Abstract
This analysis reports a quantitative modeling and simulation approach for oral dapagliflozin, a primarily uridine diphosphate‐glucuronosyltransferase (UGT)–metabolized human sodium‐glucose cotransporter 2 selective inhibitor. A mechanistic dapagliflozin physiologically based pharmacokinetic (PBPK) model was developed using in vitro metabolism and clinical pharmacokinetic (PK) data and verified for context of use (e.g., exposure predictions in pediatric subjects aged 1 month to 18 years). Dapagliflozin exposure is challenging to predict in pediatric populations owing to differences in UGT1A9 ontogeny maturation and paucity of clinical PK data in younger age groups. Based on the exposure–response relationship of dapagliflozin, twofold acceptance criteria were applied between model‐predicted and observed drug exposures and PK parameters (area under the curve and maximum drug concentration) in various scenarios, including monotherapy in healthy adults (single/multiple dose), monotherapy in hepatically or renally impaired patients, and drug–drug interactions with UGT1A9 modulators, such as mefenamic acid and rifampin. The PBPK model captured the observed exposure within twofold of the observed monotherapy data in adults and adolescents and in special population. As a guide to determining dosing regimens in pediatric studies, the verified PBPK model, along with UGT enzyme ontogeny maturation understanding, was used for predictions of dapagliflozin monotherapy exposures in pediatric subjects aged 1 month to 18 years that best matched exposure in adult patients with a 10‐mg single dose of dapagliflozin.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
Dapagliflozin is approved for the treatment of type 2 diabetes mellitus as monotherapy or in combination with other antidiabetic drugs in adults. In vitro absorption, distribution, metabolism, and excretion and disposition studies suggest that dapagliflozin is a substrate of uridine diphosphate‐glucuronosyltransferase (UGT) isoform 1A9.
WHAT QUESTION DID THIS STUDY ADDRESS?
What are the dosing recommendations for pediatric patients? Is dapagliflozin tablet exposure affected in patients with moderate/severe hepatic/renal impairment? What exposure changes occur when dapagliflozin is coadministered with inhibitors/inducers of UGT1A9?
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
A verified physiologically based pharmacokinetic (PBPK) model with UGT1A9 ontogeny was developed for prospective monotherapy exposure predictions to inform clinical trials in pediatric age groups between 1 month and 18 years by predicting the single dose that best matched exposure after a 10‐mg single dose in adult patients.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
Given the challenges in conducting pediatric studies, it is beneficial to use verified PBPK models to predict changes in drug exposure in various scenarios to guide dosing information when clinical trials are not possible, especially in pediatric subjects < 2 years old.
Dapagliflozin (Farxiga, AstraZeneca Pharmaceuticals LP, Wilmington, DE, USA) is an oral medication developed to treat type 2 diabetes mellitus (T2DM) through selective and reversible inhibition of human sodium‐glucose cotransporter 2, the major transporter for renal glucose reabsorption. 1 Dapagliflozin reduces plasma glucose by inhibiting renal glucose reabsorption in the renal proximal tubule to promote glucose urinary excretion. Dapagliflozin 10‐mg oral tablet is approved in 89 countries for the treatment of T2DM as monotherapy or in combination with other antidiabetic drugs in adults. 2
Recent evidence shows that dapagliflozin reduces the incidence of cardiovascular events, such as heart failure (HF), with clinically meaningful significance. A large, randomized, double‐blind, placebo‐controlled, phase III trial of dapagliflozin in patients with T2DM with established atherosclerotic cardiovascular disease or at risk for such disease found that dapagliflozin reduced the incidence of cardiovascular death and hospitalization from HF (NCT01730534). 3 Regulatory approval for dapagliflozin use for HF in patients with and without established atherosclerotic cardiovascular disease is being pursued. 4 Dapagliflozin metabolism to its main inactive metabolite dapagliflozin 3‐O‐glucuronide is primarily mediated by uridine diphosphate‐glucuronosyltransferase (UGT) isoform 1A9. Figure 1 shows the dapagliflozin disposition pathway based on absorption, distribution, metabolism, and excretion (ADME) studies. 5 , 6 Furthermore, Figure S1 illustrates a mass balance created using physiologically based pharmacokinetic (PBPK) modeling in pediatric subjects between 1 and 3 months old.
Figure 1.
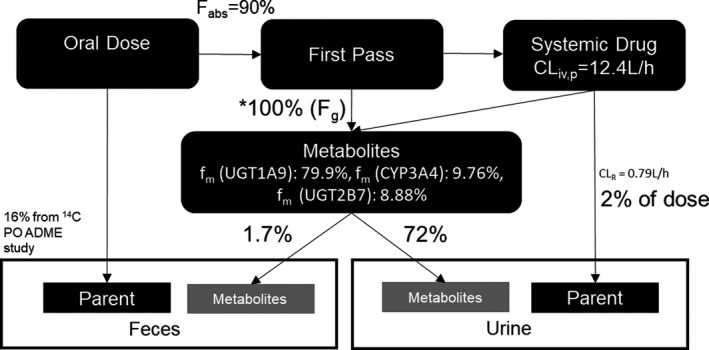
Dapagliflozin metabolism and disposition pathway based on ADME studies using oral and intravenous 14C‐dapagliflozin. A mass balance ADME study using six healthy subjects who received a single oral 10‐mg dapagliflozin dose and an 80 μg [14C]‐dapagliflozin intravenous dose containing 200 nCi radioactivity was conducted. This study showed that only ~ 16% of unchanged drug was recovered in feces and 2% of the dose was excreted in urine as unchanged dapagliflozin; enzymes metabolized the remaining drug, and the metabolites were excreted in feces and urine. *Predicted based on dapagliflozin PBPK model 10‐mg oral dose simulations. ADME, absorption, distribution, metabolism, and excretion; CLiv,p, intravenous plasma clearance; CLR, renal clearance; CYP, cytochrome P450; Fg, fraction escaping gut metabolism; fabs, fraction absorbed; fm, fractions metabolized; PBPK, physiologically based pharmacokinetics; PO, per oral; UGT, uridine diphosphate‐glucuronosyltransferase.
Age‐related changes in the abundance and functional activity of metabolizing enzymes play a major role in drug disposition and are key determinants of pharmacokinetics (PK). Pediatric populations, particularly young age groups (1 to 24 months), lack the maturation of physiological factors that affect dapagliflozin ADME, such as the ontogeny of metabolizing enzymes and renal function. Such physiological factors may have clinically meaningful safety and efficacy impacts on the dapagliflozin dosing regimen for young age groups. In addition, to promote research and development of new medicines for children, the Research to Accelerate Cures and Equity for Children Act 7 and US Food and Drug Administration mandates inclusion of children in clinical trials, where possible, to obtain regulatory approval. 7 This creates a need to predict doses and exposure that would match safe exposure in adults as a justification for dosing decisions during pediatric clinical trials.
PBPK modeling simulates a mechanistic representation of the drug ADME through the human body to calculate drug PK parameters, such as area under the curve (AUC), which shows the total plasma drug exposure, and the highest drug concentration in the plasma (Cmax). This modeling approach uses a three‐component system: system component, drug‐dependent component, and clinical trial–specific component. The system component is defined as the physiological factors affecting ADME, such as abundance of metabolizing or transporter enzymes and tissue blood flow and are specified according to relevant organs. The drug‐dependent component is described as the drug characteristics that affect ADME, such as solubility, main metabolizing enzymes, and enzyme interactions such as inhibitory or inductive activity. The clinical trial–specific component defines the scenario to be simulated, including age, gender, dosing regimen, and dosing population.
