Summary
Multiple oncogenic events are involved in the development of adult T‐cell leukaemia/lymphoma (ATL). Because CD28 plays a pivotal role in T‐cell activation, we focused on alterations of the CD28 gene in ATL. We found multiple genetic abnormalities related to CD28 among the 144 patients enrolled in the present study. These involved gene fusions with the cytotoxic T‐lymphocyte‐associated antigen 4 or the inducible T‐cell co‐stimulator in 14 patients (10%), CD28‐activating mutations in 3 (2%), and CD28 copy number variations in 34 (24%). Patients with such CD28 gene alterations were significantly younger than those without. In patients not receiving allogeneic haematopoietic stem cell transplantation, those with CD28 gene alterations tended to have a worse prognosis than those without. Finally, patients with chronic or smouldering ATL subtypes with CD28 gene alterations had a significantly worse prognosis than those without. These findings indicate that ATL, especially chronic or smouldering subtypes, have a more aggressive clinical course and are more refractory to conventional chemotherapies or mogamulizumab if they harbour CD28 gene alterations, likely because of continuous, prolonged, and enhanced CD28 activatory signalling. Novel treatment strategies to overcome the effects of these CD28 gene alterations are warranted.
Keywords: adult T‐cell leukaemia/lymphoma, CD28, fusion, mutation, copy number variation
Introduction
Adult T‐cell leukaemia/lymphoma (ATL) is a peripheral T‐cell neoplasm caused by human T‐cell lymphotropic virus type‐1 (HTLV‐1). 1 , 2 , 3 , 4 Kataoka et al. recently delineated the entire landscape of genetic aberrations in ATL and concluded that alterations to the T‐cell receptor (TCR) and related signalling pathways resulting in cell activation were frequently observed. 5 Because antigen engagement of the TCR initiates a genetic program that results in T‐cell activation, it is reasonable that these pathways might contribute to carcinogenesis. Nevertheless, TCR engagement alone is not sufficient for full activation of normal T cells, which requires a second signal, commonly via ligation of CD28. 6 , 7 , 8 In this context, T‐cell‐activating alterations to the CD28 gene have been reported not only in ATL, but also in other peripheral T‐cell neoplasms such as angioimmunoblastic T‐cell lymphoma (AITL), peripheral T‐cell lymphoma‐not otherwise specified (PTCL‐NOS), or cutaneous T‐cell lymphoma (CTCL). 9 , 10 , 11 , 12 These reports indicate that CD28 gene‐related activating alterations may play an important role in the pathogenesis of some mature T‐cell neoplasms. The aim of the present study was to determine the clinical significance of CD28 gene‐related activating alterations in ATL.
Methods
ATL patients
The study included 144 ATL patients. Tumour samples were obtained at the time of initial presentation at the participating hospitals, and we used the clinical characteristics recorded at that time. Details are available in Data S1. 2 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22
Nucleic acid extraction
Details are available in Data S1.
Detection of CD28 gene alterations
The primer pairs used are shown in Figure S1 and Table SI. For positive controls for the four fusions, we synthesized their cDNAs in vitro (Table SII). The CD28‐activating mutations were detected using a highly sensitive SNaPshot Multiplex Kit (Applied Biosystems, Foster City, CA, USA) (Figure S2). To investigate CD28 copy number variation (CNV), fluorescence in situ hybridization (FISH) was performed. Details are available in Data S1. 5 , 9 , 10 , 11 , 23 , 24 , 25 , 26 , 27
Detection of CCR4 gene mutations
Statistical analysis
The start date for assessing overall survival (OS) was the day when the tumour sample was obtained. Details are available in Data S1. 28
Results
CD28 gene alterations in ATL patients
The ATL patients enrolled in this study included 65 men and 79 women (age range, 41–90 years; median, 64 years) (Table SIII). A multiplex reverse transcription polymerase chain reaction (RT‐PCR) analysis for CD28 fusions is shown in Figure S1B. CD28 fusions were observed in the tumours of 14 patients (10%) (Table I). Sequences of the fusion boundary regions from a patient with inducible T‐cell co‐stimulator (ICOS) (ex1)–CD28 (ex2), and those from another patient with cytotoxic T‐lymphocyte‐associated antigen 4 (CTLA4) (ex3)–CD28 (ex4) are shown in Fig 1A and B, respectively. Both sequences are completely consistent with earlier reports. 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 CD28‐activating mutations were present in three patients (2%). CD28 copy number variation (CNV) was found in 34 patients (24%). Among these, three patients concurrently harboured two different types of CD28 gene alterations. Collectively, alterations of any type involving the CD28 gene were present in 48 patients (33%) (Table I). To illustrate the FISH analysis, CD28:CEP2 (centromere enumeration probe for chromosome 2) signal numbers of 7:2, 6:3, 5:2, 4:2, 3:2, and 2:2 are shown in Fig 2A–F, respectively.
Table 1.
Types and frequencies of CD28 gene alterations in ATL according to clinical subtypes.
