Abstract
Multiple myeloma (MM) is an incurable disease caused by the infiltration of malignant plasma B cells into bone marrow, whose pathogenesis remains largely unknown. Long non-coding RNAs (lncRNAs) have emerged as important factors in pathogenesis. Our previous study validated that lncRNA ST3 β-galactoside α-2,3-sialyltransferase 6 antisense RNA 1 (ST3GAL6-AS1) was upregulated markedly in MM. Therefore, the aim of the study was to investigate the molecular mechanisms of ST3GAL6-AS1 in MM cells. ST3GAL6-AS1 expression levels in MM cells was detected using reverse transcription-quantitative PCR. ST3GAL6-AS1 antisense oligonucleotides and small interfering RNAs were transfected into MM cells to downregulate expression. In vitro assays were performed to investigate the functional role of ST3GAL6-AS1 in MM cells. RNA pull-down, RNA immunoprecipitation and comprehensive identification of RNA-binding proteins using mass spectrometry assays were used to determine the mechanism of ST3GAL6-AS1-mediated regulation of underlying targets. It was reported that knockdown of ST3GAL6-AS1 suppressed the adhesion, migration and invasion ability of MM cells in vitro. Expression of ST3GAL6 was significantly reduced when ST3GAL6-AS1 was knock downed in MM cells. Moreover, mechanistic investigation showed that ST3GAL6-AS1 could suppress ST3GAL6 mRNA degradation via interacting with heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1). The present results suggested that upregulated lncRNA ST3GAL6-AS1 promotes adhesion and invasion of MM cells by binding with hnRNPA2B1 to regulate ST3GAL6 expression.
Keywords: multiple myeloma, ST3GAL6-AS1, ST3GAL6, adhesion, invasion, heterogeneous nuclear ribonucleoprotein A2B1
Introduction
Multiple myeloma (MM) is a fatal malignant proliferation of antibody-secreting plasma cells (PCs) in bone marrow (BM) (1), the second most common hematological malignancy in the world in 2019 (2-4). Driven by access to more efficacious drugs and clinical trials, an increasing number of newly diagnosed patients with MM have an overall survival of >10 years on average globally in recent years (5-7). However, MM is still an incurable disease due to drug resistance and relapse (8).
Over the past decade, studies have demonstrated that non-coding RNAs are dysregulated and play key roles in the pathogenesis of MM, similar to protein coding genes (9,10). Long non-coding RNA (lncRNA) is a sort of non-coding transcripts >200 nucleotides in length (11), which regulate gene transcription and mRNA translation through various mechanisms, such as interaction with RNA-binding proteins, epigenetic modification of gene expression and microRNA modulation (11). Therefore, lncRNAs may have potential as new therapeutic targets of MM.
In our preceding study, a landscape of differentially expressed lncRNAs and mRNAs was identified in five newly diagnosed cases of MM using microarray analysis (12). It was reported that lncRNA ST3 β-galactoside α-2,3-sialyltransferase 6 antisense RNA 1 (ST3GAL6-AS1) is upregulated in patients with newly diagnosed MM and relapse. Based on these results, it was hypothesized that ST3GAL6-AS1 may be closely related to the pathogenesis of MM. We confirmed the co-expression relationship between ST3GAL6-AS1 and ST3GAL6, and the correlation coefficient was >0.6 among pan-cancer and 0.563 in MM (12). Moreover, ST3GAL6 has been proved to be upregulated in MM and influencing homing and survival of MM cells (13). The present study investigated the role of ST3GAL6-AS1 in MM and explored the mechanisms of ST3GAL6-AS1 and ST3GAL6 interaction.
Materials and methods
MM specimens
Approval was obtained from The Ethics Committee of the Second Affiliated Hospital of Xi'an Jiaotong University (Xi'an, China; approval no. 2015186). When the written informed consent was obtained from all patients, bone marrow specimens were obtained using aspiration from 83 patients with newly diagnosed MM. All patients with MM were diagnosed according to International Myeloma Working Group (IMWG) (14) criteria after bone marrow morphology, biochemical parameters, imaging data, chromosomal and genetic examinations. Patient staging and stratification were according to 2016 IMWG staging (15) and 2013 Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) stratification (16). Patients with concomitant cardiopulmonary and other cancer types were excluded. The other related information was as previously described (12). The general clinical and laboratory features of the 83 patients with newly MM are summarized in Table I.
Table I.
Information of 83 newly diagnosed patients with multiple myeloma.
| Characteristics | value |
|---|---|
| Sex, n (%) | 39 (47.0) |
| Male | 44 (53.0) |
| Female | |
| Age, median (range) years | 57 (42-75) |
| WBC, median (range) ×109/l | 4.9 (3.1-12.3) |
| Hb, median (range) g/l | 82 (60-103) |
| PLT, median (range) ×109/l | 193 (43-367) |
| Type, n (%) | |
| IgG | 55 (66.3) |
| IgA | 15 (18.1) |
| IgD | 3 (3.6) |
| Light chain | 8 (9.6) |
| NS | 2 (2.4) |
| D-S stage, n (%) | |
| I | 27 (32.5) |
| II | 24 (28.9) |
| III | 32 (38.6) |
| ISS stage, n (%) | |
| I | 37 (44.6) |
| II | 20 (24.1) |
| III | 26 (31.3) |
| R-ISS stage, n (%) | |
| I | 35 (42.2) |
| II | 23 (27.7) |
| III | 25 (30.1) |
| mSMART risk status, n (%) | |
| Low | 46 (55.4) |
| Intermediate | 21 (25.3) |
| High | 16 (19.3) |
| Percentage of MM cells in | 37 (14-73) |
| bone marrow, median (range) | |
| Bence-Jone protein, n (%) | 59 (71.1) |
| ESR elevated | 63 (75.9) |
| LDH elevated | 41 (49.4) |
| Hypercalcemia, n (%) | 9 (10.8) |
| Renal failurea, n (%) | 27 (32.5) |
| Bone lesions | 42 (50.6) |
Serum creatinine ≥2 mg/100 ml. NS, non-secretory; D-S, Durie-Salmon; ISS, International Staging System; R-ISS, Revised International Staging System; ESR, erythrocyte sedimentation; LDH, lactate dehydrogenase; mSMART, Mayo Stratification of Myeloma and Risk-Adapted Therapy.
