Abstract
Glioblastoma (GBM) recurrence is attributed to the presence of therapy‐resistant glioblastoma stem cells. Steroid receptor coactivator‐1 (SRC‐1) acts as an oncogenic regulator in many human tumors. The relationship between SRC‐1 and GBM has not yet been studied. Herein, we investigate the role of SRC‐1 in GBM. In this study, we found that SRC‐1 expression is positively correlated with grades of glioma and inversely correlated with glioma patient’s prognosis. Steroid receptor coactivator‐1 promotes the proliferation, migration, and tumor growth of GBM cells. Notably, SRC‐1 knockdown suppresses the stemness of GBM cells. Mechanistically, long noncoding RNA X‐inactive specific transcript (XIST) is regulated by SRC‐1 at the posttranscriptional level and mediates the function of SRC‐1 in promoting stemness‐like properties of GBM. Steroid receptor coactivator‐1 can promote the expression of Kruppel‐like factor 4 (KLF4) through the XIST/microRNA (miR)‐152 axis. Additionally, arenobufagin and bufalin, SRC small molecule inhibitors, can reduce the proliferation and stemness of GBM cells. This study reveals SRC‐1 promotes the stemness of GBM by activating the long noncoding RNA XIST/miR‐152/KLF4 pathway and provides novel markers for diagnosis and therapy of GBM.
Keywords: cancer stem cell, glioblastoma, lncRNA XIST, posttranscriptional regulation, steroid receptor coactivator‐1
Steroid receptor coactivator‐1 (SRC‐1) was positively correlated with grades of glioma and inversely correlated with glioma patient’s prognosis. SRC‐1 promoted the proliferation, migration, and tumor growth. Additionally, SRC‐1 enhanced stem‐like characteristics of GBM cells. Mechanistic studies showed that SRC‐1 enhances the stemness by activating the long noncoding RNA (lncRNA) X‐inactive specific transcript (XIST)/microRNA (miR)‐152/Kruppel‐like factor 4 (KLF4) pathway in GBM cells. Arenobufagin and bufalin, SRC small molecule inhibitors, reduced the proliferation and stemness of GBM cells.
1. INTRODUCTION
Glioblastoma (GBM) has an extremely grim prognosis and is the most common aggressive brain tumor. 1 , 2 It was classified as grade IV glioma of the astrocytic lineage by the WHO. 1 , 3 Standard therapy, consisting of surgery and the combination of temozolomide (TMZ) chemotherapy and radiation, offers median survival of only 15‐18 months after initial diagnosis. 4 , 5 , 6 , 7 , 8 The poor prognosis of GBM patients is related to the oncogenic nature of the GBM cell and the existence of a small subpopulation of glioblastoma stem cells (GSCs). Glioblastoma stem cells have the capability of self‐renewal and multiple differentiation and are resistant to chemotherapy and radiotherapy. Consequently, they are likely responsible for failure of treatment and high recurrence rates in GBM. 9 , 10 , 11
Steroid receptor coactivator‐1 (SRC‐1, also called NCOA1) is a transcriptional coactivator in the SRC family that also contains SRC‐2 (TIF2 or GRIP1) and SRC‐3 (AIB1 or ACTR). These coactivators promote transcription by interacting with nuclear receptors such as estrogen and progesterone receptors, as well as other transcription factors such as activator protein‐1, nuclear factor‐κB, β‐catenin, Ets‐2, PEA3, and HOXC11. 12 , 13 A previous study showed that SRC‐1 promotes metastasis through mediating Ets‐2‐mediated human epidermal growth factor receptor‐2 (HER2) expression and by activating colony stimulating factor‐1 expression for macrophage recruitment. 14 Loss‐of‐function deletion of the SRC‐1 gene in mice reduces estrogen effect on the vascular injury response. 15 Additionally, recent studies reported that SRC‐1 is overexpressed in breast cancer and hepatocellular carcinoma, and regulates the proliferation of breast cancer, prostate cancer, and hepatocellular carcinoma. 12 , 14 , 16 , 17 , 18 , 19 , 20 , 21 , 22 Furthermore, SRC‐1 is important for cancer stem‐like cells in lung and breast cancer. 23 However, the relationship between SRC‐1 and GBM has not yet been studied in detail.
In this study, we found that SRC‐1 is positively correlated with grades of glioma and inversely correlated with glioma patient’s prognosis. Steroid receptor coactivator‐1 promoted the proliferation, migration, and tumor growth. Additionally, SRC‐1 enhanced stem‐like characteristics of GBM cells. Mechanistic studies showed that SRC‐1 enhances the stemness by activating the long noncoding RNA (lncRNA) X‐inactive specific transcript (XIST)/microRNA (miR)‐152/Kruppel‐like factor 4 (KLF4) pathway in GBM cells. Our findings show that SRC‐1 could be used as a diagnostic marker and therapeutic target for GBM.
2. MATERIALS AND METHODS
2.1. Tissue microarray
Paraffin‐embedded tumor tissue microarrays were purchased from Shanghai Outdo Biotech. Brain tumor classification was conducted according to WHO criteria. 3 There were 59 specimens on the tissue microarray, including three normal brain tissues, 20 astrocytoma, 34 glioblastoma, and two between astrocytoma and glioblastoma. The clinical characteristics of the cohort are described in Table S1. The immunohistochemical staining was scored according to the four‐point system (score 0‐3) as previously reported. 24 , 25 , 26 A score of 2 or 3 was considered high expression, and the score of 0 or 1 was low expression. The mean immunoreactive score (IRS) was considered as the final IRS (Table S1).
2.2. Cell culture
LN229, U251, U87‐MG, T98G, U118, C6, SVG‐p12 (human fetal glial cell), HEK293T, and HeLa cell lines were obtained from ATCC. All cells were incubated in DMEM (Gibco) supplemented with 10% FBS and 1% (v/v) penicillin‐streptomycin at 37°C under 5% CO2.
2.3. Lentivirus production and virus infection
The shRNA knockdown plasmids (GV248‐shSRC1‐1 and GV248‐shSRC1‐2) targeting SRC‐1 as well as a nontargeting shRNA plasmid were purchased from Genechem. Full‐length cDNA encoding human SRC‐1 was generated by PCR from the SRC‐1 recombinant plasmid from Genechem and its sequence was confirmed (NM_003743) by DNA sequencing. The fragment was subcloned into pCDH‐CMV‐MCS‐EF1‐Puro plasmid (System Biosciences). To generate recombinant lentivirus, these constructs were cotransfected with lentiviral packaging vectors (pSPAX2 and pMD2.G) into 293T cells by using Lipofectamine 2000 (Invitrogen). Then media supernatant was collected. For transduction of GBM cells, lentiviral supernatant was added into the culture medium for 24 hours. Puromycin (1 μg/mL) was used to remove noninfected GBM cells. Surviving clones were isolated and examined by real‐time PCR and western blot to determine SRC‐1 knockdown or overexpression efficiency. Primers used for PCR are shown in Table S2.
2.4. Cell proliferation assay and colony formation assay
The MTT and colony formation assays were carried out as we described previously. 27 , 28 , 29
2.5. Cell cycle analysis
Cell cycle analysis was carried out as we described previously. 29 , 30
2.6. Wound healing assay
Wound healing assay was carried out as we described previously. 29 , 31 , 32 , 33
2.7. Western blot
The western blot assay was carried out according to our previous studies. 27 , 29 Antibodies information is summarized in Table S3.
2.8. RNA extraction and RT‐quantitative PCR
The RNA extraction and RT‐quantitative PCR (qPCR) were carried out according to our previous studies. 34 Primers used for qPCR are shown in Table S4.
2.9. Immunofluorescence
Immunofluorescence (IF) was carried out as we described previously. 29 Antibody information is listed in Table S3.
2.10. Sphere formation assay
The sphere formation assay was carried out according to our previous studies. 27
2.11. CD133 marker detection by flow cytometry
Glioblastoma stem cells were cultured and enriched by sphere formation, and then GSCs were harvested, dissociated, washed with PBS, and incubated with 2 μL CD133‐PE Ab (1:50) (Miltenyi Biotec) for 30 minutes, The GSCs were then washed, suspended in PBS, and subjected to flow cytometry (BD AccuriC6) to determine the percentages of viable CD133+ cells.
2.12. RNA sequencing
The RNA sequencing (RNA‐seq) experiments were undertaken by Novogene.
