ABSTRACT
Tumor-associated macrophages contribute to cell growth, development, and metastasis in various cancers. However, the underlying mechanisms of M2 macrophage that modulate the progression of gastric cancer (GC) remain largely unknown. In this study, we detected the ratio of macrophages in GC tissues and found that the proportion of M2 macrophages was increased in GC tissues. We then co-cultured GC cells with M1 and M2 macrophages, respectively, and then assessed cell proliferation and tumorigenicity of GC cells by MTT and colony formation assay. The results indicated that M2 macrophages promoted the proliferation of GC cells, but M1 not. Besides, GW4869, an exosomes inhibitor, reduced the effects induced by M2 macrophage. Then, we isolated and identified exosomes derived from M1 and M2 macrophage, and confirmed that the exosomes could be taken up by GC cells. We demonstrated that M2 macrophage-exosomes could induce the proliferation and tumorigenesis in vitro and in vivo. Moreover, miR-487a was enriched in M2 macrophage-exosomes and further determined that miR-487a exert the functions by targeting TIA1. In conclusion, exosomal miR-487a derived from M2 macrophage promotes the proliferation and tumorigenesis in gastric cancer, and the novel findings might be helpful to the development of novel diagnostic and therapeutic methods in GC.
KEYWORDS: M2 macrophage, exosome, proliferation, tumorigenesis, microRNA, gastric cancer
Introduction
Gastric cancer (GC), the fifth most common malignant tumors in the world, is the third most lethal cancers globally [1,2]. Since the early stage of GC is asymptomatic, it is always delayed in the diagnosis and missed the best opportunities for treatment [3]. Despite advances in diagnostic methods and various treatments, the prognosis of GC patients (especially clinical stage at III and IV) remains poor, and the overall 5-year survival rate of GC less 30% [4–6]. Thus, it is necessary to understand the underlying molecular mechanisms of GC comprehensively that might be useful for the development of novel diagnostic and therapeutic methods.
Macrophages (Mφs) are the essential participators in cancer pathogenesis and they could be divided into two general species (namely M1 and M2) based on their function [7]. Specifically, M1 polarized Mφs is provided with the anti-tumor ability, while M2 tumor-associated Mφs (TAMs) possess the tumor-promoting effect [8]. Furthermore, more subdivisions exist within the general M2 Mφ category [7]. With the help of a typical scheme, M2 Mφs can also be divided into M2a, M2b, M2c, and M2d/TAM subclasses [7,9]. Generally speaking, IL-10 production is joint for all M2 subclasses [7].
Exosomes are a class of homogeneous membrane vesicles ranged from 30 to 150 nm [10], and play roles as mediators of intercellular communication by transferring complex and distinctive molecules, including mRNA, miRNA [11,12], proteins [13,14], and lipids [15]. Exosomes are characterized by specific surface proteins, including CD9, CD63, CD81 and TSG101 [16–19]. Exosomes can be released by almost all kinds of normal and cancer cells [20,21], and exist stably in almost all kinds of body fluids, including the blood [22], saliva [23], skin milk [24,25], cerebrospinal fluid [26] and urine [27]. In the past 3 years, more and more researchers focused on the roles and effects of exosomes in the microenvironment, and many studies reported that tumor-derived exosomes contributed to cancer growth, development, and metastasis. MicroRNA, as a class of vital gene expression regulators, has been received extensive attention to be intensive studied combined with exosomes. For example, Ma et al [28] reported that exosomal miR-199a-3p promotes proliferation and migration in neuroblastoma, and plasma-derived exosomal miR199a-3p may be used as an efficient, accurate, and noninvasive biomarkers clinically. Bai et al [29] reported that exosomes derived from GC cells, carrying miR-135b, down-regulates the protein level of FOXO1, and promotes angiogenesis. These findings suggested that miRNAs can be delivered to receptor cells through exosomes to be a gene expression regulator.
In this study, we identified that M2 macrophage-derived exosomes promoted tumor growth and tumorigenesis of GC. We found that exosomal miR-487a, specifically highly expressed in M2 macrophage-exosomes rather than in M1 macrophage-exosomes, were transferred to GC cells to induce the proliferation and tumorigenesis. And exosomal miR-487a regulated the expression of TIA1 negatively. In a word, exosomal miR-487a derived from M2 macrophage promotes tumorigenesis in gastric cancer, and the novel findings may help treat GC in the future.
Materials and methods
Patients and specimens
All tissues used in this study were collected from patients undergoing surgery at the Affiliated Hospital of Zunyi Medical University (Zunyi, China). None of the patients received preoperative radiotherapy or chemotherapy.
This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Zunyi Medical University (Zunyi, China). Each patient and their respective guardian signed informed consent forms.