The use of PBPK modeling for informing drug prescribing information for small molecule drugs is well established. 8 Many such drug PBPK models have been verified to reliably reproduce human clinical study PK values. 9 , 10 Verified PBPK drug models can be extended to prospective predictions when human clinical study data are not available. This includes predicting drug PK in specific populations, such as the pediatric population, by characterizing pediatric‐specific systems that affect drug ADME and thus PK predictions, such as enzyme ontogeny, body weight, and renal excretion maturity. Parkinson et al. have shown that following a single oral dose of dapagliflozin, adult and pediatric patients with T2DM had similar exposure–response relationships after accounting for significant covariates, including estimated glomerular filtration rate (GFR). 11 Figure S2 shows a flowchart outlining the development of a PBPK model for reliable PK predictions in simulated scenarios. The comprehensive and robust PBPK model demonstrates an ability to predict drug PK changes within a twofold acceptance criteria of PK data from observed clinical data in various scenarios, including drug–drug interactions (DDIs) and administration in specific populations with a different drug ADME than in the healthy adult population. The twofold acceptance criteria were also based on the exposure–response relationship of dapagliflozin (NCT00736879). Verification of PBPK models to reasonably simulate PK changes in such scenarios builds confidence for prospective simulations by these models when there are no equivalent clinical data. Verified PBPK models have previously been shown to reasonably predict PK changes in various simulated scenarios and have been well received by regulatory agencies as part of establishing safety regulations for drug applications. 12
The objectives of this analysis were as follows:
Develop a mechanistic quantitative PBPK model of the dapagliflozin tablet using Simcyp (Simcyp Limited, Sheffield, UK) with physiological parameters and human in vitro and in vivo data to simulate systemic exposure.
Reproduce dapagliflozin clinical PK data for DDI with rifampin, a cytochrome P450 (CYP) 3A inducer, and mefenamic acid, a UGT1A9 inhibitor, as agents using the PBPK model.
Use the observed clinical data of dapagliflozin exposure in adolescents to optimize the mechanistic PBPK model for prospective drug exposure predictions in pediatric (1 month to 12 years) age groups.
Use literature data to model the UGT1A9 ontogeny in the pediatric population for prospective pediatric dapagliflozin exposure and then apply UGT1A9 ontogeny models to a virtual pediatric population to determine the dapagliflozin single oral dose that matches the exposure in healthy adults with the 10‐mg single oral dose to guide dosing for pediatric studies.
Compare the observed clinical data with the mechanistic PBPK model predictions in healthy adults and populations with severe renal or hepatic impairment to verify the mechanistic PBPK model and build confidence to predict dapagliflozin exposure in specific populations.
Methods
Dapagliflozin PBPK model development
Based on current understanding of dapagliflozin drug disposition, a mechanistic PBPK model for dapagliflozin was developed using physiochemical data and data from in vitro and in vivo ADME studies. Details of the key input parameters for the dapagliflozin PBPK model are shown in Table 1 . PBPK modeling software (Simcyp version 18, Simcyp Limited) was used to build and simulate human exposure for the dapagliflozin oral tablet.
Table 1.
Dapagliflozin model input parameters used for PBPK model development
| Parameters and models | Dapagliflozin adult | Source | |
|---|---|---|---|
| Physiochemical properties | MW (g/mol) | 408.88 | Experimental data |
| Log P | 2.52 | NDA202293 | |
| pKa | Neutral | Experimental data | |
| B/P ratio | 0.88 | NDA202293 | |
| f u,plasma | 0.086 | Experimental data | |
| Absorption | Absorption model | First‐order absorption model | |
| f a | 0.90 | Clinical PK data; NCT00908271 | |
| ka (1/h) | 1.98 | Clinical PK data; NDA202293 | |
| f u,gut | 0.0247 | Simcyp predicted | |
| P eff,man | 2.536 × 10‐4 cm/s | Simcyp predicted | |
| Distribution | Distribution model | Minimal PBPK | |
| Single Adjusting Compartment Q (L/h) | 10 | NDA 209803 | |
| V ss (L/kg) | 1.19 | NDA 209803 | |
| Elimination | Clearance type | Enzyme kinetics | |
| CLiv (L/h) | 12.4 | Clinical PK data (NCT00908271) | |
| CYP3A4 CLint (µL/min/pmol of isoform) | 0.037 | Simcyp iterative approach | |
| UGT2B7 CLint (µL/min/pmol of isoform) | 0.032 | Simcyp iterative approach | |
| UGT1A9 CLint (µL/min/pmol of isoform) | 1.10 | Simcyp iterative approach | |
| Fumic/inc | 0.72 | Simcyp predicted | |
| Additional HLM CLint | 1.32 | Simcyp iterative approach | |
| Renal clearance (L/h) | 0.79 | Clinical PK data (NCT00908271) |
| Parameters and models | Dapagliflozin pediatric | Source | |
|---|---|---|---|
| Physiochemical properties | MW (g/mol) | 408.88 | Experimental data |
| Log P | 2.52 | NDA202293 | |
| pKa | Neutral | Experimental data | |
| B/P ratio | 0.88 | NDA202293 | |
| f u,plasma | 0.086 | Experimental data | |
| Absorption | Absorption model | First‐order absorption model | |
| f a | 0.78 | Estimated | |
| ka (1/h) | 1.98 | Clinical PK data; NDA202293 | |
| f u,gut | 0.0247 | Simcyp predicted | |
| P eff,man | 2.536 × 10‐4 cm/s | Simcyp predicted | |
| Distribution | Distribution model | Minimal PBPK | |
| Single Adjusting Compartment Q (L/h) | 10 | NDA 209803 | |
| V ss (L/kg) | 1.19 | NDA 209803 | |
| Elimination | Clearance type | Enzyme kinetics | |
| CLpo (L/h) | 15.20 | Parameter estimated | |
| CYP3A4 CLint (µL/min/pmol of isoform) | 0.024 | Simcyp iterative approach | |
| UGT2B7 CLint (µL/min/pmol of isoform) | 0.045 | Simcyp iterative approach | |
| UGT1A9 CLint (µL/min/pmol of isoform) | 0.854 | Simcyp iterative approach | |
| Fumic/inc | 0.72 | Simcyp predicted | |
| Additional HLM CLint | 0.00 | Simcyp iterative approach | |
| Renal clearance (L/h) | 0.79 | Clinical PK data (NCT00908271) |
| Pediatric input parameters for UGT1A9 ontogeny model development | ||
|---|---|---|
| Scenario 1 (Simcyp default UGT1A9 ontogeny model; sigmoidal Eq. 1) | Scenario 2 (literature based UGT1A9 ontogeny model) | |
| Model parameters | Sigmoidal equation with additional exponential function | |
| AdultMax | 1.041 | 1.305 |
| FBirth | 0.086 | 0 |
| Age50 | 1.16 | 0.086 |
| n | 1.2 | 1.1 |
| Sigmoidal equation age cap | 15 | 1 |
| C 0 | N.A. | 1.243 |
| C 1 | N.A. | −0.833 |
| C 2 | N.A. | 0.2 |
| C 3 | N.A. | 25 |
| Exponential equation age cap | N.A. | 20 |
| Population | All simulations were based on healthy population (“Sim‐Healthy Volunteers”) except for pediatric simulations, hepatically impaired simulations, and renally impaired simulations (“Sim‐Pediatric,” “Sim‐CirrhosisCP‐C,” and “Sim‐RenalGFR_less_30,” respectively). | |
| Clinical data |
MB102‐001; NCT00736879; MB102002: Oral monotherapy, single‐ and multiple‐dose data in healthy adults MB102‐059; NCT00908271: Oral and 14C i.v. monotherapy single dose in healthy adults MB102‐007; NCT00554450: Oral monotherapy single dose in T2DM population with renal impairment MB102‐027: Oral monotherapy single dose in hepatically impaired population MB102‐074; NCT01068756: Oral dapagliflozin drug–drug interaction with rifampin and mefenamic acid MB102‐091; NCT01525238: PKPD study of dapagliflozin in children and adolescents aged 10 to 17 years with T2DM |
|
AdultMax, maximal adult UGT activity; Age50, the age at which half the maximal fraction of adult UGT activity is reached; B/P ratio, blood‐to‐plasma ratio; C 0, baseline at birth; C 1, coefficient; C 2, Slope; C 3, adjusting age; CLint, intrinsic clearance; CLiv, intravenous clearance; CLpo, oral clearance; f a, fraction absorbed; FBirth, fraction of adult UGT activity at birth; f u,gut, fraction unbound in gut enterocytes; f u,inc, fraction unbound in the incubation; f u,mic, fraction unbound in microsomes; f u,plasma, fraction unbound in plasma; HLM, human liver microsomes; i.v., intravenous; ka, first‐order absorption rate constant; MW, molecular weight; n, hill coefficient; N.A., not available; PBPK, physiologically based pharmacokinetic; P eff,man, human intestinal effective permeability; PK, pharmacokinetic; pKa, acid dissociation constant (logarithmic scale); PKPD, pharmacokinetic and pharmacodynamic; T2DM, type 2 diabetes mellitus; V ss, volume of distribution at steady state.