| Type of gene alterations | N (%) | Total number (%) | ||||
|---|---|---|---|---|---|---|
| Clinical subtype | Acute | Lymphoma | Chronic | Smoldering | ||
| 79 (55) | 41 (28) | 11 (8) | 13 (9) | 144 (100) | ||
| CD28 fusions | 11 (14) | 2 (5) | 1 (9) | 1 (8) | 15/144 (10) | |
| CTLA4 (ex1)‐CD28 (ex2) | 0 | 0 | 0 | 0 | 0 | |
| CTLA4 (ex2)‐CD28 (ex4) | 0 | 0 | 0 | 0 | 0 | |
| CTLA4 (ex3)‐CD28 (ex4) | 1 (1) | 1 (2) | 0 | 0 | 2/144 (1) | |
| ICOS (ex1)‐CD28 (ex2) | 10 (13) | 1 (2) | 1 (9) | 1 (8) | 13/144 (9) | |
| CD28 mutations | 2 (3) | 0 | 0 | 1 (8) | 3/144 (2) | |
| F51I/V | 0 | 0 | 0 | 1 (8) | 1/144 (1) | |
| D124V/E | 2 (3) | 0 | 0 | 0 | 2/144 (1) | |
| T195I/L/P | 0 | 0 | 0 | 0 | 0 | |
| CD28 CNV | 20 (25) | 12 (29) | 0 | 2 (15) | 34/144 (24) | |
| Gain* | 9 (11) | 10 (24) | 0 | 1 (8) | 20/144 (14) | |
| Amplification † | 11 (14) | 2 (5) | 0 | 1 (8) | 14 /144 (10) | |
| Overall CD28 gene alterations | 31 (39) ‡ | 13 (32) § | 1 (9) | 3 (23) ¶ | 48/144 (33) | |
ATL, adult T‐cell leukemia/lymphoma; CNV, copy number variations; CTLA4, cytotoxic T‐lymphocyte associated antigen 4; ICOS, inducible T‐cell co‐stimulator.
Gains were all CD28:CEP2 signal number of 3:2.
Amplifications included CD28:CEP2 signal number of 7:2 in 2, 6:2 in 2, 6:3 in 1, 5:2 in 4, and 4:2 in 5 patients.
Two patients with acute‐type harbored two different types of CD28 gene alterations; one had a CTLA4 (ex3)‐CD28 (ex4) fusion and a CNV gain, a second had an ICOS (ex1)‐CD28 (ex2) fusion and a CD28 mutation (D124E).
One patient with lymphoma‐type had both ICOS (ex1)‐CD28 (ex2) and CTLA4 (ex3)‐CD28 (ex4).
One patient with smoldering‐type harbored two different types of CD28 gene alterations; an ICOS (ex1)‐CD28 (ex2) fusion and a CNV gain.
Fig 1.
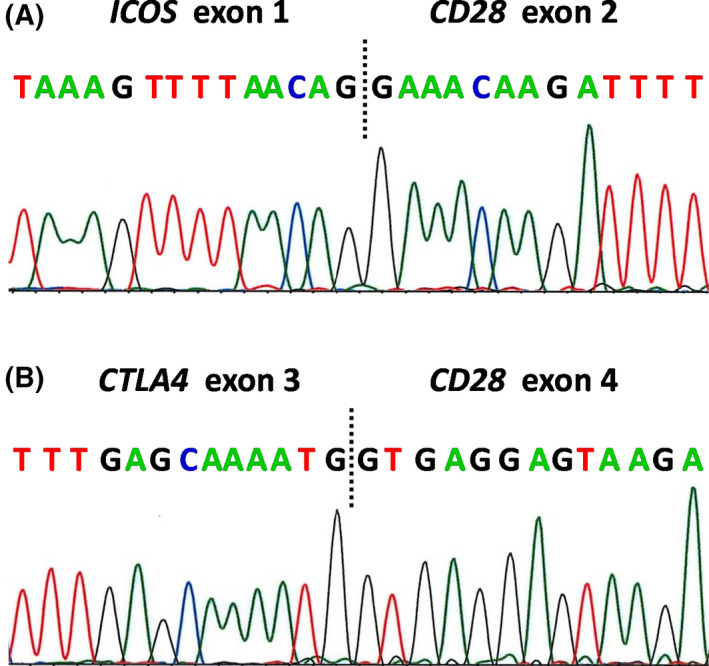
Sequences of the boundary regions of CD28 gene‐related fusions. Sequences of reverse transcription polymerase chain reaction (RT‐PCR) products of (A) inducible T‐cell co‐stimulator (ICOS) (ex1)–CD28 (ex2), and (B) cytotoxic T‐lymphocyte‐associated antigen 4 (CTLA4) (ex3)–CD28 (ex4). [Colour figure can be viewed at wileyonlinelibrary.com]
Fig 2.

CD28 copy number variation (CNV) in adult T‐cell leukaemia/lymphoma (ATL) by fluorescence in situ hybridization (FISH). FISH analyses on FFPE sections from eight individual ATL patients. CD28 signals on chromosome 2q33 are green, and centromeric signals of chromosome 2 are red. CD28 signal number: centromeric signal number ratios were 7:2 (A), 6:3 (B), 5:2 (C), 4:2 (D), 3:2 (E) and 2:2 (F). [Colour figure can be viewed at wileyonlinelibrary.com]
Clinical characteristics of ATL patients stratified by CD28 gene alterations
There were no significant differences in sex, clinical subtype, Eastern Cooperative Oncology Group (ECOG) performance status (PS), Ann Arbor stage, serum soluble interleukin‐2 receptor (sIL‐2R) level, serum‐adjusted calcium (Ca), serum albumin (Alb), white blood cell (WBC) counts, haemoglobin (Hb), or platelet (Plt) counts between patients with or without CD28 gene alterations. There were also no significant differences in the presence or absence of CC chemokine receptor 4 (CCR4) mutations. Patients with CD28 gene alterations were significantly younger than those without (Table II). In relation to the types of CD28 gene alterations, there were no significant differences in those characteristics between patients with CD28 fusions and those without any CD28 gene alterations. The same was true for the CD28 mutations or CNV (Table SIV).
Table II.
