Abstract
Viruses are omnipresent and persistent in wastewater, which poses a risk to human health. In this review, we summarise the different qualitative and quantitative methods for virus analysis in wastewater and systematically discuss the spatial distribution and temporal patterns of various viruses (i.e., enteric viruses, Caliciviridae (Noroviruses (NoVs)), Picornaviridae (Enteroviruses (EVs)), Hepatitis A virus (HAV)), and Adenoviridae (Adenoviruses (AdVs))) in wastewater systems. Then we critically review recent SARS-CoV-2 studies to understand the ongoing COVID-19 pandemic through wastewater surveillance. SARS-CoV-2 genetic material has been detected in wastewater from France, the Netherlands, Australia, Italy, Japan, Spain, Turkey, India, Pakistan, China, and the USA. We then discuss the utility of wastewater-based epidemiology (WBE) to estimate the occurrence, distribution, and genetic diversity of these viruses and generate human health risk assessment. Finally, we not only promote the prevention of viral infectious disease transmission through wastewater but also highlight the potential use of WBE as an early warning system for public health assessment.
Keywords: Viruses, SARS-CoV-2, Spatial distribution, Wastewater, Human health hazards
Graphical Abstract
1. Introduction
Viruses are omnipresent and persistent in raw and treated wastewater, which is a concern because they can pose risks to human health (Corpuz et al., 2020). A significant source of viruses in wastewater is human faecal matter, mainly from known infected persons with or without indications (Haramoto et al., 2018). Water-borne viruses, such as Noroviruses (NoVs), enteroviruses (EVs), Hepatitis A virus (HAV), and Adenoviruses (AdVs), are widely distributed in wastewater (La Rosa et al., 2020). Another example, SARS-CoV-2, is responsible for the COVID-19 pandemic that is currently affecting the world (Casanova et al., 2009, Peng et al., 2020). SARS-CoV-2 has also been detected in the faeces of infected patients (Holshue et al., 2020) and in wastewater (Holshue et al., 2020, Mallapaty, 2020). SARS-CoV-2 RNA is shed in body fluids, such as faeces, saliva, and sputum, and these fluids are often disposed of via wastewater systems (Holshue et al., 2020, Mallapaty, 2020). To date, studies have identified SARS-CoV-2 in the wastewater systems of different countries, including Australia (Ahmed et al., 2020), France (Wurtzer et al., 2020), Italy (La Rosa et al., 2020), the Netherlands (Medema et al., 2020), Spain (Randazzo et al., 2020), the Czech Republic (Mlejnkova et al., 2020), Japan (Haramoto et al., 2020), Turkey (Kocamemi et al., 2020), Israel (Or et al., 2020), India (Kumar et al., 2020), Pakistan, (Sharif,, Yaqub et al., 2020), China (Chen et al., 2020, Zhang et al., 2020) and the USA (Wu et al., 2020). Based on these studies, clear evidence of the presence of SARS-CoV-2 in sewage is available, and this transmission pathway increases the risk of human exposure to the virus.
To prevent wastewater transmission and decrease the threat to human health, there is a need to better understand the critical role of wastewater as a potential source of viruses, including SARS-CoV-2. In the current review, we first briefly introduce multiple human viruses, including HAV, EVs, NoVs, AVs, AdVs, and SARS-CoV-2, whose RNA has been detected in wastewater in recent studies. Moreover, methods for qualitative and quantitative analysis of these viruses are also discussed. We summarise the main human viruses found in wastewater and demonstrate the spatial and temporal distribution of viruses in the wastewater of various countries. We further highlight the key future approaches required to strengthen our knowledge and understanding of the existence, persistence, and possible human health risks associated with the presence of EV, NoV, AV, AdV, and SARS-CoV-2 RNA in wastewater systems. We also describe the presence of SARS-CoV-2 in wastewater and critically discuss current knowledge regarding wastewater surveillance to develop a better understanding of the epidemiology of several human viruses, including SARS-CoV-2, which causes COVID-19. The occurrence of SARS-CoV-2 in wastewater suggests wastewater analysis as a potential tool for investigating the invasion, occurrence, molecular epidemiology, and possible eradication of human viruses in human populations. Finally, our findings indicate that in addition to providing information about the transmission of infectious diseases through wastewater, wastewater analyses can also provide information that could be used to monitor viral circulation in communities and serve as a warning of potential outbreaks of contagious diseases. The abbreviations used in the current review are presented in Table S1.
2. Viruses: genomes, classification, and infection symptoms
In this section, we introduce various human viruses, such as EVs, NoVs, AVs, AdVs, and SARS-CoV-2, and detailed information about their genomes, classification, and health symptoms are listed in Table S2. Human enteric viruses are significant causes of severe acute water-borne diseases in both developed and developing countries (Prevost et al., 2015). Due to their long-term persistence in environmental water systems and their strong resistance to decontamination rather than disinfection, these enteric viruses can cause severe illness. Most respiratory viruses appear to be as infective in humans as in tissue culture. Doses < 1 TCID 50 of influenza virus, AdVs, and rhinovirus were reported to infect 50% of the tested population (Yezli and Otter, 2011). Likewise, low doses of the enteric viruses, HAV, NoV, poliovirus rotavirus, and echovirus, also caused infection in at least some of the volunteers tested (Yezli and Otter, 2011).
Human diseases caused by enteric viruses are frequently asymptomatic or paucisymptomatic; nonetheless, they can also induce numerous respiratory, intestinal, and conjunctival symptoms or hepatic infection (Kitajima et al., 2020). Human enteric viruses pose a risk of severe disease and mortality in high-risk populations, including children, older adults, and immunocompromised patients (Pavlinac et al., 2014). Enteric viruses are mainly produced inside human host cells and are substantially transmitted through the faecal-oral route (Bhunia, 2018).
The most crucial characteristic of enteric viruses is the ease with which they can be transferred from person to person; they can cause disease at low transfer doses of less than 20 particles (Warriner, 2005). Human enteric viruses are more stably resistant to environmental alterations than bacteriological endospores (Warriner, 2005). These human enteric viruses multiply inside the digestive tracts of their hosts, who then excrete them in faeces at significant quantities of up to 1011 viral particles/g stool for periods ranging from days to months. Accordingly, wastewater is likely to contain a considerable abundance of enteric viruses (Prevost et al., 2015).
These effluents are generally treated at wastewater treatment plants (WWTPs) that are not explicitly designed to remove human enteric viruses (Prado et al., 2011). Generally, treated wastewater from WWTP sewage flows directly in a riverine system that is used for diverse purposes, such as shellfish farming, agriculture, recreation, market gardening, and catchment to recharge the groundwater system and produce drinking water (Prevost et al., 2015). There are four major groups of human gastroenteritis viruses: AsVs, AdVs, Rotaviruses (RoVs), and calicivirus, which includes NoV and SaV. These viruses can be transmitted not only through the faecal-oral route but also by consumption of contaminated water and food.
AsVs are the main source of human acute gastroenteritis worldwide (Bosch et al., 2014). These non-enveloped viruses have a positive-sense single-stranded RNA, and their genomes consist primarily of open reading frames (ORFs and ORF2) that encode the capsid protein predecessor; this permits discrimination of eight different AsV genotypes, AsV-1 to AsV-8 (Prevost et al., 2015). The eight distinct serotypes of AsVs can infect individuals of all ages, including elderly people, adults, and young children; after a 3–4-day incubation period, they cause mild gastroenteritis and a wide variety of other symptoms, such as nausea, dehydration, vomiting, and diarrhoea. A considerable number of AsV viral particles are eliminated in the faeces of infected persons and circulate in wastewater systems (Prevost et al., 2015). These viruses have been found in wastewater in France (Kaas et al., 2019), Uruguay (Lizasoain et al., 2018), Spain (López-Gálvez et al., 2018), Canada (Qiu et al., 2016), Japan (Kobayashi et al., 2017), New Zealand (Gyawali et al., 2018), Egypt (Shaheen and Elmahdy, 2019), and India (Prevost et al., 2015).
AdVs with a high occurrence in water systems have been recommended as index organisms for viral pathogens since they fit most of the requirements of an ideal indicator (Kundu et al., 2013). Several studies have estimated that more than 90% of the human population worldwide is seropositive for one or more of the AdV serotypes. Human AdVs occur at significantly higher frequencies in sewage systems than other enteric viruses, and they are excreted by infected persons at higher concentrations of up to 1011 particles/g stool (Fong et al., 2010).
Outbreaks of human AdV infections primarily occur in day-care centres, hospitals, swimming pools, military quarters, and similar facilities. Although more than 51 human AdVs have been identified, only one-third of the AdV types cause human infections. These human AdV serotypes are well defined and represent six species (A-F) within the AdV genus of the family Adenoviridae.
Several AdV serotypes have been reported to have significant clinical consequences in terms of specific infections; for example, serotypes 40 and 41 of type F have been shown to be present in most cases of AdV-linked gastroenteritis in children, whereas serotypes 1, 2, and 5 of variety C cause childhood respiratory infections (Kuo et al., 2010).
In addition to replication in the respiratory tract, replication in the urinary bladder has been observed. These viruses are spread via the faecal-oral route. AdVs are among the most common viruses in untreated sewage, and their concentrations can be ten times greater than those of EVs. AdVs have been widely found in wastewater in France (Kaas et al., 2019), Uruguay (Lizasoain et al., 2018), Tunisia (Ibrahim et al., 2018), the USA (Gerba et al., 2018, Jahne et al., 2020), Brazil (Assis et al., 2018), Canada (Qiu et al., 2018), Greece (Kaliakatsos et al., 2019), and Japan (Tandukar et al., 2020).
EVs are transported in the environment to a significant degree via groundwater, seawater, river aerosols typically released from WWTPs, estuarine water, inadequately treated water, private wells, and drinking water that directly or indirectly receives either treated or untreated wastewater (Tiwari and Dhole, 2018). These EVs are frequently transmitted through the faecal-oral route and infect and replicate inside the intestinal tract of the infected host to a significant degree (Van Der Sanden et al., 2018).
The EVs include poliovirus, coxsackievirus (CB) groups A and B, and echoviruses (E) (Tseng et al., 2007). To date, more than 100 enterovirus serotypes have been identified, including more than 70 serotypes that have been discovered in humans (Tiwari and Dhole, 2018). Enteroviruses cause symptoms in humans that vary from asymptomatic contagion to severe gastroenteritis and include aseptic meningitis and myocarditis. Human coxsackievirus and echovirus show a varying pattern in terms of the serotypes most commonly found in wastewater, and clinical segregates such as echovirus 3, 6, and 19 and coxsackievirus A9, B4, and B5 have consecutively been identified as the most widespread serotypes (Tiwari and Dhole, 2018). In the environment, EVs can survive for long periods at pH values ranging from acidic to alkaline (3–10), mainly at low temperatures (Kocwa-Haluch, 2001). EVs such as coxsackievirus serotypes B-3, B-4, and B-5 and E1, E7, and E11 were significantly quantified in wastewater (Tiwari and Dhole, 2018).
Overall, EVs show vast potential for use as a water quality indicator to quantify the risk of infectious EV transmission and assess the primary source of faecal pollution in water systems (Kitajima et al., 2014). At least 100 human EV types have been identified. These viruses have been found in wastewater in Iran (Moazeni et al., 2017), the USA (Brinkman et al., 2017, Gerba et al., 2018), Italy (Pennino et al., 2018), Canada (Qiu et al., 2018) (Simhon et al., 2019), Greece (Kaliakatsos et al., 2019), and Japan (Tandukar et al., 2020). Most EVs are transmitted through the faecal-oral route, and these viruses are most frequently detected in wastewater-contaminated water.
HAV is one of the most important water-borne viruses, causes human enteric hepatitis, and is mainly transmitted through the faecal-oral route (Ouardani et al., 2016). HAV is morphologically indistinguishable from other members of its family. Nevertheless, HAV easily contaminates wastewater due to the vast number of viral particles eliminated by diseased individuals who may be symptomatic or asymptomatic; these particles pass in significant amounts through ineffective sewage treatment plants and can quickly spread to water systems, such as lakes, rivers, and oceans (Ouardani et al., 2016).
Because it is a non-enveloped virus, HAV is stable in the environment for an extended period and is resistant to wastewater treatment processes (La Rosa et al., 2010). Once excreted in faeces, HAV remains alive and can be disseminated through consumption of contaminated water and food (Vohra, 2010). HAV infection is associated with a wide range of common symptoms, including jaundice, vomiting, nausea, pale stools, fatigue, abdominal pain, and dark urine (Kaya et al., 2013). However, in some cases, mainly in teenagers and children under the age of six years, HAV can be asymptomatic (Ciocca, 2000). HAV has been detected and reported in wastewater in various countries, including South Africa (Adefisoye et al., 2016), Tunisia (Ouardani et al., 2016), France (Kaas et al., 2019), and other industrialised countries (Fenaux et al., 2019).
Hepatitis E virus (HEV) is the principal aetiologic mediator of enteric transmission through drinking water polluted with faecal matter and non-A hepatitis worldwide. HEV is the only member of the family Hepeviridae type Herpesvirus. The genome of HEV, which is approximately 7.2 kb in length, includes three open reading frames (ORF1, ORF2, and ORF3) (Long et al., 2017). ORF1 encodes non-structural proteins that are mainly involved in HEV replication, ORF2 encrypts the main capsid proteins, and ORF3 encrypts a small protein that may be involved in HEV–host interaction as well as in virion morphogenesis (Meng, 2010).
HEV infection can lead to acute viral hepatitis in young and middle-aged people (15–40 years old) in developing countries and areas. HEV presents an incubation period ranging from 2 to 9 weeks and is clinically indistinguishable from hepatitis A. This virus has been detected in wastewater in France (Miura et al., 2016, Courault et al., 2017, Kaas et al., 2019), China (Li et al., 2017), Colombia (Baez et al., 2017), Portugal (Matos et al., 2018), Italy (Di Profio et al., 2019), Germany (Beyer et al., 2020), and some other industrialised countries (Fenaux et al., 2019).
RoV-induced gastroenteritis is a self-limiting disease that can be mild to severe. Although person-to-person transmission is an important route, the main RoV transmission route is the faecal-oral route. More than 1000 RoV particles can be present in one gram of faeces, and these viruses have been detected at significant levels in wastewater in many countries, such as South Africa (Adefisoye et al., 2016, Osuolale and Okoh, 2017), Japan (Yasui et al., 2016), Canada (Qiu et al., 2016), the USA (Schmitz et al., 2016, Gerba et al., 2018), Tunisia (Ibrahim et al., 2016), Italy (Mezzanotte et al., 2016), France (Kaas et al., 2019), Japan (Kobayashi et al., 2017), Uruguay (Lizasoain et al., 2018), Egypt (Rizk et al., 2019), the UK (Adriaenssens et al., 2018), Brazil (Assis et al., 2018), New Zealand (Gyawali et al., 2018), Canada (Qiu et al., 2018), New Zealand (Clemens et al., 2020), and Tunisia (Ibrahim et al., 2020).
SARS-CoV-2 is a single-stranded positive-sense enveloped RNA virus that belongs to a group of SARS-related coronavirus species in the subgenus Sarbecovirus of the family Coronaviridae (Kitajima et al., 2020). SARS-CoV-2 contains four structural proteins: S (spike), M (membrane), E (envelope), and N (nucleocapsid); N encloses the RNA genome, and S, E, and M compose the viral envelope (Bosch et al., 2014, Fox, 2020). SARS-CoV-2 is also indistinctly linked to “classical” human CoV strains, such as HKU1, OC43, 229E, OC43, and NL63, in the genus Alphacoronavirus or Betacoronavirus; these strains were initially differentiated in the 1960s and have been recognised to cause approximately 15–30% of common cold cases worldwide (Mesel-Lemoine et al., 2012, Kitajima et al., 2020).
SARS-CoV-2 caused the recent outbreak of the zoonotic disease that is now widely known as coronavirus disease 2019 (COVID-19), a pneumonia-like sickness caused by a hitherto uncharacterised aetiologic agent (Arora et al., 2020). The coronavirus research group of the International Committee on Taxonomy of Viruses recently classified that zoonotic virus as SARS-CoV-2, a member of the Coronaviridae family, based mainly on its genetic structure, the crown- or halo-like structure of its envelope glycoprotein, its typical chemical composition, and its method of replication (Arora et al., 2020). SARS-CoV-2 virus, the aetiological agent of COVID-19, is mainly spread through respiratory droplets and human-to-human interactions.
The common symptoms of infection with SARS-CoV-2, the coronavirus responsible for COVID-19, include headache, loss of taste, sore throat, congestion, runny nose, nausea, vomiting, diarrhoea, fever, chills, cough, shortness of breath, fatigue, and body aches (Chen et al., 2020, Kitajima et al., 2020, Wurtzer et al., 2020). Detection of SARS-CoV-2 in wastewater systems represents an advantageous approach to assessing the COVID-19 epidemic in different communities. A method of detecting SARS-CoV-2 in wastewater systems was initially implemented in the Netherlands. The study reported that in the three weeks before the first COVID-19 case was reported in the Netherlands, SARS-CoV-2 viral RNA was not detected in the wastewater system; however, the amount of viral RNA began to increase over time, and the number of COVID-19 cases also increased (Medema et al., 2020).