A first‐order absorption model with a predicted human intestinal effective permeability of 2.54 × 10‐4 cm/second was calculated based on the Caco‐2 cell monolayer apparent permeability of 18.4 × 10‐6 cm/second using prediction calculations available in Simcyp. Human PK data after an intravenous dose (MB102059, NCT00908271), estimating the volume of distribution at steady state of 1.19 L/hour and intravenous clearance (CLiv) of 12.4 L/hour, were available to specify parameters and calculate enzyme kinetic clearance using the iterative model available within Simcyp. Metabolizing enzyme (CYP3A4, UGT1A9, UGT2B7) intrinsic in vitro clearance (CLint) were predicted assuming a healthy volunteer population of 18‐ to 65‐year‐olds and 0.5 proportion of females with a fraction metabolized (fm) of 0.10 by CYP3A4, 0.80 by UGT1A9, and 0.05 by UGT2B7 as measured (clinical PK data; NDA202293). Dapagliflozin does not induce significant DDI as an agent via metabolizing enzymes or transporters, including kidney transporters.
Dapagliflozin model verification with single and multiple doses in healthy adults
No dose‐limiting safety or tolerability signals were observed with up to 500‐mg dapagliflozin monotherapy in human clinical trials (MB102001, NCT00736879). Predefined acceptance criteria of 0.5‐ to 2‐fold were set to assess PBPK model predictions based on clinical relevance criteria and the wide therapeutic index of dapagliflozin.
Simulation settings
A dapagliflozin PBPK model simulated a clinical trial design in healthy adult patients following single doses between 5 and 100 mg and multiple 10‐mg once‐daily doses for 14 days using the Simcyp “healthy adult volunteer” virtual population (MB102001, NCT00736879). These simulations comprised 100 subjects (10 trials of 10 patients each) in the simulated virtual population, with matched age, range, body weight, and ratio of female to male subjects as reported in the clinical study used for verification in the fasted state. Prospective simulations for specific age groups comprised 100 patients with the target age range and a ratio of female to male subjects of 0.5 in the fasted state.
PBPK model exposure predictions in populations with severe hepatic or renal impairment
Simulations were conducted using the mechanistic dapagliflozin PBPK model to predict changes in dapagliflozin exposure in patients with severe hepatic impairment (MB102027) or T2DM with renal impairment (MB102007). Patients with Child‐Pugh class C were classified as those with severe hepatic impairment. Patients with diabetes with creatinine clearance < 30 mL/minute and not receiving dialysis were classified as severely renally impaired.
Simulations for severe hepatic and renal impairment required modified population parameters with reduced liver and kidney metabolizing enzyme abundance. The Simcyp virtual populations for severe hepatic and renal impairments, “Sim‐CirrhosisCP‐C” and “GFR < 30 mL/min,” respectively, did not incorporate the reduction in the UGT enzyme abundance found in the literature. 13 , 14 Sensitivity analysis of hepatic UGT1A9 absolute abundance for clearance with the Simcyp virtual population “Sim‐CirrhosisCP‐C” was undertaken to match the observed oral clearance, which resulted in a 46% reduction in UGT enzyme compared with the healthy population enzyme abundance. 15 Subsequently, the Simcyp virtual population “Sim‐CirrhosisCP‐C” enzyme abundance for both UGT1A9 and UGT2B7 were reduced by 46% for simulations of severe hepatic impairment, assuming impairment led to the same reduction in enzyme abundance in the organ.
The predicted clearance from the sensitivity analysis of the kidney UGT1A9 and UGT2B7 enzymes using the Simcyp virtual population “GFR < 30 mL/min” failed to match the observed clearance from the clinical study. Therefore, the Simcyp “GFR < 30 mL/min” virtual population parameters were amended to have no kidney UGT1A9 and UGT2B7 enzyme abundance for simulations of severe renal impairment to simulate the worst‐case scenario.
Simulations comprised 100 patients (10 trials of 10 patients each) using modified population parameters for the 10‐mg dapagliflozin single oral dose for severe hepatic impairment exposure and the 50‐mg dapagliflozin single oral dose for severe renal impairment exposure (highest clinical dose used in the phase II trial).
PBPK model exposure predictions in DDIs with rifampin and mefenamic acid
The dapagliflozin mechanistic PBPK model was used to simulate dapagliflozin exposure differences by simulating matching DDI studies with UGT inducers and inhibitors to further build the confidence for model predictions to calculate changes to dapagliflozin ADME appropriately. The PBPK model for DDI for the CYP3A inducer rifampin, 16 available in the compound library within the PBPK platform of verified clinical drug compounds, and the UGT1A9 inhibitor mefenamic acid, 17 developed in house, were used for simulations. Rifampin and mefenamic acid PBPK models have been previously verified for reliable DDI predictions by reproducing clinical DDI study results. Rifampin is a known metabolizing enzyme inducer of CYP3A and UGT1A9, and mefenamic acid is an UGT1A9 inhibitor. As expected, a clinical study (NCT01068756) reported a 22% decrease in dapagliflozin area under the plasma concentration‐time curve extrapolated to infinity postadministration (AUCINF) with rifampin coadministration and a 51% increase with mefenamic acid.
PBPK model simulations were performed with a matching clinical design with an adult healthy subject virtual population.