Immunohistochemical
Immunohistochemical analysis for CD68 antigen was performed on GC tissue slides. GC tissue samples were embedded in paraffin and sliced into sections at the thickness of 5 μm. After deparaffinization at 60°C temperature, slides were rehydrated by immersion in decreasing grades of ethanol. Subsequently, endogenous peroxidase activity was inhibited by immersing the tissues for 20 min in methanol containing 0.3% hydrogen peroxide (H2O2). Antigens retrieval was performed by autoclaving for 10 min in citrate buffer (pH 6). After antigen retrieval, the sections were blocked by 5% BSA at room temperature for 30 min, and then incubated with CD68 antibody (Abcam, UK) which was diluted 1:100 at 4°C overnights. The following day, the stained sections were incubated with secondary antibody biotin-conjugated goat anti-rabbit IgG (Proteintech Group, Inc./Thermo Fisher Scientific; SA00004-2; 1:200) at room temperature for 30 min in dark, and visualized with diaminobenzidine (DAB) chromogen. Finally, the nucleus was counterstained by hematoxylin. The pictures were taken by a DMi8 optical microscope (Leica).
Flow cytometry
When cell confluency reached 80 ~ 90%, cells (1 × 106 cells) were gently collected into centrifuge tubes for centrifugation and washed (1 × PBS, 0.5% bovine serum albumin, 2 mM EDTA) twice, counted, and then incubated with respective primary antibody at 4°C for 30 min in dark. Cells were washed three times with cold PBS and then stained with secondary antibody. Monoclonal antibodies for M1together with M2 macrophage are anti-CD4 and anti-CD8 (BD Biosciences, USA). Besides, Monoclonal antibodies for M2 macrophage are anti-CD206 and anti-CD163 (BD Biosciences, USA). The stained cells were analyzed with FACS Calibur (BD Biosciences, USA). The data were analyzed by FlowJo software 8.7.1 (Treestar Inc., USA).
Cell co-culture system
M1 macrophage or M2 macrophage (3 × 105/well) were seeded in the upper chamber of a co-culture system with a 0.4 µm pore membrane, and HS-746 T cells (5 × 105/well) were placed in the lower chamber. After 48 hours of culture, HS-746 T cells were collected for subsequent experiments.
Exosomes isolation and identification
Macrophages were cultured in the conditional medium supplemented with 10% exosome-free fetal bovine serum (FBS, Gibco-BRL, Grand Island, NewYork) for 48 h. When cell confluence reached 80 ~ 90%, 40 mL of used medium from different cells was collected, and exosomes were isolated through ultracentrifugation described in previous studies [30]. The collected medium was centrifuged at 4°C, 500 g for 10 min, followed by 20,000 g for 30 min at 4°C. The supernatants were penetrated through a 0.22 μm filter purchased from Millipore and ultra-centrifuged at 4°C, 100,000 g overnight. The exosome precipitate was washed with PBS and then resuspended in PBS.
Identification of exosomes: The sample was adsorbed onto a carbon-coated nickel grid and examined at an 80 kV acceleration voltage in an electron microscope (JEM-1230, Nihon Denshi, Tokyo, Japan). Particle size analysis for exosomes was conducted with the nanoparticle tracking analysis (NS300, Malvern Instruments Ltd, Worcestershire, United Kingdom). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was prepared for protein denaturation and electrophoresis, followed by the membrane transferring. The expression of exosomes specific marker proteins were detected by western blot.
Visualization of exosomes
PKH67 green fluorescent membrane dye (Sigma, USA) was used to label exosomes. Labeled exosomes were incubated with HS-746 T cells for 2 h. After incubation, cells were fixed using 4% paraformaldehyde for 30 min and following stained nucleus with DAPI.
Exosome treatment
HS-746 T cells were cultured by conditional medium supplemented with 100 µg exosomes. The control group was cultured with the fresh medium supplemented with the same amount of PBS. After 48 hours of culture, the cells were collected for subsequent experiments.
MTT assay
HS-746 T cells were seeded into 96-well plates (5 × 103 cells per well) and cultured overnight. Subsequently, 10 μL of MTT (3-(4,5-dimethylthiazol-2-yl)- 2,5-diphenyltetrazolium bromide) solution (5 mg/ml; Beyotime, China) was added into each well at 24, 48, 72 and 96 h, respectively. Following incubated for 2 h, the absorbance was measured at 450 nm using a spectrophotometer (FLX800, Bio-TEK).
Colony formation assay
HS-746 T cells were seeded into well plates (500 cells per well) and cultured for 14 days. The cells in every well were fixed with 4% paraformaldehyde for 30 min and then stained with 0.2% crystal violet at room temperature for 30 min. The number of cell colonies (more than 50 cells) was counted.
qRT-PCR
Total RNA of cells or exosomes was extracted with Trizol reagent (Invitrogen, USA) and cDNA was synthesized using Reverse Transcription Reagent Kit (Takara, Dalian, China) according to the manufacturer’s instructions. Then, qRT-PCR was performed with SYBR qPCR Mix Kit (Takara, Dalian, China). The PCR primers are listed in Table 1. U6 snRNA was used as an internal control for the expression analysis of miR-487a. The relative expression levels of target genes were evaluated using the 2-ΔΔCT method.
