Abstract
Poly(ADP‐ribose) glycohydrolase (PARG) as a main enzyme hydrolyzing poly(ADP‐ribose) in eukaryotes, and its silencing can inhibit benzo(a)pyrene (BaP)‐induced carcinogenesis. A thorough understanding of the mechanism of PARG silenced inhibition of BaP‐induced carcinogenesis provides a new therapeutic target for the prevention and treatment of environmental hazard induced lung cancer. We found that the expression of several subtypes of the histone H2B was downregulated in BaP‐induced carcinogenesis via PARG silencing as determined by label‐free proteomics and confirmed by previous cell line‐ and mouse model‐based studies. Analysis using the GEPIA2 online tool indicated that the transcription levels of H2BFS, HIST1H2BD, and HIST1H2BK in lung adenocarcinoma (LUAD) tissues and squamous cell lung carcinoma (LUSC) tissues were higher than those in normal lung tissues, while the transcription levels of HIST1H2BH in LUSC tissues were higher than those in normal lung tissues. The expression levels of HIST1H2BB, HIST1H2BH, and HIST1H2BL were significantly different in different lung cancer (LC) stages. Moreover, the expression of H2BFS, HIST1H2BD, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BO, HIST2H2BE, and HIST2H2BF was positively correlated with that of PARG in LC tissues. Analysis of the Kaplan‐Meier plotter database indicated that high H2B levels predicted low survival in all LC patients suggesting that H2B could be a new biomarker for determining the prognosis of the LC, and that its expression can be inhibited by PARG silencing in BaP‐induced carcinogenesis.
Keywords: benzo(a)pyrene, lung cancer, poly(ADP‐ribose) glycohydrolase, tumorigenesis
Study overview. After BaP‐induced malignant transformation, H2B expression was upregulated. PARG silencing inhibited the abnormally increased H2B levels and thus showed the progression of BaP‐induced LC.
1. INTRODUCTION
Poly(ADP‐ribose) glycohydrolase (PARG) is the main enzyme that hydrolyzes poly(ADP‐ribose) in eukaryotes and plays an important role in tumorigenesis. 1 , 2 In our previous study, PARG silencing inhibited the malignant transformation induced by benzo(a)pyrene (BaP), but its mechanism was unclear. 3 , 4
The histone H2B and its subtypes perform diverse biological functions, and their expression levels vary among different types of cancer. HIST1H2BA (H2B type 1‐A) maintains the integrity of testicular specific proteins and chromatin. 5 HIST1H2BJ (H2B type 1‐J) is involved in chromatin remodeling in schizophrenia. 6 HITT2H2BE (H2B type 2‐E) inhibits cell proliferation and deactivates odor sensing neurons. 7 , 8 Another study reported that HIST1H2BG, HIST1H2BJ, HIST2H2BE, HIST1H2BC, HIST1H2BH, and HIST1H2BE are highly expressed in lung adenocarcinoma (LUAD) tissues. 9 Several similar studies have shown that H2B are highly expressed in lung cancer (LC) tissues. All the above mentioned evidence indicates that H2B plays an important role in lung tumorigenesis, but its regulatory mechanism is still unknown.
In our previous study, PARG silencing effectively prevented the development of BaP‐induced LC. In this study, we found that PARG silencing prevents the development of BaP‐induced LC by stabilizing the abnormally high levels of H2B, which plays an important role in the staging and prognosis of LC. Our results provide strong evidence for elucidating the LC development. Different H2B subtypes exhibit different protein structures and sequences, indicating that they undergo different chemical modifications. Histone posttranslational modifications represent a diverse set of epigenetic marks that are involved, not only in dynamic cellular processes, such as transcription and DNA repair, but also in destabilizing chromatin. 10 , 11 , 12 The mechanism elucidated in the present study will provides a strong basis for future studies on the role of H2B‐modified pedigree expression in lung tumorigenesis.
