SUMMARY
The trachea and esophagus arise from the separation of a common foregut tube during early fetal development. Mutations in key signaling pathways such as Hedgehog (HH)/Gli can disrupt tracheoesophageal (TE) morphogenesis and cause life-threatening birth defects (TEDs), however the underlying cellular mechanisms are unknown. Here we use mouse and Xenopus to define the HH/Gli-dependent processes orchestrating TE morphogenesis. We show that downstream of Gli the Foxf1+ splanchnic mesenchyme promotes medial constriction of the foregut at the boundary between the presumptive Sox2+ esophageal and Nkx2–1+ tracheal epithelium. We identify a unique boundary epithelium co-expressing Sox2 and Nkx2–1 that fuses to form a transient septum. Septum formation and resolution into distinct trachea and esophagus requires endosome-mediated epithelial remodeling involving the small GTPase Rab11, and localized extracellular matrix degradation. These are disrupted in Gli-deficient embryos. This work provides a new mechanistic framework for TE morphogenesis and informs the cellular basis of human TEDs.
Graphical Abstract
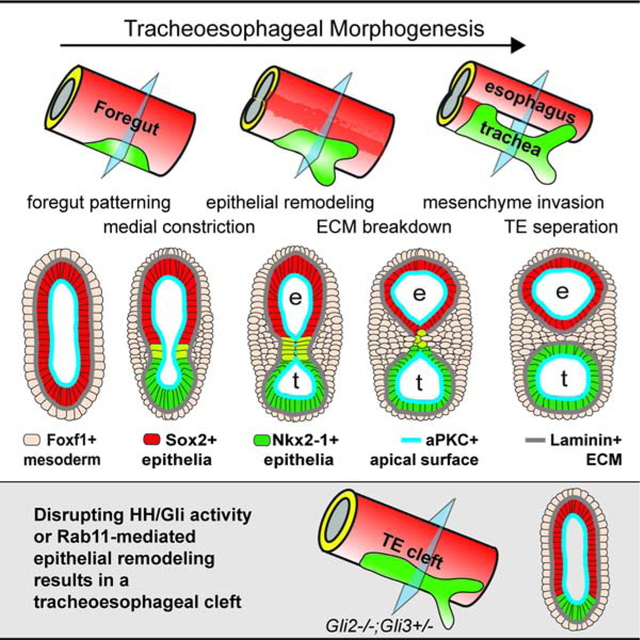
eTOC summary:
The etiology of tracheoesophageal birth defects is unknown. Nasr et al define the conserved cellular mechanisms of foregut morphogenesis in Xenopus and mouse, and show how disruption of Rab11-mediated epithelial remodeling downstream of Hedgehog/Gli signaling results in tracheoesophageal clefts similar to human patients.
INTRODUCTION
Between 25–35 days of human gestation, the fetal gut tube separates into the distinct trachea and esophagus (Billmyre et al., 2015; Que, 2015). Disruptions in this process result in life-threatening defects that impair neonatal respiration and feeding, including esophageal atresia (EA), tracheal atresia (TA), and/or tracheoesophageal fistula (TEF) (Brosens et al., 2014). The etiology of TEDs, occurring in ~1:3500 births, is poorly understood (Shaw-Smith, 2006). Even when patient genetics or animal models have revealed the genes involved, how mutations result in TEDs is unclear because the cellular basis of TE morphogenesis are poorly defined.
Studies in mouse and Xenopus have shown that TE development is initiated by a signaling cascade involving HH, WNT and other signals between the foregut endoderm and the surrounding splanchnic mesoderm. These signals pattern the epithelium into a ventral respiratory domain expressing the transcription factor Nkx2–1 and a dorsal esophageal domain expressing the transcription factor Sox2 around mouse embryonic day (E) 9.0 and Xenopus NF35, 50 hours post fertilization (hpf) (Hines and Sun, 2014; Rankin et al., 2016). Over the next few days, the foregut separates into distinct TE tubes, with paired lung buds emerging from the posterior aspect of the Nkx2–1+ domain via an Fgf10-mediated mechanism that is distinct from TE separation (Hines and Sun, 2014).
Mouse mutations in these patterning genes can result in TEDs similar to those seen in patients. For example, Sox2 and Nkx2–1 knockouts result in EA and TA, respectively (Minoo et al., 1999; Que et al., 2007; Trisno et al., 2018), while HH pathway mutations, such as in the ligand Shh or the transcription factors Gli2 and Gli3, can cause a spectrum of defects ranging from EA/TEF to laryngotracheoesophageal clefts (LTECs) (Litingtung et al., 1998; Motoyama et al., 1998; Rankin et al., 2016; Tabler et al., 2017). How these mutations result in TEDs is unclear since the cellular processes that HH regulates in this context are unknown. While a number of models for TE morphogenesis have been postulated (Billmyre et al., 2015; Que, 2015), the underlying cellular mechanisms remain to be elucidated.
Here we use Xenopus and mouse to define the conserved cellular mechanisms orchestrating TE morphogenesis. We show that HH/Gli signaling regulates multiple steps of TE separation, including dorsal-ventral (D-V) patterning, medial constriction and endosome-mediated epithelial remodeling. These results provide a mechanistic foundation for TE morphogenesis to inform the genotype-phenotype basis of human TEDs.
RESULTS and DISCUSSION
TE Morphogenesis is Conserved in Xenopus and Mouse
To define the cell biology of TE morphogenesis, we took a comparative approach using mouse and Xenopus. TE development appears to be conserved with patterning of the foregut epithelium into Sox2+ dorsal and Nkx2–1+ ventral domains by NF35 in Xenopus and E9.5 in mouse. The foregut then separates into distinct TE tubes by NF42 (80 hpf) in Xenopus and E11.5 in mice (Figures 1A–C’ and S1A–F’) (Rankin et al., 2015). Close examination of Xenopus embryos over the course of TE morphogenesis allowed us to classify the process into four major steps: 1) D-V patterning; 2) medial constriction at the Sox2-Nkx2–1 boundary; 3) epithelial fusion and remodeling of a transient septum; and 4) mesenchymal invasion separating the TE tubes. We investigated the cellular basis of each step, first using Xenopus where we could screen many potential mechanisms, followed by a comparison to mouse.
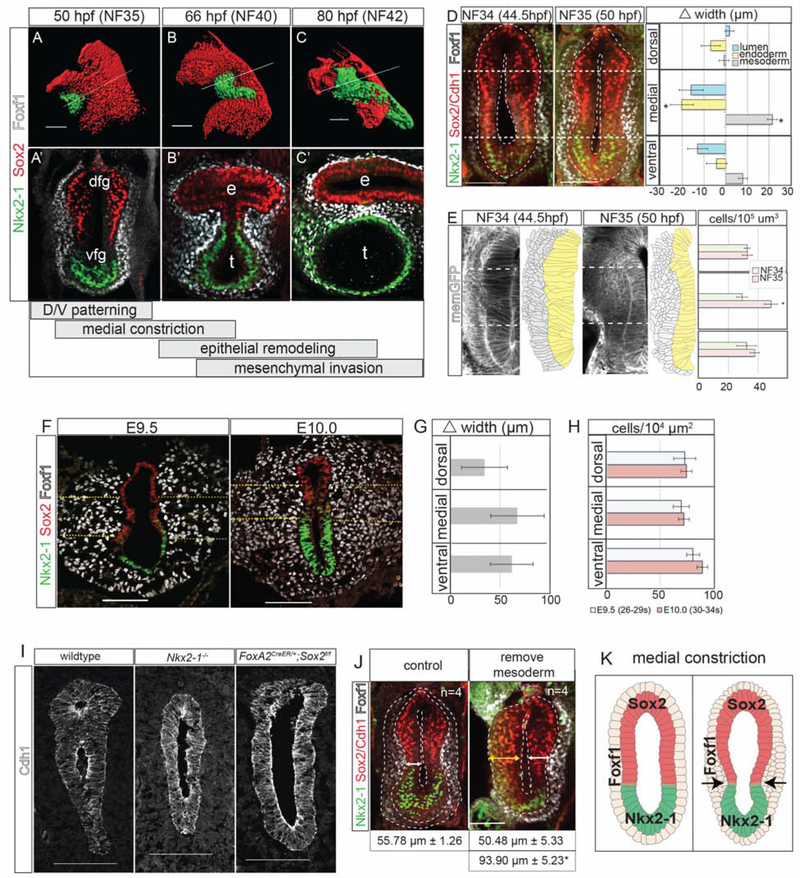
Figure 1: Foxf1+ Mesenchyme Promotes Medial Constriction at the Sox2-Nkx2–1 Boundary.
