Abstract
Protein domains are the basic units of proteins that can fold, function, and evolve independently. Knowledge of protein domains is critical for protein classification, understanding their biological functions, annotating their evolutionary mechanisms and protein design. Thus, over the past two decades, a number of protein domain identification approaches have been developed, and a variety of protein domain databases have also been constructed. This review divides protein domain prediction methods into two categories, namely sequence-based and structure-based. These methods are introduced in detail, and their advantages and limitations are compared. Furthermore, this review also provides a comprehensive overview of popular online protein domain sequence and structure databases. Finally, we discuss potential improvements of these prediction methods.
Keywords: Artificial reef, Natural reef, Stable isotopes, MixSIAR, Trophic structure, Trophic pathway
1. Introduction
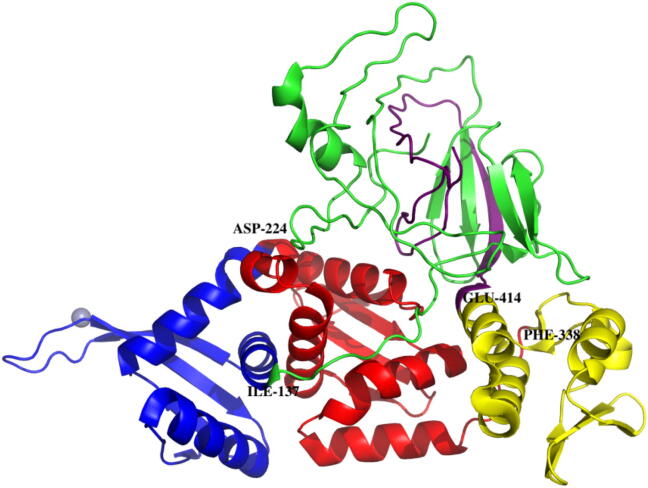
Protein domains are the fundamental units of protein structure, folding, function, evolution and design. They are considered to be homologous portions of sequences encoded in different gene contexts that have remained intact through evolution at the sequence level. They are local, compact units of structure, with hydrophobic interiors and hydrophilic exteriors forming a globular-like state that cannot be further subdivided from structural terms [1]. Small proteins are often found to be composed of only a single domain, while most large proteins consist of multiple domains for achieving various composite cellular functions. For example, the archaeal intein-encoded homing endonuclease PI-PFUI (PDB:1DQ3), which is shown in Fig. 1, includes 4 domains: [Cys1–Ile137 (green)|Gly415–Asn454(purple)], [Phe138–Asp224 (blue)], [Asn225–Phe338 (red)], and [Gly339–Glu414(yellow)]. Moreover, [Cys1–Ile137(green)] and [Gly415–Asn454(purple)] are separate in sequence but close in spatial orientation and constitute a discontinuous domain. A domain is a compact unit and often linked to others by domain linker areas. One way to identify a domain is to find the part of a target protein that has sequence or structural similarities with a template through homology alignment. Another way is to predict the domain boundaries from a protein sequence. One challenge for such methods is assembling two fragments separated in sequence into discontinuous domains. In early stages, most sequence-based methods ignore discontinuous domains. Until recently, discontinuous domains have received significant attention, and some methods have attempted to identify discontinuous domains.
Fig. 1.
Schematic diagram of structure and domain of an archaeal intein-encoded homing endonuclease PI-PFUI.
Domains can work independently or cooperate with neighbour domains, and different arrangements of domains can create proteins of different functions. Therefore, the correct detection of protein domains and their boundaries is pivotal not only for protein classification and understanding their biological functions but also for protein structure prediction and protein design. For example, DCS [2] integrated domain compositions of proteins for protein function prediction. PANDA [3] used domain architecture as input to improve protein function prediction. INGA [4] combined sequence similarity, domain architecture searches and protein–protein interaction network to predict GO functional terms of unknown protein. When predicting the structure of large multidomain proteins, the state-of-the-art protein structure prediction tools like I-TASSER [5] often first predict domain boundaries and then predict single-domain structure for reassembly. In the past two decades, many approaches have been developed to identify protein domains. Meanwhile, a variety of protein domain databases have been constructed that allow for the classification of protein sequence and structure and enable the transfer of experimentally obtained functional annotations to other proteins in the same protein domain family.