2. MATERIALS AND METHODS
2.1. Cell lines and culture selection
The cell lines used in this study was been constructed in our previous work. 4 The normal human bronchial epithelial cell line (C‐16HBE cells) and human bronchial epithelial cells with a silenced PARG gene (C‐shPARG cells) were used as the control groups and were grown in minimum essential Eagle's medium (Gibco, The United States) containing 10% fetal bovine serum (Gibco, The United States). BTC‐16HBE and BTC‐shPARG cells were treated with 40 μM BaP (Sigma, The United States) 1 day/week for 15 weeks.
2.2. Mouse model establishment
The mouse model used in this study was constructed in our previous work. 13 Wild‐type C57 mice with malignant transformation induced by BaP (C57‐female‐T) and mice with silenced PARG gene without malignant transformation under the same treatment conditions (shPARG‐female‐T) were used. C57‐female‐C and shPARG‐female‐C mouse models without BaP treatment were used as the control groups.
Treatment and maintenance of animals were performed according to the regulations of the Animal Care and Use Committee of Shenzhen CDC Experimental Animal Center. The mice were anesthetized with ether prior to experimentation, and we followed all the necessary procedures to ensure that no unnecessary pain was inflicted at any stage of the experiment.
2.3. Label‐free proteomics
An acid extraction assay was performed for histones extraction. 14 The products digested by arginase (Arg‐C) were separated by high‐performance liquid chromatography, and raw data were analyzed using Q Exactive. The raw data were analyzed using the human protein sequence database SwissProt. The software Proteome Discoverer (Thermo, The United States) was used for data comparison to obtain qualitative and quantitative information of protein or peptide segments in each group. Quantitative information was analyzed using the Perseus software.
2.4. Western blotting
Western blotting experiment based on the previous articles. 13 The relative expression of H2B (Abcam, England) was assessed relative to that of β‐actin (Santa Cruz, The United States). Histogram construction and statistical analysis of the relative expression of each group were performed using the GraphPad Prism software and Student's t test.
2.5. Immunofluorescence
Immunofluorescence analysis was performed according to a method described previously. 13
2.6. Bioinformatics analysis
GEPIA2 (http://gepia2.cancer-pku.cn/#index) was used to analyze the relative expression of H2B subtypes and their relationship with tumor stage in LC patients. The log2(TPM) was calculated simply as log2 of the transcript count per million (TPM) and log2FC was calculated as median (Tumor) − median (Normal). TPM is a normalization technique to scale the read count per gene/transcript toward the total read count of the sequencing run to compensate for different sequencing depths. The expression data were first transformed into long2(TPM + 1) values for differential analysis. Genes with higher |log2FC| values and lower q values than pre‐set thresholds were considered to be differentially expressed. Differential gene expression was analyzed using one‐way analysis of variance by considering the pathological stage as a variable. GEPIA2 was also used for performing gene correlation analysis (Pearson analysis) of PARG and H2B subtypes in LC. We used nonlog‐scale axis for calculation, and the log‐scale axis for visualization. Kaplan‐Meier Plotter (www.kmplot.com) was used to analyze the relationship between the H2B subtype and overall survival (OS), progression‐free survival (FP), and progressive postpartum survival (PPS) in LC patients.
3. RESULT
3.1. Label‐free proteomics explains the inhibition of the abnormal increase in the H2B level by PARG silencing in BaP‐induced carcinogenesis
Label‐free proteomics was used to determine the expression of histone subtypes in 16HBE and shPARG cells with or without BaP treatment. The results suggested that the relative expression of H2B subtypes increased in BTC‐16HBE cells but remained unchanged in BTC‐shPARG cells (Table 1 and Figure 1).
TABLE 1.
