Abstract
Urbanization represents a profound shift in human behavior, with significant cultural and health-associated consequences2,3. Here we investigate chemical and microbial characteristics of houses and their human occupants across an urbanization gradient in the Amazon rainforest, from a remote Peruvian Amerindian village to the Brazilian city of Manaus. Urbanization was associated with reduced microbial outdoor exposure, increased contact with housing materials, antimicrobials, and cleaning products, and increased exposure to chemical diversity. Urbanization degree correlated with changes in house bacterial and micro-eukaryotic community composition, increased house and skin fungal diversity, and increased relative abundance of human skin-associated fungi and bacteria in houses. Overall, our results indicate large-scale effects of urbanization on chemical and microbial exposures and on the human microbiota.
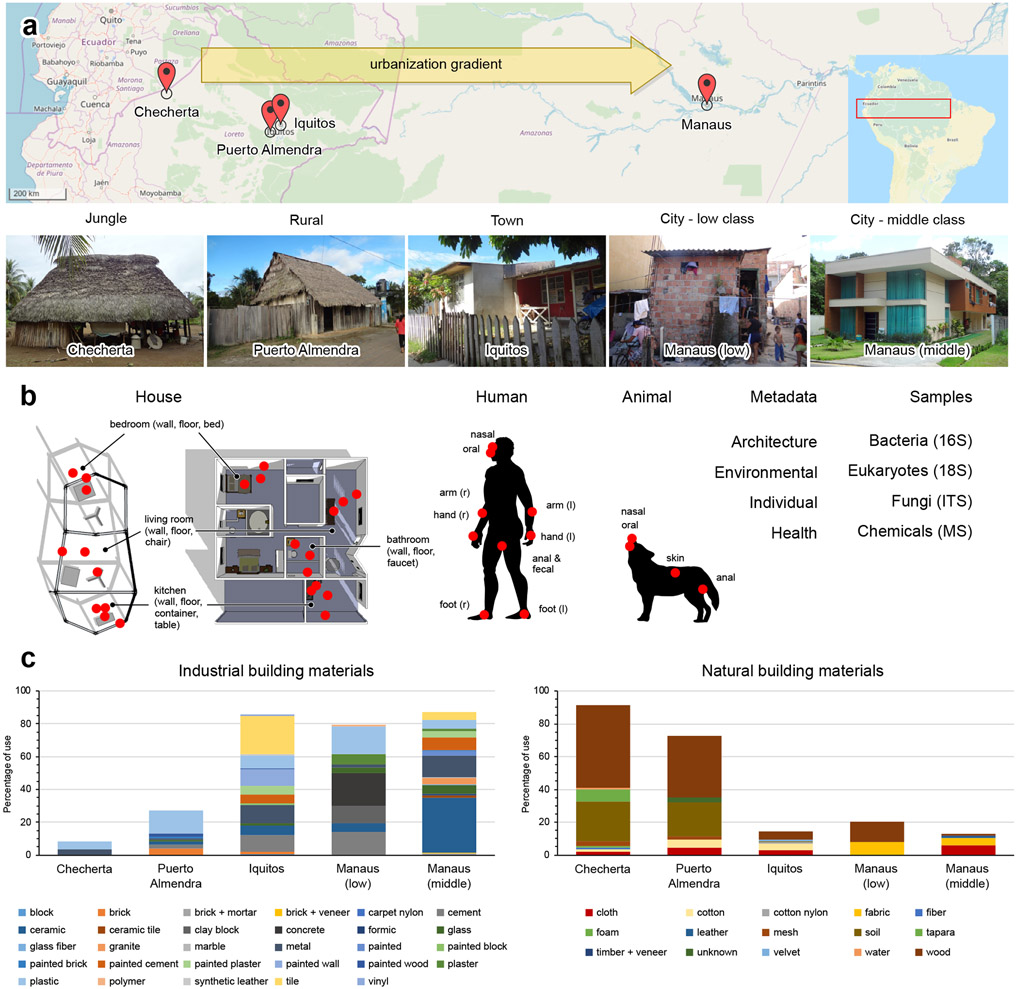
Urbanization represents a major shift from traditional lifestyles. Today, over 50% of the world population is urban, and by 2050, the proportion will exceed 66%1. Metabolic and autoimmune diseases have increased in parallel with these urbanization-associated lifestyle changes2,3, and human microbiota diversity has decreased4-7. Urbanization associated changes are numerous, and include diet4,5, urban design and density, house architecture, and exposures to environment (both outdoors and indoors8), animals, parasites, and consumer goods. Here, we expand upon prior work on household bacteria8 by performing a comprehensive chemical and pan-microbial survey spanning an urbanization gradient in the Amazon. We collected human, animal and household samples across a gradient of urbanization along a similar latitude in South America, from a remote village isolated in the Amazon rainforest (Checherta), to a rural village (Puerto Almendra), a large town (Iquitos) and a metropolis (Manaus). This urbanization gradient is associated with significant changes in house architecture, leading to reduced outdoor exposure, changes in construction materials (more industrial, less natural, p<2.2e-16, Spearman test, Fig. 1) and reduced number of inhabitants per house with increased urbanization (K-W χ2=469.09, p<2.2e-16) (Fig. 1, Supplementary Table 1). To capture differences between social strata, we sampled homes from low-income and middle class areas in the city of Manaus. Microbial and eukaryotic composition and diversity were characterized through 16S ribosomal RNA gene (16S), 18S ribosomal RNA gene (18S) and internal transcribed spacer 1 (ITS1) sequencing of extracted DNA. Chemical composition was assessed by LC-MS/MS for a subset of samples (Fig. 1).
Figure 1. Study design.
a. Samples were collected in four different locations in South America along the Amazon river across an urbanization gradient: Checherta: remote jungle village, Puerto Almendra: rural village, Iquitos: large town, Manaus: metropolis (OpenStreetMap). In Manaus two different socio-economic classes were studied: lower income and middle-class. b. At each location 10 different houses and their inhabitants (humans and pets) were sampled. Sampling locations are illustrated by red dots. Microbial samples were collected for bacterial, fungal and eukaryotic analysis, with replicate samples for LC-MS/MS-based chemical profiling. Architectural and environmental parameters were monitored. Samples from the houses included wall, floor, bed (hammock), chair handle, table, faucet (water container), countertop, cup and fireplace. Human samples included skin (arm, hand, foot); oral, nasal and anal/fecal samples. Pet samples included oral/nasal, skin and anal samples. c. Use of building materials across the five locations (right, natural building materials; left, industrial building materials; determined by visual inspection).