Characteristics of adult T‐cell leukaemia/lymphoma (ATL) patients according to CD28 gene alterations.
| Characteristics | CD28 gene alterations | P value | |
|---|---|---|---|
| Absence | Presence | ||
| N (%) | 96 (67) | 48 (33) | |
| Sex | |||
| Female | 53 (55) | 26 (54) | 1·000 |
| Male | 43 (45) | 22 (46) | |
| Clinical subtype | |||
| Chronic, smouldering | 20 (21) | 4 (8) | 0·062 |
| Acute, lymphoma | 76 (79) | 44 (92) | |
| ECOG PS † | |||
| 0, 1 | 75 (79) | 31 (65) | 0·072 |
| 2, 3, 4 | 20 (21) | 17 (35) | |
| Ann Arbor stage | |||
| I, II | 15 (16) | 4 (8) | 0·299 |
| III, IV | 81 (84) | 44 (92) | |
| Serum sIL‐2R (U/ml) ‡ | |||
| ≤20 000 | 61 (68) | 28 (61) | 0·450 |
| >20 000 | 29 (32) | 18 (39) | |
| Serum Ca (mg/dl) * , § | |||
| ≤11·0 | 80 (88) | 39 (85) | 0·603 |
| >11·0 | 11 (12) | 7 (15) | |
| Serum Alb (g/dl) ¶ | |||
| ≥3·5 | 67 (74) | 31 (66) | 0·429 |
| <3·5 | 24 (26) | 16 (33) | |
| Age (year) | |||
| Mean | 66 | 60 | 0·035 |
| Median | 66 | 61 | |
| Range | 41–90 | 41–84 | |
| WBC (/μl)** | |||
| Mean | 13 153 | 20 570 | 0·634 |
| Median | 8750 | 8410 | |
| Range | 2800–68 400 | 2500–232 100 | |
| Hb (g/l)** | |||
| Mean | 129 | 126 | 0·809 |
| Median | 130 | 132 | |
| Range | 79–160 | 88–171 | |
| Plt (×103/μl)** | |||
| Mean | 229 | 279 | 0·548 |
| Median | 215 | 205 | |
| Range | 15–622 | 60–380 | |
| CCR4 gene mutation | |||
| Absence | 65 (68) | 30 (63) | 0·578 |
| Presence | 31 (32) | 18 (37) | |
ATL, adult T‐cell leukaemia/lymphoma; ECOG, Eastern Cooperative Oncology Group; PS, performance status; sIL‐2R, soluble interleukin‐2 receptor; Ca, calcium; Alb, albumin; WBC, white blood cell count; Hb, haemoglobin; Plt, platelet count; CCR4, CC chemokine receptor 4.
When serum Alb level was <4·0 g/dl, serum Ca was adjusted by the concentration of serum Alb as follows: adjusted Ca level (mg/dl) = measured Ca level (mg/dl) + [4·0−Alb level (g/dl)].
One patient's data were unknown.
Eight patients' data were unknown.
Seven patients' data were unknown.
Six patients' data were unknown.
Five patients' data were unknown.
OS of ATL patients stratified by CD28 gene alterations
Five‐year OS of all patients enrolled in the present study was 45·6% (Fig 3A), and that of 48 patients with CD28 gene alterations and 96 patients without any alterations was 38·9% and 49·6%, respectively (not significantly different, P = 0·076) (Fig 3B). Five‐year OS of 14 patients with CD28 fusions was 38·1% (Fig 3C), that of three with CD28 mutations was 33·3% (Fig 3D), and that of 34 with CD28 CNV was 41·6% (Fig 3E). None of these were significantly different from patients without any CD28 gene alterations.
Fig 3.
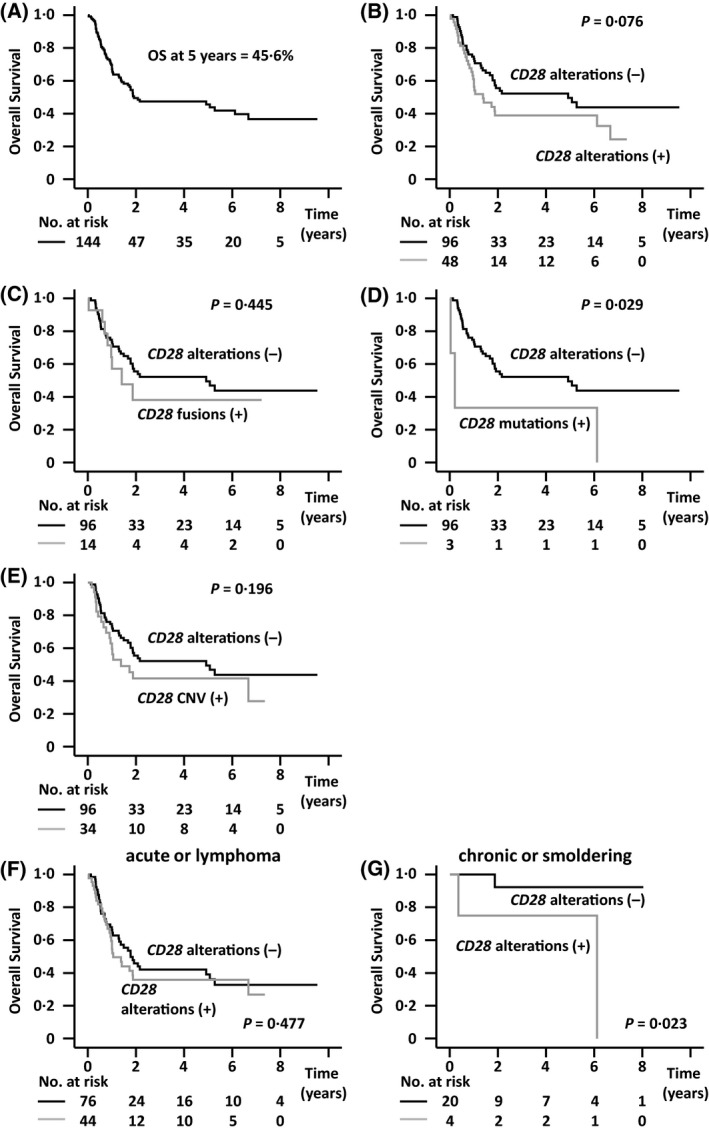
Overall survival (OS) of all adult T‐cell leukaemia/lymphoma (ATL) patients enrolled in the study, stratified according to CD28 gene alterations. (A) OS of all ATL patients enrolled in the study (n = 144). (B) OS according to CD28 gene alterations. (C) OS of the 14 ATL patients with CD28 fusions and the 96 without CD28 gene alterations. (D) OS of the 3 ATL patients with CD28 mutations and the 96 without CD28 gene alterations. (E) OS of the 34 ATL patients with CD28 CNV and the 96 without CD28 gene alterations. (B–E) P < 0·05/4 (two‐sided) was considered statistically significant after Bonferroni correction. (F) OS of ATL patients with acute or lymphoma subtypes according to CD28 gene alterations. (G) OS of ATL patients with chronic or smouldering subtypes according to CD28 gene alterations.
