SUMMARY
A fundamental challenge in immunology is to decipher the principles governing immune responses at the whole-organism scale. Here, using a comparative infection model, we observe immune signal propagation within and between organs to obtain a dynamic map of immune processes at the organism level. We uncover two inter-organ mechanisms of protective immunity mediated by soluble and cellular factors. First, analyzing ligand-receptor connectivity across tissues reveals that type I IFNs trigger a whole-body antiviral state protecting the host within hours after skin vaccination. Second, combining parabiosis, single-cell analyses and gene knockouts, we uncover a multi-organ web of tissue-resident memory T cells that functionally adapt to their environment to stop viral spread across the organism. These results have implications for manipulating tissue-resident memory T cells through vaccination, and open up new lines of inquiry for the analysis of immune responses at the organism level.
INTRODUCTION
No metazoan cell or organ is an island. Cells and organs must coordinate their actions both locally and systemically, at the whole-organism scale (Droujinine and Perrimon, 2016). For example, the mammalian immune system opposes infection through secreted and cellular factors that transcend organ boundaries. Secreted factors, such as cytokines and growth factors, can act on local and distant tissues to trigger inflammation (e.g., TNF, IL-1) (Medzhitov, 2008), antiviral responses (e.g., type I IFNs) (López and Hermesh, 2011), or to modify the cellular output of hematopoiesis (e.g., GM-CSF, IL-6) (Manz and Boettcher, 2014). In addition, cellular factors such as memory T cells can act as systemic or local safeguards against infection by patrolling across organs or residing in a previously infected tissue (Schenkel and Masopust, 2014). Yet, despite this knowledge, many basic questions about how mammalian immune responses operate at the organism level remain unanswered, such as, when and where are immune pathways activated throughout the body upon local or systemic infection, and how do these pathways contribute to host protection?
To answer these questions, we need to tackle the challenges posed by the ubiquitous nature of immune factors by developing new ways to observe and quantify immune responses at the level of the entire organism. Current approaches to study immunity in vivo mostly focus on measuring changes at the level of proteins, RNAs or chromatin in one or more immune cell types in one organ (Brandes et al., 2013; Mostafavi et al., 2016), or across multiple organs (Guilliams et al., 2016; Heng et al., 2008; Okabe and Medzhitov, 2014; Spitzer et al., 2015). However, to dissect organismal immunity, it is also critical (1) to capture the dynamic changes of an immune response, as opposed to static snapshots, (2) to include all potential players from immune to non-immune cells, and (3) to maintain the links to the rest of the system that spans across all tissues. We reasoned that gene expression analyses of whole organs can help to address these challenges as this methodology successfully found shared and tissue-specific expression patterns that vary across healthy individuals (Melé et al., 2015), and with disease or aging (Baruch et al., 2014; Dobrin et al., 2009; Huang et al., 2011; Keller et al., 2008). In addition, organ-level expression can detect immunological changes driven by cell composition or direct gene regulation, even in rare cells (Ariotti et al., 2014; Brandes et al., 2013). Based on these evidence, we hypothesized that systematically measuring gene expression over time in whole organs can track immune processes across a mammalian organism.
To test this hypothesis, we focused on immune processes leading to protective immunity at the organism level. We used a comparative mouse infection model based on two poxvirus strains with vaccinating (Modified Vaccinia Ankara, MVA) and pathogenic (Western Reserve, WR) properties. The DNA genome of WR is 85% (165,427/194,710 bp) identical to MVA, which is explained by the loss of 94 genes encoding virulence factors from the MVA genome (Figure 1A) (Meyer et al., 1991). As a result, MVA is non-virulent and non-replicative in most mammalian cells, which makes it a live attenuated vaccine and vector of choice in clinical settings (Moss, 2011).
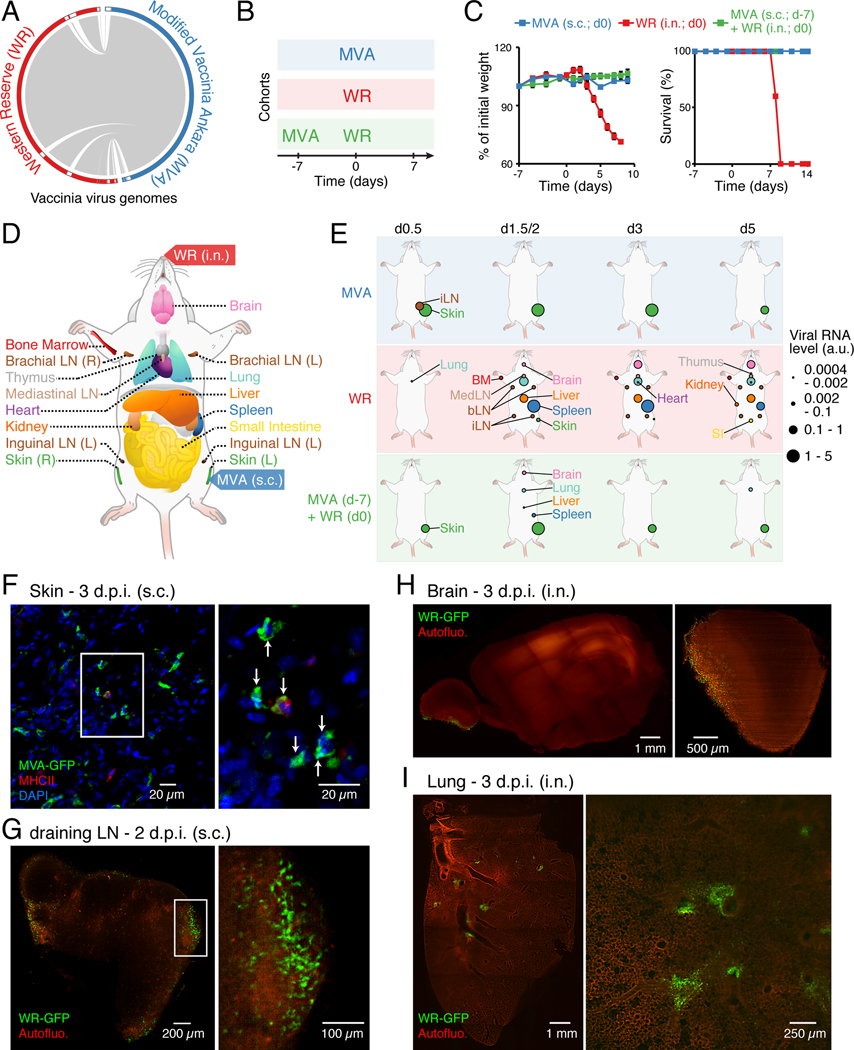
Figure 1. Dynamics of viral spread at the whole-organism scale.
(A) Viral genome alignment. Grey lines depict shared sequences. White boxes in the outer circle show genomic regions absent in the other strain.
(B-C) Cohorts of mice used to track vaccinating (MVA subcutaneously; s.c.), lethal (WR intranasally; i.n.) and protective (MVA followed by WR) responses (B), and matching weight (C, left) and survival (C, right) measurements. Error bars, SEM (n = 5).
(D) Schematic depicting the mouse tissues collected in this study (17 total including blood, not shown).
(E) Organismal viral spread for indicated cohorts and times post-infection (top). Circle sizes, normalized expression for viral gene E3L (encoded by MVA and WR).
(F-I) Whole-mount tissue imaging of skin (F), draining lymph node (inguinal; G), brain (H), and lung (I) at 2 or 3 day post-infection (d.p.i.) using indicated GFP-expressing virus strains and routes of infection. For skin (F) and dLN (G), insets indicate position of images on the right. For skin (F), MHC class II and nuclei (DAPI) were stained, and arrows indicate MVA-GFP+ cells. For brain (H) and lung (I), shown are tiled images (left), and representative sections (right). Autofluo., autofluorescence.
Here, using this comparative viral infection model, we successfully observed immune signal propagation across organs, and obtained a dynamic map of immune processes at the organism level. We discovered and functionally validated two inter-organ mechanisms mediated by secreted and cellular factors – type I IFNs and tissue-resident memory T cells, respectively – that protect the host from systemic viral pathogenesis.
RESULTS
A comparative viral infection model to study immune dynamics at the whole-organism scale
To study the dynamics of immune responses at the organism level, we used poxvirus strains MVA and WR to compare three types of immune responses in vivo: (1) vaccinating (MVA only), (2) lethal (WR only), and (3) protective (MVA followed by WR challenge) (Figures 1B). MVA was administered subcutaneously (s.c.) and WR intranasally (i.n.) to mimic routes of vaccination and natural infection, respectively. WR infection was lethal within 8 days on average, whereas MVA had no effect on animal weight and survival (Figure 1C), consistent with previous work (Wyatt et al., 2004). We collected 17 tissues per animal, including blood, at 11 time points post-infection (0.25, 0.5, 0.75, 1, 1.5, 2, 3, 4, 5, 6, 7 days) for all three cohorts and from uninfected controls – using four biological replicates per condition (Figure 1D). To track viral spread across tissues, we measured the RNA levels of the viral gene E3L – encoded in both MVA and WR genomes (Table S1). MVA-derived E3L RNAs were detected at skin and, to a lesser extent, in the inguinal draining lymph node (dLN) (Figure 1E and S1A-B). Importantly, absolute quantifications of viral DNA and RNA across tissues confirmed that MVA did not spread beyond skin and the dLN (Figure S1C-F), in agreement with previous studies (Gómez et al., 2007). By contrast, WR rapidly spread to most tissues within a time frame (36–72 h) preceding the appearance of body weight changes (>72 h) (Figures 1E and S1A-B). Interestingly, the early dissemination of WR was similar in MVA-vaccinated hosts in terms of target tissues (lung, liver, spleen, brain), but was followed by immediate viral clearance (Figure 1E and S1A-B). The signal present at skin in the MVA-WR cohort was due to remaining MVA mRNAs in infected cells.
Whole-mount tissue imaging confirmed the patterns of viral spread and revealed key aspects of viral pathogenesis that cannot be inferred from viral gene expression alone (Figure 1F-I). For example, MVA infected many skin cells, including stromal and immune cells such as MHC-II+ cells (Figures 1F), as reported by others (Mahe et al., 2009). MVA also reached the subcapsular sinus area of the dLN (Figures S1G), in agreement with previous work (Eickhoff et al., 2015). After WR entry, lungs were infected in multiple foci whereas infected brain cells were limited to the olfactory bulb, suggesting that WR’s access to the brain is likely limited to the nasal cavity (Figures 1H-I). Lastly, the presence of infectious WR virions was confirmed in whole-tissue lysates (Figure S1H).
Whole-tissue gene expression uncovers changes in local and systemic immune responses
Next, we sought to study host gene expression dynamics across the organism in lethal, vaccinating and protective contexts. We performed whole-tissue RNA-seq on 9 out of the 17 tissues collected and 5 time points (0, 0.5, 1.5, 3, 5 days), which were selected based on (1) MVA presence for skin and dLN, (2) WR presence for brain, lung, liver, spleen, bone marrow, blood, and (3) pathological signs despite little to no detectable WR presence for small intestine. All measured profiles clustered by tissue type in principal component analysis (PCA) and showed a separation between blood and solid tissues, with brain and liver being the most distinct (Figure 2A). MVA and WR did not modify the transcriptional boundaries existing between tissues, but distinct responses were readily detectable at the single-tissue level. For example, skin and liver transcriptional signatures changed within hours upon MVA immunization, while WR impacted the transcriptional outputs of these two tissues at later times (Figure 2B-C), in agreement with the timing and geography of WR propagation (Figure S1B).
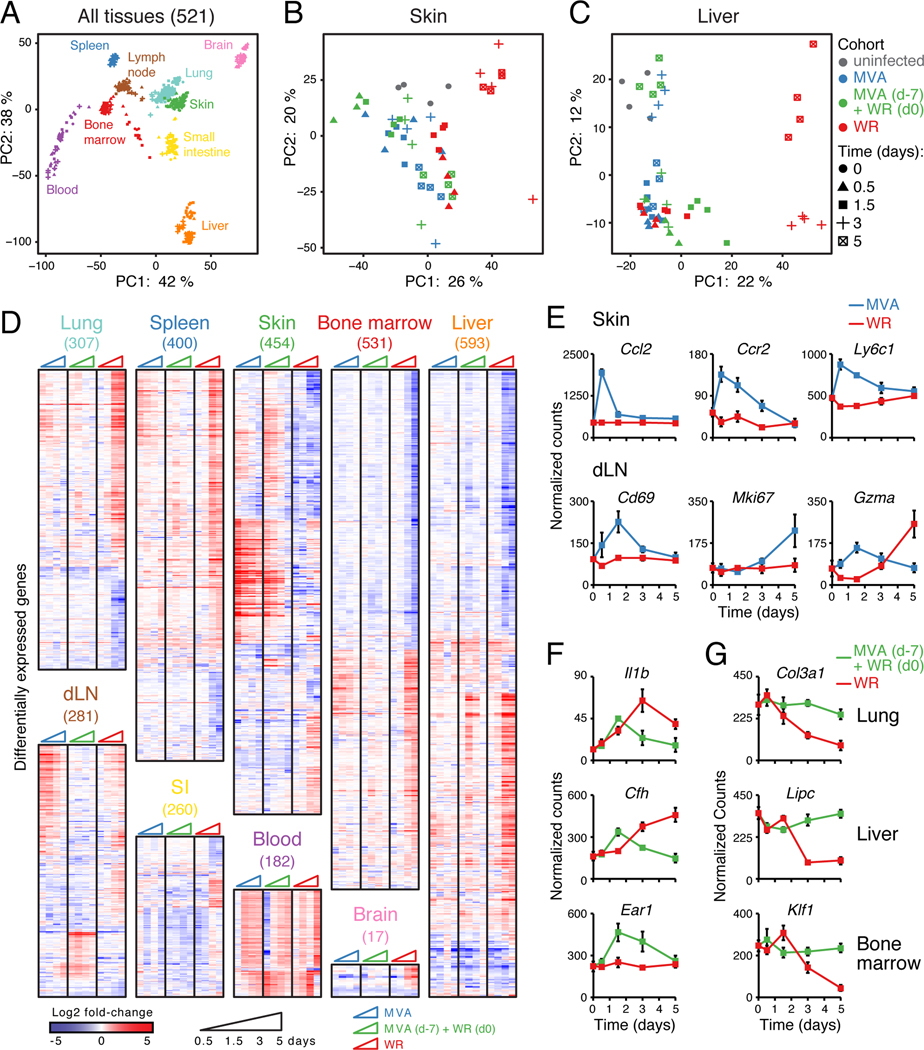
Figure 2. Whole-tissue gene expression reveals local and systemic immune responses.
(A-C) Principal component analysis (PCA) of whole-tissue mRNA profiles for 9 tissues (521 samples; A), skin (B), and liver (C). Colors, tissues (A) and cohorts (B-C); symbols, time after infection; axes, percentage of variance; lymph node, inguinal draining LN.
(D) Heatmaps of differentially expressed genes (numbers in parentheses on top) from whole-tissue mRNA profiles ordered by hierarchical clustering (Pearson’s correlation) and tissue type. Shown are log2 fold-change values relative to matching, uninfected tissues (FDR-adjusted p-value < 0.05, absolute fold change > 2, n = 4).
(E-G) Normalized read counts for indicated genes, cohorts and tissues. Error bars, SEM (n = 4).
In total, we identified 2,018 genes differentially expressed between infected and matching, uninfected samples across all 9 tissues, which were mostly tissue-specific with some exceptions (Figure 2D and S2A; Table S2). Gene ontology (GO) enrichment analysis revealed that all tissue types regulated genes associated with immune processes in at least one of the three cohorts (Figure S2B; Table S2). For example, some MVA-regulated genes pointed to well-known processes such as the early induction of the monocyte chemo-attractant Ccl2 and surface markers Ccr2 and Ly6c1 (Figure 2E), which mirrors the influx of monocytes observed at skin (Figure S2C). In addition, genes known as markers for T cell activation (Cd69), proliferation (Mki67) and effector function (Gzma) were upregulated in dLN upon MVA infection (Figure 2E), reflecting the increase in virus-specific CD8+ T cells in dLN and skin (Figure S2C-D). For the protective response (MVA/WR), we identified pro-inflammatory genes encoding secreted factors in lung (Il1b) and liver (acute phase protein Cfh), and genes involved in eosinopoiesis in bone marrow (Ear1) (Figure 2F). Lastly, WR pathogenesis in naïve mice down-regulated genes important for the extracellular matrix at lung (Col3a1), metabolism at liver (Lipc) and hematopoiesis at bone marrow (Klf1), suggesting that WR replication negatively impacts basic tissue functions likely through cell death (Figure 2G).