Cell culture and transfection
The human MM cell lines RPMI 8226 and MM.1S and human umbilical vein endothelial cells (HUVEC) were purchased from American Type Culture Collection, and the mycoplasma testing were negative. All cells were cultured in RPMI 1640 medium (HyClone; GE Healthcare Life Sciences) or Dulbecco's modified Eagle's medium (HyClone; GE Healthcare Life Sciences) supplemented with penicillin/streptomycin (100 U/ml) (Thermo Fisher Scientific, Inc.) in 10% fetal bovine serum (FBS) (Biological Industries, Inc.). RPMI 8226 and MM.1S cells were seeded in 10 cm diameter culture dishes at a density of 8×106 cells. HUVEC cells were seeded in 5-cm diameter culture dishes at a density of 3×106 cells. The cells were cultured in an incubator at 37°C with a 5% CO2 atmosphere. All transfection experiments were carried out using electroporation (CUY21 EDIT II electroporator; BEX, Co., Ltd.; http://www.bexnet.co.jp/english/product/device/in-vivo-in-vitro-electroporator/cuy21edit2.html) according to the manufacturer's instructions. Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) was used as pulsing buffer to suspend the cells in electroporation cuvette. In total, 25 μl of cell suspension (1×106 cells) containing 1 μl smart silencer or NC (20 μM) was dispensed into electric cuvettes (electrode spacing =2 mm; material aluminum electrode; cat. no. SE-202; BEX, Co., Ltd.). The electroporation conditions are shown in Table SI. After electroporation, cells were cultured in RPMI 1640 medium supplemented with 10% FBS (Biological Industries) in an incubator at 37°C with a 5% CO2 atmosphere. Cells were harvested 24 h after transfection for subsequent experiment. The target sequences of ST3GAL6-AS1 siRNA were as follows: 5′-TCCAGATCATCCAGGATCA-3′, 5′-ACCTCTCATCAGAAATCAA-3′ and 5′-GGACTAAACTATATTCACT-3′. The target sequences of ST3GAL6-AS1 antisense oligonucle- otides (ASO) were 5′-CCCAAATTCGAGGAGGAAGG-3′, 5′-GCCTGAAGATGTGGCATCTA-3′ and 5′-GAATCCTGACAAACCTCTCA-3′. The mixture of siRNA and ASO was denoted as a smart silencer (ss-ST3GAL6-AS1), which was purchased from Guangzhou RiboBio Co., Ltd. The siRNA sequence target ST3GAL6 and hnRNPA2B1 are shown in Table SII. The sequence of the negative control (NC) was forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. Actinomycin D was utilized to affect the synthesis of RNA in this study. Initial solution concentration was 10 μg/μl, and the effective density was 10 μg/ml (17).
Cell viability assay
A Cell Counting Kit-8 (CCK-8) assay was used to signify the viability and proliferation ability of MM cells. After being transfected with ST3GAL6-AS1 smart silencer, RPMI 8226 and MM.1S cells (4,000 cells/well) were seeded in three duplicates on a 96-well plate. A CCK-8 proliferation assay kit (Shanghai Qihai Futai Biotechnology Co., Ltd.) was employed according to the manufacturer's instructions. The experiments were repeated three times.
Apoptosis assay
RPMI 8226 and MM.1S cells were collected 24 h after transfection and washed twice with cold phosphate-buffered saline (PBS). Annexin V-FITC and propidium iodide (PI) (BD Biosciences) were used to mark the cells. The apoptosis rate was determined using a BD FACSCanto II flow cytometer (BD Biosciences) within 30 min. The data was analyzed by FlowJo version 10 (BD Biosciences). All assays were repeated at least three times.
Cell cycle assay
RPMI 8226 and MM.1S cells were collected 24 h after transfection and washed twice with cold PBS, and were then suspended in the cold 75% ethanol and fixed over-night at 4°C. After fixation, the cells were washed twice in cold PBS, suspended in 300 μl PBS containing 40 μg/ml RNase A and incubated for 30 min at room temperature before 300 μl staining solution (50 μg/ml PI in PBS/5 mM EDTA) was added. Cell cycle analysis was detected using a BD FACSCanto II flow cytometer (BD Biosciences) within 24 h. The data was analyzed by FlowJo version 10 (BD Biosciences). All assays were repeated at least three times.
Cell adhesion assay
Assays for adhesion of MM cells to collagen I, fibronectin and HUVEC were performed using 96-well plates. Collagen and fibronectin (both BD Biosciences) were coated in 96-well plate overnight. HUVEC cells (7,000 cells/well) were cultured to confluence overnight in 96-well plates to establish a monolayer. MM cells were labeled with 5 μM CFSE (Thermo Fisher Scientific, Inc.) for 30 min in dark at 37°C. After being prelabeled with CFSE (Thermo Fisher Scientific, Inc.) and transfection, MM cells were added to the collagen I, fibronectin and HUVEC and allowed to adhere for 60 min at 37°C. Non-adherent cells were aspirated off, 96-well plates coated with collagen I, fibronectin and HUVEC were washed with PBS for three times. Images were captured using a fluorescence microscope in three different views of each well. ImageJ version 1.8.0 (National Institutes of Health) was used to count the number of the cells. All assays were repeated at least three times.
Cell migration and invasion assay
To determine the migratory and invasive potential of cells, a 24-well culture plate and Transwell chamber (Corning, Inc.) were used. For the migration assay, MM.1S and RPMI 8226 cells (5×105) were washed once in serum free medium and seeded into the upper chamber onto a rehydrated basal layer membrane with a diameter of 8 μm. The lower chamber was filled with complete medium containing 10% FBS. For invasion analysis, the procedure was the same as above except that the upper chamber was precoated with Matrigel (BD Biosciences) overnight in 37°C. After incubation at 37°C with a 5% CO2 atmosphere for 24 h, the cells in lower chamber were collected to be counted by flow cytometry (18). The cells were counted three times and all assays were repeated at least three times.
RNA isolation and reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from bone marrow mononuclear cell (BMSCs) samples of patients with MM, healthy donors and MM cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RNA purity and concentration were determined using NanoDrop ND-1000 (Thermo Fisher Scientific, Inc.). RNA samples were reverse transcribed into cDNA using a Primescript RT master mix with Oligo(dT) primers and random primers in accordance with manufacturer's protocols (Takara Biotechnology Co., Ltd.). Then, the qPCR was performed using SYBR Premix Ex Taq™ II (TliRNaseH Plus; Takara Biotechnology Co., Ltd.) and a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. Primers of ST3GAL6-AS1, ST3GAL6 and β-actin used performed as previously described (11), and the sequence of all the primers are presented in Table SIII. The thermocycling conditions were: 10 min at 95°C, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Cq values were collected for β-actin and the genes of interest during the log phase of the cycle and results were quantified using the 2−ΔΔCq method (19).
Western blot analysis
Cultured cells were lysed with RIPA buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% Sodium Deoxycholate, 1 mM EDTA, 0.1% SDS) containing complete protease inhibitors and phosphatase inhibitor cocktail (Xi'an Hat Biotechnology Co., Ltd, http://www.fiveheart.com/) after washing with cold PBS three times. The enriched cell lysate was centrifuged at 12,000 × g for 15 min at 4°C. Protein concentration was determined using a BCA assay (Xi'an Hat Biotechnology Co., Ltd.; http://www.fiveheart.com/) according to the manufacturer's instructions. The acquired supernatant was boiled in loading buffer and separated using a 10% SDS-PAGE gel. The quantity of protein loaded per lane was 50 ng. The separated proteins were transferred to a 0.45 μm polyvinylidene fluoride (PVDF; Merck KGaA) membrane. After blocking with 5% non-fat milk in PBS containing 0.1% Tween-20 (PBST) at room temperature, the PVDF membrane was incubated with antibodies overnight at 4°C and then with HRP-conjugated goat anti-rabbit (cat. no. SA00001-2, 1:10,000 dilution) or goat anti-mouse IgG (cat. no. SA00001-1; 1:10,000 dilution) (both ProteinTech Group, Inc.) for 1 h at room temperature. All bands were detected using an ECL Western blot kit (EMD Millipore). The following antibodies were used in this study: Anti-ST3GAL6 Polyclonal antibody (cat. no. 13154-1-AP; 1:1,000 dilution; ProteinTech Group, Inc.), anti-hnRNPA2B1 monoclonal antibody (cat. no. ab6102; 1:2,000 dilution; Abcam) and anti-β-actin monoclonal antibody (cat. no. HRP-66009; 1:2,000 dilution; ProteinTech Group, Inc.).