1A–C’: Immunostaining showing TE morphogenesis in Xenopus (X.) laevis. Scale bar, 100 μm.
1D: Medial constriction in X. laevis with quantification of the difference in foregut width between NF34 and NF35. Scale bar, 100 μm. Difference of means test,*p<0.5.
1E: Transgenic membrane-GFP X. laevis show increased medial mesenchyme cell density. Scale bar, 100 μm. Student’s two-tailed t-test,*p<0.05.
1F: Medial constriction mouse embryos showing. Dashed yellow lines denote medial mesoderm. Scale bar, 100 μm.
1G: Average change in mesoderm width between E9.5 and E10.0 was not significant. Difference of means test.
1H. Average mesoderm cell density at E9.5 and E10.0 was not significantly different. Student’s two-tailed t-test.
1I: Nkx2–1 and Sox2 mouse mutants (E10.5–E11) fail to undergo medial constriction. Scale bar, 100 μm.
1J: Removal of the lateral plate mesoderm prevents medial constriction in X. laevis embryos. Student’s two-tailed t-test between side without mesoderm, and either control embryos or the contralateral side.*p<0.05. Scale bar, 100 μm.
1I: Summary of medial constriction.
Foxf1+ Mesenchyme Promotes Medial Constriction at the Sox2-Nkx2–1 Boundary
In Xenopus, the first morphological indication of TE separation was a medial constriction of the foregut at the Sox2-Nkx2–1 boundary by NF35, with a significant narrowing of the gut tube lumen coincident with thickening of the medial Foxf1+ splanchnic mesoderm (Figure 1D). Analysis of transgenic membrane-GFP embryos revealed that the mesoderm thickening was not due to increased mesodermal cell size or cell shape changes, but rather to a condensation of Foxf1+ mesodermal cells (Figures 1E and S1K). This was accompanied by the epithelium transitioning from a thick pseudostratified layer at NF34 to a thinner columnar epithelium at NF35 (Figure 1E). Phospho-Histone H3 staining indicated that the medial mesenchyme had a slightly higher proliferation rate than the dorsal or ventral regions, which may in part explain the localized increase in mesenchymal cell numbers (Figure S1G,H).
The mouse foregut also constricted at the Sox2-Nkx2–1 boundary between E9.5 and E10.0 (Figure 1F), but there was no relative increase in medial mesoderm thickness, cell density or proliferation compared to the ventral or dorsal tissue as observed in Xenopus (Figures 1G, H and S1I–L). However, in both species the Foxf1+ cells were more tightly packed against the epithelium at the constriction point (Figures 1A–F and S1I), suggesting a close association of the mesenchyme with the basement membrane in this region.
Since foregut constriction occurs at the Sox2-Nkx2–1 boundary, we re-examined mouse Nkx2–1 and Sox2 mutants where the foregut does not separate, but the underlying mechanisms have not been reported. Both Nkx2–1 and Sox2 mutants failed to undergo medial constriction (Figure 1I), indicating that proper D-V patterning and the function of these transcription factors are essential to initiate TE morphogenesis. We postulate that the Sox2-Nkx2–1 boundary might have some inherent information that regulates local mesenchyme behavior. These data help explain why human SOX2 mutations frequently cause EA; if the SOX2+ domain is too small, foregut constriction may not occur or might initiate too dorsally, resulting in insufficient esophageal tissue.
Since mesenchymal condensations can promote epithelial bending in some contexts (Walton et al., 2012), we tested whether the mesoderm was required for medial constriction in Xenopus by surgically removing the splanchnic mesenchyme from one side of the NF32 foregut. Imaging at NF35 revealed a failure of medial constriction and epithelial thinning relative to the contralateral control side (Figure 1J), suggesting that the mesoderm is required for medial constriction, possibly by exerting a pushing force on the epithelium. Together, these data indicate that medial constriction at the Sox2-Nkx2–1 boundary is the critical first step in TE morphogenesis (Figure 1K) and that disruptions in this process can lead to TEDs.
Rab11 and Endosome-Mediated Epithelial Remodeling are Required for TE Septation
As the foregut constricts, the opposing epithelial walls come into contact, forming a transient epithelial septum (Figures 2A–C). In Xenopus this septum was 5–7 cells in length, whereas in mouse the fusion point was only few cells long. Close examination revealed a unique cell population in the septum that co-expresses Nkx2–1 and Sox2 (Figures 2D and S2A). We examined whether modulators of epithelial behavior were differentially active at the Sox2+/Nkx2–1+ contact point. Before collapse of the medial lumen, the endoderm is a polarized epithelium with aPKC on the apical/luminal surface and Cdh1 enriched in adherens junctions and on the basolateral surface. As the epithelial walls touched, aPKC was specifically reduced at the contact point (Figure 2B–C), where the transient septum formed like a closing zipper between the presumptive esophagus and trachea. Concurrently, Cdh1 and Integrin became enriched on what was previously the apical surface (Figure 2B–F), suggesting that cell-cell adhesion holds the two sides together.
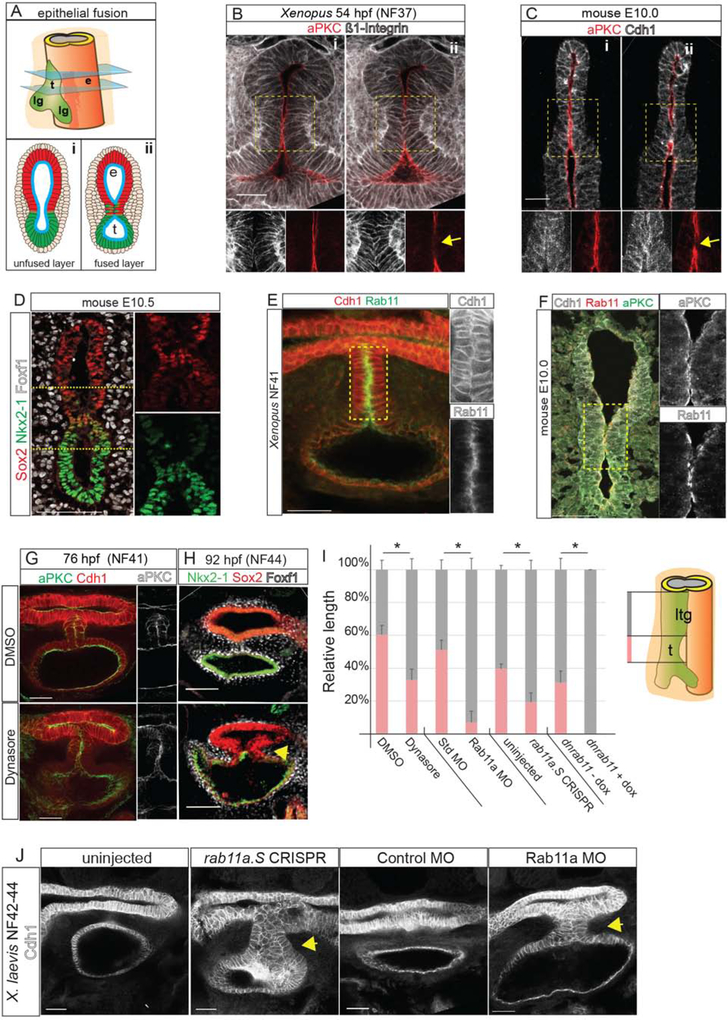
Figure 2: Endosome-Mediated Epithelial Remodeling is Required for TE Septation.
2A: Model of epithelial fusion.
2B–C: Sequential optical sections of X. laevis (B) and mouse (C) embryo immunostaining showing loss of aPKC and increased Integrin or Cdh1 at the contact point (arrow, ii). Scale bar, 50 μm.
2D: A unique population of cells co-expressing of Sox2 and Nkx2–1 in the mouse foregut. Scale bar, 100 μm.
2E: Rab11 and Cdh1 are enriched in the X. laevis septum. Scale bar, 50 μm.
2F: Rab11, aPKC and Cdh1 enriched at the fusion point in mouse. Scale bar, 50 μm.
2G–H: Inhibition of endosome recycling by Dynasore treatment of X. laevis (NF32–41) results in a failure to reduce aPKC at NF41 (G) and a TEC at NF44 (H). Scale bar, 100 μm.
2I: Quantification of reduced trachea (t) length and relative to laryngotracheal (ltg) in NF42–44 X. laevis embryos. Student’s two-tailed t-test, between manipulated and control sibling embryos *p<0.05.
2J. Rab11a CRISPR-mediated mutation or MO knockdown results in a TEC at NF42–44 in X. laevis. Scale bar, 50 μm.