To tease apart direct transcriptional changes through gene regulation from indirect effects due to cell recruitment or proliferation, we measured the proportions of 7 immune cell subsets at lung and liver (Figure S2E). While MVA led to little changes at lung and liver, WR triggered an increase in neutrophils and decrease in B cells (Figure S2F). The rapid influx of neutrophils at lung and liver in the WR cohort preceded disease symptoms (moribund state, weight loss), similar to influenza (Brandes et al., 2013). In the protective response (MVA/WR), myeloid cells such as neutrophils showed a rapid (36 h) but transient increase, while CD8+ T cells increased after day 3–5 (Figure S2F). Importantly, for MVA and MVA/WR cohorts, the changes in expression detected within the first 12 h at lung and liver are likely due to active gene regulation, given that no significant changes in cellular composition were detectable at that time (Figure S2F).
Inferring ligand-receptor connectivity across tissues reveals a whole-body antiviral state
Skin MVA vaccination led to changes in gene expression in most tissues despite the limited presence of MVA in skin and dLN (Figure 3A), suggesting that secreted factors are likely acting systemically upon local release. To test this idea, we assembled a list of 2,198 cognate, secreted ligand-receptor pairs, and examined their expression in tissues (Figure 3B). We identified 92 out of 2,198 unique ligand-receptor pairs responsible for 452 putative connections within and between tissues in all cohorts (Table S3). In the MVA cohort, most of the ligands emanated from skin and dLN (Figure 3C-D), whereas in the WR and MVA/WR cohorts, many connections originated from lung, liver and spleen (Figure S3A-B). Skin, liver and lymphoid tissues (dLN, BM, spleen) were the most interconnected tissues with responses including wound repair, inflammation, or chemotaxis (Figure S3C-D; Table S3).
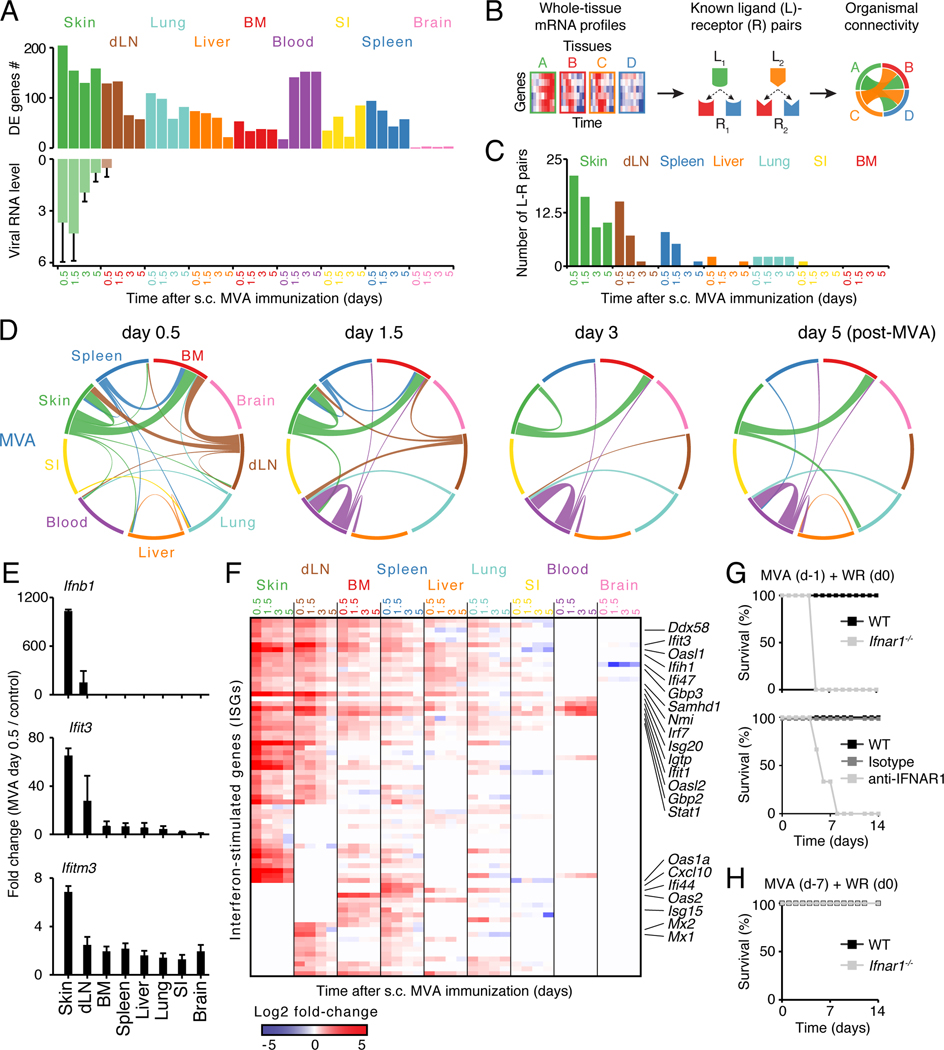
Figure 3. Analysis of ligand-receptor connectivity across tissues reveals a whole-body antiviral state.
(A) Bar graphs showing numbers of differentially expressed (DE) genes (top) and RNA levels of Vaccinia virus gene E3L (qPCR; bottom). Error bars, SD.
(B) Schematic overview of the analysis for ligand-receptor pair connectivity across tissues. Known ligand-receptor pairs were extracted from whole-tissue mRNA profiles and their potential links within and between tissues visualized using a circos plot.
(C) Bar graph showing the numbers of ligand (L)-receptor (R) pairs emanating from indicated tissues upon MVA immunization (s.c., subcutaneous).
(D) Inter-organ connectivity of ligand-receptor pairs at indicated times after MVA immunization. Line color, tissue source for ligands; line thickness, number of ligand-receptor pairs.
(E) Bar graphs showing fold changes between MVA-infected and control tissues for Ifnb1 and two ISGs: Ifit3 and Ifitm3 (qPCR). Error bars, SD (n = 4).
(F) Heatmap of all interferon-stimulated genes (ISGs) regulated across tissues upon skin MVA vaccination. Shown are log2 fold-change values relative to matching, uninfected tissues (FDR-adjusted p-value < 0.05 and absolute fold change > 2, n = 4).
(G-H) Survival analysis of wild-type (WT), anti-IFNAR1 or isotype antibody-treated, and Ifnar1−/− mice immunized subcutaneously with MVA at 1 (G) or 7 (H) days prior to intranasal WR challenge. Data are representative of three independent experiments.
Among these connections, type I interferons (IFNs) were produced only in skin and the dLN, yet induced IFN-stimulated genes (ISGs) across tissues within 12 h upon MVA vaccination (Figure 3E-F; Table S3). We asked if systemic ISG induction could confer host protection by comparing the survival of wild type versus Ifnar1−/− or anti-IFNAR1 antibody-treated mice challenged with WR at 24 hours or 7 days after skin MVA vaccination. Wild type but not knockout or antibody-treated animals were protected against WR challenge within 24 h while all groups were protected at day 7 (Figure 3G-H and S3E-F). Locally produced type I IFNs can thus trigger a whole-body antiviral state protective for the host, which helps explain the systemic effects of MVA in newborn mice (Mayr et al., 1986; Vilsmeier, 1999).
Protective memory responses to a respiratory viral challenge induce expression changes in lung, liver and spleen
Next, we investigated the organismal responses at play during the memory phase of a protective response. Mice were challenged with WR three weeks after MVA vaccination – a time frame consistent with a memory recall response as effector T cells had vanished by then (Figure 4A-B and S4A-C) (Pillai et al., 2011). We observed changes in mRNA expression in lung, liver and to a minor extent in spleen at 12 and 36 hours after WR challenge, whereas mediastinal LN, brain, small intestine, skin and bone marrow showed little to no changes (Figure 4C and S4D-E). Collectively, lung, liver and spleen displayed 178 differentially expressed genes, including ISGs across all three organs, and genes linked to leukocyte migration at lung or inflammatory and complement responses at liver (Figure 4D and S4F-G; Table S4). Interestingly, whole-lung and -liver responses were driven by T cell immunity (Figure 4E; Table S4). In addition, such rapid and organ-wide activation of antiviral genes is reminiscent of the effects of tissue-resident memory CD8+ T (TRM) cells (Ariotti et al., 2014). However, TRM cells are typically seeded locally, at the site of pathogen entry, not systemically (Mueller and Mackay, 2016) – with two exceptions of distant TRM seeding across the skin epithelia (Jiang et al., 2012) and from nasal to uterine mucosa (Stary et al., 2015). Taken together, the timing and nature of the changes observed across tissues supported a hypothetic model whereby resident memory cell activity can help protect the host from systemic WR spread.
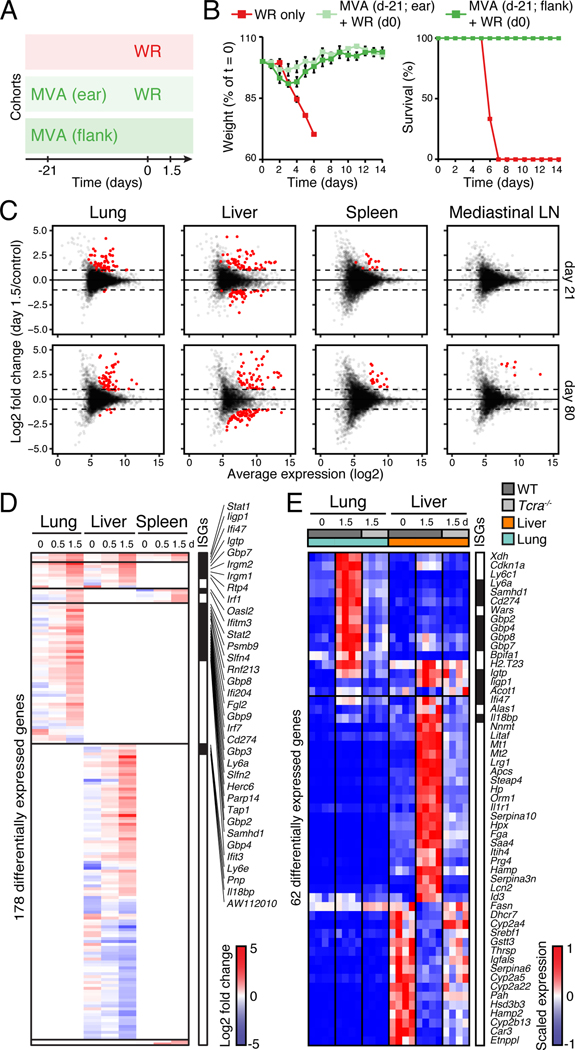
Figure 4. Protective memory responses to a respiratory viral challenge induce expression changes in lung, liver and spleen.
(A-B) Cohorts of mice used to track memory protective responses using MVA immunization at skin (ear or flank) followed by intranasal WR challenge at day 21 (A), and matching weight and survival measurements (B). Error bars, SEM (n = 4).
(C) Dot plots showing log2 fold-change in gene expression (Y axis) in tissues collected at 1.5 day post-WR challenge (on day 21 or 80 after MVA) relative to uninfected controls against log2 average expression (X axis) (n = 4). Red dots, genes with FDR < 0.05 and absolute log2 fold change > 1.
(D-E) Heatmaps showing differentially expressed genes in lung, liver, and spleen at indicated days (d) after intranasal WR challenge of wild-type (D-E) and/or Tcra−/− (E) mice immunized with MVA at flank skin 21 days earlier (FDR-adjusted p-value < 0.05 and absolute fold change > 2, n = 4). Shown are log2 fold-change values relative to matching, uninfected tissues (D), and normalized read counts scaled per row (4 replicates/condition; E). Day 0, mice immunized only. Interferon-stimulated genes (ISGs) are indicated in black on the right.
Skin vaccination seeds tissue-resident memory CD8+ T cells in multiple distant tissues to confer host protection
To test the hypothesis that CD8+ TRM cells could confer systemic host protection, we first asked if T cells were necessary for protection against WR challenge. Using Tcra−/− mice and CD8+ T cell depletion, we found that T cells are critical for host protection in our MVA/WR model (Figure 5A-B and S5A), in agreement with previous work (Salek-Ardakani et al., 2011). Second, we monitored the distribution of virus-specific CD8+ T cells across tissues and found viral B8R20–27 peptide-specific CD8+ T cells in lymphoid tissues, lung and liver at 3 weeks, 3 months and 15 months after MVA vaccination (Figures 5C-E and S5B-D). Importantly, <5% of lung memory CD8+ T cells were parenchyma-associated – similarly to previous work (Anderson et al., 2012) – at 3 weeks and 15 months after MVA vaccination, including 0.25% +/− 0.11 SD and 0.055% +/− 0.026 SD of B8R-specific cells, respectively (Figure 5F and S5E). Third, to directly test if the protective response of memory T cells was driven by tissue-resident cells, we performed parabiosis surgeries on pairs of congenic mice to establish a shared blood circulatory system (Figure S5F) (Wright et al., 2001). Mice were joined 1, 14 or 28 days after or 14 days before skin MVA immunization of one parabiont from each pair, and were separated 2 weeks later for −28, −14 and −1 groups, and two weeks after vaccination for the +14 group (Figure 5G). In groups where parabionts were joined 14 and 28 days after immunization of one partner, the naïve partners were significantly less protected when challenged with WR than the immunized ones, whereas naïve and immunized parabionts were equally protected in the −1 and +14 groups (Figure 5H). Taken together, these experiments demonstrated that CD8+ TRM cells seeded in distant tissues during the effector phase of the response are crucial for systemic host protection.
Figure 5. Skin vaccination seeds tissue-resident memory CD8+ T cells in multiple distant tissues to confer host protection.
(A-B) Relative viral DNA amount measured by qPCR in the brain (day 2 post-WR; A) and weight measurements (B) of wild-type (WT) and T-cell deficient (Tcra−/−) mice immunized subcutaneously with MVA three weeks prior to intranasal WR challenge. Arbitrary units, a.u. (WT values set to 1) (A). Error bars, SD (n = 5) in A, and SEM (n = 7) in B. *; Student’s t-test p-value < 0.05.
(C-E) Flow cytometry analysis of Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) memory CD8α+ T cells (gated on live CD3ε+CD44+CD62L− cells) from indicated tissues from mice vaccinated at skin with MVA 3 weeks (C), 3 months (E, left) or 15 months (E, right) earlier, and quantifications in percentage of parent gate (%) and absolute count per 105 live cells (cells) (D). Error bars, SD (n = 5).
(F) Flow cytometry analysis of vascular and parenchymal B8R-specific memory CD8α+ T cells (gated on live CD3ε+CD44+CD62L− cells) by intravascular staining with CD45 (CD45 i.v.). Bar graphs show quantifications in percentage of parent gate (%) and absolute count per 105 live cells (cells). Error bars, SD (n = 3). *; Student’s t-test p-value < 0.05.
(G-H) Timed parabiosis experiments show the role of tissue-resident cells in host protection. One parabiont (CD45.2) was immunized at skin with MVA at indicated days before (−28, −14, −1) or after (+14) joining with the other parabiont (CD45.1) (G). For groups −28, −14 and −1, mice were joined for 2 weeks before splitting, whereas for group +14, mice were joined for 2 weeks before immunization of one parabiont and split after another 2 weeks. Weight measurements of immunized (dark grey) and naïve (light grey) parabionts after WR challenge are shown in panel H. Data are representative of two to three independent experiments. Error bars, SEM (n = 5). *; Student’s t-test p-value < 0.05.
See also Figure S5.
Tissue-resident memory CD8+ T cells are activated across tissues as virus spreads
To investigate the mechanisms of memory T cell activation at lung and liver upon WR challenge, we analyzed 2,450 memory CD8+ T cells with single-cell (sc) RNA-seq, including 490 B8R-specific cells (Figure 6A). Differentially expressed genes between single memory CD8+ T cells from WR-challenged and control mice revealed clearly distinct phenotypic states in lung and liver (Figure 6B). We then asked if this shift in memory T cell states is due to WR antigen-specific activation and/or cytokine-mediated reactivation. To answer this, we sought to identify antigen-induced genes so as to use their expression levels for scoring the activation state of single T cells upon antigen recognition. We administered the viral B8R peptide to immunized mice intranasally, and performed scRNA-seq on B8R-specific memory CD8+ T cells. Differential expression analysis between B8R-treated and control animals identified a transcriptional signature including genes known to function in memory CD8+ T cell responses such as Ifng, Gzmb, Xcl1, and Ccl4 (Figure 6C) (Lauvau et al., 2016). Importantly, the average expression of B8R peptide-induced genes (referred to as B8R activation score) was significantly higher in tetramer positive versus negative cells upon B8R peptide challenge (p-value = 1.64 × 10−6) (Figure S6A). Thus, our scoring method identified cells directly reactivated by antigen as opposed to cytokines. In WR-challenged mice, we found significant number of memory T cells with high B8R activation scores in both lung and liver (p-value = 5.85 × 10−32 and 2.17 × 10−33, respectively; Wilcoxon rank sum test) (Figure 6D-E and S6B). Furthermore, we observed similar trends of activation based on IFN-γ production (Figure S6C), and in two independent experiments profiling 746 single lung memory CD8+ T cells with our custom scRNA-seq method (p-values = 6.79 × 10−21) (Figure S6D-E), and 7,292 cells using a droplet-based method (p-value = 6.74 × 10−46) (Figure S6F-G).