Cell nucleus/cytoplasm isolation
MM.1S and RPMI 8226 cells were cultured and washed twice with cold PBS. Nucleo-cytoplasmic fractionation was performed using a PARIS™ kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The purity of the cytoplasmic and nuclear fraction was confirmed by NanoDrop-1000. GAPDH, β-actin and U6 were used as markers of cytoplasm and nucleus in RT-qPCR and performed as aforementioned.
Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS)
ChIRP-MS, performed by KangChen Biotech, was used to search for proteins interacting with lncRNA ST3GAL6-AS1. In general, harvested cells were re-suspend with pre-cold PBS, reversibly crosslinked with UV/formaldehyde at room temperature, then hybridized with biotinylated DNA antisense and pulled down using streptavidin dynabeads (Thermo Fisher Scientific, Inc.). The mixture after elution underwent tryptic digestion and desalting into peptides before MS. For MS detection, ~1/2 peptides of each sample were separated and analyzed using a nano-UPLC (EASY-nLC1200) coupled to Q-Exactive MS (Thermo Fisher Scientific, Inc.). Raw MS files were processed using MaxQuant (version 1.5.6.0). The protein sequence database (Uniprot_ organism_2016_09) was downloaded from UNIPROT. This database and its reverse decoy were then searched against using MaxQuant software. Both peptide and protein false discovery rate (FDR) should be <0.01.
RNA pull-down
Desthiobiotinylation labeled RNAs were synthesized using a Pierce™ RNA 3′ End Desthiobiotinylation kit (cat. no. 20163; Thermo Fisher Scientific, Inc.) according to the manufacturer's instruction. The RNA pull-down assay was conducted using Pierce™ Magnetic RNA-Protein Pull-Down kit (cat. no. 20164; Thermo Fisher Scientific, Inc.) according to the manufacturer's instruction. Finally, retrieved proteins were analyzed using western blot as before mentioned. Antisense ST3GAL6-AS1 fragment (5′-UGCACCUUCCUCCUCGAAUUUGGGGGCGGCCACCGUGCAACUGGC-3′) was taken as a NC.
RNA immunoprecipitation (RIP)
RIP was performed utilizing a Magna RIP™ RNA-Binding Protein Immunoprecipitation kit (cat. no. 17-700; Sigma-Aldrich; Merck KGaA) according to the manufacturer's instruction. Equal amounts of anti-hnRNPA2B1 antibody (cat. no. ab31645; 5 μg per immunoprecipitation; Abcam) and IgG antibody (contained in aforementioned kit; 5 μg per immunoprecipitation) were used in each RIP reaction. Obtained RNA after elution was isolated by phenol-chloroformisoamyl alcohol for RT-qPCR as aforementioned.
Online software
Oncomine (https://www.oncomine.org) is an online database, integrating RNA and DNA-seq data from Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA) database and published literature. It was used to validate the expression of ST3GAL6 and hnRNPA2B1 in patients with MM in the present study. RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) online software could predict secondary structures of single stranded RNA or DNA sequences, which was used to predict secondary structures of ST3GAL6-AS1.
Statistical analysis
Each experiment was performed at least three times, and values were reported as mean ± SD. The unpaired independent t-test and Mann-Whitney U test were used to compare the two groups. For the comparison of multiple groups, Kruskal-Wallis test was used for non-parametric data, while ANOVA followed by Tukey post hoc test was used for parametric data. SPSS for Windows, version 18.0 (IBM Corp.) and GraphPad Prism version 7.0 (GraphPad Software) were used for data analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of ST3GAL6-AS1 is associated with stage of patients with MM
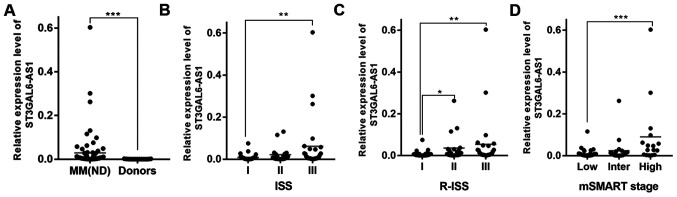
We identified that ST3GAL6-AS1 was upregulated in patients with newly diagnosed and relapse/refractory MM in previous study (12). The present study further validated that ST3GAL6-AS1 was overexpressed in MM cell lines (Fig. S1) and newly diagnosed MM by comparing cells from 83 patients with MM with normal plasma cells from healthy donors. The patient group included 39 males and 44 females with a median age of 57 (range, 42-75) years. Patients with MM were classified according to IMWG classification (14) as IgG (n=55), IgA (n=15), IgD (n=3), light chain (n=8) and non-secretory (n=2). At diagnosis, the peripheral WBC count ranged from 3.1 to 12.3×109/l, concentration of hemoglobin from 60 to 103 g/l, PLT count from 43 to 367×109/l, and myeloma cell percentage from 14 to 73%. Other basic characteristics of patients are shown in Table I, which were similar to our previous research results (12). ST3GAL6-AS1 was overexpressed in MM (P<0.0001; Fig. 1A) compared with healthy donors. Consistently, the expression of ST3GAL6-AS1 in patients with MM was also associated with ISS (P=0.288 for stage I vs. stage II, P=0.005 for stage I vs. stage III, P=0.790 for stage II vs. stage III; Fig. 1B), R-ISS (P=0.033 for stage I vs. stage II, P=0.005 for stage I vs. stage III, P=0.999 for stage II vs. stage III; Fig. 1C) and mSMART stage (P=0.772 for stage low vs. intermediate, P=0.001 for low vs. high, P=0.063 for intermediate vs. high; Fig. 1D). These data revealed that the levels of ST3GAL6-AS1 were increased at advanced stages.
Figure 1.
ST3GAL6-AS1 is overexpressed in MM and predicts poor prognosis in patients with MM. (A) lncRNA ST3GAL6-AS1 was upregulated in patients with newly diagnosed MM, compared with healthy donors. Expression level of ST3GAL6-AS1 was related to (B) ISS, (C) R-ISS and (D) mSMART stage. *P<0.05, **P<0.01, ***P<0.001 vs. respective control. MM, multiple myeloma; lnc, long non-coding; R-ISS Revised International Staging System; ISS, International Staging System; Inter, intermediate. mSMART; Mayo Stratification of Myeloma and Risk-Adapted Therapy.