Histone expression level analyzed by proteomics
| Group | Gene | Description | Fold change | P‐value |
|---|---|---|---|---|
| BTC‐16HBE/C‐16HBE | H1FV | Histone H1.0 | −1.11 | .03 |
| BTC‐16HBE/C‐16HBE | HIST1H1E | Histone H1.4 | −0.87 | .01 |
| BTC‐16HBE/C‐16HBE | HIST2H2AC | Histone H2A type 2‐C | 1.27 | .01 |
| BTC‐16HBE/C‐16HBE | H2BFS | Histone H2B type F‐S | 6.14 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BB | Histone H2B type 1‐B | 5.41 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BC | Histone H2B type 1‐C/E/F/G/I | 5.39 | .01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BD | Histone H2B type 1‐D | 4.87 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BH | Histone H2B type 1‐H | 4.9 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BJ | Histone H2B type 1‐J | 6.29 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BK | Histone H2B type 1‐K | 5.86 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BL | Histone H2B type 1‐L | 5.61 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST1H2BM | Histone H2B type 1‐M | 4.21 | .03 |
| BTC‐16HBE/C‐16HBE | HIST1H2BO | Histone H2B type 1‐O | 5.43 | <.01 |
| BTC‐16HBE/C‐16HBE | HIST2H2BE | Histone H2B type 2‐E | 4.57 | .01 |
| BTC‐16HBE/C‐16HBE | HIST2H2BF | Histone H2B type 2‐F | 4.92 | .01 |
| BTC‐16HBE/C‐16HBE | HIST1H3A | Histone H3.1 | −2.03 | <.001 |
| BTC‐16HBE/C‐16HBE | HIST3H3 | Histone H3.1t | −1.77 | <.001 |
| BTC‐16HBE/C‐16HBE | HIST2H3A | Histone H3.2 | −2.03 | <.001 |
| BTC‐16HBE/C‐16HBE | H3F3C | Histone H3.3C | −1.94 | <.001 |
| BTC‐shPARG/C‐shPARG | HIST2H2AC | Histone H2A type 2‐C | 1.11 | .42 |
FIGURE 1.
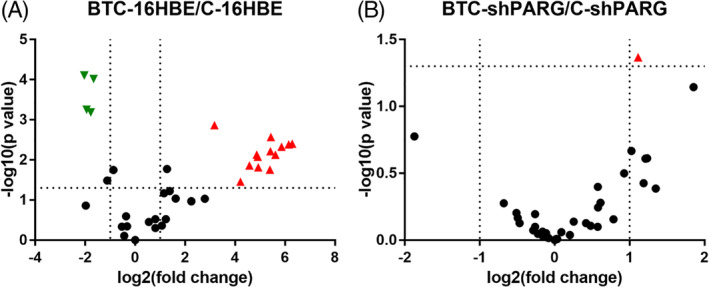
PARG silencing down‐regulates H2B levels in BaP‐induced carcinogenesis. A, A volcano plot showing histone expression in BTC‐16HBE/C‐16HBE. B, A volcano plot showing histone expression in BTC‐shPARG/C‐shPARG. The red equilateral triangle represents the protein with significantly high expression, while the green inverted triangle represents the protein with significantly low expression [Color figure can be viewed at wileyonlinelibrary.com]
3.2. PARG silencing‐mediated downregulation of H2B expression in BaP‐induced carcinogenesis was confirmed in cell lines and mouse models
Western blotting results indicated that the relative expression of H2B in BTC‐16HBE cells was higher than that in C‐16HBE cells (mean ± SD = 0.5542 ± 0.1482, P = .0038), while the relative expression of H2B in BTC‐shPARG cells was lower than in C‐shPARG cells (mean ± SD = −1.157 ± 0.3553, P = .0087) (Figure 2A,B). An increase in H2B fluorescence intensity was observed in BTC‐16HBE cells compared with that in C‐16HBE cells, that no significant difference in the H2B fluorescence intensity was observed in BTC‐shPARG cells compared with that in C‐shPARG cells (Figure 2E). In addition, in vivo studies showed an increase in the relative expression of H2B in C57‐Female‐T mice compared with that in C57‐Female‐C mice (mean ± SD = 0.2747 ± 0.02173, P < .0001), and a decrease in the relative expression of H2B in shPARG‐Female‐T mice compared with that in shPARG‐Female‐C mice (mean ± SD = −0.07096 ± 0.02788, P = .0438) (Figure 2C,D).