Table 1.
Primer sequences used for the qRT-PCR analysis
| Application | Oligonudeotides | Sequences (5’-3’) | |
|---|---|---|---|
| miR-487a | RT Forward Reverse |
GTCGTATCCAGTGCAGGGTCCGAGGTA TTCGCACTGGATACGACAACTGG CGCTGGCAATCATACAGGGACAT GTGCAGGGTCCGAGGT |
|
| U6 | RT Forward Reverse |
AACGCTTCACGAATTTGCGT CTCGCTTCGGCAGCACA AACGCTTCACGAATTTGCGT |
|
| GAPDH | Forward Reverse |
TGTGGGCATCAATGGATTTGG ACACCATGTATTCCGGGTCAAT |
|
Western blot
Total protein of cells or exosomes was extracted by RIPA lysis buffer (Beyotime, China). And the concentration was measured by the BCA kit (Beyotime, China). The protein denatured by 95 °C heating were separated by SDS-PAGE and transferred onto a PVDF membrane. After blocking by 5% skim milk at room temperature for 2 h, the membranes were incubated, respectively, with specific primary antibodies at 4°C overnight. The primary antibodies: pJAK1 (1:500, ab75599), JAK1 (1:500, ab183894), p-STAT3 (1:1000, ab76315), STAT3 (1:1000, ab68153), E-cadherin (1:500, ab231303), N-cadherin (1:500, ab245117) and vimentin (1:500, ab20346), GAPDH (1:5000, ab8245) and Lamin A (1:2000, ab108595). All of the antibodies were purchased from Abcam, USA: CD63 (1:1000, ab134045), CD9 (1:1000, ab92726), CD81 (1:1000, ab109201), TSG101 (1:1000, ab125011), Tubulin (1:2000, ab7291). The membranes were incubated with secondary antibodies combined with HRP (1: 5000, proteintech, China) at room temperature for 2 h in the next day. The bands on the membranes were visualized with ECL Western Blotting Substrate and collected photos using a Tanon 5200 system (Tanon, China).
Animal experiments
Twenty male BALB/c nude mice aged 6 weeks were randomly divided into four groups: I control (injection of HS-746 T cells alone); II M1-exosomes (injection of HS-746 T cells treated with M1 macrophage-derived exosomes); III M2-exosomes (injection of HS-746 T cells treated with M2 macrophage-derived exosomes) and IV OE-miR-487a M2-exosomes (injection of HS-746 T cells treated with exosomes derived from M2 macrophage transfected with miR-487a mimics) . There were 5 mice in each group. The infected cells (1 × 106) were resuspended in 100 µl PBS and subcutaneously inoculated into the right groin of nude mice. The tumor volume and weight of mice were measured every 2 days. After 2 weeks of injection, the tumor tissues were dissected.
Statistical analysis
Each experiment was repeated three times independently, and data were shown as mean±SEM. Differences between groups were compared by Student’s t-test and p < 0.05 was defined as significant. All statistical analyses were performed by GraphPad Prism 7.0.
Results
1. The proportion of M2 macrophage was increased in human GC tissues
To assess the vital roles of macrophages in tumor progression, we assessed the expression level of CD68 (macrophage marker) in tumor tissues and para-carcinoma tissues by immunohistochemistry (n = 5) (Figure 1a-Figure 1B). Compared with para-carcinoma tissues, the number of macrophages in GC tissues was increased in Figure 1a-Figure 1B. Then, we detected the M1/M2 macrophages percentage via flow cytometry assay in Figure 1c, and found the proportion of CD8+/CD4+ macrophage was significantly decreased in GC tissues (Figure 1c). Furthermore, the percentage of M2 macrophage in para- and GC tissues were also analyzed by Flow cytometry in Figure 1d, and the data showed that the proportion of CD163+/CD206+ macrophage was significantly increased in GC tissues (Figure 1d). Besides, mRNA expression levels of M2 phenotype markers including irf4 and Arg-1 were up-regulated in tumor-associated macrophage (TAMs) compared to normal tissues macrophage (NTM), where the mRNA expression levels of M1 phenotype markers (including TNF-α, irf-5, iNOS, IL-6) were downregulated (Figure 1e). Herein, the above results demonstrated that M2 macrophage contributed to the progression of GC.
Figure 1.