3. Virus detection methods in wastewater systems
Several studies have been performed on virus detection in the wastewater system. The virus detection accuracy significantly relies on the sample volume, the nucleic acid extraction yield (nucleic acid-base methods), and the purity (Corpuz et al., 2020). At the same time, sample processing methods can equally influence the efficacy of the subsequent detection method. Therefore, the choice of a precise method for quantification or detection also substantially relies on the ease of cultivation of the viruses under laboratory circumstances. The viruses' particular characteristics affect the genome copy (GC) concentration and detection methods used for the samples being analysed (Ibrahim et al., 2017, Corpuz et al., 2020). For instance, recent research conducted by Hjelmsø et al. (2017) indicated that the concentration of nucleic acids and the nature of the methods applied for extraction substantially influence viral metagenomic assay findings, especially those of viral community composition, viral specificness, and viral pathogen detection. Hence, the methods for viral concentration, nucleic acid extraction, and virus detection must be chosen appropriately. For further detail regarding the concentration methods used for wastewater samples, the sample processing methods used for sludge samples, and nucleic acid extraction methods, see Corpuz et al. (2020).
Additionally, this review summarises different studies in which various human viruses (i.e., HAV, EVs, NoVs, AVs, and AdVs) have been detected at a variety of sampling points, and recent evidence of SARS-CoV-2 in wastewater systems in the analysed WWTPs has been reported worldwide in these studies, which are outlined in Table 1, Table 2. Some individual studies assessed the fate of various human viruses and SARS-CoV-2 within WWTPs by analysing wastewater samples collected from secondary treatment steps (Ibrahim et al., 2017, Balboa et al., 2020, Haramoto et al., 2020, Randazzo et al., 2020). Other studies collected samples before and after treatment to evaluate virus removal efficiency (Hamza et al., 2017, Hamza and Hamza, 2018, Balboa et al., 2020). Aside from viruses in the wastewater, the occurrence of viruses in sludges generated in the examined WWTP operations has also been investigated (El-Senousy et al., 2018, Elmahdy et al., 2019). The sludge and wastewater samples contain both types of RNA and DNA (enveloped and non-enveloped) viruses. The encapsulation of their nucleic acids by capsid proteins characterises non-enveloped viruses. Enveloped viruses contain an additional lipid bilayer membrane surrounded by capsid proteins (Corpuz et al., 2020). During sample processing, lipid bilayer disruption can decrease recovery and affect the subsequent detection (Ye et al., 2016, Corpuz et al., 2020). This is of particular significance for investigations associated with coronaviruses (CoVs) (Corpuz et al., 2020). The diverse methods employed to detect and quantify viruses in the wastewater system include pulsed-field gel electrophoresis, epifluorescence microscopy, immunofluorescence assays, electronic transmission microscopy, flow cytometry, traditional cell culture, and molecular methods (Wu and Liu, 2009, Corpuz et al., 2020). These methods mainly provide distinct information about the presence of viruses (qualitative data and quantitative data) in both sludge and wastewater samples. The molecular methods primarily applied for quantifying a virus are based on determining the number of selected segments of the virus's genetic material. The virus can be detected through this method even if inactivated, i.e., the viral capsid or envelope compromised and even when the viral genetic material is incoherent. In contrast, immunological methods and cell culture-based methods are mainly used to analyse the viability of viruses. Furthermore, a description of each method type and its advantages and disadvantages are summarised in Table S3.
Table 1.
Reported diversity of human virus distribution, detection/quantification and nucleic acid extraction methods used, number of positive samples, and concentrations in wastewater systems in different countries.
| Country | Virus types | Viral detection/quantification and nucleic acid extraction method used | Concentration/Pre-treatment method | Total sample analysed, positive samples % | Concentration (range) GC/L | References |
|---|---|---|---|---|---|---|
| Africa Continent | ||||||
| Eastern Cape, South Africa | HAdV, RV, HAV | HAdV DNA extracted from 200 µL of wastewater samples using DNA extraction kits (Quick gDNATM Mini-Prep; Zymo Research, USA), RNA extraction of HAV and RV using RNA purification kits (Quick-RNATM Mini-Prep; Zymo Research, Irvine, USA), and detection using TaqMan Probe-Based qPCR quantitative assays. | Pre-filtered using glass fibre (Millipore, Ireland), AlCl3 pass through HA filter, & elution followed with Tris–EDTA (TE). | Total of 48 samples analysed. | HAdV (8.4 × 101 to 1.3 × 105) | (Adefisoye et al., 2016) |
| HAdV 16/48 (33.3%). | HAV < 1 | |||||
| HAV 3/48 (6.25%). | ||||||
| RV (ND). | ||||||
| Eastern Cape, South Africa | AiV-1 | Sewage samples analysed for the presence of AiV-1 using RT-PCR. Amplification and sequencing of 3CD and VP1 genomic regions, followed by a phylogenetic study of selected genome sequences, revealed the occurrence of AiV-1, genotype B. | Elution with glycine | Total of 12 samples analysed. | NA | (Onosi et al., 2019) |
| AiV-1 10/12 (83.3%). | ||||||
| Eastern Cape, South Africa | RoVs, HEVs | RNA extracted from 100 µL of wastewater samples using a Z.R. Viral RNA KitTM (Zymo Research Corporation, 17062 Murphy Ave., Irvine, CA 92614, USA). HEVs detected with singleplex RT-PCR assays. | Adsorption-elution-method (Al-method & Mg-method) | Total of 70 samples analysed. | RoVs (1.9 × 103 to 1.2 × 105) | (Osuolale and Okoh, 2017) |
| Adsorption-elution using electronegative membrane | RoVs (41.7%). | |||||
| HEVs (ND). | ||||||
| Tunis, Tunisia | AiV | RNA extracted from (800 µL) wastewater samples using an automatic extractor NucliSENSR Easy MagTM platform (BioMerieux, Marcy L’Etoile, France). AiV genome detected and quantified via RT-PCR using primer sets Ai6261 and Ai6779 to amplify a 519-bp fragment at the 3CD junction. | Beef extract & AlCl3 method followed by –polyethylene glycol (PEG) | Total of 102 samples analysed. | NA | (Ibrahim et al., 2017) |
| 51/102 (50%). | ||||||
| Tunisia | HAV | tRNA extracted from concentrates (150 µL) of wastewater samples using a Nucleo Spin RNA Virus Kit (Macherey-Nagel, Germany). Genotype of HAV strains detected; semi-nested RT-PCR was performed to amplify a 222-bp fragment at the VP3/VP1 junction. | Beef extract & AlCl3 method followed by PEG | Total of 271 samples of wastewater analysed, | (6.7 × 101 to 5.6 × 107) | (Ouardani et al., 2016) |
| 146/271 (53.9%). | ||||||
| Tunis, Tunisia | RVA | RNA extracted from 800 µL of wastewater samples using an automatic extractor NucliSENSR Easy MagTM platform (BioMerieux, Marcy L’Etoile, France). The RVA genome was detected and quantified by RT-PCR using various primers (Vp2-F1 to Vp2-F5, Vp2-R1, Vp2-R2) and a Vp2-P probe. | Beef extract & AlCl3 method followed by PEG | Total of 102 wastewater samples analysed, | (3.9 × 101 to 2.8 × 103) | (Ibrahim et al., 2016) |
| 51/102 (50%). | ||||||
| Greater Cairo, Egypt | HBoV | Viral nucleic acids extracted from 200 µL of wastewater suspensions using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). TaqMan probe assay used for quantification of HBoV-1, HBoV-2, 3, and 4; SYBR green qPCR assay conducted; real-time qPCR performed. | Beef extract & AlCl3 method followed by PEG | Total of 66 treated wastewater samples processed, 38/66 (57.5%). | (6.0 × 103 to 4.9 × 104) | (Hamza et al., 2017) |
| Egypt | HPV, HPyV | Viral nucleic acids extracted from 200 µL of wastewater suspensions using a QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Nested and semi-nested PCR targeting the L1 coding region employed to quantify a broad spectrum of cutaneous and mucosal HPV genotypes. Quantitative SYBR green qPCR assays performed using the primers GP5+/GP6+, which target a partial sequence of L1 genes. HPyV detected and quantified through real-time PCR targeting the VP1 capsid protein-encoding gene. | Elution with beef extract- glycine | Total of 66 treated wastewater samples processed, H HPV (30.5%), HPyV (82.4%). | HPV median (3.9 × 105) HPyV (5.1 × 1002 to 4.72 × 1003) | (Hamza and Hamza, 2018) |
| Tunis, Tunisia | HAdV | DNA extracted from 800 µL of hospital wastewater samples using an automatic extractor platform (NucliSENSR Easy MagTM, BioMerieux, Marcy L’Etoile, France). Nested PCR performed using two primer pairs (Adv-Hex1DEG/Adv-Hex2DEG and Adv-Hex3DEG/Adv-Hex4DEG) to amplify the gene segment coding for HAdV. | Beef extract & AlCl3 method followed by PEG | Total of 102 hospital wastewater samples analysed, (64%). | NA | (Ibrahim et al., 2018) |
| South Africa | NoV | Viral nucleic acid recovered from 100 µL of wastewater suspensions using a NucliSENSR Easy MagTM platform (BioMerieux, Marcy L’Etoile, France). Norovirus RNA-dependent RNA polymerase (RdRp) (region A, 326 bp) amplified from selected NoV GI- and GII-positive samples through conventional PCR using two pairs of primers (JV12Y and JV13I). | Elution with beef extract- glycine | Total of 108 wastewater samples analysed, NoV 78/108 (72.2%) | NoV (1.02 × 102 to 3.41 × 106) | (Mabasa et al., 2018) |
| NoV GI and GII (5.00 × 103 to 1.31 × 106) | ||||||
| Egypt | AiV and HBoV | Nucleic acids recovered from (240 μL) of sewage eluate using QIAamp Viral RNA and DNA kits (Qiagen, Inc., Valencia, CA, USA). The viral RNA was reverse transcribed via random primers in the presence of AiV-1 and detected through semi-nested PCR. HBoV was identified by nested PCR targeting the VP1/VP2 region to identify HBoV-2/3/4 species. PCR to amplify a 543-bp fragment was performed using the primers 234F1 and 234R1. | Adsorption-elution using electronegative membrane, method followed by PEG | Total of 24 wastewater samples analysed (12 untreated and 12 treated). | NA | (Shaheen et al., 2019) |
| AiV-1 2/12 (16.6%) untreated and 1/12 (8.3%) treated. | ||||||
| HBoV 5/12 (41.6%) untreated and 3/12 (25%) treated. | ||||||
| Egypt | Pepper mild | Nucleic acids recovered from (60 μL) of wastewater influent and effluent samples using a QIAamp DNA Blood Mini Kit (QIAGEN, Hilden, Germany). A viral nucleic acid. DNA standard for PMMoV was prepared using the primers HaPMMV2 and PM1602, targeting ~ 319 bp of the genome. The DNA standard for HAdV was produced by cloning through real-time qRT-PCR. | Virus adsorption & elution (VIRADEL) | Total of 66 wastewater influent and effluent samples analysed. | PMMoV (3.9 × 104 to 3.3 × 108) influent and (3.9 × 104 to 1.2 × 107) effluent | (Hamza et al., 2019) |
| mottle virus | ||||||
| (PMMoV) and HAdV | ||||||
| PMMoV (94%) of influent and (78%) of effluent. | HAdV (1.5 × 104 to 1.5 × 107) influent and (2.6 × 104 to 4.4 × 106) effluent | |||||
| HAdV (88%) of influent and (78%) of effluent. | ||||||
| Egypt | HAdV | Viral nucleic acid primarily extracted from (300 μL) wastewater samples using a DNA extraction kit (Patho Gene-spinTM, Korea). Real-time PCR for HAdV was performed using SYBR GREEN (Applied Biosystems Step OneTM Real-time PCR system). | Sewage sample elution with beef extract- glycine. | Total of 96 samples, including 32 raw sewage, 32 treated sewage, and 32 sewage sludge samples, analysed. | (2.02 × 106 to7.23 × 106) stool, (8.7 × 105 to 4.3 × 106) raw sewage, (1.22 × 104 to 3.7 × 106) treated sewage, and (1.48 × 106 to 1.77 × 107) sludge | (Elmahdy et al., 2019) |
| Sludge samples ultracentrifugation followed beef extract elution | ||||||
| HAdV was found in 17/32, 27/32, 16/32, and 25/32 (28.3%, 84%, 50%, and 78%) of the raw sewage, treated sewage, and sewage sludge samples, respectively. | ||||||
| Giza, Egypt | Coxsackievirus, EVs | Viral RNA recovered from concentrated (100 µL) clinical specimens of water and wastewater samples using BIOZOL Total RNA Extraction reagent (BIOFLUX—Japan). Nested RT-PCR was performed using primers in 1st- and 2nd-round PCR to amplify a 138-bp fragment. | Elution with glycine. | Total of 48 samples (12 raw Nile water, 12 drinking water, 12 raw sewage, and 12 treated sewage samples) analysed. | (9 × 101 to 7 × 105) the raw Nile, (9 × 101 to 2 × 102) drinking water, (9 × 102 to 8 × 107) raw sewage, and (9 × 101 to 7 × 103) treated sewage | (El-Senousy et al., 2018) |
| Eluted viruses were re-concentrated by PEG | ||||||
| EVs were found in 33%, 25%, 25%, and 8.3% of these samples, respectively. | ||||||
| Tunisia | SaV | RNA recovered from (800 µL) of wastewater using an automatic extractor (NucliSENS® Easy MagTM platform, BioMérieux, Marcy L'Etoile, France). SaV quantification performed through real-time RT-PCR using SaV124F and SaV1245R primers and a SaV124TP probe to amplify the segment of the gene that encodes the polymerase. | Beef extract & AlCl3 method followed by PEG | Total of 102 wastewater samples analysed, | NA | (Ibrahim et al., 2019) |
| SaV 30/102 (29.4%). | ||||||
| Tunisia | SaV | RNA extracted from (150 µL) of concentrated wastewater samples using the Nucleo Spin RNA Virus Kit (Macherey-Nagel; Germany). SaV quantification performed through real-time RT-qPCR using SaV124F and SaV1245R primers and the SaV124TP probe, which mainly target the polymerase-capsid junction in ORF-1. | Beef extract & AlCl3 method followed by PEG | Total of 218 wastewater samples analysed, SaV 87/218 (39.9%) | (4.3 × 103 to 5.3 × 108) | (Varela et al., 2018) |
| North America Continent | ||||||
| Arizona, USA | GIV NoV | RNA extracted from a concentrated (360 µL) wastewater sample spiked with murine norovirus (MNV, S7-PP3 strain) using the ZR Viral DNA/RNA Kit (Zymo Research, Irvine, CA). GIV NoV quantification performed by semi-nested PCR using COG4F, G4SKF, and G4SKR primers to amplify a 340-bp region of the GIV NoV partial capsid gene. | Ultrafiltration by using electronegative filter | Total of 50 wastewater samples analysed, 13/50 (26%). | NA | (Kitajima et al., 2016) |
| USA | ReoV | ReoV RNA recovered from cell culture homogenous lysates through the study of dsRNA segment patterns. Primers that mainly target conserved regions of the ReoV L1, L3, and S2 genes were developed and used in the molecular detection of ReoV RNA in water. These assays were performed using RT-PCR. | Adsorption-elution | Total of 30 wastewater samples analysed, 9/30 (30%). | NA | (Betancourt and Gerba, 2016) |
| Calgary, Canada | RV, AdV | RNA extracted from a concentrated (200 µL) wastewater sample. Virus mix containing NoV GII and AdV was expected from clinical stool samples and confirmed through in-house RT-qPCR assays. | VIRADEL | Total of 12 wastewater samples analysed, RV 6/6 (50%). | RV (6.6 log10) | (Qiu et al., 2016) |
| Adv (NA) | ||||||
| Adv 6/12 (50%) | ||||||
| Canada | NoV | Viral nucleic acid primarily recovered from concentrated (327 μL) wastewater samples by cell culture. Virus mixture containing NoV from clinical stool samples using real-time quantitative PCR (RT-qPCR) and confirmed by in-house RT-qPCR. | VIRADEL | Total of 12 wastewater samples analysed. | NoV most numerous (6.6 log10) | (Qiu et al., 2016) |
| Ohio, USA | EVs | Nucleic acids extracted from (10 mL) concentrated wastewater samples using a QIAamp DNA Blood Maxi Extraction kit (Qiagen, Valencia, CA). Quantitative reverse transcriptase PCR used to detect EVs. EVs counted in each sample using RT-qPCR. | Filtration and elution with beef extract-Celite | Total of 12 wastewater samples analysed. | (7.05 × 103 to 8.3 × 105) | (Brinkman et al., 2017) |
| Arizona, USA | AiV 1, SaV | Viral RNA extracted from concentrated (650 µL) wastewater samples using the ZR Viral DNA/RNA Kit (Zymo Research, Irvine, CA). Quantification of AiV 1 genotype-specific qPCR performed by real-time PCR using forward and reverse primers (AiV-AB-F and AiV-AB-R) and probes (AiV-A-TP and AiV-B-TP). | Filtration by using electronegative filter & elution followed with Tris–EDTA (TE) | Total of 26 wastewater (13 influent and 13 effluent) samples analysed | NA | (Kitajima et al., 2018) |