FIGURE 2.
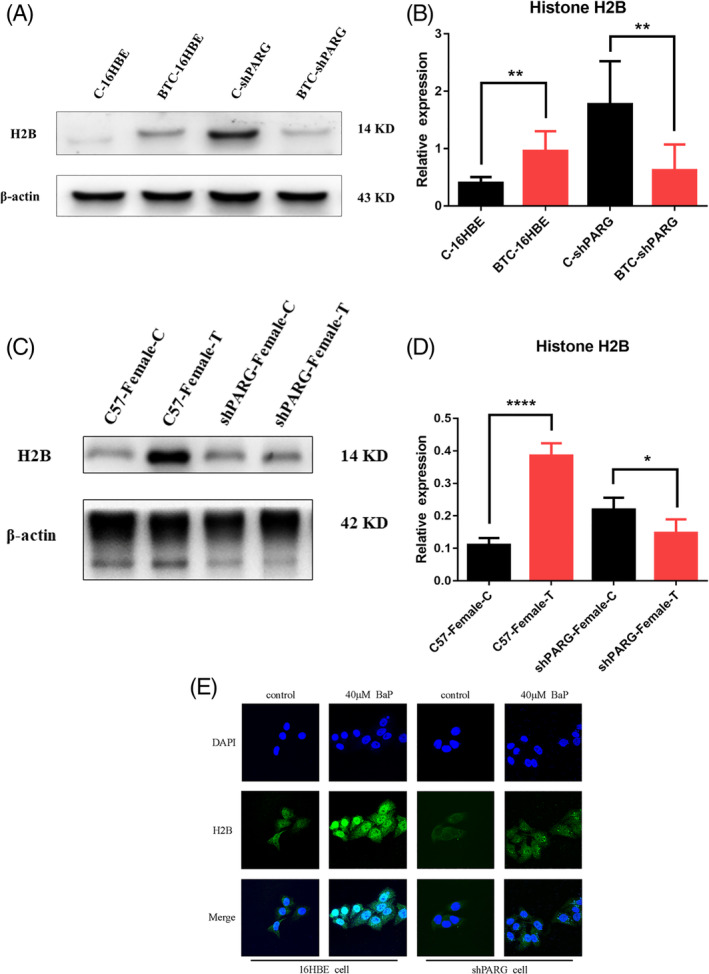
Effects of PARG on the expression of H2B in BaP‐induced carcinogenesis. A, Western blot showing H2B expression in each cell. B,Histogram of the relative expression of H2B in each cell. C,Western blot showing H2B expression in mice. D, Histogram of the relative expression of H2B in each mice. E, Fluorescence intensity of H2B in each cell [Color figure can be viewed at wileyonlinelibrary.com]
3.3. Relationship between the mRNA levels of H2B and LC
We compared the mRNA expression of H2B in LC tissues with that in normal lung tissues using the GEPIA2 dataset. The results indicated that the expression of H2BFS, HIST1H2BD, and HIST1H2BK was higher in LUAD and LUSC tissues than in normal lung tissues, whereas and the expression of HIST1H2BH was increased in LUSC tissues than in normal lung tissues (Figure 3A,B). Using the GEPIA2 dataset, we found that the expression of HIST1H2BB, HIST1H2BH, and HIST1H2BL according to the tumor stage of LC varied significantly, whereas the expression of H2BFS, HIST1H2BC, HIST1H2BD, HIST1H2BJ, HIST1H2BK, HIST1H2BM, HIST2H2BE, and HIST2H2BF according to the tumor stage of LC did not change significantly (Figure 3C). The expression of H2BFS, HIST1H2BD, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BO, HIST2H2BE, and HIST2H2BF was positively correlated with that of PARG in LC tissues (Figure 4).