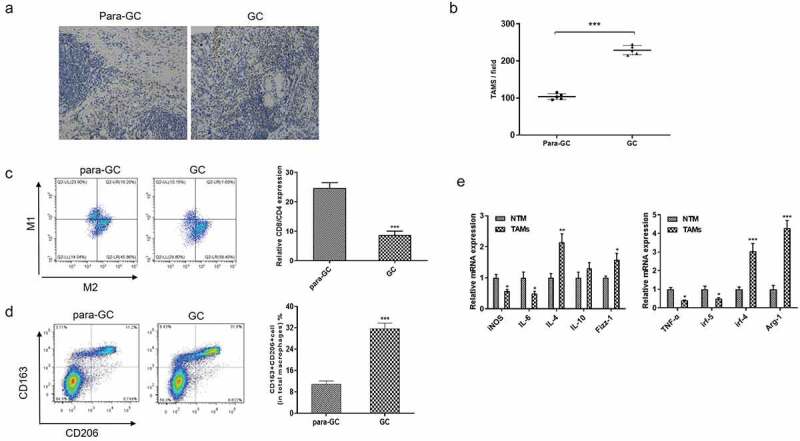
The proportion of M2 macrophage was increased in human GC tissues. (A-B). Immunohistochemistry analyze of CD68 expression level in para-GC tissues and GC tissues (n = 5). Bar scan = 100 μm. (C-D). Flow cytometry analysis of M1 and M2 macrophage markers in para-GC tissues and GC tissues. (E). qRT-PCR analyze of M1 and M2 macrophage markers in tumor-associated macrophage (TAMs) compared to normal tissues macrophage. *p < 0.05, **p < 0.01, ***p < 0.001 vs control. Data are displayed as mean ± SEM
2. M2 macrophage increased the proliferation and tumorigenesis of GC cells
To investigate the effects of M2 macrophage on GC progression, we constructed a co-culture system that consisted of two phenotypes macrophage and GC cells, HS-746 T cells. MTT assay revealed that M2 macrophage significantly promoted HS-746 T cells growth, while M1 macrophage did not affect the proliferation of HS-746 T cells (Figure 2a). Besides, HS-746 T cells co-cultured with M2 macrophage exhibited increased colony formation, compared with control and HS-746 T cells co-cultured with M1 macrophage (Figure 2b).
Figure 2.
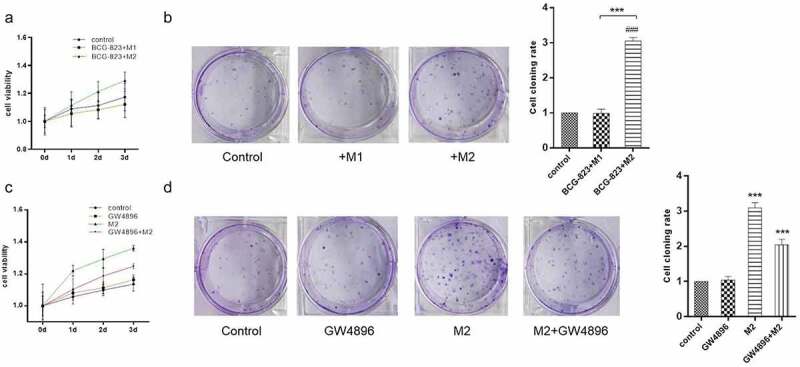
M2 macrophage increased the proliferation and tumorigenesis of GC cells through exosomes. (A). MTT assay of HS-746 T cells co-cultured with M1 macrophage, M2 macrophage or single culture. (B). colony formation assay was applied to assess the tumorigenesis ability of HS-746 T cells treated with the same conditions as (A). (C). MTT assay of HS-746 T cells treated with GW4896, M2 macrophage, GW4896 + M2 macrophage or no-treatment. (D). colony formation assay was applied to assess the tumorigenesis ability of HS-746 T cells treated with the same conditions as (C). ***p < 0.001 vs control. ###p < 0.001 vs M1 macrophage co-culture group. Data are displayed as mean ± SEM
In recent years, increasing studies have shown that exosomes, act as intercellular communication transporters, play key roles in the tumor microenvironment. To further verify whether the pro-proliferation of M2 macrophage depended on exosomes, we added GW4869 (exosome inhibitor) to reduced exosome secretion by M2 macrophage that co-cultured with GC cells. The results of MTT assay and colony formation assay showed that GW4869 inhibited the proliferation of HS-746 T cells induced by M2 macrophage (Figure 2c- D). These results suggested that M2 macrophage increased the proliferation and tumorigenesis of GC cells relied on exosome secretion.
3. Exosomes derived from M2 macrophage promoted the proliferation and tumorigenesis of GC cells
To confirm whether exosomes derived from M2 macrophage are involved in GC progression, we isolated exosomes from the conditioned medium of M2 macrophage and identified exosomes by electron microscopy and Nanoparticle tracking analysis (NTA). It could be clearly observed that typical exosomal double-membrane vesicles under the electron microscope (Figure 3a). NTA further determined that the vesicle particles were ranged from 100 to 150 nm in diameter with an enriched diameter of 130 nm (Figure 3b). To confirm the purity of exosomes, positive exosomal markers CD63, CD9, CD81, TSG101, and negative exosomal marker Tubulin were analyzed by western blot. As Figure 3c shown, the four recognized exosomal markers were abundant in particles which we extracted. These results indicated that the vesicle particles isolated from the M2 macrophage displayed typical exosomal characteristics.
Figure 3.