Knockdown of ST3GAL6-AS1 has no influence on proliferation, cell cycle or apoptosis of MM cells
To investigate the biological significance of ST3GAL6-AS1 in MM, siRNA and ASO-mediated knockdown was performed in two MM cell lines, MM.1S and RPMI 8226. The efficiency of knockdown was measured using RT-qPCR and ST3GAL6-AS1 was effectively downregulated in MM.1S and RPMI 8226 cells (P=0.0007 for MM.1S, P=0.005 for RPMI 8226; Fig. S2A). Cell proliferation was assessed using CCK-8 assays and it was demonstrated that the expression changes of ST3GAL6-AS1 did not affect MM cell proliferation (P=0.083; Fig. S2B). Furthermore, as determined by flow cytometric analysis, the expression level change of ST3GAL6-AS1 had no effect on the cell cycle (P=0.058 for MM.1S and P=0.130 for RPMI 8226 at G2/M phase in ST3GAL6-AS1-knockdown group vs. NC group; Fig. S2C) or apoptosis (P=0.071 for MM.1S, P=0.082 for RPMI 8226 in ST3GAL6-AS1-knockdown group vs. NC group; Fig. S2D).
Knockdown of ST3GAL6-AS1 decreases the adhesion, migration and invasion ability of MM cells
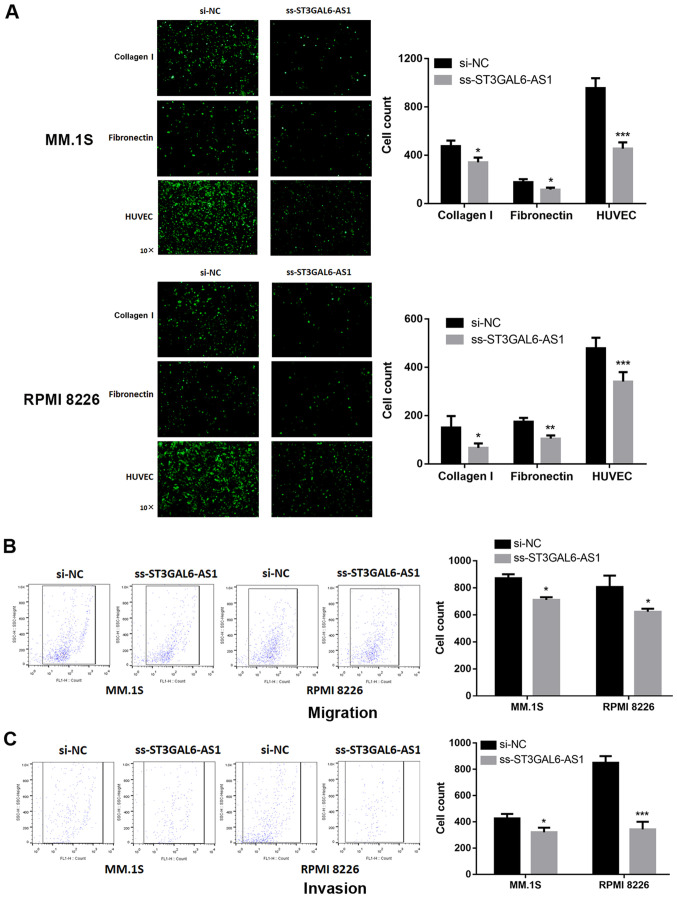
Given the evidence for strong co-expression between ST3GAL6-AS1 and ST3GAL6 (12), and knockdown of ST3GAL6 resulting in alternated MM cell adhesion and migration (13), adhesion and Transwell assays were performed. A significant reduction in adhesion to collagen I-coated and fibronectin-coated plates was reported for ST3GAL6-AS1-knockdown in MM.1S (P=0.032 for collagen I, P=0.029 for fibronectin; Fig. 2A) and RPMI 8226 cells (P=0.042 for collagen I, P=0.007 for fibronectin; Fig. 2A). The ability of ss-ST3GAL6-AS1 cells to adhere to endothelial cells was also assessed. Knockdown of ST3GAL6-AS1 in both MM.1S and RPMI 8226 cells resulted in decreased cell adhesion to HUVEC in vitro compared with control cells (P=0.0006 for MM.1S, P=0.0009 for RPMI 8226; Fig. 2A). Migration and invasion was also affected in ST3GAL6-AS1-knockdown in MM cells. As shown in Fig. 2B, a reduced number of ss-ST3GAL6-AS1 cells migrated through the bottom membrane of Transwell chamber compared with control cells (P=0.037 for MM.1S, P=0.032 for RPMI 8226; Fig. 2B). Furthermore, knockdown of ST3GAL6-AS1 also decreased the number of invading cells (P=0.041 for MM.1S, P=0.0008 for RPMI 8226; Fig. 2C). These data demonstrated that ST3GAL6-AS1 promoted MM cell adhesion, migration and invasion ability in vitro.
Figure 2.
Effect of long non-coding RNA ST3GAL6-AS1 on adhesion, migration and invasion ability of MM cells in vitro. After ST3GAL6-AS1 knockdown, the MM cell lines exhibited significant decrease in (A) cell adhesion, (B) migration and (C) invasion ability, compared with the control group. *P<0.05, **P<0.01, ***P<0.001 vs. respective control. si, small interfering; NC, negative control.