FIGURE 3.
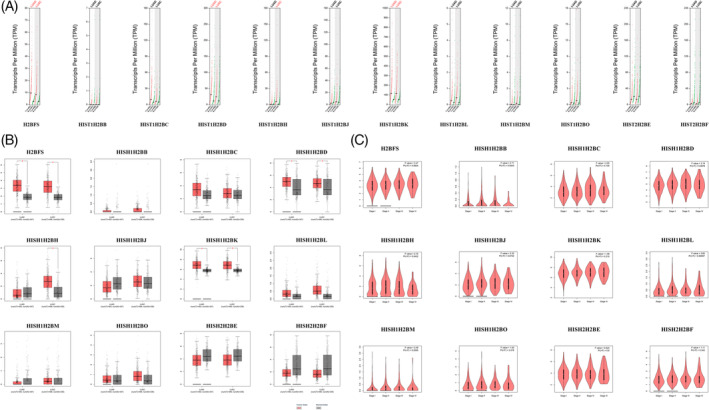
Expression of H2B in LC and that of H2B in association with LC tumor stage. A,B, The expression of H2B in LUAC and LUSC. C, The expression of H2B considering the LC tumor stage [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 4.
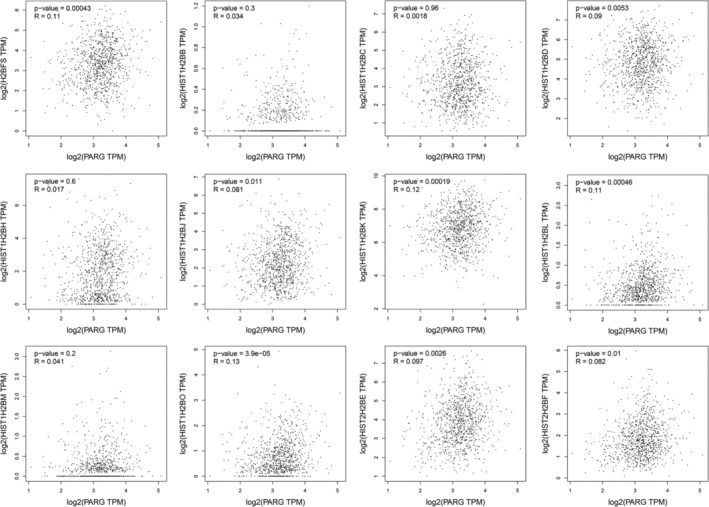
Pearson correlation analysis correlation between the expression of H2B subtypes and that of PARG in LC tissues
3.4. Correlation between H2B and improved prognosis in cirrhotic patients
We further explored the critical efficiency of H2B in the survival of patients with non‐small cell lung cancer. Kaplan‐Meier curve analysis and log‐rank test revealed that the increased HIST1H2BH mRNA level was significantly associated with the OS, FP, and PPS in all the LC patients (P < .05). Increased mRNA levels of H2BFS, HIST1H2BD, HIST1H2BK, HIST1H2BL, HIST1H2BM, and HIST1H2BO were associated with both FP and PPS in all the LC patients (P < .05) (Figure 5).
FIGURE 5.