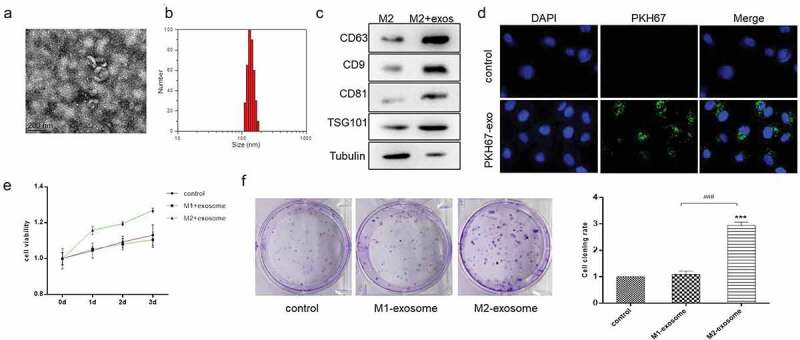
Exosomes derived from M2 macrophage promoted the proliferation and tumorigenesis of GC cells. (A). Scanning electron microscopy of exosomes isolated from conditioned medium of M2 macrophage. (B). Nanoparticle tracking analysis (NTA) of M2 macrophage-exosomes isolated by ultracentrifugation. (C). Western blot analysis of exosome markers. Tubulin was used as internal control. (D). Immunofluorescence analysis of the internalization of PKH67-labeled M2 macrophage-exosome (green) by HS-746 T cells. (E & F). HS-746 T were treated with M1 macrophage-exosomes, M2 macrophage-exosomes (100 ug/ml) or no-treatment. After 24 h, MTT was used to test cell viability of HS-746 T (E), and colony formation assay was used to detect cell proliferation (F). ***p < 0.001 vs control. ###p < 0.001 vs M1 macrophage-exosomes group. Data are displayed as mean ± SEM
To visualize exosome transfer in vitro, M2 macrophage-exosomes were labeled with PKH67, a green fluorescent dye, and incubated with HS-746 T cells. After 2 h, we observed the green fluorescence in the cytoplasm of HS-746 T (Figure 3d), which suggested that M2 macrophage-exosomes were internalized by GC cells.
To further explore the functional roles of M2 macrophage-exosomes, we treated HS-746 T cells with exosomes derived from M1 macrophage and M2 macrophage, respectively. MTT assay showed that compared with the no-treatment group and the M1-exo treatment group, the M2-exo treatment significantly induced the proliferation of HS-746 T cells (Figure 3e). And the results of clone formation indicated that M2 macrophage-exosomes promoted cell colony-forming ability of HS-746 T cells (figure 3f). Therefore, we hypothesized that the M2 macrophage-exosomes contained a unique and effective molecule that promoted GC cell growth.
4. M2 macrophage-exosomes exerted the functions in a miR-487a-dependent manner
Previously, miR-487a has been reported to be involved in the proliferation and migration of numerous types of tumors [31–34]. Particularly, it has been confirmed that miR-487a promotes the progression of gastric cancer via targeting TIA1 [34].
To investigate whether the carcinogenic effects of M2 macrophage-exosomes on GC cells are mediated by miR-487a, qRT-PCR was performed to analyze the expression level of miR-487a in exosomes derived from different phenotypes macrophage. As shown in Figure 4a, miR-487a was highly expressed in M2 macrophage-exosomes. Furthermore, we detected the expression level of miR-478a in HS-746 T cells treated with M1-exo, M2-exo, or nothing. The results demonstrated that miR-478a was markedly up-regulated by M2 exosome, and the M1 exosome treatment group showed similar effects on the non-treatment group (Figure 4b). Besides, TIA1, the direct target gene of miR-487a, exhibited the expected trend, which was down-regulated by M2 exosomes (Figure 4c). Confocal high content analysis displayed that the cy3-labeled-Mir-487a were efficiently delivered to HS-746 T cells via exosomes (Figure 4d). MiR-487a could be transferred to GC cells through M2 macrophage exosomes and promotes the progression of gastric cancer via down-regulating TIA1. These results suggested that M2 macrophage exosomes promoted GC progression in a miR-487a-dependent manner.
Figure 4.