Interaction of CD28 gene alterations with clinical subtypes in terms of OS
We investigated the interaction of CD28 gene alterations with clinical subtypes in terms of OS. When the hazard ratio (HR) for death in patients of acute or lymphoma subtypes without CD28 gene alterations was determined as 1·000, the HR in those with CD28 gene alterations, and patients of chronic or smouldering subtypes with and without CD28 gene alterations were 1·199, 0·788, and 0·072, respectively (P interaction = 0·316; Figure S3A).
OS of ATL patients stratified by clinical subtypes
The five‐year OS of patients with acute or lymphoma subtypes was 37·6% (n = 120, data not shown). Of these, the five‐year OS of 44 patients with CD28 gene alterations was 35·8% and that of 76 patients without CD28 gene alterations was 39·1% (P = 0·477; Fig 3F). HR for OS in patients with acute or lymphoma subtypes with CD28 gene alterations compared with that in patients without CD28 gene alterations was 1·197 [95% confidence interval (CI), 0·729–1·968; Figure S4A]. The five‐year OS of patients with chronic or smouldering subtypes was 89·1% (n = 24, data not shown). In this group, the five‐year OS of the four patients with CD28 gene alterations was 75·0%, which was significantly shorter than that of the 20 without CD28 gene alterations (92·3%, P = 0·023; Fig 3G). HR for OS in patients of chronic or smouldering subtypes with CD28 gene alterations compared with that without CD28 gene alterations was 9·849 (95% CI, 0·883–109·907, Figure S4A).
Interaction of CD28 gene alterations in patients who received and not received allogeneic haematopoietic stem cell transplantation treatment in terms of OS
Allogeneic haematopoietic stem cell transplantation (HSCT) is generally accepted as the only curative treatment for ATL patients. However, treatment‐related mortality for allogeneic HSCT is high compared with that for other treatments. 16 , 17 , 18 In addition, patients with CD28 gene alterations received allogeneic HSCT significantly more frequently compared with those without CD28 gene alterations (Table SV). Thus, we investigated the interaction of CD28 gene alterations in terms of OS in patients who received allogeneic HSCT and in those who did not. When the HR in patients who did not receive allogeneic HSCT without CD28 gene alterations was determined as 1·000, the HR in those with CD28 gene alterations, and in patients who received allogeneic HSCT with and without CD28 gene alterations, was 1·887, 1·010, and 0·825, respectively (P interaction = 0·241, Figure S3B).
Survival of ATL patients receiving allogeneic HSCT, stratified by CD28 gene alterations
We evaluated the impact of CD28 gene alterations on patients receiving allogeneic HSCT, separately from that in non‐transplanted patients. However, five‐year survival from the day of allogeneic HSCT in all 35 transplanted patients was 50·6% (Fig 4A), and did not differ between the 18 with and the 17 without CD28 gene alterations (47·1% and 52·4%, respectively, P = 0·748, Fig 4B). For the eight patients with CD28 fusions, five‐year survival from the day of HSCT was 37·5%, also not significantly different from that in those without CD28 gene alterations (P = 0·461, Fig 4C). No patients with CD28 mutations had received allogeneic HSCT. Finally, five‐year survival from the day of HSCT in 11 patients with CD28 CNV was 60·0%, again not significantly different from that of patients without CD28 gene alterations (P = 0·672, Fig 4D). Next, we investigated survival of the patients receiving allogeneic HSCT stratified by clinical subtype. Five‐year survival from the day of HSCT in patients with acute or lymphoma subtypes was 47·3% (n = 32, data not shown). Five‐year survival from the day of HSCT in the 15 patients without CD28 gene alterations was 48·6%, and 43·8% in the 17 with CD28 gene alterations, again not significantly different (P = 0·759, Fig 4E). HR for survival in patients of acute or lymphoma subtypes with CD28 gene alterations compared with those without CD28 gene alterations was 1·167 (95% CI, 0·434–3·142, Figure S4B). Five‐year survival from the day of HSCT in the three transplanted patients with chronic or smouldering subtypes could not be estimated. These three patients were all alive at their last follow‐up. One of the three had CD28 gene‐related alterations.
Fig 4.