2.13. Animal and tumor models
2.13.1. Tumor xenografts in nude mice
Five‐week‐old nude mice (BALB/c) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. To initiate tumors, 1 × 107 cells in 100 μL PBS : Matrigel (2:1, v/v; BD Biosciences) were injected s.c. into the dorsal flank of each nude mouse by two blinded technicians. After the tumor volume reached approximately 100 mm3, the tumor volumes (A × B2/2; A being the greatest diameter and B being the diameter perpendicular to A) were measured twice a week by digital calipers. At the end of the studies, mice were sacrificed, and tumors were dissected, weighed, stored, and fixed.
2.13.2. Rat brain orthotopic implantation model
Male Wistar rats (200‐300 g) were purchased from Liaoning Changsheng Technology Co., Ltd. The C6 GBM cell suspension (1 × 106 cells in 10 μL PBS) was injected stereotactically over a 10‐minute period using a Hamilton syringe at a depth of 5 mm. At the end of the studies, mice were sacrificed, and brains were excised and fixed. Tumor volumes were calculated using the formula A × B2/2, where A was the maximum long‐dimension tumor diameter and B was the maximum short‐dimension tumor diameter. 35
All animal procedures and animal experiments were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals under the approval of the SPF Laboratory Animal Center at Dalian Medical University.
2.14. Immunohistochemistry and H&E staining
Tumors and brains were fixed in 4% paraformaldehyde (Sigma‐Aldrich) and prepared for histological analysis. All H&E and immunohistochemical (IHC) staining was carried out as previously described. 29 , 34 , 36 Antibody information is listed in Table S3.
2.15. Promoter reporter and dual luciferase assays
Human XIST promoter was amplified from the human genomic DNA template and inserted into pGL3‐basic vector (Promega). For the luciferase reporter assays, cells were seeded in 24‐well plates and transfected with the indicated plasmids. For normalization of transfection efficiency, pRL‐TK (Renilla luciferase) reporter plasmid was added to each transfection. Forty‐eight hours after transfection, luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega).
2.16. Statistical analysis
All statistical analyses were carried out using SPSS 19.0 software. The results are represented as means ± SD. Statistical differences between two groups were analyzed by two‐tailed Student’s t test. Levene’s test was used to test variance equality. The relationship between SRC‐1 expression and clinicopathologic parameters was analyzed by the χ2 test. Statistically significant differences were considered significant when P < .05 (*P < .05, **P < .01, ***P < .001) and not significant when P> .05.
3. RESULTS
3.1. Steroid receptor coactivator‐1 expression is positively correlated with grades of glioma and inversely correlated with glioma patient's prognosis
Western blot and IF were used to detect the expression and distribution of SRC‐1 in glioma tumor tissue and GBM cell lines. The data from western blot revealed that SRC‐1 expression was upregulated in several GBM cells compared with SVG p12 (Figure 1A). Immunofluorescence revealed that SRC‐1 was a dominantly nuclear‐expressing protein of GBM tissue and glial fibrillary acidic protein expressed in cytoplasm of GBM cells (Figure 1B).
Figure 1.
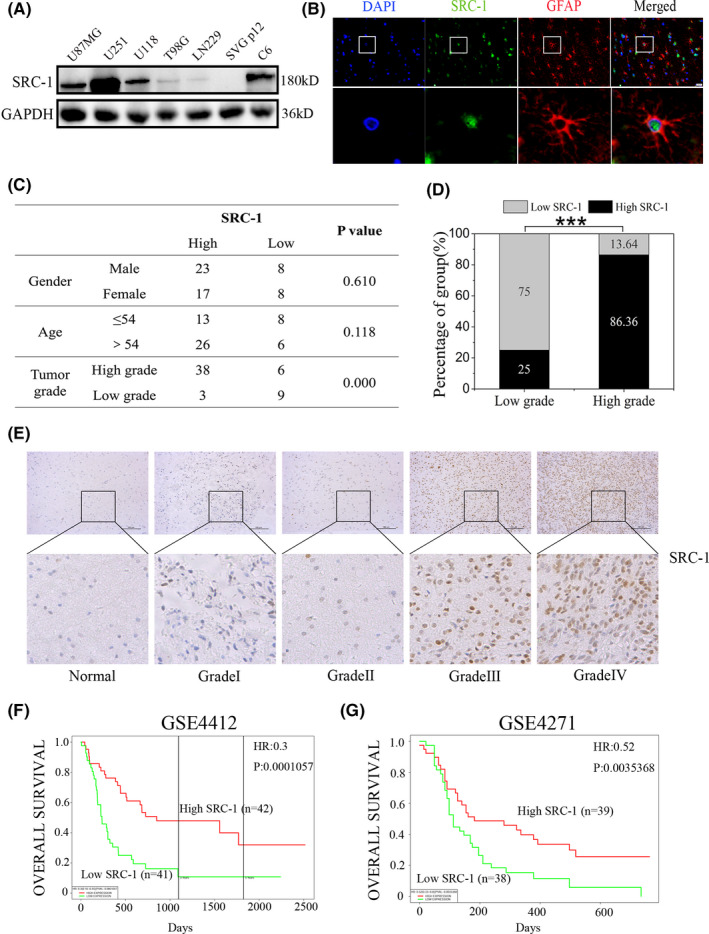
Steroid receptor coactivator‐1 (SRC‐1) expression levels positively correlated with clinical glioma grades and inversely correlated with glioma patient prognosis. A, Western blot analysis for SRC‐1 in U87MG, U251, U118, T98G, LN229, C6, and SVG p12 cells. B, Clinical glioblastoma (GBM) tissue stained for glial fibrillary acidic protein (GFAP) (red) and SRC‐1 (green) by IF analysis. Scale bar, 50 μm. C, Relationship between clinicopathologic variables and SRC‐1 expression. D, SRC‐1 protein levels of low grade (Ⅰ, Ⅱ) and high grade (Ⅲ, Ⅳ) of GBM were compared using the immunoreactive score. P values were calculated using Fisher’s exact test. ***P < .001. E, Representative images of immunohistochemical staining of SRC‐1 in glioma tissues. Scale bar, 100 μm. F, G, Overall survival of 85 glioma patients in the GSE4412 cohort and 77 glioma patients in the GSE4271 cohort
To further confirm the potential clinical implications of SRC‐1 in gliomas, IHC was carried out using a glioma tissue microarray (Table S1). No statistically significant correlations were identified between SRC‐1 expression and gender or age (Figure 1C). In low‐grade tumors (grades Ⅰ and Ⅱ), the percentage of high expression of SRC‐1 was 25.00% (3/12), whereas in high‐grade tumors (grades Ⅲ and Ⅳ), the percentage of high expression of SRC‐1 was 86.36% (38/44). According to our analysis, we found that the expression of SRC‐1 was strongly associated with the grade of the tumors (Figure 1C,D). Representative images of SRC‐1 by IHC staining in different WHO grades of glioma tissues and normal brain tissue are shown in Figure 1E. Moreover, patients with high expression of SRC‐1 showed poorer overall survival than those with low expression (GSE44412, GSE4271) (Figure 1F). Together, our findings indicate that SRC‐1 expression is positively correlated with clinical glioma malignant grade and inversely correlated with patient’s prognosis.
3.2. Steroid receptor coactivator‐1 promotes cell proliferation and migration in GBM cells
To explore the possible functions of SRC‐1 in GBM, SRC‐1 knockdown and overexpression cells were developed and named as U251‐shSRC1, U87‐shSRC1, C6‐shSRC1, and LN229‐SRC1. These cells were used to investigate the effects of SRC‐1 on cell proliferation and migration in GBM cells. First, the results of MTT and colony formation revealed that SRC‐1 knockdown significantly suppressed U251, U87, and C6 cell proliferation, whereas SRC‐1 overexpression increased cell proliferation in LN229 cells (Figure 2A‐F). SRC‐1 knockdown increased the percentage of U251 cells in the G0/G1 phase with a concomitant decrease in S and G2/M phase (Figure 2G). Furthermore, SRC‐1 knockdown suppressed the expression of proliferating cell nuclear antigen (PCNA), cyclin D1, and cyclin‐dependent kinase 4 (CDK4) (Figure 2H). In contrast, SRC‐1 overexpression significantly reduced the percentage of LN229 cells in the G0/G1 phase and upregulated the expression of PCNA, cyclin D1, and CDK4 (Figure 2I,J).
Figure 2.