Model optimization for pediatric predictions with adolescent (11‐ to 17‐year‐olds) observed data
The mechanistic PBPK model that simulated matching clinical design in adolescents with T2DM aged 11 to 17 years resulted in an overprediction of AUC and Cmax compared with the observed data from the clinical study (NCT015238). Although the PK prediction ratios were strictly within 0.5‐ to 2‐fold, because the model would be used to match PK particularly in very low doses, it was particularly important to ensure optimal recreation. Fraction absorbed (F a) was reduced from 0.9 to 0.78 as 0.9 led to overprediction of Cmax, particularly for the 2.5‐mg simulation. Sensitivity analysis of f a was done for 10‐mg simulations and led to good fit of observed Cmax for both the 2.5‐mg and 10‐mg doses. F a was changed from 0.9 to 0.78 to match the Cmax in individuals aged 12 to 17 years. Based on the literature, 18 , 19 , 20 it is expected that gut motility and absorption in children is slower and reduced compared with adults. AUC overprediction indicated that the current mechanistic PBPK model needs improvement for use in the specific context of pediatric PK predictions. To address this slight overprediction, elimination parameters, particularly those used to predict enzyme clearance in the PBPK model, were revised. A switch from CLiv to oral clearance (CLpo) was made for iterative predictions for the metabolizing enzyme intrinsic clearance (CLint). To estimate the optimal CLpo value that leads to the best match with clinical PK (NCT015238), the value of CLpo was selected based on fitting of the adolescent PK data considering the body weight of these subjects. Both the Nelder‐Mead or hybrid minimization methods, with objective function weighted least square weighted by reciprocal of prediction squared, were used to fit the data. Results from both the Nelder‐Mead method and the hybrid method agreed that a CLpo of 15.2 L/hour best matched clinical PK data from dapagliflozin monotherapy. After optimizing CLpo of the dapagliflozin PBPK model, the CYP and UGT CLint were also adjusted using the optimized CLpo values for iterative prediction using the same aforementioned fraction metabolized and population details. Details of the key input parameters for the optimized dapagliflozin PBPK model are shown in Table 1 . This optimized model then simulated the clinical design to test the ability of the model to recreate observed exposure with the 10‐mg and 2.5‐mg single doses in obese adolescents aged 11 to 17 years with T2DM. The final PBPK model for drug exposure predictions for the purpose of pediatric simulations used a “middle‐out” approach. The simulations used the Simcyp “Sim‐Pediatric” virtual population with the default ontogeny UGT1A9 model with matching age and body weight ranges found in the clinical study. Body weight was a particularly important parameter to be matched in simulations as the clinical study reported a particularly high body weight in the recruited patients with T2DM.
UGT1A9 ontogeny model development for prospective optimal‐dose predictions in pediatric age groups
The Simcyp default ontogeny models for dapagliflozin‐metabolizing enzymes used in the pediatric populations for simulations were revised against the available literature data (Figure 2 ). The CYP3A4 and UGT2B7 default ontogeny models in the Simcyp “Sim‐Pediatric” virtual population for simulations agreed with the literature data and did not require the use of a modified model for simulations, especially since CYP3A4 and UGT2B7 are relatively minor clearance pathways for oral dapagliflozin. However, there was a difference between the UGT1A9 default ontogeny model in the virtual pediatric population used for simulations and those reported in recent literature. 21 , 22 Notably, UGT1A9 maturation provides very low doses, and it is important not to underdose the pediatric population. Details of the default and changed ontogeny model parameters are shown in Table 1 and applied in sigmoidal function (Eq. 1) and added additional exponential function (Eq. 2). The additional exponential in Eq. 2 is manually activated and replaces sigmoidal Eq. 1 for ages beyond the age cap defined for Eq. 1 and within the age cap specified for Eq. 2.
Figure 2.
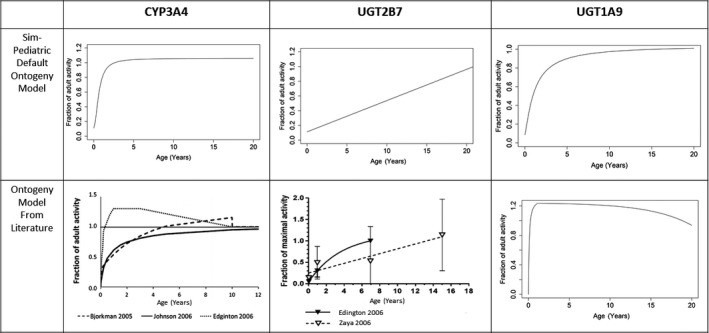
Comparison of the metabolic enzyme ontogeny models in Simcyp and in the literature. “Sim‐Pediatric” default ontogeny profiles (scenario 1) for CYP3A4, UGT2B7, and UGT1A9 and recent data based on literature (scenario 2) for CYP3A4, UGT2B7, and UGT1A9 are compared. CYP, cytochrome P450; UGT, UDP‐glucuronosyltransferase.
Sigmoidal function (Eq. 1):
| (1) |
Additional exponential component (Eq. 2):
| (2) |
The “Sim‐Pediatric” default ontogeny profiles for CYP3A4, 23 UGT2B7, 24 , 25 and UGT1A9 26 , 27 and those based on the literature for CYP3A4, 28 , 29 , 30 UGT2B7, 24 , 30 and UGT1A9 21 are compared in Figure 2 .
Pediatric age groups were classified as follows: 12 to 18 years, 6 to 12 years, 2 to 6 years, 1.5 to 2 years, 1 to 1.5 years, 0.5 to 1 years, 3 to 6 months, and 1 to 3 months. Sensitivity analyses on the doses for all pediatric age groups using both scenarios and optimized for the dapagliflozin PBPK model were done to predict the oral dose that best matches the dapagliflozin exposure from the 10‐mg oral single dose in healthy adults observed in the clinical studies (MB102001, NCT00736879).
Results
PBPK simulations of dapagliflozin exposure in healthy adults
Figure 3a,b shows the mean dapagliflozin concentration‐time profiles in plasma simulated by the mechanistic dapagliflozin PBPK model for single and multiple once‐daily oral doses of dapagliflozin between 5 and 100 mg. The simulated clinical design showed that the predicted PK parameters, including AUCINF and Cmax, were well within the predetermined 0.5‐ to 2‐fold acceptance criteria of PK parameters observed in clinical trials (Table 2 ). Simulations of single doses between 5 and 100 mg also recreated the clear linear systemic exposure found in the clinical study (MB102001) (Figure S3 ). Therefore, this mechanistic dapagliflozin PBPK model was determined sufficiently robust to continue simulations in specific populations and DDIs.
Figure 3.