This review is organized as follows: Section 2 provides a detailed summary of sequence-based and structure-based protein domain identification methods. Section 3 presents protein domain sequence and structure databases. Section 4 provides discussion and conclusion.
2. Protein domain detection methods
Domains are considered to be the structural, functional, and evolutionary units of proteins. On a sequence level, domains are homologous segments in evolution, and on a structure level, domains are units that can fold and work independently. Proteins can usually be decomposed into domains based on their similar sequence or structural characteristics.
Many methods have been developed to detect domains in protein sequences based on these characteristics over the past two decades. Table 1 lists most of the protein domain identification methods with a brief description and URL when available. In general, domain detection methods can be classified into those that are sequence-based or structure-based. A detailed introduction of these methods follows.
Table 1.
Sequence-based protein domain identification methods.
| Category | Method | Description | Year | URL | Reference |
|---|---|---|---|---|---|
| Homology-based | CHOP | Search target sequences against PDB, Pfam-A, and SWISS-PROT to find templates. | 2004 | http://www.rostlab.org/services/CHOP/ | [6] |
| DomPred | Combine homology and secondary structure element alignment to find templates. | 2005 | http://bioinf.cs.ucl.ac.uk/software.html | [15] | |
| SSEP-Domain | Based on the secondary structure elements alignment and profile-profile alignment. | 2006 | http://www.bio.ifi.lmu.de/SSEP/ | [16] | |
| ThreaDom | Deduce domain boundary locations based on multiple threading alignments. | 2013 | http://zhanglab.ccmb.med.umich.edu/ThreaDom/ | [18] | |
| CLADE | Identifie domains by a multi-source strategy which combining multiple HMMs profile. | 2016 | http://www.lcqb.upmc.fr/CLADE | [13] | |
| MetaCLADE | A multi-source domain annotation tool for metagenomic dataset. | 2018 | http://www.lcqb.upmc.fr/metaclade | [14] | |
| Ab initio methods | Domain Guess by Size | Detect domain boundaries based on the distributions of chain and domain lengths. | 2000 | [26] | |
| CHOPnet | Feed-forward neural network that uses amino acid composition and secondary structure and solvent accessibility as features. | 2004 | [27] | ||
| PPRODO | Feed-forward neural network that uses position-specific scoring matrix (PSSM) generated by PSI-BLAST as features. | 2005 | http://gene.kias.re.kr/~jlee/pprodo/ | [28] | |
| DOMpro | RNN uses secondary structure and solvent accessibility as features. | 2005 | http://www.igb.uci.edu/servers/psss.html | [31] | |
| KemaDom | Combine three SVM classifiers that use different features as inputs to predict domain boundaries. | 2006 | http://www.iipl.fudan.edu.cn/lschen/kemadom.htm | [30] | |
| DomainDiscovery | SVM uses inter-domain linker index, PSSM, secondary structural, and solvent accessibility as features. | 2006 | [33] | ||
| IGRN | An improved general regression network model that is trained by the information of PSSM, interdomain linker index, secondary structure, and solvent accessibility. | 2008 | [32] | ||
| DomSVR | Sequence is encoded by physicochemical and biological properties. SVR uses encoded sequence to predict domain boundary. | 2010 | [35] | ||
| DoBo | SVM uses evolutionary domain boundary signals embedded in homologous proteins as input features. | 2011 | http://sysbio.rnet.missouri.edu/dobo/ | [34] | |
| DROP | An SVM to predict domain linkers using 25 optimal features selected from a set of 3000 features. | 2011 | http://tuat.ac.jp/~domserv/DROP.html | [37] | |
| DomHR | Identify domain boundaries in proteins by defining the edge of domain and boundary regions as a hinge region. | 2013 | http://cal.tongji.edu.cn/domain/ | [39] | |
| PDP-CON | Combine predicted results from six single domain boundary prediction methods. | 2016 | https://cmaterju.org/cmaterbioinfo/ | [38] | |
| ConDo | Use long-range, coevolutionary features to train neural networks. | 2018 | https://github.com/gicsaw/ConDo.git | [40] | |
| DNN-Dom | Combine CNN and BGRU to predict domain boundary by combining amino acid composition information, PSSM, solvent accessibility, and secondary structure. A balanced Random Forest is used to solve the imbalance samples problem. | 2019 | http://isyslab.info/DNN-Dom/ | [42] | |