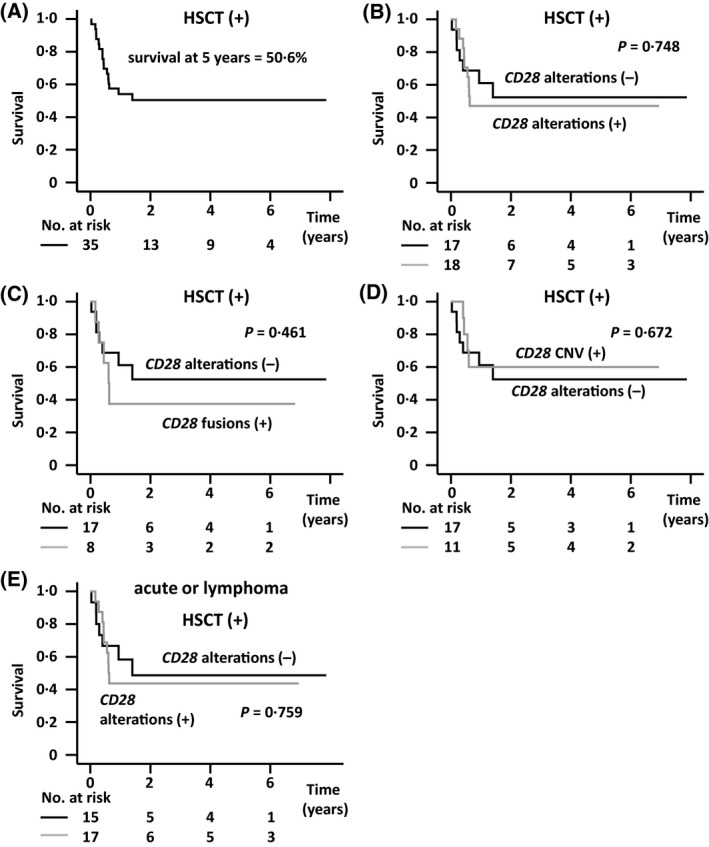
(A) Survival of adult T‐cell leukaemia/lymphoma (ATL) patients who received allogeneic haematopoietic stem cell transplantation (HSCT), stratified according to CD28 gene alterations. (B) Survival from the day of allogeneic HSCT according to CD28 gene alterations. (C) Survival from the day of allogeneic HSCT of the 8 ATL patients with CD28 fusions and the 17 without CD28 gene alterations. (D) Survival from the day of allogeneic HSCT of the 11 ATL patients with CD28 copy number variation (CNV) and the 17 without CD28 gene alterations. (B–D) P < 0·05/3 (two‐sided) was considered statistically significant after correction. (E) Survival from the day of allogeneic HSCT in patients with acute or lymphoma subtypes stratified by CD28 gene alterations.
Survival of ATL patients who did not receive allogeneic HSCT stratified by CD28 gene alterations
Five‐year OS of patients not receiving allogeneic HSCT was 43·6% (n = 109, Fig 5A). In this cohort, five‐year OS of 30 patients with CD28 gene alterations vs. 79 without was 32·8% vs. 48·4%, respectively, thus, the former tended to have a worse prognosis than the latter (P = 0·024, Fig 5B). Five‐year OS of six patients with CD28 fusions was not reached (Fig 5C), but five‐year OS of three patients with CD28 mutations was 33·3% (Fig 5D), and that of 23 patients with CD28 CNV was 31·1% (Fig 5E). The latter two tended to have worse prognoses than those without CD28 gene alterations (P = 0·044, and P = 0·039, respectively). Next, we investigated OS of the patients who did not receive allogeneic HSCT stratified according to the clinical subtypes. Of these, five‐year OS of all patients with acute or lymphoma subtypes was 33·9% (data not shown). Stratifying for CD28 gene alterations showed that five‐year OS of 61 patients without such alterations was 36·5%, compared with 29·8% for 27 with such alterations. However, this difference was not significant (P = 0·251, Fig 5F). HR for OS in patients with acute or lymphoma subtypes with CD28 gene alterations compared with those without CD28 gene alterations was 1·407 (95% CI, 0·783–2·530, Figure S4C). The five‐year OS of 21 patients with chronic or smouldering subtypes, who did not receive allogeneic HSCT, was 86·8% (data not shown). In this group, five‐year OS of the 18 patients without CD28 gene alterations was 90·9%, and that of the three patients with CD28 gene alterations was 66·7%. Despite the small number, this difference was significant (P = 0·010, Fig 5G). HR for OS in patients with chronic or smouldering subtypes with CD28 gene alterations compared with those without CD28 gene alterations was 12·688 (95% CI, 1·094–147·217, Figure S4C).
Fig 5.

Overall survival (OS) of adult T‐cell leukaemia/lymphoma (ATL) patients who did not receive allogeneic HSCT, stratified according to CD28 gene alterations. (A) OS of all ATL patients who did not receive allogeneic HSCT (n = 109). (B) OS of the ATL patients who did not receive allogeneic HSCT according to CD28 gene alterations. (C) OS of the 6 ATL patients with CD28 fusions and the 79 without CD28 gene alterations. (D) OS of the 3 ATL patients with CD28 mutations and the 79 without CD28 gene alterations. (E) OS of the 23 ATL patients with CD28 copy number variation (CNV) and the 79 without CD28 gene alterations. (B–E) P < 0·05/4 (two‐sided) was considered statistically significant after Bonferroni correction. (F) OS of the ATL patients with acute or lymphoma subtypes, who did not receive allogeneic HSCT, stratified by CD28 gene alterations. (G) OS of the ATL patients with chronic or smouldering subtypes, who did not receive allogeneic HSCT, according to CD28 gene alterations.
Survival of ATL patients who did not receive allogeneic HSCT, but did receive mogamulizumab, stratified by CD28 gene alterations
Five‐year survival from the first dose of mogamulizumab in 53 patients who did not receive allogeneic HSCT, but were on mogamulizumab‐containing regimens, was 40·0% (data not shown). In this cohort, five‐year survival from the first dose of antibody in patients with CD28 gene alterations was 34·2% (n = 17), compared to 46·5% (n = 36) in those without CD28 gene alterations (P = 0·078; Figure S5A). Five‐year survival from the first dose of antibody in patients with CD28 gene fusions was not reached (n = 3), and was not significantly different from that in those without CD28 gene alterations (n = 36; P = 0·931) (Figure S5B). The same was true for the single patient with CD28 mutations who died 3·5 years after starting mogamulizumab treatment, not different compared to survival without CD28 gene alterations (n = 36, P = 0·819; Figure S5C). Five‐year survival from the first dose of antibody in patients with CD28 CNV was 40·1% (n = 14), again not significantly different from that in those without CD28 gene alterations (n = 36, P = 0·054; Figure S5D). We next investigated survival of patients treated with mogamulizumab stratified by clinical subtype. Five‐year survival from the first dose of mogamulizumab in 46 patients with acute or lymphoma subtypes was 40·4% (data not shown). In this cohort, five‐year survival from the first dose of antibody in patients with CD28 gene alterations was 32·5% (n = 14), compared to 43·2% (n = 32) in those without CD28 gene alterations (P = 0·203; Figure S5E). The median survival from the first dose of mogamulizumab in seven patients with chronic or smouldering subtypes, who did not receive allogeneic HSCT, was 3·5 years (data not shown). Here also, there was no significant difference between patients with (n = 3) or without (n = 4) CD28 gene alterations (P = 0·320; Figure S5F).