Remodeling of polarized epithelium is regulated by endosome recycling, where aPKC and cadherins are removed from the cell surface by endocytosis and shuttled to other membrane domains (Ivanov et al., 2004; Wang et al., 2018). Immunostaining for endosomal protein Rab11 revealed abundant apical puncta where the epithelium fuses in both Xenopus and in mouse, coincident with loss of aPKC and Cdh1 relocalization (Figure 2E–F). To test whether endosome recycling is required for TE separation, we treated Xenopus embryos at NF32 with Dynasore, a dynamin inhibitor that blocks endocytosis (Macia et al., 2006). In Dynasore-treated embryos, aPKC remained aberrantly localized on the apical surface of the epithelial contact site at NF41, resulting in a tracheoesophageal cleft (TEC) at NF44 (Figure 2G–I).
To corroborate the pharmacological inhibition, we performed F0 CRISPR/Cas9-mediated indel mutation of rab11a in Xenopus, as well as knockdown with a well-characterized Rab11a antisense morpholino oligo (MO) (Kim et al., 2012). Both of these resulted in 25–30% of the embryos having gastrulation defects, as expected (Kim et al., 2012; Ossipova et al., 2015); however ~50% of the remaining embryos (which were grossly normal) had disrupted epithelial remodeling with a failure of the transient epithelial septum to resolve at NF42–43 resulting in a TEC (Figures 2I,J and S2E). immunostaining confirmed the reduced Rab11 protein in rab11a-CRISPRs and Rab11a-MO embryos (Figure S2B). To overcome the earlier role of Rab11 in gastrulation, we generated transgenic Xenopus embryos expressing a dominant negative Rab11 in the foregut upon induction with doxycycline [Tg(hhex:trTA;TRE:dnRab11a-GFP)]. These also had disrupted epithelial fusion with enrichment of the dnRab11a-GFP on the basal-lateral epithelium rather than the normal apical surface (Figure S2C–E). Together, these data demonstrate that Rab11a-dependent endosome-mediated epithelial remodeling is essential for the foregut walls to fuse properly and that disruptions in this process can lead to TEDs.
Localized ECM Degradation and Mesenchymal Invasion Resolve the TE Septum
Between NF40–42 in Xenopus and E10.5–11.5 in mice, the length of the Sox2-Nkx2–1 boundary decreased as the length of the separated trachea and esophagus increased (Figure 3A). The presumptive trachea and esophagus had comparable proliferation in both species (data not shown), indicating that differential growth is unlikely to account for TE separation (Ioannides et al., 2010). This is consistent with a “splitting and extension” model (Que, 2015), where active septation occurs along with relatively equal growth of the separated trachea and esophagus.
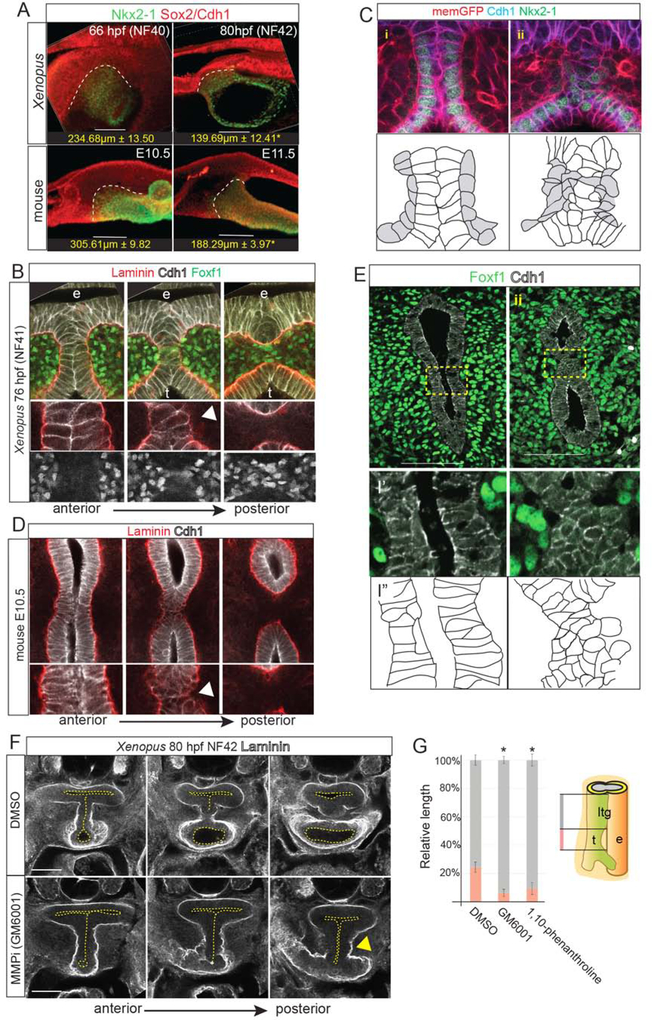
Figure 3: Localized ECM Degradation and Mesenchymal Invasion Resolve the TE Septum.
3A: Wholemount immunostaining of septum resolution in X. laevis and mouse embryos, quantifying the length of the Sox2+/Nkx2–1+ boundary. Student’s two-tailed t-test,*p<0.05.
3B: Serial optical sections showing Laminin, Cdh1 and Foxf1 during TE septum resolution in NF41 X. laevis embryos. Arrowhead indicates localized Laminin breakdown.
3C: Immunostaining of transgenic membrane GFP NF41 Xenopus embryo (i) anterior to and (ii) at the septation point showing Cdh1+ epithelial (white in schematic) round up as mesenchymal cells (grey) invade. Scale bar, 100 μm.
3D: Serial optical sections showing Laminin and Cdh1 during TE septum resolution in E10.5 mouse embryos. Arrowhead indicates localized Laminin breakdown.
3E: Immunostaining of Foxf1 and Cdh1 (schematic below) in an E10.5 mouse embryo (i) anterior to and (ii) at the septation point. Scale bar, 100 μm.
3F: Inhibition of MMP activity in Xenopus with GM6001 (from NF32–42) results in impaired Laminin breakdown (arrowhead) and a TEC.
3G: Quantification of relative lengths of the laryngotracheal groove (ltg) and trachea (t) in DMSO, GM6001, or 1,10-phenanthroline-treated NF41 X. laevis embryos. One-way ANOVA, *p<0.05.
Wholemount staining indicate that septation occurs in a posterior to anterior wave starting at the lung buds (Figures 3A–E, S3 and Supplemental Movies). In both Xenopus and mouse, the anterior septum was composed of Cdh1+ columnar cells and Laminin-rich basement membrane surrounded the entire foregut epithelium. However, in more posterior optical sections, Cdh1 and Laminin levels became reduced as basement membrane broke down and epithelial cells in the septum rounded up and lost adhesion to one another (Figure 3B–E and Supplementary Movies). Cdh1+ puncta indicative of new adherens junctions were observed between opposing epithelial cells to seal the presumptive tracheal and esophageal lumens (Figures 3B–D). Inhibiting ECM remodeling matrix metalloproteinases (MMPs) (Vu and Werb, 2000) with either GM6001 or 1,10-phenanthroline impaired Laminin breakdown in Xenopus, resulting in a TEC and shorter trachea (Figure 3F–G). As the septum resolved, fibronectin (Fn1)-rich Foxf1+ mesoderm cells, with enriched cortical actin, were observed between the separating trachea and esophagus, suggesting that mesenchymal cells actively migrate across the midline on the Fn1+ matrix (Figure S3C, D and F).
Cleaved caspase-3 staining revealed a few dying cells in the resolving mouse septum (Figure S3G) (Ioannides et al., 2010), but little septum cell death in Xenopus (data not shown). Rather, when the septum cells lost adhesion to one another, they appeared to incorporate into the esophageal or tracheal epithelium. This is consistent with reports in Kim et. al, 2019 and our observation that, immediately after TE separation it is not uncommon to see Nkx2–1+ cells in the ventral esophagus and Sox2+ cells in the dorsal trachea (Figure S1E). Thus, in both Xenopus and mouse, the transient septum is resolved by epithelial remodeling, localized ECM degradation, and mesenchymal cell invasion to separate the foregut into a distinct esophagus and trachea (Figure S3H). Disruption of this process at any point along the anterior-posterior axis could result in a TEF.