| SaV 8/13(62%) influent and 1/13(8%) effluent. | ||||||
| AiV 7/13 (54%) influent and 7/13 (54%) effluent | ||||||
| South America Continent | ||||||
| Rio De Janeiro, Brazil | NoV | Viral nucleic acid recovered from concentrated (140 μL) wastewater samples using the QIAamp Viral RNATM Mini Kit (QIAGEN, Valencia, CA, USA). NoV quantification by semi-nested PCR targeting the 50-end ORF2 region. | NA | Total of 156 wastewater samples analysed | (104 to 106) | (Fioretti et al., 2018) |
| (52%). | ||||||
| Rio de Janeiro, Brazil | NoV | Viral RNA recovered from concentrated (140 μL) wastewater samples using the QIAamp Viral RNA Mini Kit (QIAGEN, CA, USA) qPCR) method. | Adsorption of viruses to pre-flocculated skimmed-milk proteins followed centrifugation | Total of 156 wastewater samples analysed, NoV GI and GII 38.5% and 96.1%, respectively. | NoV GI and GII (4 to 6.2 log10) and (4.4 to 7.3 log10), respectively | (Fumian et al., 2019) |
| Rio De Janeiro, Brazil | HSaV | Viral RNA recovered from concentrated (140 μL) wastewater and faecal samples using the QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA, USA). HSaV quantitative detection performed using the TaqManTM-based real-time PCR through the polymerase-capsid junction localised in ORF1 of HSaV (GI, GII, GIV, and GV) in a single reaction. | Adsorption of viruses to pre-flocculated skimmed-milk proteins followed centrifugation | Total of 156 wastewater samples analysed, 51/156 (33.0%). | (104 to 106) | (Fioretti et al., 2016) |
| Antioquia, Colombia | HEV | Viral RNA extracted from concentrated wastewater samples using a commercial kit (QIAamp Viral RNA Mini, QIAgen, Netherlands). RT-nested PCR method used to target the ORF2/3 region (nt 5258–5394) to detect the HEV genome. | Filtration & tangential ultrafiltration | Total of 30 wastewater and drinking water samples analysed, Wastewater 5/30 (16.7%). | NA | (Baez et al., 2017) |
| Drinking water 7/30 (23.3%). | ||||||
| Uruguay | AiV-1 | Nested PCR method used for amplification of the 3CD junction section by Taq DNA polymerase (5 U/ll) with primers 6261/6779, which amplifies a 519-bp region, in the 1st-round PCR and primers C94b/246 k to amplify a 266-bp region in the 2nd-round PCR. | Ultracentrifugation | Total of 96 wastewater samples analysed, 54/96 (56%). | NA | (Burutarán et al., 2016) |
| Santiago, Chile | JCPyV | Viral nucleic acid obtained from concentrated wastewater samples using the High Pure Viral Nucleic Acid Kit (Roche Diagnostics GmbH, Germany). JCPyV detection and quantification performed through real-time qPCR. An 89-bp fragment of the large T antigen (LTAg) coding region of JCPyV (positions 4251 to 4339 in reference sequence NC_001699.1) was amplified using forward and reverse JE3 (Mad-1) primers and a 6-FAM/BHQ1 JE3 (Mad1) probe. | Ultracentrifugation | Total of 72 (36 influents and 36 effluents) wastewater samples analysed, JCPyV 29/36 (80.56%) influent and 18/36 (50%) effluent. | NA | (Levican et al., 2019) |
| Germany | HEV | Viral nucleic acid obtained from concentrated (5 mL) wastewater samples using NucliSENS easy MAG (BioMérieux, Germany). Nested RT-PCR used to amplify a 332-bp product from HEV (ORF1). | Ultracentrifugation followed PEG | Total of 184 (111 influent and 83 effluent) wastewater samples analysed, 93/111 (84%) influents and 26/83 (31%) effluents. | Median 3× 103 (influent) and 1× 103 (effluent) | (Beyer et al., 2020) |
| Mexico | Poliovirus | Viral nucleic acid obtained from concentrated wastewater samples. Poliovirus serotype detected by qRT-PCR. Sabin isolates screened through real-time PCR assay for VDPVs by sequencing in VP1. | Elution with beef extract- glycine | Total of 125 wastewater samples analysed, poliovirus 37/125 (29.6%). | NA | (Estívariz et al., 2019) |
| Asia Continent | ||||||
| India | EVs (polio) | EVs (polio) found in wastewater from clinically isolated samples using pan-EV primers (CDC, Atlanta, GA). Tissue culture used for EV isolation, and serotype confirmed by EV neutralisation tests. | Centrifugation followed PEG | Total of 109 wastewater samples analysed, (50.0%) | NA | (Tiwari and Dhole, 2018) |
| Pakistan | EVs (polio) | EV RNA content of wastewater concentrates estimated by real-time RT-PCR using the qScript XLT qPCR Toughmix system (Quantabio) in a Rotor-Gene Q instrument (Qiagen). Nucleotide sequence of the VP1 coding region of EV strains analysed using the Sanger method. | Separation-inoculated on rhabdomyosarcoma cell culture flasks. | NA | (7.0 to 7.5 log10) | (Majumdar et al., 2018) |
| Pakistan | HEV | RNA extracted from the virus in concentrated wastewater using the QIAamp RNA extraction kit (Qiagen, Hamburg, Germany). | Centrifugation followed PEG | Total of 86 wastewater samples analysed | NA | (Ahmad et al., 2010) |
| 35/86 (40.7%) | ||||||
| Japan | NoV | RNA extracted from the virus in concentrated wastewater using the QIAamp Viral RNA Mini QIAcube Kit (Qiagen, Hilden, Germany) and QIAcube (Qiagen, Hilden, Germany). Genotypes and variants quantified through amplification of the partial capsid protein (VP1) and RNA-dependent RNA polymerase genes of NoV GI and GII by single-round PCR, nested PCR, and sequencing using the primers p290, COG1F, COG2F, G1SKR, and G2SKR. | Centrifugation followed PEG | Total of 147 (70 wastewater and 77 stool) samples analysed, NOV GII (77%) stool. | NA | (Masago et al., 2016) |
| NOV GII (81%) | ||||||
| Miyagi, Japan | HPeV | Viral RNA recovered from concentrated (140 μL) clinical wastewater samples using the QIAamp Viral RNA Mini Kit (Qiagen) and complementary DNA (cDNA) synthesised via RT using superscript II-RT. HPeV directly verified by PCR targeting the VP1 region and purified PCR products of the VP1 region. | Centrifugation followed PEG | Total of 79 wastewater samples analysed, HPeV 14/79 (18%), | NA | (Abe et al., 2016) |
| Japan | NoV GI | RNA recovered from concentrated (140 μL) wastewater samples using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany) with QIAcube (Qiagen) and cDNA produced using the iScript Advanced cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). NoV GI quantified through qPCR using the CFX96 real-time PCR quantification system with primers and probes COG1F, COG1R, RING1(a)-TP, and RING1(b)-TP for GI and COG2F, COG2R, and RING2AL-TP for GII. | Centrifugation followed PEG | Total of 17 wastewater samples analysed, NoV GI 17/17(100%). | Up to (8.7 × 104) | (Kazama et al., 2016) |
| Japan | NoV GII | Viral RNA recovered from concentrate (100 µL) using NucliSENS ® miniMAG ® (BioMerieux, Tokyo, Japan). Matching DNA (cDNA) prepared using the Prime Script RT reagent kit (Takara Bio, Japan). TaqMan qPCR assays used in MF-qPCR for quantification of NoV GI, NoV GII, NoV GIV, MgV, MNV, and IAC plasmid DNA. | Centrifugation followed PEG | Total of 26 wastewater samples analysed, 12/26 (46%). | NA | (Ito et al., 2017) |
| Shen Zhen, China | HEV | RNA recovered from concentrated wastewater samples using the UltraPureTM RNA Kit (CWBIO, Beijing, China). Recovered RNA further used for matching DNA (cDNA) synthesis using the HiFiScript 1st strand cDNA Synthesis Kit (CWBIO, Beijing, China). Nested PCR used to amplify a fragment of ORF2 (nt 5,983-6,349) of the HEV genome. | Centrifugation followed PEG | Total of 152 wastewater samples analysed. | NA | (Li et al., 2017) |
| 2/152(1.32%). | ||||||
| Karaj, Iran | SAFV | RNA extracted from concentrated (100 µL) river and wastewater samples using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA). RT-PCR and RT-qPCR method used a gold standard to quantify SAFV through targeting the 5′UTR region of the genome. | Centrifugation followed Dextran-PEG | Total of 50 (28 river, 12 treated, and 10 untreated) wastewater samples analysed, 10/28 (35.7%) river, 4/12 (33.3%) treated, 4/10 (40%) untreated. | (2 × 106 to 6.4 × 106) river | (Aminipour et al., 2020) |
| (1.2 × 106 to 5.2 × 106) treated 2.4× 106 to 6.8× 106) untreated | ||||||
| Tehran, Iran | TTV | Viral nucleic acids recovered from concentrated (140 μL) final wastewater elute samples using the QIAamp RNA Mini Kit (Qiagen, Germany). Nested PCR used to detect the presence of TTV using 1st-round primers (NG054 and NG147) and 2nd- round amplification using (NG132 and NG133). | Elution with beef extract- glycine | Total of 13 wastewater samples analysed, (76.9%). | NA | (Tavakoli Nick et al., 2019) |
| Vietnam | NoV | Viral RNA recovered from concentrate (300 µL) from wastewater samples using the Direct-zol RNA MiniPrep kit (Zymo Research, Irvine, CA, USA). cDNA obtained via RT using the iScript Advanced cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qPCR assays performed to quantitatively detect NoV GI and GII using SsoFast Probes Supermix with primers COG1F and COG1R and probes RING1-TP(a) and RING1-TP(b) for NoV GI and primers COG2F and ALPF and probes COG2R and RING2AL-TP for NoV GII quantification. | Filtration with mixed cellulose ester | Total of 39 wastewater samples analysed, positive | NoV GI maximum (5.6 × 102) | (Nguyen et al., 2018) |
| membrane | rates of NoV GI and GII were 87% and 95%, respectively. | NoV GII (1.3 × 101 to 3.1 × 103) | ||||
| Europe Continent | ||||||
| Italy | HSaV | Nucleic acid recovered from concentrated (100 µL) wastewater samples and purified using the NucliSens extraction kit (BioMerieux, Paris, France). Nested RT-PCR assay targeting the capsid region (VP1) that detects all HSaV genotypes was performed using three forward primers (SaV124F, SaV1F, and SaV5F) and two reverse primers (SV-R13 and SV-R14) in the 1st round of PCR and primers, 1245Rfwd and SV-R2, in the nested PCR. | Polyethylene glycol-dextran separation | Total of 166 wastewater samples analysed, 56/166 (33.7%). | NA | (Mancini et al., 2019) |
| Portugal | HEV | Nucleic acid recovered from concentrated (140 µL) wastewater samples using the QIAmp® Viral RNA Mini Kit (QIAGEN, Hilden, Germany). HEV quantification was done using an RT-qPCR TaqMan probe assay targeting the open reading frame (ORF) 2 region of the HEV. | Ultracentrifugation | Total of 60 wastewater samples analysed, 2/60 (3.3%) | NA | (Matos et al., 2018) |
| French Polynesia | HEVs, NOV, SaV, RoV, HAdV, HPyV | Viral nucleic acids recovered from concentrated (200 µL) wastewater samples using the High Pure Viral Nucleic Acid Kit (Roche Molecular Biochemicals Ltd, Mannheim, Germany). | Flocculation with PEG | Total of 6 wastewater samples analysed. | HEVs average (2.3 × 105) and SaV (8.3 × 106). average (7.2 × 102) and GII (1.7 × 107) | (Kaas et al., 2019) |
| NoV GI 2/6(33.3%) and GII 5/6 (83.3%). | ||||||
| HAdV 5/6 (83.3%). | HAdV average (1.7 × 105). HEV (LOD) | |||||
| Viral RNA genomes (NoV GI and GII, AsV, SaV, EV, HAV, HEVs, HPyV, and RoV) detected using RT-qPCR or qPCR assays. | ||||||
| HEVs 4/6 (66.7%). | ||||||
| RoV and AsV 3/6 (50%). | RoV and AsV average (3.3 × 104 2.6 × 103) | |||||
| HAV (ND). | ||||||
| HPyV 6/6 (100%) | HPyV average (3.4 × 107). | |||||
| Gothenburg, Sweden | NoV | RNA recovered from concentrated (300 µL) wastewater samples using Biorobot EZ1 (Qiagen) with the EZ1 Virus Mini Kit v.2.0 (Qiagen). Real-time qPCR method used as a duplex PCR for NoV GGI and GGII with primers (NV-G1-fwd1b, NV-G1-rev, NV-G2fwd, and COG2R) at regions of the ORF1-ORF2 junction for NoV GGI and GGII quantification. | Fluctuations in concentration | Total of 160 wastewater samples analysed, NoV GI 22/26 (84.6%) (warm) and 25/28 (89.2%) (cold) untreated wastewater. | Average NoV GI and GII (6.2 and 6.8 log10) genome equivalents (g.e.)/L cold (5.3 and 5.9 log10) warm g.e./L untreated | (Dienus et al., 2016) |
| NoV GII 15/26 (57.69%) (warm) and 26/28 (92.85%) (cold) untreated wastewater. | Treated detection limit (BDL) warm to average (3.8) log10 g.e./L cold | |||||
| NoV GI 16/26 (61.5%) warm, 17/28 (60.7%) cold and NoV GII 15/26(57.69%) warm, 26/28 (92.85%) cold in treated water. | ||||||
| Western France | HEV | Viral RNA extracted from concentrated wastewater using a NucliSENS kit (BioMerieux, Lyon, France). HEV quantification using an Ultrasens QRT-PCR kit (Invitrogen, France). RNA assessed based on a standard curve obtained from in vitro transcription of a plasmid comprising a fragment of the HEV genotype 3 f strain. | Centrifugation followed PEG | Total of 32 (18 influent and 14 effluent) samples analysed, collected from 4 WWTP (A, B, C, and D) | NA | (Miura et al., 2016) |
| sites | ||||||
| HEV (10%, 11%, 13%, and 12% influent) and (8%, 5%, 9%, and 13% effluent) WWTP A, B, C and D positive, respectively. | ||||||
| Southern Italy | HEV | RNA extracted from concentrated wastewater using the TRIzol LS (Invitrogen, Ltd., Paisley, UK) method and studied through HEV-specific RT-qPCR, focusing on a conserved 68-nucleotide region of the ORF3 genome. | Elution with phosphate buffered saline | Total of 56 wastewater samples analysed, 13/56 (23.2%). | (6.1 × 102 to 5.8 × 105) GC/mL | (Di Profio et al., 2019) |
| Southern Italy | HEVs | RNA extracted from concentrated wastewater. HEV quantified through PCR, and virus typing performed by seroneutralization. The presence of EVs in the CPE-positive samples was measured through RT-nested PCR using primers focused on VP1 and AN89. | Two-phase separation method by Dextran-PEG | Total of 731 wastewater samples analysed, 161/731 (22.0%). | NA | (Pennino et al., 2018) |
| North Wales, UK | EVs | RNA obtained from concentrated wastewater. RNA viruses (NoVGI and GII, SaVGI), along with potential EVs (HAdVs and PyV strains BK and JC), were measured by RT-qPCR. | Beef extract elution followed PEG | Total of 91 (52 influent and 39 effluent) wastewater samples analysed. | AdV and JCV (104 & 6 × 105) | (Farkas et al., 2018) |
| NoVGI, GII and SaVGI (NA) | ||||||
| AdV and JCV (100%). | ||||||
| NoVGI, GII and SaVGI (35%, 62% and 27%) influent and (38%, 49% | ||||||
| and 10%), respectively. | ||||||
| Italy | HBoVs | Viral nucleic acids recovered from concentrated (10 mL) chloroform-treated wastewater samples using NucliSENS easyMAG (BioMerieux, Marcy L’Etoile, France). Viral DNA was used as a template for nested PCR using a broad-range pair of primers targeting the VP1/VP2 region of HBoV. | Centrifugation | Total of 134 wastewater samples analysed, 106 (79.1%). | (5.51E+03 to 1.84E+05) | (Iaconelli et al., 2016) |
| Catalonia, Spain | HAdV, NoV | Viral nucleic acids extracted from concentrated wastewater samples using the QIAmp Viral RNA kit (Qiagen, Inc., Valencia, CA). HAdV and NoV GII quantitated using real-time qPCR and RT-qPCR. | Skimmed milk flocculation | Total of 12 wastewater samples analysed, HAdV and NoV GII 12/12 (100%) samples. | HAdV and NoV GII (1.98 × 105) and (5.17 × 106), respectively | (Gonzales-Gustavson et al., 2019) |
| Australia Continent | ||||||
| Sydney and Melbourne, Australia | HAdVs | Viral DNA obtained from concentrated wastewater samples using the QIAamp Viral Mini Kit (Qiagen, Hilden, Germany). HAdV quantification was performed using qPCR. The standard for quantification was obtained as a 301-bp fragment in 1st-round PCR through the product of the hexon gene cloned into pET26b (Gen Script) and then measured by spectrophotometry. | Ultracentrifugation | Total of 68 wastewater samples analysed. | Average (1.8 × 107) | (Lun et al., 2019) |
| Queensland, Australia | HPyV | qPCR amplification method used for EC H8 and HF183 and qPCR amplification in a (20 μL) reaction mixture using (10 μL) of SsoFast EvaGreen supermix (Bio-Rad Laboratories, CA, USA), 400 nM each primer (EC H8 assay), 300 nM each primer (HF183 assay), and 3 μL of template DNA. | Filtration by using electronegative filter followed Tenfold serial dilutions | NA | HPyV average (2.56 × 105) GC/mL | (Hughes et al., 2017) |
Not Detected = (ND); Not Available = (NA); Low Detection = (LOD).
Table 2.
