCAM, and KRT19. While further studies are needed to validate our results, we suggest that CTC‐based multigene profiling may be a useful strategy for diagnosing metastatic prostate cancer and guiding related treatments.
CONFLICT OF INTEREST
The authors have no conflicts of interest.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF), which is funded by the Korean government (MSIT) (NRF‐2018R1C1B6004574 and NRF‐2019R1A2B5B01070170).
Cho H, Chung JI, Kim J, et al. Multigene model for predicting metastatic prostate cancer using circulating tumor cells by microfluidic magnetophoresis. Cancer Sci. 2021;112:859–870. 10.1111/cas.14745
Hyungseok Cho and Jae Il Chung have contributed equally to this study.
[Correction added on 23 December 2020, after first online publication: The affiliation details of Seok‐Soo Byun and Jae‐Seung Chung have been updated]
Contributor Information
Jae‐Seung Chung, Email: biogen@hanmail.net.
Ki‐Ho Han, Email: mems@inje.ac.kr.
REFERENCES
- 1. Shen MM, Rubin MA. Prostate cancer research at the crossroads. Cold Spring Harb Perspect Med. 2019;9:a036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394‐424. [DOI] [PubMed] [Google Scholar]
- 3. Broncy L, Paterlini‐Bréchot P. Clinical impact of circulating tumor cells in patients with localized prostate cancer. Cells. 2019;8:676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nuhn P, De Bono JS, Fizazi K, et al. Update on systemic prostate cancer therapies: management of metastatic castration‐resistant prostate cancer in the era of precision oncology. Eur Urol. 2019;75:88‐99. [DOI] [PubMed] [Google Scholar]
- 5. Pantel K, Hille C, Scher HI. Circulating tumor cells in prostate cancer: from discovery to clinical utility. Clin Chem. 2019;65:87‐99. [DOI] [PubMed] [Google Scholar]
- 6. Miyamoto DT, Sequist LV, Lee RJ. Circulating tumour cells—monitoring treatment response in prostate cancer. Nat Rev Clin oncol. 2014;11:401‐412. [DOI] [PubMed] [Google Scholar]
- 7. Danila DC, Fleisher M, Scher HI. Circulating tumor cells as biomarkers in prostate cancer. Clin Cancer Res. 2011;17:3903‐3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration‐resistant prostate cancer. Clin Cancer Res. 2008;14:6302‐6309. [DOI] [PubMed] [Google Scholar]
- 9. Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration‐resistant prostate cancer. Clin Cancer Res. 2007;13:7053‐7058. [DOI] [PubMed] [Google Scholar]
- 10. Sciarra A, Gentilucci A, Silvestri I, et al. Androgen receptor variant 7 (AR‐V7) in sequencing therapeutic agents for castratrion resistant prostate cancer: A critical review. Medicine. 2019;98:e15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Antonarakis ES, Lu C, Wang H, et al. AR‐V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028‐1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Okegawa T, Ninomiya N, Masuda K, et al. AR‐V7 in circulating tumor cells cluster as a predictive biomarker of abiraterone acetate and enzalutamide treatment in castration‐resistant prostate cancer patients. Prostate. 2018;78:576‐582. [DOI] [PubMed] [Google Scholar]
- 13. Scher HI, Lu D, Schreiber NA, et al. Association of AR‐V7 on circulating tumor cells as a treatment‐specific biomarker with outcomes and survival in castration‐resistant prostate cancer. JAMA oncology. 2016;2:1441‐1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meyer CP, Pantel K, Tennstedt P, et al. Limited prognostic value of preoperative circulating tumor cells for early biochemical recurrence in patients with localized prostate cancer. Urol Oncol. 2016;34(235):e11‐e16. [DOI] [PubMed] [Google Scholar]
- 15. Thalgott M, Rack B, Horn T, et al. Detection of circulating tumor cells in locally advanced high‐risk prostate cancer during neoadjuvant chemotherapy and radical prostatectomy. Anticancer Res. 2015;35:5679‐5685. [PubMed] [Google Scholar]
- 16. Davis JW, Nakanishi H, Kumar VS, et al. Circulating tumor cells in peripheral blood samples from patients with increased serum prostate specific antigen: initial results in early prostate cancer. J Urol. 2008;179:2187‐2191. [DOI] [PubMed] [Google Scholar]