HH/Gli Activity is Required for D-V Patterning, Medial Constriction and Epithelial Remodeling
Having defined the cellular events controlling TE morphogenesis, we next asked how disruptions in the HH signaling pathway cause TEDs. HH ligands are expressed in the foregut epithelium and signal to the mesoderm to activate Gli2 and Gli3 (Ioannides et al., 2003; Rankin et al., 2016). In the absence of a HH signal, Gli2 is degraded and Gli3 is proteolytically cleaved into a transcriptional repressor (Gli3R). When the HH pathway is activated, Gli2 and Gli3 become transcriptional activators (Briscoe and Therond, 2013). Multiple human TEDs have been associated with mutations in SHH, GLI2, and GLI3, including heterozygous truncating mutations in GLI3 (Johnston et al., 2005).
We examined an allelic series of HH/Gli mouse mutants to determine which steps in TE morphogenesis were disrupted. Consistent with previous reports (Litingtung et al., 1998), Shh−/− embryos exhibited TA with a single hypomorphic gut tube at E15.5 (Figure 4A). Gli2−/−;Gli3+/− mutants also failed to undergo TE separation, displaying a TEC along the length of the foregut (Figure 4A) (Motoyama et al., 1998). In contrast, Gli2+/−;Gli3−/−, Gli3−/− and Gli2−/− (Figure 4A and data not shown) mutants were indistinguishable from wildtype littermates. We also examined transgenic embryos ectopically expressing a truncated repressor form of Gli3 (Vokes et al., 2008) in the foregut mesendoderm (Foxg1Cre;Gli3T), which mimics the GLI3 mutation in Pallister-Hall Syndrome (PHS) [OMIM# 151560] (Johnston et al., 2005). Like PHS patients, Foxg1;Gli3T embryos had LTECs (Figure 4A). These results indicate that increasing levels of Gli3-repressor relative to Gli activator cause increasing severity of TEDs.
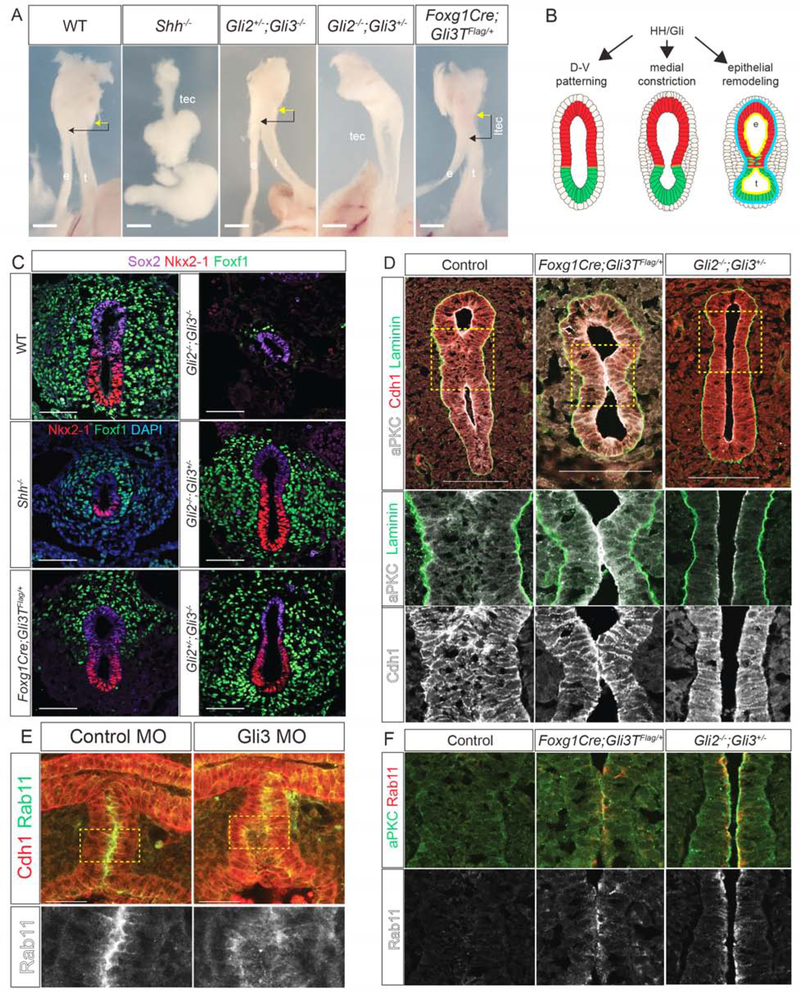
Figure 4: HH/Gli Activity Is Required for D-V Patterning, Medial Constriction and Epithelial Remodeling.
4A: E15.5 mouse foregut in Shh/Gli mutant embryos. Esophagus, e; trachea, t; trachea-esophageal cleft, tec; laryngotracheal-esophageal cleft, ltec. Arrows denote distance between cricoid cartilage (yellow) and TE septation point (black). Scale bar, 6.35 mm.
4B: Summary of HH/Gli-regulated events.
4C: Nkx2–1, Foxf1 and Sox2 (or DAPI) immunostaining in E10.0Shh/Gli mutants. Scale bar, 50 μm.
4D: aPKC, Laminin and Cdh1 immunostaining in E11.0 Foxg1Cre;Gli3T and Gli2−/−;Gli3+/− mutants showing a failure epithelial fusion and persistent aPKC. Scale bar, 100 μm.
4E: Immunostaining of control MO and Gli3 MO injected NF41 X. laevis embryos showing mislocalized Rab11 in Gli3 morphants. Scale bar, 50 μm.
4F: Immunostaining of aPKC and Rab11 in Foxg1Cre;Gli3T and Gli2−/−;Gli3+/− E11.0 mutants showing a failure of Rab11 reduction compared to controls.
Nasr et al. Endosome-Mediated Epithelial Remodeling Downstream of Hedgehog/Gli Is Required for Tracheoesophageal Separation DEVELOPMENTAL-CELL-D-19–00386R1
To understand the cellular basis of these defects (Figure 4B), we examined earlier developmental stages. At E10.0, Gli2−/−;Gli3−/− embryos, which lack all HH activity, failed to undergo D-V patterning, with no Nkx2–1+ respiratory progenitors and very little Foxf1+ mesenchyme (Rankin et al., 2016) (Figure 4C). In contrast, the foregut was properly patterned in all other genotypes with ventral Nkx2–1 and dorsal Sox2. Even though Shh−/− mutants were correctly patterned the foregut was smaller with fewer Nkx2–1+ and Foxf1+ cells and failed to constrict, similar to NF37 Xenopus embryos treated with HH antagonist cyclopamine (Figures 4C and S4B). Unlike Shh−/− mutants, the foreguts of both Gli2−/−;Gli3+/− and Foxg1Cre;Gli3T constricted at E10.0, but Foxg1Cre;Gli3T embryos had fewer Foxf1+ cells in the ventral region (Figure 4C). Thus high levels of HH activity are needed for D-V patterning, medial constriction and to maintain Foxf1+ mesoderm, consistent with Foxf1 being a direct Gli transcriptional target (Mahlapuu et al., 2001).
We next assessed Gli2−/−;Gli3+/− and Foxg1Cre;Gli3T mutants at E11.0 for defects in epithelial remodeling and septation (Figure 4D). In Gli2−/−;Gli3+/− mutants the foregut lumen remained open, suggesting that although medial constriction was initiated, the process was insufficient to bring the epithelial walls into contact. Alternatively, disrupted epithelial remodeling might result in failure of cell adhesion at the constriction point, followed by a relaxation and a TEC. Indeed, in Foxg1Cre;Gli3T embryos, aPKC and Rab11 persisted at the anterior contact point where the epithelium touched but failed to fuse, whereas in wild type embryos aPKC, Rab11 and Cdh1 were rapidly downregulated as the opposite walls of the epithelium fused (Figure 4F). This was similar to Xenopus embryos with disrupted endosome recycling, suggesting that the LTEC was caused by impaired epithelial remodeling in the septum.
Gli3 MO knockdown in X. laevis and CRISPR/Cas9-mediated gli3 mutation in X. tropicalis resulted in phenotypes similar to Gli2−/−;Gli3+/− and Foxg1Cre;Gli3T mouse mutants, with a delay in medial constriction and failure of TE separation resulting in TECs (Figure S4). In most cases, the transient septum formed but failed to resolve, with the lumen eventually reopening to form a cleft. CRISPR-mediated indel mutations of the gli3 C-terminus, predicted to result in a Gli3R form like the PHS patient mutation, resulted in persistent aPKC in the septum, like Foxg1Cre;Gli3T mouse embryos. Immunostaining of Gli3 MO X. laevis embryos showed a mislocalization of Rab11, which is normally enriched at the point of apical membrane fusion (Figure 4E). These results collectively indicate that HH/Gli-regulated epithelial remodeling is required for TE morphogenesis in both Xenopus and mouse, and that this is compromised in Gli3 mutant models of PHS.