HSCT‐censored OS of ATL patients stratified by CD28 gene alterations
We estimated the survival of all patients enrolled in the present study, from which the transplanted patients had been censored at the day of allogeneic HSCT, in order to reduce the impact of allogeneic HSCT on survival. In this way, five‐year HSCT‐censored survival was 46·6% (Figure S6A); survival of the 48 patients with CD28 gene alterations and of the 96 patients without any alterations was 38·1% and 50·5%, respectively (P = 0·063; Figure S6B). Thus, the former tended to have a worse survival than the latter. Next, we investigated survival of the patients stratified by clinical subtype. The five‐year HSCT‐censored OS of patients with acute or lymphoma subtypes was 36·6% (n = 120, data not shown). Of these, five‐year OS of the 76 patients without CD28 gene alterations was 38·1%, and that of the 44 patients with CD28 gene alterations 34·5%, again not significantly different (P = 0·444; Figure S6C). The OS of patients with chronic or smouldering subtypes was 88·1% (n = 24, data not shown). Of these, five‐year OS of the four patients with CD28 gene alterations was 75·0%, which was significantly less than for the 20 patients without CD28 gene alterations (91·7%, P = 0·012; Figure S6D).
Discussion
Multiple oncogenic events are required for the development of ATL in HTLV‐1‐infected cells after a long latency. Of the many genetic alterations, the present study focused on CD28, finding that compared with other types of peripheral T‐cell neoplasms such as AITL, PTCL‐NOS, and CTCL, CD28 gene alterations are more frequent in ATL. This implies that they may play an important role in ATL tumorigenesis, just as CD28 signalling plays an important role in non‐neoplastic T‐cell activation.
In the entire cohort of ATL patients studied here, those with various different CD28 gene alterations showed no statistically significant differences in OS compared to those without any CD28 alterations. However, there was a trend for the former to have a poorer survival. For some subgroup comparisons, although not statistically significant, it was likely that of the CD28 alterations, especially mutations contributed to the trend towards poorer survival. Notably, however, the impact of CD28 gene alterations on OS did achieve statistical significance in patients with chronic or smouldering subtypes (but not in acute or lymphoma subtypes).
In the cohort of ATL patients receiving allogeneic HSCT, survival of those with or without CD28 gene alterations was not significantly different. Regarding the observation that, in this cohort, the proportion of patients with CD28 gene alterations was relatively high (51%, 18/35), we feel that this is likely to reflect the younger age of the patients with these alterations.
In contrast, among patients who did not receive allogeneic HSCT, those with CD28 gene alterations tended to have worse OS than those without. Of the different CD28 gene alterations, patients with CD28 mutations or CD28 CNV had a worse prognosis relative to patients without any CD28 gene alterations. These findings suggest that ATL with CD28 gene alterations, especially CD28 mutations and CNV, have a more aggressive clinical course and are refractory to conventional chemotherapies, likely because of continuous, prolonged, and enhanced CD28 signaling. 9 , 29 , 30 , 31 , 32 Notably, again, the impact of CD28 gene alterations on OS reached statistical significance in patients with chronic or smouldering subtypes, but not in those with acute or lymphoma subtypes. In addition, patients with CD28 gene alterations were significantly younger than those without, and this characteristic was especially noticeable in the case of CD28 fusions, in agreement with an earlier study. 33 Thus, CD28 gene alterations may be relatively early events among the genetic alterations required for the development of ATL. Collectively all these data suggest that, for patients with chronic or smouldering subtypes, CD28 gene alterations are likely to be critical risk factors for progression to acute or lymphoma subtypes.
In the field of ATL treatment, several novel agents such as mogamulizumab 19 , 20 , 21 and lenalidomide 22 have become available in the clinic. On the other hand, the present study suggests that mogamulizumab treatment does not overcome the aggressiveness associated with CD28 gene alterations, similar to other conventional chemotherapies. Consistent with all these findings, CD28 gene alterations tended to be associated with worse HSCT‐censored OS, and among patients with chronic or smouldering subtypes, this difference achieved statistical significance.
In conclusion, we found that CD28 gene alterations such as fusions, mutations, and CNV were frequent in ATL patients. Among patients not receiving allogeneic HSCT, those with CD28 gene alterations tended to have a worse prognosis than those without, and this difference achieved statistical significance in chronic or smouldering subtypes. These findings indicate that ATL harbouring CD28 gene alterations, especially chronic or smouldering ATL, have a more aggressive clinical course and are refractory to conventional chemotherapies or mogamulizumab, likely because of continuous, prolonged, and enhanced CD28 signalling. Novel treatment strategies to overcome the effects of these CD28 gene alterations are warranted.
Funding
This work was supported by grants‐in‐aid for Early‐Career Scientists (20K16177 to Y. Sakamoto), or scientific research (Aichi Cancer Research Foundation to Y. Sakamoto, and the Nitto Foundation to Y. Sakamoto), grants‐in‐aid from the Japan Agency for Medical Research and Development (Nos. 19cm0106301h0004, & 20cm0106301h0005 to T Ishida), and grants‐in‐aid from the National Cancer Research and Development Fund (29‐A‐3 to S. Iida).
