Abstract
New strategies to efficiently treat bacterial infections are crucial to circumvent the increase of resistant strains and to mitigate side effects during treatment. Skin and soft tissue infections represent one of the areas suffering the most from these resistant strains. We developed a new drug delivery system composed of the green algae, Chlamydomonas reinhardtii, which is generally recognized as safe, to target specifically skin diseases. A two‐step functionalization strategy was used to chemically modify the algae with the antibiotic vancomycin. Chlamydomonas reinhardtii was found to mask vancomycin and the insertion of a photocleavable linker was used for the release of the antibiotic. This living drug carrier was evaluated in presence of Bacillus subtilis and, only upon UVA1‐mediated release, growth inhibition of bacteria was observed. These results represent one of the first examples of a living organism used as a drug delivery system for the release of an antibiotic by UVA1‐irradiation.
Keywords: antibiotics, controlled release, drug delivery system, photocleavage, vancomycin
Algae control the release: A new approach for antibiotic caging and controlled release was developed using Chlamydomonas reinhardtii. The algae were covalently modified with vancomycin, and using a photocleavable linker, light‐mediated release of an antibacterial agent from the cell surface and inhibited the growth of Bacillus subtilis (see figure).
Due to the steady increase in the number of pathogenic bacteria, including resistant strains, [1] there is an urgent need for the development of new treatment strategies. [2] One of the most important burdens to our health system is the treatment of skin and soft tissue infections (SSTIs). [3] This disease affected up to 1 million people in the United States alone in 2010. [4] The most identified pathogen responsible for SSTI is Staphylococcus aureus (S. aureus) and the microorganism was found to be methicillin resistant (MRSA) in around half of the cases. [5] Fortunately, these strains are still sensitive to last resort antibiotics such as vancomycin, however, resistant mechanisms have already been identified. [6] Several approaches have been developed during the last decades in order to mitigate the development of resistant strains by using drug delivery systems. The advantage of such methodology is to efficiently release the antibiotic at a precise time and location, thereby avoiding the development of resistance and lowering the side effects of the drugs. [7]
Currently, the main technology relies on the use of nanoparticles (NPs), so‐called “nanoantibiotics”, which can exhibit intrinsic antibacterial activity and can be further functionalized. [8] These systems can be classified in six main categories based on their composition: [9] An ideal drug delivery system would control the release of the drug using an external stimulus.[ 7 , 10 ] This strategy was applied for the release of vancomycin by well‐designed NP systems using different stimuli, such as bacterial enzymes, [11] pH variation, [12] presence of bacterial cells, [13] near‐infrared light, [14] and temperature change. [15] The main issue that was raised with the use of NPs is their potential toxicity,[ 8b , 16 ] burst effects, [17] or limited drug loadings. [18] Moreover, exposure to nanoparticles may promote bacterial resistance in certain situations. [19]
In this study, we aimed to develop a new drug delivery system that is optimized for bacterial skin infections with triggered release of antibiotics. We selected living cells as a carrier based on promising results observed for such systems. [20] In particular the algae Chlamydomonas reinhardtii (C. reinhardtii) would represent the ideal candidate carrier due to its property to be biodegradable, [21] generally recognized as safe (GRAS), [22] not triggering an immune response, [23] antibiotic producer, [24] and for its potential for further genetic modifications. [25] Furthermore, promising results have already been demonstrated for chemical surface engineering of this organism. [26]
In this study, we aimed to functionalize the surface of C. reinhardtii with an antibiotic and to release the active compound by light irradiation. This specific release method was chosen due to the fact that phototherapy already showed promising results for skin treatment. [27] As a proof of concept, vancomycin was selected as the antibiotic (Figure 1).
Figure 1.