Conclusion
We have defined the HH/Gli-regulated cellular mechanisms orchestrating TE morphogenesis: 1) D-V foregut patterning; 2) mesenchymal medial constriction at the Sox2-Nkx2–1 boundary; 3) epithelial fusion to form a transient septum; and 4) Rab11-dependent endosome-mediated epithelial remodeling and localized ECM breakdown to resolve the septum. These results provide a cellular explanation for the previous mouse models of TE separation, and help explain the genotype-phenotype association of TEDs in patients with SHH and GLI mutations. Complete loss of HH/Gli results in TA, while one copy of Gli3 appears to be sufficient to support D-V patterning and constriction, but not septation, resulting in a TEC. We predict that partial or transient disruption in epithelial remodeling would result in an interruption in progressive septation and a TEF. Our data also suggest that a balance between Gli activator and repressor function is critical, with excessive Gli3R resulting in TEDs. Further studies will focus on identifying mesenchymal Gli targets and how these regulate TE morphogenesis. Candidates include proteins that modulate cell adhesion, endosome-mediated epithelial remodeling, and ECM degradation. Consistent with this prediction, human TEDs have been linked to mutations in Integrin Alpha 6 (ITGA6), Filamin A (FLNA), and Fraser extracellular matrix subunit 1 (FRAS1) (Brosens et al., 2014).
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Aaron Zorn (aaron.zorn@cchmc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Xenopus and all mouse lines described were housed at Cincinnati Children’s Hospital Medical Center (CCHMC) and maintained according to the NIH Guidelines for the Care and Use of Laboratory Animals. All animals were housed in a 12-hour light-dark cycle with standard chow and water, and only healthy animals were used for experiments. Both mouse and Xenopus embryos were collected to analyze tracheoesophageal morphogenesis, and the sex of embryos was not recorded as there is no obvious sex-dependent penetrance of TECs (Shaw-Smith, 2006). All experiments were performed using guidelines approved by the CCHMC Institutional Animal Care and Use Committee (IACUC2019–0006 and IACUC2019–0053).
METHOD DETAILS
Experimental Design
All Xenopus experiments were performed at least two times. In the case of Xenopus experiments involving small molecules or induced mutations, mutant embryos were compared with untreated or vehicle-treated sibling embryos as controls. In the case of mouse experiments, at least three mouse mutants, age-matched by somite number if collected at E11.5 or earlier, were used for analysis in each experiment, with age-matched littermate controls. For mouse experiments only examining normal TE development, at least three age-matched controls were used. The sex of each embryo was not deliberately determined. Only healthy animals were used for this study. For most of the analysis, samples were not blinded except the determination of TEC phenotypes in Xenopus embryos, which were scored blind. No data was excluded from analysis.
Xenopus experiments
Xenopus laevis and Xenopus tropicalis were purchased from Nasco or the National Xenopus Resource Center (X. laevis memGFP) and housed according to established CCHMC IACUC protocols. X. tropicalis “Superman” animals were generously provided by Mustafa Khokha (Yale University). Generation of Xenopus embryos was performed using ovulation and in vitro fertilization techniques previously described.
For X. laevis microsurgery experiments the ectoderm and lateral plate mesoderm surrounding the foregut were removed from one side of NF32/33 embryos and cultured in 0.5XMBS with gentamicin until NF35 when they were fixed along with unmanipulated siblings. For morpholino (MO) knockdowns we injected previously validated Gli3-MOs (5 ng), Gli2-MO (5 ng) (Rankin et al., 2016) and Rab11a-MO (50 ng) (Kim et al., 2012), or equal concentrations of standard control morpholinos, into the dorsal-ventral region of 2–8 cell stage X. laevis embryos to target the future foregut region.
For transient F0 mosaic CRISPR/Cas9 mutagenesis, guide RNAs (gRNAs) were designed using CRISPRScan or CHOPCHOP and synthesized by IDT. For gli2 and gli3 loss of function, gRNAs targeting the X. tropicalis gli2 exon 4 or gli3 exon 2 were designed to produce truncating indels or missense mutations resulting in premature stop codon prior to the DNA-binding domains. A gRNA targeting the X. tropicalis gli3 in exon13 corresponding to the human Pallister Hall Syndrome GLI3d699 mutation was designed such that a premature stop codon would prevent translation of the Gli3 activator domain resulting in a constitutive repressor form. For the rab11a LOF mutations, we designed gRNAs targeting a region in exon 2 that has identical sequence identity between and X. tropicalis rab11a and X. laevis rab11a.S (X.laevis rab11a.L has a 2-nucleotide mismatch and is predicted not to be targeted). X. laevis embryos were used for rab11a CRISPR/Cas9 mutagenesis because the Rab11 antibodies (to verify loss of Rab11) work in laevis but not tropicalis. RNA-seq data in Xenbase.org indicated that rab11a.S transcripts are expressed two-fold higher than rab11a.L in X. laevis tadpoles. Consistent with the mRNA levels, Rab11 immunofluorescence showed that rab11a.S CRISPR mutants (with wildtype rab11a.L), have dramatically reduced Rab11a protein levels (Figure S2B). To generate F0 mosaic gli2, gli3, gli3R and rab11a.S mutant embryos, the gRNAs (750 pg) were then injected with Cas9 protein (1–1.5 ng; PNA Biosciences) into X. tropicalis or X. laevis embryos at the one-cell stage or the eight-cell stage targeted to the foregut endoderm. For negative controls, tyrosinase (tyr) gRNAs were injected. These resulted in a high frequency of F0 embryos lacking pigment indicating effective indel mutations, but no detectable TED.
To genotype embryos, genomic DNA was extracted from tails and the genomic regions target by the gRNAs were PCR amplified with 2X Phusion Master Mix (Thermo Fisher Scientific F531) according to manufacturer’s instructions. Successful indel mutation was initially verified with a T7E1 (New England BioLabs Incorporated M0302S) digest of the PCR product, followed by, Sanger sequencing. Overall efficiency and allele frequency of specific mutations were determined by TIDE sequencing decomposition analyses (https://tide.deskgen.com). The table below shows the efficiency of truncating indel mutations for each gRNA.
For small molecule treatments, Xenopus laevis embryos were cultured at room temperature, covered in aluminum foil, in 0.1XMBS in the presence of small molecules that were refreshed once every 12–24 hours beginning at NF28/29–32/33. Control sibling embryos were treated with equal volumes of the small molecule solvent (e.g., EtOH or DMSO). Small molecule treatment concentrations were: cyclopamine (Selleck or Tocris, 80μM), Dynasore (Millipore Sigma, 50μM), GM6001 (Millipore Sigma, 150μM) and 1,10-phenanthroline (Millipore Sigma, 5 μM).
For Xenopus transgenesis, the I-SceI meganuclease method was used as described (Sterner et al., 2019). The Tg(hhex;rtTA;TRE:dnRab11a-GFP) construct was generated as follows. The −1.2Kb X.laevis hhex.L promoter (Rankin et al., 2011) and the dNRab11a-GFP fusion coding sequence (Kim et al., 2012) were PCR amplified gel purified and TOPO-TA cloned into the pCR8 Gateway entry vectors (Thermo Scientific #K250020). Gateway LR Clonase II Plus enzyme (Thermo Scientific #12538120) was used in standard recombination reactions according to manufacturer’s instructions to transfer the hhex.L promoter into the pDXTP and dNRab11-GFP into the pDXTR transgenesis plasmids (Sterner et al., 2019).
Transgenic embryos were generated as follows: 200pg of pDXTP-hhex promoter and 200pg of pDXTR-dNRab11-GFP were incubate in a 25uL reaction containing 2.5uL of I-SceI meganuclease enzyme (New England Biolabs #R0694S) in 0.5X I-SceI buffer. The reaction was incubated at 37°C for 30 to 40 minutes and then immediately injected into 1-cell embryos near the sperm entry point within the first 45 to 60 minutes after fertilization. Embryos were cultured at 13°C for the first two hours after injection and subsequently at 18°C to 23°C degrees thereafter. Transgenic embryos were selected based on GFP fluorescence in the eye, which becomes visible during early tailbud stages (Sterner et al., 2019). In half of the embryos the transgenes were activated by the addition of Doxycycline hyclate (Sigma #D9891) at a final concentration of 50ug/mL culture buffer. The addition of Dox alone has no effect on the embryos and did not cause TEDs.