Author contributions
YS, TI and HIn designed the research. YS, TI, AM, MT, HIw, KY, YT, AI, SK, AU and SI performed the experiments. TI, RU and HIn analysed and interpreted data. All authors wrote and approved the manuscript.
Conflicts of interest
YS, AM, MT, HIw, YT, AI, SK and HIn have no conflicts of interest to disclose. TI received honoraria from Kyowa Kirin Co., Ltd., and Celgene K.K. KY received honoraria from Kyowa Kirin Co., Ltd., and Celgene. A.U received honoraria from Kyowa Kirin Co., Ltd., and Celgene. SI received research founding from Kyowa‐Hakko Kirin, Chugai, Takeda, Ono, Celgene, Janssen, Bristol‐Myers Squibb, MSD, Gilead, Abbvie, Sanofi, and Daiichi Sankyo, and honoraria from Takeda, Ono, Celgene, Janssen, Bristol‐Myers Squibb, and Daiichi Sankyo. RU received research funding from Kyowa Kirin Co., Ltd., Chugai Pharmaceutical Co., Ltd., and Ono Pharmaceutical Co.
Supporting information
Data S1. Supplementary methods.
Table SI. Primer sequences.
Table SII. Control cDNA sequences.
Table SIII. Characteristics of ATL patients.
Table SIV. Characteristics of ATL patients according to types of CD28 gene alterations.
Table SV. Characteristics of ATL patients who subsequently received allogeneic HSCT or not.
Fig S1. Detection of CD28 gene fusions.
Fig S2. Detection of CD28 gene mutations.
Fig S3. Interaction of CD28 gene alterations with clinical subtype and treatment.
Fig S4. Forest plots showing the effect of CD28 gene alterations on survival in each patient category.
Fig S5. Survival of ATL patients who did not receive allogeneic HSCT, but received mogamulizumab, stratified according to CD28 gene alterations.
Fig S6. HSCT‐censored OS of all ATL patients enrolled in the study, stratified according to CD28 gene alterations.
Acknowledgements
We thank the Japan Institute of Statistical Technology (Tokyo, Japan) for their critical review of the statistical analyses throughout the manuscript.
Contributor Information
Takashi Ishida, Email: itakashi@med.nagoya-u.ac.jp.
Hiroshi Inagaki, Email: hinagaki@med.nagoya-cu.ac.jp.
References
- 1. Uchiyama T, Yodoi J, Sagawa K, Takatsuki K, Uchino H. Adult T‐cell leukemia: clinical and hematologic features of 16 cases. Blood. 1977;50:481–92. [PubMed] [Google Scholar]
- 2. Shimoyama M. Diagnostic criteria and classification of clinical subtypes of adult T‐cell leukemia‐lymphoma. A report from the Lymphoma Study Group (1984–1987). Br J Haematol. 1991;79:428–37. [DOI] [PubMed] [Google Scholar]
- 3. Matsuoka M, Jeang KT. Human T‐cell leukaemia virus type 1 (HTLV‐1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–80. [DOI] [PubMed] [Google Scholar]
- 4. Ishitsuka K, Tamura K. Human T‐cell leukaemia virus type I and adult T‐cell leukaemia‐lymphoma. Lancet Oncol. 2014;15:e517–e526. [DOI] [PubMed] [Google Scholar]
- 5. Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304–15. [DOI] [PubMed] [Google Scholar]
- 6. Chen L, Flies DB. Molecular mechanisms of T cell co‐stimulation and co‐inhibition. Nat Rev Immunol. 2013;13:227–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Esensten JH, Helou YA, Chopra G, Weiss A, Bluestone JA. CD28 costimulation: from mechanism to therapy. Immunity. 2016;44:973–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. [DOI] [PubMed] [Google Scholar]
- 9. Ungewickell A, Bhaduri A, Rios E, Reuter J, Lee CS, Mah A, et al. Genomic analysis of mycosis fungoides and Sezary syndrome identifies recurrent alterations in TNFR2. Nat Genet. 2015;47:1056–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vallois D, Dupuy A, Lemonnier F, Allen G, Missiaglia E, Fataccioli V, et al. RNA fusions involving CD28 are rare in peripheral T‐cell lymphomas and concentrate mainly in those derived from follicular helper T cells. Haematologica. 2018;103:e360–e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang L, Ni X, Covington KR, Yang BY, Shiu J, Zhang X, et al. Genomic profiling of Sezary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat Genet. 2015;47:1426–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, et al. Molecular heterogeneity in peripheral T‐cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia. 2019;33:2867–83. [DOI] [PubMed] [Google Scholar]
- 13. Yamada Y, Tomonaga M, Fukuda H, Hanada S, Utsunomiya A, Tara M, et al. A new G‐CSF‐supported combination chemotherapy, LSG15, for adult T‐cell leukaemia‐lymphoma: Japan Clinical Oncology Group Study 9303. Br J Haematol. 2001;113:375–82. [DOI] [PubMed] [Google Scholar]
- 14. Tsukasaki K, Utsunomiya A, Fukuda H, Shibata T, Fukushima T, Takatsuka Y, et al. VCAP‐AMPVECP compared with biweekly CHOP for adult T‐cell leukemia‐lymphoma: Japan Clinical Oncology Group Study JCOG9801. J Clin Oncol. 2007;25:5458–64. [DOI] [PubMed] [Google Scholar]
- 15. Cook LB, Fuji S, Hermine O, Bazarbachi A, Ramos JC, Ratner L, et al. Revised adult T‐cell leukemia‐lymphoma International Consensus Meeting Report. J Clin Oncol. 2019;37:677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ishida T, Hishizawa M, Kato K, Tanosaki R, Fukuda T, Taniguchi S, et al. Allogeneic hematopoietic stem cell transplantation for adult T‐cell leukemia‐lymphoma with special emphasis on preconditioning regimen: a nationwide retrospective study. Blood. 2012a;120:1734–41. [DOI] [PubMed] [Google Scholar]