The reported SARS-CoV-2 distribution, nucleic acid extraction and detection/quantification methods used, concentration/pre-treatment methods used, target genes, number of positive samples, and concentrations in wastewater systems in different countries.
| Country & Location | Nucleic acid extraction & Detection/quantification method used | Concentration/Pre-treatment method | Target gens | Total sample analysed & water type | Positive samples | Concentration range (GC/L) | References |
|---|---|---|---|---|---|---|---|
| France | Viral particles and genomes extracted from concentrated (11 mL) ultracentrifugation wastewater samples using an optimised protocol (Power Faecal Pro kit in a QIA symphony extractor, QIAGEN). SARS-CoV2 quantitative analysis done by RT-qPCR. | Ultracentrifugation | RNA-dependent RNA polymerase gene (RdRp) | Total of 31 wastewater samples analysed | 23/23 (100%) untreated & 6/8 (75%) treated samples detected positive | Max: > 106.5 untreated | (Wurtzer et al., 2020) |
| Paris | |||||||
| (23 untreated wastewater, 8 treated wastewater). | |||||||
| Max: ~105 treated | |||||||
| Netherlands | Viral RNA genome recovered from concentrated sewage samples using the RNeasy Power Microbiome Kit (Qiagen, Hilden, Germany). SARS-CoV2 quantitative analysis done by real-time RT-PCR. | Centrifugation | Envelope protein gene (E) | Total of 24 untreated wastewater samples analysed. | 14/24 (58%) samples detected positive | NA | (Medema et al., 2020) |
| Australia Queensland | Viral RNA genome recovered from concentrated wastewater samples using a combination of two kits (RNeasy PowerWater Kit and RNeasy PowerMicrobiome Kit; Qiagen, Hilden, Germany). SARS-CoV2 quantitative analysis done by RT-qPCR. | Filtration (0.4 μm pore size) | Nucleocapsid gene (N) | Total of 9 untreated wastewater samples analysed. | 2/9 (22%) samples detected positive | Max: 1.2 × 102 | (Ahmed et al., 2020) |
| ItalyRome | Viral nucleic acids recovered from concentrated sewage samples using the NucliSENS miniMAG semi-automated extraction system. SARS-CoV2 quantitative analysis done by nested RT-PCR | Pasteurzation | Open reading frame | Total of 12 treated wastewater samples analysed | 6/12 (50%) samples detected positive | NA | (La Rosa et al., 2020) |
| (57 °C, 30 min) | |||||||
| 1ab | |||||||
| (ORF1ab) | |||||||
| USA Massachusetts | Viral RNA genome recovered from concentrated wastewater samples using polyethylene glycol 8000 (PEG) & by reverse transcriptase NEB & qPCR (TaqMan fast advanced master mix, Thermo Fisher). SARS-CoV2 quantitative analysis done by RT-qPCR. | Pasteurisation | Spike protein gene (S) | Total of 14 untreated wastewater samples analysed. | 10/14 (71%) samples detected positive | Max: > 2× 104 | (Wu et al., 2020) |
| (60 °C, 90 min) & Filtration (0.2μm pore size) | |||||||
| USA | Viral RNA genome recovered from concentrated wastewater samples. SARS-CoV2 quantitative analysis done by RT-qPCR. | Filtration (5 μm & 0.4 μm pore size) | Nucleocapsid gene (N) | Total of 7 untreated wastewater samples analysed. | 7/7 (100%) samples detected positive | Max: > 3× 105 | (Nemudryi et al., 2020) |
| Bozeman | |||||||
| USA | Viral RNA genome recovered from 2.5 mL of primary well mixed sludge using the RNeasy Power Soil Total RNA kit (Qiagen). SARS-CoV2 quantitative analysis done by one-step qRT-PCR. | NA | Nucleocapsid gene (N) | Total of 44 untreated primary sewage samples analysed. | 44/44 (100%) samples detected positive | 1.7 × 106 to 4.6 × 108 | (Peccia et al., 2020) |
| New Haven | |||||||
| USA | Viral RNA genome recovered from wastewater samples using (NucliSENS easyMag, bioMerieux, Inc., Durham, NC, USA). SARS-CoV2 quantitative analysis done by reverse transcription droplet digital PCR (RT-ddPCR). | Centrifugation & filtration | Nucleocapsid gene (N) | Total of 198 raw wastewater samples analysed | 98/198 (49.5%) samples detected positive | 101 to 104 | (Gonzalez et al., 2020) |
| Southeast Virginia | |||||||
| Spain | Viral RNA genome recovered from concentrated (150 µL) wastewater samples using the NucleoSpin RNA virus kit (Macherey-Nagel GmbH and Co., Düren, Germany). SARS-CoV2 quantitative analysis done by RT-qPCR. | pH adjustment at 6; | Nucleocapsid gene (N) | Total of 72 samples analysed | 35/42 (83%) influent & 2/18 (11%) secondary treated samples detected positive. | NA | (Randazzo et al., 2020) |
| Murcia | 42 influent samples, 18 secondary treated &12 tertiary treated wastewater | ||||||
| Spain | Viral RNA genome recovered from concentrates (150 µL) wastewater samples using the NucleoSpin RNA virus kit (Macherey-Nagel GmbH and Co., Düren, Germany). SARS-CoV-2 RNA was detected using the One Step RT-PCR Kit & by RT-qPCR. | pH adjustment at 6; | Nucleocapsid gene (N) | Total of 24 wastewater samples analysed | Only 12/15 (80%) untreated samples detected positive | 105 to106 untreated | (Randazzo et al., 2020) |
| Valencia | |||||||
| (15 untreated & | |||||||
| 9 treated) | |||||||
| Spain | Viral RNA genome recovered from concentrated wastewater samples by (Seegene, Seoul, South Korea) SARS-CoV-2 RNA quantified by one-step multiplex RT-qPCR. | Ultrafiltration | E, N, ORF1ab, RdRp & S | Total 35 samples analysed | Only 2/18 (11.1%) secondary & | < 2.5 × 105 secondary & untreated NA | (Balboa et al., 2020) |
| Ourense | |||||||
| 5 untreated wastewaters | |||||||
| 5/5 (100%) untreated samples detected positive | |||||||
| 25 treated (secondary 18 & tertiary 12) | |||||||
| Czech Republic | Viral RNA genome isolated from concentrated wastewater samples using the NucliSENSfi miniMAGfi system (BioMérieux, Marcy l’Etoile, France). SARS-CoV2 quantitative analysis done by RT-qPCR. | Direct flocculation using beef extract- glycine & centrifugation | NA | Total 112 untreated wastewater samples analysed | 13/112 (11.6%) samples detected positive | NA | (Mlejnkova et al., 2020) |
| Israel | Viral RNA recovered from concentrated wastewater samples using the RNeasy mini kit (QIAGEN) & Easy MAG (bioMerieux, France). SARS-CoV2 quantitative analysis done by RT-qPCR. | Centrifugation | Envelope protein gene | Total 26 untreated wastewater samples analysed | 10/26 (38.5%) samples detected positive | NA | (Or et al., 2020) |
| Various locations | |||||||
| (E) | |||||||
| Turkey | RNA genome extracted from concentrated wastewater samples using the QIAamp Cador Pathogen | Centrifugation; filtration (0.45 μm & 0.2 μm pore size); pH adjustment at 7.0 to 7.2 | NA | Total 7 untreated wastewater samples analysed | 5/7 (71.4%) samples detected positive | ND to 9.33 × 104 | (Kocamemi et al., 2020) |
| Mini Kit (Qiagen, Hilden, Germany). SARS-CoV2 quantitative analysis done by RT-qPCR | |||||||
| Istanbul | |||||||
| Japan | Viral RNA genome recovered from concentrated (140 µL) sewage samples using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). SARS-CoV2 quantitative analysis done by RT-qPCR. | NA | Nucleocapsid gene (N) | Total of 13 samples analysed | Only 1/5 (20%) secondary-treated samples detected positive | 2.4 × 103 | (Haramoto et al., 2020) |
| Yamanashi | |||||||
| 5 secondary-treated wastewater samples | |||||||
| 5 influents samples | |||||||
| 3 river water samples | |||||||
| India | Viral RNA genome recovered from concentrated wastewater samples using the Allplex™ 2019-nCoV | Filtration (0.45 μm) & PEG centrifugation (4 °C for 30 minutes) | Spike protein gene (S) | Total 6 untreated wastewater samples analysed | 2/6 (33.3%) samples detected positive | NA | (Arora et al., 2020) |
| Jaipur | |||||||
| Assay kit (RP10244Y RP10243X). SARS-CoV2 quantitative analysis done by RT-qPCR. | |||||||
| India | Viral RNA genome recovered from concentrated wastewater samples using the NucleoSpin® RNA Virus Kit (Macherey-Nagel GmbH & Co. KG, Germany). SARS-CoV2 quantitative analysis done by RT-qPCR. | Filtration (0.22 μm) & PEG centrifugation | ORF1ab, N & S | 20 untreated & treated | 20/20 (100%) untreated | 5.6× 10 to | (Kumar et al., 2020) |
| Ahmedabad | wastewater samples | 3.5× 102 untreated | |||||
| 20/20 (100%) treated samples detected positive | |||||||
| analysed | |||||||
| Pakistan | Viral RNA genome recovered from concentrated wastewater samples using the Spin star viral nucleic acid kit 1.0 (ADT Biotech, Phileo Damansara 1, Petaling Jaya Part No.811803). SARS-CoV2 quantitative analysis done by RT-qPCR | Filtration & PEG centrifugation | ORF1ab | Total of 78 untreated wastewater samples analysed | 21/78 (26.9%) samples detected positive | NA | (Sharif) |
| Various locations | |||||||
| Pakistan Lahore | Viral RNA genome recovered from concentrated wastewater samples using BSL-3 of IM, UVAS. SARS-CoV2 quantitative analysis done by RT-qPCR | Centrifugation (4 °C for 15 minutes) | ORF1ab | Total of 28 untreated wastewater samples analysed | 22/28 (78.6%) samples detected positive | 2.67 × 102 to 3.60 × 104 | (Yaqub et al., 2020) |
| ChinaWuhan | SARS-CoV2 quantitative analysis done by RT-qPCR | NA | NA | Total of 42 untreated stool samples analysed | 28/42 (66.67%) samples detected positive | NA | (Chen et al., 2020) |
| China | SARS-CoV2 quantitative analysis done by RT-qPCR | Centrifugation (56 °C for 30 minutes inactivation) | ORF1ab | Total of 15 untreated stool samples analysed | 4/15 (26.7%) samples detected positive | 0.05 to 1.87 × 105 | (Zhang et al., 2020) |
| Wuhan |
Not Detected = (ND); Not Available = (NA).
4. Occurrence of viruses worldwide in wastewater
4.1. Spatial distribution of human viruses in wastewater systems
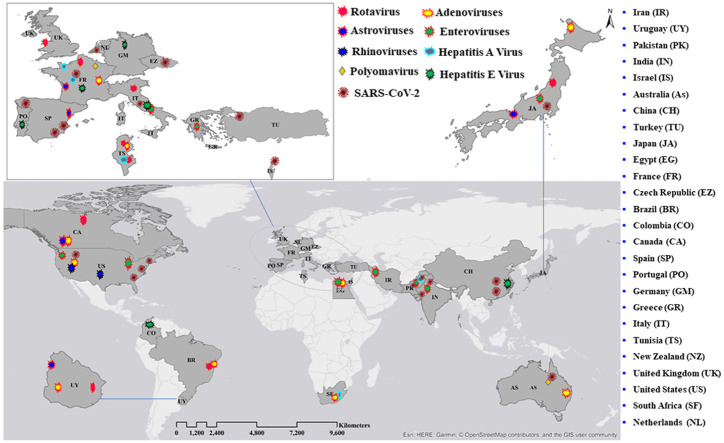
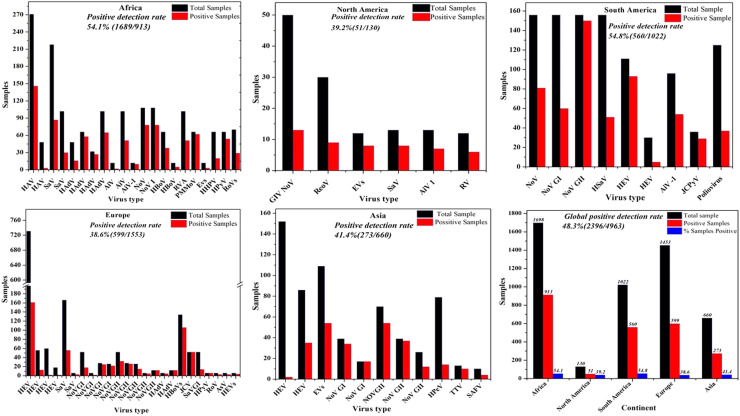
Here, we discuss the detection of various viruses in wastewater in multiple countries worldwide. According to the collected data, wastewater on all inhabited continents contains detectable viruses. Detailed information on each virus found in wastewater in each country is presented in this section (see Fig. 1). Further details showing the diversity of human virus continent distributions, quantification methods used, concentration/pre-treatment method, total sample analysed, positive samples, and virus concentrations (range GC/L) in wastewater systems worldwide are summarised in Table 1. We also describe in detail the viruses in wastewater in different areas in Part 1 of Supporting information.
Fig. 1.
Spatial patterns and diversity of human viruses in wastewater systems worldwide (Points on the map do not correspond to precise geographical locations).
4.2. Temporal patterns of human viruses in wastewater systems
Although most studies have indicated that changes in the levels of viral pathogens in sewage over time are not governed by predictable rules (Farkas et al., 2018), some studies have found that the levels of specific viral pathogens in sewage follow a pattern of imperceptible daily changes and obvious seasonal changes (Abe et al., 2016, Kitajima et al., 2018). Farkas et al. detected diurnal patterns during sampling periods; however, obvious changes in viral titres were not found, apart from slight fluctuations in raw wastewater (Farkas et al., 2018).
These results showed that the viral level in sewage is not affected by daily chemical fluctuations. Therefore, one sample taken during the day may be adequate for enumerating the viral load of treated wastewater within an order of magnitude, while collection of four samples per day is recommended for testing wastewater influent samples. Regarding seasonal changes, Farkas et al. also found that AdV titres were high and relatively constant in different seasons. In contrast, high concentrations of NoV GI and GII and SaV GI titres were demonstrated in winter and fall, and low counts were observed in summer (Farkas et al., 2018).
AiV-1 was also frequently detected in the colder months of the year in Fray Bentos, Bella Unión, Paysandú, and Salto, Uruguay (Burutarán et al., 2016). Similarly, Ouardani et al. demonstrated that HAV circulates throughout the year in Tunisia, with high concentrations in winter and autumn. In contrast, in coastal areas, the highest rates occur in summer and fall (Ouardani et al., 2016). Ibrahim et al. (2017) and Abe et al. (2016) demonstrated a clear difference in the monthly and seasonal distributions, respectively, of viruses in wastewater.
Brinkman et al. (2017) collected monthly municipal wastewater samples for 1 year and quantified EVs in wastewater. Sequence analysis and principal component analysis demonstrated that EV A and EV B were present, with EV A comprising over 45% of detections in the spring and EV B accounting for more than 80% of detections during the summer and autumn. EV C was detected throughout the year, while EV D was observed occasionally. The analysis of human EV in wastewater has provided novel insights into seasonal trends in EVs at the community level and could help elucidate the EV disease burden.
Nguyen et al. (2018) demonstrated NoV contamination in oysters from Hue city, Vietnam. The concentration of NoV GII was lower in the dry season than in the flood season, suggesting that seasonal flooding and sewage cause NoV contamination of oysters. However, the temporal patterns of SARS-COV-2 in wastewater are still unclear and require more research. Nonetheless, a recent study conducted by Vallejo et al. applied a wastewater detection approach in the urban area of Coruña (Spain), and investigative sampling, analysis, and monitoring were conducted on April 15th. The initial results indicated that a substantial level of SARS-COV-2 viral RNA was present in the wastewater system of the Bens WWTP. In addition, starting on April 19th, 24 h composite wastewater samples were continually analysed until early June, and surveillance will continue at the Bens WWTP until the viral genetic material disappears. The results of this study identified a decrease in COVID-19 incidence and further confirmed that the time course of the quantifiable discovery of SARS-CoV-2 in wastewater was inversely correlated with the number of confirmed COVID-19 cases (Vallejo et al., 2020).
4.3. Respiratory viruses in wastewater systems and associated human risks
In the field of ecological virology, studies of water-borne transmission have focused mainly on EVs. Respiratory viruses such as AdVs, CoVs, and SARS-CoV-2 have been reported in wastewater systems (Ahmed et al., 2020, Kitajima et al., 2020). According to early descriptions, these human viruses cause severe diarrhoea as well as respiratory illness.
4.4. Conventional respiratory viruses
Respiratory diseases are the most common human diseases worldwide, and most of them are caused by viruses such as the influenza virus, coronaviruses, rhinoviruses, AdV, and respiratory syncytial virus (Bruning et al., 2017, Corpuz et al., 2020, Zhang et al., 2020). These viruses can infect the upper and lower respiratory tract, leading to acute viral rhinitis and pharyngitis, bronchitis, and pneumonia. Respiratory infection usually leads to self-inoculation because virus-contaminated fingers or hands rub the eyes or cause viral transmission through the nose or mouth. Inhalation of contaminated aerosols is another important route of transmission. Although faecal-oral transmission is not the main route of transmission of respiratory infectious diseases, some viruses that cause contagious respiratory diseases are extensively detected in faeces and wastewater.