All of the confocal analysis in Xenopus embryos was performed in wholemount. Embryos were cut transversely just anterior to the pharynx and posterior to the lung buds in 100% MeOH. After serial rehydrations into 1XPBS with 0.1% Triton X-100 (1XPBSTr), embryos underwent antigen retrieval for 45 minutes in 1X citrate buffer at 65°C. Following three washes in 1XPBSTr, embryos were blocked in a 1XPBSTr solution of 1% bovine serum albumin (BSA) and 5% DMSO overnight at 4°C on a rocker for at least sixteen hours. Primary antibodies were added to embryos for at least sixteen hours, again rocking overnight at 4°C. Following five thirty-minute washes in 1XPBSTr, secondary antibodies were added in a 1XPBSTr solution containing 0.1% BSA overnight, rocking at 4°C for a t least sixteen hours. After five thirty-minute washes in 1XPBSTr, embryos were serially dehydrated into 100% methanol, and stored at 4°C until imaging in Murray’s Clear solution.
Mouse experiments
All mouse experiments performed were approved by the CCHMC Institutional Animal Care and Use Committee (IACUC). Gli2tm2.1Alj/J mice were obtained from The Jackson Laboratory. Foxg1Cre, mTmG, Dermo1Cre, and ShhCreGFP mice were obtained from Debora Sinner’s lab. Gli3TFlag mice were provided by Rolf Stottmann and Joo-Seop Park. Gli3XtJ mice were obtained from Rolf Stottmann’s lab. Nkx2–1GFP mice were provided by Jeffrey Whitsett and John Shannon. FoxA2tm2.1(cre/Esr*)Moon/J and Sox2tm1.1Lan/J mice were provided by James Wells (tamoxifen administered via oral gavage at E6.5). Mice were maintained in the CCHMC animal facility. Timed matings and somite counting were used to obtain embryos at the stages described. Genotyping was performed using Phusion Hot Start Flex 2x Master Mix or Quickload 2x Master Mix (New England BioLabs, Incorporated). After collection, all mouse embryos were washed in 4% paraformaldehyde, rocking overnight at 4°C, before two five-minute washes in 1XPBS.
Mouse embryos used for cryosectioning were washed for one to two days, rocking at 4°C, in 30% sucrose in 1XPBS. Embryos were then emb edded in OCT (Sakura, VWR) and stored at −80°C until sectioning at 8 μm thickness or 60 μm thickness. For section immunohistochemistry, sections were washed in 1XPBS, then 1XPBS with 0.1% Triton X-100 for permeabilization before blocking for one hour in 5% normal donkey serum in 1XPBS. After incubation in primary antibodies in 1XPBS at 4°C ov ernight, slides were washed in 1XPBS before incubation in secondary antibodies in 1XPBS at room temperature for one hour, followed by 1XPBS washes and mounting using ProLong Gold Antifade (Thermo Fisher Scientific).
Mouse embryos used for wholemount immunostaining were incubated in 100% methanol at −20°C until dissection and staining of the foregut. Briefly, embryos were washed for two hours at room temperature in Dent’s Bleach solution before serial rehydration into 1XPBS from methanol. Embryos were blocked in 5% normal donkey serum in 1XPBS with 0.2% Triton X-100 for two hours, rocking at room temperature. Embryos were then placed in primary antibodies in blocking solution, rocking overnight at 4°C. After a series of washes in 1XPBS with 0.1% Triton X-100, embryos were incubated with secondary antibodies in blocking solution, rocking overnight at 4°C. After three washes in 1XP BS with 0.1% Triton X-100, embryos underwent serial dehydration into 100% methanol before overnight storage at 4°C. Embryos were removed from methanol and placed into Murray’s Clear for at least fifteen minutes before confocal imaging.
QUANTIFICATION AND STATISTICAL ANALYSIS
NIS Elements software was used to obtain image quantifications, and the indicated tests appear in corresponding figure legends. For cell proliferation and cell death counts, the number of pHH3+ or CC3+/Cdh1+ and pHH3+ or CC3+/Foxf1+ cells were counted, as well as pHH3+ or CC3+/DAPI+ cells when possible. In Xenopus, the mesoderm thickness in the dorsal, medial or ventral regions was measured as the distance between the lateral ectoderm and endoderm. In mouse, the mesoderm thickness in the dorsal, medial or ventral regions was measured as the distance between the endoderm and the lateral boundary of Foxf1+ mesoderm. Mesoderm density in Xenopus was calculated as the number of Foxf1+ cells within a confocal Z-stack ~105 μm3, whereas the cell density in mouse was calculated based on DAPI staining from one optical section of a 30–60μm confocal Z-stack.
For small molecule treatments, the laryngotracheal groove was calculated as the distance between the start of the groove and the distinct trachea, marked by absent cytoskeletal staining between the trachea and esophagus. The tracheal length was calculated as the distance between the start of the distinct trachea, described above, and the start of the lung buds. Data shown is from one representative experiment.
For all experiments, n = at least 3 embryos per condition per replicate experiment. Unpaired Student’s two-tailed t-test and difference of means calculations were performed in Microsoft Excel. All other significance calculations were performed using one-way ANOVA (for one control with multiple conditions), two-way ANOVA (for two controls conditions with equal numbers within conditions), or mixed effects analysis (for two control conditions with unequal numbers within conditions) in Prism. For two-way ANOVA and mixed effects analysis calculations, the Geisser-Greenhouse correction and Tukey’s multiple comparisons test were applied. p < 0.05 (*) indicated significance for all tests, and the mean and SEM were included in all calculations.
DATA AND CODE AVAILABILITY
This study did not generate or analyze any datasets or codes.
Supplementary Material
Supplemental movie 1 related to Fig. 3 Xenopus Septum Resolution.
Confocal Z-stack of cross sections through a NF41 Xenopus foregut (anterior to posterior) immunostained for Cdh1(red) epithelium, Foxf1 (white) mesenchyme and the Laminin (green) basement membrane showing that cells in the transient septum round up as the Laminin breaks down and mesenchymal cells invade to separate the nascent trachea and esophagus.
Supplemental movie 2 related to Fig. 3 Xenopus NF41 foregut surface rendering
Surface rendering of Confocal Z-stack of NF 41 Xenopus foregut.
Supplemental movie 3 related to Fig. 3 Mouse E10.5 Septum Resolution.
Confocal Z-stack of cross sections (from anterior to posterior) through an E10.5 mouse foregut immunostained for Cdh1(red) epithelium and Laminin (red) basement membrane showing that epithelial cells at the contact point round up and Laminin breaks down as the nascent trachea and esophagus separate.