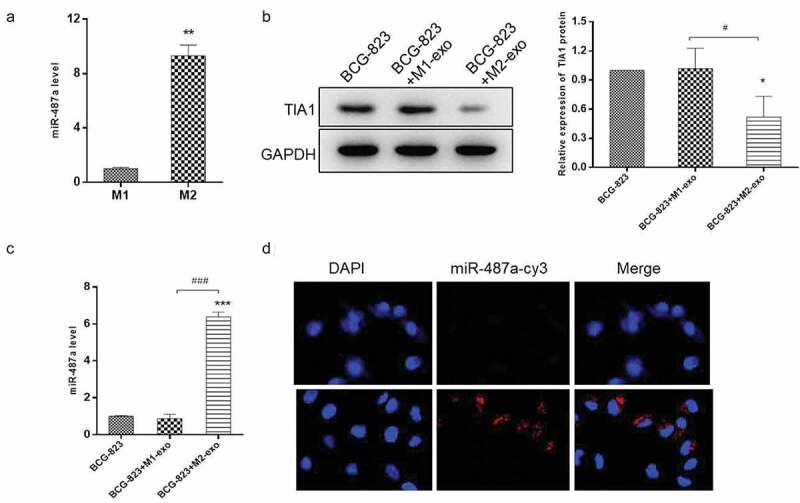
miR-487a is highly expressed in M2 macrophage-exosomes and can be transferred to GC cells via exosomes. (A). qRT-PCR were performed to detect the expression level of miR-487a in M1 and M2 macrophage-exosomes. (B & C). HS-746 T cells were treated with M1 macrophage-exosomes, M2 macrophage-exosomes or no-treatment. After 24 h, the expression level of miR-487a were detected by qRT-PCR (B), and the protein level of TIA1 were analyzed by western blot (C). GAPDH were used as the internal control. (D). Immunofluorescence analysis of the internalization of Cy3-labaled-miR-487a mimic (red) by HS-746 T cells. *p < 0.05, ***p < 0.001 vs control. #p < 0.05, ###p < 0.001 vs M1 macrophage-exosomes group. Data are displayed as mean ± SEM
5. Exosomal miR-487a induced the proliferation and tumorigenesis of GC cells in vivo
To determine whether exosomal miR-487a could induce tumorigenesis effectively in vivo, BGC823 cells mixed with exosomes derived from M1 macrophage, M2 macrophage, or M2 macrophage transfected with miR-487a mimics were subcutaneously injected into nude mice, tumor volume and mice weight were measured and recorded every 2 days. The control group was injected by HS-746 T cells alone. After 15 days, we observed that tumors of BGC823 cells mixed with miR-487a mimics M2 exosomes developed larger and faster than those produced by M2 macrophage exosomes, while no significant difference was observed between the control and M1 exosomes group (Figure 5a & B). Importantly, there was no significant difference in the weight of these mice (Figure 5c). These results demonstrated that M2 macrophage exosomal miR-478a can promote the proliferation and tumorigenesis of GC cells in vivo.
Figure 5.
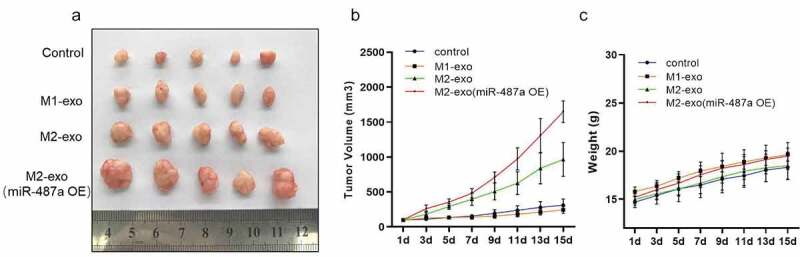
Exosomal miR-487a induced the proliferation and tumorigenesis of GC cells in vivo. (A). Representative images of tumors in nude mice that subcutaneous injection of HS-746 T cells alone or co-injected with exosomes derived from M1 macrophage, M2 macrophage or M2 macrophage transfected with miR-487a mimics. n = 5. (B & C). Tumor volumes were measured by growth curve every 2 days (B), and weights of nude mice were monitored at one time (C). Data are displayed as mean ± SEM
Discussion
Macrophages are a population of phagocytic immune cells present in almost all tissues [35]. Tumor-associated macrophage (TAMs) are defined as macrophages developed from peripheral blood monocytes infiltrating into solid tumor tissues. Macrophages are characterized by specific markers, including CD16, CD68, CD163 [36]. As a critical part of tumor microenvironment, TAMs play multiple roles in tumorigenesis and growth [37,38], immune regulation [39], metastasis [40], and chemo-resistance [41]. TAMs can be activated and polarized to different phenotypes in response to the tumor microenvironment. Activated macrophages are usually classified into M1(classical activated macrophage) and M2(alternating activated macrophage) phenotypes [42]. Generally, M1 macrophages have an inflammatory response to invading pathogens and tumor cells, while M2 macrophages have an immunosuppressive phenotype, which is conducive to tissue repair and tumor progression [43]. In recent years, increasing researches suggested that M2 macrophages exert a strong pro-tumor effect by producing anti-inflammatory cytokines such as IL-10, IL-13, and CD206. For instance, Lailler et al [39] revealed that the activated ERK1/2 signaling increased infiltration of TAMs with the polarization of M2 phenotype in glioblastoma.
However, these studies mainly concentrated on the functions and mechanisms biological process “macrophage polarization”, little researchers explored how the polarized M2 macrophage to regulate microenvironment and the development of tumor. In our research, we demonstrated that exosomes are the critical message transmission mediators from M2 macrophage to GC cells. Moreover, exosomes derived from M1 macrophage have no regulatory effects. It indicated that M2 macrophage-exosomes carry certain particular regulators that M1 macrophage-exosomes don’t have.