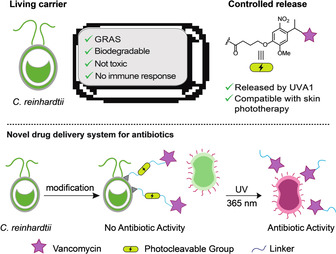
Design of a novel drug delivery system to potentially treat SSTIs. The carrier is the green algae C. reinhardtii selected as a generally recognized as safe organism (GRAS), [22] biodegradable, [21] and not immunogenic. [23] A photocleavable linker was chosen as the controlled release system of choice due to its compatibility with existing phototherapy using UV light to treat bacterial skin infection.
The new drug delivery system consists of living functionalized algae, which possess the advantage to produce beneficial compounds such as O2 and could be genetically engineered. In the first part of our study, we focused on a methodology to attach vancomycin covalently on the surface of C. reinhardtii and we hypothesized that the algae would mask the antibiotic from the bacteria. The synthetic route should include bioorthogonal reactions and the chemical steps should not affect the viability of C. reinhardtii. We decided to combine our reported strategy using N‐hydroxysuccinimide (NHS) ester for the functionalization of the algae [26c] with the Cu free azide‐alkyne cycloaddition used by Cooper and co‐workers (Figure 2 A). [28] This strategy would provide the bioorthogonal anchor dibenzocyclooctyne (DBCO) 1 on the algae surface and we hypothesized that the Cu free azide‐alkyne cycloaddition would not affect the viability of the organism.
Figure 2.
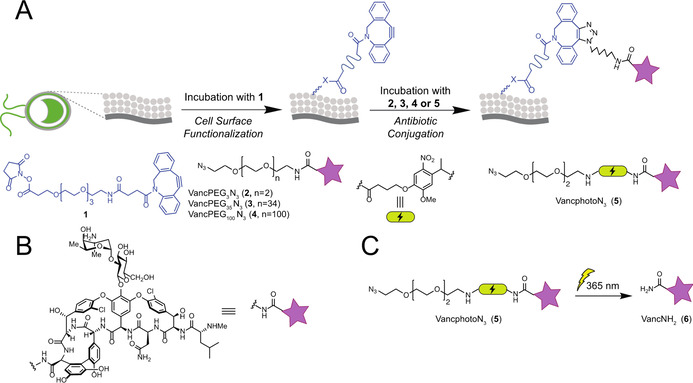
A) General two‐step procedure for the algal surface modification using dibenzocyclooctyne and vancomycin azide derivatives. B) Representation of vancomycin derivatives C) Photolysis reaction of VancphotoN3 (5) and its degradation product VancNH2 (6).
We started our investigation with the surface functionalization of C. reinhardtii using a DBCO anchor. The algae were incubated with the NHS‐PEG4‐DBCO (1) for one hour and the cells were centrifuged and washed with PBS buffer. The labeling strategy was evaluated using an azide‐containing fluorescent derivatives of vancomycin, which was synthesized and attached covalently using a three ethylene glycol (PEG) chain as linker. We were pleased to observe by microscope experiments the presence of the labelled antibiotic on the surface of the organism (see Supporting Information).