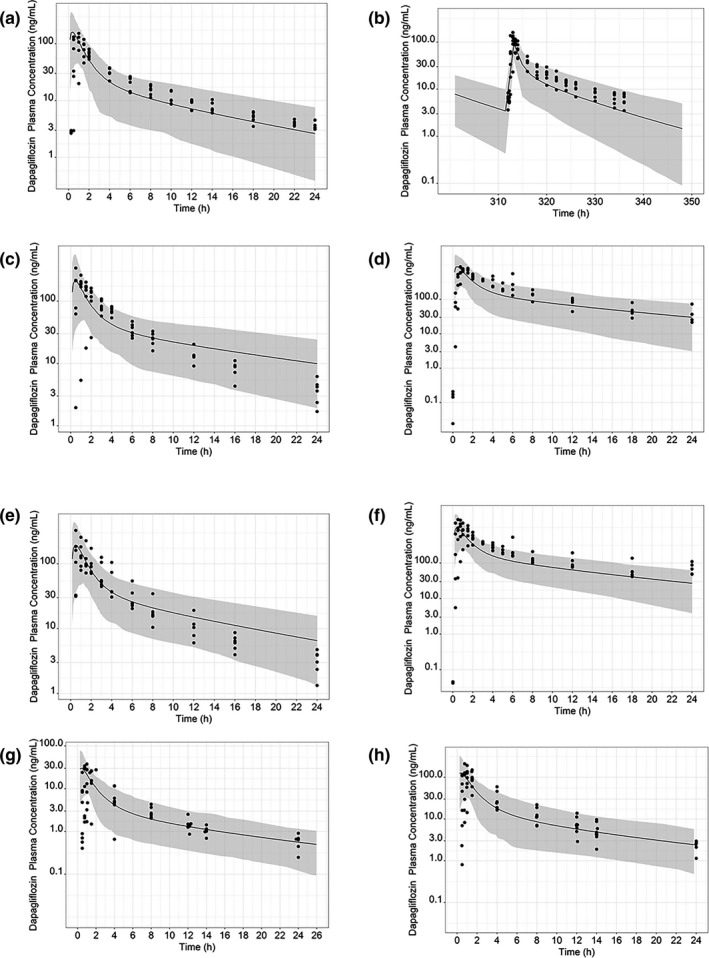
Simulated plasma concentration‐time profile for dapagliflozin after (a) a single 10‐mg dose and (b) multiple 10‐mg doses once daily in healthy adults; (c) a single 10‐mg dose in a population with severe hepatic impairment; (d) a single 50‐mg dose in a population with severe renal impairment; (e) a single 10‐mg dose in a population with moderate hepatic impairment; (f) a single 50‐mg dose in a population with moderate renal impairment; (g) a single 2.5‐mg dose in adolescents aged 12 to 17 years with type 2 diabetes mellitus; and (h) a single 10‐mg dose in adolescents aged 11 to 17 years with type 2 diabetes mellitus. The continuous black line represents the predicted mean dapagliflozin plasma concentration; the shaded area represents 95% prediction intervals. Closed circles are the observed data points from dapagliflozin clinical trials of a single dose (MB102001), multiple doses (MB102002), MB102007 for renal impairment dosing, and MB102027 for hepatic impairment dosing. Closed circles are the observed data points from dapagliflozin clinical trials in adolescents (NCT01525238).
Table 2.
Geometric mean AUC and Cmax from PBPK model simulations compared with observed clinical data of dapagliflozin exposure following single or multiple doses
| Population and dose | Parameters from observed data | Parameters from simulation | Mean ratio of simulated/observed | |||
|---|---|---|---|---|---|---|
| AUCINF, h·ng/mL (95% CI) | Cmax, ng/mL (95% CI) | AUCINF, h·ng/mL (95% CI) | Cmax, ng/mL (95% CI) | AUC | Cmax | |
| Dapagliflozin exposure following a single dose in healthy volunteers | ||||||
| Healthy volunteers, 5 mg | 185 (157–219) | 67 (57–80) | 290.25 (266.49–316.14) | 94.49 (85.09–104.93) | 1.57 | 1.41 |
| Healthy volunteers, 10 mg | 368 (316–433) | 116 (83–135) | 411.49 (377.24–448.85) | 146.23 (128.08–166.94) | 1.12 | 1.26 |
| Healthy volunteers, 20 mg | 977 (813–1177) | 269 (233–310) | 822.9 (754.49–897.70) | 292.45 (256.17–333.87) | 0.84 | 1.09 |
| Healthy volunteers, 100 mg | 4363 (3867–494) | 1112 (867–1468) | 5881.00 (5400.66–6404.06) | 1899.21 (1712.82–2105.89) | 1.35 | 1.71 |
| Dapagliflozin exposure following multiple doses in healthy volunteers | ||||||
| Healthy volunteers, 10 mg q.d. | 506 (438–613) | 119 (87–168) | 430.89 (393.31–472.07) | 150.71 (132.70–171.15) | 0.85 | 1.27 |
| Dapagliflozin exposure following a single dose in severely renal and hepatic impaired adults | ||||||
| Moderate hepatic impairment, 10 mg | 632 (1129–2575) | 153 (200–492) | 710.59 (650.26–776.52) | 168.64 (145.93–194.90) | 1.12 | 1.10 |
| Severe hepatic impairment, 10 mg | 776 (834–1419) | 190 (119–242) | 1023.38 (936.15–1118.74) | 175.42 (152.94–201.21) | 1.32 | 0.92 |
| Moderate renal impairment, 50 mg | 5182 (4191–6681) | 897 (696–1250) | 2930.48 (2635.97–3257.91) | 848.92 (732.98–983.20) | 0.57 | 0.95 |
| Severe renal impairment, 50 mg | 4884 (4409–5358) | 772 (692–857) | 2757.40 (2443.21–311.98) | 788.90 (685.35–908.08) | 0.56 | 1.02 |
| Mean AUC and Cmax from PBPK model simulations with default virtual pediatric population (scenario 1) compared with observed clinical data of dapagliflozin exposure following single doses in adolescents with T2DM | ||||||
|---|---|---|---|---|---|---|
| Dapagliflozin single dose | AUCINF, h·ng/mL (95% CI) | Cmax, ng/mL (95% CI) | AUCINF, h·ng/mL (95% CI) | Cmax, ng/mL (95% CI) | AUC | Cmax |
| 2.5 mg | 101.0 (83.7–120) | 24.8 (14–31) | 79.82 (73.51–86.67) | 26.76 (23.57–30.38) | 0.79 | 1.08 |
| 10 mg | 427.0 (346–535) | 118.0 (94–155) | 335.24 (307.46–365.54) | 112.85 (99.35–128.18) | 0.79 | 0.96 |
Data are expressed as geometric means. Single‐dose and multiple‐dose PK analyses are from clinical studies MB102001 and MB102002, respectively. Single‐dose 10‐mg oral dapagliflozin in a hepatically impaired population and single‐dose 50‐mg oral dapagliflozin in a renally impaired population PK analysis are from clinical studies MB102007, and MB102027, respectively. Single‐dose PK is from clinical study NCT01525238. Oral and 14C intravenous monotherapy single dose in healthy adults is from clinical study NCT00908271. AUC, area under the curve; AUCINF, extrapolated area under the plasma concentration‐time curve from time zero to infinity; CI, confidence interval; Cmax, highest drug concentration in the plasma; PBPK, physiologically based pharmacokinetic; q.d., once daily; T2DM, type 2 diabetes mellitus.
PBPK simulations of dapagliflozin exposure in an adult population with severe renal or hepatic impairment
Certain patients may have physiological factors affected by morbidities that influence ADME and, by extension, dapagliflozin exposure. For example, patients with cancer 31 or T2DM 15 may suffer from varying degrees of hepatic or renal impairment, and this may affect dapagliflozin clearance and, therefore, drug exposure. Mean dapagliflozin concentration‐time profiles in plasma simulated by the mechanistic dapagliflozin PBPK model following single 10‐mg and 50‐mg oral doses using modified virtual population parameters for severe hepatic and renal impaired populations are shown in Figure 3c,d and those for moderate hepatic and renal impaired populations are shown in Figure 3e,f . A single 10‐mg oral dose of dapagliflozin in patients with the Child‐Pugh class C hepatic function classification led to drug exposure increases in AUC and Cmax that were 67% and 40% higher, respectively, compared with those patients with normal hepatic function. A single 50‐mg oral dose in patients with diabetes with severe renal impairment led to a 70% increase in AUC. PBPK model simulations reasonably captured observed exposures from renal and hepatic impaired populations (both moderate and severe) within the twofold acceptance criteria (Table 2 ). Since dapagliflozin metabolism is predominantly performed by the kidney and liver UGT enzymes, which is also reflected by clearance predictions using the PBPK model, this model reflects the drug exposure changes from physiological factors well.