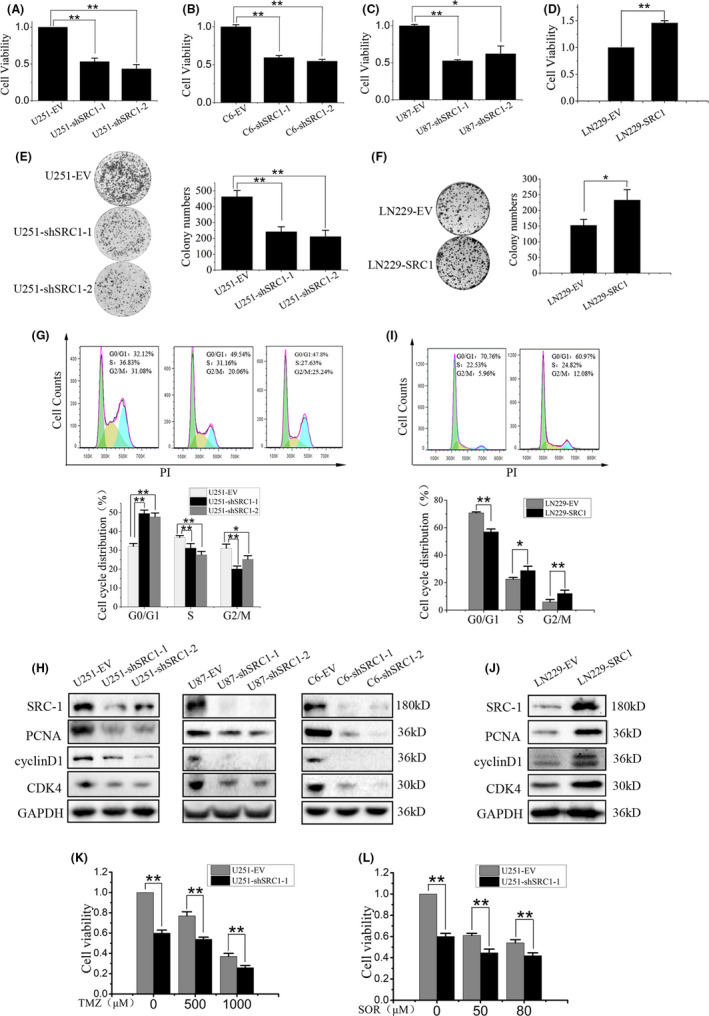
Steroid receptor coactivator‐1 (SRC‐1) promotes the cell proliferation of glioblastoma cells. A‐D, Cell viability was examined by MTT assay for U251, U87, C6, and LN229 cells in 48 h (n = 3). E, F, Cell colony formation ability was examined in U251 and LN229 cells (n = 3). G, I, Flow cytometry analysis was used to examine cell cycles in U251 and LN229 cells (n = 3). PI, propidium iodide. H, J, Western blot analysis for SRC‐1, proliferating cell nuclear antigen (PCNA), cyclin D1, and cyclin‐dependent kinase 4 (CDK4) in U251, U87, C6, and LN229 cells. K, L, Cell viability was detected in U251 cells, which were treated with temozolomide (TMZ; 500 and 1000 μmol/L) and sorafenib (SOR; 50 and 80 μmol/L). *P < .05, **P < .01
To further evaluate the functional significance of SRC‐1 in the chemoresistance of GBM, cell viability was examined in the U251‐EV and U251‐shSRC1 cells under treatment with increasing concentrations of TMZ and sorafenib (SOR). Silencing SRC‐1 remarkably reduced the proliferation of U251 cells treated with TMZ or SOR (Figure 2K,L).
Next, migration rate was examined in U251, U87, C6, and LN229 cells by wound healing assay. As shown in Figure 3A‐D, SRC‐1 knockdown significantly reduced U251, U87, and C6 cell migration rates, and the reverse was shown in LN229‐SRC1 and LN229‐EV cells. To verify the roles of SRC‐1 on the epithelial‐mesenchymal transition (EMT) process, the protein expression of N‐cadherin, E‐cadherin, Vimentin, and TWIST1 was detected by western blot and IF. Our data confirmed that SRC‐1 plays an important role in promoting EMT of GBM cells (Figure 3E‐H). Together, these data suggest that SRC‐1 promotes GBM cell proliferation and migration.
Figure 3.
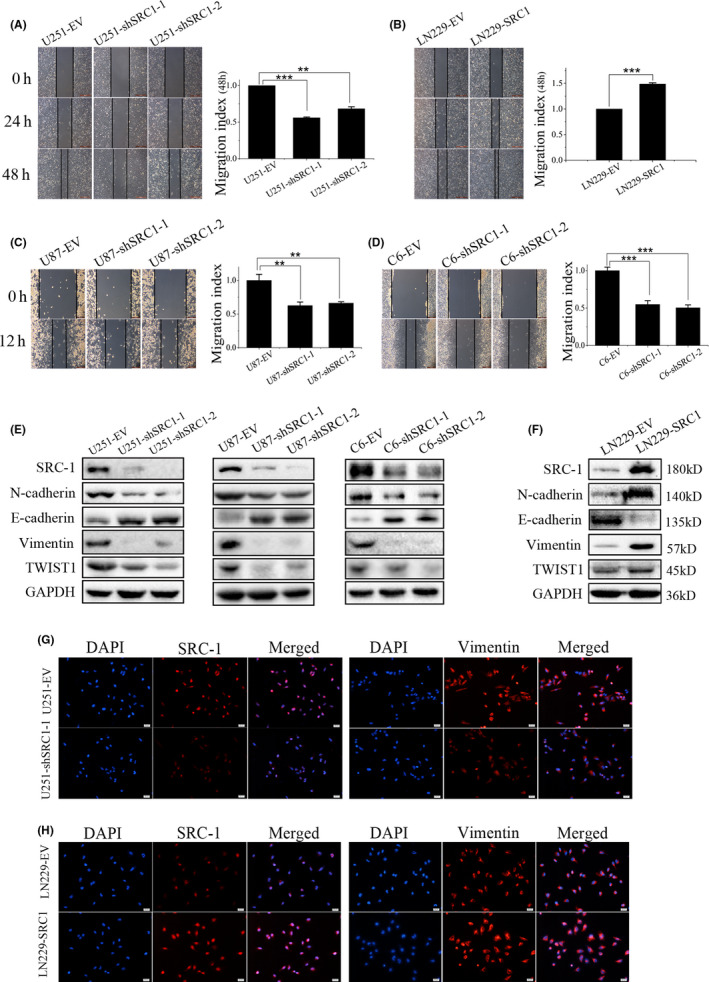
Steroid receptor coactivator‐1 (SRC‐1) promotes the cell migration of glioblastoma cells. A‐D, Migration ability was detected in U251, U87, C6, and LN229 cells by wound healing assay (n = 3). Scale bar, 500 and 200 μm. E, F, Western blot analysis for SRC‐1, N‐cadherin, E‐cadherin, Vimentin, and TWIST1 in U251, U87, C6, and LN229 cells. G, H, Representative immunofluorescence images of U251 and LN229 cells stained for Vimentin (red) and DAPI (blue). Scale bar, 50 μm. **P < .01, ***P < .001
3.3. Steroid receptor coactivator‐1 enhances stem‐like characteristics in GBM cells
To determine the roles of SRC‐1 in GSCs, the self‐renewal capacities of U251, U87, C6, and LN229 cells was detected by sphere formation assay. The results showed that SRC‐1 knockdown significantly decreased sphere initiation and growth, and opposite results were shown in LN229‐SRC1 cells compared with LN229‐EV cells (Figure 4A‐D). To further clarify the roles of SRC‐1 on the GSCs at the molecular level, the expression levels of CD133 and KLF4 were examined by flow cytometry and western blot. The results of flow cytometry showed that the percentage of CD133+ cell counts were significantly increased in LN229‐SRC1 cell spheres when compared with LN229‐EV cell spheres (Figure 4E). The expression of CD133 and KLF4 was decreased in SRC‐1 knockdown cells compared to control cells and was increased in LN229‐SRC1 cells compared to LN229‐EV cells (Figure 4F,G). These results indicate that SRC‐1 can enhance the self‐renewal capacity and stem‐like characteristics in GBM cells.
Figure 4.