The photocaging properties were investigated in the next experiment. Recently, the research group of Cooper and collaborators demonstrated that the immobilization of vancomycin derivative (VancPEG3N3 (2)) on the surface of magnetic particles via NHS‐PEG4‐DBCO linker (1) improved the activity of the antibiotic toward the sensitive S. aureus and also against resistant strains. [28] Taking advantage of this result, the C. reinhardtii surface was modified using vancomycin azide derivatives possessing a PEG3 linker chain. Additionally, to investigate the chain size of the linker to the effectiveness of the photocaging, Vancomycin derivatives with 35 and approximately 100 ehtylene glycol units (VancPEG35N3 (3), VancPEG100N3 (4)) were also synthesized. Using our labeling strategy (Figure 2 A), the compounds were attached on the surface of the algae and the antibacterial activity against Bacillus subtilis (B. subtilis) of the construct was investigated. As expected, independent of the PEG chain size, the modified C. reinhardtii did not exhibit antibacterial activity (see the Supporting Information). Similar effects were observed by the group of Simard, where the nanoparticle size influenced the efficiency of vancomycin [29] and by the groups of Wong and Choi during their study of photocaging properties of ciprofloxacin attached to NPs. [30]
The next step focused on the development of a method to control the release of the active agent from the carrier by an external stimulus. An ideal photocleavable linker would absorb the light above 340 nm in order to be compatible with existing skin phototherapy and not affecting C. reinhardtii viability. It has been shown that treatment with ultraviolet A1 light (UVA1, 340–400 nm), did not trigger serious side effects and was effective against several skin diseases.[ 27 , 31 ] Disadvantages of this approach include (1) duration of treatment resulting in undesired UV damage, and (2) the treatment has to be repeated several times in order to be efficient. [32] To increase the efficacy of this method, a photocleavable linker o‐nitrobenzyl (o‐NB) was selected for its favorable absorption properties at 365 nm [33] and was attached to vancomycin, via a three step strategy, to obtain the vancomycin derivative (VancphotoN3 (5)). The first step included the synthesis of the modified linker using the amino PEG3 azide and 4‐{4‐[1‐(9‐Fluorenylmethyloxycarbonyl‐amino)ethyl]‐2‐methoxy‐5‐nitrophenoxy}butanoic acid. A Fmoc deprotection following by the coupling with vancomycin in presence of propylphosphonic anhydride (T3P) furnished the desired product 5. Using our photocaged vancomycin 5, we optimized the parameters of the labeling strategy. An increase in the concentration of the DBCO linker by a factor of 4 raised the labeling efficiency by approximately 20 % (see Supporting Information). The data obtained from these screens indicated that using a VancphotoN3 (5) concentration of 5 μm and 100 μm of DBCO linker would be ideal for the modification of the algae surface. The impact of the labeling strategy was evaluated on the viability of C. reinhardtii and we were pleased to observe that the algae were not significantly affected by the treatment (see Supporting Information).
To test the best photocleavable conditions, the amide derivative (VancNH2 (6)), resulting from the UVA1 cleavage of 5 (Figure 2 C), was synthesized. Pleasingly, the biological activity of 6 was evaluated against B. subtilis and an encouraging minimal inhibitory concentration (MIC) value of 0.06 μg mL−1 was determined (MIC of vancomycin: 0.125–0.25 μg mL−1). The conversion from 5 to 6 by UVA1 light was determined via analyzing the formation of the amide derivative 6 by UHPLC‐MS measurements in SIM mode (applied for the limit detection units) using the synthetic compound as analytic standard. The irradiation time was varied from 5 min to 15 min and the results demonstrate that the concentration of the product increased quickly and reached a maximum of 70 % conversion after 5 min. The experiment was also tested in the presence of the algae and a slight decrease of 10 % in conversion was observed (see Supporting Information). Based on these results, 5 min of UVA1‐irradiation was chosen in order to preserve algal viability. Using our optimal conditions, we covalently attached construct 5 on the surface of the algae. Gratefully, after 5 min of UVA1‐irradiation, a conversion of 20–25 % of VancNH2 (5) was determined by UHPLC‐MS measurements. The lower yield, in comparison of the previous results, could be explained by the proximity of vancomycin with the surface of the algae, which could mask the linker from the light, retention of the compound on the algae, or other factors. Interestingly, the measured concentration (0.27 μg mL−1) of released VancNH2 (6) is higher that the MIC value determined for 6 (0.06 μg mL−1) and, consequently, would be sufficient to inhibit the growth of B. subtilis (See Supporting Information). Furthermore, the device selected for our UVA1 experiments, SI MA 032 from Sina, is commercially available and inexpensive, is compatible with other skin treatments (365 m) and is already used in the cosmetic industry.