For instance, rhinoviruses have been identified in sewage in the USA (Brinkman et al., 2017). According to Fong et al. (2010), AdVs (40 and 41 type F), both respiratory AdVs (2 and 3 types C and B) and AdVs (12 types) that cause meningoencephalitis with early replication in the digestive or respiratory tract, are found in wastewater systems and in drain overflows and waterways that receive these releases (Fong et al., 2010). While it is possible that swimming in sewage-polluted waters is also linked to respiratory illness, aetiological agents associated with respiratory diseases are not frequently detected in swimming areas (Wade et al., 2006, Kitajima et al., 2020).
4.5. SARS-CoV-2 in wastewater systems
According to previous studies, the occurrence of CoVs in wastewater systems is limited; however, few ecological research studies have focused on CoVs. It has been assumed but not well confirmed that enveloped CoVs are primarily spread by human-to-human contact rather than by the faecal-oral route; therefore, the occurrence of CoVs in faeces requires further nuanced clarification (Kitajima et al., 2020). Although few studies have used the appearance of CoV genetic RNA in wastewater systems as a main disease surveillance tool, use of the method for this purpose is gaining traction (Ahmed et al., 2020, Kitajima et al., 2020). During the SARS epidemic in China in 2004, SARS RNA was first detected in 10/10 (100%) untreated and 3/10 (30%) disinfected hospital wastewater samples collected in Beijing, China, which was used to identify the presence of SARS patients (Wang et al., 2005).
In December 2019, cases of SARS-COV-2, the aetiological agent of the ongoing COVID-19 pandemic, were first reported (Haramoto et al., 2020). To date, the available studies on the surrogate SARS-COV-2 viruses recommend assuming that the strain that causes COVID-19 might be less persistent in wastewater systems, primarily due to the occurrence of either carbon-based matter or matrix autochthonous flora in such systems; such influences are certainly able to trigger metabolic pathways that hasten the die-off of viruses (Carraturo et al., 2020). However, recent cases of SARS-CoV-2 have been accompanied by insistent shedding of RNA viruses in faecal samples in 27–89% of patients at concentrations ranging from 0.8 to 7.5 log10 genome copies/g (Ahmed et al., 2020, Wölfel et al., 2020, Zhang et al., 2020). Thus, it is clear that SARS-CoV-2 is also present in wastewater systems (Lodder and de Roda Husman, 2020).
Actually, SARS-CoV-2 has been detected in wastewater systems in several countries, and this could be very informative for risk measurement. Respiratory symptoms are frequently reported in patients with COVID-19, and several ongoing studies have revealed that the intestinal tract can also be affected by SARS-CoV-2 (Wong et al., 2020). A meta-analysis confirmed that approximately 15% of COVID-19 patients experienced mostly intestinal symptoms and that approximately 10% of patients had intestinal symptoms but not respiratory symptoms (Ali et al., 2020). Correspondingly, SARS-CoV-2 RNA was frequently detected in the faeces of COVID-19 patients who did not experience intestinal symptoms (Vallejo et al., 2020, Yeo et al., 2020). Several studies reported that approximately 53.9% of patients were positive for faecal viral RNA (Vallejo et al., 2020), and some studies indicated that viruses were excreted in significant amounts in the patients’ stools for an extended period of time (Chen et al., 2020); moreover, in a few cases, the individual was confirmed to be negative based on respiratory samples after a month or more (Vallejo et al., 2020, Wölfel et al., 2020). In addition, the virus can significantly infect enterocytes in the human small intestine (Lamers et al., 2020). The occurrence of the infectious virus in human faeces highlights the potential for viral replication in the intestinal epithelium of infected individuals (Zhou et al., 2020).
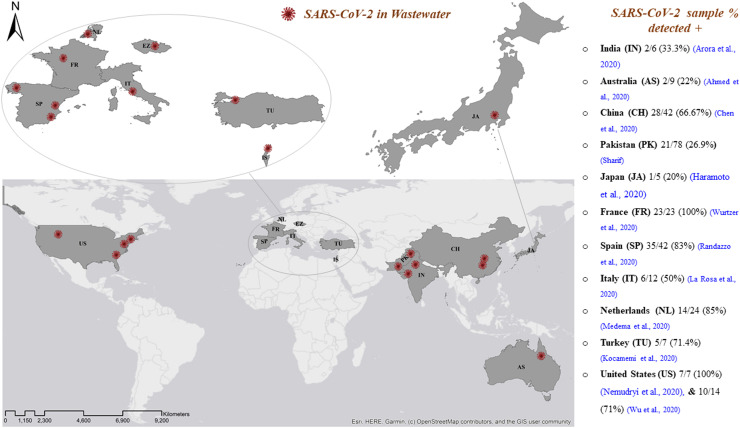
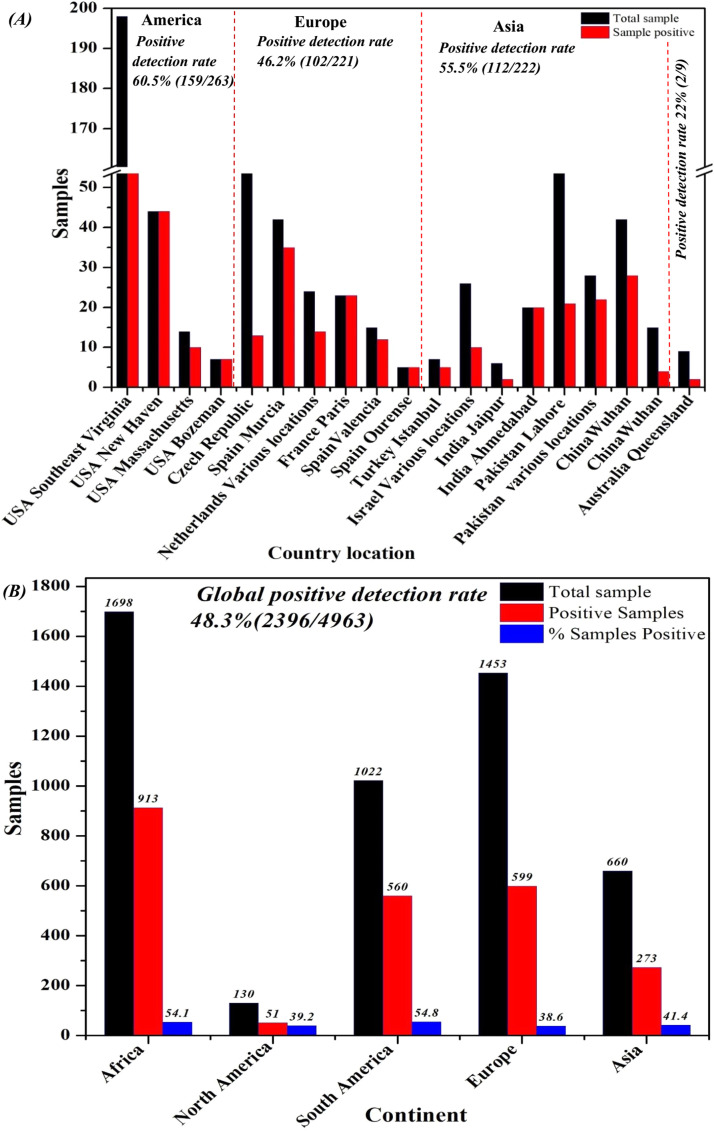
As shown in Fig. 3, recent studies have identified SARS-CoV-2 in wastewater systems in many countries on different continents (Europe, Australia, Asia, and America), including the Netherlands in a sample collected in February 2020. Of the samples, 14/24 (58%) that were positive (Medema et al., 2020) were from Rome, Italy, collected between February 3 and April 2, 2020. There were 6/12 (50%) positive (La Rosa et al., 2020) samples from Yamanashi, Japan collected between March 17 and May 7, 2020. We found that 1/5 (20%) secondarily treated positive samples (Haramoto et al., 2020) were from Israel, collected from various locations at the end of March to April 2020. Additionally, 10/26 (38.5%) of the samples (Or et al., 2020) from Istanbul, Turkey, collected on April 21, 2020 were positive; 5/7 (71.4%) (Kocamemi et al., 2020) samples from Paris, France, collected from March 5 to April 23, 2020 were positive; 23/23 (100%) samples (Wurtzer et al., 2020) from Murcia, Spain, and Valencia collected from March 12 to April 14, 2020 were positive, of which 35/42 (83%) influent samples were positive, 2/18 (11%) secondarily treated samples were positive (Randazzo et al., 2020), and 12/15 (80%) were positive (Randazzo et al., 2020). Ourense samples were collected twice a week from April 6–21 2020. The 2/18 (11.1%) secondary and 5/5 (100%) untreated positive samples (Balboa et al., 2020) from the Czech Republic were collected from April to June 2020. Among the 13/112 (11.6%) positive samples (Mlejnkova et al., 2020) from Queensland, Australia, 2/9 (22%) were positive (Ahmed et al., 2020). Among the Jaipur and Ahmedabad samples collected from India between May 3 and June 14, 2020 (Arora et al., 2020) and May 8 and 27, 2020 (Kumar et al., 2020), 2/6 (33.3%) and 20/20 (100%) were positive, respectively. From March 20 to April 9, 2020, among samples collected from various locations in 17 districts, 21/78 (26.9%) were positive (Sharif), and Lahore samples were collected on alternate days between July 13–25, 2020. We found that 22/28 (78.6%) of the positive samples (Yaqub et al., 2020) from Wuhan Pulmonary Hospital, China, were collected after the initial identification of 2019-NCoV. Moreover, 4/15 (26.7%) (Zhang et al., 2020) samples collected from Zhongnan Hospital of Wuhan University from January 20 to February 9, 2020, were positive. We found that 28/42 (66.67%) (Chen et al., 2020) samples from the USA were positive. The samples were collected from Massachusetts (Wu et al., 2020), Bozeman (Nemudryi et al., 2020), New Haven (Peccia et al., 2020), and Southeast Virginia (Gonzalez et al., 2020) from March 18–25, 2020, March 27, 2020, March 19 to May 1, 2020, and March 11 to July 27, 2020, respectively, and 10/14 (71%), 7/7 (100%), 44/44 (100%), and 98/198 (49.5%) samples were positive, respectively.( Fig. 4).
Fig. 3.
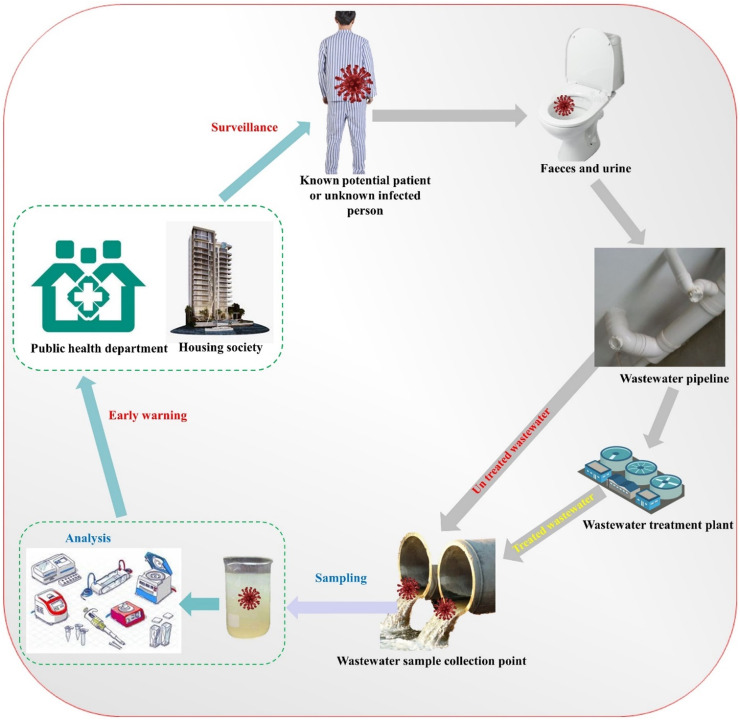
Conceptual model of the wastewater-based epidemiology (WBE) surveillance and early warning system for infectious disease epidemics caused by SARS-CoV-2 pathogens (Mao et al., 2020, Mao et al., 2020).
Fig. 4.
Spatial distribution of SARS-CoV-2 in wastewater systems worldwide.
Medema et al. (2020) first reported detection of SARS-CoV-2 in wastewater system samples collected from WWTPs in the Netherlands; 14/24 (58%) untreated wastewater samples were positive. In this first study, we used two different commercial systems, the RNeasy Power Microbiome Kit and the Nuclisens kit, in combination with the semi-automated KingFisher mL purification system to extract RNA from SARS-CoV-2 viral particles in wastewater samples. For identification, the one-step RT-qPCR method was used with primers specific for three regions (N1-N3) of the nucleocapsid protein gene (N) and envelope protein gene (E). A quantitative culture assay for F-specific RNA phage was additionally performed in this study to indirectly evaluate the efficacy of SARS-CoV-2 recovery through the purification and concentration treatment steps, and the effect of these sample processing steps on the viability of the viruses was examined.
Wurtzer et al. (2020) investigated 23/23 (100%) untreated wastewater samples that were found to be positive; the samples were collected from three main WWTPs in Paris and analysed using RT-qPCR with primers mainly targeting the RNA-dependent RNA polymerase gene (RdRP) and envelope protein gene (E). Following concentration via ultracentrifugation, the viral genome was extracted using an optimized protocol from the commercial PowerFecal Pro kit on a QIAsymphony extractor (Qiagen).
Through RT-PCR, La Rosa et al. (2020) assessed 6/12 (50%) untreated wastewater samples from an Italian WWTP that were SARS-CoV-2 positive. This study used a group of primers for SARS-CoV-2: a newly built set specifically targeting ORF1ab and an available set mainly intended for the pharyngeal swab, specific for the spike protein gene (S). In this study, wide-range primers, which were primarily developed before quantifying the new coronavirus strain, were also examined, amplifying a conserved region of the ORF1ab of Coronaviridae members. In contrast, these specific primers did not give signals due to nucleotide variations in this region discovered with subsequent sequencing of the recent SARS-CoV-2 strain.
Using RT-qPCR, Ahmed et al. (2020) found that 2/9 (22%), with a maximum concentration of 1.2 × 102 GC/L, wastewater samples were SARS-CoV-2 positive in Australia, Queensland. In this study, RT-qPCR was used with the primers N_Sarbeco and NIID_2019-nCOV_N specific for the nucleocapsid protein gene (N). A one-step kit RT-qPCR was adopted, and reverse transcription and qPCR occurred properly in the identical reaction.
Based on RT-qPCR analysis, Kumaret et al. (2020) reported that 20/20 (22%) wastewater samples were positive for SARS-CoV-2, with a concentration ranging from 5.6 × 10–3.5 × 102 GC/L, in Ahmedabad, India. In this study, the viral RNA genome was recovered using a NucleoSpin® RNA Virus Kit (Macherey-Nagel GmbH and Co. KG, Germany). RT-qPCR was employed with primers targeting the Spike protein gene (S), nucleocapsid protein gene (N), and ORF1ab.
Yaqub et al. (2020) reported that 21/78 (26.9%) wastewater samples from Lahore, Pakistan, were positive for SARS-CoV-2 at concentrations ranging from 2.67 × 102 to 3.60 × 104 GC/L, and the quantitative analysis was performed using RT-qPCR. The viral RNA genome was recovered in BSL-3 of IM, UVAS, using RT-qPCR with primers targeting the ORF1ab.
Or et al. (2020) reported 10/26 (38.5%) wastewater samples collected from various SARS-CoV-2-positive samples detected in Israel, and the quantitative analysis was conducted using RT-qPCR. This study examined the viral RNA genome isolated using a Spin star viral nucleic acid kit 1.0 (ADT Biotech, Phileo Damansara 1, Petaling Jaya Part No.811803). RT-qPCR with primers targeting the Envelope protein gene (E) was also employed.
Using RT-qPCR, Nemudryi et al. (2020) found that 7/7 (22%) wastewater samples from Bozeman, USA, were positive for SARS-CoV-2, with a maximum concentration > 3 × 105 GC/L. In this study, Nemudryi et al. (2020) also assessed the phylogenetic relationship of the isolated SARS-CoV-2 genome with the other global sequences by using ten available primer pairs for amplification together with non-quantitative RT-PCR and sequenced several polymorphous regions diffused in the genome. In contrast, Wu et al. (2020) assessed 7/7 (22%) samples with a maximum concentration > 2 × 104 GC/L, Peccia et al. (2020) assessed 44/44 (100%) with a concentration ranging from 1.7 × 106 to 4.6 × 108 GC/L, and Gonzalez et al. (2020) evaluated 98/198 (49.5%) samples with a concentration ranging from 101 to 104 GC/L among SARS-CoV-2-positive wastewater samples detected in Massachusetts, New Haven, and Southeast Virginia, USA, respectively.
Similarly, Randazzo et al. (2020a) evaluated 35/42 (83%), Randazzo et al. (2020b) reported 12/15 (80%) samples with concentrations ranging from 105 to 106 GC/L, and Balboa et al. (2020) assessed 2/18 (11.1%) secondary and 5/5 (100%) untreated wastewater samples that were SARS-CoV-2-positive in Murcia, Valencia, and Ourense, Spain, respectively. Furthermore, a study conducted by Vallejo et al. applied a wastewater detection approach in the urban area of A Coruña (Spain) with the key objective of developing novel statistical regression models that could be used to assess the dynamics of the COVID-19 epidemic among the population of 369,098. The results of the regression models suggested that the real number of individuals infected with COVID-19 was determined with a reliability of approximately 90% (Vallejo et al., 2020).