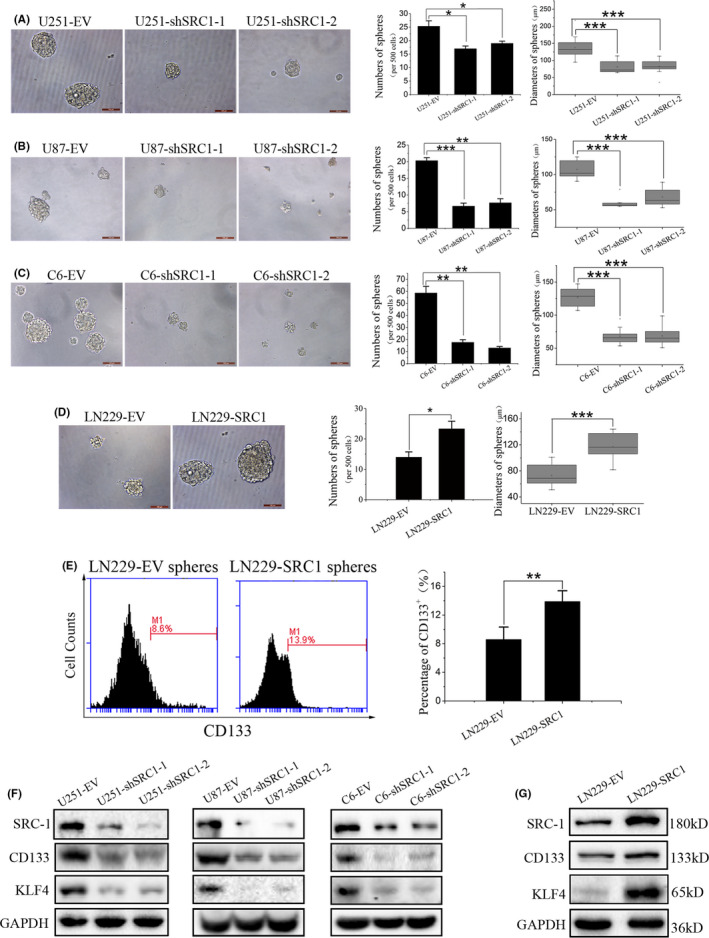
Steroid receptor coactivator‐1 (SRC‐1) enhances the self‐renewal ability of glioblastoma stem cells. A‐D, Self‐renewal ability was detected in U251, U87, C6, and LN229 cells by sphere formation assay. Scale bar, 100 μm. E, Representative images and quantitative assessment of CD133 were examined in LN229‐SRC1 spheres compared with LN229‐EV spheres using flow cytometry. F, G, Expression of SRC‐1, CD133, and Kruppel‐like factor 4 (KLF4) were detected by western blot analysis in U251, U87, C6, and LN229 cells. *P < .05, **P < .01, ***P < .001
3.4. Steroid receptor coactivator‐1 knockdown inhibits GBM growth in vivo
To further verify the in vivo tumorigenesis‐promoting effect of SRC‐1, a xenograft tumor growth assay was carried out on nude mice. LN229‐EV and LN229‐SRC1 cells were injected s.c. into the dorsal flanks of nude mice. Tumor volume, weight, and protein expression were evaluated in these mice. Tumors derived from LN229‐SRC1 cells grew faster than those derived from the control cells (Figure 5A,B). The results of western blot and IHC showed that the expression of SRC‐1, PCNA, KLF4, and Vimentin of the SRC‐1 overexpression group was distinctly higher than in the control group (Figure 5C,D). These findings indicate that overexpression of SRC‐1 promotes tumor growth and development of GBM.
Figure 5.
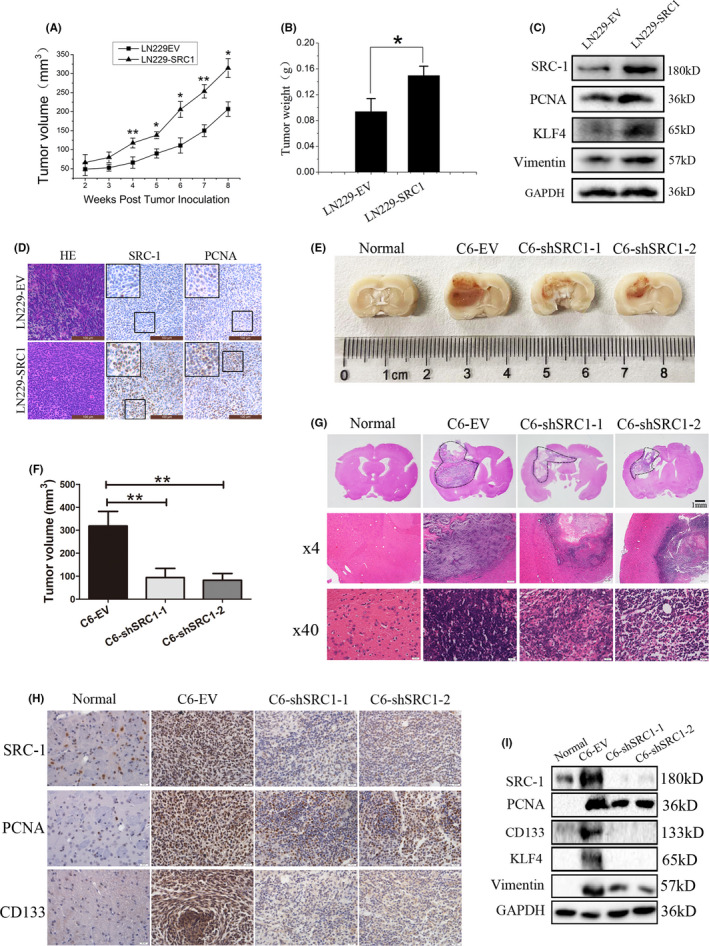
Overexpression of steroid receptor coactivator‐1 (SRC‐1) increases glioblastoma (GBM) tumorigenesis and SRC‐1 knockdown inhibits orthotopic GBM growth in vivo. A, Tumor volume was measured and calculated. n = 6. B, Tumor weight was measured in 8 weeks. n = 6. C, Western blot analysis for SRC‐1, Kruppel‐like factor 4 (KLF4), Vimentin, and proliferating cell nuclear antigen (PCNA) in LN229‐EV and LN229‐SRC1 tumors. D, H&E staining showing the tissue morphology and immunohistochemical (IHC) staining showing the SRC‐1 and PCNA expression pattern in xenograft tumors of nude mice. Scale bar, 100 μm. E, Pathological picture of orthotopic gliomas after 28 days. F, Glioma volume was measured and calculated after 28 days. n = 6. G, Representative H&E staining showing the tissue morphology. Scale bar, 1 mm and 20 μm. H, IHC staining showing the SRC‐1, PCNA, and CD133 expression patterns in rat brain orthotopic glioma. I, Western blot analysis for SRC‐1, KLF4, Vimentin, PCNA, and CD133 in normal brain and glioma tissues. *P < .05, **P < .01
The rat brain orthotopic implantation model of C6 cells was established to further investigate the effect on glioma growth of SRC‐1. The results shown by pathological pictures, glioma volume, and H&E indicated that SRC‐1 knockdown efficiently suppressed GBM growth (Figure 5E‐G). Furthermore, SRC‐1 knockdown inhibited the expression of PCNA, CD133, KLF4, and Vimentin of GBM (Figure 5H,I). Together, these data suggest that SRC‐1 promotes tumorigenesis of GBM and the downregulation of SRC‐1 reduced intracranial GBM growth in vivo.
