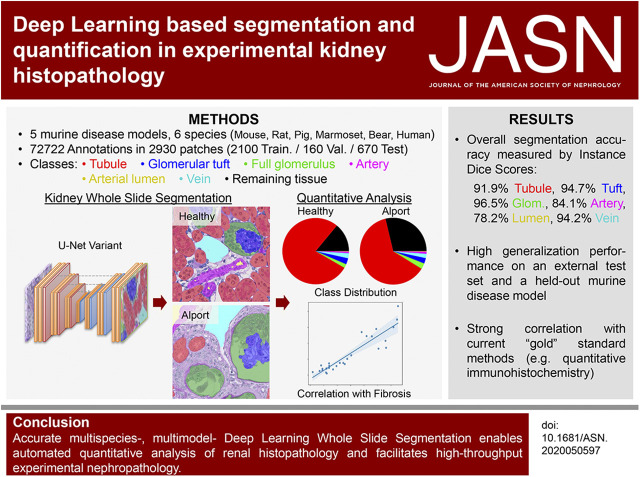
Significance Statement
Nephropathologic analyses provide important outcomes-related data in the animal model studies that are essential to understanding kidney disease pathophysiology. In this work, the authors used a deep learning technique, the convolutional neural network, as a multiclass histology segmentation tool to evaluate kidney disease in animal models. This enabled a rapid, automated, high-performance segmentation of digital whole-slide images of periodic acid–Schiff–stained kidney tissues, allowing high-throughput quantitative and comparative analyses in multiple murine disease models and other species. The convolutional neural network also performed well in evaluating patient samples, providing a translational bridge between preclinical and clinical research. Extracted quantitative morphologic features closely correlated with standard morphometric measurements. Deep learning–based segmentation in experimental renal pathology is a promising step toward reproducible, unbiased, and high-throughput quantitative digital nephropathology.
Keywords: digital pathology, segmentation, histopathology, animal model
Visual Abstract
Abstract
Background
Nephropathologic analyses provide important outcomes-related data in experiments with the animal models that are essential for understanding kidney disease pathophysiology. Precision medicine increases the demand for quantitative, unbiased, reproducible, and efficient histopathologic analyses, which will require novel high-throughput tools. A deep learning technique, the convolutional neural network, is increasingly applied in pathology because of its high performance in tasks like histology segmentation.
Methods
We investigated use of a convolutional neural network architecture for accurate segmentation of periodic acid–Schiff-stained kidney tissue from healthy mice and five murine disease models and from other species used in preclinical research. We trained the convolutional neural network to segment six major renal structures: glomerular tuft, glomerulus including Bowman’s capsule, tubules, arteries, arterial lumina, and veins. To achieve high accuracy, we performed a large number of expert-based annotations, 72,722 in total.
Results
Multiclass segmentation performance was very high in all disease models. The convolutional neural network allowed high-throughput and large-scale, quantitative and comparative analyses of various models. In disease models, computational feature extraction revealed interstitial expansion, tubular dilation and atrophy, and glomerular size variability. Validation showed a high correlation of findings with current standard morphometric analysis. The convolutional neural network also showed high performance in other species used in research—including rats, pigs, bears, and marmosets—as well as in humans, providing a translational bridge between preclinical and clinical studies.
Conclusions
We developed a deep learning algorithm for accurate multiclass segmentation of digital whole-slide images of periodic acid–Schiff-stained kidneys from various species and renal disease models. This enables reproducible quantitative histopathologic analyses in preclinical models that also might be applicable to clinical studies.
Many basic science and preclinical studies require experiments in animals with histopathologic assessment representing a major readout. The demands on robust but at the same time objective, precise, and quantitative data steadily increase. In both clinical practice and research, histopathologic evaluations are often performed manually. This is both time-consuming and not seldom poorly reproducible, particularly if not performed by experts. The projected decrease in pathologist workforce, which is particularly noticeable in highly specialized fields like nephropathology, and heavy engagement in clinical duties further complicate the situation.1
High-throughput digitization of histologic slides, generating so-called whole-slide images (WSIs), enables the effective use of computer-assisted histopathologic analysis. Deep learning (DL) is a subset of artificial intelligence that applies computer algorithms to find meaningful representations of raw data through multiple layers of abstraction.2 DL’s most popular technique, the convolutional neural network (CNN), is increasingly applied in pathology3 due to its high performance in tasks like detection of nuclei,4 histology segmentation,5 or prediction of molecular alterations from hematoxylin-and-eosin–stained sections.6 We have previously shown that ML- and DL-based techniques can facilitate glomerulus detection and segmentation in WSIs.7–10 Recently, two other groups reported the feasibility of the DL-based segmentation of human kidney WSIs,11,12 and glomerulus segmentation was already successfully used for subsequent analysis of glomerulosclerosis in periodic acid–Schiff (PAS)–13,14 or trichrome-stained biopsy specimens.15 The usefulness of DL in animal models with broad histopathologic injury patterns was not yet analyzed.
Our main aim was to develop a CNN for multiclass segmentation of mouse kidney PAS-stained histology, focusing on five commonly used models of kidney diseases. We demonstrate the applicability of our CNN for large-scale histopathologic segmentation followed by quantitative data extraction and confirm the performance by correlation with traditional image analysis tools. We also show the applicability for other species used in research, and for patient kidney samples.
Methods
Histology Samples
We used paraffin-embedded kidney tissue fixed in formalin or methyl Carnoy’s solution. Sections of 1–2-µm thickness were stained with PAS and counterstained with hematoxylin. Slides were digitalized using the whole-slide scanners NanoZoomer HT2 with ×20 objective (Hamamatsu Photonics, Hamamatsu, Japan) or Aperio AT2 with ×20 or ×40 objective (Leica Biosystems, Wetzlar, Germany).
All samples from mice, rats, and pigs came from already published studies and were retrospectively analyzed.16–21 All animal experiments were approved by the local government authorities: mouse, rats, pigs: Landesamt für Umwelt und Verbraucherschutz Nordrhein Westfalen; marmosets: institutional animal welfare committee and subsequently by the Lower Saxony State Office for Consumer Protection and Food Safety (LAVES) (reference number 33.19‐42502‐04‐17/2496); and bears: bear samples were obtained by hunters during the hunting seasons in Maine. Hunters were asked to participate on a voluntary basis and no bears were killed for the specific purpose of this study. All methods were carried out in accordance with relevant guidelines and regulations.
Mouse Models
We reanalyzed healthy male 10–12-week-old C57BL/6N mice (n=41) and five widely used murine models of kidney diseases with different causes, i.e., unilateral ureteral obstruction (UUO, n=15),16,17 adenine-induced nephropathy (adenine, n=15),18 Col4a3 knock out (Alport, n=15),16 unilateral ischemia-reperfusion injury (IRI, n=15),16,17 and nephrotoxic serum nephritis (NTN, n=15),19 and an additional sixth model used only for testing, the diabetic/metabolic nephropathy (db/db, n=3).20 The surgical UUO and IRI models were conducted in male 10–12-week-old C57BL/6N mice as previously described.16,17 An additional UUO day 10 cohort of three male C57BL/6J mice was contributed by R. Kramann and S. Menzel and used as an external control cohort. For the adenine model, male 10–12-week-old mice on C57BL/6N background were fed with 0.2% adenine-enriched diet as previously described.22 For the NTN model, kidneys from male 12–14-week-old 129×1/SvJ mice were harvested 10 days after intravenous injection of a sheep-anti-mouse glomerulus antiserum.19 Col4a3 knockout mice were bred on a 129×1/SvJ genetic background and euthanized at 8 weeks of age. The db/db mice (BKS.Cg-Dock7m+/+Leprdb/J) were fed a high-fat Western diet for 9 weeks and a normal diet for another 5 weeks before euthanasia.20
In the UUO (sham, day 5, day 10 samples), IRI (sham, day 14, day 21 samples), and adenine (day 1, day 14, day 21 samples) models, additional immunostainings and quantifications were performed as previously described17,22 for comparison with network-based automated segmentation results from PAS stainings. In short, sections were deparaffinized and endogenous peroxidase was blocked with 3% H2O2. Slides were incubated with a primary antibody against α-SMA (α-smooth muscle actin, Dako/Agilent, M085101–2; Santa Clara, CA), followed by colorimetric detection using DAB and nuclear counterstain with methyl green. The stainings were digitalized and further processed using the viewing software NDP.view (Hamamatsu Photonics, Hamamatsu, Japan). The percentage of positively stained area was analyzed in whole cortices at ×20 magnification using ImageJ software by measuring DAB-positive pixels in 8-Bit images (National Institutes of Health, Bethesda, MD) as previously described.16,18 All analyses were performed in a blinded manner.
Patient Samples
Sixteen PAS-stained sections from formalin-fixed and paraffin-embedded human kidney specimens (nine tumor nephrectomies and seven biopsy specimens [two minimal change disease, one pauci-immune GN, four acute tubular injury]) were anonymously obtained from the archive of the Institute for Pathology of the RWTH Aachen University. In the case of tumor nephrectomies, healthy tissue far away from the tumors was used. Patient characteristics were: M/F=7:9; age=63.13±11.86 years. The study was approved by the local ethical committee of the RWTH University (No. EK315/19).
Further Species
For an extended analysis across different species, we used healthy kidney tissue from rats, pigs, common marmosets, and black bears. We used renal tissue from male Wistar rats (n=8) and German landrace pigs (n=6). Renal tissue from male (n=2) and female (n=6) common marmosets was provided by the German Primate Center, Goettingen. Kidney tissue from black bears (n=8) was provided by the Jackson Laboratory and collected by local huntsmen from male animals at different ages all across Maine, US. Hunters were provided with detailed collection directions and provided datasheets voluntarily about deviations to requested timing in sample collection and fixation and metadata about the bears.
Dataset and Ground Truth
All technical terms used in the following sections are described in a glossary in Supplemental Table 1. The WSIs (n=168 in total) were split into training, validation, and test sets as follows: the 41 healthy mouse WSIs—30 training, three validation, eight test; the 15 WSIs from each mouse model—11 training, one validation, three test; the three db/db and three external UUO were only used for the test; the six pig WSIs—five training, one test; the eight marmoset, bear and rat WSIs—each split to five training, three test; and the 16 human WSIs—ten training, six test slides: two test WSIs for performance quantifications and all four slides of acute tubular injury to visually show transferability to human disease.