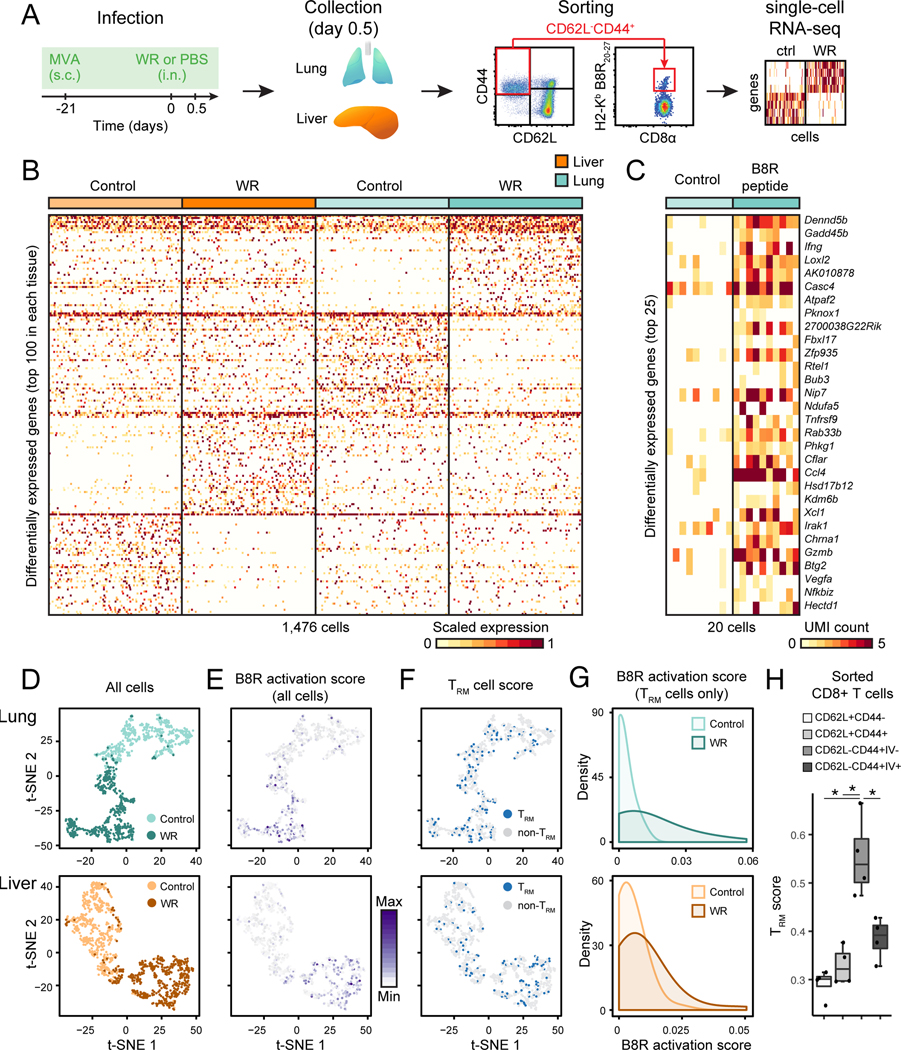
Figure 6. Tissue-resident memory CD8+ T cells are activated in lung and liver as virus spreads.
(A) Experimental workflow: mice were immunized with MVA at skin 21 days prior to intranasal WR challenge (or PBS as control) for 0.5 day, and single lung and liver memory CD8+ T cells (live CD3ε+CD44+CD62L− cells), including virus-specific cells were sorted by FACS prior to single-cell (sc) RNA-seq.
(B) Heatmap of 1,476 single memory CD8+ T cells (columns) from lung and liver showing the top 100 differentially expressed genes (rows) between control (PBS only) and WR-challenged mice in each tissue type (FDR < 0.01, expression fold change > 3).
(C) Heatmap of 20 single virus-specific (H2-Kb B8R20–27) CD8+ T cells (columns) showing the top 25 differentially expressed genes (rows) between cells from lungs of mice challenged with B8R20–27 peptide (20 μg) or saline as control (FDR < 0.01, expression fold change > 3). UMI, unique molecular identifiers.
(D-E) Impact of WR challenge on single CD8+ memory T cell states. Visualization of single cells from lung (top) and liver (bottom) from control (MVA vaccination only) and WR-challenged mice using t-SNE (D). t-SNE plots from D are shown in E and colored based on the B8R activation score (scaled average expression of genes differentially expressed in both WR (B) and peptide B8R20–27 (C) challenges).
(F-G) Tissue-resident memory CD8+ T cells (TRM) are activated in both lung and liver upon intranasal WR challenge. t-SNE plots from D are shown in F and colored based on the TRM cell score (scaled average expression of genes associated with the TRM phenotype). Blue dots indicate a TRM score > mean TRM score of all cells + 1 SD. In panel G are shown the distributions of B8R activation scores (E) in TRM cells (F).
(H) TRM scores for indicated CD8+ T cell populations sorted from lung after intravascular (IV) immunostaining. Error bars, SD (n = 4). *; Student’s t-test p-value < 0.05.
Next, given the critical role of resident memory cells for host protection in our model, we investigated single TRM cell responses. To do so, we identified TRM cells from single memory CD8+ T cell profiles by computing the average expression of 84 genes (referred to as the TRM cell score) shared by CD8+ TRM cells across tissues (Table S5) (Mackay et al., 2016). We found TRM cells in lung (13.5%, 50/370, in control and 21.4%, 79/369, in WR) and liver (9.8%, 36/368, in control and 16.5%, 61/369, in WR) exhibiting a high TRM score (Figure 6F and S6E-G), which represents TRM cell proportions in agreement with quantifications based on intravascular (IV) staining for lung (Figure 5F), and two commonly used, yet imperfect, TRM markers CD69 and CD103 for lung and liver (Figure S6H-I) (Steinert et al., 2015). As an additional validation for our TRM scoring approach, we found that IV-negative (parenchyma-associated) memory CD8+ T cells displayed a significantly higher TRM score than IV-positive (vasculature-associated), or CD44+CD62L+ and CD44−CD62L+ CD8+ T cells at lung (Figure 6G). Using TRM scores, we found a fraction of memory CD8+ T cells displaying a TRM state and being reactivated in an antigen-specific manner as measured by high B8R activation scores (p-value = 1.07 × 10−7 for lung and 2.2 × 10−3 for liver; Wilcoxon rank sum test) (Figure 6H and S6E-G). Altogether, these results demonstrated that TRM (high TRM score) and non-TRM (low TRM score) memory CD8+ T cells at lung and liver were activated at least in part in an antigen-dependent manner (high B8R score) within hours of a respiratory WR challenge.
Local tissue environments shape the functional abilities of memory CD8+ T cells to bolster organ-specific responses
Two observations from our experiments suggested that tissue-specific adaptations help shape the phenotypic states of memory CD8+ T cells at steady state and upon reactivation. First, the classical TRM markers CD69 and CD103 were skewed in expression on memory T cells at lung (mostly CD103+) and liver (mostly CD69+) (Figure S6H-I), in agreement with others (Fernandez-Ruiz et al., 2016; Laidlaw et al., 2014; Takamura et al., 2016; Wakim et al., 2013). CD69 and CD103 have been shown to vary in expression based on tissue and infectious contexts (Steinert et al., 2015), but it remains unclear whether such variations reflect tissue-specific cellular states or that these molecules are poor markers of TRM cells. Second, our single-cell analyses supported a role for tissue adaptations in memory CD8+ T cells by hinting at shared and tissue-specific gene modules in lung and liver.
To test this hypothesis, we tested for differential gene expression between lung and liver single-cell profiles, which revealed tissue-specific genes in memory CD8+ T cells – including in cells with the same antigen specificity (B8R) (Figure 7A-B). We validated these results at the protein level for: CCL5 – well-described effector molecule in memory CD8+ T cells (Lauvau et al., 2016), and present in nearly all lung and liver cells; CX3CR1 which resolved two subpopulations in lung and liver; and CXCR6 and ITGA1 showing liver-specific expression in subsets of cells (Figure S7A-B) – as recently shown for CXCR6 (Fernandez-Ruiz et al., 2016). In addition, a number of other surface molecules (e.g., Fcer1g), transcriptional regulators (e.g., Junb, Tnfaip3), and effector cytokines (e.g., Xcl1) displayed variations in their expression based on tissue type (Figure 7C). We further confirmed that the expression of these genes and others varied according to the tissue of residence by measuring gene expression in pools of 5,000 memory CD8+ T cells (CD44+CD62L−) (Figure S7C-F; Table S6).
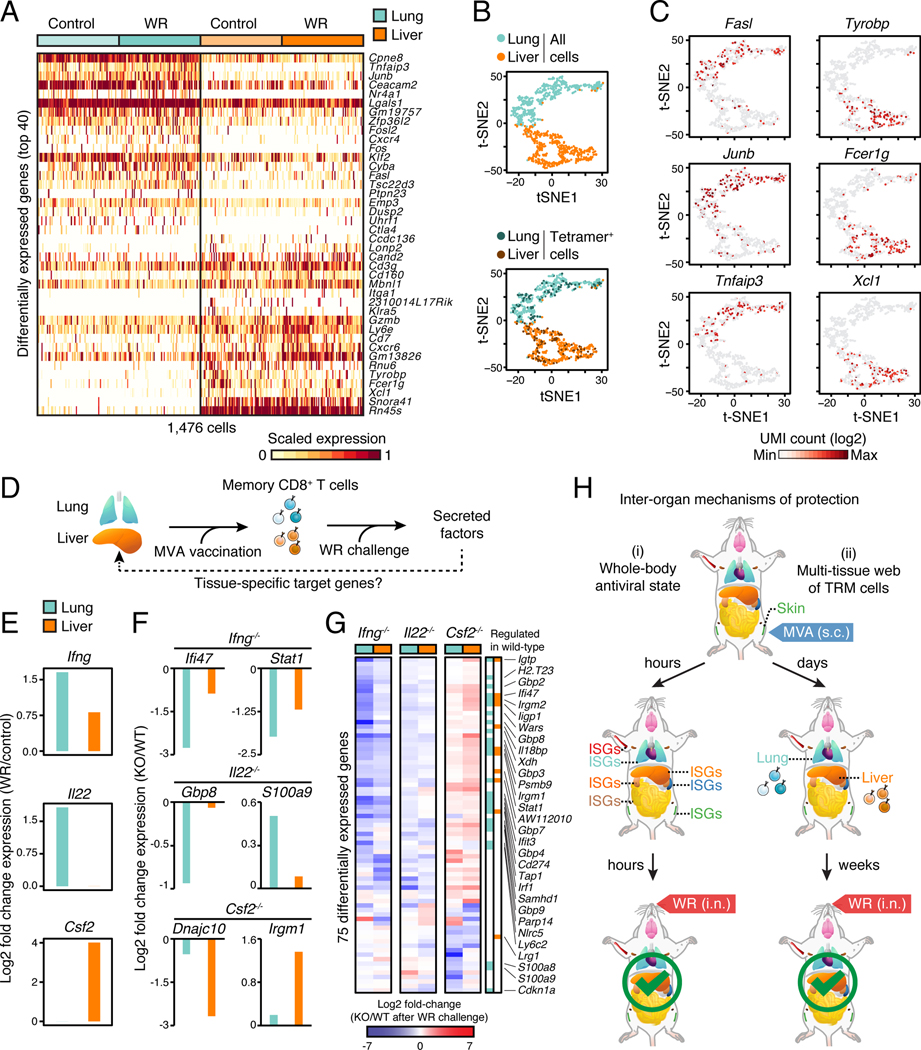
Figure 7. Local tissue environments shape the functional abilities of memory CD8+ T cells to bolster organ-specific responses.
(A) Heatmap of 1,476 single memory CD8+ T cells (columns) showing the top 40 differentially expressed genes (rows) between lung and liver of control (MVA vaccination only) and WR-challenged mice (FDR < 0.01, expression fold change > 3).
(B) Visualization of single cells from lung and liver in MVA-vaccinated mice (Control) using t-SNE. Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells (Tetramer+) are labeled in the bottom plot for lung (25.4%, 94/370 cells) and liver (25.3%, 93/368 cells).
(C) Expression levels of tissue-specific genes in single cells. t-SNE plot from B colored based on expression levels (UMI, unique molecular identifiers) of indicated genes.
(D) Illustration of the impact of tissue-adapted memory CD8+ T cells on their respective tissue of residence. From left to right, vaccination at skin seeds memory CD8+ T cells in lung and liver, which secrete factors controlling tissue responses upon WR challenge.
(E) Secreted factor induction in lung and liver at day 0.5 after WR challenge of MVA-vaccinated mice. Fold change values were calculated using single-cell RNA-seq profiles from WR-challenged versus vaccinated only mice as control.
(F-G) Tissue-level expression changes for target genes of secreted factors produced in memory CD8+ T cells. Mice were immunized at skin with MVA and challenged 3 weeks later with WR intranasally for 1.5 days. Shown are log2 fold-change values of knockout (KO) relative to wild-type (WT) mice for selected (F) and all differentially expressed genes (G) (FDR-adjusted p-value < 0.05, absolute fold change > 2, n = 4).
(H) Schematic depicting the inter-organ mechanisms of protection reported in this study. Upon skin MVA vaccination, type I IFNs produced locally trigger a whole-body antiviral state within hours (i; left), and tissue-resident memory CD8+ T cells (TRM) seeded in distant tissues help block viral spread (ii; right).
Lastly, given that tissue-specific differences were maintained upon WR challenge (Figure S7G), we hypothesized that tissue-driven adaptations can shape the processes targeted by memory T cells in their host tissue. To test this idea, we reasoned that combining single-cell and whole-tissue transcriptional profiles from challenged hosts would reveal the molecular links, if any, between memory T cells and surrounding parenchyma, stroma and/or immune cells (Figure 7D). First, we mined our transcriptional data from single memory CD8+ T cells for secreted factors produced in lung or liver upon WR challenge, and focused on three factors: Ifng (lung and liver), Il22 (lung-specific), and Csf2 (liver-specific) (Figure 7E). Second, to test the functional effects of these factors on lung and liver, we measured the impact of genetic deletions on whole-tissue mRNA responses upon WR challenge (Table S6). Deleting Ifng decreased ISG induction in both lung and liver (e.g., Ifi47, Stat1) (Figure 7F-G), albeit to a lesser extent in liver, which is consistent with the lower induction of Ifng in liver compared to lung (Figure S6C and 7E). Il22−/− mice displayed a broader downregulation in target genes in lung compared to liver (e.g., Gbp8, S100a9) (Figure 7F-G), consistent with the effects of the IL-22 cytokine on epithelia (Sabat et al., 2014). On the contrary, knocking out Csf2 impacted liver target genes more than lung ones, with the majority of genes being upregulated in Csf2−/− mice compared to wild-type (Figure 7F-G). Overall, 40% (30/75) of the genes impacted by knocking out Ifng, Il22 or Csf2 (Figure 7G) were regulated in wild-type tissues in a T cell-dependent manner upon WR challenge (Figure 4D-E), suggesting that knockout effects were, at least in part, not due to indirect effects linked to deleting these pleiotropic cytokines. In light of these results, we propose the existence of a tissue-specific coupling between the factors secreted by memory T cells and the processes these factors trigger in their environment of residence.