MiR-487a have been reported as an oncogenic miRNA in various other cancers [31,33,44]. In our previous study [34], miR-487a was highly expressed in GC tissues and negatively correlated with TIA1. miR-487a promoted GC growth both in vitro and in vivo through targeting TIA1. Thus, we hypothesized that miR-487a might be the key regulator of M2 macrophage-exosomes. Subsequent experiments proved this hypothesis. The expression of miR-478a in M2 macrophage-exosomes was higher in M1. MiR-487a could be transferred to GC cells through M2 macrophage-exosomes and promotes progression of gastric cancer via down-regulating TIA1. Finally, in-vivo experiments indicated that M2 macrophage exosomal miR-487a induced the proliferation and tumorigenesis of GC cells, M2 macrophage exosomes promoted the proliferation and tumorigenesis of GC cells in a miR-487a-dependent manner.
Conclusion
In conclusion, the above-mentioned results demonstrated that M2 macrophage-exosomes transfer miR-487a into GC cells, and miR-487a promotes the progression of gastric cancer via down-regulating TIA1. Therefore, exosomes from M2 macrophages with up-regulated levels of miR-487a could be a potential therapeutic pathway for GC. However, due to the insufficiency in the investigation on the role and mechanism of exosomal miR-487a in the tumorigenesis and microenvironment of GC, further experiments are required to further elucidate the intrinsic mechanisms of exosomal miR-487a.
Acknowledgments
All mice were treated according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication, 8th edition, 2011), and approved by the Institutional Laboratory Animal Care and Use Committee of Zunyi Medical University.
Funding Statement
This research was funded by Science and Technology Foundation of Guizhou Province (QKHJC[2020]1Y335 and Word J [2014] 2185).
Author Contributions
Conceptualization, Xuefeng Yang and Shouyang Yu; Data curation, Xuefeng Yang, Shuang Cai, Yue Shu, Aichen Tang, Xun Deng, Nian He and Xu Chen; Formal analysis, Yuanwei Zhang and Yan Qu; Investigation, Shouyang Yu; Methodology, Lei Wan; Writing – original draft, Shuang Cai; Writing – review & editing, Shouyang Yu.
Disclosure statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- [1].Bray F. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Ferlay J. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. [DOI] [PubMed] [Google Scholar]
- [3].Fu M. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol Cancer. 2019;18(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wagner AD. Chemotherapy for advanced gastric cancer. Cochrane Database Syst Rev. 2017;8:CD004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang W, Tan Y, Ma H. Combined aspirin and apatinib treatment suppresses gastric cancer cell proliferation. Oncol Lett. 2017;14(5):5409–5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zubarayev M, Min EK, Son T. Clinical and molecular prognostic markers of survival after surgery for gastric cancer: tumor-node-metastasis staging system and beyond. Transl Gastroenterol Hepatol. 2019;4:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rőszer T. Understanding the mysterious M2 macrophage through activation markers and effector mechanisms. Mediators Inflamm. 2015;2015:816460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Owen JL, Mohamadzadeh M. Macrophages and chemokines as mediators of angiogenesis. Front Physiol. 2013;4:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Duluc D. Interferon-gamma reverses the immunosuppressive and protumoral properties and prevents the generation of human tumor-associated macrophages. Int J Cancer. 2009;125(2):367–373. [DOI] [PubMed] [Google Scholar]
- [10].Zhang X. Exosomes in cancer: small particle, big player. J Hematol Oncol. 2015;8(1):83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Salas-Huetos A. The role of miRNAs in male human reproduction: a systematic review. Andrology. 2019. 10.1111/andr.12714. [DOI] [PubMed] [Google Scholar]
- [12].Sadri Nahand J. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int J Cancer. 2019. 10.1002/ijc.32688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kuang M. FGB and FGG derived from plasma exosomes as potential biomarkers to distinguish benign from malignant pulmonary nodules. Clin Exp Med. 2019. 10.1007/s10238-019-00581-8. [DOI] [PubMed] [Google Scholar]
- [14].Li H. S100-A9 protein in exosomes derived from follicular fluid promotes inflammation via activation of NF-kappaB pathway in polycystic ovary syndrome. J Cell Mol Med. 2019;130(6):777–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Meurer S. Endoglin trafficking/exosomal targeting in liver cells depends on N-glycosylation. Cells. 2019;8(9). 10.3390/cells8090997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Alvarez-Rodriguez M. Exosomes in specific fractions of the boar ejaculate contain CD44: a marker for epididymosomes?. Theriogenology. 2019;140:143–152. [DOI] [PubMed] [Google Scholar]