- 17. Beltran H, Jendrisak A, Landers M, et al. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22:1510‐1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dago AE, Stepansky A, Carlsson A, et al. Rapid phenotypic and genomic change in response to therapeutic pressure in prostate cancer inferred by high content analysis of single circulating tumor cells. PLoS One. 2014;9:e101777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steinestel J, Luedeke M, Arndt A, et al. Detecting predictive androgen receptor modifications in circulating prostate cancer cells. Oncotarget. 2015;10:4213‐4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Magnusson C, Augustsson P, Lenshof A, et al. Clinical‐scale cell‐surface‐marker independent acoustic microfluidic enrichment of tumor cells from blood. Anal Chem. 2017;89:11954‐11961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen–dependent and–independent sorting of rare circulating tumor cells. Sci Trans Med. 2013;5:179ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li P, Mao Z, Peng Z, et al. Acoustic separation of circulating tumor cells. Proc Natl Acad Sci. 2015;112:4970‐4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Augustsson P, Magnusson C, Nordin M, et al. Microfluidic, label‐free enrichment of prostate cancer cells in blood based on acoustophoresis. Anal Chem. 2012;84:7954‐7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stott SL, Hsu C‐H, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex‐generating herringbone‐chip. Proc Natl Acad Sci. 2010;107:18392‐18397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cho H, Kim J, Jeon C‐W, et al. A disposable microfluidic device with a reusable magnetophoretic functional substrate for isolation of circulating tumor cells. Lab Chip. 2017;17:4113‐4123. [DOI] [PubMed] [Google Scholar]
- 26. Ma Y, Luk A, Young FP, et al. Droplet digital PCR based androgen receptor variant 7 (AR‐V7) detection from prostate cancer patient blood biopsies. Int J Mol Sci. 2016;17:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361‐387. [DOI] [PubMed] [Google Scholar]
- 28. Wintrobe MM. Examiniation of the blood and bone marrow In: Wintrobe's clinical hematology. Vol. 1, 12th ed. Lippincott Williams & Wilkins; 2008:2–7. [Google Scholar]
- 29. Tryfonidis K, Kafousi M, Perraki M, et al. Detection of circulating cytokeratin‐19 mRNA‐positive cells in the blood and the mitotic index of the primary tumor have independent prognostic value in early breast cancer. Clin Breast Cancer. 2014;14:442‐450. [DOI] [PubMed] [Google Scholar]
- 30. Markou A, Strati A, Malamos N, et al. Molecular characterization of circulating tumor cells in breast cancer by a liquid bead array hybridization assay. Clin Chem. 2011;57:421‐430. [DOI] [PubMed] [Google Scholar]
- 31. Tsongalis GJ, Peterson JD, de Abreu FB, et al. Routine use of the Ion Torrent AmpliSeq™ Cancer Hotspot Panel for identification of clinically actionable somatic mutations. Clin chem Lab Med. 2014;52:707‐714. [DOI] [PubMed] [Google Scholar]
- 32. Chin EL, da Silva C, Hegde M. Assessment of clinical analytical sensitivity and specificity of next‐generation sequencing for detection of simple and complex mutations. BMC Genet. 2013;14:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration‐resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233‐239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual‐level surrogate for survival in metastatic castration‐resistant prostate cancer. J Clin Oncol. 2015;33:1348‐1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chung J‐S, Wang Y, Henderson J, et al. Circulating tumor cell‐based molecular classifier for predicting resistance to abiraterone and enzalutamide in metastatic castration‐resistant prostate cancer. Neoplasia. 2019;21:802‐809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Azad AA, Volik SV, Wyatt AW, et al. Androgen receptor gene aberrations in circulating cell‐free DNA: biomarkers of therapeutic resistance in castration‐resistant prostate cancer. Clin Cancer Res. 2015;21:2315‐2324. [DOI] [PubMed] [Google Scholar]