DISCUSSION
By revisiting a comparative poxvirus infection model, we validated the central hypothesis of this work that monitoring gene expression changes across tissues can uncover immune processes operating at the whole-organism scale. What are some of the advantages and caveats associated with our whole-tissue profiling approach? First, at steady state, our results on the variations in transcript abundance between organs agree with recent reports in humans (Melé et al., 2015), which provides an independent validation of our custom RNA-seq method. On the basis of these whole-tissue mRNA profiles, our approach successfully captured the dynamic changes of many molecular and cellular immune processes that can be linked within and between organs through, for example, ligand-receptor connectivity or cell migration events such as recruitment to inflamed tissues. Second, the power of our strategy rests on the systematic monitoring of intact organs, which avoids potential sampling biases, and more importantly, preserves the native organization of inter-cellular communications between immune and non-immune cells across the body. Thus, our data captured information about cell-intrinsic and -extrinsic activities to reveal the net output of tissue-level responses, which cannot be obtained by processing cells isolated from tissues, and yet, is critical to shed light on organismal immune mechanisms that would otherwise not be detectable. Third, several processes are likely missed by our approach, such as changes in expression that cannot be detected at the RNA (e.g., small variations in few cells) and protein levels (e.g., pre-formed cytokine precursors). Future work is required to capture the many additional facets of organismal immune processes by, for example, tracking changes at the protein level in tissues and selected cells, combining single-cell and whole-tissue profiles, and using targeted RNA-seq strategies or higher sequencing depth to capture more subtle changes.
Our whole-tissue datasets can be mined in many ways and, as a proof-of-principle, we analyzed the expression of known pairs of secreted factors and matching receptors to draw organism-level maps of connectivity within and between tissues. In doing so, we uncovered that type I IFNs produced mostly at skin can prompt a whole-body antiviral conditioning through inter-organ signaling, leading to host protection within hours (Figure 7H). The antiviral effects of type I IFNs through ISGs have been largely studied for their cell-intrinsic effects (McNab et al., 2015; Schneider et al., 2014), whereas inter-organ signaling of type I IFNs remains poorly understood. One exception comes from recent work on the capacity of type I IFNs to signal from lungs infected with influenza virus to the bone marrow (Hermesh et al., 2010). Thus, our finding that type I IFNs can prompt an antiviral state at the organism level brings a new perspective on the biology of these cytokines by extending their perimeter of action upon localized synthesis and secretion – perhaps as a means for the host to anticipate systemic viral spread by preemptively arming distant tissues with antiviral properties. Moreover, the organismal effects of type I IFNs provide a model to probe fundamental questions about the organization and evolution of inter-organ communications through secreted factors (Droujinine and Perrimon, 2016). Moving beyond type I IFNs, our results lay the foundation for future analyses of inter-organ communication through secreted factors, and for functional investigations of the various ligand-receptor connections predicted in this work.
We discovered mechanisms of inter-organ protection mediated by memory CD8+ T cells, shedding new light on how the memory T cell compartment is organized spatially and functionally to protect the host against systemic viral spread. First, we found that skin vaccination with MVA generated TRM cells which protected the host by seeding multiple distant tissues such as lung or liver (Figure 7H). TRM cells are thought to only seed the site of pathogen entry, where they can trigger a rapid antimicrobial state during reinfection (Mueller and Mackay, 2016; Schenkel and Masopust, 2014). Despite this predominant view of a local seeding for TRM cells, our findings corroborate recent work showing that repeated skin infections can lead to some degree of protection in distant skin epithelia (Jiang et al., 2012), and that intranasal vaccination can seed TRM cells in distant mucosa (Stary et al., 2015). Thus, our data reinforce the notion that, upon local vaccination, TRM cells can be seeded systemically within a time frame – during the effector phase of the response – in agreement with previous work (Hofmann and Pircher, 2011; Masopust et al., 2010; Stary et al., 2015). The nature of the pathogen, the dose and the tissue of entry are likely to be key drivers for the seeding of TRM cells locally and/or across distant organs.
Second, memory CD8+ T cells, including TRM cells, expressed tissue-specific genes encoding surface and effector molecules as well as transcriptional regulators. While tissue-specific adaptations are well documented for macrophages throughout development and beyond (Okabe and Medzhitov, 2016), examples of such adaptations are only beginning to emerge for resident lymphocytes such as T regulatory cells (Fan and Rudensky, 2016; Panduro et al., 2016). While it is clear that local adaptations of immune cells are key for tissue homeostasis (Okabe and Medzhitov, 2016), our results show that these local adaptations can also dictate the communication conduits between immune and parenchymal cells so as to cope effectively with an infection given local tissue constraints. Indeed, by functionally adapting to their tissue of residence, memory CD8+ T cells were empowered to trigger tissue-specific responses upon reactivation. While pre-established differences in memory T cell states across tissues are likely to drive tissue-specific responses, it is also important to consider that varying reactivation conditions might play a role in this process as well (Richer et al., 2013). Future work will be critical to test whether shaping tissue-level communications through local adaptation is a universal feature of immune cells that is important for host protection and homeostasis.
In conclusion, this study opens up new lines of inquiry that pave the way for systematic analyses of immune responses at the whole-organism scale. Future studies based on additional vaccine and infection models will have implications for our understanding of how protective immunity works at the organism level, and thus, how to manipulate the immune system against disease.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Nicolas Chevrier (nchevrier@uchicago.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
We used female C57BL/6J mice, B6.129S7-Ifngtm1Ts/J (Stock 002287), B6.129S-Csf2tm1Mlg/J (Stock 026812), and C57BL/6-Il22tm1.1(icre)Stck/J (Stock 027524) (Jackson Laboratories) which were 5- to 8-weeks old for all experiments. Knockout mice were kindly provided by Christophe Benoist, Diane Mathis, and Mei X. Wu (Ifnar1−/−) and Arlene Sharpe (Tcra−/−). Animals were housed in specific pathogen free and BSL2 conditions at Harvard University, and all experiments were performed in accordance with the US National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Harvard University Institutional Animal Care and Use Committee.
Viruses
For Vaccinia virus work, we followed procedures previously described (Earl and Moss, 2001; Earl et al., 2001) with modifications.
1). Original stocks.
Vaccinia virus strain Western Reserve (WR; NR-55, Lot 3579605), strain WR Expressing Enhanced Green Fluorescent Protein (WR-eGFP; NR-624, Lot 3925477), and strain Modified Vaccinia Ankara (MVA; NR-726, Lot 4225252) were obtained from the Biodefense and Emerging Infections Research Resources Repository (BEI Resources). MVA-eGFP was kindly provided by Bernard Verrier. For each strain, initial plaque purification was conducted under agarose overlay in 6 well-plates using Vero (ATCC CCL-81) and BHK-21 (ATCC CCL-10) cells for WR and MVA strains, respectively. Several well isolated plaques were picked with a pre-wet P-1000 tip, placed into 0.5 mL DMEM containing 2.5% FBS, and frozen at −80°C. One plaque from each strain was selected to prepare stocks for all experiments.
2). Propagation.
HeLa S3 (ATCC CCL-2.2) and BHK-21 cells were infected with WR or MVA, respectively, and harvested 2–4 days later upon visual inspection of cytopathic effects. Cells were centrifuged at 1,800 g for 6 min at 4°C, supernatants discarded, and cell pellets resuspended in 10 mM Tris-HCl buffer (pH 9.0). Because Vaccinia virus is strongly cell-associated, cell pellets went through 4 freeze/thaw cycles using a dry ice/ethanol bath and a 37 °C water batch. After each freeze/thaw cycle, lysates were centrifuged at 500 g for 6 min at 4°C, and virus-containing supernatants pooled.
3). Purification.
Prior to titering and in vivo use, virus preparations were purified using four consecutive ultracentrifugation steps. First, crude stocks were concentrated using a 36% sucrose cushion (w/v in 1 mM Tris-HCl buffer at pH 9.0) by centrifuging at ~32,900 g for 90 min at 4°C. Second, supernatants were discarded and pellets resuspended in 1 mM Tris-HCl buffer, pH 9.0. Concentrated virus was sonicated on ice using a cup horn sonicator (3 cycles of 20 seconds at power 1–2), and purified twice consecutively using 10–50% continuous sucrose gradients prepared with a Gradient Master instrument (Biocomp 153-001) and centrifuged at ~26,000 g for 20 min at 4°C. Third, virus bands from both sucrose gradients were pooled and centrifuged at ~32,900 g for 60 min at 4°C. Virus-containing pellets were resuspended in 1 mM Tris-HCl buffer (pH 9.0), sonicated as described above, and split into single use aliquots stored at −80°C.
4). Molecular characterization.
For WR strains, we performed PCR analysis of the cytokine response modifier B (crmB) conserved sequence and crmB-NlaIII RFLP analysis using the following primer pair: VL2N (5’-ACATGCATGCCAGGAC-3’) and VL33 (5’-ACCATTACAAACATTATCC-3’) (Loparev et al., 2001). For MVA, we performed PCR using primers flanking the 6 major deletion sites compared to the parent strain Chorioallantois Vaccinia virus Ankara (CVA). Primers were selected to produce PCR fragments ranging from 366 to 698 bp (NCBI U94848), compared to 3194 to 7159 bp for CVA (NCBI AM501482) (Meisinger-Henschel et al., 2007).
5). Viral titers.
We performed serial 10-fold dilutions in DMEM without additives followed by inoculation of BS-C-1 (ATCC CCL-26) or BHK-21 cells for WR and MVA, respectively, in 24 or 6 well-plates. Two days after inoculation, plaque-forming units (PFUs) were quantified using (1) crystal violet staining for WR, and (2) immunostaining for MVA using rabbit polyclonal anti-vaccinia virus primary antibody (ThermoFisher Scientific PA1-7258) and HRP-Goat anti-rabbit secondary antibody (ThermoFisher Scientific 656120) diluted at 1:1000 in PBS containing 3% FBS. Peroxidase activity was then detected using o-Phenylenediamine dihydrochloride (OPD; Sigma P9187) by incubating for 30 min.
METHOD DETAILS
Infection
Mice were infected with 107 PFUs intranasally (i.n.) for WR and subcutaneously (s.c.; flank skin) for MVA, unless mentioned otherwise. Body weights were measured daily, and animals were sacrificed upon losing more than 30% of their initial weight. For CD8 T cell depletion studies, mice were injected intraperitoneally with 200 μg of anti-CD8β antibody (Clone 53-5.8: BioXCell BE0223) or isotype control (Clone HRPN: BioXCell BE0088) at day −6 and −3 and 100 μg at day 0, 4 and 8 of WR challenge. For IFNAR1 inhibition, 400 μg of anti-IFNAR-1 antibody (Clone MAR1-5A3: BioXCell BE0241) or isotype control (mouse IgG1: BioXCell BE0083) was administered intraperitoneally at 6 h before and after both MVA and WR infection (4 times in total).
Intravascular immunostaining
For staining of vasculature-associated leukocytes, mice were injected intravenously with 3 μg of anti-CD45-PE antibody (clone 30-F11; Biolegend) in 200 μL saline. After 3 min, mice were sacrificed, and tissues were harvested and processed for flow cytometry analysis (see sections on Tissue Harvest and Flow Cytometry below).
Parabiosis
Surgery was performed as described in Wright et al., 2001. Mice were anesthetized by intraperitoneal injection of ketamine HCl (100 mg/kg body weight) and xylazine (10 mg/kg body weight), or with avertin (500 mg/kg body weight). After shaving the corresponding lateral flank skin of each mouse, matching skin incisions were made from the elbow to the knee joint. The elbow and knee joints were attached by a single 4-0 silk suture, and the dorsal and ventral skins were approximated by staples or continuous suture. After an interval of at least two weeks, parabiosed mice were surgically separated by a reversal of the above procedure, and challenged with WR after a two-week period of recovery after separation.
Tissue harvest
Mice were anesthesized with avertin (250–500 mg/kg) and perfused transcardially with PBS containing 10 mM EDTA (to avoid signal contamination from blood in tissues). Prior to perfusion, blood was collected by cardiac puncture and stored on ice, and immediately after perfusion, tissues were rinsed in PBS, placed in RNA later solution (Sigma R0901), and kept at 4°C overnight prior to transfer at −80°C for storage. For each mouse, we harvested 17 tissues total: five lymph nodes (2 brachial, 2 inguinal, and mediastinal), left and right flank skin, thymus, heart, lung, spleen, kidney, small intestine, liver, brain, bone marrow (BM) and peripheral blood mononuclear cells (PBMCs). Small intestine was cut longitudinally and washed extensively in PBS to completely remove feces contamination. Blood was used for extraction of peripheral blood mononuclear cells (PBMCs) using Lymphocyte Separation Medium (Corning 25-072), and sera using BD Microtainer tubes with serum separator (BD 365956). Bone marrow cells were collected from femora and tibiae, stored overnight in RNA later at 4°C, centrifuged at 5,000 g for 5 min at 4°C, and cell pellets stored at −80°C.
Whole-mount tissue imaging
Tissue clearing of brain, lung and dLN was performed with slight modifications of the original CLARITY protocol (Chung et al., 2013). For brain, mice were perfused with PBS containing 1 mM EDTA and then with hydrogel monomer (HM) solution containing 2% acrylamide, 0.025% bisacrylamide, 4% paraformaldehyde (PFA), and 0.125% VA-044 in PBS. The whole brain was further incubated in HM solution for 3 days at 4°C, followed by polymerization for 3 hours at 37°C. Clearing of whole brains was performed for 2 days using the X-CLARITY Electrophoretic Tissue Clearing System (Logos Biosystems). For lung and dLN, mice were perfused with 4% PFA in PBS, and tissues were incubated in HM solution without bisacrylamide for 3 days at 4°C. After polymerization for 3 hours at 37°C, tissue clearing was performed passively by incubation in clearing solution (Logos Biosystems C13001) at 37°C for 16 days (lung) or 7 days (dLN). Cleared tissues were washed in PBS containing 0.5% Triton X-100 for 1 (brain and dLN) or 2 days (lung) at room temperature, and incubated in refractive index matching solution containing 88% w/v Histodenz (Sigma D2158) and 0.01% sodium azide in 1X PBS at pH 7.5 (Treweek et al., 2015), for 2 days at room temperature. Images were acquired using a Lightsheet Z.1 microscope (Zeiss). A 488-nm laser was used to excite GFP and a 561-nm laser was used to produce autofluorescence. For whole-mount skin imaging, mice were perfused with 4% PFA in PBS and hair removed with hair removal cream (Nair). Skin samples were dissected, fat removed and incubated overnight at 4ºC in Zamboni’s fixative (paraformaldehyde/picric acid). Samples with washed in PBS with 1% Triton X-100 and immunostained with primary (anti-GFP, anti-IA/IE) and fluorophore-labeled secondary antibodies for 2–3 days each. Skin was dehydrated in methanol, and mounted in Benzyl Alcohol (Sigma 402834) and Benzyl Benzoate (Sigma B6630) mixed 1:2 for clearing followed by imaging using an LSM 880 confocal microscope (Zeiss).
Flow Cytometry
Lung, liver and skin were minced with scissors and digested at 37°C for 40–60 min in RPMI1640 medium containing 2% FBS, 32 mM HEPES, 100 U/mL Penicillin, 100 μg/mL Streptomycin, 0.5 mg/mL DNase, and 1.66 or 0.5 mg/mL of Collagenase IV (Worthington Biochemical) for lung and liver, respectively, or 0.25 mg/mL of Liberase TL (Sigma 5401020001) for skin. Tissues were then mashed and filtered to obtain single-cell suspensions. For lung and liver, lymphocytes were enriched using a gradient of 40/80% Percoll PLUS (GE Healthcare Bio-Sciences 17-5445-02). Spleen and LNs were mashed on 70 μm filter. Peripheral blood was collected by puncturing heart or cheek and mononuclear cells were isolated with Lymphocyte Separation Medium (Corning). Single-cell suspensions were stained in the presence of Fc receptor-blocking antibodies (anti-mouse CD16/32, clone 93) using the following antibodies (Biolegend): CCL5 (2E9/CCL5), CCR2 (475301), CD103 (2E7), CD11b (M1/70), CD11c (N418), CD19 (6D5), CD24 (M1/69), CD3e (145-2c11), CD44 (IM7), CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD49a/ITGA1 (HMα1), CD49b (DX5), CD62L (MEL-14), CD64 (X54-5/7.1), CD69 (H1.2F3), CD8a (53-6.7), CD8b (53-5.8), CX3CR1 (SA011F11), CXCR6 (SA051D1), I-A/I-E (M5/114.15.2), IFN-γ (XMG1.2), KLRG1 (2F1/KLRG1), Ly6C (HK1.4), Ly6G (1A8). Intracellular staining was performed after surface staining using the BD Cytofix/Cytoperm Kit (BD Biosciences 554714). H-2Kb-restricted and Brilliant Violet 421-labeled MHC class I tetramers for the VACV B8R20–27 peptide (TSYKFESV) were provided by the NIH Tetramer Core Facility.