A study in Wuhan, China, conducted by Zhang et al. (2020) revealed that SARS-CoV-2 genomes were not detected in the influent of a hospital septic tank (after a primary disinfection tank). Nonetheless, 4/15 (26.7%) in effluent samples were found to be positive, with concentrations ranging from 0.50 × 103 to 1.87 × 105 GC/L, after disinfection with sodium hypochlorite. The SARS-CoV-2 viral genomes in the septic tank effluent were detected after the free chlorine concentration had dropped to a non-detectable level. The presence in the influent and absence in the effluent indicated the release of embedded SARS-CoV-2 in human stool particles, which provided protection from the treatment process. Though a viability analysis was not performed in the latter research, the results suggested the need for more research to verify that the wastewater or sewage treatment systems are not SARSCoV-2-spreading pathways.
Additionally, details regarding the distribution of these reports, the nucleic acid extraction and detection/quantification methods used, the concentration/pre-treatment methods used, the target genes, the total number of samples analysed, the water type, the number of positive samples, and the concentration range (GC/L) of SARS-CoV-2 in wastewater systems worldwide are presented in Table 2.
Overall, our results showed that the total positive SARS-CoV-2 detection rate in effluent wastewater systems worldwide is approximately 52.5% (375/715). The positive detection rates measured in America, Asia, Europe, and Australia were 60.5% (159/263), > 55.5% (112/222), > 46.2% (102/221) and > 22% (2/9), respectively ( Fig. 5A and B). These results indicate that the incidence of SARS-CoV-2 in effluent wastewater systems poses a severe epidemiological health threat in America, Asia, and Europe.
Fig. 5.
SARS-CoV-2 detected in effluent wastewater systems in different countries located on the American, Asian, and European continents and the positive detection rates (A). Global positive detection rate (B).
Nevertheless, despite various studies showing that transmission of the virus can occur via the faecal-oral axis (Ding and Liang, 2020, Vallejo et al., 2020), inadequate evidence is available to confirm this route of transmission (Vallejo et al., 2020, Ronchi et al., 2020). Moreover, no evidence is available to demonstrate infection via wastewater, which might lead to uncertainty regarding the presence of SARS-CoV-2 in wastewater systems (Cha et al., 2020, Chin et al., 2020, Yeo et al., 2020). Considering that several respiratory viruses other than EV, including SARS-CoV-2, have been identified in wastewater systems, other respiratory infectious disease-causing viruses in sewage still require attention to reduce the risk of environmental transmission through avenues such as aerosol inhalation.
5. Understanding the ongoing COVID-19 pandemic via wastewater surveillance
With respect to understanding ongoing infectious diseases via wastewater surveillance, WBE is an important approach that uses wastewater to identify the transmission of viruses to the public, and it provides an opportunity to assess the occurrence, distribution, and genetic diversity of viruses (Choi et al., 2018, Kitajima et al., 2020). In comparison to patient testing, WBE has advantages; for instance, it is a cost-efficient method of acquiring extensive population data and of early detection (Hart and Halden, 2020). WBE can now be used to detect and manage SARS-CoV-2, which is associated with severe infectious disease transmission among communities (Bivins et al., 2020). A conceptual model of a WBE and early warning system for epidemics of infectious diseases caused by SARS-CoV-2 pathogens is presented in Fig. 2 (Mao et al., 2020, Mao et al., 2020).
Fig. 2.
Positive detection rates of different human viruses in effluent wastewater systems worldwide.
One study has shown that SARS-CoV-2 viral RNA is detectable in human faeces from days to a week prior to the start of symptoms (Chen et al., 2020). Similarly, another study suggested that monitoring SARS-CoV-2 RNA concentrations in wastewater might predict COVID-19 epidemics seven days before they are revealed by individual patient testing and three days before they are revealed by hospital admission (Peccia et al., 2020). Thus, these studies conclude that the presence of viral DNA in wastewater is a significant indicator that can be used to detect hotspots in local community regions. Therefore, while the tool has a comparatively low cost, it offers an early warning system that can be used to identify new epidemics, trends in existing outbreaks, and the occurrence of contagions.
SARS-CoV-2 is known to primarily cause asymptomatic/paucisymptomatic infections (Prezioso et al., 2020), and it is difficult to assess the actual degree of viral spread/circulation among the public and make comparisons among diverse countries that have differing clinical analytic testing competencies and may even use different diagnostic approaches or assays (Kitajima et al., 2020). Several uncertainties associated with the use of WBE have been noted; these include changes in the rate of viral excretion by infected persons (Choi et al., 2018), temporal delays, spatial inconsistencies due to travel and time, precipitation dilution, inactivation during sample transport, infrequent clinical testing, the stability of the viral genome in wastewater, sampling variability, and the lack of sensitive detection assays that can detect low virus loads, and these uncertainties limit the technique’s ability to detect and quantify viruses (Kitajima et al., 2020). Therefore, in the future, additional work should be performed in this research field, including efficient standardisation of sampling, improvements in quality control, and development of analysis methods. For example, Bivins et al. called for a global collaborative to maximise contributions in the fight against COVID-19 using wastewater analysis (Bivins et al., 2020).
Despite these limitations, several efforts to develop wastewater surveillance methods for SARS-CoV-2 are ongoing (Hart and Halden, 2020, Daughton, 2020). As mentioned above, there are several preliminary reports on the molecular detection of SARS-CoV-2 viral RNA in wastewater in the Netherlands, Turkey, India, Pakistan, Australia, France, and the USA. The findings of these reports and other ongoing efforts worldwide might contribute to epidemiological modelling of the occurrence of SARS-CoV-2 in communities and might serve as an indicator of its threat to communities endeavouring to slow the spread of the contagion. To increase human acceptance of wastewater scrutiny, an agenda highlighting the moral issues associated with primary access to hygiene, privacy, and privileges might be required (Kitajima et al., 2020). WBE has the advantage of providing epidemiological knowledge regarding public infection/disease occurrence without identifying the affected individuals, which occurs when the outcomes of medical diagnoses during current COVID-19 epidemics are tabulated (Hirano and Murakami, 2020).
6. Conclusions and future outlook
A variety of viral pathogens are frequently reported in sewage in different countries on diverse continents, including Africa, North America, Asia, Europe, South America, and Australia. Based on individual studies reporting the number of samples positive for viruses, our results showed that the total positive detection rate for various human viruses such as NoVs, EVs, HAV, and AdVs in effluent wastewater systems worldwide is approximately 48.3% (2396/4963). Detection rates of 54.8% (560/1022), 54.1% (1689/913), 41.4% (273/660), 39.2% (51/130), and 38.6% (599/1553) were measured in South America, Africa, Asia, North America, and Europe, respectively.
Although some viral pathogens change seasonally, most are irregularly detected. In South Africa, Tunisia, and Egypt, the effluents of some sewage treatment plants do not meet standards and represented a potential threat to public health through environmental transmission when HAdV entered the aqueous environment. Moreover, municipal wastewater effluents that contain a variety of human viruses can circulate in the environment when the water is used for agricultural irrigation or for wastewater reclamation and reuse. SARS-CoV-2 has been detected in wastewater from France, the Netherlands, the Czech Republic, Australia, Italy, Israel, Japan, Spain, Turkey, India, Pakistan, China, and the USA. Worldwide, the positive detection rate for SARS-CoV-2 in effluent wastewater systems is approximately 52.5% (375/715); this is obviously linked to the current pandemic. The rank order of SARS-CoV-2 positive detection rates is America > Asia > Europe > Australia. Based on our findings, the levels of SARS-CoV-2 and other viruses in effluent wastewater systems in America are comparatively higher than those in the rest of the world. The high incidence of SARS-CoV-2, the causative agent of the COVID-19 epidemic, and other viruses in American wastewater systems indicates that adverse human health effects may be higher in America than on other continents. Wastewater may also pose a risk to environmental and public health, which indicates that the fate of SARS-CoV-2 and its transfer in the environment must be better understood.
The foregoing considerations show that it is essential to take adequate measures to monitor the risk of sewage-mediated transmission. Although viral pathogens in sewage represent a risk, sewage can also provide efficient information and an early warning of the presence of infectious viral diseases, and this information can be used to improve public security in certain areas, such as in the community. In recent years, the use of WBE to assess drug abuse in community populations has produced initial results.
Some experts have attempted to use sewage analysis to screen for potential viral carriers and asymptomatic patients, and this approach has developed into an early warning system. Several efforts to develop WBE surveillance methods for SARS-CoV-2 via wastewater are ongoing. WBE provides an efficient, low-cost surveillance tool and an early warning system for identification of new epidemics, evaluation of trends in existing outbreaks and prediction of the occurrence of contagions.
These global reports might contribute to epidemiological modelling of the occurrence of SARS-CoV-2 in communities and help investigators determine how to use wastewater information as a threat indicator for communities endeavouring to slow the spread of the contagion.
Knowledge gaps remain regarding the possible role of wastewater systems in the transmission of SARS-CoV-2 viral RNA. The presence of SARS-CoV-2 viral RNA in different environmental matrices, including wastewater systems, is unknown.
Ongoing studies have suggested that the stability of SARS-CoV-2 viral RNA in water is similar to its stability in aerosols and on surfaces. In future research, similar tools should be used to measure the stability of SARS-CoV-2 viral RNA in different water systems.
At present, RT-qPCR assays are mainly designed for medical specimen testing and are being used to detect SARS-CoV-2 viral RNA in wastewater samples. The application of different assays to different water matrices might produce conflicting results. Current WBE surveillance methods must focus more attention on improving the sensitivity of SARS-CoV-2 viral RNA detection in wastewater systems.
In the future, individuals belonging to communities in some countries may face some degree of health risk from various viruses present in wastewater systems. On the one hand, adequate measures must be taken to prevent the risk of wastewater transmission; on the other hand, the presence of viruses in wastewater can be used to provide new perspectives. For example, we can make full use of the information available from sewage to evaluate public health and to further prevent the outbreak of large-scale infectious diseases while preventing the spread of viral pathogens in wastewater systems.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was funded by the Science and Technology Program of Guizhou Province ([Grant No. (2019)5618; (2019)4428] and Qiankehe Zhicheng [2020]4Y190), STS of CAS (KFJ-STS-QYZD-185), Chinese Academy of Sciences, President’s International Fellowship Initiative (CAS, PIFI) for 2021 (2021PC0058; [2020]4Y190), STS of CAS Sciences, President’s International Fellowship Initiative (CAS, PIFI) for 2021; ZY thnaks UK NERC Fellowship grant (NE/R013349/2). Dr. Nabeel Khan Niazi is thankful to the University of Agriculture in Faisalabad, Pakistan. The authors would also like to thank all the anonymous reviewers for their constructive comments that significantly improved the quality of the manuscript.
Editor: Dr. R Teresa
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jhazmat.2021.125439.
Appendix A. Supplementary material
Supplementary material.
.
References
- Abe M., Ueki Y., Miura T., Kimura S., Suzuki Y., Sugawara N., Masago Y., Omura T., Watanabe S. Detection of human parechoviruses in clinical and municipal wastewater samples in Miyagi, Japan, in 2012–2014. Jpn. J. Infect. Dis. 2016;69:414–417. doi: 10.7883/yoken.JJID.2015.551. [DOI] [PubMed] [Google Scholar]
- Adefisoye M.A., Nwodo U.U., Green E., Okoh A.I. Quantitative PCR detection and characterisation of human adenovirus, rotavirus and hepatitis A virus in discharged effluents of two wastewater treatment facilities in the Eastern Cape, South Africa. Food Environ. Virol. 2016;8:262–274. doi: 10.1007/s12560-016-9246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens E.M., Farkas K., Harrison C., Jones D.L., Allison H.E., McCarthy A.J. Viromic analysis of wastewater input to a river catchment reveals a diverse assemblage of RNA viruses. mSystems. 2018;3:e00025–00018. doi: 10.1128/mSystems.00025-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad T., Waheed Y., Tahir S., Safi S.Z., Fatima K., Afzal M.S., Qadri I. Frequency of HEV contamination in sewerage waters in Pakistan. J. Infect. Dev. Ctries. 2010;4:842–845. doi: 10.3855/jidc.612. [DOI] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali W., Mao K., Zhang H., Junaid M., Xu N., Rasool A., Feng X., Yang Z. Comprehensive review of the basic chemical behaviours, sources, processes, and endpoints of trace element contamination in paddy soil-rice systems in rice-growing countries. J. Hazard. Mater. 2020;397 doi: 10.1016/j.jhazmat.2020.122720. [DOI] [PubMed] [Google Scholar]
- Aminipour M., Ghaderi M., Harzandi N. First occurrence of saffold virus in sewage and river water samples in Karaj, Iran. Food Environ. Virol. 2020;12:75–80. doi: 10.1007/s12560-019-09415-y. [DOI] [PubMed] [Google Scholar]
- Arora S., Nag A., Sethi J., Rajvanshi J., Saxena S., Shrivastava S.K., Gupta A.B. Sewage surveillance for the presence of SARS-CoV-2 genome as a useful wastewater based epidemiology (WBE) tracking tool in India. MedRxiv. 2020;82:2823–2836. doi: 10.2166/wst.2020.540. [DOI] [PubMed] [Google Scholar]
- Assis A.S.F., Fumian T.M., Miagostovich M.P., Drumond B.P., da Rosa e Silva M.L. Adenovirus and rotavirus recovery from a treated effluent through an optimized skimmed-milk flocculation method. Environ. Sci. Pollut. Res. 2018;25:17025–17032. doi: 10.1007/s11356-018-1873-x. [DOI] [PubMed] [Google Scholar]
- Baez P.A., Lopez M.C., Duque-Jaramillo A., Pelaez D., Molina F., Navas M.-C. First evidence of the hepatitis E virus in environmental waters in Colombia. PLoS One. 2017;12 doi: 10.1371/journal.pone.0177525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodriguez S., Martínez-Lamas L., Vasallo F., Regueiro B., Lema J. The fate of SARS-COV-2 in WWTPs points out the sludge line as a suitable spot for monitoring. MedRxiv. 2020 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboa S., Mauricio-Iglesias M., Rodríguez S., Martínez-Lamas L., Vasallo F.J., Regueiro B., Lema J.M. The fate of SARS-CoV-2 in WWTPs points out the sludge line as a suitable spot for monitoring. medRxiv. 2020 doi: 10.1016/j.scitotenv.2021.145268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.Q., Gerba C.P. Rethinking the significance of reovirus in water and wastewater. Food Environ. Virol. 2016;8:161–173. doi: 10.1007/s12560-016-9250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer S., Szewzyk R., Gnirss R., Johne R., Selinka H.-C. Detection and Characterization of hepatitis E Virus Genotype 3 in Wastewater and Urban Surface Waters in Germany. Food Environ. Virol. 2020;12:137–147. doi: 10.1007/s12560-020-09424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhunia A.K. Foodborne Microbial Pathogens. Springer; 2018. Biology of Microbial Pathogens; pp. 25–42. [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J. ACS Publications; 2020. Wastewater-Based Epidemiology: Global Collaborative to Maximize Contributions in the Fight Against COVID-19, in. [DOI] [PubMed] [Google Scholar]
- Bosch A., Pintó R.M., Guix S. Human astroviruses. Clin. Microbiol. Rev. 2014;27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman N.E., Fout G.S., Keely S.P. Retrospective surveillance of wastewater to examine seasonal dynamics of enterovirus infections. mSphere. 2017;2:e00099–00017. doi: 10.1128/mSphere.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruning A.H., Leeflang M.M., Vos J.M., Spijker R., de Jong M.D., Wolthers K.C., Pajkrt D. Rapid tests for influenza, respiratory syncytial virus, and other respiratory viruses: a systematic review and meta-analysis. Clin. Infect. Dis. 2017;65:1026–1032. doi: 10.1093/cid/cix461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burutarán L., Lizasoain A., García M., Tort L.F.L., Colina R., Victoria M. Detection and molecular characterization of aichivirus 1 in wastewater samples from Uruguay. Food Environ. Virol. 2016;8:13–17. doi: 10.1007/s12560-015-9217-1. [DOI] [PubMed] [Google Scholar]