We were ready to investigate our system with the optimized parameters and to determine the antibacterial activity on B. subtilis of the modified algae before and after photocleavage. The algae were modified with VancphotoN3 (5, 5 μm) according to the general protocol (Figure 2 A) and were added to a suspension of B. subtilis in 24‐well plates. The system was irradiated at 365 nm for 5 min. As a negative control experiment, the modified algae were added after irradiation, and after 12 h of incubation, the bacterial growth inhibition was found only in the wells, which were irradiated in presence of the modified algae (Figure 3). Pleasingly, the antibacterial activity was even observed in the system using a loading concentration of VancphotoN3 (5) as low as 2.5 μm (see Supporting Information). Furthermore, control experiments provided evidence that neither VancphotoN3 (5) incubated with algae nor the by‐products after UVA1‐irradiation significantly influenced bacterial growth (see Supporting Information). The effect of UVA1‐irradiation on the algae was also tested and we did not observe a significant decrease on cell viability (see Supporting Information).
Figure 3.
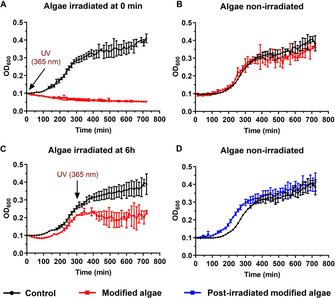
B. subtilis growth curves in the presence of functionalized algae with VancphotoN3 (5, red curve) or in the absence of the algae as control experiment (black curve). DBCO (1) concentration for the algae functionalization was 100 μm and VancphotoN3 (5) was 5 μm. A) The samples were UVA1‐irradiated at a wavelength of 365 nm for 5 min. B) The samples were not exposed to a UVA1 irradiation. C) The samples were UVA1‐irradiated at a wavelength of 365 nm for 5 min after 5 h of incubation. D) The functionalized algae with VancphotoN3, were UVA1‐irradiated at a wavelength of 365 nm for 5 min, the algae cells were washed with PBS (3x) and the resulting cells were incubated with B. subtilis. The growth curve of this experiment is displayed in blue. Data points adjusted to initial value 0.1 and represent mean value ±SD (n=3).
In order to demonstrate the advantage of this method, the experiment using the modified algae with VancphotoN3 (5) was repeated with an UVA1‐irradiation step during the exponential phase of the bacterial growth (Figure 3 C). After incubating the bacteria with the modified algae for 5 h in 24‐well plates, the system was UVA1‐irradiated at 365 nm for 5 min and the bacterial concentration was analyzed. A clear inhibition of the bacterial growth was observed for the samples containing the modified algae compare to the negative control. However, a lower loading concentration (2.5 μm) of 5 was not sufficient, in this case, to inhibit bacterial growth (see Supporting Information). This might be explained by a higher concentration of bacteria present after several hours of incubation.
In summary, a new and efficient drug delivery system was designed and optimized. The developed system relies on the green algae C. reinhardtii, an organism generally recognized as safe. [22] In a first step, the chemical anchor DBCO was installed providing a platform for the functionalization of the C. reinhardtii surface. Vancomycin was covalently attached, and, as predicted, no antibacterial activity of the system was observed. Consequently, using a photocleavable linker, we were able to release the active vancomycin derivative, VancNH2 (6), in a highly‐controlled manner by light irradiation. The release experiments could even be performed in presence of B. subtilis and a strong growth inhibition by released antibiotics was observed after UV‐irradiation (365 nm, 5 min). Control experiments demonstrated that the bacteria were not affected by the UVA1‐irradiation step. Potentially, the controlled release strategy is compatible with therapeutic approaches to threat SSTIs and other skin treatments, such as phototherapy using UVA1. Furthermore, we could demonstrate that the functionalization steps and the UVA1 irradiation did not affect significantly the cell viability of C. reinhardtii. In summary, these results represent one of the first reports where a living organism was chemically modified and used for the controlled release of an antibiotic. Based on this living organism, the system could be further modified either by chemistry or by biotechnology, harvesting the advantages of both fields. This methodology opens new ways for drug delivery and might be critical in the development of strategies for skin and soft tissue infections,
Experimental Section
The experimental procedures and the characterization of the compounds are found in the Supporting Information.