Plaque assay
Tissues were collected and weighed at day 3 after intranasal WR infection, homogenized in PBS containing 1% FBS by adding 5-mm stainless steel beads and running 3 × 4 min cycles at 30 Hz on the Tissue Lyzer II. For liver samples, tissues were homogenized using M tubes and running 3 cycles of the RNA_02.01 program on the gentle MACS octo dissociator (Miltenyi Biotec). Tissue lysates underwent 3 freeze/thaw cycles using dry ice/ethanol and 37 °C water baths. Lysates were centrifuged at 3,200 x g for 5 min at 4°C and supernatants were used for plaque assay using BS-C-1 cells seeded in T25 flasks. Cells were inoculated with tissue lysates diluted in DMEM containing 10% FBS and incubated for 1 h at 37°C/5% CO2. Cells were then washed with PBS 3 times and incubated for 36 h in DMEM containing 10% FBS, and stained with crystal violet for quantification of plaque-forming units.
Viral DNA qPCR
Tissues were homogenized in 10 mM Tris-Cl (pH 9.0) by adding 5-mm stainless steel beads and running 3 × 4 min cycles at 30 Hz on the Tissue Lyzer II (Qiagen). Tissue homogenates were lysed at 60°C for 1 h in 50 mM Tris-HCl containing 100 mM EDTA, 0.05% SDS, and 0.5 mg/mL Proteinase K. Total DNA was purified directly from tissue lysates using magnetic beads Agencourt AMPure XP (Beckman Coulter A63880). Viral DNA was quantified by qPCR using 30 ng of total DNA as input, and primers targeting loci within the Vaccinia virus genes H5R (5’-gcgacattgtagaaagcgtg-3’, 5’-ccagcttcaacttgtaccatagg-3’) and F7L (5’-gctcgtcatgggatcctg-3’, 5’-tgcttcggattcatccagatc-3’), and the host gene Ifnb1 (5’-ctggcttccatcatgaacaa-3’, 5’-agagggctgtggtggagaa-3’) as an internal control. The CFX384 Real-Time PCR Detection System (Bio-Rad Laboratories) was used with LightCycler 480 SYBR Green I Master mix (Roche) and 0.5 μM of each primer in a final volume of 10 μL with 45 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 40 seconds. Amplification products were subjected to melting curve analysis using the CFX Manager System (Bio-Rad Laboratories) to exclude the amplification of non-specific products. To generate standard curves for H5R and F7L primers, known PFU numbers of MVA were spiked into 30 ng of mouse genomic DNA. PFU numbers were determined in two ways using plaque assay and qPCR measurements on known plasmid copy numbers that contained the viral gene I4L (5’-ctggcggctagaatggcata-3’, 5’-gacactctggcagccgaaat-3’) (Liu et al., 2006).
Whole-tissue RNA extraction
Tissues stored in RNAlater were thawed and transferred to 2.0 mL Safe-Lock Tubes (Eppendorf 022363344) containing 700–1500 μL of Isol-RNA Lysis Reagent (5Prime 2302700) depending on the tissue size and type. Tissues were lysed by adding 5-mm stainless steel beads (Qiagen 69989) and running 1–3 cycles of 4 min at 30 Hz on the Tissue Lyzer II (Qiagen). For liver and small intestine samples, tissues were lysed using M tubes (Miltenyi biotec 130-096-335) and running 1–4 cycles of the RNA_02.01 program on the gentleMACS octo dissociator (Miltenyi biotec). Next, for high throughput RNA extraction, lysates were processed in deep 96-well plates (USA Scientific 1896–2000) by adding chloroform for phase separation by centrifugation, followed by precipitation of total RNA in the aqueous phase using magnetic beads coated with silane (Dynabeads MyOne Silane; ThermoFisher Scientific 37002D) and buffer RLT (Qiagen 79216). Genomic DNA contamination was removed by on-bead DNase I (ThermoFisher Scientific AM2239) treatment at 37°C for 20 min. After washing steps with 80% ethanol, RNA was eluted from beads and sample concentrations were measured using the Quant-iT RiboGreen RNA Assay Kit (ThermoFisher Scientific R11490). RNA quality was confirmed using a 2100 Bioanalyzer (Agilent Technologies).
RT-qPCR
Total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific 4368813) with both random nonamers and oligo(dT) primers. Real-time quantitative PCR reactions were performed on the CFX384 Real-Time PCR Detection System (Bio-Rad Laboratories) with LightCycler 480 SYBR Green I Master mix (Roche) and 0.5 μM of each primer in a final volume of 10 μL with 40 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 40 seconds. The following forward-reverse primer pairs were used to measure levels of Vaccinia virus genes E3L (5’-gcctccgttgtcataaacca-3’, 5’-cgtagtgctatggtgtacagctc-3’) and F7L (5’-gctcgtcatgggatcctg-3’, 5’-tgcttcggattcatccagatc-3’), and mouse genes Gapdh (5’-ggcaaattcaacggcacagt-3’, 5’-agatggtgatgggcttccc-3’), Ifnb1 (5’-caggcaacctttaagcatcag-3’, 5’-cctttgacctttcaaatgcag-3’), Ifit3 (5’-tgaactgctcagcccaca-3’, 5’-tcccggttgacctcactc-3’), and Ifitm3 (5’-cccccaaactacgaaagaatca-3’, 5’-accatcttccgatccctagac-3’). Amplification products were subjected to melting curve analysis using the CFX Manager System (Bio-Rad Laboratories) to exclude the amplification of non-specific products. Gapdh levels were used as an endogenous control for normalization. To generate standard curves for viral E3L and F7L primers, RNA was extracted from sorted GFP+ BHK-21 cells that were infected with MVA-eGFP – to ensure that all cells were productively infected – and used as a template in indicated concentrations.
RNA sequencing
We developed a protocol to generate multiplexed RNA-seq libraries using the following workflow: (1) oligo(dT)-primed reverse transcription (RT) with sample barcoding followed by pooling; (2) 3’-end extension of pooled, barcoded cDNAs using the Klenow fragment; and (3) PCR amplification followed by Illumina sequencing.
First, for each tissue sample, 100 ng of total RNA was heat-fragmented at 94°C for 2.5 min in a 10-μL reaction volume in the presence of 1X AffinityScript RT Buffer (AffinityScript Multi-Temp RT kit; Agilent Technologies 600109) and 1 pmole of a custom RT primer containing sequences from 5’ to 3’ for the Illumina P7 adaptor, an 8-bp sample barcode, the Illumina read 2 primer, and an anchored oligo(dT)22 for priming (Table S7). Fragmentation conditions were optimized to generate RNA fragments ranging in size from 400 to 600 bp on average. Samples were cooled on ice and RT performed by adding 10 μL of 4 mM dNTPs, 10 mM DTT, 20 units of RNase Block (Agilent Technologies 300151), and 0.8 μL of AffinityScript Multiple Temperature RT enzyme, and incubating at 42°C for 90 min. After RT, barcoded cDNA samples were pooled (up to 96 samples per library) using RNA Clean & Concentrator columns (Zymo Research R1015), and cleaned up from residual primers and RNA using exonuclease I (New England Biolabs M0293) treatment and alkaline hydrolysis with NaOH.
Second, single-stranded cDNA samples were cleaned up using Oligo Clean & Concentrator columns (Zymo Research D4060), and extended in 3’ with Klenow Fragment 3’ -> 5’ exo- (New England Biolabs M0212) using a random nonamer primer coupled with the Illumina read 1 primer sequence and blocked in 3’ end with a modified base (ddC) to avoid extension of the primer itself by the Klenow fragment (Table S7).
Third, cDNAs were purified using magnetic beads Agencourt AMPure XP (Beckman Coulter A63880) and amplified with 8–10 cycles of PCR using Q5 Hot Start High-Fidelity 2X Master Mix (New England Biolabs M0494) (Table S7). Libraries were gel purified using E-Gel EX Agarose Gels, 2% (ThermoFisher Scientific G402002), quantified with a Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific Q32851), and sequenced using an Illumina HiSeq 2500 (single read 50 bp).
Single-cell RNA sequencing
1). Library preparation.
We used a plate-based protocol and sequencing libraries from single CD8+ memory T cells (see sorting scheme in Figure S5B) were prepared based on the SCRB-seq protocol (Soumillon et al., 2014) with modifications using the following workflow: (1) oligo(dT)-primed RT reaction with cell and molecular barcoding followed by pooling of 384 cells; (2) single primer PCR amplication; and (3) full-length cDNA tagmentation and amplication by PCR.
First, single lung and liver lymphocytes were isolated by FACS into 96 or 384 well-plates containing 2 μL/well of lysis buffer containing 50 mM Tris-HCl pH8.0, 0.5% NP-40, 10 mM DTT, 0.8 U/μL RiboLock RNase Inhibitor (ThermoFisher Scientific EO0381), and 6.25% PEG8000. cDNA was prepared directly from single cell lysates in a 4-μL final reaction volume containing the following: (1) 2 μL of lysate; (2) 1 μL containing 2 pmoles of a custom RT primer biotinylated in 5’ and containing sequences from 5’ to 3’ for the Illumina read 1 primer, a 6-bp cell barcode, a 10-bp unique molecular identifier (UMI) (Islam et al., 2013), and an anchored oligo(dT)30 for priming (Table S7); and (3) 1 μL of RT mix containing 0.4 μL of 5X RT buffer, 0.2 μL of 10 mM dNTPs, 2 pmoles of template switching oligo (Table S7), and 0.125 μL Maxima H Minus Reverse Transcriptase (ThermoFisher Scientific EP0753), and. First, barcoded RT primers (384 total) were added to lysates, which were denatured at 72ºC for 1 min and snap cooled on ice. Second, the RT mix was added and plates were incubated at 42ºC for 120 min. For each library, double stranded cDNA from 384 single cells were pooled using DNA Clean & Concentrator-5 columns (Zymo Research D4013), and residual RT primers were removed using exonuclease I (New England Biolabs M0293).
Second, full length cDNA was amplified with 18 cycles of single primer PCR (Table S7) using the Advantage 2 PCR Kit (Clontech 639206), and cleaned up using magnetic beads Agencourt AMPure XP (Beckman Coulter A63880). cDNA was quantified with a Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific Q32851).
Third, 1 ng of cDNA per library was tagmented and amplified by PCR using the Nextera XT Kit (Illumina) (Table S7). Libraries were gel purified using E-Gel EX Agarose Gels, 2% (ThermoFisher Scientific G402002), quantified with a Qubit dsDNA High Sensitivity Assay Kit (ThermoFisher Scientific Q32851), and sequenced on an Illumina NextSeq 500 by pooling up to 4 libraries of 384 cells each (1,536 cells) to avoid batch effects.
For droplet-based analysis (Figure S6F-G), we used the inDrops method for single cell encapsulation, library preparation and UMI count table generation as described (Zilionis et al., 2017).
QUANTIFICATION AND STATISTICAL ANALYSIS
Genome alignment
Alignment of MVA (177,922 bp; GenBank ID U94848.1) and WR (194,710 bp; Genbank ID NC_006998.1) genomes was performed using MAFFT (Katoh and Standley, 2013) and plotted using Circos (Krzywinski et al., 2009).
RNA sequencing data analysis
1). Raw read processing.
Sequencing read files were processed using the RNA-seq pipeline in the bcbio-nextgen project version 0.8.9a0-6c9bb01 (https://bcbio-nextgen.readthedocs.org/en/latest/). Reads were aligned to the mm10 genome augmented with transcripts from Ensembl release 78 with STAR version 2.5.2b (Dobin et al., 2013). Quality control metrics were compiled with a combination of FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), Qualimap (Garcia-Alcalde et al., 2012), MultiQC (https://github.com/ewels/MultiQC) and custom metrics (5 million mapped reads were obtained on average per sample). Expression quantification was performed using both featureCounts version 1.4.4 (Liao et al., 2014) with multi-mapping reads excluded and Sailfish version 0.9.2 (Patro et al., 2014) with a kmer size of 31 with 30 bootstrap samples. Furthermore, by mapping RNA-seq reads onto both viral and mouse transcriptomes, we found that 29 out of 601 whole-tissue RNA-seq samples had >1,000 reads mapping to viral transcripts from WR or MVA. These 29/601 samples had on average 4,173 viral reads +/− 5,190 SD for 4.75 million mouse reads +/− 1.5M SD (i.e., reads mapping unambiguously to mouse transcripts and not viral ones). Thus, only 0.1% +/− 0.21 SD of the total reads mapping to either mouse or viral transcriptome were of viral origin, with a min = 0.014% and a max = 1.16% (at skin of MVA-vaccinated animals), which did not affect downstream analyses of host gene expression.
2). Differential expression analysis.
Using custom scripts in R (http://www.R-project.org), for each tissue type, we filtered the read count matrix to keep genes with at least 50 cpm in 2 samples, and normalized cpm values across samples using the calcNormFactor function in edgeR (Robinson et al., 2010). Batch effects were corrected using limma (Ritchie et al., 2015) for skin and spleen samples for the MVA (d-7) + WR (d0) cohort. We identified genes with at least two-fold expression difference and a Benjamini and Hochberg FDR adjusted p-value < 0.05 (0.01 for skin and spleen) by comparing tissues from all three cohorts (MVA, WR, and MVA+WR) and matching uninfected tissues using limma. Besides the MVA (d-7) + WR (d0) cohort, all other whole-tissue RNA-seq measurements were processed similarly with FDR and fold-change cutoffs as indicated.
3). Gene Ontology (GO) enrichment.
GO term enrichment analysis was done on differentially expressed genes from each tissue using DAVID (Huang et al., 2009).
4). Visualization.
For principal component analysis (PCA), normalized count values were log2 transformed, scaled to unit variance, and centered by subtracting the mean before applying the prcomp function in R. Heatmaps were generated using Gene-E (https://software.broadinstitute.org/GENE-E/).
5). Ligand-receptor pair connectivity mapping.
We assembled a list of known ligand-receptor pairs from mining the literature, KEGG (Kanehisa et al., 2016), and a recent study (Ramilowski et al., 2015). Only known non-membrane bound ligands were selected for further analysis, which yielded a total of 2,198 ligand-receptor pairs (617 receptors and 562 ligands) that were used as inputs for further analysis. Using the list of differentially expressed genes defined in section 3 above, we selected all ligands being upregulated at any time point (FDR adjusted p-value < 0.05) and their corresponding receptors having at least 50 cpm in expression value in any tissue but at the same time point. Ligand-receptor connectivity plots (Figure 3 and S3) were drawn using Circos (Krzywinski et al., 2009).
Single-cell RNA sequencing data analysis
1). Data preprocessing and normalization.
We mapped read 2 sequences onto RefSeq mRNAs using BWA, parsed the output based on cell barcodes stored in read 1 (first 6 bp), and computed gene expression using UMIs stored in read 1 (base 7 to 16) to produce raw single-cell gene expression matrices. For initial filtering, cells with total UMI counts between 100 and 5000 were kept, together with cells in which at least 100 genes were expressed. Furthermore, genes expressed in at least 10 cells were kept, which led to a total of 2,450 cells used for downstream analyses, including 1,476/1,536 cells for experiment 1 (Figure 6B) and 947/1,152 cells for experiment 2 (Figure S6D-E). Furthermore, each experiment included virus-specific memory CD8+ T cells (based on staining with MHC class I tetramers for the VACV B8R20–27 peptide): 375 and 115 for experiments 1 and 2, respectively. Resulting UMI count tables were normalized by dividing UMI counts by the total number of UMIs in that cell, and multiplying by the median of total UMIs across all cells (Grün et al., 2015). To address the issue of low sensitivity in single-cell RNA-seq datasets due to dropout events, weighted pseudocounts were added to genes with missing values by calculating the ratio of the average expression of the gene in all cells and the number of genes detected in that cell (Bailey, 2012; Kharchenko et al., 2014; Tirosh et al., 2016).
2). Differential expression analysis.
To identify genes regulated in single memory CD8+ T cells by WR challenge in MVA-vaccinated mice at lung and liver, we compared control and WR cells within and between both tissue types (i.e., lung control vs lung WR; liver control vs liver WR; lung control vs liver control; lung WR vs liver WR). We used thresholds of at least 3-fold average expression difference and Benjamini and Hochberg FDR adjusted p-value < 0.01 using signal-to-noise ratio test and confirmed by t-test and using SCDE (Kharchenko et al., 2014). For inDrops data, we used SCDE.
3). Score calculations.
Normalized UMI counts were scaled between 0 and 1 by dividing the UMI counts of each gene in each cell by the maximum UMI count of that gene across all cells.