3.5. Steroid receptor coactivator‐1 promotes stem‐like characteristics through the lncRNA XIST/miR‐152/KLF4 pathway
In further efforts to investigate the downstream target genes for SRC‐1 in GBM, we extracted total RNA from both U251‐EV and U251‐shSRC1 cells and generated the global gene expression profile by RNA‐seq (Figure 6A). The RNA expression level of lncRNA XIST was less than eightfold in U251‐shSRC1 cells compared with U251‐EV cells (Figure 6B and Table S5). Furthermore, XIST was recently determined to be a key factor for maintenance of GBM stemness. We thus hypothesized that XIST might act as a downstream target gene for SRC‐1 mediated glioma stemness. By analyzing GEO databases (GSE2223 and GSE4058), we found that XIST was notably elevated in human glioma and GBM tissues (Figure 6C,D). Furthermore, XIST expression was significantly higher in high‐grade glioma and low‐grade glioma tissues than in normal tissues, but no significant difference in XIST expression levels was observed between high‐ and low‐grade glioma tissues (Figure 6E). We validated the RNA‐seq data by using RT‐qPCR analysis. The results confirmed that XIST expression was robustly inhibited following SRC‐1 knockdown and that XIST expression was increased following SRC‐1 overexpression (Figure 6F,G). MicroRNA‐152 acts as a competing endogenous RNA of XIST and KLF4 is a downstream target of miR‐152. 37 , 38 By analyzing a GEO database (GSE2223), we found that KLF4 was upregulated in human GBM tissues, but KLF4 expression was not significantly higher in glioma tissues than in normal tissues (Figure 6H‐J). The expression of KLF4 and miR‐152 was detected by RT‐qPCR and western blot. The results revealed that KLF4 expression was inhibited and miR‐152 expression was upregulated by SRC‐1 knockdown, whereas overexpression of SRC‐1 increased KLF4 expression and decreased miR‐152 expression (Figure 6K,L). Then XIST expression was reverse rescued by transient siRNA transfection in LN229‐SRC1 cells. The RT‐qPCR results revealed that XIST reversed the expression of miR‐152 but not KLF4 at RNA level, and western blot analysis showed that KLF4 protein expression was significantly decreased (Figure 7A,B). Furthermore, knockdown of XIST by siRNA abolished the induced sphere‐forming ability of SRC‐1 in LN229‐SRC1 cells (Figure 7C). Next, to further confirm the role of miR‐152 for SRC‐1‐regulated KLF4 and SRC‐1‐mediated stem‐like characteristics in GBM cells, miR‐152 expression was rescued by transient transfection of miR‐152 mimic in LN229‐SRC1 cell. The results revealed that miR‐152 reversed the expression of KLF4 and XIST, and that miR‐152 mimic abolished the induced sphere‐forming ability of SRC‐1/XIST in LN229‐SRC1 cells (Figure 7D‐F).
Figure 6.

Steroid receptor coactivator‐1 (SRC‐1) promotes the expression of X‐inactive specific transcript (XIST) and Kruppel‐like factor 4 (KLF4) and suppresses microRNA (miR)‐152 expression. A, An unbiased transcriptome expression profiling heat map between U251‐EV and U251‐shSRC1 cells. B, All differentially expressed genes are listed in the column graph according to their fold change. Right panel, enlarged section showing log2 fold change <−6. C‐E, XIST levels were analyzed in glioma tissues of GSE2223 and GSE4058 databases. F, G, RT‐quantitative PCR (qPCR) analysis for SRC‐1 and XIST in U251 and LN229 cells. H‐J, KLF4 levels were analyzed in glioma tissues of GSE2223 databases. K, L, RT‐qPCR analysis for miR‐152 and KLF4 in U251 and LN229 cells. *P < .05, **P < .01, ***P < .001. GBM, glioblastoma; NS, not significant
Figure 7.
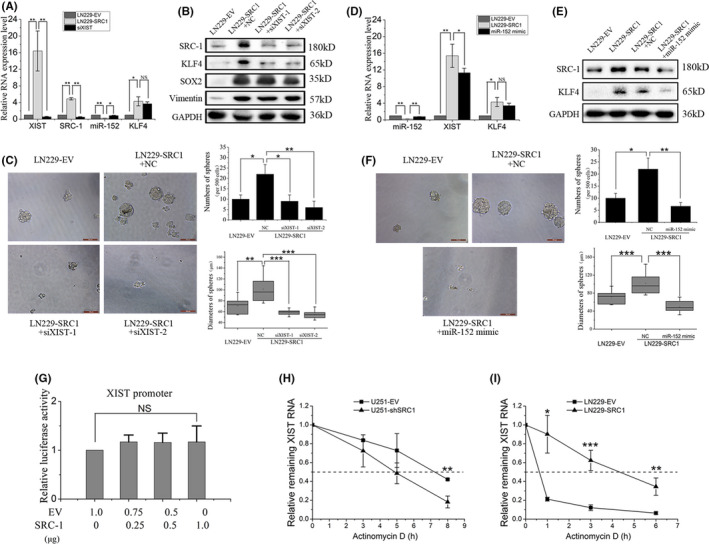
Steroid receptor coactivator‐1 (SRC‐1) increases Kruppel‐like factor 4 (KLF4) expression and stemness of glioblastoma cells through upregulating the X‐inactive specific transcript (XIST)/microRNA (miR)‐152 pathway. A, B, RT‐quantitative PCR (qPCR) and western blot analysis for SRC‐1, XIST, miR‐152, KLF4, and Vimentin in LN229‐EV and LN229‐SRC1 cells transfected with XIST‐siRNA (siXIST) or control siRNA (NC). C, Sphere‐forming ability was detected in LN229‐EV and LN229‐SRC1 cells transfected with siXIST or NC. Scale bar, 100 μm. D, E, RT‐qPCR and western blot analysis for SRC‐1, XIST, miR‐152, and KLF4 in LN229‐EV and LN229‐SRC1 cells transfected with miR‐152 mimic or control RNA (NC). F, Sphere‐forming ability was detected in LN229‐EV and LN229‐SRC1 cells transfected with miR‐152 mimic or NC. Scale bare, 100 μm. G, Promoter activities of 293T cells were determined by a dual‐luciferase assay kit. H, I, Total RNA was analyzed by RT‐qPCR to examine the mRNA half‐life of XIST. *P < .05, **P < .01, ***P < .001. NS, not significant
To further investigate how SRC‐1 affects XIST expression, we first examined whether SRC‐1 regulates the XIST RNA transcription process. To this end, we cloned the sequence of XIST promoter (‒1982, +219) by PCR and inserted the sequence into luciferase reporter pGL3‐basic plasmid. Dual‐luciferase reporter results showed that firefly luciferase did not significantly change after SRC‐1 overexpression in 293T cells, which suggested that SRC‐1 does not influence the proximal XIST promoter activity (Figure 7G). These results implied that SRC‐1 might posttranscriptionally regulate XIST. U251 and LN229 cells were treated with actinomycin D (Act D), which blocks de novo RNA synthesis, and total RNA was isolated at indicated time points after Act D application. Relative XIST RNA levels were measured by RT‐qPCR normalized to that at 0 hour. Knockdown of SRC‐1 resulted in a decrease of the half‐life of XIST from 7 to 5 hours in U251 cells, whereas overexpression of SRC‐1 increased its half‐life from 0.5 to 4.5 hours in LN229 cells, indicating that SRC‐1 promotes the stability of XIST RNA after transcription (Figure 7H,I). Taken together, these data confirm that SRC‐1 stabilizes XIST RNA and enhances stem‐like characteristics, at least partly, through upregulating the XIST/miR‐152/KLF4 pathway.
3.6. Arenobufagin and bufalin inhibit proliferation and stemness of GBM cells
Our previous study reported that arenobufagin (ARE) and bufalin (BFU; SRC inhibitor), homogeneous bufadienolides, could inhibit SRC‐1 expression in HER2‐overexpressing breast cancer cells. 39 Thus, in this study, ARE and BFU were used to treat LN229‐EV, LN229‐SRC1, and U251 cells at 50 nmol/L for 48 hours. The results showed that the expression of SRC‐1, XIST, and KLF4 had all obviously declined in control and SRC‐1 overexpression cells (Figure 8A‐C). We then tested the efficacy of ARE and BFU on proliferation of GBM cell lines. After treatment with different concentrations (0.01‐10 μmol/L) of ARE and BFU for 48 hours, the proliferation of cells was significantly suppressed in a dose‐dependent manner (Figure 8D,E). Furthermore, ARE and BFU decreased the sphere‐forming ability of SRC‐1 overexpressed cells (Figure 8F). Collectively, these data indicate that bufadienolides could inhibit proliferation and stemness through suppressing SRC‐1 expression in GBM cells.
Figure 8.