- Carraturo F., Del Giudice C., Morelli M., Cerullo V., Libralato G., Galdiero E., Guida M. Persistence of SARS-CoV-2 in the environment and COVID-19 transmission risk from environmental matrices and surfaces. Environ. Pollut. 2020;265 doi: 10.1016/j.envpol.2020.115010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova L., Rutala W.A., Weber D.J., Sobsey M.D. Survival of surrogate coronaviruses in water. Water Res. 2009;43:1893–1898. doi: 10.1016/j.watres.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha M.H., Regueiro M., Sandhu D.S. Gastrointestinal and hepatic manifestations of COVID-19: a comprehensive review. World J. Gastroenterol. 2020;26:2323–2331. doi: 10.3748/wjg.v26.i19.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen L., Deng Q., Zhang G., Wu K., Ni L., Yang Y., Liu B., Wang W., Wei C. The presence of SARS‐CoV‐2 RNA in the feces of COVID‐19 patients. J. Med. Virol. 2020;92:833–840. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- Chin A., Chu J., Perera M., Hui K., Yen H.-L., Chan M., Peiris M., Poon L. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1 doi: 10.1016/S2666-5247(20)30003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P.M., Tscharke B.J., Donner E., O'Brien J.W., Grant S.C., Kaserzon S.L., Mackie R., O'Malley E., Crosbie N.D., Thomas K.V. Wastewater-based epidemiology biomarkers: past, present and future. TrAC Trends Anal. Chem. 2018;105:453–469. [Google Scholar]
- Ciocca M. Clinical course and consequences of hepatitis A infection. Vaccine. 2000;18:S71–S74. doi: 10.1016/s0264-410x(99)00470-3. [DOI] [PubMed] [Google Scholar]
- Clemens H., Pang L., Morgan L.K., Weaver L. Attenuation of rotavirus, MS2 bacteriophage and biomolecule-modified silica nanoparticles in undisturbed silt loam over gravels dosed with onsite wastewater. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115272. [DOI] [PubMed] [Google Scholar]
- Corpuz M.V.A., Buonerba A., Vigliotta G., Zarra T., Ballesteros F., Jr, Campiglia P., Belgiorno V., Korshin G., Naddeo V. Viruses in wastewater: occurrence, abundance and detection methods. Sci. Total Environ. 2020;745 doi: 10.1016/j.scitotenv.2020.140910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courault D., Albert I., Perelle S., Fraisse A., Renault P., Salemkour A., Amato P. Assessment and risk modeling of airborne enteric viruses emitted from wastewater reused for irrigation. Sci. Total Environ. 2017;592:512–526. doi: 10.1016/j.scitotenv.2017.03.105. [DOI] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Profio F., Melegari I., Palombieri A., Sarchese V., Arbuatti A., Fruci P., Marsilio F., Martella V., Di Martino B. High prevalence of hepatitis E virus in raw sewage in Southern Italy. Virus Res. 2019;272 doi: 10.1016/j.virusres.2019.197710. [DOI] [PubMed] [Google Scholar]
- Dienus O., Sokolova E., Nyström F., Matussek A., Löfgren S., Blom L., Pettersson T.J.R., Lindgren P.-E. Norovirus dynamics in wastewater discharges and in the recipient drinking water source: long-term monitoring and hydrodynamic modeling. Environ. Sci. Technol. 2016;50:10851–10858. doi: 10.1021/acs.est.6b02110. [DOI] [PubMed] [Google Scholar]
- Ding S., Liang T.J. Is SARS-CoV-2 also an enteric pathogen with potential fecal–oral transmission? A COVID-19 virological and clinical review. Gastroenterology. 2020;159:53–61. doi: 10.1053/j.gastro.2020.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmahdy E.M., Ahmed N.I., Shaheen M.N.F., Mohamed E.-C.B., Loutfy S.A. Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health. 2019;17:287–294. doi: 10.2166/wh.2019.303. [DOI] [PubMed] [Google Scholar]
- Elmahdy E.M., Ahmed N.I., Shaheen M.N., Mohamed E.-C.B., Loutfy S.A. Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health. 2019;17:287–294. doi: 10.2166/wh.2019.303. [DOI] [PubMed] [Google Scholar]
- El-Senousy W.M., Abdel-Moneim A., Abdel-Latif M., El-Hefnawy M.H., Khalil R.G. Coxsackievirus B4 as a causative agent of diabetes mellitus Type 1: is there a role of inefficiently treated drinking water and sewage in virus spreading? Food Environ. Virol. 2018;10:89–98. doi: 10.1007/s12560-017-9322-4. [DOI] [PubMed] [Google Scholar]
- Estívariz C.F., Pérez-Sánchez E.E., Bahena A., Burns C.C., Gary H.E., García-Lozano H., Rey-Benito G., Peñaranda S., Castillo-Montufar K.V., Nava-Acosta R.S., Meschke J.S., Oberste M.S., Lopez-Martínez I., Díaz-Quiñonez J.A. Field performance of two methods for detection of poliovirus in wastewater samples, Mexico 2016–2017. Food Environ. Virol. 2019;11:364–373. doi: 10.1007/s12560-019-09399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Marshall M., Cooper D., McDonald J.E., Malham S.K., Peters D.E., Maloney J.D., Jones D.L. Seasonal and diurnal surveillance of treated and untreated wastewater for human enteric viruses. Environ. Sci. Pollut. Res. 2018;25:33391–33401. doi: 10.1007/s11356-018-3261-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Cooper D.M., McDonald J.E., Malham S.K., de Rougemont A., Jones D.L. Seasonal and spatial dynamics of enteric viruses in wastewater and in riverine and estuarine receiving waters. Sci. Total Environ. 2018;634:1174–1183. doi: 10.1016/j.scitotenv.2018.04.038. [DOI] [PubMed] [Google Scholar]
- Fenaux H., Chassaing M., Berger S., Gantzer C., Bertrand I., Schvoerer E. Transmission of hepatitis E virus by water: an issue still pending in industrialized countries. Water Res. 2019;151:144–157. doi: 10.1016/j.watres.2018.12.014. [DOI] [PubMed] [Google Scholar]
- Fioretti J.M., Rocha M.S., Fumian T.M., Ginuino A., da Silva T.P., de Assis M.R., Rodrigues Jd.S., Carvalho-Costa F.A., Miagostovich M.P. Occurrence of human sapoviruses in wastewater and stool samples in Rio De Janeiro, Brazil. J. Appl. Microbiol. 2016;121:855–862. doi: 10.1111/jam.13205. [DOI] [PubMed] [Google Scholar]
- Fioretti J.M., Fumian T.M., Rocha M.S., dos Santos Id.A.L., Carvalho-Costa F.A., de Assis M.R., Rodrigues Jd.S., Leite J.P.G., Miagostovich M.P. Surveillance of noroviruses in Rio De Janeiro, Brazil: occurrence of new GIV genotype in clinical and wastewater samples. Food Environ. Virol. 2018;10:1–6. doi: 10.1007/s12560-017-9308-2. [DOI] [PubMed] [Google Scholar]
- Fong T.-T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong T.-T., Phanikumar M.S., Xagoraraki I., Rose J.B. Quantitative detection of human adenoviruses in wastewater and combined sewer overflows influencing a Michigan river. Appl. Environ. Microbiol. 2010;76:715–723. doi: 10.1128/AEM.01316-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. What you need to know about the novel coronavirus. Nature. 2020 doi: 10.1038/d41586-020-00209-y. [DOI] [PubMed] [Google Scholar]
- Fumian T.M., Fioretti J.M., Lun J.H., dos Santos I.A.L., White P.A., Miagostovich M.P. Detection of norovirus epidemic genotypes in raw sewage using next generation sequencing. Environ. Int. 2019;123:282–291. doi: 10.1016/j.envint.2018.11.054. [DOI] [PubMed] [Google Scholar]
- Gerba C.P., Betancourt W.Q., Kitajima M., Rock C.M. Reducing uncertainty in estimating virus reduction by advanced water treatment processes. Water Res. 2018;133:282–288. doi: 10.1016/j.watres.2018.01.044. [DOI] [PubMed] [Google Scholar]
- Gonzales-Gustavson E., Rusiñol M., Medema G., Calvo M., Girones R. Quantitative risk assessment of norovirus and adenovirus for the use of reclaimed water to irrigate lettuce in Catalonia. Water Res. 2019;153:91–99. doi: 10.1016/j.watres.2018.12.070. [DOI] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M., Yetka K., Thompson H., Keeling D., Mitchell J., Gonzalez D. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyawali P., Croucher D., Hewitt J. Preliminary evaluation of BioFire FilmArray® Gastrointestinal Panel for the detection of noroviruses and other enteric viruses from wastewater and shellfish. Environ. Sci. Pollut. Res. 2018;25:27657–27661. doi: 10.1007/s11356-018-2869-2. [DOI] [PubMed] [Google Scholar]
- Hamza H., Hamza I.A. Oncogenic papillomavirus and polyomavirus in urban sewage in Egypt. Sci. Total Environ. 2018;610:1413–1420. doi: 10.1016/j.scitotenv.2017.08.218. [DOI] [PubMed] [Google Scholar]
- Hamza H., Leifels M., Wilhelm M., Hamza I.A. Relative abundance of human bocaviruses in urban sewage in Greater Cairo, Egypt. Food Environ. Virol. 2017;9:304–313. doi: 10.1007/s12560-017-9287-3. [DOI] [PubMed] [Google Scholar]
- Hamza H., Rizk N.M., Gad M.A., Hamza I.A. Pepper mild mottle virus in wastewater in Egypt: a potential indicator of wastewater pollution and the efficiency of the treatment process. Arch. Virol. 2019;164:2707–2713. doi: 10.1007/s00705-019-04383-x. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Kitajima M., Hata A., Torrey J.R., Masago Y., Sano D., Katayama H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018;135:168–186. doi: 10.1016/j.watres.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Haramoto E., Malla B., Thakali O., Kitajima M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. medRxiv. 2020 doi: 10.1016/j.scitotenv.2020.140405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart O.E., Halden R.U. Computational analysis of SARS-CoV-2/COVID-19 surveillance by wastewater-based epidemiology locally and globally: feasibility, economy, opportunities and challenges. Sci. Total Environ. 2020;730 doi: 10.1016/j.scitotenv.2020.138875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Murakami M. COVID-19: a new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52:731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelmsø M.H., Hellmér M., Fernandez-Cassi X., Timoneda N., Lukjancenko O., Seidel M., Elsässer D., Aarestrup F.M., Löfström C., Bofill-Mas S. Evaluation of methods for the concentration and extraction of viruses from sewage in the context of metagenomic sequencing. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State-nCo V.C.I. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes B., Beale D.J., Dennis P.G., Cook S., Ahmed W. Cross-comparison of human wastewater-associated molecular markers in relation to fecal indicator bacteria and enteric viruses in recreational beach waters. Appl. Environ. Microbiol. 2017;83:e00028–00017. doi: 10.1128/AEM.00028-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaconelli M., Divizia M., Della Libera S., Di Bonito P., Rosa G. La. Frequent detection and genetic diversity of human bocavirus in Urban Sewage Samples. Food Environ. Virol. 2016;8:289–295. doi: 10.1007/s12560-016-9251-7. [DOI] [PubMed] [Google Scholar]
- Ibrahim C., Cherif N., Hammami S., Pothier P., Hassen A. Quantification and genotyping of rotavirus A within two wastewater treatment processes. Clean Soil Air Water. 2016;44:393–401. [Google Scholar]
- Ibrahim C., Hammami S., Mejri S., Mehri I., Pothier P., Hassen A. Detection of Aichi virus genotype B in two lines of wastewater treatment processes. Microb. Pathog. 2017;109:305–312. doi: 10.1016/j.micpath.2017.06.001. [DOI] [PubMed] [Google Scholar]
- Ibrahim C., Hassen A., Pothier P., Mejri S., Hammami S. Molecular detection and genotypic characterization of enteric adenoviruses in a hospital wastewater. Environ. Sci. Pollut. Res. 2018;25:10977–10987. doi: 10.1007/s11356-018-1399-2. [DOI] [PubMed] [Google Scholar]
- Ibrahim C., Hammami S., Chérif N., Mejri S., Pothier P., Hassen A. Detection of Sapoviruses in two biological lines of Tunisian hospital wastewater treatment. Int. J. Environ. Health Res. 2019;29:400–413. doi: 10.1080/09603123.2018.1546835. [DOI] [PubMed] [Google Scholar]
- Ibrahim C., Hammami S., Pothier P., Khelifi N., Hassen A. The performance of biological and tertiary wastewater treatment procedures for rotaviruses A removal. Environ. Sci. Pollut. Res. 2020;27:5718–5729. doi: 10.1007/s11356-019-05487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Kitajima M., Kato T., Ishii S., Segawa T., Okabe S., Sano D. Target virus log10 reduction values determined for two reclaimed wastewater irrigation scenarios in Japan based on tolerable annual disease burden. Water Res. 2017;125:438–448. doi: 10.1016/j.watres.2017.08.057. [DOI] [PubMed] [Google Scholar]
- Jahne M.A., Brinkman N.E., Keely S.P., Zimmerman B.D., Wheaton E.A., Garland J.L. Droplet digital PCR quantification of norovirus and adenovirus in decentralized wastewater and graywater collections: implications for onsite reuse. Water Res. 2020;169 doi: 10.1016/j.watres.2019.115213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas L., Ogorzaly L., Lecellier G., Berteaux-Lecellier V., Cauchie H.-M., Langlet J. Detection of human enteric viruses in french polynesian wastewaters, environmental waters and giant clams. Food Environ. Virol. 2019;11:52–64. doi: 10.1007/s12560-018-9358-0. [DOI] [PubMed] [Google Scholar]
- Kaliakatsos A., Kalogerakis N., Manios T., Venieri D. Efficiency of two constructed wetland systems for wastewater treatment: removal of bacterial indicators and enteric viruses. J. Chem. Technol. Biotechnol. 2019;94:2123–2130. [Google Scholar]
- S. Kaya, A.E. Eskazan, N. Ay, B. Baysal, M.V. Bahadir, A. Onur, R. Duymus, Acute acalculous cholecystitis due to viral hepatitis A, Case reports in infectious diseases, 2013 (2013). [DOI] [PMC free article] [PubMed]
- Kazama S., Masago Y., Tohma K., Souma N., Imagawa T., Suzuki A., Liu X., Saito M., Oshitani H., Omura T. Temporal dynamics of norovirus determined through monitoring of municipal wastewater by pyrosequencing and virological surveillance of gastroenteritis cases. Water Res. 2016;92:244–253. doi: 10.1016/j.watres.2015.10.024. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Iker B.C., Pepper I.L., Gerba C.P. Relative abundance and treatment reduction of viruses during wastewater treatment processes—identification of potential viral indicators. Sci. Total Environ. 2014;488:290–296. doi: 10.1016/j.scitotenv.2014.04.087. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Gerba C.P. Genetically distinct genogroup IV norovirus strains identified in wastewater. Arch. Virol. 2016;161:3521–3525. doi: 10.1007/s00705-016-3036-z. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Rachmadi A.T., Iker B.C., Haramoto E., Gerba C.P. Temporal variations in genotype distribution of human sapoviruses and Aichi virus 1 in wastewater in Southern Arizona, United States. J. Appl. Microbiol. 2018;124:1324–1332. doi: 10.1111/jam.13712. [DOI] [PubMed] [Google Scholar]
- Kitajima M., Ahmed W., Bibby K., Carducci A., Gerba C.P., Hamilton K.A., Haramoto E., Rose J.B. SARS-CoV-2 in wastewater: state of the knowledge and research needs. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N., Oshiki M., Ito T., Segawa T., Hatamoto M., Kato T., Yamaguchi T., Kubota K., Takahashi M., Iguchi A., Tagawa T., Okubo T., Uemura S., Harada H., Motoyama T., Araki N., Sano D. Removal of human pathogenic viruses in a down-flow hanging sponge (DHS) reactor treating municipal wastewater and health risks associated with utilization of the effluent for agricultural irrigation. Water Res. 2017;110:389–398. doi: 10.1016/j.watres.2016.10.054. [DOI] [PubMed] [Google Scholar]
- Kocamemi B.A., Kurt H., Hacioglu S., Yarali C., Saatci A.M., Pakdemirli B. First Data-Set on SARS-CoV-2 Detection for Istanbul Wastewaters in Turkey. MedRxiv. 2020 [Google Scholar]
- Kocwa-Haluch R. Waterborne enteroviruses as a hazard for human health. Pol. J. Environ. Stud. 2001;10:485–488. [Google Scholar]
- Kumar M., Patel A.K., Shah A.V., Raval J., Rajpara N., Joshi M., Joshi C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020;746 doi: 10.1016/j.scitotenv.2020.141326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu A., McBride G., Wuertz S. Adenovirus-associated health risks for recreational activities in a multi-use coastal watershed based on site-specific quantitative microbial risk assessment. Water Res. 2013;47:6309–6325. doi: 10.1016/j.watres.2013.08.002. [DOI] [PubMed] [Google Scholar]
- Kuo D.H.-W., Simmons F.J., Blair S., Hart E., Rose J.B., Xagoraraki I. Assessment of human adenovirus removal in a full-scale membrane bioreactor treating municipal wastewater. Water Res. 2010;44:1520–1530. doi: 10.1016/j.watres.2009.10.039. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Pourshaban M., Iaconelli M., Muscillo M. Quantitative real-time PCR of enteric viruses in influent and effluent samples from wastewater treatment plants in Italy. Ann. dell'Ist. Super. di Sanita. 2010;46:266–273. doi: 10.4415/ANN_10_03_07. [DOI] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods-A scoping review. Water Res. 2020;179 doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Iaconelli M., Mancini P., Ferraro G.B., Veneri C., Bonadonna L., Lucentini L., Suffredini E. First detection of SARS-CoV-2 in untreated wastewaters in Italy. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B., van Schayck J.P., Mykytyn A.Z., Duimel H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levican J., Levican A., Ampuero M., Gaggero A. JC polyomavirus circulation in one-year surveillance in wastewater in Santiago, Chile. Infect. Genet. Evol. 2019;71:151–158. doi: 10.1016/j.meegid.2019.03.017. [DOI] [PubMed] [Google Scholar]