First, to identify TRM cells from single-cell RNA-seq profiles of memory CD8+ T cells extracted from lung and liver, we used a set of 84 genes recently reported as TRM-specific using various infection models (Mackay et al., 2016). We defined as TRM score the average expression of these 84 genes within each cell using normalized, scaled UMI counts. The TRM score threshold used to distinguish between TRM and non-TRM cells was set as one standard deviation over the mean TRM score across all cells, which identified approximately 15% TRM cells. These numbers agreed with the proportion of TRM cells identified using standard flow cytometry analyses for the surface markers CD69 and CD103 (Figure S6H-I), and using intravascular staining (Figure 5F and S5E-F). In addition, to directly measure the potential contribution of differentially regulated genes on TRM scoring, we asked if removing these genes from the TRM-specific gene set would impact our results. We found 2 genes in lung (i.e., Gsg2, Xcl1) and 5 genes in liver (i.e., Cxcr6, Fos, Gpr55, Litaf, Rnf149) that were both present in the TRM 84-gene set and differentially regulated upon WR challenge. Excluding these genes from further analyses had no significant effects on (1) the number of TRM cells identified in both tissues, and (2) the activation score of TRM cells upon WR activation.
Second, to identify cells active in an antigen-specific manner using our single-cell data, we used a set of activation genes by comparing B8R-specific cells in control versus B8R peptide-challenged vaccinated mice (Figure 6C). To select this activation gene set, we used thresholds of at least 3-fold upregulation and Benjamini and Hochberg FDR adjusted p-value < 0.1 using signal-to-noise ratio test and t-test. Next, we intersected this activation gene set with differentially expressed genes upon WR challenge in lung and liver separately, and the resulting set of overlapping genes was used to compute the activation score as the average expression within each cell using normalized, scaled UMI counts.
Lastly, the distributions of TRM and activation scores in control and WR cells for lung and liver were plotted as density plots, and differences between their means across conditions were tested using the Wilcoxon Rank Sum test.
4). Visualization.
To visualize cell-to-cell variations, we first performed PCA on the differentially expressed genes identified above using normalized UMI counts with weighted zero values (Bailey, 2012), which were then log2 transformed, scaled to unit variance, and centered by subtracting the mean before applying the prcomp function in R. Next, to plot cell-to-cell distances in two dimensions, we applied the t-distributed stochastic neighbor embedding (t-SNE) algorithm (Maaten and Hinton, 2008) on scores from the first 4 principal components, which showed maximum variance. t-SNE plots were colored based on gene expression (normalized UMI counts) for indicated genes, or based on TRM and activation scores computed above. Lastly, for heatmap generation in GENE-E, we used normalized UMI counts directly (Figure 6C), or, to ease the visualization of large number of cells (Figure 6B and 7A), we first computed a rolling mean with a window of 5 cells (Paul et al., 2015).
DATA AND SOFTWARE AVAILABILITY
The data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE87633 (whole-tissue RNA-seq) and GSE90697 (single-cell RNA-seq).
Supplementary Material
Table S1. Expression level of Vaccinia virus gene E3L as measured by qPCR, Related to Figure 1 and S1
Table S4. Differentially expressed genes identified from whole-tissue RNA-seq in mice challenged with WR during the memory phase of the response, Related to Figure 4 and S4
Table S6. Whole-tissue and bulk population RNA-seq analysis from wild-type and Ifng−/−, Il22−/− and Csf2−/− mice, Related to Figure 7 and S7
Table S7. List of primers used in this study, Related to STAR Methods
(A) Heatmap showing average transcript abundance for the viral gene E3L (present in both MVA and WR genomes) as measured by qPCR in indicated tissues (n = 2–4).
(B) Bar graph representation of data from (A) for brain, spleen, lung, liver, skin and draining LN (inguinal; dLN). Error bars, SD.
(C-F) Quantification of Vaccinia virus MVA RNA (C-D) and DNA (E-F) in indicated tissues after subcutaneous (s.c.: light gray) or intravenous (i.v.: dark gray) infection with 107 PFUs/mouse. qPCR measurements were performed on viral genomic DNA (H5R and F7L genes) and mRNA (E3L and F7L genes). Standard curves for each primer pair are shown for known quantities of MVA-infected cells (C) and MVA DNA (E), with PCR efficiency and correlation coefficients indicated on each plot. Error bars, SD.
(G) Whole-mount tissue imaging of draining lymph node (inguinal) at 2 days post-infection (d.p.i.) with GFP-expressing MVA administered subcutaneously (s.c.). Inset indicates position of the image on the right. Autofluo., autofluorescence.
(H) Plaque assay images from whole-tissue homogenates (left), and quantification plot (right).
(A) Heatmap showing all 2,018 differentially expressed genes from Figure 2D, and ordered by k-means clustering (k = 12). Values represent log2 fold-change relative to matching, uninfected tissues (FDR-adjusted p-value < 0.05, absolute fold change > 2, n = 4).
(B) Heatmap visualization of gene enrichment analyses for all differentially expressed genes in each tissue type and cohort (combined across all 4 time points). Shown are enrichment p-values for indicated Gene Ontology (GO) terms (rows) (FDR-adjusted p-value < 0.05).
(C-F) Flow cytometry analysis of indicated immune cell subsets in skin (C) and dLN (inguinal; D) upon subcutaneous MVA infection, and in lung and liver for all three cohorts (E-F). For panels C and D, numbers in dot plots indicate percentages relative to parent gate, and bar graphs at the bottom show absolute cell counts per 105 live cells (nd; non-draining LN). Line graphs in panel F show percentages in live cells, corresponding to the composition in whole tissues. VACV, Vaccinia virus. Error bars, SD (n = 2–3).
(A) Inter-organ connectivity of ligand-receptor pairs upon skin vaccination shown as circos plots for indicated times. Line color and thickness reflect the tissue source for upregulated ligands and the number of ligand-receptor (L-R) pairs, respectively.
(B) Bar graphs showing the numbers of L-R pairs emanating from indicated tissues in the MVA + WR (top) or WR (bottom) cohorts.
(C-D) Heatmaps showing the number of L-R pairs found for indicated tissue pairs (C), and the number of occurrences for indicated L-R pairs (D) across all times and cohorts.
(E-F) Weight measurements of wild-type (WT), anti-IFNAR1 or isotype antibody-treated, and Ifnar1−/− mice immunized subcutaneously with MVA at 1 (E) or 7 (F) days prior to intranasal challenge with WR. Error bars, SEM (n = 4). Data are representative of three independent experiments.
(A) Weight (top) and survival (bottom) measurements for indicated mouse cohorts (legend). MVA vaccination routes were subcutaneous at ear or flank skin (s.c.), intra-muscular (i.m.), oral (per os; p.o.), or intranasal (i.n.). Error bars, SEM (n = 4).
(B-C) Flow cytometry analysis of the CD8+ T cell response to MVA vaccination at skin. Shown are dots plots (left) and matching quantifications (right) for live CD3ε+CD8α+ CD44+CD62L− cells that are KLRG1+ (top), or CD62L− and tetramer-positive (bottom), for lung (A) and liver (B). Numbers in dot plots indicate percentages relative to parent gate. Line graphs on the right show percentages relative to parent gate (%) and absolute cell counts per 105 live cells (cells). Error bars, SD (n = 3).
(D) Dot plots showing average log2 fold-change in gene expression in tissues collected at 1.5 day post-WR challenge following 21 days (X axis) or 80 days (Y axis) of MVA vaccination relative to uninfected controls (n = 4). Colored dots represent genes with FDR < 0.05 and absolute log2 fold change > 1 in both day 21 and day 80 (red), day 21 only (green), and day 80 only (blue).
(E) Dot plots showing average log2 fold-change in gene expression (Y axis) in tissues collected at 1.5 day post-WR challenge following 21 days of MVA vaccination relative to uninfected controls against log2 average expression (X axis) (n = 4). Red dots, gene with FDR < 0.05 and absolute log2 fold change > 1.
(F) Gene enrichment analysis for differentially expressed genes in lung and liver after WR challenge 21 days post-MVA vaccination at skin. Shown are enrichment p-values (X axis) for indicated Gene Ontology (GO) terms (Y axis) (FDR-adjusted p-value < 0.05).
(G) Line graphs for selected genes from lung, liver and spleen from indicated cohorts and tissues. Shown are average normalized read counts. Error bars, SEM (n = 4).
(A) Weight measurements of MVA-vaccinated mice treated with isotype control (light grey) or anti-CD8β (dark grey) antibodies, and challenged with WR intranasally (2 weeks after vaccination). Error bars, SEM (n = 4). *; Student’s t-test p-value < 0.05.
(B) Gating strategy for flow cytometry analysis of memory CD8+ T cells. Shown are dissociated lung cells. Numbers in red in bottom plots (1–4) indicate matching gates from the CD44/CD62L gate (bottom left).
(C) Quantification of flow cytometry data from panel A for Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells in percentage of parent gates based on CD44 and CD62L expression (top) and absolute counts per 105 live cells (bottom). Error bars, SD (n = 5).
(D-F) Flow cytometry analysis of Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) memory CD8α+ T cells (gated on live CD3ε+CD44+CD62L− cells) from indicated tissues of mice vaccinated at skin with MVA 3 months (D, left) or 15 months (D, right, and F) earlier. In panels E and F, vascular and parenchymal cells are measured by intravascular staining with CD45 (CD45 i.v.), and bar graphs show quantifications in percentage of parent gate (%) and absolute count per 105 live cells (cells). Error bars, SD (n = 3).
(G) Chimerism measurements after parabiosis surgery. Flow cytometry analysis of CD45.1+ and CD45.2+ cells in peripheral blood mononuclear cells (PBMCs) in indicated parabionts, and bar graph showing quantifications in percentage (%) of parent gate (i.e., all CD45+ cells). Ratio of partner-derived to endogenous blood immune cells reached 50:50 within two weeks after surgery. Error bars, SD (n = 5).
(A-B) Distributions of B8R activation scores in single cells that are tetramer-positive or -negative (tet+/−) from mice immunized with MVA and challenged with B8R peptide (A); and from all single cells shown in Figure 6D for lung (left) and liver (right) (WR challenge; B).
(C) Flow cytometry analysis of intracellular interferon gamma (IFN-γ) protein abundance in memory CD8+ T cells in mice vaccinated with MVA and challenged with WR 21 days later (WR), or left unchallenged as control (Ctrl). Shown are percentages of CD3ε+CD44+CD62L− cells that express IFNg in lung (left) and liver (right) at day 0.5 post-challenge. Error bars, SD (n = 3). p; Student’s t-test p-value.
(D-G) Independent validations of single-cell RNA-seq measurements from lung memory CD8+ T cells. 746 (D-E) and 7,292 (F-G) single cells were processed with a plate-based scRNA-seq protocol (as in Figure 6A) or a droplet-based protocol, respectively. t-SNE plots from both protocols are colored based on treatment (MVA vaccination only vs vaccination + challenge; D and F, left), B8R activation scoring (from D and F, right), and TRM scoring (from E and G, left). Panels on the right in E and G depict the distributions of B8R activation scores (from D and F, right) in TRM cells (from E and G, left).
(H-I) Flow cytometry analysis of TRM markers CD69 and CD103 at lung (top) and liver (bottom) on total (H) and Vaccinia virus peptide B8R-specific (I) memory CD8+ T cells. Matching quantifications in percentage of parent gate and absolute count per 105 live cells are shown on the right of each plot. Error bars, SD (n = 4). *; Student’s t-test p-value < 0.05.
(A) Expression levels of tissue-specific genes in single cells. t-SNE plot from Figure 7B colored based on expression levels (UMI, unique molecular identifiers) of indicated genes.
(B) Flow cytometry analysis of surface protein expression for all 4 markers from panel A. Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells are gated on CD3ε+CD44+CD62L− memory cells. Bar graphs showing quantifications in percentage from indicated gates. Error bars, SD (n = 3–5). *; Student’s t-test p-value < 0.05.
(C) Visualization of transcriptional profiles of 5,000 pooled, sorted CD44−CD62L+CD8+ T cells from lung and liver after MVA vaccination by principal component analysis (PCA). Axes indicate percentage (%) of variance for PC1 and 2.
(D) Expression of tissue-specific memory CD8+ T cell markers shown in Figure 7C in sorted lung and liver CD44−CD62L+ CD8+ T cells. Shown are the fold-change in expression for the indicated genes between memory CD8+ T cells from lung and liver.
(E-F) Heatmap showing genes that are expressed in a tissue-specific manner in memory CD8+ T cells after s.c. MVA vaccination for 3 weeks. Shown are scaled, normalized read counts (rows) for 4 independent replicate mice (columns; n = 4). Colors on the right indicate genes found to be differentially expressed between lung (blue) and liver (orange) memory CD8+ T cells based on scRNA-seq measurements (from Figure 7A).
(G) Tissue-specific expression patterns in single memory CD8+ T cells are preserved upon WR challenge in vaccinated mice. t-SNE plots of single cells from lung (369) and liver (369) in MVA-vaccinated mice challenged with WR (top). Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells (tet+) are labeled in the top right plot for lung (94/369 cells) and liver (94/369 cells). The bottom 3 rows of plots are colored based on expression levels (UMI, unique molecular identifiers) of indicated genes.