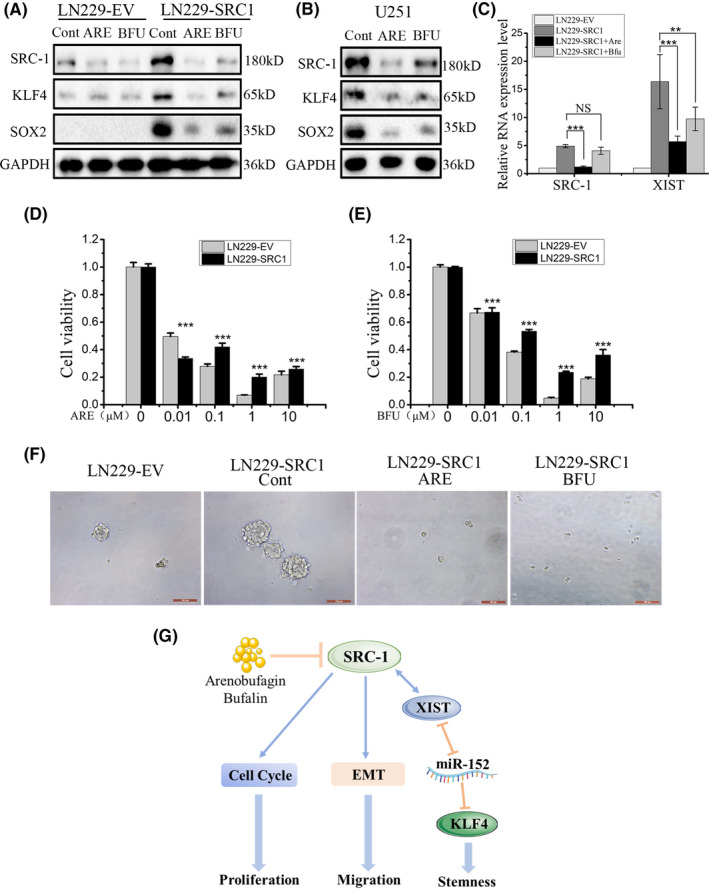
Arenobufagin (ARE) and bufalin (BFU) suppressed proliferation and stemness of glioblastoma cells. A‐C, Western blot and RT‐quantitative PCR analysis for steroid receptor coactivator‐1 (SRC‐1), X‐inactive specific transcript (XIST), and Kruppel‐like factor 4 (KLF4) in LN229 and U251 cells. D, E, Cell viability was detected in LN229 cells. F, Sphere‐forming ability was detected in LN229 cells treated with ARE (100 nmol/L) and BUF (100 nmol/L). Scale bar, 100 μm. G, Schematic diagram of mechanism of this research. SRC‐1 promoted proliferation and migration of GBM cells. SRC‐1 enhanced the stemness by activating the XIST/microRNA (miR)‐152/ KLF4 pathway in GBM cells. Additionally, ARE and BFU reduced the proliferation and stemness of GBM cells. **P < .01, ***P < .001. Cont, control; NS, not significant
4. DISCUSSION
It has been reported that SRC‐1 is overexpressed and plays a crucial promoting role in several human cancers, including breast cancer, liver cancer, lung cancer, and prostate cancer. 20 , 23 , 40 , 41 Accumulated evidence has revealed the pivotal roles that sex steroid hormones and steroid receptor coactivators play in the regulation of brain structure and function. 42 Steroid receptor coactivator‐1 also participates in the regulation of vascular endothelial growth factor expression and its expression was upregulated by progesterone in D54 cells. 43 , 44 However, there were no in‐depth or detailed studies about the expression and roles of SRC‐1 in GBM.
Accumulated studies have revealed that SRC‐1 is the predominant p160 family member in the brain and mostly localized in the neurons, but some astrocytes are also positive for SRC‐1. 45 , 46 , 47 , 48 Our data of brain tissue microarray and analysis of GEO databases indicated that SRC‐1 expression is positively correlated with clinical glioma malignant grade and inversely correlated with glioma patient’s prognosis, which implied that SRC‐1 might be involved in tumorigenesis of GBM. We also determined the possible function of SRC‐1 in the procession of GBM. The results showed that SRC‐1 knockdown induces G2/M arrest and suppresses proliferation of GBM cells. Our data also revealed that SRC‐1 promotes migration of GBM cells by upregulating the EMT process; this is consistent with previous reports that SRC‐1 promotes breast cancer metastasis by upregulating the expression of transcription factor TWIST1, which could increase the expression of N‐cadherin and Vimentin. 12 Furthermore, in our study, SRC‐1 also promoted tumorigenesis in a xenograft mouse model and SRC‐1 knockdown inhibited GBM growth in a rat intracranial brain tumor model.
Our data primarily indicate that SRC‐1 promotes stem‐like characteristics and the self‐renewal ability of GBM cells, consistent with a recent report that SRC‐1 is important for the proliferation of breast and lung cancer stem‐like cells. 23 A number of other reports have indicated possible associated roles for GSCs in GBM initiation, propagation, and maintenance, as well as for recurrence and chemoresistance. 49 , 50 , 51 , 52 , 53 , 54 , 55 , 56 , 57 There are many stemness factors, ALDH1, CD44, CD133, c‐Kit, KLF4, Nanog, Nestin, Oct4, and Sox2, that promote self‐renewal and survival pathways in GBM. 58 , 59 , 60 , 61 , 62 Therefore, to combat the growth of glioblastoma, identifying and targeting a key stemness factor that drives the most aggressive cell proliferation is crucial. The data from Figure 4 reveal that SRC‐1 promotes stem‐like characteristics and the self‐renewal ability of GBM cells. Furthermore, SRC‐1 upregulates the expression of KLF4, which is a master regulator of GSCs. Collectively, our data strongly suggest SRC‐1 as a potential target to GBM stemness, thus overcoming GSCs mediated drug resistance. U251 cells were treated with SRC‐1 knockdown or GBM chemotherapy drug (TMZ and SOR), and the results revealed that knockdown of SRC‐1 enhanced the sensitivity of GBM cells to chemotherapy drugs, which could be of great significance in the clinical treatment of GBM.
Next, we investigated the mechanism of SRC‐1 promoting GBM stemness. From the results of RNA‐seq, we obtained a series of differentially expressed genes, including the XIST gene (−8.3378‐fold), which was recently revealed to be a key factor associated with GSCs and transcriptional silencing of the X chromosome in mammals. 37 , 63 Previous studies have also confirmed the function of lncRNA XIST as a tumor regulator in various human cancers, including breast cancer, ovarian cancer, and hepatocellular carcinoma. 64 , 65 , 66 A recent study showed that the lncRNA XIST is essential for long‐term survival of hematopoietic stem cells. 67 In addition, knockdown of lncRNA XIST exerts tumor‐suppressive functions by upregulating miR‐152, which functions as a tumor suppressor by targeting KLF4 in human GSCs. 37 , 38 Furthermore, our data from GEO database analyses indicated that XIST and KLF4 are upregulated in human GBM samples. Our results showed that SRC‐1 promotes the expression of XIST and KLF4 and suppresses miR‐152 expression. Knockdown of XIST abolishes the induced sphere‐forming ability of SRC‐1 and rescues the expression of miR‐152 and KLF4. Similarly, miR‐152 mimic restores the stemness of SRC‐1. Overall, these data confirm that SRC‐1 increases KLF4 expression and stem‐like characteristics, at least in part, through upregulating the XIST/miR‐152 pathway. Further studies assessed how SRC‐1 regulates lncRNA XIST. We found that SRC‐1 enhances XIST RNA expression at the posttranscriptional level rather than through promoting its transcriptional activity. A previous study showed that JPX and TSIX could activate transcription of XIST by evicting CTCF protein or in a sex‐specific manner; however, two earlier studies used nuclear run‐on analysis and measurements of steady‐state levels to argue that XIST is posttranscriptionally regulated by RNA stabilization rather than by increased transcriptional rate. 68 , 69 , 70 , 71
Recent studies have shown that altering coactivator function is a new entrée into the treatment of cancer. For instance, our previous study showed that ARE and BFU can inhibit SRC‐1 and SRC‐3 expression in HER2‐positive breast cancer cells. Additionally, ARE and BFU can block breast cancer, glioma, and lung cancer cell proliferation. 39 , 72 , 73 , 74 In our study, ARE and BFU suppressed the expression of SRC‐1 in GBM cells. Even though SRC‐1 was overexpressed, ARE and BFU restrained the cell proliferation and sphere‐forming ability of GBM cells. These data confirm that inhibiting SRC‐1 expression with ARE and BFU can block proliferation and stemness of GBM cells. From these data, we can also conclude that ARE is more effective in inhibiting proliferation and stemness of GBM cells than BFU.
In summary, our study indicates that SRC‐1 is overexpressed in both GBM cells and tissue as well as being positively correlated with clinical glioma malignant grade. The SRC‐1 expression level is inversely correlated with glioma patient’s prognosis. In addition, SRC‐1 is a key regulator of the cell cycle, EMT, stemness, and tumorigenesis of GBM. Furthermore, our results show that SRC‐1 enhances stemness of GBM through the XIST/miR‐152/KLF4 regulatory pathway (Figure 8G). Knockdown of SRC‐1 could enhance the sensitivity of clinical chemotherapy drugs. In conclusion, our findings suggest that SRC‐1 could be a therapeutic target as well as a diagnostic marker in GBM.