Ground truth annotations were generated for patches of size 174×174 µm2 (resampled into 516×516-pixel integer label images) by eight qualified annotators as outlined in the section “Data Quality and Quantity” using QuPath.23 All annotations were corrected by a nephropathologist and researcher with long experience in nephrologic basic research. Six predefined classes (i.e., renal structures) were annotated: (1) full glomerulus, (2) glomerular tuft, (3) tubule, (4) artery, (5) arterial lumen, and (6) vein including renal pelvis and large nontissue areas. Classes and annotation procedure are defined in detail in Supplemental Figure 1, A–G and Supplemental Table 2. The remaining tissue comprising capillaries, adventitia of arteries, interstitial cells and matrix, and urothelium was defined as the “interstitium.” For annotations, we mostly selected 20 random patches per slide. An overview of our annotations is provided in Supplemental Table 3. In total, we performed 2930 annotated patches and 72,722 annotated structures and split the annotated patches into 2100 training (600 murine healthy, 220 each murine model, 200 human, 50 each remaining species), 160 validation (60 murine healthy, 20 each murine model), and 670 test patches (160 murine healthy, 60 each murine model, 30 murine db/db, 30 external murine UUO, 30 each remaining species including human) for the development of our CNN (Figure 1, Supplemental Table 4).
Figure 1.
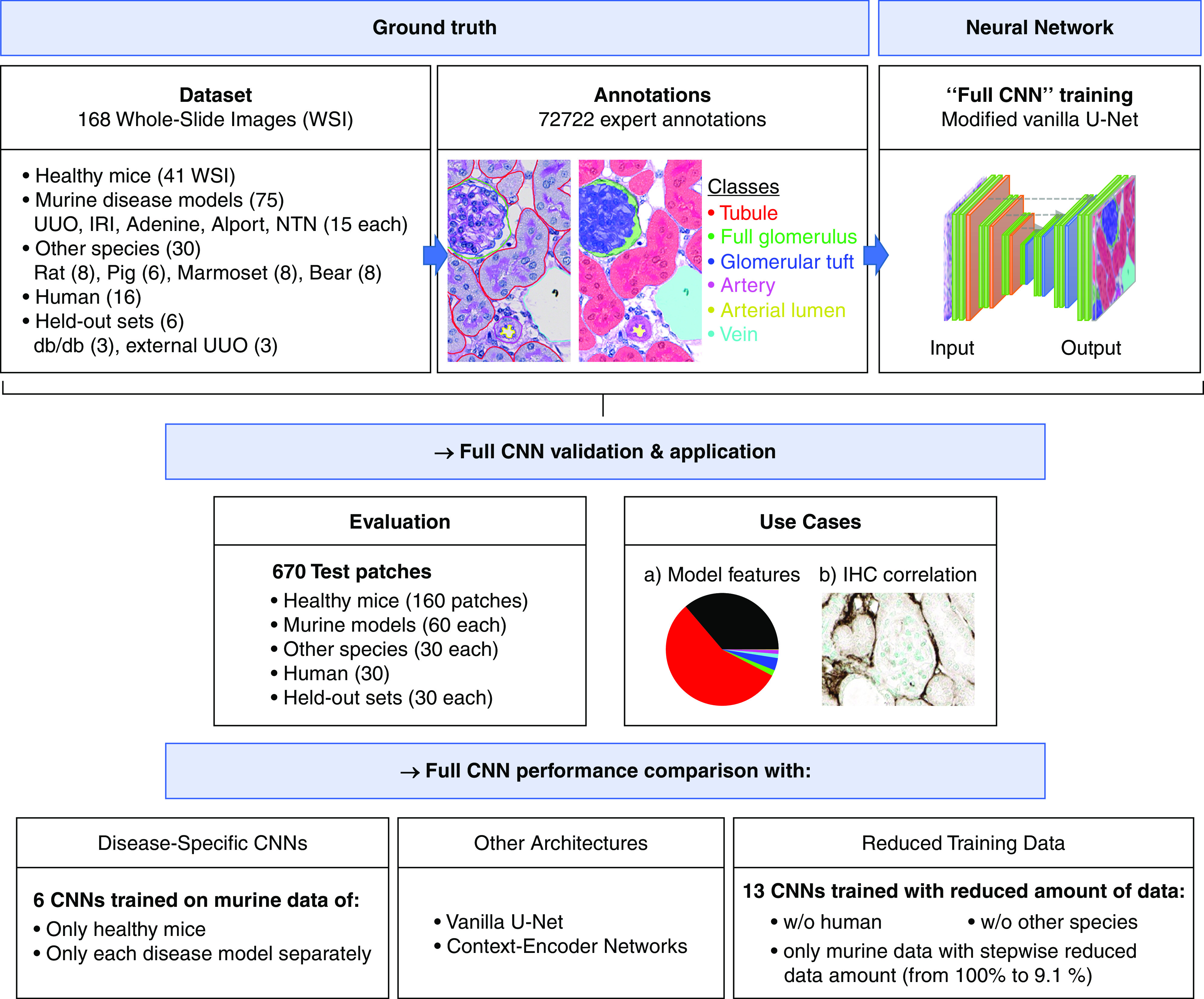
Overview of experimental design. Our DL model (here: Full CNN) was trained with annotations from healthy and diseased murine kidneys and with annotations from five different species including humans. A total of 72,722 single instance annotations comprised six different renal structures: “tubule,” “full glomerulus,” “glomerular tuft,” “artery,” “arterial lumen,” and “vein.” The model was tested on healthy and diseased murine kidneys, on five different other species, on a held-out murine disease model, and on an external UUO cohort. We used the automatically segmented kidneys to perform quantitative feature analysis and correlations with IHC. Further experiments included an ablation study on varying training dataset sizes to analyze its effect on model performance, and we also compared the full CNN with its variants solely trained on single murine models and with different state-of-the-art segmentation networks including the vanilla U-Net and context-encoder networks. IHC, immunohistochemistry; w/o, without.
Data Quality and Quantity
The most crucial prerequisite for high performance of a DL system is the optimization of data quality and quantity. We performed the following optimization techniques: (1) the expert annotators were instructed and coached to precisely comply with the developed structure definitions (Supplemental Figure 1, Supplemental Table 2) to reduce interannotator variability, thus yielding consistent annotations. (2) After manual annotation of about 20% of all annotations, we used these to train an initial segmentation network. We then used its predictions as preannotations facilitating the annotation effort for the annotators. These predictions were loaded into QuPath, converting the manual annotation task into a prediction correction task, reducing the annotation effort (Supplemental Figure 1H). (3) We applied the concept of active learning24 to optimize the selection of image patches for annotation. We used the initial segmentation network to compute whole-slide segmentation results and visually selected patches with the highest prediction errors most often showing complex or rare structures. We have repeated steps (2) and (3) when about 60% of all annotations have been performed. This concept yields an extremely high degree of sample efficiency to ensure that the network will learn and improve in an optimal way.
CNN Development
CNN Model
Our employed DL model was on the basis of the U-Net architecture25 (for details see Supplemental Table 4). The U-Net was initially developed for biomedical image segmentation and represents one of the most popular and powerful segmentation techniques nowadays. We applied the following changes to the original architecture: (1) we increased its depth by one to increase its receptive field, (2) we then used half channel numbers on each architectural level to reduce the risk of overfitting, (3) we did not halve feature channel numbers when upsampling via transposed convolutions to effectively increase its capacity, and (4) we empirically applied instance normalization and leaky ReLU activation due to its empirically shown superiority over batch normalization and ReLU activation,26 overall resulting in about 37 million learnable parameters in our CNN. As network inputs, we extracted bigger image slide patches of 216×216 µm2, resampled into 640×640-pixel RGB images, around the annotated patches of 174×174 µm2, to improve prediction accuracy close at borders due to the resulting context awareness.27
Border Class
To ensure the separation of different, touching instances of the same class, we introduced a new border class following28 by performing dilation on all tubules using a ball-shaped structuring element of radius three pixels. Considering arteries and glomeruli, only the overlap between their dilated versions, employing a radius of seven pixels, was also assigned to the border class. This way, the network was able to maintain a continuous label transition prediction from afferent and efferent arteriole to the glomerulus, thereby greatly improving the prediction accuracy of small afferent and efferent arterioles. The border class mainly represented the tubular basement membranes.
Training Routines
We trained our CNN using the optimizer RAdam29 on random mini-batches of size six and applied weight decay with a factor of 1E−5 for regularization. We further scheduled the learning rate in a reduce-on-plateau fashion to reduce overfitting as follows: it was initially set to 0.001 and was divided by 3 when the validation loss had not fallen for 15 epochs. When the learning rate fell below 4E−6, training terminated and the network configuration providing the lowest validation error was chosen as the final model. Also, our data augmentation pipeline consisted of spatial (i.e., affine, piecewise affine, elastic, flipping, 90-degree rotation) and color transformations (i.e., hue and saturation shifting, gamma contrast, normalization) to improve the CNN’s generalizability by simulating variance in tissue morphology and staining. The weighted categoric crossentropy and the Dice-loss30 were applied as equally weighted loss functions measuring the dissimilarity between prediction and ground truth for network optimization. Using weighted categoric crossentropy, we gave the border class a ten-times-greater weight than other classes to strongly enforce the separation of different instances from the same class. Overall, three-channel inputs (RGB) of spatial resolution 640×640 pixels were being forwarded through the network producing eight class probability maps, i.e., full glomerulus, glomerular tuft, tubule, vein including nontissue background and renal pelvis, artery, arterial lumen, tubular border, and remaining tissue representing our interstitium class, of spatial size 516×516 pixels. For each pixel, the class with the highest probability was assigned as the predicted label. To account for reproducibility, our code is publicly available (at https://github.com/NBouteldja/KidneySegmentation_Histology).