- 17. Ishida T, Hishizawa M, Kato K, Tanosaki R, Fukuda T, Takatsuka Y, et al. Impact of graft‐versus‐host disease on allogeneic hematopoietic cell transplantation for adult T cell leukemia‐lymphoma focusing on preconditioning regimens: nationwide retrospective study. Biol Blood Marrow Transplant. 2013;19:1731–9. [DOI] [PubMed] [Google Scholar]
- 18. Utsunomiya A. Progress in allogeneic hematopoietic cell transplantation in adult T‐cell leukemia‐lymphoma. Front Microbiol. 2019;10:2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishii T, Ishida T, Utsunomiya A, Inagaki A, Yano H, Komatsu H, et al. Defucosylated humanized anti‐CCR4 monoclonal antibody KW‐0761 as a novel immunotherapeutic agent for adult T‐cell leukemia/lymphoma. Clin Cancer Res. 2010;16:1520–31. [DOI] [PubMed] [Google Scholar]
- 20. Ishida T, Joh T, Uike N, Yamamoto K, Utsunomiya A, Yoshida S, et al. Defucosylated anti‐CCR4 monoclonal antibody (KW‐0761) for relapsed adult T‐cell leukemia‐lymphoma: a multicenter phase II study. J Clin Oncol. 2012b;30:837–42. [DOI] [PubMed] [Google Scholar]
- 21. Ishida T, Jo T, Takemoto S, Suzushima H, Uozumi K, Yamamoto K, et al. Dose‐intensified chemotherapy alone or in combination with mogamulizumab in newly diagnosed aggressive adult T‐cell leukaemialymphoma: a randomized phase II study. Br J Haematol. 2015;169:672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishida T, Fujiwara H, Nosaka K, Taira N, Abe Y, Imaizumi Y, et al. Multicenter phase II study of lenalidomide in relapsed or recurrent adult T‐cell leukemia/lymphoma: ATLL‐002. J Clin Oncol. 2016;34:4086–93. [DOI] [PubMed] [Google Scholar]
- 23. Inagaki H, Okabe M, Seto M, Nakamura S, Ueda R, Eimoto T. API2‐MALT1 fusion transcripts involved in mucosa‐associated lymphoid tissue lymphoma: multiplex RT‐PCR detection using formalin‐fixed paraffin‐embedded specimens. Am J Pathol. 2001;158:699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nakayama T, Miyabe S, Okabe M, Sakuma H, Ijichi K, Hasegawa Y, et al. Clinicopathological significance of the CRTC3‐MAML2 fusion transcript in mucoepidermoid carcinoma. Mod Pathol. 2009;22:1575–81. [DOI] [PubMed] [Google Scholar]
- 25. Sakamoto Y, Ishida T, Masaki A, Murase T, Yonekura K, Tashiro Y, et al. CCR4 mutations associated with superior outcome of adult T‐cell leukemia/lymphoma under mogamulizumab treatment. Blood. 2018;132:758–61. [DOI] [PubMed] [Google Scholar]
- 26. Xia H, Nakayama T, Sakuma H, Yamada S, Sato F, Takino H, et al. Analysis of API2‐MALT1 fusion, trisomies, and immunoglobulin VH genes in pulmonary mucosa‐associated lymphoid tissue lymphoma. Hum Pathol. 2011;42:1297–304. [DOI] [PubMed] [Google Scholar]
- 27. Mossafa H, Damotte D, Jenabian A, Delarue R, Vincenneau A, Amouroux I, et al. Non‐Hodgkin's lymphomas with Burkitt‐like cells are associated with c‐Myc amplification and poor prognosis. Leuk Lymphoma. 2006;47:1885–93. [DOI] [PubMed] [Google Scholar]
- 28. Schemper M, Smith TL. A note on quantifying follow‐up in studies of failure time. Control Clin Trials. 1996;17:343–6. [DOI] [PubMed] [Google Scholar]
- 29. Rohr J, Guo S, Huo J, Bouska A, Lachel C, Li Y, et al. Recurrent activating mutations of CD28 in peripheral T‐cell lymphomas. Leukemia. 2016;30:1062–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gmyrek GB, Pingel J, Choi J, Green JM. Functional analysis of acquired CD28 mutations identified in cutaneous T cell lymphoma. Cell Immunol. 2017;319:28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015;47:1011–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vallois D, Dobay MPD, Morin RD, Lemonnier F, Missiaglia E, Juilland M, et al. Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T‐cell‐derived lymphomas. Blood. 2016;128:1490–502. [DOI] [PubMed] [Google Scholar]
- 33. Yoshida N, Shigemori K, Donaldson N, Trevisani C, Cordero NA, Stevenson KE, et al. Genomic landscape of young ATLL patients identifies frequent targetable CD28 fusions. Blood. 2020;135:1467–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplementary methods.
Table SI. Primer sequences.
Table SII. Control cDNA sequences.
Table SIII. Characteristics of ATL patients.
Table SIV. Characteristics of ATL patients according to types of CD28 gene alterations.
Table SV. Characteristics of ATL patients who subsequently received allogeneic HSCT or not.
Fig S1. Detection of CD28 gene fusions.
Fig S2. Detection of CD28 gene mutations.
Fig S3. Interaction of CD28 gene alterations with clinical subtype and treatment.
Fig S4. Forest plots showing the effect of CD28 gene alterations on survival in each patient category.
Fig S5. Survival of ATL patients who did not receive allogeneic HSCT, but received mogamulizumab, stratified according to CD28 gene alterations.
Fig S6. HSCT‐censored OS of all ATL patients enrolled in the study, stratified according to CD28 gene alterations.