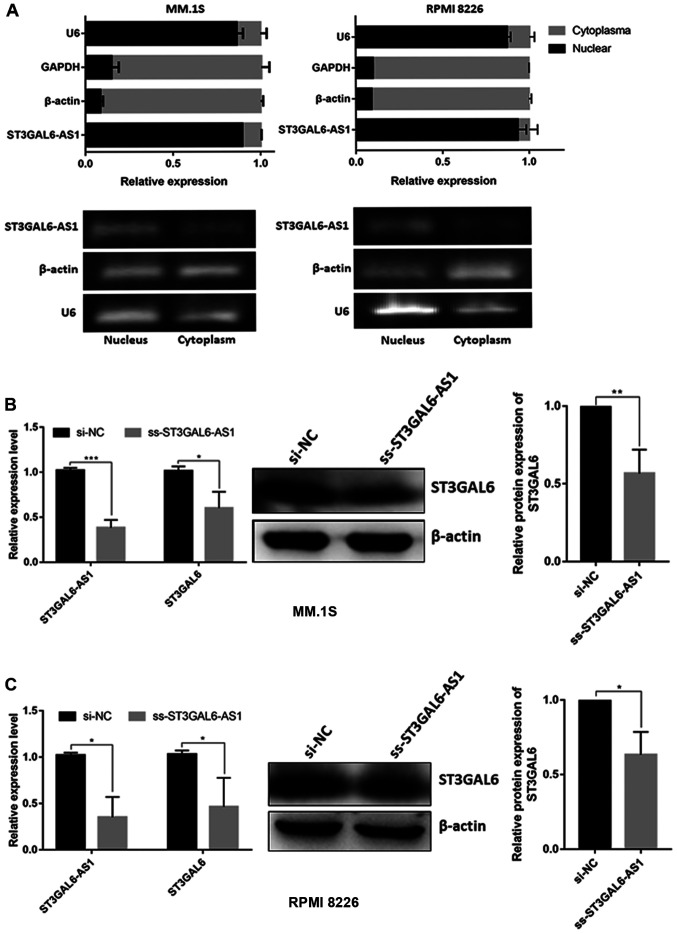
ST3GAL6-AS1 is a nuclear lncRNA and regulates the expression of ST3GAL6
By isolating nuclear and cytoplasmic RNA, it was confirmed that the ST3GAL6-AS1 transcript was mainly present in the nucleus in MM.1S and RPMI 8226 cell (Fig. 3A). Since nuclear lncRNAs usually coordinate the transcriptional or translational regulation of nearby protein-coding genes (11), and we previously confirmed the co-expression relationship between ST3GAL6-AS1 and ST3GAL6 among pan-cancer with correlation coefficient >0.6 (12), it was investigated whether ST3GAL6 expression was mediated by lncRNA ST3GAL6-AS1. After knocking down ST3GAL6-AS1 with ss-ST3GAL6-AS1, ST3GAL6 expression was significantly reduced in MM cells at both the mRNA and protein levels in MM.1S (P=0.034 for RNA level, P=0.0083 for protein level; Fig. 3B) and RPMI 8226 cells (P=0.029 for RNA level, P=0.030 for protein level; Fig. 3C). Our previous study indicated that ST3GAL6 was upregulated in MM (11), which is entirely consistent with the data from Oncomine database (https://www.oncomine.org) (P= 0.021; Fig. S3). So, the study further explored the role of ST3GAL6 in MM cells by downregulating its expression using siRNA. Following ST3GAL6-knockdown, a significant reduction in adhesion to collagen I-coated, fibronectin-coated plates and HUVEC was observed in MM.1S (P=0.002 for collagen I, P=0.007 for fibronectin, P<0.001 for HUVEC; Fig. S4A) and RPMI 8226 (P=0.033 for collagen I, P<0.001 for fibronectin, P=0.002 for HUVEC; Fig. S4B). Migration ability was also affected in ST3GAL6-knockdown MM cells (P=0.031 for MM.1S, P=0.016 for RPMI 8226; Fig. S4C), as well as invasion ability (P=0.029 for MM.1S, P<0.001 for RPMI 8226; Fig. S4D). Cell proliferation, cell cycle and apoptosis were not changed when ST3GAL6 downregulated (P>0.05; Fig. S5A-D). These data showed that nuclear lncRNA ST3GAL6-AS1 increases the expression of its downstream target gene, ST3GAL6.
Figure 3.
Cellular localization of ST3GAL6-AS1 and its downstream target analysis. (A) Cell nucleus/cytoplasm isolation following by RT-qPCR of MM cell lines indicated that ST3GAL6-AS1 located in cell nucleus. (B) RT-qPCR and (C) western blot analysis of ST3GAL6 expression after ST3GAL6-AS1-knockdown in MM cell lines. *P<0.05, **P<0.01, ***P<0.001 vs. respective control. RT-q, reverse transcription-quantitative; MM, multiple myeloma; si, small interfering; NC, negative control.
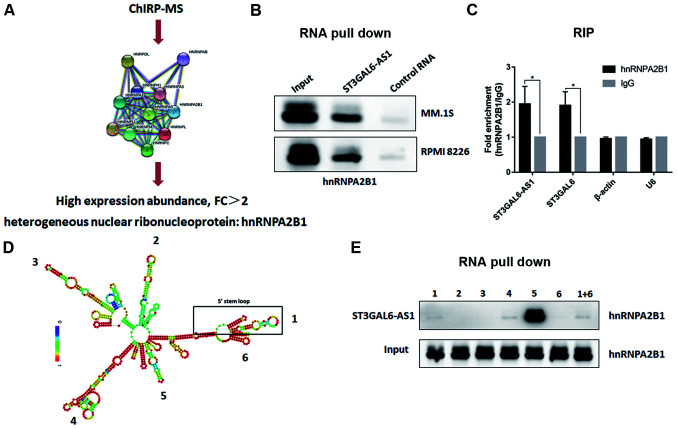
ST3GAL6-AS1 interacts with heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1) protein
To determine the precise mechanism underlying ST3GAL6-AS1-guided ST3GAL6 regulation, a ChIRP assay with biotin-labeled oligos was performed and ChIRP-purified proteins were identified using MS. Through screening and analyzing the peptide signals, hnRNPA2B1 protein was found to be enriched in the ChIRP lysate (Fig. 4A). hnRNPA2B1 was upregulated in patients with MM (P=0.048) based on the Oncomine database (Fig. S6). To confirm that hnRNPA2B1 protein interacted with ST3GAL6-AS1, RNA pull-down was performed with biotinylated ST3GAL6-AS1, having taken antisense ST3GAL6-AS1 as a NC. Finally, hnRNPA2B1 was identified as a direct interacting protein of lncRNA ST3GAL6-AS1 using an RNA pull-down assay (Fig. 4B). Moreover, RIP verified the interaction between hnRNPA2B1 and ST3GAL6-AS1 (P= 0.026; Fig. 4C) in vitro, and hnRNPA2B1 also bound to ST3GAL6 mRNA (P=0.019; Fig. 4C). To further clarify the binding area of ST3GAL6-AS1 to hnRNPA2B1, its secondary construction was calculated using the RNAfold web server. ST3GAL6-AS1 had five stem loops, which were coded by Arabic number from 5′ stem loop (Fig. 4D). Fragments of its six stem loops from 5′ to 3′ were synthesized separately, which were utilized to conduct RNA pull-down after biotinylated (Fig. 4D). As shown in Fig. 4E, fragment 5 was confirmed to interact with hnRNPA2B1.
Figure 4.
ST3GAL6-AS1 interacts with hnRNPA2B1 by stem loop 5. (A) ChIRP-MS revealed that hnRNPA2B1 protein binds to ST3GAL6-AS1. (B) RNA pull-down assay confirmed that hnRNPA2B1 protein bind to ST3GAL6-AS1. (C) RIP method was used to verify the interaction of hnRNPA2B1 and ST3GAL6-AS1 reversely. (D) Secondary structure prediction of ST3GAL6-AS1. (E) RNA pull-down assay showed that hnRNPA2B1 protein specifically bind to ST3GAL6-AS1 stem loop 5. *P<0.05 vs. IgG. hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; RIP, RNA immunoprecipitation.