Supplemental Mouse 4 related to Fig. 3 mouse E10.5 foregut surface rendering
Surface rendering of Confocal Z-stack of an E10.5 mouse foregut.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Sox2 | Abcam | ab79351 RRID: AB_10710406 |
| Goat polyclonal anti-Sox2 (Y-17) | Santa Cruz Biotechnology | sc-17320 RRID: AB_2286684 |
| Rabbit polyclonal anti-Nkx2–1 (H-190) | Santa Cruz Biotechnology | sc-13040X RRID: AB_793532 |
| Goat polyclonal anti-Foxf1 | R&D | AF4798 RRID: AB_2105588 |
| Rat monoclonal anti-Cdh1 | R&D | MAB7481 RRID: AB_2076679 |
| Rabbit polyclonal anti-laminin | Sigma-Aldrich | L9393 RRID: AB_477163 |
| Mouse monoclonal anti-ß1 integrin | Developmental Studies Hybridoma Bank | 8C8 (deposited to the DSHB by Hausen, P./ Gawantka, V.) RRID: AB_528309 |
| Rabbit polyclonal anti-ß-catenin (H-102) | Santa Cruz Biotechnology | sc-7199 RRID: AB_634603 |
| Rabbit polyclonal anti-phospho-histone H3 | Cell Signaling | 9701L RRID: AB_331535 |
| Rabbit polyclonal anti-cleaved caspase 3 | Cell Signaling | 9661 RRID: AB_2341188 |
| Mouse monoclonal anti-Cdh1 (5D3) | Developmental Studies Hybridoma Bank | 5D3 (deposited to the DSHB by Gumbiner, B.M.) RRID: AB_528116 |
| Mouse monoclonal anti-Cdh3 (6B6) | Developmental Studies Hybridoma Bank | 6B6 (deposited to the DSHB by Gumbiner, B.M.) RRID: AB_528113 |
| Rabbit polyclonal anti-aPKC | Abcam | ab59364 RRID: AB_944858 |
| Goat polyclonal anti-p-PKC ζ (C-20) | Santa Cruz Biotechnology | sc-216-G RRID: AB_632241 |
| Rabbit monoclonal anti-PKCμ (C-20) | Santa Cruz Biotechnology | sc-639 RRID: AB_2172392 |
| Rabbit monoclonal anti-Rab11 (3H18L5) | Invitrogen | 700184 RRID: AB_2532295 |
| Rabbit polyclonal anti-Rab11 | Cell Signaling | 5589 RRID: AB_10693925 |
| Mouse monoclonal anti-Fibronectin 4H2 | De Simone Lab | Available from DSHB #4H2 RRID: AB_2721949 |
| Chicken polyclonal anti-ß-galactosidase | Abcam | ab9361 RRID: AB_307210 |
| Chicken polyclonal anti-GFP | Aves Labs | GFP-1020 RRID: AB_10000240 |
| Mouse monoclonal anti-DsRed | Takara Bio | 632392 RRID: AB_2801258 |
| Donkey anti-goat 488 | Jackson ImmunoResearch Laboratories | 705-546-147 RRID: AB_2340430 |
| Donkey anti-goat Cy3 | Jackson ImmunoResearch Laboratories | 705-166-147 RRID: AB_2340413 |
| Donkey anti-rabbit Cy3 | Jackson ImmunoResearch Laboratories | 711-165-152 RRID: AB_2307443 |
| Donkey anti-rat 647 | Jackson ImmunoResearch Laboratories | 712-606-153 RRID: AB_2340696 |
| Donkey anti-mouse Cy3 | Jackson ImmunoResearch Laboratories | 715-165-151 RRID: AB_2315777 |
| Donkey anti-mouse 647 | Jackson ImmunoResearch Laboratories | 715-606-151 RRID: AB_2340866 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Cyclopamine | Tocris | 1623 |
| Cyclopamine | Selleck Chemicals | S1146 |
| GM6001 | Millipore Sigma | 364205 |
| 1,10-phenanthroline | Millipore Sigma | 131377–25G |
| Dynasore | Millipore Sigma | 324414 |
| Hydroxyurea | Alfa Aesar | A10831 |
| T7E1 | New England BioLabs, Incorporated | M0302S |
| Cas9 | PNA Biosciences | CP01–50 |
| 2x Phusion Master Mix | Thermo Fisher Scientific | F531 |
| Phusion Hot Start Flex 2X Master Mix | New England BioLabs, Incorporated | M0536S |
| QuickLoad 2X Master Mix | New England BioLabs, Incorporated | M0271L |
| I-Sce1 | New England Biolabs | R0694S |
| Doxycycline hyclate | Sigma | D9891 |
| pCR8/GW/TOPO TA Cloning Kit with One Shot TOP10 E.coli | Thermo Fisher Scientific | K250020 |
| LR Clonase II Plus enzyme | Thermo Fisher Scientific | 12538120 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| Normal donkey serum | Jackson ImmunoResearch Laboratories | 017-000-121 |
| ProLong Gold Antifade | Thermo Fisher Scientific | P36930 |
| 10X Citrate Buffer | Abcam | ab64214 |
| Alexa Fluor Phalloidin 488 | Life Technologies | A12379 |
| DAPI | Thermo Fisher Scientific | D1306 RRID: AB_2629482 |
| Experimental Models: Organisms/Strains | ||
| Xenopus laevis: Xla.Tg(CMV:memGFP,cryga:mCherry)NXR | National Xenopus Resource | NXR_0.0012 |
| Xenopus laevis females | NASCO | LM00531 |
| Xenopus laevis males | NASCO | LM00715 |
| Xenopus tropicalis females | NASCO | LM00823 |
| Xenopus tropicalis males | NASCO | LM00822 |
| Xenopus tropicalis Superman males | Mustafa Khokha | N/A |
| Xenopus tropicalis Superman females | Mustafa Khokha | N/A |
| Xenopus laevis Tg(hhex;rtTA;TRE:dnRab11a-GFP) | This study | This study |
| Mouse: Gli2tm2.1Alj/J | The Jackson Laboratory | JAX: 007922 |
| Mouse; Gli3XtJ | Rolf Stottmann | (JAX: 000026) |
| Mouse: Gt(ROSA)26Sortm3(Gli3)Amc/J | Rolf Stottmann & Joo-Seop Park | (JAX: 013124) |
| Mouse: Shhtm1(EGFP/cre)Cjt/J | Debora Sinner | (JAX: 005622) |
| Mouse: Foxg1tm1(cre)Skm | Debora Sinner | (JAX: 006084) |
| Mouse: Nkx2–1GFP | John Shannon & Jeffrey Whitsett | (Longmire et al., 2012) |
| Mouse: FoxA2tm2.1(cre/Esr*)Moon/J | James Wells | (JAX: 008464) |
| Mouse: Sox2tm1.1Lan/J | James Wells | (JAX: 013093) |
| Mouse: B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | Debora Sinner | (JAX: 007676) |
| Mouse: B6.129X1-Twist2tm1.1(cre)Dor/J | Debora Sinner | (JAX: 008712) |
| Oligonucleotides | ||
| Control Morpholino: CCTCTTACCTCAGTTACAATTTATA | GeneTools | (Nguyen et al., 2005) |
| X. laevis Gli2 Morpholino: GCACAGAACGCAGGTAATGCTCCAT | GeneTools | (Nguyen et al., 2005) |
| X. laevis Gli3 Morpholino: TAGTGCTACGGGACTGGGCTTCCAT | GeneTools | (Nguyen et al., 2005) |
| X. laevis Rab11a Morpholino: TACCCATCGTCGCGGCACTTCTGAC | GeneTools | (Kim et al., 2012) |
| X. tropicalis CRISPR gRNA tyr: GGAACTGGCCCCTGCAAACATGG | IDTDNA | (Blitz et al., 2013) |
| X. tropicalis CRISPR gRNA gli3 exon 2 LOF: CGGTAGGGAACTGAGGGTTCAGG | IDTDNA | This study |
| Genotyping, X. tropicalis gli3 LOF CRISPR forward: AAGAAACGCCATCACCATGC | IDTDNA | This study |
| Genotyping, X. tropicalis gli3 LOF CRISPR reverse: TCCAAGCAGTCCCTAATAGCA | IDTDNA | This study |
| X. tropicalis CRISPR gRNA gli2 LOF: GGGCTACCGCTGAGAGTTGGGGG | IDTDNA | This study |
| Genotyping, X. tropicalis gli2 LOF CRISPR forward: CTGTGCTAATAACCCACATTTCTC | IDTDNA | This study |
| Genotyping, X. tropicalis gli2 LOF CRISPR reverse: TGCAGACCCCCACTATCCA | IDTDNA | This study |
| X. tropicalis CRISPR gRNA gli3R PHS: GTGCTCTATGAGGTGGAACTGGG | IDTDNA | This study |
| Genotyping, X. tropicalis gli3R CRISPR forward: GCATTGAGTGCATGACATTG | IDTDNA | This study |
| Genotyping, X. tropicalis gli3R CRISPR reverse: CATTCCCATGACAACACAGC | IDTDNA | This study |
| X. laevis CRISPR gRNA rab11.a exon 1 LOF: GAGATTACTCTTCCCCACAC CGG | IDTDNA | This study |
| Genotyping, X. laevis rab11a.S CRISPR forward: ACAGTGTGCATAGAATTTGGCG | IDTDNA | This study |
| Genotyping, X. laevis rab11a.S CRISPR forward: CGTGTGTTGCCTAGAGGGAC | IDTDNA | This study |
| X. laevis pCS2-EGFP-dnRab11a | Sergei Sokol | (Kim et al., 2012) |
| Amplification of EGFP-dnRab11a from provided pCS2-EGFP-dnRab11, forward: ATGGTGAGCAAGGGCGAGGAGCT | IDTDNA | This study |
| Amplification of EGFP-dnRab11a from provided pCS2-EGFP-dnRab11, reverse: TTAGATGTTCTGACAGCATTGCAT | IDTDNA | This study |
| Amplification of X. laevis hhex promoter, forward: GCATACAGACCCATGCCAGTG | IDTDNA | (Rankin et al., 2011) |
| Amplification of X. laevis hhex promoter, reverse: GTCGATTCCTCTTTCCACACTCAG | IDTDNA | (Rankin et al., 2011) |
| Software and Algorithms | ||
| NIS Elements | Nikon | N/A |
| IMARIS | Bitplane | N/A |
| Excel | Microsoft | N/A |
| Prism | GraphPad | N/A |
| TIDE | N/A | https://tide.deskgen.com |
| CRISPRScan | N/A | http://www.crisprscan.org |
| CHOPCHOP | N/A | http://chopchop.cbu.uib.no |
Highlight bullet points:
The Sox2+ esophagus and Nkx2–1+ trachea arise from the separation of the foregut
HH/Gli-dependent medial constriction of the foregut initiates morphogenesis
Rab11-dependent epithelial remodeling and ECM degradation separates the foregut
HH/Gli mutations reveal the cellular basis of tracheoesophageal birth defects
ACKNOWLEDGMENTS
The authors would like to thank the Zorn and Wells labs for their feedback. We acknowledge the CCHMC Confocal Imaging Core. This project was funded by NIH P01HD093363 and was supported in part by NIH P30 DL0778392 to the Digestive Diseases Research Core Center in Cincinnati. T.N. is supported by NIH F30HL142201 and T32 GM063483-14 to the University of Cincinnati Medical Scientist Training Program. We are grateful to Sergei Sokol for the dnRab11 constructs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
There are no conflicts of interest to declare.