Author Contribution
I. S. S., S. S and K. G. designed the study. I. S. S. carried out the synthesis, characterization of the derivatives and biological experiments. I. S. S., S. S. and K. G. analyzed and discussed the results. I. S. S., S. S. and K. G. wrote the manuscript.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Acknowledgements
We acknowledge the Swiss National Science Foundation (SNSF, No. 182043) and a Bundesstipendium (to I.S.S.) for financial support. We acknowledge the NMR and Mass spectrometry facilities, the Center for Microscopy and Image Analysis (ZMB) and the Flow Cytometry Facility of the University of Zurich for training and maintenance of the instruments. Dr. Katja Zerbe and Myriam Gwerder are thanked for support and helpful discussions.
I. S. Shchelik, S. Sieber, K. Gademann, Chem. Eur. J. 2020, 26, 16644.
References
- 1.CDC, Antibiotic Resistance Threats in the United States, 2019 Atlanta, GA: U.S. Department of Health and Human Services, CDC, 2019.
- 2. Santajit S., Indrawattana N., BioMed Res. Int. 2016, 2016, 2475067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walsh T. L., Chan L., Konopka C. I., Burkitt M. J., Moffa M. A., Bremmer D. N., Murillo M. A., Watson C., Chan-Tompkins N. H., BMC Infect. Dis. 2016, 16, 721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lam P. L., Lee K. K. H., Wong R. S. M., Cheng G. Y. M., Bian Z. X., Chui C. H., Gambari R., Crit. Rev. Microbiol. 2018, 44, 40–78. [DOI] [PubMed] [Google Scholar]
- 5. Ray G. T., Suaya J. A., Baxter R., BMC Infect. Dis. 2013, 13, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chambers H. F., DeLeo F. R., Nat. Rev. Microbiol. 2009, 7, 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gao S., Tang G., Hua D., Xiong R., Han J., Jiang S., Zhang Q., Huang C., J. Mater. Chem. B 2019, 7, 709–729. [DOI] [PubMed] [Google Scholar]
- 8.Reviews:
- 8a. Gupta A., Mumtaz S., Li C.-H., Hussain I., Rotello V. M., Chem. Soc. Rev. 2019, 48, 415–427; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8b. Huh A. J., Kwon Y. J., J. Controlled Release 2011, 156, 128–145. [DOI] [PubMed] [Google Scholar]
- 9.
- 9a. Raza A., Sime F. B., Cabot P. J., Maqbool F., Roberts J. A., Falconer J. R., Drug Discovery Today 2019, 24, 858–866; [DOI] [PubMed] [Google Scholar]
- 9b. Liu Y., Shi L., Su L., van der Mei H. C., Jutte P. C., Ren Y., Busscher H. J., Chem. Soc. Rev. 2019, 48, 428–446; [DOI] [PubMed] [Google Scholar]
- 9c. Lai H.-Z., Chen W.-Y., Wu C.-Y., Chen Y.-C., ACS Appl. Mater. Interfaces 2015, 7, 2046–2054; [DOI] [PubMed] [Google Scholar]
- 9d. Sun J., Li J., Fan H., Ai S., J. Mater. Chem. B 2013, 1, 5436–5442; [DOI] [PubMed] [Google Scholar]
- 9e. Gu H., Ho P. L., Tong E., Wang L., Xu B., Nano Lett. 2003, 3, 1261–1263. [Google Scholar]
- 10.Reviews:
- 10a. Mura S., Nicolas J., Couvreur P., Nat. Mater. 2013, 12, 991–1003; [DOI] [PubMed] [Google Scholar]
- 10b. Norris M. D., Seidel K., Kirschning A., Adv. Ther. 2019, 2, 1800092; [Google Scholar]
- 10c. Wong P. T., Choi S. K., Chem. Rev. 2015, 115, 3388–3432; Photoactivation: [DOI] [PubMed] [Google Scholar]
- 10d. Li Y., Zhang Y., Wang W., Nano Res. 2018, 11, 5424–5438; [Google Scholar]
- 10e. Zhang Y., Xu C., Yang X., Pu K., Adv. Mater. 2020, 32, 2002661; [DOI] [PubMed] [Google Scholar]
- 10f. Choi S. K., Adv. Ther. 2020, 13, 2000117. [Google Scholar]
- 11.