ST3GAL6-AS1 recruits hnRNPA2B1 to promote ST3GAL6 translation
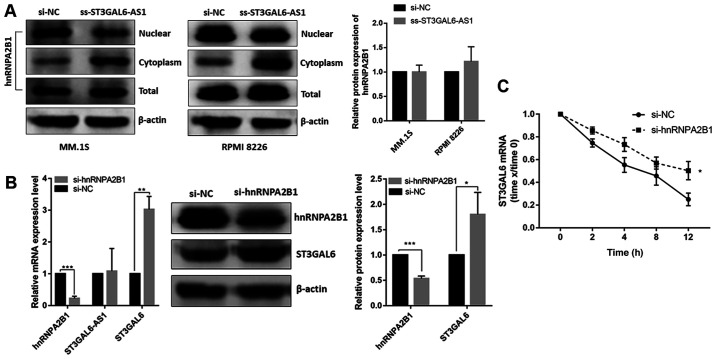
After confirming that ST3GAL6-AS1 could bind to hnRNPA2B1 as demonstrated by ChIRP and RNA pull-down assays, and hnRNPA2B1 also bound to ST3GAL6 mRNA in the RIP assay, it was necessary to determine whether ST3GAL6-AS1-mediated regulation of ST3GAL6 expression was induced by hnRNPA2B1. In ST3GAL6-AS1-knockdown RPMI 8226 cells, the expression of hnRNPA2B1 protein increased in the cytoplasm and decreased in the nucleus, but not in the total expression level (Fig. 5A). This phenomenon indicated that hnRNPA2B1 protein translocated to cell cytoplasm when ST3GAL6-AS1 was downregulated, without the total expression change. Furthermore, following knock down of hnRNPA2B1 both on mRNA and protein levels (P<0.05; Fig. 5B), ST3GAL6 expression was upregulated both at the mRNA (P=0.008; Fig. 5B) and protein levels (P=0.039; Fig. 5B), but ST3GAL6-AS1 was unchanged (P= 0.812; Fig. 5B). hnRNPA2B1-downregulated RPMI 8226 cells were treated with actinomycin D to block the synthesis of new RNA and then mRNA levels of ST3GAL6 at different time points were measured. Knockdown of hnRNPA2B1 elongated the mRNA half-life of ST3GAL6 (P=0.024; Fig. 5C).
Figure 5.
ST3GAL6-AS1 recruits hnRNPA2B1 in nucleus to promote ST3GAL6 translation in cytoplasm. (A) hnRNPA2B1 protein translocated to cell cytoplasm when ST3GAL6-AS1 knockdown in MM cell lines. (B) Efficiency of hnRNPA2B1-knockdown measured by reverse transcription quantitative PCR and western blotting. (C) Knockdown of hnRNPA2B1 significantly decreased the degradation of ST3GAL6 mRNA when MM cells were treated with acti- nomycin D. 18S rRNA was the normalizer. *P<0.05, **P<0.01, ***P<0.001 vs. respective control. hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2B1; MM, multiple myeloma; si, small interfering; NC, negative control.
Discussion
Research has revealed the dysregulation of lncRNAs in MM (20). Our previous study reported a landscape of differentially expressed lncRNAs in newly diagnosed MM using microarray and bioinformatics analyses. These previous results showed that lncRNA ST3GAL6-AS1 is significantly overexpressed in BMSCs of patients with MM and associated with ST3GAL6 expression (12). Furthermore, the present study reported that upregulated-ST3GAL6-AS1 promoted MM cell adhesion, migration and invasion ability in vitro, via interacting with hnRNPA2B1 protein to decrease the degradation of ST3GAL6 mRNA. These findings suggested that ST3GAL6-AS1 exerts an oncogenic function in MM, and its overexpression contributes to MM pathogenesis and progression.
Studies have shown that lncRNAs contribute to MM pathogenesis and progression, such as MALAT1 (9,21-25), UCA1 (26) and NEAT1 (27). However, until now, the function and molecular mechanism of lncRNA ST3GAL6-AS1 in MM were far from being well understood. One recent study showed ST3GAL6-AS1 expression was low in tumor tissues and cell lines of colorectal cancer (CRC), and inhibited CRC cell proliferation, migration and promoted apoptosis. The group also demonstrated that ST3GAL6-AS1 activated ST3GAL6 transcription by recruiting histone methyltransferase MLL1 to the ST3GAL6 promoter region (28). On the contrary, our previous study confirmed that ST3GAL6-AS1 is significantly upregulated in patients with MM and MM cell lines (12). The aforementioned studies results suggested that ST3GAL6-AS1 may be a novel multifunctional regulator in different cancer types. There are some lncRNAs in similar situations. For example, H19 is upregulated and acts as an oncogene in gastric (29) and bladder cancer (30) and CRC (31). High expression of H19 can also significantly promote tumor proliferation, invasion and migration (29,30). Conversely, H19 acts as a tumor suppressor in prostate (32) and liver cancer (33), and its high expression can inhibit tumor proliferation and migration and reduce the sensitivity of liver cancer cells to chemotherapeutic drugs (34). The present study confirmed that overexpressed ST3GAL6-AS1 in MM predicted poor prognosis. After identifying the importance of ST3GAL6-AS1 in MM, knockdown experiments were conducted to elucidate the specific role of ST3GAL6-AS1. Knockdown of ST3GAL6-AS1 decreased ST3GAL6 expression and inhibited the adhesion, migration and invasion ability of MM cells.
Sialyltransferases (STs) are a group of enzymes that mediate protein sialylation by conjugating sialic acids to other sugar molecules in glycans. ST family of proteins is divided into three subfamilies: α-2,3- (ST3GAL I-VI), α-2,6- (ST6GAL I-II And ST6GALNAc I-VI) and α-2,8-STs (ST8SIA I-VI) (29), according to the type of linkage form (35). Alterations in the protein sialylation process have been associated with malignant transformation and metastasis (36). Up to now, only a small number of studies investigating ST3GAL6 in cancer progression were published. ST3GAL6 is upregulated in hepatocellular carcinoma cell lines and tissue samples, and negatively regulated by miR-26a through Akt/mTOR signaling pathway to promote cell proliferation, migration and invasion (36). Another study identified that ST3GAL6 is highly expressed in MM cell lines and patients, influenced MM cell adhesion and migration in vitro and homing to bone marrow in vivo (13). In an effort to understand the molecular mechanism of how ST3GAL6-AS1 regulates the expression of ST3GAL6, the present study conducted a ChIRP-MS assay to identify the possible interacting proteins with ST3GAL6-AS1, since numerous existing studies have demonstrated that lncRNA in nucleus might regulate the gene expression at the transcription level by interacting with some regulatory factors (11,20,37,38).
Most lncRNAs display strong nuclear localization (39), including ST3GAL6-AS1 based on the cell nucleus/cytoplasm isolation assay of the present study. It was demonstrated that lncRNA ST3GAL6-AS1 interacted with hnRNPA2B1 protein directly with its stem loop 5. Heterogeneous nuclear ribonucleoproteins (hnRNPs), predominantly expressed in the nucleus, are a large family of RNA-binding proteins that contribute to multiple aspects of nucleic acid metabolism, including alternative splicing, mRNA stabilization and transcriptional and translational regulation (40). hnRNPA2B1 is among the most abundant hnRNPs, with a multitude of functions such as telomere maintenance (41,42), DNA replication and repair, transcription, pre-mRNA splicing and mRNA nucleo-cytoplasmic export, via interacting with DNA or, more commonly, RNA (43,44). An increasing number of studies have demonstrated the vital role of lncRNA and hnRNP interactions in regulating gene expression (39,45). It has been reported that hnRNPA2B1 could bind with lncRNA lnc-HC to regulate cholesterol metabolism in hepatocytes (46), and interacts with LNMAT2 to promote lymphatic metastasis in bladder cancer (47).