Postprocessing
In contrast to network ensembling, we applied the regularization technique test-time augmentation to improve the CNN’s robustness at low cost. During inference, test-time augmentation forward flipped versions of the input and averages their respectively back-flipped predictions to reduce prediction variance by considering multiple estimations. We also performed the following postprocessing techniques to all classes except the interstitium: (1) we removed too-small instance predictions and assigned them to the remaining interstitium class, except for respective glomerular tuft and arterial lumen predictions that were assigned to their superior classes glomerulus and artery; (2) we performed hole filling; and (3) we performed dilated tubular instance predictions due to their thicker border predictions.
Evaluation
Quantitative Evaluation
We quantitatively evaluated network performance using instance-level Dice scores, i.e., in all image/ground truth pairs, we computed regular Dice scores between each ground truth instance and its maximally overlapping prediction (0 for false negatives), and vice versa for each prediction instance to also account for false positives. These Dice scores were averaged over all instances in all images, resulting in the instance-level Dice score. This metric accurately denoted the mean detected area coverage per instance. We also employed the commonly used average precision (AP) as a detection metric. After counting and summing all true positives (TPs), false positives (FPs), and false negatives (FN) across all images, the AP was calculated as follows:
A prediction was considered a TP when it overlapped with at least 50% of a ground-truth instance. Both metrics range from 0 (maximal discordance: no overlap/TP) to 1 (maximal agreement: perfect overlap/detections).
Performance versus Amount of Training Data
A key unresolved issue regarding DL systems is the specification of the minimum amount of training data necessary to reach satisfactory performances for a given task. Therefore, we performed an ablation study on performance differences when training on different training set sizes. In total, we trained another 13 CNNs from scratch using the following training sets: From all 2100 training patches (representing our full CNN), we removed human patches or other species patches, or used murine patches only and in a stepwise manner removed randomly 9.1% of the patches (i.e., using only 90.9%, 81% … 9.1% of the murine patches, but always including patches from healthy and each model). The validation and test sets as employed for our full CNN always remained the same.
Full CNN versus Specialized Single Models
We examined the effect on network performance when jointly training on data from different domains, i.e., different species and murine disease models. We compared our full CNN trained on all training data (including murine models and species) with six networks, each solely trained and tested on a particular single murine model, i.e., healthy, UUO, adenine, Alport, IRI, NTN, to analyze (1) whether the network benefits from shared multidomain information by potentially learning more specialized class features, (2) whether the network can learn the same domain-specific features maintaining equal segmentation performance, or (3) whether the heterogeneity of multidomain information might perturb the network, resulting in lower prediction accuracies.
State-of-the-Art Model Comparison
We compared our model with its unmodified variant, the vanilla U-Net,27 to explore whether our technical modifications to the standard network architecture had an effect on performance. We also compared our network with the context-encoder network,31 another novel state-of-the-art segmentation network particularly suitable for the segmentation of structures with different sizes that was shown to outperform the vanilla U-Net. For all comparisons, the same training and test sets were used.
Comparative Feature Extraction
On the basis of the CNN segmentation results, we extracted the following histologic features from cortical areas: (1) relative proportions of tissue area covered by each class, (2) single class instance sizes (including sizes of Bowman’s space by subtracting the glomerular tuft area from each full glomerulus), and (3) tubular diameters. We included all instances independent of the plane on which they were cut. We used data from four individual mice at each of the following model time points: UUO day 10, adenine day 14, Alport mice at 8 weeks of age, IRI day 14, NTN day 10, and randomly chosen healthy mice. In each WSI, we extracted ten cortical patches of size 700×700 µm2 for feature computation. We defined the maximum tubular diameter as the diameter of the largest circle fully fitting inside the tubules, a feature that can represent both tubular dilation and atrophy. Tubular diameter computation was performed by employing the distance transform function and extracting its maximum value. For class instance size and tubular diameter computation, only instances fully inside our selected patches were considered.
Correlation with Immunohistochemical Analysis
Next to qualitative and quantitative performance evaluation, we correlated our results with standard morphometric analyses, to assess the capabilities of facilitating relevant histopathologic applications. We employed data from the three different murine models UUO, adenine, and IRI. We extracted five cortical patches of size 700×700 µm2 in each WSI and correlated the remaining interstitial area coverage predicted by our automated approach with results from a computer-assisted morphometric analysis of immunohistochemical stainings for α-SMA from the same kidneys, in which big vessels were always excluded.16,18
Statistical Analyses
To measure the strength of the (linear) correlation between immunohistochemical fibrosis quantifications and network-based interstitial area estimations, we employed the Pearson correlation coefficient and the Spearman correlation coefficient and computed respective P values on the basis of the t-distribution. We used t-tests for comparison between CNN, the vanilla U-Net, and the context-encoder by comparing respective Dice score distributions of each class across all models, and to compare pairwise class instance sizes from healthy and all disease models (P<0.05 was considered statistically significant).
Results
Ground Truth
For the training and evaluation of our full CNN, we performed 72,722 annotations of six classes, i.e., renal structures, selected on the basis of the most commonly performed compartment-specific quantifications in animal models: tubule, full glomerulus, glomerular tuft, artery (including intima and media but excluding adventitia), arterial lumen, and vein (including renal pelvis and nontissue slide background). We used kidneys from murine disease models, different species, and humans (Figure 1, Supplemental Figure 1, Supplemental Table 3). Inclusion of renal pelvis and large nontissue areas in the “vein” instead of our “interstitium” class improved predictions of such large white structures due to their great local similarities and was an important prerequisite for more precise quantitative analyses, particularly of the interstitium. We have not distinguished different tubular segments, particularly due to the difficult distinction of injured tubules in the disease models. The tubular class did not include tubular basement membranes, to allow a very specific analysis of tubular cells. Both cortex and medulla were annotated, whereas perirenal tissues were not included. We recognized some obstacles in generating annotations, outlined in detail in Supplemental Figure 2. All annotations were ultimately corrected by two experts in nephropathology and structures that were not feasible to assign to a class on the basis of our class definitions with sufficient certainty and consensus were not included in annotations (altogether representing only very few instances).
Accurate Multiclass Segmentation of Murine Kidney Sections
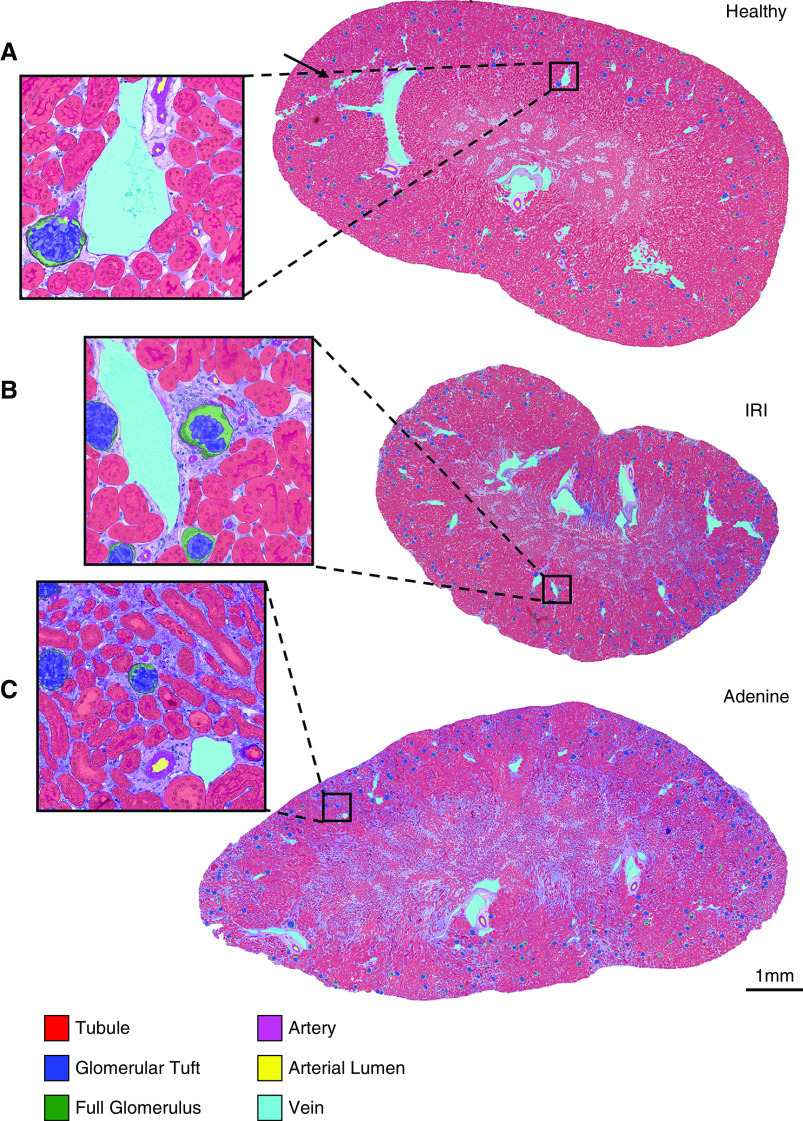
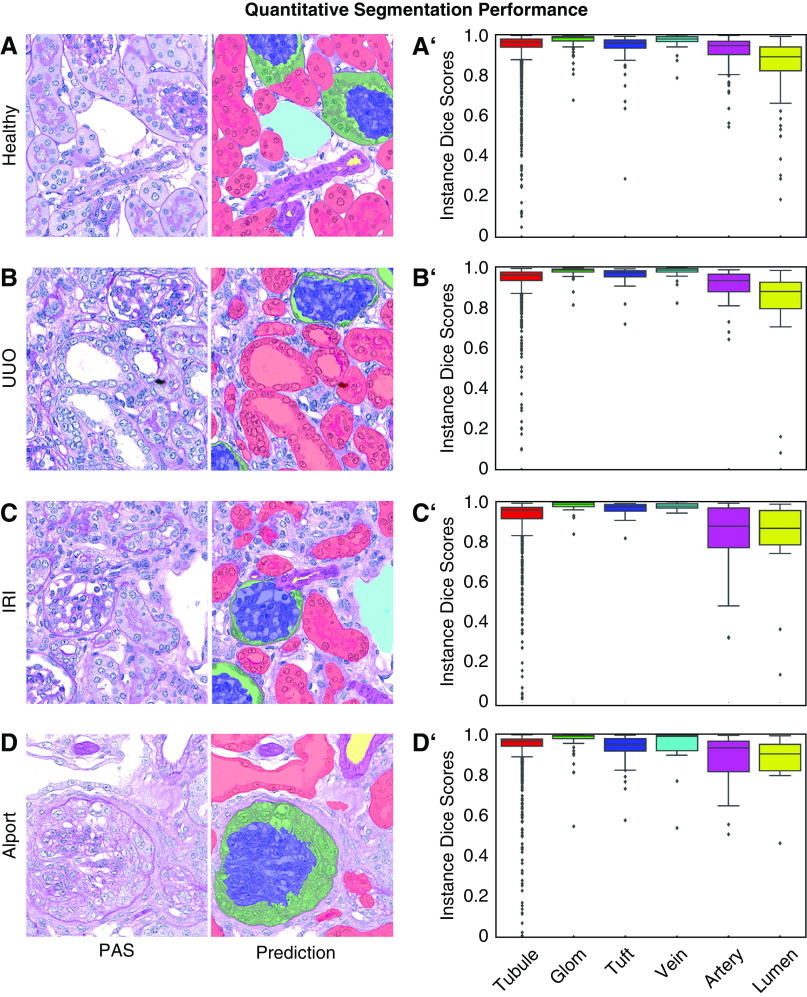