DISCLOSURE
The authors have no conflict of interest to declare.
Supporting information
Table S1
Table S2
Table S3
Table S4
Table S5
ACKNOWLEDGMENTS
This work was supported by National Natural Science Foundation of China (NO. 81472492), Dalian Medical University Talents Innovation and Entrepreneurship Projects (NO. 201092), Liaoning Provincial Natural Science Foundation of China (No. 2019‐BS‐056).
Gong M, Wang X, Mu L, et al. Steroid receptor coactivator‐1 enhances the stemness of glioblastoma by activating long noncoding RNA XIST/miR‐152/KLF4 pathway. Cancer Sci. 2021;112:604–618. 10.1111/cas.14685
Funding information
National Natural Science Foundation of China, Grant/Award Number: 81472492; Dalian Medical University Talents Innovation and Entrepreneurship Projects, Grant/Award Number: 201092; Liaoning Provincial Natural Science Foundation of China, Grant/Award Number: 2019‐BS‐056.
Gong and Wang contributed equally to this study.
Contributor Information
Liang Liu, Email: dy7liuliang@163.com.
Yongshun Zhao, Email: zhaoyongshun_2005@aliyun.com.
Yuhui Yuan, Email: yuhuiyuan@hotmail.com.
REFERENCES
- 1. Eder K, Kalman B. Molecular heterogeneity of glioblastoma and its clinical relevance. Pathol Oncol Res. 2014;20(4):777‐787. [DOI] [PubMed] [Google Scholar]
- 2. Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro‐oncology. 2012;14(suppl 5):v1‐49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thomas AA, Brennan CW, DeAngelis LM, Omuro AM. Emerging therapies for glioblastoma. JAMA Neurol. 2014;71(11):1437‐1444. [DOI] [PubMed] [Google Scholar]
- 5. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842‐1850. [DOI] [PubMed] [Google Scholar]
- 6. Li QJ, Cai JQ, Liu CY. Evolving molecular genetics of glioblastoma. Chin Med J (Engl). 2016;129(4):464‐471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683‐2710. [DOI] [PubMed] [Google Scholar]
- 8. Noda SE, El‐Jawahri A, Patel D, Lautenschlaeger T, Siedow M, Chakravarti A. Molecular advances of brain tumors in radiation oncology. Semin Radiat Oncol. 2009;19(3):171‐178. [DOI] [PubMed] [Google Scholar]
- 9. Liebelt BD, Shingu T, Zhou X, Ren J, Shin SA, Hu J. Glioma Stem Cells: Signaling, Microenvironment, and Therapy. Stem Cells Int. 2016;2016:7849890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Piccirillo SG, Vescovi AL. Brain tumour stem cells: possibilities of new therapeutic strategies. Expert Opin Biol Ther. 2007;7(8):1129‐1135. [DOI] [PubMed] [Google Scholar]
- 11. Yuan X, Curtin J, Xiong Y, et al. Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene. 2004;23(58):9392‐9400. [DOI] [PubMed] [Google Scholar]
- 12. Qin L, Liu Z, Chen H, Xu J. The steroid receptor coactivator‐1 regulates twist expression and promotes breast cancer metastasis. Can Res. 2009;69(9):3819‐3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McIlroy M, McCartan D, Early S, et al. Interaction of developmental transcription factor HOXC11 with steroid receptor coactivator SRC‐1 mediates resistance to endocrine therapy in breast cancer [corrected]. Can Res. 2010;70(4):1585‐1594. [DOI] [PubMed] [Google Scholar]
- 14. Wang S, Yuan Y, Liao L, et al. Disruption of the SRC‐1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci USA. 2009;106(1):151‐156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan Y, Xu J. Loss‐of‐function deletion of the steroid receptor coactivator‐1 gene in mice reduces estrogen effect on the vascular injury response. Arterioscler Thromb Vasc Biol. 2007;27(7):1521‐1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fleming FJ, Hill AD, McDermott EW, O'Higgins NJ, Young LS. Differential recruitment of coregulator proteins steroid receptor coactivator‐1 and silencing mediator for retinoid and thyroid receptors to the estrogen receptor‐estrogen response element by beta‐estradiol and 4‐hydroxytamoxifen in human breast cancer. J Clin Endocrinol Metab. 2004;89(1):375‐383. [DOI] [PubMed] [Google Scholar]
- 17. McBryan J, Theissen SM, Byrne C, et al. Metastatic progression with resistance to aromatase inhibitors is driven by the steroid receptor coactivator SRC‐1. Can Res. 2012;72(2):548‐559. [DOI] [PubMed] [Google Scholar]
- 18. Tai H, Kubota N, Kato S. Involvement of nuclear receptor coactivator SRC‐1 in estrogen‐dependent cell growth of MCF‐7 cells. Biochem Biophys Res Commun. 2000;267(1):311‐316. [DOI] [PubMed] [Google Scholar]
- 19. Kishimoto H, Wang Z, Bhat‐Nakshatri P, Chang D, Clarke R, Nakshatri H. The p160 family coactivators regulate breast cancer cell proliferation and invasion through autocrine/paracrine activity of SDF‐1alpha/CXCL12. Carcinogenesis. 2005;26(10):1706‐1715. [DOI] [PubMed] [Google Scholar]
- 20. Tong Z, Li M, Wang W, et al. Steroid receptor coactivator 1 promotes human hepatocellular carcinoma progression by enhancing wnt/beta‐catenin signaling. J Biol Chem. 2015;290(30):18596‐18608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh CA, Qin L, Tien JC, Young LS, Xu J. The function of steroid receptor coactivator‐1 in normal tissues and cancer. International journal of biological sciences. 2012;8(4):470‐485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang L, Lonard DM, O'Malley BW. The role of steroid receptor coactivators in hormone dependent cancers and their potential as therapeutic targets. Hormones Cancer. 2016;7(4):229‐235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rohira AD, Yan F, Wang L, et al. Targeting SRC coactivators blocks the tumor‐initiating capacity of cancer stem‐like cells. Can Res. 2017;77(16):4293‐4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER‐ICA) in breast cancer tissue. Pathologe. 1987;8(3):138‐140. [PubMed] [Google Scholar]
- 25. Horvai AE, Roy R, Borys D, O'Donnell RJ. Regulators of skeletal development: a cluster analysis of 206 bone tumors reveals diagnostically useful markers. Mod Pathol. 2012;25(11):1452‐1461. [DOI] [PubMed] [Google Scholar]
- 26. Adams EJ, Green JA, Clark AH, Youngson JH. Comparison of different scoring systems for immunohistochemical staining. J Clin Pathol. 1999;52(1):75‐77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang P, Chen Y, Gong M, et al. Gab2 ablation reverses the stemness of HER2‐overexpressing breast cancer cells. Cell Physiol Biochem. 2018;50(1):52‐65. [DOI] [PubMed] [Google Scholar]
- 28. Wang T, Liu B, Guan Y, et al. Melatonin inhibits the proliferation of breast cancer cells induced by bisphenol A via targeting estrogen receptor‐related pathways. Thorac. Cancer. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu M, Liu Q, Pei Y, et al. Aqp‐1 Gene Knockout Attenuates Hypoxic Pulmonary Hypertension of Mice. Arterioscler Thromb Vasc Biol. 2019;39(1):48‐62. [DOI] [PubMed] [Google Scholar]
- 30. Wang T, Zhuang Z, Zhang P, et al. Effect of arenobufagin on human pancreatic carcinoma cells. Oncol Lett. 2017;14(4):4971‐4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang S, Li G, Liu C, et al. miR‐30e‐5p represses angiogenesis and metastasis by directly targeting AEG‐1 in squamous cell carcinoma of the head and neck. Cancer Sci. 2020;111(2):356‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gong R, Lin W, Gao A, et al. Forkhead box C1 promotes metastasis and invasion of non‐small cell lung cancer by binding directly to the lysyl oxidase promoter. Cancer Sci. 2019;110(12):3663‐3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y, Yang H, Du Y, et al. Long noncoding RNA TP53TG1 promotes pancreatic ductal adenocarcinoma development by acting as a molecular sponge of microRNA‐96. Cancer Sci. 2019;110(9):2760‐2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jin H, Wang Y, Zhou L, et al. Melatonin attenuates hypoxic pulmonary hypertension by inhibiting the inflammation and the proliferation of pulmonary arterial smooth muscle cells. J Pineal Res. 2014;57(4):442‐450. [DOI] [PubMed] [Google Scholar]