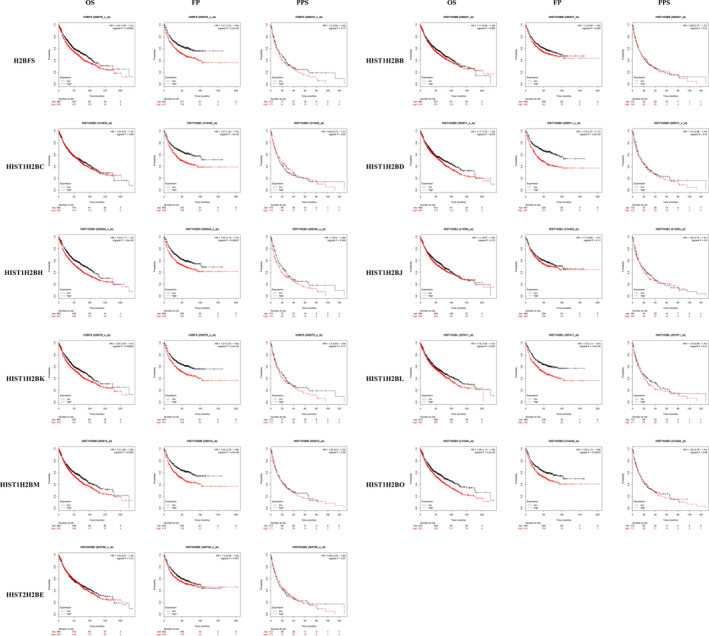
Prognostic value of the H2B mRNA level in LC patients. The overall survival (OS), progression‐free survival (FP), and postprogression survival (PPS) of all LC patients with respect to H2B expression [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
A mutation in the H2B gene represents a new class of oncogenic driver. 15 Histones are the basic substrates for chromatin modification and recombinant enzymes, and H2B have been shown to undergo mutation in a variety of tumors. 16 Our previous study showed that PARG silencing delay BaP‐induced carcinogenesis. The present study is the first to demonstrate the association between PARG silencing and H2B downregulation in BaP‐induced carcinogenesis. Because of the differences in the sequences and structures of H2B subtypes, we found changes in the expression of H2B subtypes and the modified lineage of H2B after translation. The role of H2B posttranslational modifications in tumorigenesis has been identified. For example, H2BK5ac has been reported to regulate the transformation of epithelial cells into mesenchymal cells. 17 We believe that our findings will contribute to the existing knowledge and provide a theoretical basis for the better prognosis and treatment of LC.
In this study, we found that the levels of several H2B subtypes increased after BaP‐induced malignant cell transformation, but PARG silencing inhibited this phenomenon. Our in vivo experiments confirmed that PARG silencing inhibited BaP‐induced malignant transformation by inhibiting the expression of H2B. H2B expression analysis using the online tumor database indicated that the increased expression of H2BFS, HIST1H2BD, HIST1H2BH, and HIST1H2BK in LC tissues might play an important role in LC oncogenesis. The transcription products of H2BFS, HIST1H2BD, HIST1H2BH, HIST1H2BK, HIST1H2BL, HIST1H2BM, and HIST1H2BO are potential prognostic markers that can be used for improving prognostic accuracy, thereby improving the survival rate of LC patients.
In summary, we have determined here for the first time, that PARG silencing reduces BaP‐induced carcinogenesis by suppressing H2B expression. This finding provides a theoretical basis of our next study on the role PARG silencing in posttranslational modification pedigree changes in H2B in the context of lung tumorigenesis.
ACKNOWLEDGMENTS
Thanks for the support of Natural Science Foundation of Guangdong Province (Project No. 2019A1515011080), Shenzhen Basic Research Project (Project No. JCYJ20190807102803567), Shenzhen Basic Research Discipline Layout (Project no. JCYJ20160328161613864) and Guangdong Medical Science and Technology Research Fund (Project No. A2019554).
Zeng Z, Lu J, Wu D, et al. Poly(ADP‐ribose) glycohydrolase silencing‐mediated H2B expression inhibits benzo(a)pyrene‐induced carcinogenesis. Environmental Toxicology. 2021;36:291–297. 10.1002/tox.23034
Funding information Guangdong Medical Science and Technology Research Fund, Grant/Award Number: A2019554; Guangdong natural science foundation general project, Grant/Award Number: 2019A1515011080; Shenzhen basic research discipline layout project, Grant/Award Number: JCYJ20160328161613864; Shenzhen Basic Research Project, Grant/Award Number: JCYJ20190807102803567
Contributor Information
Haiyan Huang, Email: hhy424@126.com.