The present study observed that hnRNPA2B1 also bound to ST3GAL6 mRNA using a RIP assay. The expression of ST3GAL6 in mRNA and protein level was increased when hnRNPA2B1 was downregulated. Following ST3GAL6-AS1 knockdown, hnRNPA2B1 translocated to the cytoplasm where it bound to ST3GAL6 mRNA, leading its degradation. These results indicated that ST3GAL6-AS1 increased hnRNPA2B1 protein localization in the nucleus, thereby decreasing the degradation of ST3GAL6 via hnRNPA2B1 in the cytoplasm.
There are some limitations of the present study. First, in vivo experiments were not performed to verify the function of ST3GAL6-AS1 in MM. In further studies, we plan to construct a ST3GAL6-AS1 knockout model using CRISPER/Cas9 technology to verify the in vitro experiment results. Second, we did not conduct an analysis of overall survival of patients with MM based on ST3GAL6-AS1 expression level due to the short follow-up period. The significance of ST3GAL6-AS1 on survival of patients should be clarified in further investigations.
Taken together, the present study confirmed that ST3GAL6-AS1, located in the nucleus, promotes adhesion, migration and invasion of MM cells by binding with hnRNPA2B1 to regulate ST3GAL6 expression. These findings indicate that ST3GAL6-AS1 may help us improve the understanding of the mechanism of MM invasion, which may imply new strategies and targets for MM treatment.
Supplementary Data
Acknowledgments
Not applicable.
Funding Statement
This work was supported by The Science and Technology Coordinating Innovative Engineering Projects of Shaanxi Province (grant no. 2018ZDXM-SF-039).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YS contributed significantly to experiment performing and manuscript writing. YF helped perform the data analysis and interpretation. WZ helped collected the clinical specimens. FL, YJ and YP performed the experiment and confirmed the authenticity of all raw data. AH and JH contributed to the conception of the study and constructive discussions.
Ethics approval and consent to participate
Approval for this study was obtained from The Ethics Committee of the Second Affiliated Hospital of Xian Jiaotong University (Xi'an, China; approval no. 2015186). Written informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors confirm that there were no competing interests.
References
- 1.Rajkumar SV. Multiple myeloma: 2020 update on diagnosis, risk-stratification and management. Am J Hematol. 2020;95:548–567. doi: 10.1002/ajh.25791. [DOI] [PubMed] [Google Scholar]
- 2.Nobili L, Ronchetti D, Agnelli L, Taiana E, Vinci C, Neri A. Long non-coding RNAs in multiple myeloma. Genes (Basel) 2018;9:69. doi: 10.3390/genes9020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar SK, Rajkumar V, Kyle RA, van Duin M, Sonneveld P, Mateos MV, Gay F, Anderson KC. Multiple myeloma. Nat Rev Dis Primers. 2017;3:17046. doi: 10.1038/nrdp.2017.46. [DOI] [PubMed] [Google Scholar]
- 4.Fitzmaurice C, Abate D, Abbasi N, Abbastabar H, Abd-Allah F, Abdel-Rahman O, Abdelalim A, Abdoli A, Abdollahpour I, Abdulle AS, et al. Global Burden of Disease Cancer Collaboration Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2019;5:1749–1768. doi: 10.1001/jamaoncol.2019.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA. Pathogenesis of bone disease in multiple myeloma: From bench to bedside. Blood Cancer J. 2018;8:7. doi: 10.1038/s41408-017-0037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chim CS, Kumar SK, Orlowski RZ, Cook G, Richardson PG, Gertz MA, Giralt S, Mateos MV, Leleu X, Anderson KC. Management of relapsed and refractory multiple myeloma: Novel agents, antibodies, immunotherapies and beyond. Leukemia. 2018;32:252–262. doi: 10.1038/leu.2017.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soekojo CY, Ooi M, de Mel S, Chng WJ. Immunotherapy in multiple myeloma. Cells. 2020;9:9. doi: 10.3390/cells9030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robak P, Drozdz I, Szemraj J, Robak T. Drug resistance in multiple myeloma. Cancer Treat Rev. 2018;70:199–208. doi: 10.1016/j.ctrv.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, Taiana E, Agnelli L, Cantafio ME, Romeo E, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018;32:1948–1957. doi: 10.1038/s41375-018-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Zhao H, Wang Z, Cheng L, Yang L, Shi H, Yang H, Sun J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp F, Mendell JT. Functional classification and experi- mental dissection of long noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen Y, Feng Y, Chen H, Huang L, Wang F, Bai J, Yang Y, Wang J, Zhao W, Jia Y, et al. Focusing on long non-coding RNA dysregulation in newly diagnosed multiple myeloma. Life Sci. 2018;196:133–142. doi: 10.1016/j.lfs.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 13.Glavey SV, Manier S, Natoni A, Sacco A, Moschetta M, Reagan MR, Murillo LS, Sahin I, Wu P, Mishima Y, et al. The sialyltransferase ST3GAL6 influences homing and survival in multiple myeloma. Blood. 2014;124:1765–1776. doi: 10.1182/blood-2014-03-560862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rajkumar SV, Dimopoulos MA, Palumbo A, Blade J, Merlini G, Mateos MV, Kumar S, Hillengass J, Kastritis E, Richardson P, et al. International Myeloma Working Group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 2014;15:e538–e548. doi: 10.1016/S1470-2045(14)70442-5. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Paiva B, Anderson KC, Durie B, Landgren O, Moreau P, Munshi N, Lonial S, Bladé J, Mateos MV, et al. International Myeloma Working Group consensus criteria for response and minimal residual disease assessment in multiple myeloma. Lancet Oncol. 2016;17:e328–e346. doi: 10.1016/S1470-2045(16)30206-6. [DOI] [PubMed] [Google Scholar]