- 35. Lu J, Xu Z, Duan H, et al. Tumor‐associated macrophage interleukin‐beta promotes glycerol‐3‐phosphate dehydrogenase activation, glycolysis and tumorigenesis in glioma cells. Cancer Sci. 2020;111(6):1979‐1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yuan Y, Liao L, Tulis DA, Xu J. Steroid receptor coactivator‐3 is required for inhibition of neointima formation by estrogen. Circulation. 2002;105(22):2653‐2659. [DOI] [PubMed] [Google Scholar]
- 37. Yao Y, Ma J, Xue Y, et al. Knockdown of long non‐coding RNA XIST exerts tumor‐suppressive functions in human glioblastoma stem cells by up‐regulating miR‐152. Cancer Lett. 2015;359(1):75‐86. [DOI] [PubMed] [Google Scholar]
- 38. Ma J, Yao Y, Wang P, et al. MiR‐152 functions as a tumor suppressor in glioblastoma stem cells by targeting Kruppel‐like factor 4. Cancer Lett. 2014;355(1):85‐95. [DOI] [PubMed] [Google Scholar]
- 39. Wang T, Mu L, Jin H, et al. The effects of bufadienolides on HER2 overexpressing breast cancer cells. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2016;37(6):7155‐7163. [DOI] [PubMed] [Google Scholar]
- 40. Fleming FJ, Myers E, Kelly G, et al. Expression of SRC‐1, AIB1, and PEA3 in HER2 mediated endocrine resistant breast cancer; a predictive role for SRC‐1. J Clin Pathol. 2004;57(10):1069‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gregory CW, He B, Johnson RT, et al. A mechanism for androgen receptor‐mediated prostate cancer recurrence after androgen deprivation therapy. Can Res. 2001;61(11):4315‐4319. [PubMed] [Google Scholar]
- 42. Chung YG, Kim HK, Lee HK, Lee KC. Expression of androgen receptors in astrocytoma. J Korean Med Sci. 1996;11(6):517‐521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hernandez‐Hernandez OT, Gonzalez‐Garcia TK, Camacho‐Arroyo I. Progesterone receptor and SRC‐1 participate in the regulation of VEGF, EGFR and Cyclin D1 expression in human astrocytoma cell lines. J Steroid Biochem Mol Biol. 2012;132(1–2):127‐134. [DOI] [PubMed] [Google Scholar]
- 44. Hernandez‐Hernandez OT, Rodriguez‐Dorantes M, Gonzalez‐Arenas A, Camacho‐Arroyo I. Progesterone and estradiol effects on SRC‐1 and SRC‐3 expression in human astrocytoma cell lines. Endocrine. 2010;37(1):194‐200. [DOI] [PubMed] [Google Scholar]
- 45. Zhang D, Guo Q, Bian C, Zhang J, Lin S, Su B. Alterations of steroid receptor coactivator‐1 (SRC‐1) immunoreactivities in specific brain regions of young and middle‐aged female Sprague‐Dawley rats. Brain Res. 2011;1382:88‐97. [DOI] [PubMed] [Google Scholar]
- 46. Zhang D, Guo Q, Bian C, Zhang J, Cai W, Su B. Expression of steroid receptor coactivator‐1 was regulated by postnatal development but not ovariectomy in the hippocampus of rats. Dev Neurosci. 2011;33(1):57‐63. [DOI] [PubMed] [Google Scholar]
- 47. Bian C, Zhang D, Guo Q, Cai W, Zhang J. Localization and sex‐difference of steroid receptor coactivator‐1 immunoreactivities in the brain of adult female and male mice. Steroids. 2011;76(3):269‐279. [DOI] [PubMed] [Google Scholar]
- 48. Ogawa H, Nishi M, Kawata M. Localization of nuclear coactivators p300 and steroid receptor coactivator 1 in the rat hippocampus. Brain Res. 2001;890(2):197‐202. [DOI] [PubMed] [Google Scholar]
- 49. Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem‐like neural precursors from human glioblastoma. Can Res. 2004;64(19):7011‐7021. [DOI] [PubMed] [Google Scholar]
- 50. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396‐401. [DOI] [PubMed] [Google Scholar]
- 51. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756‐760. [DOI] [PubMed] [Google Scholar]
- 52. Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522‐526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Garros‐Regulez L, Aldaz P, Arrizabalaga O, et al. mTOR inhibition decreases SOX2‐SOX9 mediated glioma stem cell activity and temozolomide resistance. Expert Opin Ther Targets. 2016;20(4):393‐405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ben‐Porath I, Thomson MW, Carey VJ, et al. An embryonic stem cell‐like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40(5):499‐507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sathyan P, Zinn PO, Marisetty AL, et al. Mir‐21‐Sox2 Axis delineates glioblastoma subtypes with prognostic impact. J Neurosci. 2015;35(45):15097‐15112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gangemi RM, Griffero F, Marubbi D, et al. SOX2 silencing in glioblastoma tumor‐initiating cells causes stop of proliferation and loss of tumorigenicity. Stem Cells. 2009;27(1):40‐48. [DOI] [PubMed] [Google Scholar]
- 57. Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF‐beta signaling maintains tumorigenicity of glioma‐initiating cells through Sry‐related HMG‐box factors. Cell Stem Cell. 2009;5(5):504‐514. [DOI] [PubMed] [Google Scholar]
- 58. Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA‐1 is an enrichment marker for tumor‐initiating cells in human glioblastoma. Cell Stem Cell. 2009;4(5):440‐452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Holmberg J, He X, Peredo I, et al. Activation of neural and pluripotent stem cell signatures correlates with increased malignancy in human glioma. PLoS One. 2011;6(3):e18454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chakrabarti M, Ray SK. Synergistic anti‐tumor actions of luteolin and silibinin prevented cell migration and invasion and induced apoptosis in glioblastoma SNB19 cells and glioblastoma stem cells. Brain Res. 2015;1629:85‐93. [DOI] [PubMed] [Google Scholar]
- 62. Wang Z, Yang J, Xu G, et al. Targeting miR‐381‐NEFL axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget. 2015;6(5):3147‐3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Brown CJ, Ballabio A, Rupert JL, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38‐44. [DOI] [PubMed] [Google Scholar]
- 64. Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108(12):2419‐2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ren C, Li X, Wang T, et al. Functions and mechanisms of long noncoding RNAs in ovarian cancer. Int J Gynecol Cancer. 2015;25(4):566‐569. [DOI] [PubMed] [Google Scholar]
- 66. Kong Q, Zhang S, Liang C, et al. LncRNA XIST functions as a molecular sponge of miR‐194‐5p to regulate MAPK1 expression in hepatocellular carcinoma cell. J Cell Biochem. 2018;119(6):4458‐4468. [DOI] [PubMed] [Google Scholar]
- 67. Yildirim E, Kirby JE, Brown DE, et al. Xist RNA is a potent suppressor of hematologic cancer in mice. Cell. 2013;152(4):727‐742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, Lee JT. Jpx RNA activates Xist by evicting CTCF. Cell. 2013;153(7):1537‐1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21(5):617‐628. [DOI] [PubMed] [Google Scholar]
- 70. Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379(6561):131‐137. [DOI] [PubMed] [Google Scholar]
- 71. Panning B, Dausman J, Jaenisch R. X chromosome inactivation is mediated by Xist RNA stabilization. Cell. 1997;90(5):907‐916. [DOI] [PubMed] [Google Scholar]
- 72. Wang Y, Lonard DM, Yu Y, et al. Bufalin is a potent small‐molecule inhibitor of the steroid receptor coactivators SRC‐3 and SRC‐1. Can Res. 2014;74(5):1506‐1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Han L, Yuan B, Shimada R, et al. Cytocidal effects of arenobufagin and hellebrigenin, two active bufadienolide compounds, against human glioblastoma cell line U‐87. Int J Oncol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu T, Wu C, Weng G, et al. Bufalin inhibits cellular proliferation and cancer stem cell‐like phenotypes via upregulation of MiR‐203 in glioma. Cell Physiol Biochem. 2017;44(2):671‐681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4
Table S5