- 11a. Pornpattananangkul D., Zhang L., Olson S., Aryal S., Obonyo M., Vecchio K., Huang C.-M., Zhang L., J. Am. Chem. Soc. 2011, 133, 4132–4139; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11b. Xiong M. H., Bao Y., Yang X. Z., Wang Y.-C., Sun B., Wang J., J. Am. Chem. Soc. 2012, 134, 4355–4362; [DOI] [PubMed] [Google Scholar]
- 11c. Xiong M. H., Li Y. J., Bao Y., Yang X. Z., Hu B., Wang J., Adv. Mater. 2012, 24, 6175–6180; [DOI] [PubMed] [Google Scholar]
- 11d. Li L.-L., Xu J.-H., Qi G.-B., Zhao X., Yu F., Wang H., ACS Nano 2014, 8, 4975–4983; [DOI] [PubMed] [Google Scholar]
- 11e. Li Y., Liu G., Wang X., Hu J., Liu S., Angew. Chem. Int. Ed. 2016, 55, 1760–1764; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2016, 128, 1792–1796; [Google Scholar]
- 11f. Hajiahmadi F., Alikhani M. Y., Shariatifar H., Arabestani M. R., Ahmadvand D., Int. J. Nanomed. 2019, 14, 5943–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.
- 12a. Radovic-Moreno A. F., Lu T. K., Puscasu V. A., Yoon C. J., Langer R., Farokhzad O. C., ACS Nano 2012, 6, 4279–4287; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12b. Chu L., Gao H., Cheng T., Zhang Y., Liu J., Huang F., Yang C., Shi L., Liu J., Chem. Commun. 2016, 52, 6265–6268; [DOI] [PubMed] [Google Scholar]
- 12c. Chen M., Xie S., Wei J., Song X., Ding Z., Li X., ACS Appl. Mater. Interfaces 2018, 10, 36814–36823; [DOI] [PubMed] [Google Scholar]
- 12d. Makhathini S. S., Kalhapure R. S., Jadhav M., Waddad A. Y., Gannimani R., Omolo C. A., Rambharose S., Mocktar C., Govender T., J. Drug Targeting 2019, 27, 1094–1107; [DOI] [PubMed] [Google Scholar]
- 12e. Jadhav M., Kalhapure R. S., Rambharose S., Mocktar C., Singh S., Kodama T., Govender T., Chem. Phys. Lipids 2018, 212, 12–25. [DOI] [PubMed] [Google Scholar]
- 13. Mas N., Galiana I., Mondragón L., Aznar E., Climent E., Cabedo N., Sancenón F., Murguía J. R., Máñez R. M., Marcos M. D., Amorós P., Chem. Eur. J. 2013, 19, 11167–11171. [DOI] [PubMed] [Google Scholar]
- 14. Chiang W.-L., Lin T.-T., Sureshbabu R., Chia W.-T., Hsiao H.-C., Liu H.-Y., Yang C.-M., Sung H.-W., J. Controlled Release 2015, 199, 53–62. [DOI] [PubMed] [Google Scholar]
- 15. Zhang W., Taheri-Ledari R., Hajizadeh Z., Zolfaghari E., Ahghari M. R., Maleki A., Hamblin M. R., Tian Y., Nanoscale 2020, 12, 3855–3870. [DOI] [PubMed] [Google Scholar]
- 16. Elsaesser A., Howard C. V., Adv. Drug Delivery Rev. 2012, 64, 129–137. [DOI] [PubMed] [Google Scholar]
- 17. Huang X., Brazel C. S., J. Controlled Release 2001, 73, 121–136. [DOI] [PubMed] [Google Scholar]
- 18. Ghasemiyeh P., Mohammadi-Samani S., Res. Pharm. Sci. 2018, 13, 288–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.