- 16.Mikhael JR, Dingli D, Roy V, Reeder CB, Buadi FK, Hayman SR, Dispenzieri A, Fonseca R, Sher T, Kyle RA, et al. Mayo Clinic Management of newly diagnosed symptomatic multiple myeloma: Updated Mayo Stratification of Myeloma and Risk-Adapted Therapy (mSMART) consensus guidelines 2013. Mayo Clin Proc. 2013;88:360–376. doi: 10.1016/j.mayocp.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Grammatikakis I, Zhang P, Panda AC, Kim J, Maudsley S, Abdelmohsen K, Yang X, Martindale JL, Motiño O, Hutchison ER, et al. Alternative splicing of neuronal differentiation factor TRF2 regulated by HNRNPH1/H2. Cell Rep. 2016;15:926–934. doi: 10.1016/j.celrep.2016.03.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng Y, Li F, Zhang P, Wang X, Shen Y, Feng Y, Jia Y, Zhang R, Hu J, He A. IGF-1 promotes multiple myeloma progression through PI3K/Akt-mediated epithelial-mesenchymal transition. Life Sci. 2020;249:117503. doi: 10.1016/j.lfs.2020.117503. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho SF, Chang YC, Chang CS, Lin SF, Liu YC, Hsiao HH, Chang JG, Liu TC. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014;14:809. doi: 10.1186/1471-2407-14-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handa H, Kuroda Y, Kimura K, Masuda Y, Hattori H, Alkebsi L, Matsumoto M, Kasamatsu T, Kobayashi N, Tahara KI, et al. Long non-coding RNA MALAT1 is an inducible stress response gene associated with extramedullary spread and poor prognosis of multiple myeloma. Br J Haematol. 2017;179:449–460. doi: 10.1111/bjh.14882. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Lin J, Fang H, Fang J, Li C, Chen W, Liu S, Ondrejka S, Gong Z, Reu F, et al. Targeting the MALAT1/PARP1/LIG3 complex induces DNA damage and apoptosis in multiple myeloma. Leukemia. 2018;32:2250–2262. doi: 10.1038/s41375-018-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Chen P, Qu J, Shi L, Zhuang W, Fu J, Li J, Zhang X, Sun Y, Zhuang W. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J Biol Chem. 2014;289:29365–29375. doi: 10.1074/jbc.M114.572693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Wang H, Wu B, Yao K, Liao A, Miao M, Li Y, Yang W. Down-regulation of long non-coding RNA MALAT1 by RNA interference inhibits proliferation and induces apoptosis in multiple myeloma. Clin Exp Pharmacol Physiol. 2017;44:1032–1041. doi: 10.1111/1440-1681.12804. [DOI] [PubMed] [Google Scholar]
- 26.Sedlarikova L, Gromesova B, Kubaczkova V, Radova L, Filipova J, Jarkovsky J, Brozova L, Velichova R, Almasi M, Penka M, et al. Deregulated expression of long non-coding RNA UCA1 in multiple myeloma. Eur J Haematol. 2017;99:223–33. doi: 10.1111/ejh.12908. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Wang H. LncRNA NEAT1 promotes dexamethasone resistance in multiple myeloma by targeting miR-193a/MCL1 pathway. J Biochem Mol Toxicol. 2018;32:e22008. doi: 10.1002/jbt.22008. [DOI] [PubMed] [Google Scholar]
- 28.Hu J, Shan Y, Ma J, Pan Y, Zhou H, Jiang L, Jia L. LncRNA ST3Gal6-AS1/ST3Gal6 axis mediates colorectal cancer progression by regulating α-2,3 sialylation via PI3K/Akt signaling. Int J Cancer. 2019;145:450–460. doi: 10.1002/ijc.32103. [DOI] [PubMed] [Google Scholar]
- 29.Gan L, Lv L, Liao S. Long non coding RNA H19 regulates cell growth and metastasis via the miR 22 3p/Snail1 axis in gastric cancer. Int J Oncol. 2019;54:2157–2168. doi: 10.3892/ijo.2019.4773. [DOI] [PubMed] [Google Scholar]
- 30.Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013;333:213–221. doi: 10.1016/j.canlet.2013.01.033. [DOI] [PubMed] [Google Scholar]
- 31.Chen SW, Zhu J, Ma J, Zhang JL, Zuo S, Chen GW, Wang X, Pan YS, Liu YC, Wang PY. Overexpression of long non-coding RNA H19 is associated with unfavorable prognosis in patients with colorectal cancer and increased proliferation and migration in colon cancer cells. Oncol Lett. 2017;14:2446–2452. doi: 10.3892/ol.2017.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng R, Wang Y, Huang J, Xu M, Yan J, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 33.Ding K, Liao Y, Gong D, Zhao X, Ji W. Effect of long non-coding RNA H19 on oxidative stress and chemotherapy resistance of CD133+ cancer stem cells via the MAPK/ERK signaling pathway in hepatocellular carcinoma. Biochem Biophys Res Commun. 2018;502:194–201. doi: 10.1016/j.bbrc.2018.05.143. [DOI] [PubMed] [Google Scholar]
- 34.Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–4881. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 35.Teppa RE, Petit D, Plechakova O, Cogez V, Harduin-Lepers A. Phylogenetic-derived insights into the evolution of sialylation in eukaryotes: Comprehensive analysis of vertebrate β-galactoside α2,3/6-sialyltransferases (ST3Gal and ST6Gal) Int J Mol Sci. 2016;17:17. doi: 10.3390/ijms17081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun M, Zhao X, Liang L, Pan X, Lv H, Zhao Y. Sialyltransferase ST3GAL6 mediates the effect of microRNA-26a on cell growth, migration, and invasion in hepatocellular carcinoma through the protein kinase B/mammalian target of rapamycin pathway. Cancer Sci. 2017;108:267–276. doi: 10.1111/cas.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi J, Li YM, Fang XD. The mechanism and clinical significance of long noncoding RNA-mediated gene expression via nuclear architecture. Yi Chuan. 2017;39:189–199. doi: 10.16288/j.yczz.16-385. [DOI] [PubMed] [Google Scholar]
- 38.Jandura A, Krause HM. The new RNA world: Growing evidence for long noncoding RNA functionality. Trends Genet. 2017;33:665–676. doi: 10.1016/j.tig.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez-Dominguez JR, Lodish HF. Emerging mechanisms of long noncoding RNA function during normal and malignant hematopoiesis. Blood. 2017;130:1965–1975. doi: 10.1182/blood-2017-06-788695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geuens T, Bouhy D, Timmerman V. The hnRNP family: Insights into their role in health and disease. Hum Genet. 2016;135:851–867. doi: 10.1007/s00439-016-1683-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ford LP, Wright WE, Shay JW. A model for heterogeneous nuclear ribonucleoproteins in telomere and telomerase regulation. Oncogene. 2002;21:580–583. doi: 10.1038/sj.onc.1205086. [DOI] [PubMed] [Google Scholar]
- 42.Shishkin SS, Kovalev LI, Pashintseva NV, Kovaleva MA, Lisitskaya K. Heterogeneous nuclear ribonucleoproteins involved in the functioning of telomeres in malignant cells. Int J Mol Sci. 2019;20:20. doi: 10.3390/ijms20030745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He Y, Smith R. Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B. Cell Mol Life Sci. 2009;66:1239–1256. doi: 10.1007/s00018-008-8532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L, Wen M, Cao X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science. 2019;365:365. doi: 10.1126/science.aav0758. [DOI] [PubMed] [Google Scholar]
- 45.Qin G, Tu X, Li H, Cao P, Chen X, Song J, Han H, Li Y, Guo B, Yang L, et al. Long Noncoding RNA p53-stabilizing and activating RNA promotes p53 signaling by inhibiting heterogeneous nuclear ribonucleoprotein K deSUMOylation and suppresses hepatocellular carcinoma. Hepatology. 2020;71:112–129. doi: 10.1002/hep.30793. [DOI] [PubMed] [Google Scholar]
- 46.Lan X, Yan J, Ren J, Zhong B, Li J, Yue L, Liu L, Yi J, Sun Q, Yang X, et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology. 2016;64:58–72. doi: 10.1002/hep.28391. [DOI] [PubMed] [Google Scholar]
- 47.Chen C, Luo Y, He W, Zhao Y, Kong Y, Liu H, Zhong G, Li Y, Li J, Huang J, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2019;130:404–421. doi: 10.1172/JCI130892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.