Although network training took about 8.5 hours on the graphics processing unit (GPU) RTX2080Ti and required approximately 10 GB of GPU memory, automated segmentation of a whole murine kidney longitudinal cross-section was performed in <5 minutes on the same GPU. Qualitative segmentation results of representative WSIs from healthy and diseased kidneys showed high accuracy for all six classes (Figure 2, A–C, Supplemental Figure 3, A–C). In a healthy kidney, an accidental scratch was correctly assigned to the vein class including nontissue areas (Figure 2A, arrow). In healthy murine kidneys, our CNN was able to detect almost 95% of all tubular structures with an instance segmentation accuracy of 93.2% (Table 1). Almost all glomeruli were correctly detected and segmented, although detection and segmentation accuracies were lowest for arteries and arterial lumina (Figure 3, A and A’). Segmentation performances in UUO (Figure 3, B and B’) and IRI (Figure 3, C and C’) were similar to healthy kidneys for tubules, glomeruli, and vein classes (all >90%). Alport mice represented the most complex model, with correct segmentation of 91% of all tubules and 95% of all glomeruli, including those with severe and global pathologic alterations such as extracapillary proliferates (cellular crescents) or FSGS (Figure 3, D and D’). Detection and segmentation results for arteries and their lumina were the lowest, ranging from 79.1% (segmentation of artery in IRI) to 88.1% (segmentation of artery in healthy) and from 73.5% (segmentation of arterial lumen in IRI) to 81.1% (segmentation of arterial lumen in Alport), respectively. The CNN was able to correctly detect and segment disease-specific pathologies, e.g., dilated tubules in UUO (Figure 3B), atrophic tubules in IRI (Figure 3C), glomerular crescents and FSGS in Alport mice and NTN (Figure 3D, Supplemental Figure 4A, arrows), and tubules with renal crystals in the adenine model (Supplemental Figure 4B, arrows). Medullary structures were also accurately segmented in all models (Supplemental Figure 5, A–F’’). Almost every segmented item, e.g., one tubular cross-section, was recognized as an individual instance despite potentially touching other class instances and could be therefore further analyzed separately on instance level (Supplemental Figure 5, A’’–F’’).
Figure 2.
Automated segmentation on WSIs of murine kidneys. (A) The CNN generates segmentation predictions on a WSI of a healthy mouse kidney. All six classes, i.e., tubule, glomerulus, glomerular tuft, artery, arterial lumen, and vein, are precisely segmented. Even tissue damage in the form of an artificial scratch (arrow) is correctly assigned to the vein class including the background. Similar segmentation predictions are generated for WSIs of (B) IRI and (C) adenine kidneys.
Table 1.
Quantitative segmentation and detection performance of six classes in murine kidneys
| Mouse Models | Detection | Segmentation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full Glomerulus | Glomerular Tuft | Tubule | Artery | Arterial Lumen | Vein | Full Glomerulus | Glomerular Tuft | Tubule | Artery | Arterial Lumen | Vein | |
| Healthy mouse | 98.7 | 96.5 | 94.9 | 87.4 | 76.2 | 93.9 | 96.5 | 93.7 | 93.2 | 88.1 | 80.3 | 94.3 |
| UUO | 100 | 100 | 91.0 | 78.2 | 73.3 | 100 | 97.5 | 95.6 | 90.9 | 82.3 | 75.0 | 97.6 |
| IRI | 95.7 | 97.7 | 89.3 | 73.3 | 67.6 | 100 | 96.0 | 95.4 | 90.2 | 79.1 | 73.5 | 97.7 |
| Adenine | 100 | 100 | 93.0 | 82.4 | 80.3 | 90.3 | 98.8 | 97.2 | 93.0 | 87.9 | 80.9 | 93.5 |
| Alport | 92.5 | 93.4 | 88.6 | 73.2 | 79.2 | 80.0 | 94.7 | 91.4 | 90.6 | 80.3 | 81.1 | 89.2 |
| NTN | 96.2 | 98 | 93.5 | 86.1 | 74.0 | 89.2 | 95.5 | 94.8 | 93.2 | 86.8 | 78.2 | 92.8 |
Segmentation performance was calculated by averaging all instance Dice scores from each instance in all test images denoting the mean detected area coverage per instance. We employed an average precision metric to measure detection performance.
Figure 3.
Quantitative segmentation performance in murine kidney disease models. Representative PAS pictures and corresponding segmentation predictions generated by the CNN for murine (A) healthy, (B) UUO, (C) IRI, and (D) Alport kidneys. Instance segmentation accuracy is shown by instance Dice scores for each class in all four models (A’–D’). Data are presented in box plots with median, quartiles, and whiskers. Glom, glomerulus; Tuft, glomerular tuft.
A very small fraction of structures were not correctly detected or not precisely segmented (Supplemental Figure 6). These included glomeruli with a direct connection to the proximal tubule, in which either a part of the glomerulus was identified as tubule or tubular cells were marked as part of the glomerulus (Supplemental Figure 6, A and A’, arrow). Those examples also included special instances, e.g., fibrin within crescents (Supplemental Figure 6, B and B’, arrow), which was missing in the training dataset. We also observed some incorrectly detected tubules, mostly if severely injured, present as denuded basement membrane (Supplemental Figure 6, C and C’ arrow), massively dilated (Supplemental Figure 6, D and D’, arrowhead), or atrophic (Supplemental Figure 6, D and D’ arrow).
Detection rates were improved in all models by providing more training data (Supplemental Figure 7). In all models and almost all classes (except arteries and arterial lumina), approximately 35% of ground truth data were already sufficient to obtain 90% or higher detection rates. Especially for more complex structures such as arteries or very small structures like arterial lumina, detection performance could be substantially improved by integrating more training data, indicating that further improvement of segmentation accuracy for some classes is feasible (Supplemental Figure 7). For other classes, especially tubules, the performance was high and stable even in the case of only about 9% training data.
We compared our CNN with its variants, which have been solely trained and tested on single murine models (healthy, UUO, adenine, Alport, IRI, NTN). In almost all models and classes, especially arteries and lumina, our full CNN trained on all domains provided higher segmentation performances compared with the variants (Supplemental Figure 8, A–F).
We next compared our CNN with its unmodified variant, the vanilla U-Net, and with a context-encoder, a novel state-of-the-art segmentation framework which was shown to outperform the U-Net.31 Our modified CNN significantly outperformed the unmodified vanilla U-Net (Supplemental Table 5) and the context-encoder (Supplemental Table 5) in the majority of classes and models, including arterial structures. Thus, our modified architecture was suitable for the specific task of kidney histology segmentation.
Multiclass Segmentation in External UUO Test Set and Held-Out db/db Model
We next examined performance of our full CNN on PAS slides from an external UUO cohort and also in a completely different disease model, i.e., the db/db mice on a high-fat diet,20 both not included in the training. Quantitative evaluation confirmed very high segmentation accuracies of at least 95% area coverage with the ground truth for glomeruli, tufts, and tubules in both experiments (Supplemental Figure 9, A–D’’, Table 2). As in other models, the segmentation of arteries and their lumina was less accurate (both approximately 80%). Overall, these results are comparable to the other models included in training, indicating strong generalization capabilities of our CNN across different laboratories and models.
Table 2.
Quantitative segmentation and detection performance in kidneys from different species, held-out murine disease model db/db, and external UUO
| Kidney Type | Detection | Segmentation | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Full Glomerulus | Glomerular Tuft | Tubule | Artery | Arterial Lumen | Vein | Full Glomerulus | Glomerular Tuft | Tubule | Artery | Arterial Lumen | Vein | |
| Rat | 100 | 82.1 | 94.7 | 85.7 | 81.0 | 92.9 | 99.5 | 88.9 | 96.5 | 91.6 | 89.5 | 93.9 |
| Pig | 93.8 | 100 | 95.6 | 100 | 95.2 | 84.6 | 96.5 | 99.0 | 97.9 | 96.9 | 96.3 | 91.6 |
| Black bear | 88.3 | 85.7 | 96.8 | 94.3 | 89.2 | 100 | 87.5 | 91.5 | 97.3 | 91.8 | 94.3 | 99.7 |
| Marmoset | 100 | 100 | 95.1 | 82.7 | 73.5 | 92.9 | 98.9 | 95.9 | 96.8 | 86.0 | 86.8 | 96.2 |
| Human | 88.2 | 72.5 | 91.8 | 66.7 | 68.4 | 72.7 | 93.4 | 76.6 | 95.2 | 79.1 | 77.6 | 85.1 |
| db/db mice | 93.1 | 96.3 | 90.5 | 60.6 | 58.3 | 100 | 95.9 | 97.5 | 94.9 | 81.0 | 79.1 | 99.0 |
| External UUO | 93.6 | 97.7 | 94.8 | 68.2 | 69.6 | 87.5 | 96.6 | 98.5 | 97.0 | 78.2 | 81.4 | 93.3 |
Segmentation performance was calculated by averaging all instance Dice scores from each instance in all test images denoting the mean detected area coverage per instance. We employed an average precision metric to measure detection performance.