PBPK simulations of dapagliflozin exposure in adolescents (11‐ to 17‐year‐olds)
The optimized model with CLpo after parameter estimation in the default virtual pediatric population was used for simulation of clinical trials with adolescents aged 11 to 18 years. The optimized model reasonably recreated the PK parameters observed in clinical trials (NCT01525238) within the twofold acceptance criteria for both 2.5 mg and 10 mg, which are the lowest and highest doses used in the clinical trial. The default UGT1A9 ontogeny model (scenario 1) in the virtual “Sim‐Pediatric” population was used, as by age 11, which is the youngest age at recruitment for the study, UGT1A9 would have achieved full maturation. This has been agreed to in both the literature used to build the default model 26 , 27 and in recently published results. 21 Full details of the predictions are shown in Figure 3g,h and Table 2 .
PBPK simulations of dapagliflozin exposure change owing to DDIs of rifampin and mefenamic acid
Simulations for dapagliflozin DDIs with CYP3A and UGT modulators were performed to further build the confidence for mechanistic dapagliflozin PBPK predictions and verify clinical PK data reproducibility. As detailed in Table S1 , DDI simulations using the dapagliflozin PBPK model well replicated (within twofold) the changes in dapagliflozin exposure with coadministration of rifampin and mefenamic acid observed in clinical studies (NCT01068756). As expected, the single dapagliflozin dose (10 mg) with the UGT inducer rifampin (600 mg) induced a moderate decrease in dapagliflozin exposure in both the predictions and observations from clinical DDI studies. Conversely, when the single dapagliflozin dose (10 mg) was coadministered with the UGT inhibitor mefenamic acid (250 mg), an increase in dapagliflozin exposure was demonstrated in both the PBPK modeling predictions and clinical DDI study. This demonstrates that enzyme interactions produced by inhibitor or inducer drugs in coadministrations are accurately incorporated in the mechanistic PBPK model for dapagliflozin leading to robust dapagliflozin exposure predictions.
Prospective predictions of the optimal dapagliflozin absolute dose in pediatric age groups using PBPK modeling and simulations
The optimized “middle‐out” PBPK model was determined as sufficiently robust to make prospective predictions to determine the oral dose in pediatric age groups that best matches the drug exposure in healthy adults after a 10‐mg oral dose. Oral dapagliflozin dose predictions using both UGT1A9 ontogeny models are detailed in Table 3 . More details of predicted plasma dapagliflozin exposure using both ontogeny models are shown in Figure 4 .
Table 3.
Observed dapagliflozin exposure in adult and adolescent patients and predicted dapagliflozin exposure in different pediatric age groups
| Age group | Ontogeny model | Dapagliflozin dose, mg, SD | AUCINF, h·ng/mL (95% CI) | Cmax, ng/mL (95% CI) |
|---|---|---|---|---|
| Healthy adults (observed, MB102001) | NA | 10 | 368.00 | 116.00 |
| T2DM adolescents (observed, NCT01525238) | NA | 10 | 427.00 | 118.00 |
| Healthy adults (predicted) | NA | 10 | 383.01 (353.58–414.90) | 146.23 (128.08–166.94) |
| 12 to 18 years (predicted) | Scenario 1 | 10 | 512.49 (465.25–564.53) | 170.87 (149.39–195.44) |
| Scenario 2 | 10 | 567.95 (516.33–624.74) | 177.32 (154.81–203.10) | |
| 6 to 12 years (predicted) | Scenario 1 | 5 | 510.92 (460.30–567.11) | 141.88 (124.88–161.21) |
| Scenario 2 | 5 | 427.04 (383.65–475.34) | 134.17 (118.21–152.29) | |
| 2 to 6 years (predicted) | Scenario 1 | 1.70 | 339.05 (305.85–375.85) | 71.47 (63.68–80.21) |
| Scenario 2 | 2.50 | 374.18 (336.18–416.47) | 97.48 (86.94–109.30) | |
| 1.5 to 2 years (predicted) | Scenario 1 | 1.20 | 392.25 (156.28–431.85) | 62.42 (56.08–69.48) |
| Scenario 2 | 2.00 | 407.47 (366.11–453.50) | 93.08 (83.66–103.57) | |
| 1 to 1.5 years (predicted) | Scenario 1 | 1.00 | 411.80 (374.90–431.85) | 56.77 (51.13–63.04) |
| Scenario 2 | 1.60 | 365.36 (328.33–406.56) | 79.55 (71.67–88.31) | |
| 0.5 to 1 year (predicted) | Scenario 1 | 0.50 | 316.71 (287.57–348.80) | 33.22 (30.02–36.76) |
| Scenario 2 | 1.25 | 360.51 (322.60–402.88) | 70.63 (63.86–78.13) | |
| 3 to 6 months (predicted) | Scenario 1 | 0.40 | 427.00 (389.32–469.92) | 29.63 (26.92–32.61) |
| Scenario 2 | 0.90 | 377.30 (336.23–423.40) | 62.85 (57.13–69.14) | |
| 1 to 3 months (predicted) | Scenario 1 | 0.15 | 363.62 (332.40–339.17) | 16.72 (15.23–18.36) |
| Scenario 2 | 0.60 | 402.28 (356.85–453.48) | 54.64 (49.85–89.90) |
Data are expressed as geometric means. Scenario 1 ontogeny model is default uridine diphosphate‐glucuronosyltransferase (UGT) 1A9 model found in the virtual population, Sim‐Pediatric; scenario 2 ontogeny model is a modified UGT1A9 model based on Badee et al. 22 . AUCINF, extrapolated area under the plasma concentration‐time curve from time zero to infinity; CI, confidence interval; Cmax, highest drug concentration in the plasma; NA, not applicable; SD, single dose; T2DM, type 2 diabetes mellitus.
Figure 4.
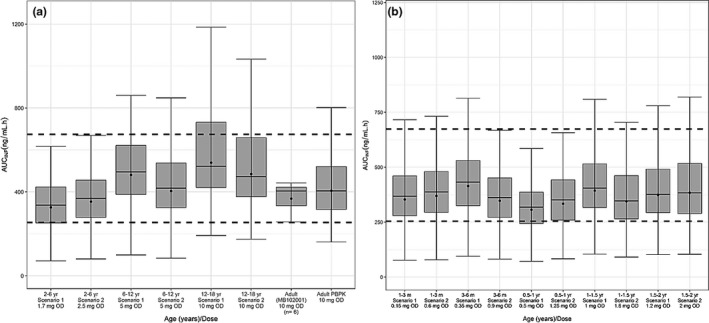
Boxplot of dapagliflozin plasma exposure from predicted optimal OD using a PBPK model in virtual pediatric populations using two different uridine diphosphate‐glucuronosyltransferase 1A9 ontogeny models in pediatric age groups (a) 2 to 18 years old and (b) 1 month to 2 years old. The dotted black lines indicate lower 2.5% and upper 97.5% of exposure reported in healthy adults after a 10‐mg oral dapagliflozin. Closed circles indicate geometric means; line indicates median. AUCINF, area under the curve extrapolated from administration to infinity; m, months old; OD, oral dose; PBPK, physiologically based pharmacokinetic; yr, years old.