- 37. Kohli M, Li J, Du M, et al. Prognostic association of plasma cell‐free DNA‐based androgen receptor amplification and circulating tumor cells in pre‐chemotherapy metastatic castration‐resistant prostate cancer patients. Prostate Cancer and Prostatic Dis. 2018;21:411‐418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kantoff PW, Halabi S, Farmer DA, et al. Prognostic significance of reverse transcriptase polymerase chain reaction for prostate‐specific antigen in men with hormone‐refractory prostate cancer. J Clin Oncol. 2001;19:3025‐3028. [DOI] [PubMed] [Google Scholar]
- 39. Ross RW, Manola J, Hennessy K, et al. Prognostic significance of baseline reverse transcriptase‐PCR for prostate‐specific antigen in men with hormone‐refractory prostate cancer treated with chemotherapy. Clin Cancer Res. 2005;11:5195‐5198. [DOI] [PubMed] [Google Scholar]
- 40. Autio KA, Dreicer R, Anderson J, et al. Safety and efficacy of BIND‐014, a docetaxel nanoparticle targeting prostate‐specific membrane antigen for patients with metastatic castration‐resistant prostate cancer: a phase 2 clinical trial. JAMA Oncol. 2018;4:1344‐1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sweat SD, Pacelli A, Murphy GP, et al. Prostate‐specific membrane antigen expression is greatest in prostate adenocarcinoma and lymph node metastases. Urology. 1998;52:637‐640. [DOI] [PubMed] [Google Scholar]
- 42. Lawal IO, Bruchertseifer F, Vorster M, et al. Prostate‐specific membrane antigen‐targeted endoradiotherapy in metastatic prostate cancer. Curr Opin Urol. 2020;30:98‐105. [DOI] [PubMed] [Google Scholar]
- 43. Udovicich C, Perera M, Hofman MS, et al. 68Ga‐prostate‐specific membrane antigen‐positron emission tomography/computed tomography in advanced prostate cancer: current state and future trends. Prostate Int. 2017;5:125‐129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hofman MS, Lawrentschuk N, Francis RJ, et al. Prostate‐specific membrane antigen PET‐CT in patients with high‐risk prostate cancer before curative‐intent surgery or radiotherapy (proPSMA): a prospective, randomised, multi‐centre study. Lancet. 2020;395:1208‐1216. [DOI] [PubMed] [Google Scholar]
- 45. Fendler WP, Calais J, Eiber M, et al. Assessment of 68Ga‐PSMA‐11 PET accuracy in localizing recurrent prostate cancer: a prospective single‐arm clinical trial. JAMA Oncol. 2019;5:856‐863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Perera M, Papa N, Roberts M, et al. Gallium‐68 prostate‐specific membrane antigen positron emission tomography in advanced prostate cancer—updated diagnostic utility, sensitivity, specificity, and distribution of prostate‐specific membrane antigen‐avid lesions: a systematic review and meta‐analysis. Eur Urol. 2019;77:403‐417. [DOI] [PubMed] [Google Scholar]
- 47. Calais J, Ceci F, Eiber M, et al. 18F‐fluciclovine PET‐CT and 68Ga‐PSMA‐11 PET‐CT in patients with early biochemical recurrence after prostatectomy: a prospective, single‐centre, single‐arm, comparative imaging trial. Lancet Oncol. 2019;20:1286‐1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Singhal U, Wang Y, Henderson J, et al. Multigene profiling of CTCs in mCRPC identifies a clinically relevant prognostic signature. Mol Cancer Res. 2018;16(4):643‐654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kozminsky M, Fouladdel S, Chung J‐S, et al. Detection of CTC Clusters and a Dedifferentiated RNA‐Expression Survival Signature in Prostate Cancer. Adv Sci (Weinh). 2018;6:1801254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parker CC, James ND, Brawley CD, et al. Radiotherapy to the primary tumour for newly diagnosed, metastatic prostate cancer (STAMPEDE): a randomised controlled phase 3 trial. Lancet. 2018;392:2353‐2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