- 19a. Kaweeteerawat C., Ubol P. N., Sangmuang S., Aueviriyavit S., Maniratanachote R., J. Toxicol. Environ. Health Sci. 2017, 80, 1276–1289; [DOI] [PubMed] [Google Scholar]
- 19b. Hachicho N., Hoffmann P., Ahlert K., Heipieper H. J., FEMS Microbiol. Lett. 2014, 355, 71–77; [DOI] [PubMed] [Google Scholar]
- 19c. Graves J. L. J., Tajkarimi M., Cunningham Q., Campbell A., Nonga H., Harrison S. H., Barrick J. E., Front. Genet. 2015, 6, 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reviews and summaries:
- 20a. Hosseinidoust Z., Mostaghaci B., Yasa O., Park B.-W., Singh A. V., Sitti M., Adv. Drug Delivery Rev. 2016, 106, 27–44; [DOI] [PubMed] [Google Scholar]
- 20b. Luo M., Feng Y., Wang T., Guan J., Adv. Funct. Mater. 2018, 28, 1706100; [Google Scholar]
- 20c. Erkoc P., Yasa I. C., Ceylan H., Yasa O., Alapan Y., Sitti M., Adv. Ther. 2019, 2, 1800064; [Google Scholar]
- 20d. Chu D., Dong X., Shi X., Zhang C., Wang Z., Adv. Mater. 2018, 30, 1706245; Recent reports using bacteria: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20e. Felfoul O., Mohammadi M., Taherkhani S., de Lanauze D., Xu Y. Z., Loghin D., Essa S., Jancik S., Houle D., Lafleur M., Gaboury L., Tabrizian M., Kaou N., Atkin M., Vuong T., et al., Nat. Nanotechnol. 2016, 11, 941–947; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20f. Park B.-W., Zhuang J., Yasa O., Sitti M., ACS Nano 2017, 11, 8910–8923; [DOI] [PubMed] [Google Scholar]
- 20g. Singh A. V., Hosseinidoust Z., Park B.-W., Yasa O., Sitti M., ACS Nano 2017, 11, 9759–9769; [DOI] [PubMed] [Google Scholar]
- 20h. Alapan Y., Yasa O., Schauer O., Giltinan J., Tabak A. F., Sourjik V., Sitti M., Sci. Robot. 2018, 3, eaar4423; [DOI] [PubMed] [Google Scholar]
- 20i. Suh S., Jo A., Traore M. A., Zhan Y., Ott S. L. C., Scaia V. M. R., Allen I. C., Davis R. M., Behkam B., Adv. Sci. 2019, 6, 1801309; using sperm cells: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20j. Xu H., Medina-Sánchez M., Magdanz V., Schwarz L., Hebenstreit F., Schmidt O. G., ACS Nano 2018, 12, 327–337; [DOI] [PubMed] [Google Scholar]
- 20k. Xu H., Medina-Sánchez M., Maitz M. F., Werner C., Schmidt O. G., ACS Nano 2020, 14, 2982–2993; using macrophage cells: [DOI] [PubMed] [Google Scholar]
- 20l. Han J., Zhen J., Van Du Nguyen, Go G., Choi Y., Ko S. Y., Park J.-O., Park S., Sci. Rep. 2016, 6, 28717; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20m. Ullah S., Seidel K., Türkkan S., Warwas D. P., Dubich T., Rohde M., Hauser H., Behrens P., Kirschning A., Köster M., Wirth D., J. Controlled Release 2019, 294, 327–336. [DOI] [PubMed] [Google Scholar]
- 21. Mahdy A., Mendez L., Ballesteros M., González-Fernández C., Energy Convers. Manage. 2014, 85, 551–557. [Google Scholar]
- 22.FDA. GRN. No. 773. Available online: https://www.accessdata.fda.gov/scripts/fdcc/index.cfm?set=GRASNotices&id=773, (accessed on 17 July 2020).