Since UGT1A9 is the main metabolizing enzyme for dapagliflozin, accurate modeling of the UGT1A9 ontogeny is particularly significant for robust predictions in younger pediatric age groups between 1 month and 2 years. Predicted dapagliflozin clearance at steady state in children is shown in Figure S4 . The scenario 2 UGT1A9 ontogeny model predicted faster enzyme activity maturation, with the enzyme activity achieving the equivalent of that in adults at 4 months of age, leading to higher clearance, and thus presented higher dose predictions than scenario 1.
Discussion
This work outlines the development of an oral dapagliflozin PBPK model and successfully reproduced observed clinical PK data in obese adolescents with T2DM, such as AUC and Cmax, within the 0.5‐ to 2‐fold acceptance criteria in various scenarios to determine robustness and prediction confidence, including in specific populations and DDIs. The model also demonstrated applicability for these scenarios for prospective predictions when clinical PK is unavailable. This model was then used to determine the absolute single oral dose in pediatric age groups between 1 month and 18 years that best matches the exposure from a single 10‐mg oral dose through prospective PK predictions.
The PBPK model for oral dapagliflozin was developed using a mechanistic approach and well predicted the exposure in a virtual healthy adult population receiving a 5‐ to 100‐mg single oral dose and 10‐mg multiple doses within the predetermined twofold acceptance criteria of clinical PK data after dapagliflozin monotherapy. Clinical DDI studies with a UGT inhibitor, mefenamic acid, and a CYP3A/UGT inducer, rifampin, were also used to verify the oral dapagliflozin PBPK model prediction confidence. The plasma concentration‐time profiles for dapagliflozin from PBPK predictions were within the determined 0.5‐ to 2‐fold acceptance criteria of observed data from adults. Having a verified PBPK model using adult and adolescent clinical data supported by literature on ontology instills confidence in predicting dapagliflozin pediatric exposure.
Continuing with the dapagliflozin PBPK model verification, simulations for oral dapagliflozin administration in patients with severe hepatic and renal impairment were performed to verify the PBPK model robustness in predicting the effects of physiological factors on dapagliflozin plasma exposure. Clinical studies of dapagliflozin monotherapy in patients with severe hepatic or renal impairment reported a > 50% increase in dapagliflozin plasma exposure owing to reduced clearance abilities. The use of virtual populations developed by Simcyp for severe renal impairment (GFR < 30 mL/minute) and severe hepatic impairment (Child‐Pugh class C) initially failed to reproduce the observed clinical PK within twofold. This was attributed to the default virtual population failing to account for the reduction in liver and kidney UGT enzyme activities in severe hepatic impairment and reduced kidney UGT enzyme activity in severe renal impairment, although there was a reduction in CYP metabolizing enzymes in both virtual populations. Dapagliflozin is a primarily UGT1A9‐metabolized oral drug; thus, accurate representation of the UGT‐metabolizing activity for both virtual populations is significant for exposure predictions. Such reductions in the UGT‐metabolizing activity have been well documented in the literature for patients with severe renal 15 and hepatic impairment. 13 , 32 The virtual population physiological parameters for UGT abundance in both liver and kidney were reduced 46% in the Simcyp virtual population “GFR < 30 mL/min” to simulate severe hepatic impairment. The UGT abundance in the kidney was eliminated in the Simcyp virtual population “Child‐Pugh class C” to match the observed clearance from clinical studies after sensitivity analysis, while other physiological parameters remained unchanged. After modifying both virtual populations, dapagliflozin exposure predictions after matching clinical design corresponded to the observed clinical PK within the twofold acceptance criteria, verifying the model’s capability for exposure predictions with significant physiological factors.
The dapagliflozin PBPK model also well predicted dapagliflozin exposure observed in clinical studies within 0.5‐ to 2‐fold for monotherapy in obese adolescents aged 11 to 17 years with T2DM after optimizing CLpo from the clinical PK data from NCT01525238. The literature used to build the default UGT1A9 ontogeny model, 26 , 27 which determines the UGT1A9‐metabolizing activity as a fraction of the metabolism in adults, was not consistent with other, more recent, literature reports. 22 Oral dapagliflozin is primarily metabolized by UGT1A9, and thus exposure predictions are expected to be significantly affected by UGT1A9 ontogeny, particularly in pediatric age groups between 1 month and 1 year. The default UGT1A9 ontogeny model in Simcyp’s pediatric virtual population was kept as “scenario 1” and a different literature source‐based UGT1A9 ontogeny model 22 was created as “scenario 2”. An optimized “middle‐out” dapagliflozin PBPK model was used for dapagliflozin exposure in pediatric age groups between 1 month and 18 years using both UGT1A9 ontogeny models.
Since the “scenario 2” UGT1A9 model reports a quicker maturation and, by extension, a higher drug clearance than “scenario 1,” dosing recommendations using “scenario 2” consistently predict a higher absolute oral dose for pediatric age groups from 1 month to 6 years to match the observed dapagliflozin exposure from a 10‐mg single dose in healthy adults. Dosing recommendations should be verified and revised when more clinical PK data are available in patients < 11 years old.
In conclusion, a mechanistic oral dapagliflozin PBPK model was developed that adequately predicted the results observed in dapagliflozin monotherapy studies in healthy and impaired populations as well as DDIs with UGT inhibitors and inducers. A CLpo‐optimized PBPK model was developed for exposure predictions in pediatric age groups and for dose recommendations to best match the exposure found in healthy adults after a 10‐mg single dose of dapagliflozin.
Funding
This study was supported by AstraZeneca.
Conflict of Interest
Venkatesh Pilla Reddy, Joanna Parkinson, David W. Boulton, and Weifeng Tang are full‐time employees of AstraZeneca. Heeseung Jo was a placement student at AstraZeneca when this study was conducted.
Author Contributions
V.P.R. and H.J. wrote the manuscript. V.P.R., D.B., J.P., and W.T designed the research. V.P.R. and H.J. performed the research. V.P.R. and H.J. analyzed the data.
Supporting information
Supplementary Material
Acknowledgments
The authors would like to thank the patients, their families, and the clinical teams who worked on the studies. Heeseung Jo was a placement student at AstraZeneca, UK, at the time the study was conducted. The authors thank Cactus Life Sciences (part of Cactus Communications, Mumbai, India) for proofreading.
Data Availability Statement
The authors confirm that the clinical data supporting the physiologically based pharmacokinetic modeling are listed in ClinicalTrials.gov, and details of these clinical studies have been published elsewhere.
References
- 1. Plosker, G.L. Dapagliflozin: a review of its use in patients with type 2 diabetes. Drugs 74, 2191–2209 (2014). [DOI] [PubMed] [Google Scholar]
- 2. Goldenberg, M.M. Pharmaceutical approval update. P & T. 39, 619–620, 26 (2014). [PMC free article] [PubMed] [Google Scholar]
- 3. Wiviott, S.D. et al Dapagliflozin and cardiovascular outcomes in type 2 diabetes. N. Engl. J. Med. 380, 347–357 (2019). [DOI] [PubMed] [Google Scholar]
- 4. Kemp, A. Farxiga granted FDA Priority Review for patients with heart failure with reduced ejection fraction (AstraZeneca, 2020). [Google Scholar]
- 5. Boulton, D.W. et al Simultaneous oral therapeutic and intravenous (1)(4)C‐microdoses to determine the absolute oral bioavailability of saxagliptin and dapagliflozin. Br. J. Clin. Pharmacol. 75, 763–768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Callegari, E. , Lin, J. , Tse, S. , Goosen, T.C. , Sahasrabudhe, V. & Kumar, V. Physiologically based pharmacokinetic modeling of the drug drug interaction between rifampin and SGLT2 inhibitors: the contribution of CYP induction to the observed interaction. Drug Metab. Pharmacokinet. 33, S48–S49 (2018). [Google Scholar]
- 7. RACE for Children Act <https://www.govtrack.us/congress/bills/115/s456> (2017). Accessed March 10, 2020. S. 456.
- 8. Shebley, M. et al Physiologically based pharmacokinetic model qualification and reporting procedures for regulatory submissions: a consortium perspective. Clin. Pharmacol. Ther. 104, 88–110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pilla Reddy, V. , Walker, M. , Sharma, P. , Ballard, P. & Vishwanathan, K. Development, verification, and prediction of osimertinib drug‐drug interactions using PBPK modeling approach to inform drug label. CPT Pharmacometrics Syst. Pharmacol. 7, 321–330 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pilla Reddy, V. , Bui, K. , Scarfe, G. , Zhou, D. & Learoyd, M. Physiologically based pharmacokinetic modeling for olaparib dosing recommendations: bridging formulations, drug interactions, and patient populations. Clin. Pharmacol. Ther. 105, 229–241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parkinson, J. et al Comparison of the exposure‐response relationship of dapagliflozin in adult and paediatric patients with type 2 diabetes mellitus. Diabetes Obes. Metab. 18, 685–692 (2016). [DOI] [PubMed] [Google Scholar]
- 12. Jamei, M. Recent advances in development and application of physiologically‐based pharmacokinetic (PBPK) models: a transition from academic curiosity to regulatory acceptance. Curr. Pharmacol. Rep. 2, 161–169 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye, L. et al Sorafenib metabolism is significantly altered in the liver tumor tissue of hepatocellular carcinoma patient. PLoS One 9, e96664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Prasad, B. et al Abundance of phase 1 and 2 drug‐metabolizing enzymes in alcoholic and hepatitis C cirrhotic livers: a quantitative targeted proteomics study. Drug Metab. Dispos. 46, 943–952 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sayama, H. , Takubo, H. , Komura, H. , Kogayu, M. & Iwaki, M. Application of a physiologically based pharmacokinetic model informed by a top‐down approach for the prediction of pharmacokinetics in chronic kidney disease patients. AAPS J. 16, 1018–1028 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soars, M.G. , Petullo, D.M. , Eckstein, J.A. , Kasper, S.C. & Wrighton, S.A. An assessment of udp‐glucuronosyltransferase induction using primary human hepatocytes. Drug Metab. Dispos. 32, 140–148 (2004). [DOI] [PubMed] [Google Scholar]
- 17. Vietri, M. , Pietrabissa, A. , Mosca, F. & Pacifici, G.M. Mycophenolic acid glucuronidation and its inhibition by non‐steroidal anti‐inflammatory drugs in human liver and kidney. Eur. J. Clin. Pharmacol. 56, 659–664 (2000). [DOI] [PubMed] [Google Scholar]
- 18. Anderson, B.J. et al Acetaminophen developmental pharmacokinetics in premature neonates and infants: a pooled population analysis. Anesthesiology 96, 1336–1345 (2002). [DOI] [PubMed] [Google Scholar]
- 19. van den Anker, J.N. et al Insufficient ketoconazole concentrations in preterm infants with fungal infections. Eur. J. Pediatr. 152, 538 (1993). [DOI] [PubMed] [Google Scholar]
- 20. de Wildt, S.N. et al Pharmacokinetics and metabolism of oral midazolam in preterm infants. Br. J. Clin. Pharmacol. 53, 390–392 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bhatt, D.K. et al Age‐ and genotype‐dependent variability in the protein abundance and activity of six major uridine diphosphate‐glucuronosyltransferases in human liver. Clin. Pharmacol. Ther. 105, 131–141 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Badee, J. , Fowler, S. , de Wildt, S.N. , Collier, A.C. , Schmidt, S. & Parrott, N. The ontogeny of UDP‐glucuronosyltransferase enzymes, recommendations for future profiling studies and application through physiologically based pharmacokinetic modelling. Clin. Pharmacokinet. 58, 189–211 (2019). [DOI] [PubMed] [Google Scholar]
- 23. Salem, F. , Johnson, T.N. , Abduljalil, K. , Tucker, G.T. & Rostami‐Hodjegan, A. A re‐evaluation and validation of ontogeny functions for cytochrome P450 1A2 and 3A4 based on in vivo data. Clin. Pharmacokinet. 53, 625–636 (2014). [DOI] [PubMed] [Google Scholar]
- 24. Zaya, M.J. , Hines, R.N. & Stevens, J.C. Epirubicin glucuronidation and UGT2B7 developmental expression. Drug Metab. Dispos. 34, 2097–2101 (2006). [DOI] [PubMed] [Google Scholar]
- 25. Emoto, C. , Fukuda, T. , Johnson, T.N. , Neuhoff, S. , Sadhasivam, S. & Vinks, A.A. Characterization of contributing factors to variability in morphine clearance through PBPK modeling implemented with OCT1 transporter. CPT Pharmacometrics Syst. Pharmacol. 6, 110–119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Strassburg, C.P. et al Developmental aspects of human hepatic drug glucuronidation in young children and adults. Gut 50, 259–265 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leakey, J.E. , Hume, R. & Burchell, B. Development of multiple activities of UDP‐glucuronyltransferase in human liver. Biochem. J. 243, 859–861 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bjorkman, S. Prediction of drug disposition in infants and children by means of physiologically based pharmacokinetic (PBPK) modelling: theophylline and midazolam as model drugs. Br. J. Clin. Pharmacol. 59, 691–704 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson, T.N. , Rostami‐Hodjegan, A. & Tucker, G.T. Prediction of the clearance of eleven drugs and associated variability in neonates, infants and children. Clin. Pharmacokinet. 45, 931–956 (2006). [DOI] [PubMed] [Google Scholar]
- 30. Edginton, A.N. , Schmitt, W. , Voith, B. & Willmann, S. A mechanistic approach for the scaling of clearance in children. Clin. Pharmacokinet. 45, 683–704 (2006). [DOI] [PubMed] [Google Scholar]
- 31. Schwenger, E. et al Harnessing meta‐analysis to refine an oncology patient population for physiology‐based pharmacokinetic modeling of drugs. Clin. Pharmacol. Ther. 103, 271–280 (2018). [DOI] [PubMed] [Google Scholar]
- 32. Hardwick, R.N. et al Altered UDP‐glucuronos yltransferase and sulfotransferase expression and function during progressive stages of human nonalcoholic fatty liver disease. Drug Metab. Dispos. 41, 554–561 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
The authors confirm that the clinical data supporting the physiologically based pharmacokinetic modeling are listed in ClinicalTrials.gov, and details of these clinical studies have been published elsewhere.