Multiclass Segmentation of Murine Kidney Sections Enables Feature Extraction and Analysis
The CNN-based segmentation made it possible to extract quantitative histologic features on a large scale. We analyzed each of the six classes in all disease models (Figure 4, A–F), overall analyzing 70,311 cortical instances. We compared healthy kidneys, UUO day 10, adenine day 14, Alport at 8 weeks of age, IRI day 14, and NTN day 10. The glomerular area significantly increased in all models, particularly in those with primary glomerular damage, i.e., Alport and NTN. This expansion of glomeruli reached areas of above 14,000 µm2 in NTN, compared with 6000 µm2 as the largest measured glomerular area in healthy mice. We observed similar findings for glomerular tufts, except for Alport mice, in which the tuft size was significantly reduced due to sclerosis (Figure 4B). Specific analyses of the area of Bowman’s space confirmed its expansion in the two models with known glomerular damage, i.e., NTN and Alport. In addition, the Bowman’s space was also significantly increased in the adenine model but decreased in the IRI model (Figure 4H). Healthy tubules exhibited two major groups with peak areas of 900 µm2 and 400 µm2, likely representing different tubular segments. In all disease models, tubular area distributions converged to a single peak at about 400–500 µm2, in line with tubular damage and simplification. Tubular dilation was found in several disease models, and prominently increased tubular sizes were detected in NTN (maximum tubular size: 20,000 µm2), Alport (17,000 µm2), and UUO (15,000 µm2), compared with healthy (11,000 µm2) (Figure 4C).
Figure 4.
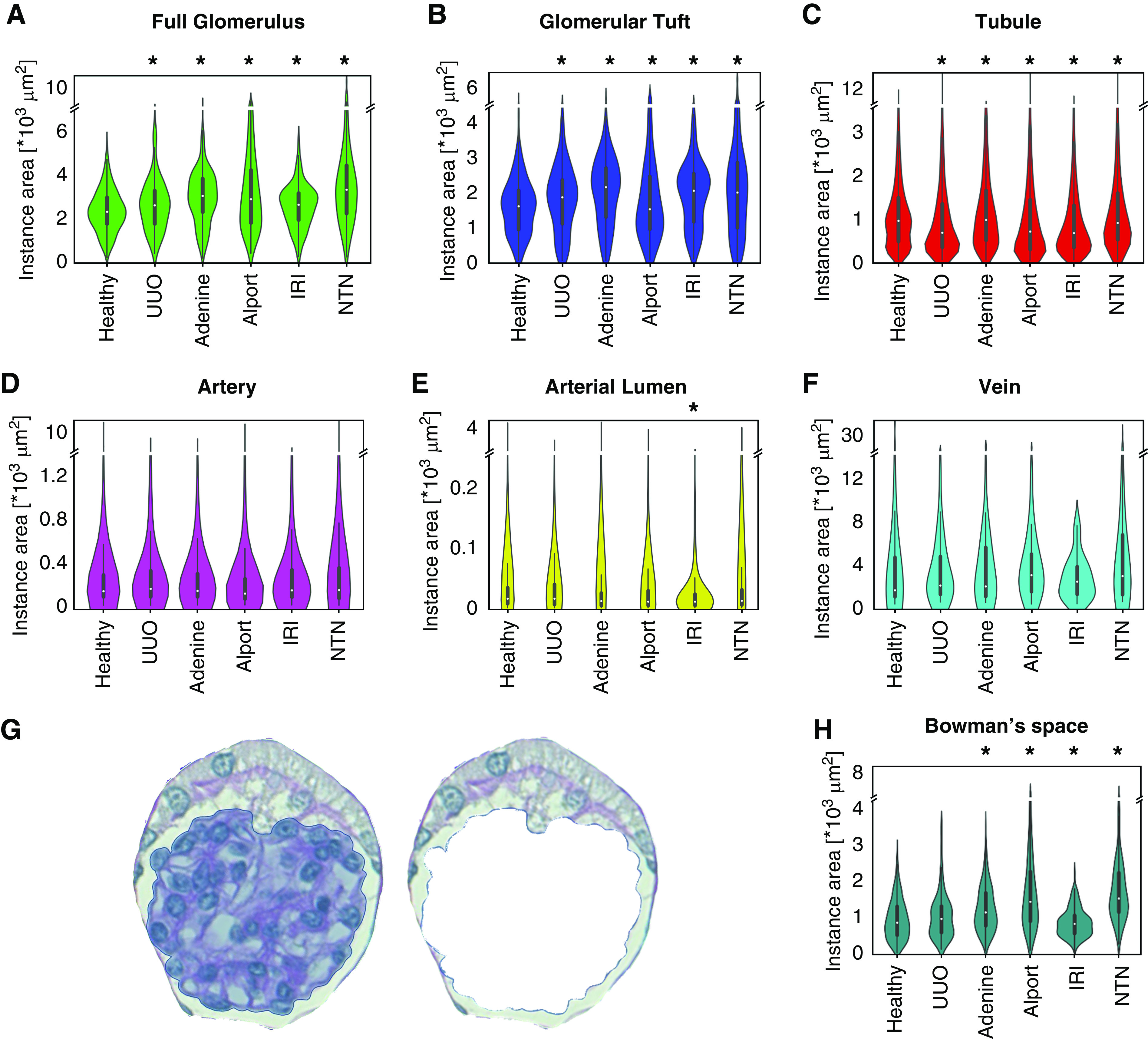
Instance sizes of each class. Violin plots show the distribution pattern of cross-sectional instance sizes for each of the six automatically segmented classes: (A) full glomerulus, (B) glomerular tuft, (C) tubule, (D) artery, (E) arterial lumen, and (F) vein in healthy, UUO, IRI, adenine, Alport, and NTN kidneys. In addition, we subtracted the glomerular tuft area from each glomerulus (G) to analyze size distribution of Bowman’s space (H). *P<0.05 versus healthy.
The maximum cross-sectional area of arteries was not changed, whereas the arterial lumen was slightly reduced in disease models compared with healthy kidneys and significantly decreased in the IRI model (Figure 4, D and E).
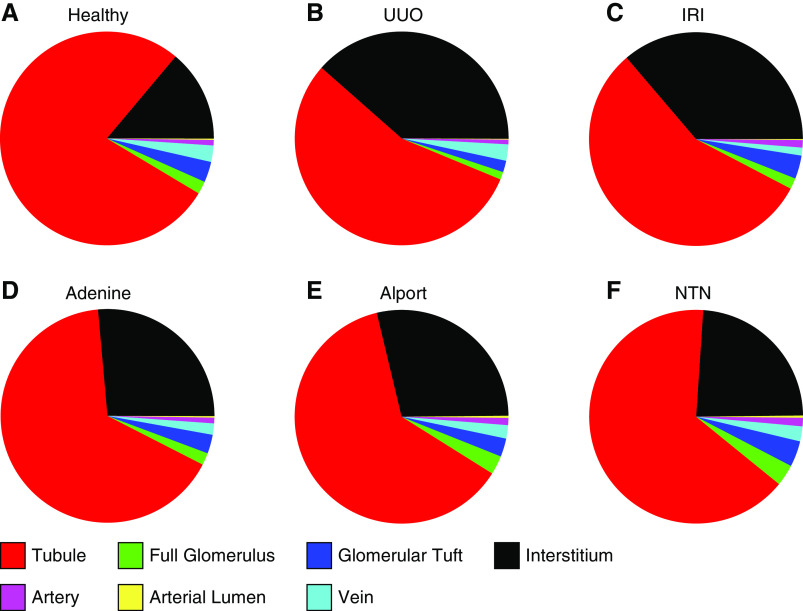
The segmentation also allowed us to analyze changes in the relative proportions of tissue area coverage of all classes in all models (Figure 5, A–F). Compared with the interstitial area in healthy kidneys (mean 14%), it increased in all disease models by two- to three-fold (UUO: 38.6%; adenine: 26.3%; Alport: 28%; IRI: 36.5%; NTN: 23.9%). Conversely, the tubular area decreased in all models by 15%–30% (from 78% in healthy to 55.3%–66.3% in disease). We found no differences in the areas occupied by arteries or their lumina.
Figure 5.
Relative area distributions of automatically segmented classes. The relative area distributions in percentages in (A) healthy, (B) UUO, (C) IRI, (D) adenine, (E) Alport, and (F) NTN kidneys additionally give information on the proportion of remaining nonclassified tubulointerstitial area (shown in black).
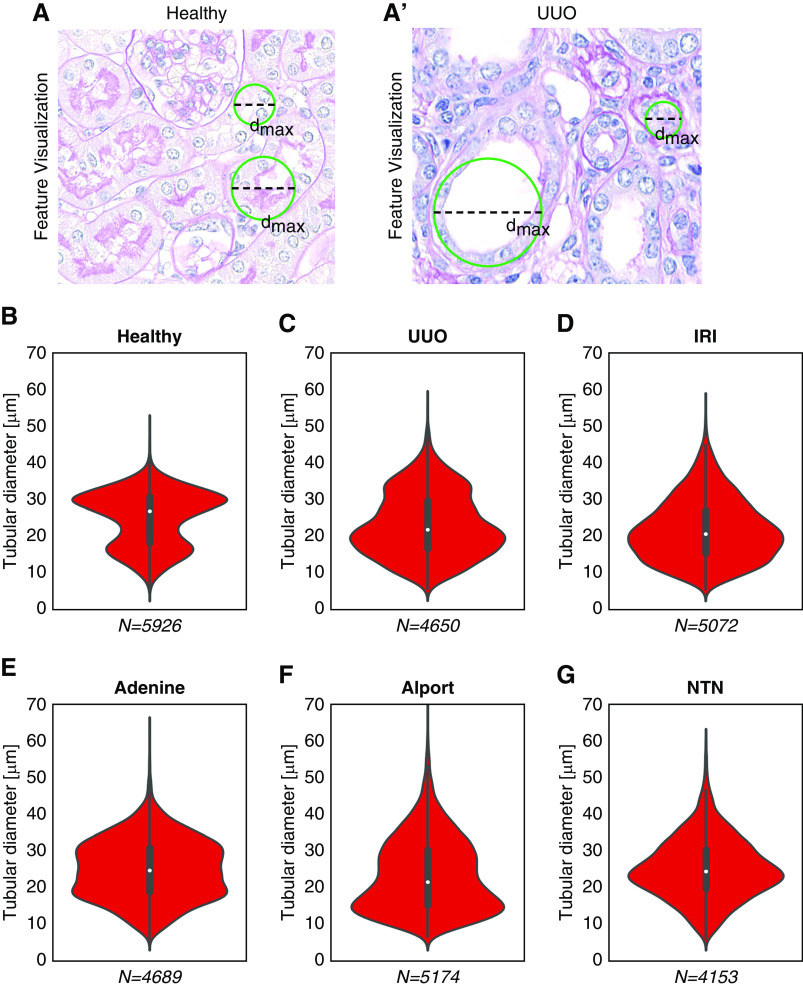
To analyze tubular changes in more detail, we measured the maximum tubular diameter in cortical tubular cross-sections. This was defined as the diameter of the largest circle completely fitting into a segmentation of a single tubular cross-section (Figure 6, A and A’). In line with tubular size (Figure 4C), diameter distribution in healthy kidneys showed two major groups with approximately 15- and 30-µm diameter, likely representing proximal and distal tubules versus collecting ducts (Figure 6A). In all disease models, the maximum diameter of tubules was higher than in healthy kidneys (means of healthy: 49 µm; UUO: 56 µm; adenine: 63 µm; Alport: 83 µm; IRI: 56 µm; NTN: 67 µm) (Figure 6, B–G). However, in UUO, IRI, and Alport, the number of small tubules also increased, representing tubular atrophy and being in line with the results of significantly decreased tubular instance sizes (Figure 4C). In the adenine model, the number of medium-sized tubules increased due to intratubular adherent or obstructing crystals. The NTN model contained the most tubules with a maximum diameter of 20 µm.