Table S2. Differentially expressed genes identified from whole-tissue RNA-seq upon Vaccinia virus vaccination and/or infection, Related to Figure 2 and S2
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Vaccinia Virus (polycolonal) | Thermo Fisher Scientific | Cat#PA1-7258 |
| Anti-Rabbit IgG (H+L), HRP | Thermo Fisher Scientific | Cat#656120 |
| InVivoMAb anti-mouse CD8β (Clone 53-5.8) | BioXCell | Cat#BE0223 |
| InVivoMAb anti-mouse IFNAR-1 (Clone MAR1-5A3) | BioXCell | Cat#BE0241 |
| InVivoMAb Rat IgG1 (Clone HRPN) | BioXCell | Cat#BE0088 |
| InVivoMAb Mouse IgG1 Isotype control | BioXCell | Cat#BE0083 |
| TruStain fcX (anti-mouse CD16/32) (Clone 93) | Biolegend | Cat#101320 |
| Anti-mouse CCL5 PE (Clone 2E9/CCL5) | Biolegend | Cat#149103 |
| Anti-mouse CCR2 APC (Clone 475301) | R&D Systems | Cat#FAB5538A |
| Anti-mouse CD103 Brilliant Violet 510 (Clone 2E7) | Biolegend | Cat#121423 |
| Anti-mouse CD11b Alexa Fluor 700 (Clone M1/70) | Biolegend | Cat#101222 |
| Anti-mouse CD11b Brilliant Violet 421 (Clone M1/70) | Biolegend | Cat#101251 |
| Anti-mouse CD11c APC (Clone N418) | Biolegend | Cat#117310 |
| Anti-mouse CD11c PE/Cy7 (Clone N418) | Biolegend | Cat#117317 |
| Anti-mouse CD19 APC/Cy7 (Clone 6D5) | Biolegend | Cat#115529 |
| Anti-mouse CD19 FITC (Clone 6D5) | Biolegend | Cat#115506 |
| Anti-mouse CD24 PE/Cy7 (Clone M1/69) | Biolegend | Cat#101822 |
| Anti-mouse CD3ε APC (Clone 145-2C11) | Biolegend | Cat#100312 |
| Anti-mouse CD3ε APC/Cy7 (Clone 145-2C11) | Biolegend | Cat#100330 |
| Anti-mouse CD3ε FITC (Clone 145-2C11) | Biolegend | Cat#100306 |
| Anti-mouse CD44 PE/Cy7 (Clone IM7) | Biolegend | Cat#103030 |
| Anti-mouse CD45 Alexa Fluor 700 (Clone 30-F11) | Biolegend | Cat#103128 |
| Anti-mouse CD45 Brilliant Violet 510 (Clone 30-F11) | Biolegend | Cat#103137 |
| Anti-mouse CD45 PE (Clone 30-F11) | Biolegend | Cat#103106 |
| Anti-mouse CD45 PerCP/Cy5.5 (Clone 30-F11) | Biolegend | Cat#103132 |
| Anti-mouse CD45.1 PE (Clone A20) | Biolegend | Cat#110708 |
| Anti-mouse CD45.2 PerCP/Cy5.5 (Clone 104) | Biolegend | Cat#109828 |
| Anti-mouse CD49a PE (Clone HMα1) | Biolegend | Cat#142603 |
| Anti-mouse CD49b FITC (Clone DX5) | Biolegend | Cat#108905 |
| Anti-mouse CD62L APC (Clone MEL-14) | Biolegend | Cat#104412 |
| Anti-mouse CD64 PE (Clone X54-5/7.1) | Biolegend | Cat#139303 |
| Anti-mouse CD69 PE (Clone H1.2F3) | Biolegend | Cat#104508 |
| Anti-mouse CD8α Alexa Fluor 488 (Clone 53-6.7) | Biolegend | Cat#100723 |
| Anti-mouse CD8α PerCP (Clone 53-6.7) | Biolegend | Cat#100731 |
| Anti-mouse CD8β.2 FITC (Clone 53-5.8) | Biolegend | Cat#140403 |
| Anti-mouse CX3CR1 PE (Clone SA011F11) | Biolegend | Cat#149005 |
| Anti-mouse CXCR6 PE (Clone SA051D1) | Biolegend | Cat#151103 |
| Anti-mouse I-A/I-E Brilliant Violet 510 (Clone M5/114.15.2) | Biolegend | Cat#107635 |
| Anti-mouse I-A/I-E PE/Cy7 (Clone M5/114.15.2) | Biolegend | Cat#107630 |
| Anti-mouse IFN-γ FITC (Clone XMG1.2) | Biolegend | Cat#505806 |
| Anti-mouse/human KLRG1 PE (Clone 2F1/KLRG1) | Biolegend | Cat#138407 |
| Anti-mouse Ly6C APC/Cy7 (Clone HK1.4) | Biolegend | Cat#128026 |
| Anti-mouse Ly6C PE (Clone HK1.4) | Biolegend | Cat#128007 |
| Anti-mouse Ly6G FITC (Clone 1A8) | Biolegend | Cat#127606 |
| Antimouse I-A/I-E purified (Clone M5/114.15.2) | Biolegend | Cat#107602 |
| Anti-GFP (Clone GFP-G1) | DSHB | Cat#GFP-G1 |
| Anti-rat IgG (H+L) Alexa Fluor 555 | Thermo Fisher Scientific | Cat#A21434 |
| Anti-mouse IgG (H+L) Alexa Fluor 647 | Thermo Fisher Scientific | Cat#A31571 |
| Bacterial and Virus Strains | ||
| Vaccinia virus strain Western Reserve | BEI Resources | NR-55, Lot 3579605 |
| Vaccinia virus strain Western Reserve expressing eGFP | BEI Resources | NR-624, Lot 3925477 |
| Vaccinia virus strain Modified Vaccinia Ankara | BEI Resources | NR-726, Lot 4225252 |
| Vaccinia virus strain Modified Vaccinia Ankara expressing GFP | Bernard Verrier | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| H-2Kb VACV B8R20–27 (TSYKFESV) tetramer | NIH Tetramer Core Facility | N/A |
| SIGMAFAST OPD (o-Phenylenediamine dihydrochloride) | Sigma-Aldrich | Cat#P9187 |
| Gram Crystal Violet | BD | Cat#212525 |
| FTY720 | Sigma-Aldrich | Cat#SML0700; CAS: 162359-56-0 |
| Lymphocyte Separation Medium | Corning | Cat#25-072 |
| DNase I | Sigma-Aldrich | Cat#D4527 |
| Collagenase, Type IV | Worthington Biochemical Corporation | Cat#LS004188 |
| Liberase TL | Sigma-Aldrich | Cat#5401020001 |
| Percoll PLUS | GE Healthcare Life Sciences | Cat#17-5445-02 |
| RNAlater | Sigma-Aldrich | Cat#R0901 |
| VA-044 | Wako Pure Chemical Industries | Cat#VA-044; CAS: 27776-21-2 |
| Histodenz | Sigma-Aldrich | Cat#D2158; CAS: 66108-95-0 |
| Electrophoretic Tissue Clearing Solution | Logos Biosystems | Cat#C13001 |
| Zamboni Fixative | Newcomer Supply | Cat#1459A |
| Benzyl alcohol | Sigma-Aldrich | Cat#402834; CAS: 100-51-6 |
| Benzyl benzoate | Sigma-Aldrich | Cat#B6630; CAS: 120-51-4 |
| RNase Block Ribonuclease Inhibitor | Agilent Technologies | Cat#300151 |
| Klenow Fragment (3’->5’ exo-) | New England Biolabs | Cat#M0212 |
| TURBO DNase | Thermo Fisher Scientific | Cat#AM2239 |
| Exonuclease I | New England Biolabs | Cat#M0293 |
| Critical Commercial Assays | ||
| BD Cytofix/Cytoperm Fixation/Permeabilization Kit | BD | Cat#554714 |
| High-Capacity cDNA Reverese Transcription Kit | Thermo Fisher Scientific | Cat#4368813 |
| LightCycler 480 SYBR Green I Master | Roche | Cat#04887352001 |
| Stainless Steel Bead, 5 mm | QIAGEN | Cat#69989 |
| Isol-RNA Lysis Reagent | 5Prime | Cat#2302700 |
| Dynabeads MyOne Silane | Thermo Fisher Scientific | Cat#37002D |
| Buffer RLT | QIAGEN | Cat#79216 |
| Quanti-iT RiboGreen RNA Assay Kit | Thermo Fisher Scientific | Cat#R11490 |
| AffinityScript Multiple Temperature Reverse Transcriptase | Agilent Technologies | Cat#600109 |
| RNA Clean & Concentrator-5 | Zymo Research | Cat#R1015 |
| Oligo Clean & Concentrator | Zymo Research | Cat#D4060 |
| Agencourt AMPure XP | Beckman Coulter | Cat#A63880 |
| Q5 Hot Start High-Fidelity 2X Master Mix | New England Biolabs | Cat#M0494 |
| E-Gel EX Agarose Gels, 2% | Thermo Fisher Scientific | Cat#G402002 |
| Qubit dsDNA High Sensitivity Assay Kit | Thermo Fisher Scientific | Cat#Q32854 |
| RiboLock RNase Inhibitor | Thermo Fisher Scientific | Cat#EO0381 |
| Maxima H Minus Reverse Transcriptase | Thermo Fisher Scientific | Cat#EP0753 |
| DNA Clean & Concentrator-5 | Zymo Research | Cat#D4013 |
| Advantage 2 PCR Kit | Clontech | Cat#639206 |
| Nextera XT DNA Library Preparation Kit | Illumina | Cat#FC-131-1024 |
| Deposited Data | ||
| Whole-tissue and bulk sort RNA-seq data sets | This paper | GEO: GSE87633 |
| Single-cell RNA-seq data sets | This paper | GEO: GSE90697 |
| Experimental Models: Cell Lines | ||
| African green monkey: Vero | ATCC | Cat#CCL-81 |
| Syrian golden hamster: BHK-21 | ATCC | Cat#CCL-10 |
| Human: Hela S3 | ATCC | Cat#CCL-2.2 |
| African green monkey: BS-C-1 | ATCC | Cat#CCL-26 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | JAX | stock 000664 |
| Mouse: Csf2−/− (B6.129S-Csf2tm1Mlg/J) | JAX | stock 026812 |
| Mouse: Ifng−/− (B6.129S7-Ifngtm1Ts/J) | JAX | stock 002287 |
| Mouse: Il22−/− (C57BL/6-Il22tm1.1(icre)Stck/J) | JAX | stock 027524 |
| Mouse: Ifnar1−/− | Christophe Benoist, Diane Mathis and Mei X. Wu | N/A |
| Mouse: Tcra−/− | Arlene Sharpe | N/A |
| Oligonucleotides | ||
| Primers for qPCR, custom RNA-seq and single-cell RNA-seq, see Table S7 | This paper | N/A |
| Software and Algorithms | ||
| R version 3.3.2 | The R Project | https://www.r-project.org |
| RStudio Version 1.0.136 | N/A | https://www.rstudio.com |
| CFX Manager | Bio-Rad | http://www.bio-rad.com/en-us/product/cfx-manager-software |
| Fiji version 2.0.0 | NIH | https://fiji.sc |
| ZEN microscope software | ZEISS | https://www.zeiss.com/microscopy/us/products/microscope-software/zen/image-analysis.html |
| MAFFT | Katoh and Standley, 2013 | http://mafft.cbrc.jp/alignment/software/ |
| Circos | Krzywinski et al., 2009 | http://circos.ca/ |
| STAR version 2.5.2b | Dobin et al., 2013 | https://code.google.com/archive/p/rna-star/ |
| FastQC | N/A | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Qualimap | Garcia-Alcalde et al., 2012 | http://qualimap.bioinfo.cipf.es/ |
| MultiQC | N/A | https://github.com/ewels/MultiQC |
| featureCounts version 1.4.4 | Liao et al., 2014 | http://bioinf.wehi.edu.au/featureCounts/ |
| Sailfish version 0.9.2 | Patro et al., 2014 | https://www.cs.cmu.edu/~ckingsf/software/sailfish/ |
| Limma | Ritchie et al., 2015 | http://bioinf.wehi.edu.au/limma/ |
| DAVID | Huang et al., 2009 | https://david.ncifcrf.gov/ |
| GENE-E | The Broad Institute | https://software.broadinstitute.org/GENE-E/ |
ACKNOWLEDGMENTS
We thank the members of the FAS Center for Systems Biology, Diane Mathis, Douglas Melton, David Knipe, William Heath, Laura Mackay and Akiko Iwasaki for valuable discussions; Andrew Murray, Philippe Cluzel, Sean Eddy and Peter Sage for critical reading of the manuscript; the New England Regional Center of Excellence for Biodefense and Emerging Infectious Diseases (NERCE) core, Christine Anderson and John Connor for help with virus handling; Joyce LaVecchio, Silvia Ionescu with FACS; Christian Daly, Claire Reardon with sequencing; Andreas Sjodin, Rory Kirchner, John Hutchinson, Shannan Ho Sui with bioinformatics; David Ginty, Emily Kuehn with whole-mount skin immunostaining; Douglas Richardson with imaging; Allon Klein, Alex Ratner with inDrops; Tian Tian, Thomas Kupper with viral DNA qPCR; and MCB graphics and Sigrid Knemeyer with art work. This work was supported by a Grant-in-Aid for Young Scientists by the Japan Society for the Promotion of Science (A.P.); and the Bauer Fellows Program, the William F. Milton Fund, and the Harvard University Center for AIDS Research (N.C).
REFERENCES
- Anderson KG, Sung H, Skon CN, Lefrancois L, Deisinger A, Vezys V, and Masopust D (2012). Cutting Edge: Intravascular Staining Redefines Lung CD8 T Cell Responses. The Journal of Immunology 189, 2702–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song J-Y, Jacobs H, Haanen JB, and Schumacher TN (2014). T cell memory. Skin-resident memory CD8⁺ T cells trigger a state of tissue-wide pathogen alert. Science 346, 101–105. [DOI] [PubMed] [Google Scholar]
- Bailey S (2012). Principal Component Analysis with Noisy and/or Missing Data. arXiv 1208.4122 [Google Scholar]
- Baruch K, Deczkowska A, David E, Castellano JM, Miller O, Kertser A, Berkutzki T, Barnett-Itzhaki Z, Bezalel D, Wyss-Coray T, et al. (2014). Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 346, 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes M, Klauschen F, Kuchen S, and Germain RN (2013). A systems analysis identifies a feedforward inflammatory circuit leading to lethal influenza infection. Cell 154, 197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K, Wallace J, Kim S-Y, Kalyanasundaram S, Andalman AS, Davidson TJ, Mirzabekov JJ, Zalocusky KA, Mattis J, Denisin AK, et al. (2013). Structural and molecular interrogation of intact biological systems. Nature 497, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrin R, Zhu J, Molony C, Argman C, Parrish ML, Carlson S, Allan MF, Pomp D, and Schadt EE (2009). Multi-tissue coexpression networks reveal unexpected subnetworks associated with disease. Genome Biol 10, R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droujinine IA, and Perrimon N (2016). Interorgan Communication Pathways in Physiology: Focus on Drosophila. Annu. Rev. Genet. 50, 539–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, and Moss B (2001). Characterization of recombinant vaccinia viruses and their products. Curr Protoc Mol Biol Chapter 16, Unit16.18–16.18.11. [DOI] [PubMed] [Google Scholar]
- Earl PL, Cooper N, Wyatt LS, Moss B, and Carroll MW (2001). Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Mol Biol Chapter 16, Unit16.16. [DOI] [PubMed] [Google Scholar]
- Eickhoff S, Brewitz A, Gerner MY, Klauschen F, Komander K, Hemmi H, Garbi N, Kaisho T, Germain RN, and Kastenmüller W (2015). Robust Anti-viral Immunity Requires Multiple Distinct T Cell-Dendritic Cell Interactions. Cell 162, 1322–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, and Rudensky AY (2016). Hallmarks of Tissue-Resident Lymphocytes. Cell 164, 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V, Cozijnsen A, Collins N, et al. (2016). Liver-Resident Memory CD8(+) T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity 45, 889–902. [DOI] [PubMed] [Google Scholar]
- Garcia-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Gotz S, Tarazona S, Dopazo J, Meyer TF, and Conesa A (2012). Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28, 2678–2679. [DOI] [PubMed] [Google Scholar]
- Gómez CE, Nájera JL, Domingo-Gil E, Ochoa-Callejero L, Gonzalez-Aseguinolaza G, and Esteban M (2007). Virus distribution of the attenuated MVA and NYVAC poxvirus strains in mice. J. Gen. Virol. 88, 2473–2478. [DOI] [PubMed] [Google Scholar]
- Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, and van Oudenaarden A (2015). Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 1–23. [DOI] [PubMed] [Google Scholar]
- Guilliams M, Dutertre C-A, Scott CL, McGovern N, Sichien D, Chakarov S, Van Gassen S, Chen J, Poidinger M, De Prijck S, et al. (2016). Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 45, 669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng TSP, Painter MW, Immunological Genome Project Consortium (2008). The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Hermesh T, Moltedo B, Moran TM, and López CB (2010). Antiviral Instruction of Bone Marrow Leukocytes during Respiratory Viral Infections. Cell Host and Microbe 7, 343–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M, and Pircher H (2011). E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc. Natl. Acad. Sci. U.S.a. 108, 16741–16746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Huang T, Zhang J, Xie L, Dong X, Zhang L, Cai Y-D, and Li Y-X (2011). Crosstissue coexpression network of aging. Omics 15, 665–671. [DOI] [PubMed] [Google Scholar]
- Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, and Linnarsson S (2013). Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Meth 11, 163–166. [DOI] [PubMed] [Google Scholar]
- Jiang X, Clark RA, Liu L, Wagers AJ, Fuhlbrigge RC, and Kupper TS (2012). Skin infection generates non-migratory memory CD8+ TRM cells providing global skin immunity. Nature 483, 227–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, and Tanabe M (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research 44, D457–D462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Choi Y, Wang P, Belt Davis D, Rabaglia ME, Oler AT, Stapleton DS, Argmann C, Schueler KL, Edwards S, et al. (2008). A gene expression network model of type 2 diabetes links cell cycle regulation in islets with diabetes susceptibility. Genome Research 18, 706–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nat Meth 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, and Marra MA (2009). Circos: An information aesthetic for comparative genomics. Genome Research 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw BJ, Zhang N, Marshall HD, Staron MM, Guan T, Hu Y, Cauley LS, Craft J, and Kaech SM (2014). CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity 41, 633–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvau G, Boutet M, Williams TM, Chin SS, and Chorro L (2016). Memory CD8+ T Cells: Innate-Like Sensors and Orchestrators of Protection. Trends in Immunology 37, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu L, Fuhlbrigge RC, Karibian K, Tian T, and Kupper TS (2006). Dynamic programming of CD8+ T cell trafficking after live viral immunization. Immunity 25, 511–520. [DOI] [PubMed] [Google Scholar]
- Loparev VN, Massung RF, Esposito JJ, and Meyer H (2001). Detection and differentiation of old world orthopoxviruses: restriction fragment length polymorphism of the crmB gene region. J. Clin. Microbiol. 39, 94–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López CB, and Hermesh T (2011). Systemic responses during local viral infections: type I IFNs sound the alarm. Current Opinion in Immunology 23, 495–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maaten LVD, and Hinton G (2008). Visualizing Data using t-SNE. Journal of Machine Learning Research 9, 2579–2605. [Google Scholar]
- Mackay LK, Minnich M, Kragten NAM, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. (2016). Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 352, 459–463. [DOI] [PubMed] [Google Scholar]
- Mahe B, Vogt A, Liard C, Duffy D, Abadie V, Bonduelle O, Boissonnas A, Sterry W, Verrier B, Blume-Peytavi U, et al. (2009). Nanoparticle-based targeting of vaccine compounds to skin antigen-presenting cells by hair follicles and their transport in mice. J. Invest. Dermatol. 129, 1156–1164. [DOI] [PubMed] [Google Scholar]
- Manz MG, and Boettcher S (2014). Emergency granulopoiesis. Nature Reviews Immunology 14, 302–314. [DOI] [PubMed] [Google Scholar]
- Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey KA, Barber DL, Kawamura KS, et al. (2010). Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr A, Büttner M, Pawlas S, Erfle V, Mayr B, Brunner R, and Osterkorn K (1986). [Comparative studies of the immunostimulating (paramunizing) effectiveness of BCG, levamisole, Corynebacterium parvum and preparations of pock viruses in various in vivo and in vitro tests]. Zentralblatt Veterinarmedizin Reihe B 33, 321–339. [PubMed] [Google Scholar]