- Li H., Li W., She R., Yu L., Wu Q., Yang J., Hu F., Soomro M.H., Shi R., Hao W., Zhao Y., Mao J. Hepatitis E virus genotype 4 sequences detected in sewage from treatment plants of China. Food Environ. Virol. 2017;9:230–233. doi: 10.1007/s12560-016-9276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizasoain A., Tort L.F.L., García M., Gillman L., Alberti A., Leite J.P.G., Miagostovich M.P., Pou S.A., Cagiao A., Razsap A., Huertas J., Berois M., Victoria M., Colina R. Human enteric viruses in a wastewater treatment plant: evaluation of activated sludge combined with UV disinfection process reveals different removal performances for viruses with different features. Lett. Appl. Microbiol. 2018;66:215–221. doi: 10.1111/lam.12839. [DOI] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S.S., Prober C.G., Fischer M. Elsevier Health Sciences; 2017. Principles and Practice of Pediatric Infectious Diseases E-Book. [Google Scholar]
- López-Gálvez F., Randazzo W., Vásquez A., Sánchez G., Decol L.T., Aznar R., Gil M.I., Allende A. Irrigating lettuce with wastewater effluent: does disinfection with chlorine dioxide inactivate viruses? J. Environ. Qual. 2018;47:1139–1145. doi: 10.2134/jeq2017.12.0485. [DOI] [PubMed] [Google Scholar]
- Lun J.H., Crosbie N.D., White P.A. Genetic diversity and quantification of human mastadenoviruses in wastewater from Sydney and Melbourne, Australia. Sci. Total Environ. 2019;675:305–312. doi: 10.1016/j.scitotenv.2019.04.162. [DOI] [PubMed] [Google Scholar]
- Mabasa V.V., Meno K.D., Taylor M.B., Mans J. Environmental surveillance for noroviruses in selected South African Wastewaters 2015–2016: emergence of the Novel GII.17. Food Environ. Virol. 2018;10:16–28. doi: 10.1007/s12560-017-9316-2. [DOI] [PubMed] [Google Scholar]
- Majumdar M., Sharif S., Klapsa D., Wilton T., Alam M.M., Fernandez-Garcia M.D., Rehman L., Mujtaba G., McAllister G., Harvala H. Open Forum Infectious Diseases. Oxford University Press; US: 2018. Environmental surveillance reveals complex enterovirus circulation patterns in human populations; p. ofy250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallapaty S. How sewage could reveal true scale of coronavirus outbreak. Nature. 2020;580:176–177. doi: 10.1038/d41586-020-00973-x. [DOI] [PubMed] [Google Scholar]
- Mancini P., Bonanno Ferraro G., Iaconelli M., Suffredini E., Valdazo-González B., Della Libera S., Divizia M., La G. Rosa, molecular characterization of human Sapovirus in untreated sewage in Italy by amplicon-based Sanger and next-generation sequencing. J. Appl. Microbiol. 2019;126:324–331. doi: 10.1111/jam.14129. [DOI] [PubMed] [Google Scholar]
- Mao K., Zhang K., Du W., Ali W., Feng X., Zhang H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health. 2020;17:1–7. doi: 10.1016/j.coesh.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Zhang H., Yang Z. Can a paper-based device trace COVID-19 sources with wastewater-based epidemiology? Environ. Sci. Technol. 2020;54:3733–3735. doi: 10.1021/acs.est.0c01174. [DOI] [PubMed] [Google Scholar]
- Masago Y., Konta Y., Kazama S., Inaba M., Imagawa T., Tohma K., Saito M., Suzuki A., Oshitani H., Omura T. Comparative evaluation of real-time PCR methods for human noroviruses in wastewater and human stool. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos A., Mesquita J.R., Gonçalves D., Abreu-Silva J., Luxo C., Nascimento M.S. First detection and molecular characterization of hepatitis E virus in water from wastewater treatment plants in Portugal. Ann. Agric. Environ. Med. 2018;25:364–367. doi: 10.26444/aaem/90497. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. MedRxiv. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Meng X. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 2010;140:256–265. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M., Millet J., Vidalain P.-O., Law H., Vabret A., Lorin V., Escriou N., Albert M.L., Nal B., Tangy F. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J. Virol. 2012;86:7577–7587. doi: 10.1128/JVI.00269-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzanotte V., Marazzi F., Bissa M., Pacchioni S., Binelli A., Parolini M., Magni S., Ruggeri F.M., De Giuli Morghen C., Zanotto C., Radaelli A. Removal of enteric viruses and Escherichia coli from municipal treated effluent by zebra mussels. Sci. Total Environ. 2016;539:395–400. doi: 10.1016/j.scitotenv.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Miura T., Lhomme S., Le Saux J.-C., Le Mehaute P., Guillois Y., Couturier E., Izopet J., Abranavel F., Le Guyader F.S. Detection of Hepatitis E virus in sewage after an outbreak on a French Island. Food Environ. Virol. 2016;8:194–199. doi: 10.1007/s12560-016-9241-9. [DOI] [PubMed] [Google Scholar]
- Mlejnkova H., Sovova K., Vasickova P., Ocenaskova V., Jasikova L., Juranova E. Preliminary study of Sars-Cov-2 occurrence in wastewater in the Czech Republic. Int. J. Environ. Res. Public Health. 2020;17:5508. doi: 10.3390/ijerph17155508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazeni M., Nikaeen M., Hadi M., Moghim S., Mouhebat L., Hatamzadeh M., Hassanzadeh A. Estimation of health risks caused by exposure to enteroviruses from agricultural application of wastewater effluents. Water Res. 2017;125:104–113. doi: 10.1016/j.watres.2017.08.028. [DOI] [PubMed] [Google Scholar]
- Nemudryi A., Nemudraia A., Surya K., Wiegand T., Buyukyoruk M., Wilkinson R., Wiedenheft B. Temporal detection and phylogenetic assessment of SARS-CoV-2 in municipal wastewater. MedRxiv. 2020 doi: 10.1016/j.xcrm.2020.100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen G.T., Pu J., Miura T., Ito H., Kazama S., Konta Y., Van Le A., Watanabe T. Oyster contamination with Human Noroviruses Impacted by Urban Drainage and Seasonal Flooding in Vietnam. Food Environ. Virol. 2018;10:61–71. doi: 10.1007/s12560-017-9325-1. [DOI] [PubMed] [Google Scholar]
- Onosi O., Upfold N.S., Jukes M.D., Luke G.A., Knox C. The first molecular detection of Aichi Virus 1 in raw sewage and mussels collected in South Africa. Food Environ. Virol. 2019;11:96–100. doi: 10.1007/s12560-018-9362-4. [DOI] [PubMed] [Google Scholar]
- Or I.B., Yaniv K., Shagan M., Ozer E., Erster O., Mendelson E., Mannasse B., Shirazi R., Kramarsky-Winter E., Nir O. Regressing SARS-CoV-2 sewage measurements onto COVID-19 burden in the population: a proof-of-concept for quantitative environmental surveillance. medRxiv. 2020 doi: 10.3389/fpubh.2021.561710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuolale O., Okoh A. Human enteric bacteria and viruses in five wastewater treatment plants in the Eastern Cape, South Africa. J. Infect. Public Health. 2017;10:541–547. doi: 10.1016/j.jiph.2016.11.012. [DOI] [PubMed] [Google Scholar]
- Ouardani I., Turki S., Aouni M., Romalde J.L. Detection and molecular characterization of Hepatitis A virus from tunisian wastewater treatment plants with different secondary treatments. Appl. Environ. Microbiol. 2016;82:3834–3845. doi: 10.1128/AEM.00619-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- P. Vohra, HEPATITISA INFECTION, Tropical Hepato-Gastroenterology, 2010, 277.
- Pavlinac P.B., John-Stewart G.C., Naulikha J.M., Onchiri F.M., Denno D.M., Odundo E.A., Singa B.O., Richardson B.A., Walson J.L. High-risk enteric pathogens associated with HIV infection and HIV exposure in Kenyan children with acute diarrhoea. AIDS (Lond. Engl. ) 2014;28:2287–2296. doi: 10.1097/QAD.0000000000000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M. SARS-CoV-2 RNA concentrations in primary municipal sewage sludge as a leading indicator of COVID-19 outbreak dynamics. medRxiv. 2020 [Google Scholar]
- Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral. Sci. 2020;12:1–6. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennino F., Nardone A., Montuori P., Aurino S., Torre I., Battistone A., Delogu R., Buttinelli G., Fiore S., Amato C., Triassi M. Large-scale survey of human enteroviruses in wastewater treatment plants of a metropolitan area of southern Italy, food and environmental. Virology. 2018;10:187–192. doi: 10.1007/s12560-017-9331-3. [DOI] [PubMed] [Google Scholar]
- Prado T., Silva D.M., Guilayn W.C., Rose T.L., Gaspar A.M.C., Miagostovich M.P. Quantification and molecular characterization of enteric viruses detected in effluents from two hospital wastewater treatment plants. Water Res. 2011;45:1287–1297. doi: 10.1016/j.watres.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Prevost B., Lucas F., Goncalves A., Richard F., Moulin L., Wurtzer S. Large scale survey of enteric viruses in river and waste water underlines the health status of the local population. Environ. Int. 2015;79:42–50. doi: 10.1016/j.envint.2015.03.004. [DOI] [PubMed] [Google Scholar]
- Prevost B., Lucas F.S., Ambert-Balay K., Pothier P., Moulin L., Wurtzer S. Deciphering the diversities of astroviruses and noroviruses in wastewater treatment plant effluents by a high-throughput sequencing method. Appl. Environ. Microbiol. 2015;81:7215–7222. doi: 10.1128/AEM.02076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezioso C., Marcocci M.E., Palamara A.T., De Chiara G., Pietropaolo V. The “Three Italy” of the COVID-19 epidemic and the possible involvement of SARS-CoV-2 in triggering complications other than pneumonia. J. Neurovirol. 2020;26:311–323. doi: 10.1007/s13365-020-00862-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Lee B.E., Ruecker N.J., Neumann N., Ashbolt N., Pang X. A one-step centrifugal ultrafiltration method to concentrate enteric viruses from wastewater. J. Virol. Methods. 2016;237:150–153. doi: 10.1016/j.jviromet.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Qiu Y., Li Q., Lee B.E., Ruecker N.J., Neumann N.F., Ashbolt N.J., Pang X. UV inactivation of human infectious viruses at two full-scale wastewater treatment plants in Canada. Water Res. 2018;147:73–81. doi: 10.1016/j.watres.2018.09.057. [DOI] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Ferrando E.C., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. medRxiv. 2020 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Cuevas-Ferrando E., Simón P., Allende A., Sánchez G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk N., Herrawy A.A.-, Gad M., Shaheen M., Elmahdy E. Existence and removal of rotaviruses group A and cryptosporidium species in a wastewater treatment plant. Pol. J. Environ. Stud. 2019;28:4331–4339. [Google Scholar]
- Ronchi A., Pagliuca F., Zito Marino F., Montella M., Franco R., Cozzolino I. Interventional cytopathology in the COVID‐19 era. Cytopathology. 2020;31:509–513. doi: 10.1111/cyt.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Kitajima M., Campillo M.E., Gerba C.P., Pepper I.L. Virus reduction during advanced bardenpho and conventional wastewater treatment processes. Environ. Sci. Technol. 2016;50:9524–9532. doi: 10.1021/acs.est.6b01384. [DOI] [PubMed] [Google Scholar]
- Shaheen M.N., Elmahdy E.M. Environmental monitoring of astrovirus and norovirus in the Rosetta branch of the River Nile and the El-Rahawy drain, Egypt. Water Supply. 2019;19:1381–1387. [Google Scholar]
- Shaheen M.N.F., Abd El-Daim S.E., Ahmed N.I., Elmahdy E.M. Environmental monitoring of Aichi virus and human bocavirus in samples from wastewater treatment plant, drain, and River Nile in Egypt. J. Water Health. 2019;18:30–37. doi: 10.2166/wh.2019.075. [DOI] [PubMed] [Google Scholar]
- S. Sharif, Detection of SARs-CoV-2 in wastewater, using the existing environmental surveillance network: An epidemiological gateway to an early warning for COVID-19 in communities.
- Simhon A., Pileggi V., Flemming C.A., Bicudo J.R., Lai G., Manoharan M. Enteric viruses in municipal wastewater effluent before and after disinfection with chlorine and ultraviolet light. J. Water Health. 2019;17:670–682. doi: 10.2166/wh.2019.111. [DOI] [PubMed] [Google Scholar]
- Tandukar S., Sherchan S.P., Haramoto E. Applicability of crAssphage, pepper mild mottle virus, and tobacco mosaic virus as indicators of reduction of enteric viruses during wastewater treatment. Sci. Rep. 2020;10:3616. doi: 10.1038/s41598-020-60547-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakoli Nick S., Mohebbi S.R., Hosseini S.M., Mirjalali H., Alebouyeh M. Occurrence and molecular characterization of Torque teno virus (TTV) in a wastewater treatment plant in Tehran. J. Water Health. 2019;17:971–977. doi: 10.2166/wh.2019.137. [DOI] [PubMed] [Google Scholar]
- Tiwari S., Dhole T.N. Assessment of enteroviruses from sewage water and clinical samples during eradication phase of polio in North India. Virol. J. 2018;15:157. doi: 10.1186/s12985-018-1075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Dhole T.N. Assessment of enteroviruses from sewage water and clinical samples during eradication phase of polio in North India. Virol. J. 2018;15:1–8. doi: 10.1186/s12985-018-1075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng F.C., Huang H.C., Chi C.Y., Lin T.L., Liu C.C., Jian J.W., Hsu L.C., Wu H.S., Yang J.Y., Chang Y.W. Epidemiological survey of enterovirus infections occurring in Taiwan between 2000 and 2005: analysis of sentinel physician surveillance data. J. Med. Virol. 2007;79:1850–1860. doi: 10.1002/jmv.21006. [DOI] [PubMed] [Google Scholar]
- Vallejo J.A., Rumbo-Feal S., Conde-Perez K., Lopez-Oriona A., Tarrio J., Reif R., Ladra S., Rodino-Janeiro B.K., Nasser M., Cid A. Highly predictive regression model of active cases of COVID-19 in a population by screening wastewater viral load. medRxiv. 2020 [Google Scholar]
- Van Der Sanden S.M., Sachs N., Koekkoek S.M., Koen G., Pajkrt D., Clevers H., Wolthers K.C. Enterovirus 71 infection of human airway organoids reveals VP1-145 as a viral infectivity determinant. Emerg. Microbes Infect. 2018;7:1–9. doi: 10.1038/s41426-018-0077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela M.F., Ouardani I., Kato T., Kadoya S., Aouni M., Sano D., Romalde J.L. Sapovirus in wastewater treatment plants in Tunisia: prevalence, removal, and genetic characterization. Appl. Environ. Microbiol. 2018;84:e02093–02017. doi: 10.1128/AEM.02093-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade T.J., Calderon R.L., Sams E., Beach M., Brenner K.P., Williams A.H., Dufour A.P. Rapidly measured indicators of recreational water quality are predictive of swimming-associated gastrointestinal illness. Environ. Health Perspect. 2006;114:24–28. doi: 10.1289/ehp.8273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Li J., Guo T., Zhen B., Kong Q., Yi B., Li Z., Song N., Jin M., Xiao W. Concentration and detection of SARS coronavirus in sewage from Xiao Tang Shan Hospital and the 309th Hospital of the Chinese People's Liberation Army. Water Sci. Technol. 2005;52:213–221. [PubMed] [Google Scholar]
- Warriner K. Improving the Safety of Fresh Fruit And Vegetables. Elsevier; 2005. Pathogens in vegetables; pp. 3–43. [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Wong S.H., Lui R.N., Sung J.J. Covid‐19 and the digestive system. J. Gastroenterol. Hepatol. 2020;35:744–748. doi: 10.1111/jgh.15047. [DOI] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Gu X., Lee W.L., Kauffman K., Hanage W., Matus M., Ghaeli N., Endo N. SARS-CoV-2 titers in wastewater are higher than expected from clinically confirmed cases. mSystems. 2020;5 doi: 10.1128/mSystems.00614-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Liu W.-T. Determination of virus abundance, diversity and distribution in a municipal wastewater treatment plant. Water Res. 2009;43:1101–1109. doi: 10.1016/j.watres.2008.11.039. [DOI] [PubMed] [Google Scholar]
- Wurtzer S., Marechal V., Mouchel J.-M., Moulin L. Time course quantitative detection of SARS-CoV-2 in Parisian wastewaters correlates with COVID-19 confirmed cases. MedRxiv. 2020 [Google Scholar]
- Yaqub T., Nawaz M., Shabbir M.Z., Ali M.A., Altaf I., Raza S., Shabbir M.A.B., Ashraf M.A., Aziz S.Z., Cheema S.Q. A longitudinal survey for genome-based identification of SARS-CoV-2 in sewage water in selected lockdown areas of Lahore city, Pakistan; a potential approach for future smart lockdown strategy. medRxiv. 2020 doi: 10.3967/bes2021.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui N., Suwa M., Sakurai K., Suzuki Y., Tsumori J., Kobayashi K., Takabatake H., Lee S.T., Yamashita N., Tanaka H. Removal characteristics and fluctuation of norovirus in a pilot-plant by an ultrafiltration membrane for the reclamation of treated sewage. Environ. Technol. 2016;37:2793–2801. doi: 10.1080/09593330.2016.1164760. [DOI] [PubMed] [Google Scholar]
- Ye Y., Ellenberg R.M., Graham K.E., Wigginton K.R. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ. Sci. Technol. 2016;50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- Yeo C., Kaushal S., Yeo D. Enteric involvement of coronaviruses: is faecal–oral transmission of SARS-CoV-2 possible? Lancet Gastroenterol. Hepatol. 2020;5:335–337. doi: 10.1016/S2468-1253(20)30048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yezli S., Otter J.A. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 2011;3:1–30. doi: 10.1007/s12560-011-9056-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., Wang L., Deng X., Liang R., Su M., He C., Hu L., Su Y., Ren J., Yu F. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020;92:408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Du R.-H., Li B., Zheng X.-S., Yang X.-L., Hu B., Wang Y.-Y., Xiao G.-F., Yan B., Shi Z.-L., Zhou P. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg. Microbes Infect. 2020;9:386–389. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., Wei Y., Lee A., Zhang A.J., Chu H. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med. 2020:1–7. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.