Figure 6.
Quantitative analysis of tubular dilation. An exemplary illustration of automated analysis of tubular dilation in PAS stainings of (A) healthy and (A’) UUO mouse kidneys (top). The maximum tubular diameter is defined as the diameter of the maximum-sized circle that fits into a tubule segmentation. Violin plots show the distribution of the analyzed tubular diameter within each model, i.e., for (B) healthy, (C) UUO, (D) IRI, (E) adenine, (F) Alport mice, and (G) NTN. dmax, maximum diameter; N, number of analyzed tubule instances.
Segmentation-Based Feature Correlates with Standard Morphometric Analyses
Our interstitium class includes several histologic compartments, namely the true interstitium, capillaries, and adventitia of arteries. To understand whether this class can still provide useful quantitative information, we compared the interstitial area of the cortex with computer-assisted morphometric analyses of the same kidneys of three selected models. We used immunohistochemical stainings for α-SMA, a widely used marker for the expansion of interstitial myofibroblasts, which is highly upregulated in the UUO, IRI, and adenine models.16,18 Representative segmentation showed that compared with healthy kidneys (Figure 2), the nonclassified interstitial areas increased in all renal disease models (Figure 7, A–C). Interstitial area estimated by our CNN strongly correlated with the expression of the myofibroblast marker α-SMA in all models (Figure 7, A’–C’).
Figure 7.
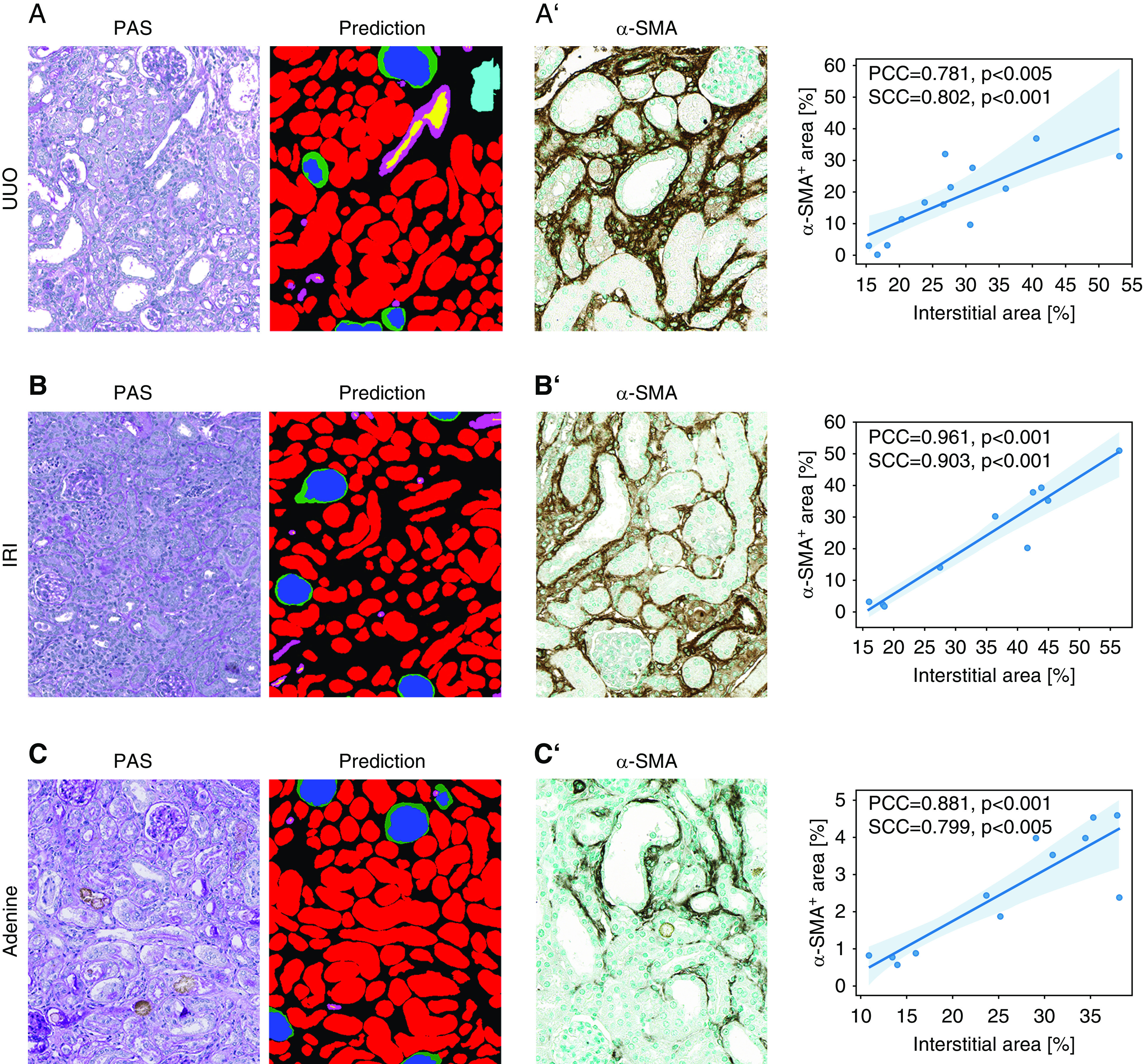
Correlation between segmentation and standard computer-assisted morphometric analyses. (A) Representative picture of the automated segmentation prediction in a murine UUO kidney section. The nonclassified remaining tissue (black) correlates with α-SMA+ area (A’) quantified in immunostainings of the same kidneys. (B) Representative picture of the automated segmentation prediction on a murine IRI kidney section. The nonclassified remaining tissue (black) correlates with α-SMA+ area (B’) quantified in immunostainings from the same kidneys. (C) Representative picture of the automated segmentation prediction on a murine adenine kidney section. The nonclassified remaining tissue (black) correlates with α-SMA+ area (C’) quantified in immunostainings from the same kidneys. PCC, Pearson correlation coefficient; SCC, Spearman correlation coefficient.
Translation of Multiclass Segmentation to Kidneys from Different Species and Humans
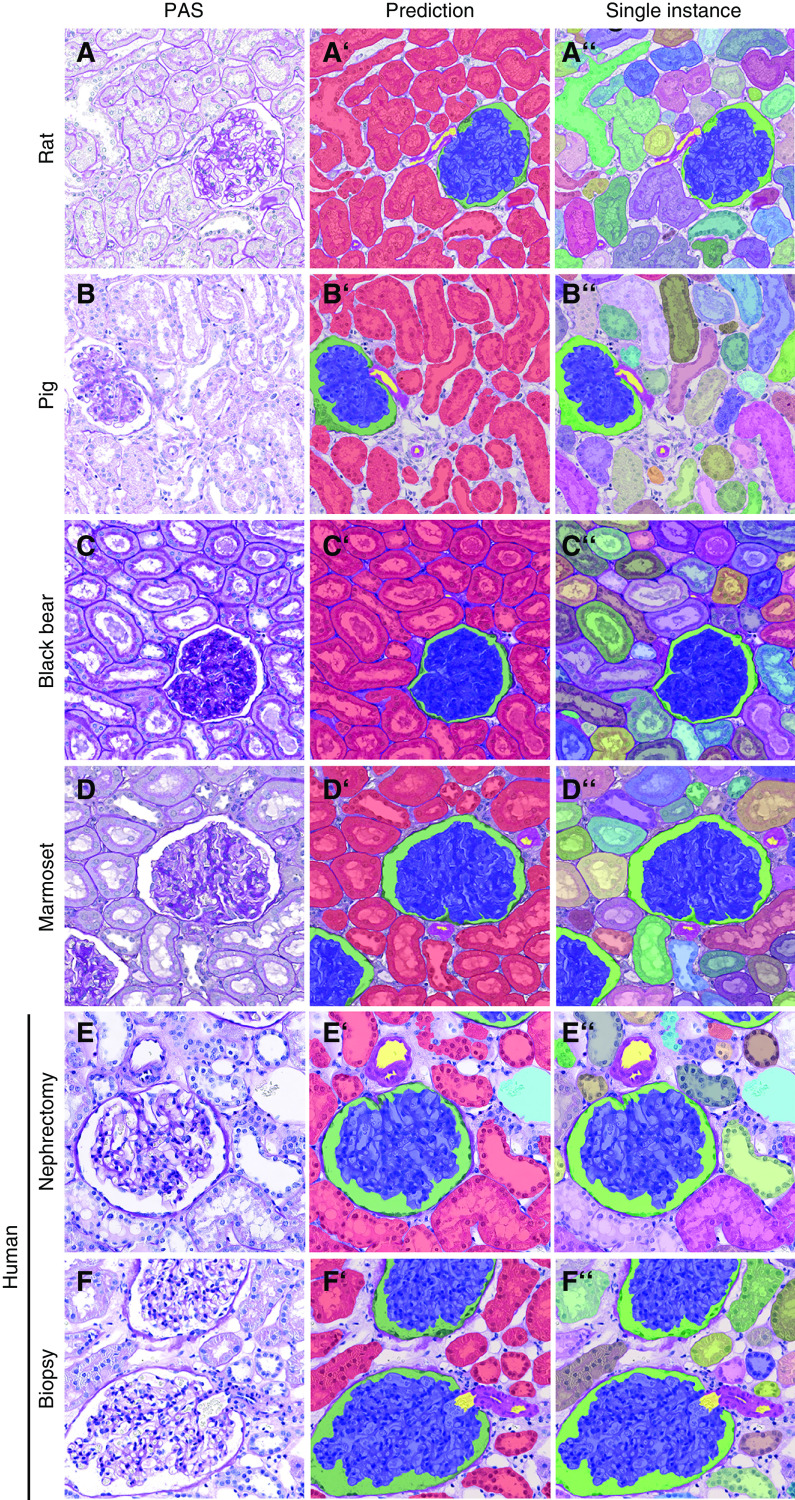
To show the broader applicability of our CNN, we applied it to kidneys of other species, including rats, pigs, black bears, and marmosets. With only a few additional training sets per species, i.e., 50 annotated patches each, the CNN was able to detect and segment all classes in the cortex (Figure 8, A–D’’) and medulla (Supplemental Figure 10, A–D’’) in all species, overall providing very high detection and segmentation accuracies of all classes (Table 2).