SUPPLEMENTAL ITEMS
Supplemental figures and legends Pdf.
REFERENCES
- Billmyre KK, Hutson M, and Klingensmith J (2015). One shall become two: Separation of the esophagus and trachea from the common foregut tube. Developmental dynamics : an official publication of the American Association of Anatomists 244, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitz IL, Biesinger J, Xie X, and Cho KW (2013). Biallelic genome modification in F(0) Xenopus tropicalis embryos using the CRISPR/Cas system. Genesis 51, 827–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, and Therond PP (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 14, 416–429. [DOI] [PubMed] [Google Scholar]
- Brosens E, Ploeg M, van Bever Y, Koopmans AE, H IJ, Rottier RJ, Wijnen R, Tibboel D, and de Klein A (2014). Clinical and etiological heterogeneity in patients with tracheo-esophageal malformations and associated anomalies. European journal of medical genetics 57, 440–452. [DOI] [PubMed] [Google Scholar]
- Hines EA, and Sun X (2014). Tissue crosstalk in lung development. J Cell Biochem 115, 1469–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannides AS, Henderson DJ, Spitz L, and Copp AJ (2003). Role of Sonic hedgehog in the development of the trachea and oesophagus. Journal of pediatric surgery 38, 29–36; discussion 29–36. [DOI] [PubMed] [Google Scholar]
- Ioannides AS, Massa V, Ferraro E, Cecconi F, Spitz L, Henderson DJ, and Copp AJ (2010). Foregut separation and tracheo-oesophageal malformations: the role of tracheal outgrowth, dorso-ventral patterning and programmed cell death. Developmental biology 337, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AI, Nusrat A, and Parkos CA (2004). Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Molecular biology of the cell 15, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Olivos-Glander I, Killoran C, Elson E, Turner JT, Peters KF, Abbott MH, Aughton DJ, Aylsworth AS, Bamshad MJ, et al. (2005). Molecular and clinical analyses of Greig cephalopolysyndactyly and Pallister-Hall syndromes: robust phenotype prediction from the type and position of GLI3 mutations. American journal of human genetics 76, 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Lake BB, Haremaki T, Weinstein DC, and Sokol SY (2012). Rab11 regulates planar polarity and migratory behavior of multiciliated cells in Xenopus embryonic epidermis. Developmental dynamics : an official publication of the American Association of Anatomists 241, 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, and Chiang C (1998). Sonic hedgehog is essential to foregut development. Nature genetics 20, 58–61. [DOI] [PubMed] [Google Scholar]
- Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, et al. (2012). Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, and Kirchhausen T (2006). Dynasore, a cell-permeable inhibitor of dynamin. Developmental cell 10, 839–850. [DOI] [PubMed] [Google Scholar]
- Mahlapuu M, Enerback S, and Carlsson P (2001). Haploinsufficiency of the forkhead gene Foxf1, a target for sonic hedgehog signaling, causes lung and foregut malformations. Development 128, 2397–2406. [DOI] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, and Kimura S (1999). Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Developmental biology 209, 60–71. [DOI] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, and Hui CC (1998). Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nature genetics 20, 54–57. [DOI] [PubMed] [Google Scholar]
- Nguyen V, Chokas AL, Stecca B, and Ruiz i Altaba A (2005). Cooperative requirement of the Gli proteins in neurogenesis. Development 132, 3267–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Chuykin I, Chu CW, and Sokol SY (2015). Vangl2 cooperates with Rab11 and Myosin V to regulate apical constriction during vertebrate gastrulation. Development 142, 99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J (2015). The initial establishment and epithelial morphogenesis of the esophagus: a new model of tracheal-esophageal separation and transition of simple columnar into stratified squamous epithelium in the developing esophagus. Wiley interdisciplinary reviews Developmental biology 4, 419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, and Hogan BL (2007). Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development 134, 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Han L, McCracken KW, Kenny AP, Anglin CT, Grigg EA, Crawford CM, Wells JM, Shannon JM, and Zorn AM (2016). A Retinoic Acid-Hedgehog Cascade Coordinates Mesoderm-Inducing Signals and Endoderm Competence during Lung Specification. Cell reports 16, 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Kormish J, Kofron M, Jegga A, and Zorn AM (2011). A gene regulatory network controlling hhex transcription in the anterior endoderm of the organizer. Developmental biology 351, 297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Thi Tran H, Wlizla M, Mancini P, Shifley ET, Bloor SD, Han L, Vleminckx K, Wert SE, and Zorn AM (2015). A Molecular atlas of Xenopus respiratory system development. Developmental dynamics : an official publication of the American Association of Anatomists 244, 69–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C (2006). Oesophageal atresia, tracheo-oesophageal fistula, and the VACTERL association: review of genetics and epidemiology. Journal of medical genetics 43, 545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner ZR, Rankin SA, Wlizla M, Choi JA, Luedeke DM, Zorn AM, and Buchholz DR (2019). Novel vectors for functional interrogation of Xenopus ORFeome coding sequences. Genesis, e23329. [DOI] [PubMed] [Google Scholar]
- Tabler JM, Rigney MM, Berman GJ, Gopalakrishnan S, Heude E, Al-Lami HA, Yannakoudakis BZ, Fitch RD, Carter C, Vokes S, et al. (2017). Cilia-mediated Hedgehog signaling controls form and function in the mammalian larynx. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trisno SL, Philo KED, McCracken KW, Cata EM, Ruiz-Torres S, Rankin SA, Han L, Nasr T, Chaturvedi P, Rothenberg ME, et al. (2018). Esophageal Organoids from Human Pluripotent Stem Cells Delineate Sox2 Functions during Esophageal Specification. Cell Stem Cell 23, 501–515 e507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vokes SA, Ji H, Wong WH, and McMahon AP (2008). A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes & development 22, 2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu TH, and Werb Z (2000). Matrix metalloproteinases: effectors of development and normal physiology. Genes & development 14, 2123–2133. [DOI] [PubMed] [Google Scholar]
- Walton KD, Kolterud A, Czerwinski MJ, Bell MJ, Prakash A, Kushwaha J, Grosse AS, Schnell S, and Gumucio DL (2012). Hedgehog-responsive mesenchymal clusters direct patterning and emergence of intestinal villi. Proc Natl Acad Sci U S A 109, 15817–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qiu Z, Xu Z, Chen SJ, Luo J, Wang X, and Chen J (2018). aPKC is a key polarity determinant in coordinating the function of three distinct cell polarities during collective migration. Development 145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental movie 1 related to Fig. 3 Xenopus Septum Resolution.
Confocal Z-stack of cross sections through a NF41 Xenopus foregut (anterior to posterior) immunostained for Cdh1(red) epithelium, Foxf1 (white) mesenchyme and the Laminin (green) basement membrane showing that cells in the transient septum round up as the Laminin breaks down and mesenchymal cells invade to separate the nascent trachea and esophagus.
Supplemental movie 2 related to Fig. 3 Xenopus NF41 foregut surface rendering
Surface rendering of Confocal Z-stack of NF 41 Xenopus foregut.
Supplemental movie 3 related to Fig. 3 Mouse E10.5 Septum Resolution.
Confocal Z-stack of cross sections (from anterior to posterior) through an E10.5 mouse foregut immunostained for Cdh1(red) epithelium and Laminin (red) basement membrane showing that epithelial cells at the contact point round up and Laminin breaks down as the nascent trachea and esophagus separate.
Supplemental Mouse 4 related to Fig. 3 mouse E10.5 foregut surface rendering
Surface rendering of Confocal Z-stack of an E10.5 mouse foregut.
Data Availability Statement
This study did not generate or analyze any datasets or codes.