Jianhui Yuan, Email: jianhui_yuan@126.com.
Zhangli Hu, Email: huzl@szu.edu.cn.
REFERENCES
- 1. Ahel I, Ahel D, Matsusaka T, et al. Poly(ADP‐ribose)‐binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451(7174):81‐85. [DOI] [PubMed] [Google Scholar]
- 2. Beneke S. Regulation of chromatin structure by poly(ADP‐ribosyl)ation. Front Genet. 2012;3:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang H, Li X, Hu G, et al. Poly(ADP‐ribose) glycohydrolase silencing down‐regulates TCTP and Cofilin‐1 associated with metastasis in benzo(a)pyrene carcinogenesis. Am J Cancer Res. 2015;5(1):155‐167. [PMC free article] [PubMed] [Google Scholar]
- 4. Li X, Li X, Zhu Z, et al. Poly(ADP‐ribose) Glycohydrolase (PARG) silencing suppresses Benzo(a)pyrene induced cell transformation. PLoS One. 2016;11(3):e0151172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maselli J, Hales BF, Chan P, Robaire B. Exposure to bleomycin, etoposide, and cis‐platinum alters rat sperm chromatin integrity and sperm head protein profile. Biol Reprod. 2012;86(5):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loe‐Mie Y, Lepagnol‐Bestel AM, Maussion G, et al. SMARCA2 and other genome‐wide supported schizophrenia‐associated genes: regulation by REST/NRSF, network organization and primate‐specific evolution. Hum Mol Genet. 2010;19(14):2841‐2857. [DOI] [PubMed] [Google Scholar]
- 7. Guo X, Liu W, Pan Y, et al. Homeobox gene IRX1 is a tumor suppressor gene in gastric carcinoma. Oncogene. 2010;29(27):3908‐3920. [DOI] [PubMed] [Google Scholar]
- 8. Santoro SW, Dulac C. The activity‐dependent histone variant H2BE modulates the life span of olfactory neurons. Elife. 2012;1:e00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beer DG, Kardia SL, Huang CC, et al. Gene‐expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816‐824. [DOI] [PubMed] [Google Scholar]
- 10. Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Esteller M. Cancer epigenomics: DNA methylomes and histone‐modification maps. Nat Rev Genet. 2007;8(4):286‐298. [DOI] [PubMed] [Google Scholar]
- 12. Wang R, Xin M, Li Y, Zhang P, Zhang M. The functions of histone modification enzymes in cancer. Curr Protein Pept Sci. 2016;17(5):438‐445. [DOI] [PubMed] [Google Scholar]
- 13. Zeng Z, Liu H, Yuan J, et al. Poly(ADP‐ribose) glycohydrolase silencing‐mediated maintenance of H2A and downregulation of H2AK9me protect human bronchial epithelial cells from benzo(a)pyrene‐induced carcinogenesis. Toxicol Lett. 2018;295:270‐276. [DOI] [PubMed] [Google Scholar]
- 14. Shechter D, Dormann HL, Allis CD, Hake SB. Extraction, purification and analysis of histones. Nat Protoc. 2007;2(6):1445‐1457. [DOI] [PubMed] [Google Scholar]
- 15. Bennett RL, Bele A, Small EC, et al. A mutation in histone H2B represents a new class of oncogenic driver. Cancer Discov. 2019;9(10):1438‐1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nacev BA, Feng L, Bagert JD, et al. The expanding landscape of 'oncohistone' mutations in human cancers. Nature. 2019;567(7749):473‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mobley RJ, Abell AN. Controlling epithelial to mesenchymal transition through acetylation of histone H2BK5. J Nat Sci. 2017;3(9):e432. [PMC free article] [PubMed] [Google Scholar]