- McNab F, Mayer-Barber K, Sher A, Wack A, and O’Garra A (2015). Type I interferons in infectious disease. Nature Reviews Immunology 15, 87–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R (2008). Origin and physiological roles of inflammation. Nature 454, 428–435. [DOI] [PubMed] [Google Scholar]
- Meisinger-Henschel C, Schmidt M, Lukassen S, Linke B, Krause L, Konietzny S, Goesmann A, Howley P, Chaplin P, Suter M, et al. (2007). Genomic sequence of chorioallantois vaccinia virus Ankara, the ancestor of modified vaccinia virus Ankara. Journal of General Virology 88, 3249–3259. [DOI] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. (2015). The human transcriptome across tissues and individuals. Science 348, 660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Sutter G, and Mayr A (1991). Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72 (Pt 5), 1031–1038. [DOI] [PubMed] [Google Scholar]
- Moss B (2011). Smallpox vaccines: targets of protective immunity. Immunol. Rev. 239, 8–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafavi S, Yoshida H, Moodley D, LeBoité H, Rothamel K, Raj T, Ye CJ, Chevrier N, Zhang S-Y, Feng T, et al. (2016). Parsing the Interferon Transcriptional Network and Its Disease Associations. Cell 1–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SN, and Mackay LK (2016). Tissue-resident memory T cells: local specialists in immune defence. Nature Reviews Immunology 16, 79–89. [DOI] [PubMed] [Google Scholar]
- Okabe Y, and Medzhitov R (2014). Tissue-Specific Signals Control Reversible Program of Localization and Functional Polarization of Macrophages. Cell 157, 832–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, and Medzhitov R (2016). Tissue biology perspective on macrophages. Nat Immunol 17, 9–17. [DOI] [PubMed] [Google Scholar]
- Panduro M, Benoist C, and Mathis D (2016). Tissue Tregs. Annu. Rev. Immunol. 34, 609–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Mount SM, and Kingsford C (2014). Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nature Biotechnology 32, 462–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F, Arkin Y, Giladi A, Jaitin DA, Kenigsberg E, Keren-Shaul H, Winter D, Lara-Astiaso D, Gury M, Weiner A, et al. (2015). Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell 163, 1663–1677. [DOI] [PubMed] [Google Scholar]
- Pillai VKB, Kannanganat S, Penaloza-MacMaster P, Chennareddi L, Robinson HL, Blackwell J, and Amara RR (2011). Different patterns of expansion, contraction and memory differentiation of HIV-1 Gag-specific CD8 T cells elicited by adenovirus type 5 and modified vaccinia Ankara vaccines. Vaccine 29, 5399–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, Kloppmann E, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, et al. (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nature Communications 6, 7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer MJ, Nolz JC, and Harty JT (2013). Pathogen-Specific Inflammatory Milieux Tune the Antigen Sensitivity of CD8. Immunity 38, 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK (2015). limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43, e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, and Smyth GK (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat R, Ouyang W, and Wolk K (2014). Therapeutic opportunities of the IL-22-IL-22R1 system. Nat Rev Drug Discov 13, 21–38. [DOI] [PubMed] [Google Scholar]
- Salek-Ardakani S, Moutaftsi M, Sette A, and Croft M (2011). Targeting OX40 promotes lung-resident memory CD8 T cell populations that protect against respiratory poxvirus infection. Journal of Virology 85, 9051–9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel JM, and Masopust D (2014). Tissue-resident memory T cells. Immunity 41, 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, and Rice CM (2014). Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Cacchiarelli D, Semrau S, van Oudenaarden A, and Mikkelsen TS (2014). Characterization of directed differentiation by high-throughput single-cell RNA-Seq. bioRxiv 003236 [Google Scholar]
- Spitzer MH, Gherardini PF, Fragiadakis GK, Bhattacharya N, Yuan RT, Hotson AN, Finck R, Carmi Y, Zunder ER, Fantl WJ, et al. (2015). An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stary G, Olive A, Radovic-Moreno AF, Gondek D, Alvarez D, Basto PA, Perro M, Vrbanac VD, Tager AM, Shi J, et al. (2015). VACCINES. A mucosal vaccine against Chlamydia trachomatis generates two waves of protective memory T cells. Science 348, aaa8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, Southern PJ, and Masopust D (2015). Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 161, 737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S, Yagi H, Hakata Y, Motozono C, McMaster SR, Masumoto T, Fujisawa M, Chikaishi T, Komeda J, Itoh J, et al. (2016). Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med. 213, 3057–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treweek JB, Chan KY, Flytzanis NC, Yang B, Deverman BE, Greenbaum A, Lignell A, Xiao C, Cai L, Ladinsky MS, et al. (2015). Whole-body tissue stabilization and selective extractions via tissue-hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat Protoc 10, 1860–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilsmeier B (1999). Paramunity-inducing effects of vaccinia strain MVA. Berl. Munch. Tierarztl. Wochenschr. 112, 329–333. [PubMed] [Google Scholar]
- Wakim LM, Gupta N, Mintern JD, and Villadangos JA (2013). Enhanced survival of lung tissue-resident memory CD8⁺ T cells during infection with influenza virus due to selective expression of IFITM3. Nat Immunol 14, 238–245. [DOI] [PubMed] [Google Scholar]
- Wright DE, Wagers AJ, Gulati AP, Johnson FL, and Weissman IL (2001). Physiological migration of hematopoietic stem and progenitor cells. Science (New York, NY) 294, 1933–1936. [DOI] [PubMed] [Google Scholar]
- Wyatt LS, Earl PL, Eller LA, and Moss B (2004). Highly attenuated smallpox vaccine protects mice with and without immune deficiencies against pathogenic vaccinia virus challenge. Proceedings of the National Academy of Sciences 101, 4590–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilionis R, Nainys J, Veres A, Savova V, Zemmour D, Klein AM, and Mazutis L (2017). Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 12, 44–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Expression level of Vaccinia virus gene E3L as measured by qPCR, Related to Figure 1 and S1
Table S4. Differentially expressed genes identified from whole-tissue RNA-seq in mice challenged with WR during the memory phase of the response, Related to Figure 4 and S4
Table S6. Whole-tissue and bulk population RNA-seq analysis from wild-type and Ifng−/−, Il22−/− and Csf2−/− mice, Related to Figure 7 and S7
Table S7. List of primers used in this study, Related to STAR Methods
(A) Heatmap showing average transcript abundance for the viral gene E3L (present in both MVA and WR genomes) as measured by qPCR in indicated tissues (n = 2–4).
(B) Bar graph representation of data from (A) for brain, spleen, lung, liver, skin and draining LN (inguinal; dLN). Error bars, SD.
(C-F) Quantification of Vaccinia virus MVA RNA (C-D) and DNA (E-F) in indicated tissues after subcutaneous (s.c.: light gray) or intravenous (i.v.: dark gray) infection with 107 PFUs/mouse. qPCR measurements were performed on viral genomic DNA (H5R and F7L genes) and mRNA (E3L and F7L genes). Standard curves for each primer pair are shown for known quantities of MVA-infected cells (C) and MVA DNA (E), with PCR efficiency and correlation coefficients indicated on each plot. Error bars, SD.
(G) Whole-mount tissue imaging of draining lymph node (inguinal) at 2 days post-infection (d.p.i.) with GFP-expressing MVA administered subcutaneously (s.c.). Inset indicates position of the image on the right. Autofluo., autofluorescence.
(H) Plaque assay images from whole-tissue homogenates (left), and quantification plot (right).
(A) Heatmap showing all 2,018 differentially expressed genes from Figure 2D, and ordered by k-means clustering (k = 12). Values represent log2 fold-change relative to matching, uninfected tissues (FDR-adjusted p-value < 0.05, absolute fold change > 2, n = 4).
(B) Heatmap visualization of gene enrichment analyses for all differentially expressed genes in each tissue type and cohort (combined across all 4 time points). Shown are enrichment p-values for indicated Gene Ontology (GO) terms (rows) (FDR-adjusted p-value < 0.05).
(C-F) Flow cytometry analysis of indicated immune cell subsets in skin (C) and dLN (inguinal; D) upon subcutaneous MVA infection, and in lung and liver for all three cohorts (E-F). For panels C and D, numbers in dot plots indicate percentages relative to parent gate, and bar graphs at the bottom show absolute cell counts per 105 live cells (nd; non-draining LN). Line graphs in panel F show percentages in live cells, corresponding to the composition in whole tissues. VACV, Vaccinia virus. Error bars, SD (n = 2–3).
(A) Inter-organ connectivity of ligand-receptor pairs upon skin vaccination shown as circos plots for indicated times. Line color and thickness reflect the tissue source for upregulated ligands and the number of ligand-receptor (L-R) pairs, respectively.
(B) Bar graphs showing the numbers of L-R pairs emanating from indicated tissues in the MVA + WR (top) or WR (bottom) cohorts.
(C-D) Heatmaps showing the number of L-R pairs found for indicated tissue pairs (C), and the number of occurrences for indicated L-R pairs (D) across all times and cohorts.
(E-F) Weight measurements of wild-type (WT), anti-IFNAR1 or isotype antibody-treated, and Ifnar1−/− mice immunized subcutaneously with MVA at 1 (E) or 7 (F) days prior to intranasal challenge with WR. Error bars, SEM (n = 4). Data are representative of three independent experiments.
(A) Weight (top) and survival (bottom) measurements for indicated mouse cohorts (legend). MVA vaccination routes were subcutaneous at ear or flank skin (s.c.), intra-muscular (i.m.), oral (per os; p.o.), or intranasal (i.n.). Error bars, SEM (n = 4).
(B-C) Flow cytometry analysis of the CD8+ T cell response to MVA vaccination at skin. Shown are dots plots (left) and matching quantifications (right) for live CD3ε+CD8α+ CD44+CD62L− cells that are KLRG1+ (top), or CD62L− and tetramer-positive (bottom), for lung (A) and liver (B). Numbers in dot plots indicate percentages relative to parent gate. Line graphs on the right show percentages relative to parent gate (%) and absolute cell counts per 105 live cells (cells). Error bars, SD (n = 3).
(D) Dot plots showing average log2 fold-change in gene expression in tissues collected at 1.5 day post-WR challenge following 21 days (X axis) or 80 days (Y axis) of MVA vaccination relative to uninfected controls (n = 4). Colored dots represent genes with FDR < 0.05 and absolute log2 fold change > 1 in both day 21 and day 80 (red), day 21 only (green), and day 80 only (blue).
(E) Dot plots showing average log2 fold-change in gene expression (Y axis) in tissues collected at 1.5 day post-WR challenge following 21 days of MVA vaccination relative to uninfected controls against log2 average expression (X axis) (n = 4). Red dots, gene with FDR < 0.05 and absolute log2 fold change > 1.
(F) Gene enrichment analysis for differentially expressed genes in lung and liver after WR challenge 21 days post-MVA vaccination at skin. Shown are enrichment p-values (X axis) for indicated Gene Ontology (GO) terms (Y axis) (FDR-adjusted p-value < 0.05).
(G) Line graphs for selected genes from lung, liver and spleen from indicated cohorts and tissues. Shown are average normalized read counts. Error bars, SEM (n = 4).
(A) Weight measurements of MVA-vaccinated mice treated with isotype control (light grey) or anti-CD8β (dark grey) antibodies, and challenged with WR intranasally (2 weeks after vaccination). Error bars, SEM (n = 4). *; Student’s t-test p-value < 0.05.
(B) Gating strategy for flow cytometry analysis of memory CD8+ T cells. Shown are dissociated lung cells. Numbers in red in bottom plots (1–4) indicate matching gates from the CD44/CD62L gate (bottom left).
(C) Quantification of flow cytometry data from panel A for Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells in percentage of parent gates based on CD44 and CD62L expression (top) and absolute counts per 105 live cells (bottom). Error bars, SD (n = 5).
(D-F) Flow cytometry analysis of Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) memory CD8α+ T cells (gated on live CD3ε+CD44+CD62L− cells) from indicated tissues of mice vaccinated at skin with MVA 3 months (D, left) or 15 months (D, right, and F) earlier. In panels E and F, vascular and parenchymal cells are measured by intravascular staining with CD45 (CD45 i.v.), and bar graphs show quantifications in percentage of parent gate (%) and absolute count per 105 live cells (cells). Error bars, SD (n = 3).
(G) Chimerism measurements after parabiosis surgery. Flow cytometry analysis of CD45.1+ and CD45.2+ cells in peripheral blood mononuclear cells (PBMCs) in indicated parabionts, and bar graph showing quantifications in percentage (%) of parent gate (i.e., all CD45+ cells). Ratio of partner-derived to endogenous blood immune cells reached 50:50 within two weeks after surgery. Error bars, SD (n = 5).
(A-B) Distributions of B8R activation scores in single cells that are tetramer-positive or -negative (tet+/−) from mice immunized with MVA and challenged with B8R peptide (A); and from all single cells shown in Figure 6D for lung (left) and liver (right) (WR challenge; B).
(C) Flow cytometry analysis of intracellular interferon gamma (IFN-γ) protein abundance in memory CD8+ T cells in mice vaccinated with MVA and challenged with WR 21 days later (WR), or left unchallenged as control (Ctrl). Shown are percentages of CD3ε+CD44+CD62L− cells that express IFNg in lung (left) and liver (right) at day 0.5 post-challenge. Error bars, SD (n = 3). p; Student’s t-test p-value.
(D-G) Independent validations of single-cell RNA-seq measurements from lung memory CD8+ T cells. 746 (D-E) and 7,292 (F-G) single cells were processed with a plate-based scRNA-seq protocol (as in Figure 6A) or a droplet-based protocol, respectively. t-SNE plots from both protocols are colored based on treatment (MVA vaccination only vs vaccination + challenge; D and F, left), B8R activation scoring (from D and F, right), and TRM scoring (from E and G, left). Panels on the right in E and G depict the distributions of B8R activation scores (from D and F, right) in TRM cells (from E and G, left).
(H-I) Flow cytometry analysis of TRM markers CD69 and CD103 at lung (top) and liver (bottom) on total (H) and Vaccinia virus peptide B8R-specific (I) memory CD8+ T cells. Matching quantifications in percentage of parent gate and absolute count per 105 live cells are shown on the right of each plot. Error bars, SD (n = 4). *; Student’s t-test p-value < 0.05.
(A) Expression levels of tissue-specific genes in single cells. t-SNE plot from Figure 7B colored based on expression levels (UMI, unique molecular identifiers) of indicated genes.
(B) Flow cytometry analysis of surface protein expression for all 4 markers from panel A. Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells are gated on CD3ε+CD44+CD62L− memory cells. Bar graphs showing quantifications in percentage from indicated gates. Error bars, SD (n = 3–5). *; Student’s t-test p-value < 0.05.
(C) Visualization of transcriptional profiles of 5,000 pooled, sorted CD44−CD62L+CD8+ T cells from lung and liver after MVA vaccination by principal component analysis (PCA). Axes indicate percentage (%) of variance for PC1 and 2.
(D) Expression of tissue-specific memory CD8+ T cell markers shown in Figure 7C in sorted lung and liver CD44−CD62L+ CD8+ T cells. Shown are the fold-change in expression for the indicated genes between memory CD8+ T cells from lung and liver.
(E-F) Heatmap showing genes that are expressed in a tissue-specific manner in memory CD8+ T cells after s.c. MVA vaccination for 3 weeks. Shown are scaled, normalized read counts (rows) for 4 independent replicate mice (columns; n = 4). Colors on the right indicate genes found to be differentially expressed between lung (blue) and liver (orange) memory CD8+ T cells based on scRNA-seq measurements (from Figure 7A).
(G) Tissue-specific expression patterns in single memory CD8+ T cells are preserved upon WR challenge in vaccinated mice. t-SNE plots of single cells from lung (369) and liver (369) in MVA-vaccinated mice challenged with WR (top). Vaccinia virus peptide B8R-specific (H2-Kb B8R20–27) CD8+ T cells (tet+) are labeled in the top right plot for lung (94/369 cells) and liver (94/369 cells). The bottom 3 rows of plots are colored based on expression levels (UMI, unique molecular identifiers) of indicated genes.
Table S2. Differentially expressed genes identified from whole-tissue RNA-seq upon Vaccinia virus vaccination and/or infection, Related to Figure 2 and S2
Data Availability Statement
The data generated in this paper has been deposited in the Gene Expression Omnibus (GEO) under accession numbers GSE87633 (whole-tissue RNA-seq) and GSE90697 (single-cell RNA-seq).