- [17].Patton MC. Hypoxia alters the release and size distribution of extracellular vesicles in pancreatic cancer cells to support their adaptive survival. J Cell Biochem. 2019;121:828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Johnson TK. Exosomes derived from induced vascular progenitor cells promote angiogenesis in vitro and in an in vivo rat hindlimb ischemia model. Am J Physiol Heart Circ Physiol. 2019;317(4):H765–H776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Tang J. [Peripheral blood exosomes from patients with multiple myeloma mediate bortezomib resistance in cultured multiple myeloma cells]. Nan Fang Yi Ke Da Xue Xue Bao. 2019;39(4):485–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ni J. Exosomes in cancer radioresistance. Front Oncol. 2019;9:869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Skog J. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vafaei S. Common molecular markers between circulating tumor cells and blood exosomes in colorectal cancer: a systematic and analytical review. Cancer Manag Res. 2019;11:8669–8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cecchettini A. Salivary extracellular vesicles versus whole saliva: new perspectives for the identification of proteomic biomarkers in sjogren’s syndrome. Clin Exp Rheumatol. 2019;37 Suppl 118(3):240–248. [PubMed] [Google Scholar]
- [24].Reif S, Elbaum Shiff Y, Golan-Gerstl R. Milk-derived exosomes (MDEs) have a different biological effect on normal fetal colon epithelial cells compared to colon tumor cells in a miRNA-dependent manner. J Transl Med. 2019;17(1):325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gao R. A comparison of exosomes derived from different periods breast milk on protecting against intestinal organoid injury. Pediatr Surg Int. 2019. 10.1007/s00383-019-04562-6. [DOI] [PubMed] [Google Scholar]
- [26].Sindeeva OA. New frontiers in diagnosis and therapy of circulating tumor markers in cerebrospinal fluid in vitro and in vivo. Cells. 2019;8(10). 10.3390/cells8101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ando W. Novel breast cancer screening: combined expression of miR-21 and MMP-1 in urinary exosomes detects 95% of breast cancer without metastasis. Sci Rep. 2019;9(1):13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ma J. Exosomal hsa-miR199a-3p promotes proliferation and migration in neuroblastoma. Front Oncol. 2019;9:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Bai M. miR-135b delivered by gastric tumor exosomes inhibits FOXO1 expression in endothelial cells and promotes angiogenesis. Mol Ther. 2019;27(10):1772–1783. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [30].Zeng Z. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ma M. MiR-487a promotes TGF-beta1-induced EMT, the migration and invasion of breast cancer cells by directly targeting MAGI2. Int J Biol Sci. 2016;12(4):397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma MT. MiR-487a resensitizes mitoxantrone (MX)-resistant breast cancer cells (MCF-7/MX) to MX by targeting breast cancer resistance protein (BCRP/ABCG2). Cancer Lett. 2013;339(1):107–115. [DOI] [PubMed] [Google Scholar]
- [33].Chang RM. miRNA-487a promotes proliferation and metastasis in hepatocellular carcinoma. Clin Cancer Res. 2017;23(10):2593–2604. [DOI] [PubMed] [Google Scholar]
- [34].Yang X. miR-487a promotes progression of gastric cancer by targeting TIA1. Biochimie. 2018;154:119–126. [DOI] [PubMed] [Google Scholar]
- [35].Sharifi L. A review on the role of M2 macrophages in bladder cancer; pathophysiology and targeting. Int Immunopharmacol. 2019;76:105880. [DOI] [PubMed] [Google Scholar]
- [36].Blessing MM. Desmoplastic infantile ganglioglioma: a MAPK pathway-driven and microglia/macrophage-rich neuroepithelial tumor. J Neuropathol Exp Neurol. 2019. 10.1093/jnen/nlz086. [DOI] [PubMed] [Google Scholar]
- [37].Yin S. SI-CLP inhibits the growth of mouse mammary adenocarcinoma by preventing recruitment of tumor-associated macrophages. Int J Cancer. 2020;146:1396–1408. [DOI] [PubMed] [Google Scholar]
- [38].Liu Y. An miR-340-5p-macrophage feedback loop modulates the progression and tumor microenvironment of glioblastoma multiforme. Oncogene. 2019. 10.1038/s41388-019-0952-x. [DOI] [PubMed] [Google Scholar]
- [39].Lailler C. ERK1/2 signaling regulates the immune microenvironment and macrophage recruitment in glioblastoma. Biosci Rep. 2019;39(9). 10.1042/BSR20191433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang J. Kruppel like factor 6 splice variant 1 (KLF6-SV1) overexpression recruits macrophages to participate in lung cancer metastasis by up-regulating TWIST1. Cancer Biol Ther. 2019;20(5):680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Larionova I. Interaction of tumor-associated macrophages and cancer chemotherapy. Oncoimmunology. 2019;8(7):1596004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol. 2010;11(10):889–896. [DOI] [PubMed] [Google Scholar]
- [43].Salmaninejad A. Tumor-associated macrophages: role in cancer development and therapeutic implications. Cell Oncol (Dordr). 2019;42(5):591–608. [DOI] [PubMed] [Google Scholar]
- [44].Gonzalez-Vallinas M. Epigenetically regulated chromosome 14q32 miRNA cluster induces metastasis and predicts poor prognosis in lung adenocarcinoma patients. Mol Cancer Res. 2018;16(3):390–402. [DOI] [PubMed] [Google Scholar]