Figure 8.
Automated segmentation of kidneys from various species. Representative pictures illustrate the segmentation quality of the CNN in kidney tissue from (A–A’’) rat, (B–B’’) pig, (C–C’’) black bear, and (D–D’’) marmoset. Predictions (A’–D’) depict different classes, whereas (A’’–D’’) display predictions on instance level for tubules. All classes are also correctly detected and segmented on human nephrectomy (E–E’’) and smaller human biopsy (F–F’’) specimens.
Finally, we tested the CNN on normal human renal biopsy specimens and nephrectomy samples. Our full CNN segmented all classes in both cortex and medulla and was applicable to large tissue specimens from nephrectomies and renal biopsy specimens (Figure 8, E–F’’, Supplemental Figure 10, E–F’’). Quantitative validation confirmed high detection segmentation accuracies of all classes. However, as compared with other species, performance was lower for glomerular tuft, arteries, and their lumina (Table 2). As a proof of concept, we additionally provided visual segmentation results in human biopsy specimens showing acute tubular damage, a feature that is also common in many animal models, yielding promising segmentation results (Supplemental Figure 11).
Discussion
We developed a CNN for automated multiclass segmentation of renal histology of different mammalian species and different experimental disease models with broad pathologic alterations. In comparison, the currently available multiclass segmentation model was developed on patient samples only and focused on transplant specimens.10 Compared with the previous work,10 we also technically extended the segmentation pipeline by employing suitable task-specific modifications to network architecture, novel approaches for data quality and quantity improvement, modern network training and regularization routines, and network performance quantification on the basis of novel and precise evaluation metrics. As a proof of concept, we used the segmentation results to provide quantitative metrics for efficient, comparative, high-throughput histopathologic analyses.
To standardize the annotation procedure, we first developed precise class definitions and performed several training sessions with all expert annotators. This step was also used in difficult radiologic segmentation tasks, in which experts underwent a period of training of up to several months, until they had reached a defined reproducibility ensuring sufficient quality of manual annotations.32 These definitions can also guide future training for further model improvement. The annotation process is highly time-consuming, which is a major limiting factor. In order to facilitate the process, we loaded predictions into QuPath, which served as preannotations and reduced manual annotation effort by up to 90%. This made it possible to perform an exceedingly large number of expert-based annotations (72,722 in total), representing the largest study to date for histopathologic structure segmentation. We also applied active learning for patch selection, i.e., we visually selected patches with the largest prediction errors and corrected them, which further strongly improved the CNN performance while reducing the number of required annotations as described by others.33 Furthermore, QuPath currently represents the most widely used open-source and freely available software for digital pathology, enabling broad, vendor-independent applicability.
We have chosen six different murine models broadly used in nephrology research. The models provide a wide variety of distinct causes and histopathologic alterations, i.e., obstructive nephropathy, IRI, crystal-induced nephropathy, immune-mediated GN, genetic glomerulopathy, and metabolic (diabetic) nephropathy. Despite the broad differences in histopathology, our CNN was able to segment all structures in all models with high accuracy. Our results suggest that a single comprehensive CNN might perform better compared with specific CNNs trained for each model, and that performance can be further improved by integrating data from different species, including humans. This follows from the partial class similarities across all models and species, effectively yielding more useful training data and thus contributing to learning more generalizable class features.
Only one-third of the training data were sufficient to reach approximately 90% accuracy in all classes, except for arteries and their lumina. For both latter classes, performance improved continuously as training datasets increased, indicating options for further improvements. Because of the amount of training data, strong color augmentations, and active learning, our CNN yielded accurate segmentation of an external UUO dataset and db/db mice, a model with distinct pathology that the network had never seen before. Our data also showed that it is possible to achieve promising segmentation accuracy in different species or models with relatively low additional annotation effort by experts. This might allow rapid adaptation of the algorithm to samples from various laboratories and translation to additional models and pathologies. This is an important prerequisite for high-throughput and reproducible analyses and will be essential to reduce the workload while at the same time increasing the quantitative precision in experimental and potentially also clinical histopathology. As a proof of concept, we applied our model to human biopsy specimens with acute tubular damage with promising segmentation accuracy. However, further studies will be needed to develop a model that is capable of efficiently segmenting the broad spectrum of human renal pathology.
We describe the applicability of implementing basic feature extraction on top of the segmentation results, providing compartment-specific quantifications. Using a handcrafted feature, tubular diameters on an entire slide could be analyzed within minutes, a task that would be impossible to perform manually. Such basic analyses can provide valuable quantitative information about healthy renal morphology and novel insights into experimental disease models and human kidney diseases, while saving an enormous amount of time. We found that the mean instance size of glomeruli was increased in all of our disease models. This was expected for models with primary glomerular damage and crescent formation, i.e., Alport and NTN, which both also exhibited larger Bowman’s space, but was surprising for models with primary tubulointerstitial damage. Possible explanations are compensatory glomerular hypertrophy with loss of nephrons and enlargement of Bowman’s space due to obstruction of the associated tubule, e.g., in the adenine model and the IRI model. An exception was the Alport model, which exhibited significantly smaller glomerular tuft sizes due to pronounced glomerulosclerosis. For tubules, we found a significant decrease in tubular size in all disease models but at the same time an increase of the maximum tubular instances in UUO, Alport, and NTN. These data provide quantitative evidence for tubular injury and atrophy in all models and model-specific cystic tubular dilation, which was confirmed by the direct analysis of tubular dilation. Overall, these large-scale, precise quantitative data provide novel read-outs for interventional studies and potentially also lead to reduced numbers of animals required for research.
Our study has several limitations. First, in our current CNN, the nonsegmented area comprises a collection of various histologic structures, including peritubular capillaries, interstitium, arterial adventitia, tubular basement membranes, and all other nonrecognized structures. Although we found a high correlation with the expression of the fibrosis marker α-SMA, our “interstitial area” does not specifically reflect fibroblasts or fibrosis. Further annotations and training of the specific subclasses, e.g., capillaries, immune cells, adventitia, and tubular basement membranes, will enable us to refine the segmentation. Second, we have not differentiated between the various tubular segments. Although automated differentiation between tubular segments would allow a more comprehensive study of tubular injury, we recognized that manual annotations of tubular segments on PAS stainings were not possible in some disease models with reasonable certainty. An automated differentiation between cortex and medulla could be the first step toward this direction. Third, our study is descriptive and does not allow to draw mechanistic implications. Fourth, human renal diseases show a multitude of different histopathologic alterations, some of which, e.g., membranous or membranoproliferative glomerular changes, are not well reflected in our animal models. Further studies, expert annotations, consensus, and technical improvements will be required for a holistic segmentation model that comprehensively covers all (human) renal diseases. Finally, although our network showed promising results on external, held-out data from a different laboratory, multicenter studies will be required to assess the full generalization capability of the network.
In conclusion, our DL algorithm for segmentation of kidney histology for multiple murine disease models and species provides a first step toward fully automated high-throughput quantitative computational experimental nephropathology.
Disclosures
P. Bankhead reports other from Philips Digital Pathology Solutions, outside the submitted work; and is the primary inventor and maintainer of the QuPath open source software platform. J. Floege reports other from Amgen, Bayer, Calliditas, Fresenius, Omeros, Retrophin, and Vifor, outside the submitted work. R. Kramann reports grants from Chugai, outside the submitted work. M. Lehrke reports consultancy agreements with Amgen, Bayer, Boehringer Ingelheim, Lilly, MSD, Novartis, and Novo Nordisk; research funding from Boehringer Ingelheim, MSD, and Novo Nordisk; honoraria from Amgen, Bayer, Boehringer Ingelheim, Lilly, MSD, Novartis, and Novo Nordisk; and being a scientific advisor to or membership with Amgen, Bayer, Boehringer Ingelheim, Lilly, MSD, Novartis, and Novo Nordisk. All remaining authors have nothing to disclose.
Funding
This study was funded by German Research Foundation (Deutsche Forschungsgemeinschaft [DFG]; grants SFB/TRR57, SFB/TRR219, BO3755/3-1, and BO3755/6-1), the German Federal Ministry of Education and Research (Bundesministerium für Bildung und Forschung [BMBF]: STOP-FSGS-01GM1901A), the German Federal Ministry of Economic Affairs and Energy (Bundesministerium für Wirtschaft und Energie [BMWi]: EMPAIA project), and the RWTH Aachen Exploratory Research Space (ERS Seed Fund: OPSF585).