- 23. Schenck T. L., Hopfner U., Chávez M. N., Machens H.-G., Somlai-Schweiger I., Giunta R. E., Bohne A. V., Nickelsen J., Allende M. L., Egaña J. T., Acta Biomater. 2015, 15, 39–47. [DOI] [PubMed] [Google Scholar]
- 24.
- 24a. Renukadevi K. P., Saravana P. S., Angayarkanni J., Int. J. Pharm. Sci. Res. 2011, 2, 1467–1472; [Google Scholar]
- 24b. Vishwakarma J., Vavilala S. L., J. Appl. Microbiol. 2019, 127, 1004–1017; [DOI] [PubMed] [Google Scholar]
- 24c. Bhowmick S., Mazumdar A., Moulick A., Adam V., Biotechnol. Adv. 2020, 43, 107571. [DOI] [PubMed] [Google Scholar]
- 25.
- 25a. Stoffels L., Taunt H. N., Charalambous B., Purton S., Plant Biotechnol. J. 2017, 15, 1130–1140; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25b. Quezada-Rivera J. J., Soria-Guerra R. E., Pérez-Juárez F. S., Martínez-González L., Valdés-Rodríguez S. E., Vasco-Méndez N. L., Morales-Domínguez J. F., Phyton 2019, 88, 25–35; [Google Scholar]
- 25c. Molino J. V. D., de Carvalho J. C. M., Mayfield S. P., PLoS ONE 2018, 13, e0192433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.
- 26a. Kerschgens I. P., Gademann K., ChemBioChem 2018, 19, 439–443; [DOI] [PubMed] [Google Scholar]
- 26b. Weibel D. B., Garstecki P., Ryan D., DiLuzio W. R., Mayer M., Seto J. E., Whitesides G. M., Proc. Natl. Acad. Sci. USA 2005, 102, 11963–11967; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26c. Szponarski M., Schwizer F., Ward T. R., Gademann K., Commun. Chem. 2018, 1, 84; [Google Scholar]
- 26d. Yasa O., Erkoc P., Alapan Y., Sitti M., Adv. Mater. 2018, 30, 1804130; [DOI] [PubMed] [Google Scholar]
- 26e. Akolpoglu M. B., Dogan N. O., Bozuyuk U., Ceylan H., Kizilel S., Sitti M., Adv. Sci. 2020, 7, 2001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gupta A., Avci P., Dai T., Huang Y.-Y., Hamblin M. R., Adv. Wound Care 2013, 2, 422–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hassan M. M., Ranzoni A., Phetsang W., Blaskovich M. A. T., Cooper M. A., Bioconjugate Chem. 2017, 28, 353–361. [DOI] [PubMed] [Google Scholar]
- 29. Kell A. J., Stewart G., Ryan S., Peytavi R., Boissinot M., Huletsky A., Bergeron M. G., Simard B., ACS Nano 2008, 2, 1777–1788. [DOI] [PubMed] [Google Scholar]
- 30. Wong P. T., Tang S., Mukherjee J., Tang K., Gam K., Isham D., Murat C., Sun R., Baker J. R., Choi S. K., Chem. Commun. 2016, 52, 10357–10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.
- 31a. Rodenbeck D. L., Silverberg J. I., Silverberg N. B., Clin. Dermatol. 2016, 34, 607–613; [DOI] [PubMed] [Google Scholar]
- 31b. Vangipuram R., Feldman S. R., Oral Dis. 2016, 22, 253–259; [DOI] [PubMed] [Google Scholar]
- 31c. Zandi S., Kalia S., Lui H., Skin Therapy Lett. 2012, 17, 1–4. [PubMed] [Google Scholar]
- 32. Gambichler T., Terras S., Kreuter A., Clin. Dermatol. 2013, 31, 438–454. [DOI] [PubMed] [Google Scholar]
- 33. Hoecker J., Liffert R., Burch P., Wehlauch R., Gademann K., Org. Biomol. Chem. 2013, 11, 3314–3321; [DOI] [PubMed] [Google Scholar]; Wehlauch R., Hoecker J., Gademann K., ChemPlusChem 2012, 77, 1071–1074. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary