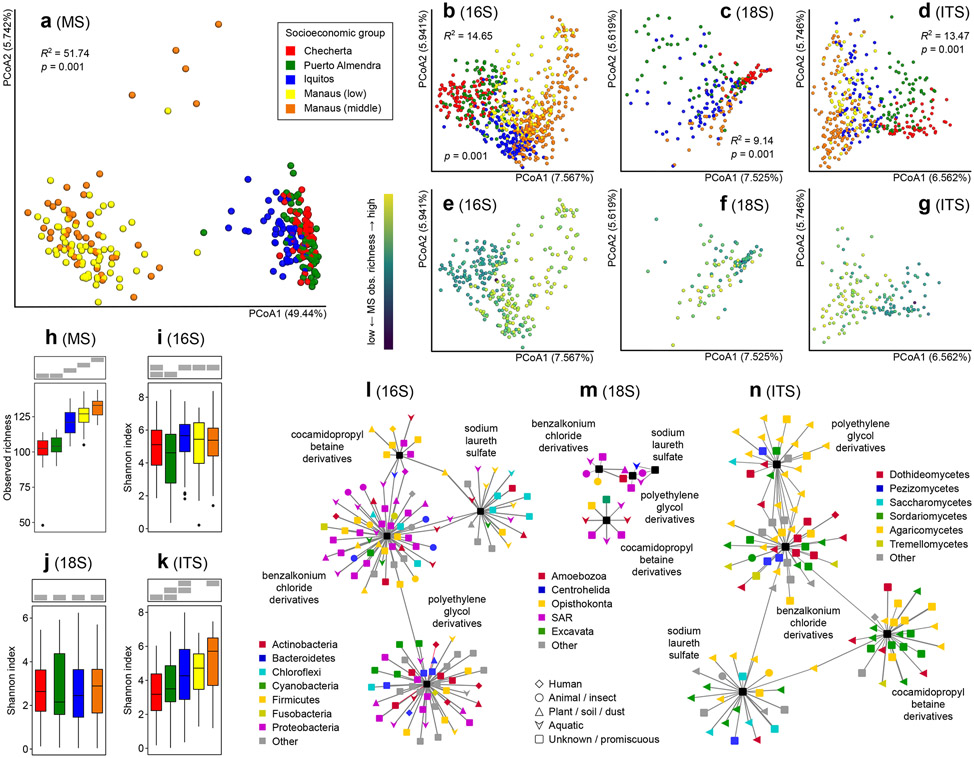
The number of distinct chemicals in each sample increased dramatically with urbanization (K-W χ2=275.94, p<2.2e-16, Fig. 2h). House surface chemical profiles from the city of Manaus (regardless of socioeconomic group) were remarkably different from the other locations (Fig. 2a PERMANOVA R2=51.74%, p=0.001, Supplementary Figs. 1 and 2). Medication-derived chemicals were only detected in the more urbanized settings of Manaus and Iquitos. These include the beta-blocker metoprolol and the antifungal ketoconazole in Manaus, and the antifungal clotrimazole in Iquitos and Manaus (Supplementary Table 4, Supplementary Figs. 1 and 3). Mass spectral molecular networking analysis identified several molecular families from personal care, cleaning products and detergents in Manaus (both lower- and middle-class homes), including sodium laureth sulfate, cocamidopropyl betaine-related molecules, polyethylene glycol derivatives, and benzalkonium chloride family members (Supplementary Table 4, Supplementary Figs. 1 and 3). These results are further supported by increased self-reported cleaning frequency in urban settings (K-W χ2=907.29, p<2.2e-16, Supplementary Table 1). In silico structure prediction using Network Annotation Propagation (NAP)9 revealed that mass spectral features putatively identified as lipids or lipid-like molecules are predominantly found in samples from Manaus, whereas organic nitrogen compounds are more abundant in the less urbanized areas (Supplementary Fig. 2). Many mass spectral features derived from cleaning products fall within the class of lipid-like molecules (detergents), thus the predominance of this chemical class in urban settings likely derives from differential cleaning habits.
Figure 2. House chemical and microbial diversity is altered by urbanization.
a. Principal coordinate analysis (PCoA) of chemical compounds (n=270), showing strong clustering by community, but not by socioeconomic group within one community (Bray-Curtis distance metric; PERMANOVA R2 and p-values are displayed). b-d. PCoA of bacterial (n=681), eukaryotic (n=215) and fungal (n=401) composition showing significant segregation among locations in the urbanization gradient (Jaccard distance metric; PERMANOVA R2 and p-values are displayed within each panel). e-g. Correlation between chemical richness and diversity of bacteria (e), eukaryotes (f) and fungi (g). h-k. Diversity of chemicals (h, observed richness, n=270) and microbes (i-k, Shannon diversity) (i, n=681, j, n=215, k, n=401) across the urbanization gradient. Boxplots display median and interquartile range, with boxplot whiskers extending to the most extreme data point within 1.5 times the interquartile range of the first (lower whisker) or third (upper whisker) quartile. Grey boxes on top of each panel indicate sample groupings (one group per row) by the M-W test. Samples sharing a grey bar at the same position are not significantly different (two-sided M-W p>0.05). Chemical and fungal diversity increased with urbanization. No significant differences were observed in 16S or 18S alpha diversity across the urbanization gradient. Chemical diversity analyses were based on all detected small molecule features in our dataset. l-n. Correlation analysis of cleaning and personal care product abundance and house bacteria (l, n=256), microeukaryotes (m, n=82) or fungi (n, n=140). Only correlations with an FDR-corrected Pearson correlation p-value greater than 0.05 are displayed. Outer nodes represent microorganisms (colored by phylum for bacteria, clade for eukaryotes and class for fungi; shaped by source) and central rectangles represent cleaning/personal care products. Names indicate the cleaning/personal care product at the center of each correlation cluster. Edge length is proportional to Pearson correlation p-value (FDR-corrected); edge thickness is proportional to Pearson correlation coefficient (independent scale for each panel). Negative correlations are indicated by red edges.
Overall microbial profiles varied significantly between sampling locations. House fungal profiles differed significantly between Checherta, Puerto Almendra, Iquitos and Manaus (Fig. 2d, PERMANOVA R2=13.47%, p=0.001). Most notably, the relative abundance of yeasts, such as Saccharomyces (K-W χ2=131.96, p<2.2e-16) and Debaryomyces (K-W χ2=103.88, p<2.2e-16), were lower in urban than in rural houses. Malassezia (K-W χ2=43.408, p=2.016e-09), Aspergillus (K-W χ2=103.12, p<2.2e-16) and Candida (K-W χ2=69.526, p=2.858e-14) clades increased with urbanization (Supplementary Table 5, Supplementary Figs. 4 and 5); while most members of these clades are non-pathogenic, these clades also include human pathogens such as Aspergillus fumigatus or Candida albicans. House fungal alpha diversity (K-W χ2=60.752, p=2.428e-12) and house fungal biomass (K-W χ2=44.915, p=4.141e-09) increased with urbanization, despite greater use of cleaning products and antimicrobials (Fig. 2k, Fig. 3, Supplementary Figs. 12, Supplementary Table 4). The more urbanized houses (Manaus low and middle, 97% and 71%) had more samples with detectable ITS sequences by qPCR, compared to less urbanized houses (Checherta 13%, Puerto Almendra 43%, Iquitos 43%, Supplementary Fig. 13a, Fisher’s exact test, p=1.71e-12). In addition, house floor fungal biomass was positively correlated to alpha diversity (Spearman rho=0.4461, p<0.0001, Supplementary Fig. 13b). Increased use of antifungals has been tied to increases in antifungal resistance in both medical and agricultural settings10; our results may reflect a similar trend on housing surfaces, in which increased antimicrobial and cleaning product usage in urban settings lead to reduced fungal sensitivity to these agents and increased fungal loads. Alternatively, our observations could reflect increased exposure to resistant strains in urban settings or a disruption in the succession dynamics of urban microbial communities, such that stable states of communities are disrupted and communities associated with random dispersal and assortment take hold. Lower alpha diversity in remote areas could be due to a higher proportion of taxa not covered by current databases, which would not be mapped and therefore would be missed in subsequent analyses. However, no significant differences in the proportion of sequence matches to the reference database were observed among the four locations (K-W χ2=1.482, p=0.83, Supplementary Fig. 14e), indicating that this is not the case. In relation to rainforest huts, urban houses experience higher temperature (K-W χ2=187.72, p=2.2e-16), less indoor natural luminous intensity (K-W χ2=75.146, p=3.331e-16), elevated indoor CO2 levels (K-W χ2=182.89, p=2.2e-16), and contain more surfaces for fungi to deposit and grow, all of which likely contribute to the urban increase in fungal alpha diversity. However, urbanization degree alone explained a larger proportion of the variation in beta diversity (RDA R2=23.03%, p=0.002) than temperature, luminous intensity in the house and CO2 levels combined (RDA R2=0.502%, non-significant).
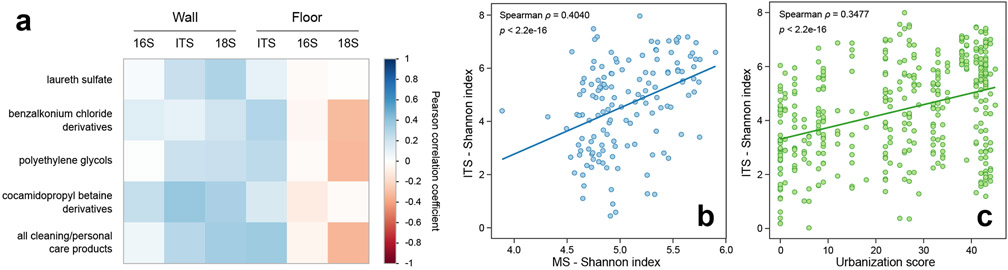
Figure 3. House fungal diversity is correlated to cleaning/personal care product abundance and overall chemical diversity.
a. Correlation plot of house microbial diversity (16S, ITS and 18S) with cleaning/personal care product derivatives (MS) (n=270). Colored according to Pearson correlation coefficient (blue, positive correlation; red, negative correlation; scale displayed right). b. Correlation of fungal diversity (ITS) with chemical diversity (MS) (n=141) (p<2.2e-16, Spearman test). c. Correlation of fungal diversity (ITS) in houses with urbanization score (n=671) (p<2.2e-16, Spearman test).
Human-derived samples revealed unexpected patterns in fungal composition across body vs geographic locations. The strongest differentiating factor for fungal communities was urbanization level (PERMANOVA R2=16.32%, p=0.001; RDA R2=37.31%, p=0.002) rather than body site (PERMANOVA R2=5.34%, p=0.001; RDA R2=7.61%, p=0.002) (Supplementary Fig. 15). This pattern contrasts starkly with typical patterns of bacterial community composition11. The fungal composition of human feet and floor samples strongly clustered by village (PERMANOVA R2=22.14%, p=0.001; RDA R2=50.05%, p=0.002) and not by sampling site (PERMANOVA R2=1.54%, p=0.001; RDA R2=0.899%, p=0.002) (Supplementary Fig. 16), with parallel increases in alpha diversity with urbanization (K-W χ2=13.349, p=0.0058 for foot and χ2=50.02, p<2.2e-16 for floor, Supplementary Fig. 12). The relative abundance of Trichosporon (23.47% in Checherta vs 1.87% in Manaus middle class), Debaryomyces (22.97% in Checherta vs 0.14% in Manaus middle class) and Saccharomyces (5.63% in Checherta vs 0.05% in Manaus middle class) decreased, while the relative abundance of Candida (0.66% in Checherta vs 8.24% in Manaus low-income) and Aspergillus (0.23% in Checherta vs 6.03% in Manaus low-income) increased on feet with urbanization (Supplementary Fig. 6, K-W χ2>58, p<0.0001). In contrast, the gut fungal diversity decreased with urbanization (fecal and anal combined, K-W χ2=18.057, p=0.0004, Supplementary Fig. 12). With urbanization, the relative abundance of Candida (2.23% in Checherta vs 9.41% in Manaus low), and Aspergillus (0.37% in Checherta vs 6.27% in Manaus middle) increased in the gut, while the relative abundance of Debaryomyces (17.93% in Checherta vs 0.51% in Manaus middle), Saccharomyces (9.17% in Checherta vs 0.11% in Manaus middle), Trichosporon (20.98% in Checherta vs 1.15% in Manaus low) and Fusarium (4.73% in Checherta vs 0.36% in Manaus middle) decreased (Supplementary Fig. 6, K-W χ2>50, p<0.0001). Malassezia and other fungi commonly associated with human skin were underrepresented in this study compared to prior reports (e.g. 12,13). To control for primer choice effects, we assessed skin fungal abundance in 18S data, trimming for fungi only. Indeed, in this analysis, a higher relative abundance of Malassezia was observed with urbanization (K-W χ2=43.408, p=2.016e-09). Otherwise, similar results were obtained for other fungal genera between ITS sequencing and 18S sequencing data trimming for fungi (e.g. urbanization-associated increases in Candida (K-W χ2=66.092, p=2.93e-14), Aspergillus (K-W χ2=41.635, p=4.795e-09) and Polyporaceae (K-W χ2= 33.704, p=2.287e-07) in 18S data; urbanization-associated decreases in relative abundance in homes with urbanization for Trichosporon (K-W χ2=17.488, p=0.0005607) and Saccharomycetaceae (K-W χ2=85.985, p<2.2e-16) in 18S data). Bacterial composition of the house surfaces segregated by settlement (Fig. 2b, PERMANOVA R2=14.65%, p=0.001), with urban homes showing a significantly higher relative abundance of skin-associated bacteria (e.g. Corynebacterium, Micrococcus and Enhydrobacter) and a lower abundance of microbes normally associated with the environment (Supplementary Figs. 10 and 11) than their rural counterparts, as previously reported8. For example, relative abundance of Actinobacteria decreased from 44.4% in Checherta to 18.5% in Manaus middle class homes (K-W χ2=126.52, p<2.2e-16) while Proteobacteria relative abundance doubled with urbanization (22.9% in Checherta huts vs 51.0% in Manaus middle class houses) (K-W χ2=168.36, p<2.2e-16). This was mirrored by increases in skin-associated Proteobacteria in urban areas (27.4% in Checherta vs 38.7% in Manaus middle class) (K-W χ2=106.23, p<2.2e-16) (Supplementary Fig. 11). Strikingly, by comparing the molecules detected in our study to bacterial datasets in the Global Natural Product Social Molecular Networking resource (GNPS)14, we found a higher proportion of molecules matched to bacterial datasets in rural locations than in urban sites (Fisher’s exact test, p=9.426e-13, Supplementary Table 6). Matched datasets include metabolomic studies of a number of environmental bacteria, including 11 different actinomycete datasets. These results further indicate a concordance between our 16S and LC-MS datasets.
Effects of urbanization on micro-eukaryotic communities remain largely unknown. We complemented our ITS and 16S analyses with 18S rRNA gene sequencing, filtering out animal, plant and fungal reads. The house micro-eukaryotic composition also differed by location (Fig. 2c, PERMANOVA R2=9.14%, p=0.001), with decreased alpha diversity for floors only (K-W χ2=11.4, p=0.009749, Supplementary Fig. 12), but not for all house sampling locations combined (Fig. 2J), paralleled by decreased alpha diversity in human foot and gut samples (K-W χ2=9.79, p=0.02, Supplementary Fig. 12). These decreases could be due to reduced inputs from the environment, and parasite treatment associated with greater access to medical care. Indeed, microscopic examination of fecal samples did not detect any parasites in Manaus, unlike in rural settings (p<2.2e-16, Spearman test; Fig. 1, Supplementary Table 2 and Figs. 17 and 18). Likewise, we observed an urbanization-associated decrease in the relative abundance (18S sequencing-based) of the common human parasite order Trypanosomatida, which includes human pathogenic genera such as Trypanosoma and Leishmania15 (0.064% in Checherta and absent in Manaus, K-W χ2=11.679, p=0.008566).
To further explore the connection between microbial and chemical exposures, we performed Partial Least Squares Singular Value Decomposition (PLSSVD) analyses of LC-MS/MS, 16S, 18S and ITS data. Results showed that both chemicals and microbes cluster by village, and that the clustering of chemical and microbes per village, respectively, are highly overlapping (Supplementary Fig. 20). Procrustes analysis of beta-diversity results for chemicals and microorganisms also showed modest correlation between datasets (Monte Carlo simulation p<0.0001 and M2=0.797 for 16S-MS Procrustes analysis; p<0.0001 and M2=0.782 for ITS-MS Procrustes analysis). Tighter clustering of 16S and MS samples for the two extremes of our urbanization gradient, Checherta and Manaus middle class, was observed (Supplementary Fig. 21a, c, M-W p=9.59e-5 Manaus middle class vs Manaus lower income; p=3.08e-5 Checherta vs Puerto Almendra). The most likely explanation for these observations is a close connection between the microbiome and its environmental chemistry in those settings, although confounding variables influencing both the microbiome and small molecule profile could also account for such results. In contrast, microbiome composition is more divergent than chemical composition between Manaus middle and Manaus lower income homes (Fig. 2).
Given the expected impact of cleaning product usage on house surface microorganisms, we analyzed the relationship between cleaning product levels and microbial communities in greater detail (Fig. 2l-n, Fig. 3, Supplementary Data Files 1-4). Overall cleaning product usage, as well as each individual cleaning product, were positively correlated with fungal diversity (Fig. 3, p<2.2e-16, Spearman test). Very few taxa were correlated with multiple cleaning products, suggesting specific effects of the different cleaning products (Fig. 2l-n). Members of the Proteobacteria phylum correlated with benzalkonium chloride derivatives, while most fungal genera of the Agaricomycetes class correlated with polyethylene glycol derivatives (p<0.05, Pearson, FDR-corrected). Many of the organisms that correlated with cleaning products occur in multiple environments or were of unknown source. Strikingly however, most of the remaining organisms were of environmental origin (aquatic for bacteria and micro-eukaryotes; plant-derived for fungi). These correlated microorganisms could either be resistant to cleaning products or colonizers filling a niche opened up by cleaning practices. Very few correlated organisms were exclusively human-derived, suggesting that the resistant or recolonizing organisms are primarily environmentally-derived. Although correlation does not mean causation, these patterns support future avenues of investigation.
Increased urbanization is linked to increased social stratification, and one might expect low-income neighborhoods in a large metropolis to resemble rural areas due to reduced access to services, commodities and goods compared to higher-income groups. Indeed, the urbanization score for lower income houses in Manaus was intermediate between rural areas and Manaus middle class. However, there were only minor differences in house bacterial composition and alpha diversity between the two groups (Fig. 2b, PERMANOVA of the two groups (same below): R2=3.71%, p=0.001; Fig. 2i, M-W p=0.1835), fungal composition (Fig. 2d, PERMANOVA R2=5.75%, p=0.001) and chemical composition (Fig. 2a, PERMANOVA R2=3.25%, p=0.001). However, reduced human bacterial alpha diversity was observed in the higher socio-economic level compared to samples from low-income neighbourhoods (M-W p<0.0001, Supplementary Fig. 12). These findings correlate well with recent data showing reduced bacterial fecal alpha diversity in populations experiencing economic development16. Interestingly, feet samples from people living in low-income areas were more similar in terms of bacterial composition to those from Checherta and Puerto Almendra than those from Iquitos and Manaus middle-class neighborhoods (Supplementary Fig. 11), consistent with walking barefoot. Increasing relative abundance of foot Staphylococcus was associated with shoe use (4.76% in Checherta, 6.64% in Manaus low-income and 29.67% in Manaus middle class) (Spearman rho=0.4627, p=1.212e-12). Similarly, more Staphylococcus were found in the floors of homes in middle-class neighbourhoods (10.79%) compared to those of low-income homes (2.94%) (M-W p=0.012). Overall, these results suggest that the differentiating effects we observed between Checherta, Puerto Almendra, Iquitos and Manaus are related to factors shared by all in the settlements, more than to individual lifestyles.
This work is an important advance from previous research on the built environment that focused on Western urban environments17 or only on bacteria8. While not necessarily extrapolatable to all locations, given that many environmental and cultural variables affect the microbiome and small molecule profile, these observations provide expanded insights into the joint chemical and microbial changes associated with changing cultural practices, and their relationship to the transition from infectious to noncommunicable diseases. Generalizability is further supported by the fact that we observed common alterations between samples collected in Manaus in 2012 and 2013, even though they were collected on two separate sampling expeditions (PERMANOVA R2=3.39% for 16S, 3.21% for ITS, p=0.001 for both) (Supplementary Fig. 22). Our results provide a comprehensive view of microbes in homes and people (bacteria, fungi, other micro-eukaryotes and chemical profile) across urbanization, enabling the identification of cross-domain changes correlated with disturbances caused by changes in urban design, architecture and human behavior. These results will inform future studies into the functional connection between urban lifestyle, microbiota and health.
Methods
Sample collection
Researchers collected samples in 2012 in the Amazon, along the same latitude, starting in the jungle (Checherta), where no epidemics were present, to the researchers’ best knowledge. They then moved to more urbanized societies with the emergence of a lower socioeconomic class (Puerto Almendra), to a large town (Iquitos) and to a metropolis (Manaus). All samples in Checherta, Puerto Almendra and Iquitos were collected in July-August 2012. Manaus 2012 samples were collected August-October 2012. Additional samples were collected in Manaus in December 2013, including samples from two different socio-economic classes (Fig. 1; sampling stratification by neighbourhood). The average temperature during 2012 and 2013 sampling was between 25 and 28°C (Supplementary Table 1). With urbanization, more industrial and less natural building materials were used (p<2.2e-16, Spearman test, Fig. 1c). Checherta houses were mainly constructed from wood, soil, cotton and cloth; Puerto Almendra houses from soil, wood, plastic, bricks, and cloth; Iquitos houses from cement, metal, plastic, tiles, ceramic, wood, cotton and paint; Manaus low-income houses from cement, ceramic, clay blocks, concrete, fabric and glass; and Manaus middle-class houses from ceramic, glass, granite, metal, plaster, tiles, plastic, brick and mortar (Fig. 1c). In more rural settings, more people were living in one house or room, while in more urbanized areas, the inhabitants had more privacy, with less inhabitants per house and more rooms available per inhabitant (χ2=469.09, p<2.2e-16, Kruskal-Wallis (K-W) test) (Supplementary Table 1). A higher cleaning frequency was noted with urbanization (χ2=907.29, p<2.2e-16, K-W test). In Iquitos, Manaus low income and Manaus middle class, the house was cleaned much more frequent (almost every day), as compared to Checherta (never) and Puerto Almendra (every week to every month) (Supplementary Table 1). The average natural luminous intensity in the house (χ2=75.146, p=3.331e-16, K-W test), CO2 inside (χ2=182.89, p<2.2e-16, K-W test) and air exchange rate inside the house decreased in more urban locations (Supplementary Table 1). With urbanization, the inhabitants had higher incidence of self-reported allergy (p=6.6e-14, Spearman test), self-reported asthma (p=0.00013, Spearman test), other self-reported autoimmune diseases, self-reported cardiovascular diseases, self-reported epilepsy, self-reported thyroid conditions and self-reported tumors. In less urban settings, the inhabitants had increasingly more parasites (p<2.2e-16, Spearman test) (Supplementary Figs. 17 and 18 and Table 1). Frequently encountered parasites included Giardia lamblia, Ascaris lumbricoides, Trichuris trichiura, Strongyloides, Ancylostoma duodenale and Hymenolepis nana.
Per location, 8 to 10 representative houses were selected and sampled, as well as their inhabitants (33 to 53 humans and their pets) (Supplementary Table 3). Six body sites were sampled per subject, six objects and eight surfaces were sampled per home, and three body samples per pet. All wall samples were collected 1.5 meters above the floor. The total number of samples were 107 to 273 house samples, 199 to 320 human samples and 21 to 48 animal samples per location (Supplementary Table 3). House sites and participants were distributed throughout the village/city, except for Manaus lower income vs middle income stratification, where samples were collected from within the shanty areas vs middle class neighbourhoods of the city, respectively. The first two of the Peruvian villages had a population size of 120–200 subjects, therefore our sampling of 40 houses represent 20–33% the village as a whole. For the two latter communities, Iquitos has a population of ~400,000 and Manaus of 2 million, so we only sampled a small transect of these locations.
An urbanization score was calculated based on a series of parameters that are introduced when rural areas become cities, i.e. the level of education, access to health care, Western practices such as use of shoes, clothes and processed food, roads and traveling time, materials used for house construction (determined by visual inspection), and the introduction of electrical appliances (fans, air conditioning, washer, dryer, wireless internet, kitchen exhaust). The degree of urbanization was determined based on the urbanization score (Supplementary Data File 5). Although there are some clear cultural differences between Manaus and Peru, these locations were selected because they were all within the same latitude, which for this particular study was necessary to keep similar environmental conditions (Supplementary Fig. 2).
Over 300 blank samples were obtained throughout the sampling expeditions. Samples were taken for bacterial, fungal, eukaryotic and LC-MS/MS analysis, and metadata collected concerning the houses (temperature, relative humidity, luminous intensity in the house, CO2, etc.) and their inhabitants (age, gender, length, weight, parasite detection, self-reported health status, footwear, pets, etc.).
Researchers complied with all relevant ethical regulations; participants provided informed consent for all sample collection. The work was performed under IRB approval from the University of Puerto Rico (UPR, # 112–172), from the Ministry of Health of Peru (#001–2013-CIEI/INS), and from the Federal University of Amazonas in Manaus, Brazil (UFAM, #46532). South America maps were obtained from OpenStreetMap (https://www.openstreetmap.org/), under the Open Data Commons Open Database License (ODbL). Human and dog silhouettes were obtained from PhyloPic (http://phylopic.org/), under the Public Domain Dedication 1.0 license.
DNA sequencing
DNA extraction and PCR amplification of the 16S, 18S, and ITS genes were performed following the protocols described in18 and the Earth Microbiome Project (EMP)19. 16S amplicons were sequenced on an Illumina HiSeq platform at the Biofrontiers Sequencing Facility located at the University of Colorado (CU) Boulder and the Genomics Core at CU Denver. 18S and ITS amplicons were sequenced on an Illumina HiSeq functioning in rapid run mode at the Institute for Genomic Medicine located at the University of California San Diego. All DNA samples were stored at −80°C.
Blank sample analysis
A total of 2,572 samples from 16S rRNA sequencing were obtained after Deblur. The 2,572 samples contained 119,655,350 sequences in total, with a minimum of two sequences per sample, a maximum of 452,208 sequences per sample, a median of 45,488 sequences per sample and an average of 53,754 sequences per sample. A total of 343 blank samples were taken during the Amazon missions and 50 blank samples were retrieved with sequences. The blanks had an average of 16,628 sequences per sample and a median of 11,284 sequences per sample. Seventeen blank samples had less than 1,000 sequences per samples and were not withheld after rarefaction. A total of 33 blanks were still present after rarefaction, with more than 1,000 sequences per sample. These remaining blank samples contained predominantly Pseudomonas spp. (62.67% on average), which were likely contamination20. Therefore, no claims or conclusions were taken with Pseudomonas counts. This taxon was however not filtered out to prevent skewing the data.
Fungal biomass quantification
To evaluate fungal biomass, ITS quantitation was performed on a subset of samples (n=204) using qPCR. Specifically, 204 floor samples from Checherta (n=23), Puerto Almendra (n=30), Iquitos (n=40), Manaus low (n=36), and Manaus middle (n=75) were processed with qPCR (Roche LightCycler 480 II, Cat# 05015243001) in triplicate and 40% replication using 10 μl reaction volumes (ThermoFisher, PowerUp SYBR green mastermix, Cat# A25742), 1 μl of gDNA, and the same ITS primers and amplification metrics as used for sequencing (EMP protocol) in a 384 well format. For samples which were sequenced but below sampling depth cutoff (n=51), high concordance between ITS sequencing and qPCR data were observed, with only 6 of these samples showing mean ITS copies by qPCR >10. Likewise, of the sequenced samples with mean ITS copies by qPCR >10 (n=88), all but 6 of them had ITS sequencing data that passed our filters. Effective limit of detection for these mixed samples is 100–1000 ITS copies, as previously reported21,22. Discrepancies between fungal biomass quantification and ITS sequencing results may also be explained by the necessary use of different cycling conditions and enzyme mixes between qPCR and preparation for sequencing, the fact that qPCR was performed close to five years after ITS sequencing, and the higher sensitivity of fluorescence-based assays to PCR inhibitors compared to sequencing approaches23.
Chemical extraction and UPLC-Q-TOF MS/MS analysis
To avoid possible biased small molecule recovery due to different swab types, we only performed LC-MS/MS analysis on swabs from Fisher (cat# 23–400-119), for which we had representation across all study locations (n=270). Swabs were extracted and analyzed using a previously validated workflow described in24,25. Swabs were extracted in 300 μL of 50:50 ethanol/water solution for 2 hours on ice then overnight at −20°C. Swab sample extractions were concentrated in a centrifugal evaporator, and redissolved in 100 μL of 50:50 ethanol/water with internal standard (fluconazole 1μM). The ethanol/water extracts were then analyzed using a previously validated UPLC-Q-TOF MS/MS method24,25. Liquid chromatography separation was performed on a Thermo Fisher Scientific UltiMate 3000 UPLC system using a 1.7 μm C18 (50 × 2.1 mm) UHPLC Column (Phenomenex). UPLC conditions of analysis were set as follow: column temperature: 40 °C, flow rate 0.5 mL/min, mobile phase A 98 % water / 2 % acetonitrile / 0.1 % formic acid (v/v), mobile phase B 98 % acetonitrile / 2 % water / 0.1 % formic acid (v/v). A linear gradient was used for the chromatographic separation: 0–2 min 0–20 % B, 2–8 min 20–99 % B, 8–9 min 99–99% B, 9–10 min 0% B.
MS/MS analysis was performed on a Maxis Q-TOF (Quadrupole-Time-of-Flight) mass spectrometer (Bruker Daltonics), controlled by the Otof Control 4.0 and Hystar 3.2 software packages (Bruker Daltonics) and equipped with ESI source. Full-scan MS spectra (m/z 80–2000) were acquired in a data-dependent positive ion mode. Instrument parameters were set as follows: nebulizer gas (nitrogen) pressure: 2 Bar, capillary voltage: 4,500 V, ion source temperature: 180°C, dry gas flow: 9 L/min, spectra rate acquisition: 10 spectra/s. MS/MS fragmentation of 10 most intense selected ions per spectrum was performed using ramped collision induced dissociation energy, ranged from 10 to 50 eV to get diverse fragmentation patterns. MS/MS active exclusion was set after 4 spectra and released after 30 seconds.
MS, 16S, 18S and ITS data analysis
MS/MS data were processed using Optimus software26 (v0.1, downloaded Jan. 3 2017) with default parameters, except as follows: m/z tolerance 20 ppm, retention time tolerance 30s, noise threshold 3,000. To control for swab characteristics and swab-derived molecules, features were only retained if they had a minimum intensity ratio of 3.0 compared to blank swabs. Feature intensity was normalized using total ion current (TIC). Beta diversity was assessed using the Bray-Curtis distance metric, and visualized with principal coordinates analysis (PCoA) using QIIME v1.9.127. The correlation between the distance matrix and certain metadata categories was tested with permutational multivariate analysis of variance (PERMANOVA)28, which reports an R2 value indicating the proportion of variation explained by this category, and a p-value representing the statistical significance. Putative feature identification was performed by performing mass spectral molecular networking and spectral library matching using the Global Natural Products Social Molecular Networking (GNPS) platform14,29. Data was filtered by removing all MS/MS peaks within ±17 Da of the precursor m/z, followed by data clustering with MS-Cluster, with a parent mass tolerance of 1 Da and a MS/MS fragment ion tolerance of 0.5 Da (these settings were used as reference spectra contributed by the GNP community also contain low resolution spectra); consensus spectra containing less than 3 spectra were discarded. Networking parameters were as follows and can be accessed publicly at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=549cfafcdaef4a7496768f45bb90771c#: cosine score above 0.65; more than 4 matched peaks; nodes in each other’s respective top 10 most similar nodes. Analog search was enabled against the library with a maximum mass shift of 100.0 Da. The spectra in the network were then searched against GNPS spectral libraries and GNPS datasets, with the same parameters. Such parameters have been associated with 1–5% false discovery rates on comparable datasets30 and all putative identification hits were manually inspected (Supplementary Fig. 3). To further enhance putative structure identification we performed in silico structure annotation through Network Annotation Propagation (NAP)9. Parameters were set to 10 first candidates for consensus score, fusion result for consensus enabled, 15 ppm accuracy for exact mass candidate search, cosine value to subselect inside a cluster: 0.8, [M+H] adduct, maximum 10 candidate structures in the graph, parent mass selection enabled, structure databases: GNPS, CHEBI, HMDB, SUPNAT. NAP output is publicly available at: https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=af425ada55d54adca9c7b28a823af54c. A major challenge of in silico annotation is the uncertainty around the correct structure among a predicted candidate list 9. Predicted structures cannot be verified through the comparison of a reference library spectrum, but fragmentation patterns need to be manually confirmed, a task infeasible for the over 10,000 mass spectral features found in our mass spectral molecular network. To get an overview of putative identifications we retrieved through GNPS library matching and in silico structure annotation, we calculated most predominant chemical classes per mass spectral molecular family (two or more connected components of a graph) by submitting in silico structures and structures retrieved through library matching to automated chemical classification using ClassyFire31,32. Subsequently, to evaluate the consistency of structure annotation, we calculated a ClassyFire score (Supplementary Fig. 2). A ClassyFire score of 1 represents a scenario where all predicted structures within the mass spectral molecular family fall within the same chemical class, whereas a score close to 0 represents a scenario where no predominant chemical class could be identified within the mass spectral molecular family, possibly resulting from false annotations. All annotations are level 2/3 according to the 2007 metabolomics consortium standard initiative33. Molecular networks were visualized using Cytoscape version 3.4.034. Chemical structures were drawn with ChemDraw Professional 16.0. Venn diagrams were built using http://bioinformatics.psb.ugent.be/webtools/Venn/.
All three types of DNA sequence data: 16S (n=2178), 18S (n=525), and ITS (n=758) were quality-filtered to discard sequences with a quality score of <20. 16S sequence data were trimmed to 100 nt, while 18S and ITS sequence data were trimmed to 150 nt. Quality filtering and trimming was done in QIITA (https://qiita.ucsd.edu/)35. Sequences were denoised using Deblur 1.0.236 and assigned taxonomy using SortMeRNA 2.037. The reference sequence databases used for these two steps were: 16S: Greengenes v13_838, 18S: SILVA v12339, and ITS: UNITE v7.140. OTU tables generated in the primary processing step were rarefied to 1,000 sequences per sample for all marker types. After this operation, 90.12% of DNA samples (16S: n=2,120, 18S: n=328, ITS: n=671) were retained. Alpha diversity for each sample and distances between samples were calculated using QIIME27. Alpha diversity as measured by Observed OTU, Chao1, Gini evenness and Shannon were calculated with QIIME. Beta diversity was measured by the abundance weighted Jaccard metric, visualized with principal coordinates analysis (PCoA) and tested with PERMANOVA28 in QIIME. The contributions of multiple factors to the community variation were compared using the cumulative effect sizes computed from the redundancy analysis (RDA)41 implemented in vegan 2.5.4 42. This analysis also reports R2 and p-values, in which R2 values of different factors are directly comparable. Blank samples were included for all sample types, at all locations and were analyzed as discussed above.
Multi-omics analysis
LC-MS/MS and microbiome data were analyzed together using two strategies: First, the beta diversity PCoAs of the two data types were transformed and overlaid using Procrustes analysis43 implemented in QIIME. Top ten dimensions were retained, with 1,000 Monte Carlo permutations performed to assess the statistical significance of correlation, as represented by the M2 metric (larger is less correlated) and its p-value. The distribution of per-sample pairwise distances weighted by the loadings of the corresponding axes in the Euclidean space was compared using the M-W test. Second, we used the Partial Least Squares Singular Value Decomposition (PLSSVD) method44 to infer the correlation within and between the two data types. This was performed using the PLSSVD function in scikit-learn, which calculates a singular value decomposition (SVD) on the covariance matrix between log transformed chemical and microbial abundances. A pseudocount was added to both datasets to avoid taking logs of zero. The loadings calculated from PLS were visualized using a biplot, where the points represent microbes, and the arrows represent chemicals. The angle between arrows provide information about correlations between chemicals. Arrows pointing the same direction indicate positive correlation, whereas arrows pointing in opposite directions indicate negative correlations. The longer the arrows are, the larger the variance is within the specific small molecule. The distance between arrows provides information about correlation between chemicals–smaller distances indicates higher correlation between chemicals. Indicator value analysis was performed using the labdsv45 package to identify which microbes and chemicals are likely to be uniquely associated to specific socio-economic classes.
The correlation between the relative abundance of MS and DNA was assessed using the Pearson correlation analysis, as implemented in QIIME. For MS, we calculated the sums of the MS1 feature abundances of four categories of chemicals that are frequently associated with cleaning and personal care products: sodium laureth sulfate, benzalkonium chloride derivatives, polyethylene glycols, and cocamidopropyl betaine derivatives. For each type of microorganism, we calculated the sums of the relative abundances of OTUs assigned to each taxonomic rank (phylum, class, order, family and genus). The sums of MS and DNA features were subject to the Pearson correlation analysis, with 1000 permutations. The p-values after Benjamini-Hochberg FDR correction were reported.
Statistics and reproducibility
We used PERMANOVA to determine whether diversity levels correlated with a categorical metadata column. Cumulative effect sizes of multiple metadata columns were calculated using the redundancy analysis (RDA). Rank-based differences among the settlements were tested using non-parametric Kruskal-Wallis tests. Pairwise differences between two groups were tested using non-parametric Mann-Whitney U tests. Monotonic relationships between two variables were tested using non-parametric Spearman correlation test. The alternative hypothesis is by default two-sided. Boxplots were made with R default code: box length = interquartile range (IQR), Q3 - Q1; upper whisker = min(max(x), Q3 + 1.5 * IQR) and lower whisker = max(min(x), Q1 – 1.5 * IQR). To compare the relationship between ITS biomass (copies per μl) and ITS richness (Chao1 index), both values of successfully quantified samples were log transformed and compared using Spearman correlation implemented in Prism v.8.0.0. Reproducibility of findings is supported by the fact that beta diversity was comparable for microbiome samples for Manaus middle class collected on two separate trips (2012 and 2013). Samples collected in Manaus in 2012 and in 2013 group together by principal coordinate analysis and are both distinct from the other settlements, further confirming reproducibility of our findings.
Data availability
Mass spectrometry data have been deposited in MassIVE (accession number MSV000082924). Molecular networking jobs can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=549cfafcdaef4a7496768f45bb90771c (full housing dataset). GNPS molecular networking jobs for dataset matching can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f1bc8144b7f648dc94215a34b94537df (Checherta), https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=044f5c1437e4453eb9d47afafadb7cfb (Puerto Almendra), https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c3736e2379d1470f8b9e990a1afeb682 (Iquitos), https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8799f335311540bdb5af75da896bd87c (Manaus low-income) and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=1070c36112c940a186fcf4454f845f08 (Manaus middle class; searches performed August 19 2018). In silico structure annotation using NAP can be accessed here: https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=af425ada55d54adca9c7b28a823af54c. The raw sequencing data and processed BIOM tables are available at Qiita (https://qiita.ucsd.edu/) under study ID 10333, and also at EMBL-EBI under submission number ERP107551.
Code availability
Instructions and source codes for replicating the bioinformatic analyses are available at: https://github.com/knightlab-analyses/amazon-urbanization.
Supplementary Material
Acknowledgements
We acknowledge the collaborators in Peru, the late Father Luigi Bocca and the interpreter J. J. Semu for their support and information sources. In Manaus, we had the valuable help of A. Vasconcelos and J. Machado with the fieldwork, and the support of the community leader Ms. Maria Aparecida Lisboa, Director of Association Fazendo Amigos (AFA), Manaus. We thank the nurses that accompanied the researchers in the jungle. Irene Fajardo Neddermann helped us in the architectural work. Daniela Vargas and Magda Magris helped prepare the urbanization score survey. Daniel McDonald, James Morton, Ricardo da Silva, and Lingjing Jiang helped with DNA and MS data analysis. Alexander Cai helped determine the sources of microorganisms correlated with cleaning products. In Peru, we had the support of staff and community members participating within the Malaria Immunology and Genetics in the Amazon Project with the Ministry of Health of Peru.
Funding: This work was supported by the Sloan Foundation (to M.G.D.-B., R.K, & P.C.D), C&D Fund, and Emch Fund for Microbial Diversity (to M.G.D.-B.). Partial support was also provided by the NIH Research Initiative for Scientific Enhancement Program 2R25GM061151–13 (to J.F.R.-C.). LIM was partially supported by a fellowship from the Canadian Institutes of Health Research (338511, http://www.cihr-irsc.gc.ca/). CC was supported by the Belgian American Educational Foundation and the Research Foundation Flanders. We acknowledge the NIH for providing the MS and MS data analysis infrastructure P41-GM103484 and GMS10RR029121 (P.C.D.).
Footnotes
Competing interests
The authors declare no competing interests.
Materials & Correspondence
Correspondence and material requests should be addressed to Maria G. Dominguez-Bello (mg.dominguez-bello@rutgers.edu - study design and sequencing), Rob Knight (robknight@ucsd.edu - sequencing and bioinformatics) or Pieter C. Dorrestein (pdorrestein@ucsd.edu - LC-MS/MS analysis).
References
- 1.United Nations Publications. World Population Prospects, the 2015 Revision. (World Population Prospects, 2016). [Google Scholar]
- 2.Oyebode O et al. Rural, urban and migrant differences in non-communicable disease risk-factors in middle income countries: a cross-sectional study of WHO-SAGE data. PLoS One 10, e0122747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ege MJ et al. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med 364, 701–709 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Yatsunenko T et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Filippo C et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U. S. A 107, 14691–14696 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemente JC et al. The microbiome of uncontacted Amerindians. Sci Adv 1, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Obregon-Tito AJ et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat. Commun 6, 6505 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruiz-Calderon JF et al. Walls talk: Microbial biogeography of homes spanning urbanization. Sci Adv 2, e1501061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.da Silva RR et al. Propagating annotations of molecular networks using in silico fragmentation. PLoS Comput. Biol 14, e1006089 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlin DS, Rautemaa-Richardson R & Alastruey-Izquierdo A The global problem of antifungal resistance: prevalence, mechanisms, and management. Lancet Infect. Dis 17, e383–e392 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Findley K et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh J et al. Biogeography and individuality shape function in the human skin metagenome. Nature 514, 59–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang M et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol 34, 828–837 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCall L-I & McKerrow JH Determinants of disease phenotype in trypanosomatid parasites. Trends Parasitol. 30, 342–349 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Stagaman K et al. Market Integration Predicts Human Gut Microbiome Attributes across a Gradient of Economic Development. mSystems 3, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams RI et al. Ten questions concerning the microbiomes of buildings. Build. Environ 109, 224–234 (2016). [Google Scholar]
- 18.Caporaso JG et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A 108 Suppl 1, 4516–4522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson LR et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salter SJ et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CM et al. BactQuant: an enhanced broad-coverage bacterial quantitative real-time PCR assay. BMC Microbiol. 12, 56 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil-Serna J, González-Salgado A, González-Jaén MAT, Vázquez C & Patiño B ITS-based detection and quantification of Aspergillus ochraceus and Aspergillus westerdijkiae in grapes and green coffee beans by real-time quantitative PCR. Int. J. Food Microbiol 131, 162–167 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Schrader C, Schielke A, Ellerbroek L & Johne R PCR inhibitors - occurrence, properties and removal. J. Appl. Microbiol 113, 1014–1026 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Bouslimani A et al. Molecular cartography of the human skin surface in 3D. Proc. Natl. Acad. Sci. U. S. A 112, E2120–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bouslimani A et al. Lifestyle chemistries from phones for individual profiling. Proc. Natl. Acad. Sci. U. S. A 113, E7645–E7654 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Protsyuk I et al. 3D molecular cartography using LC-MS facilitated by Optimus and ‘ili software. Nat. Protoc 13, 134–154 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Caporaso JG et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson MJ A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46 (2001). [Google Scholar]
- 29.Watrous J et al. Mass spectral molecular networking of living microbial colonies. Proc. Natl. Acad. Sci. U. S. A 109, E1743–52 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheubert K et al. Significance estimation for large scale metabolomics annotations by spectral matching. Nat. Commun 8, 1494 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Djoumbou Feunang Y et al. ClassyFire: automated chemical classification with a comprehensive, computable taxonomy. J. Cheminform 8, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ernst M et al. MolNetEnhancer: Enhanced Molecular Networks by Integrating Metabolome Mining and Annotation Tools. Metabolites 9, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumner LW et al. Proposed minimum reporting standards for chemical analysis. Metabolomics 3, 211–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez A et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat. Methods 15, 796–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amir A et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kopylova E, Noé L & Touzet H SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28, 3211–3217 (2012). [DOI] [PubMed] [Google Scholar]
- 38.McDonald D et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quast C et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kõljalg U et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol 22, 5271–5277 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Falony G et al. Population-level analysis of gut microbiome variation. Science 352, 560–564 (2016). [DOI] [PubMed] [Google Scholar]
- 42.CRAN - Package vegan. Available at: https://CRAN.R-project.org/package=vegan. (Accessed: 3rd June 2019)
- 43.Gower JC Generalized procrustes analysis. Psychometrika 40, 33–51 (1975). [Google Scholar]
- 44.Kapono CA et al. Creating a 3D microbial and chemical snapshot of a human habitat. Sci. Rep 8, 3669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts DW labdsv: Ordination and multivariate analysis for ecology. R package version (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited in MassIVE (accession number MSV000082924). Molecular networking jobs can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=549cfafcdaef4a7496768f45bb90771c (full housing dataset). GNPS molecular networking jobs for dataset matching can be accessed here: https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=f1bc8144b7f648dc94215a34b94537df (Checherta), https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=044f5c1437e4453eb9d47afafadb7cfb (Puerto Almendra), https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=c3736e2379d1470f8b9e990a1afeb682 (Iquitos), https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=8799f335311540bdb5af75da896bd87c (Manaus low-income) and https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=1070c36112c940a186fcf4454f845f08 (Manaus middle class; searches performed August 19 2018). In silico structure annotation using NAP can be accessed here: https://proteomics2.ucsd.edu/ProteoSAFe/status.jsp?task=af425ada55d54adca9c7b28a823af54c. The raw sequencing data and processed BIOM tables are available at Qiita (https://qiita.ucsd.edu/) under study ID 10333, and also at EMBL-EBI under submission number ERP107551.