Supplementary Material
Acknowledgments
The support for manual annotations from Felicitas Weiß, Timo Horstmann, and the whole LaBooratory is gratefully acknowledged.
Mr. Nassim Bouteldja, Dr. Barbara M. Klinkhammer, Dr. Roman D. Bülow, Prof. Dorit Merhof, and Prof. Peter Boor planned and oversaw the study. Mr. Nassim Bouteldja, Dr. Barbara M. Klinkhammer, and Dr. Roman D. Bülow planned and conducted experiments. Mr. Nassim Bouteldja, Dr. Barbara M. Klinkhammer, Dr. Roman D. Bülow, Mr. Patrick Droste, Mr. Simon W. Otten, and Dr. Saskia Freifrau von Stillfried performed annotations. Dr. Barbara M. Klinkhammer and Dr. Roman D. Bülow corrected annotations. Mr. Nassim Bouteldja performed statistical analyses. Ms. Susan M. Sheehan, Dr. Ron Korstanje, Dr. Julia Moellmann, Prof. Michael Lehrke, Ms. Sylvia Menzel, Dr. Matthias Mietsch, Dr. Charis Drummer, Prof. Rafael Kramann, and Prof. Peter Boor provided samples. Mr. Nassim Bouteldja, Dr. Barbara M. Klinkhammer, and Dr. Roman D. Bülow wrote the first draft of the manuscript and arranged the figures. Prof. Jürgen Floege, Prof. Peter Boor, and Prof. Dorit Merhof critically reviewed the manuscript and figures. All authors read and approved the final version of the article.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020050597/-/DCSupplemental.
Supplemental Table 1. Glossary of technical terms.
Supplemental Table 2. Criteria for definition of classes.
Supplemental Table 3. Quantitative information on ground truth data.
Supplemental Table 4. Architecture of our full CNN.
Supplemental Table 5. Performance comparison of our model, its unmodified variant vanilla U-Net, and state-of-the-art context-encoder.
Supplemental Figure 1. Annotation procedure.
Supplemental Figure 2. Challenging morphology for manual and automated annotations.
Supplemental Figure 3. Segmentation on whole-slide images of UUO, Alport, and NTN kidneys.
Supplemental Figure 4. Quantitative segmentation performance in murine NTN and adenine kidneys.
Supplemental Figure 5. Automated segmentation in the medulla of murine kidney sections.
Supplemental Figure 6. Examples of missclassifications.
Supplemental Figure 7. Relation between amount of training data and detection performance.
Supplemental Figure 8. Comparison between our full CNN and its variants independently trained on single models.
Supplemental Figure 9. Segmentation of nontrained and external murine kidney slides.
Supplemental Figure 10. Automated segmentation of renal medulla in different species.
Supplemental Figure 11. Automated segmentation of human biopsy specimens presenting with acute tubular damage.
References
- 1.Robboy SJ, Weintraub S, Horvath AE, Jensen BW, Alexander CB, Fody EP, et al.: Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch Pathol Lab Med 137: 1723–1732, 2013 [DOI] [PubMed] [Google Scholar]
- 2.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521: 436–444, 2015 [DOI] [PubMed] [Google Scholar]
- 3.Boor P: Artificial intelligence in nephropathology. Nat Rev Nephrol 16: 4–6, 2020 [DOI] [PubMed] [Google Scholar]
- 4.Sirinukunwattana K, Ahmed Raza SE, Yee-Wah Tsang, Snead DRJ, Cree IA, Rajpoot NM: Locality sensitive deep learning for detection and classification of nuclei in routine colon cancer histology images. IEEE Trans Med Imaging 35: 1196–1206, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Jia Z, Wang LB, Ai Y, Zhang F, Lai M, et al.: Large scale tissue histopathology image classification, segmentation, and visualization via deep convolutional activation features. BMC Bioinformatics 18: 281, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kather JN, Pearson AT, Halama N, Jäger D, Krause J, Loosen SH, et al. : Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 25: 1054–1056, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadermayr M, Eschweiler D, Jeevanesan A, Klinkhammer BM, Boor P, Merhof D: Segmenting renal whole slide images virtually without training data. Comput Biol Med 90: 88–97, 2017 [DOI] [PubMed] [Google Scholar]
- 8.Gadermayr M, Gupta L, Appel V, Boor P, Klinkhammer BM, Merhof D: Generative adversarial networks for facilitating stain-independent supervised and unsupervised segmentation: A study on kidney histology. IEEE Trans Med Imaging 38: 2293–2302, 2019 [DOI] [PubMed] [Google Scholar]
- 9.Gupta L, Klinkhammer BM, Boor P, Merhof D, Gadermayr M: Stain independent segmentation of whole slide images: A case study in renal histology. Proceedings from the 2018 IEEE 15th International Symposium on Biomedical Imaging, Washington, DC, April 4–7, 2018, pp 1360–1364
- 10.Sheehan SM, Korstanje R: Automatic glomerular identification and quantification of histological phenotypes using image analysis and machine learning. Am J Physiol Renal Physiol 315: F1644–F1651, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermsen M, de Bel T, den Boer M, Steenbergen EJ, Kers J, Florquin S, et al.: Deep learning-based histopathologic assessment of kidney tissue. J Am Soc Nephrol 30: 1968–1979, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginley B, Lutnick B, Jen KY, Fogo AB, Jain S, Rosenberg A, et al.: Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 30: 1953–1967, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bueno G, Fernandez-Carrobles MM, Gonzalez-Lopez L, Deniz O: Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput Methods Programs Biomed 184: 105273, 2020 [DOI] [PubMed] [Google Scholar]
- 14.Bueno G, Gonzalez-Lopez L, Garcia-Rojo M, Laurinavicius A, Deniz O: Data for glomeruli characterization in histopathological images. Data Brief 29: 105314, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kannan S, Morgan LA, Liang B, Cheung MG, Lin CQ, Mun D, et al.: Segmentation of glomeruli within Trichrome images using deep learning. Kidney Int Rep 4: 955–962, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehling J, Bábíčková J, Gremse F, Klinkhammer BM, Baetke S, Knuechel R, et al.: Quantitative micro-computed tomography imaging of vascular dysfunction in progressive kidney diseases. J Am Soc Nephrol 27: 520–532, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Djudjaj S, Papasotiriou M, Bülow RD, Wagnerova A, Lindenmeyer MT, Cohen CD, et al.: Keratins are novel markers of renal epithelial cell injury. Kidney Int 89: 792–808, 2016 [DOI] [PubMed] [Google Scholar]
- 18.Baues M, Klinkhammer BM, Ehling J, Gremse F, van Zandvoort MAMJ, Reutelingsperger CPM, et al.: A collagen-binding protein enables molecular imaging of kidney fibrosis in vivo. Kidney Int 97: 609–614, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djudjaj S, Lue H, Rong S, Papasotiriou M, Klinkhammer BM, Zok S, et al.: Macrophage migration inhibitory factor mediates proliferative GN via CD74. J Am Soc Nephrol 27: 1650–1664, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moellmann J, Klinkhammer BM, Onstein J, Stöhr R, Jankowski V, Jankowski J, et al.: Glucagon-like peptide 1 and its cleavage products are renoprotective in murine diabetic nephropathy. Diabetes 67: 2410–2419, 2018 [DOI] [PubMed] [Google Scholar]
- 21.Mancina E, Kalenski J, Paschenda P, Beckers C, Bleilevens C, Boor P, et al.: Determination of the preferred conditions for the isolated perfusion of porcine kidneys. Eur Surg Res 54: 44–54, 2015 [DOI] [PubMed] [Google Scholar]
- 22.Klinkhammer BM, Djudjaj S, Kunter U, Palsson R, Edvardsson VO, Wiech T, et al. : Cellular and molecular mechanisms of kidney injury in 2,8-dihydroxyadenine nephropathy. J Am Soc Nephrol 31: 799–816, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al.: QuPath: Open source software for digital pathology image analysis. Sci Rep 7: 16878, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Settles B: Active Learning Literature Survey. Computer Sciences Technical Report 1648 University of Wisconsin-Madison, 2009 [Google Scholar]
- 25.Falk T, Mai D, Bensch R, Çiçek Ö, Abdulkadir A, Marrakchi Y, et al.: U-Net: Deep learning for cell counting, detection, and morphometry. Nat Methods 16: 67–70, 2019 [DOI] [PubMed] [Google Scholar]
- 26.Isensee F, Petersen J, Kohl SAA, Jäger PF, Maier-Hein KH: nnU-Net: Breaking the spell on successful medical image segmentation. arXiv 2019. arXiv:1904.08128
- 27.Ronneberger O, Fischer P, Brox T: U-Net: Convolutional Networks for Biomedical Image Segmentation, Cham, Springer International Publishing, 2015, pp 234–241 [Google Scholar]
- 28.Chen H, Qi X, Yu L, Dou Q, Qin J, Heng P-A: DCAN: Deep contour-aware networks for object instance segmentation from histology images. Med Image Anal 36: 135–146, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Liu L, Jiang H, He P, Chen W, Liu X, Gao J, et al. : On the variance of the adaptive learning rate and beyond. Proceedings from the 2020 International Conference on Learning Representations (ICLR), 2020
- 30.Milletari F, Navab N, Ahmadi S-A: V-Net: Fully convolutional neural networks for volumetric medical image segmentation. Proceedings from the 2016 Forth International Conference on 3D Vision (3DV), Stanford, CA, October 25–28, 2016, 565–571
- 31.Gu Z, Cheng J, Fu H, Zhou K, Hao H, Zhao Y, et al. : CE-Net: Context encoder network for 2D medical image segmentation. IEEE Trans Med Imaging 38: 2281–2292, 2019 [DOI] [PubMed] [Google Scholar]
- 32.Kennedy DN, Filipek PA, Caviness VR: Anatomic segmentation and volumetric calculations in nuclear magnetic resonance imaging. IEEE Trans Med Imaging 8: 1–7, 1989 [DOI] [PubMed] [Google Scholar]
- 33.Lutnick B, Ginley B, Govind D, McGarry SD, LaViolette PS, Yacoub R, et al.: An integrated iterative annotation technique for easing neural network training in medical image analysis. Nat Mach Intell 1: 112–119, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.