| DeepDom | Use sequences information encoded by physical–chemical properties to train a bidirectional LSTM model to predict domain boundaries. | 2019 | https://github.com/yuexujiang/DeepDom | [41] | |
| FuPred | Predict protein domain boundaries using predicted contact maps generated by ANN. | 2020 | https://zhanglab.ccmb.med.umich.edu/FUpred | [44] |
2.1. Sequence-based domain identification methods
Compared with structural information, protein sequence information is easier to obtain. Moreover, with the development of sequencing technology, the amount of protein sequence data has grown rapidly, which facilitates sequence-based methods to identify domains. Based on the observation that similar domains often occur in different proteins, homology-based methods have been developed to detect domains by comparing them with homologous sequences with known annotated domains. Homology-based methods can achieve good accuracy when sequences with domain information can be identified. However, their prediction accuracy decreases sharply for targets lacking homologous templates. Ab initio methods have been developed to overcome this limitation. Ab initio methods assume that domain boundaries have some features that are different from other regions in a protein. Statistical methods and machine learning methods are often used to learn these features and identify domain boundaries. With the development of machine learning technology and the growing number of protein sequences in databases, these ab initio methods have progressed significantly in recent years. Table 1 lists most of the sequence-based protein domain identification methods with a brief description and URL when available.
2.1.1. Homology-based methods
The basic principle of homology-based methods is finding the homologous segments in different protein sequences through sequence alignment. Homology-based methods need templates with known domain annotations and efficient sequence alignment algorithms to find templates that match a target sequence. For example, CHOP [6] implements three hierarchical steps to predict domain boundaries. Target sequences are aligned with data from PDB [7], Pfam-A [8], and SWISS-PROT [9] to find homologous sequences with domain annotations.
In some cases, homologous templates cannot be identified by simple alignment algorithms. Then, more advanced algorithms are needed to find remote homologous templates. Profiles are widely used in homologous sequence searching because these can represent domain families rather than single domain sequences and allow greater residue divergence in matched sequences to find remote templates. A profile describes the frequency of different amino acids at each position in a sequence that belongs to a given domain family. For example, several advanced alignment tools, such as HMMer [10], HHblits [11], HHsearch [12], are widely used to identify domains. Based on the observation that many proteins are not globally conserved but might be locally conserved in separate phylogenetic clades, CLADE [13] modified Pfam HMMs profile library and proposed a multi-source strategy that combines multiple HMMs profile to identify a domain. MetaCLADE [14] was further proposed to annotate metagenomic dataset also based on a multi-source domain annotation strategy. Predicted Secondary structure information is another item that can provide additional information to help identify remote homologous templates. For example, DomPred [15] combines secondary structure element alignment and multiple sequence alignment to find homologous sequences. Then, the domain boundaries of homologous sequences are used to predict the boundary of a target sequence. SSEP-Domain [16] identifies potential boundaries based on secondary structure element alignment and profile–profile alignments (PPA) [17].
ThreaDom [18], developed recently, adopts a threading-based algorithm to improve remote homologous templates detection [19], [5]. It first uses eight LOMETS [20] programs to thread a target sequence through PDB [7] to find homologous templates and then constructs a multiple sequence alignment based on the target sequence. According to these multiple sequence alignments, a domain conservation score (DCS) is calculated to measure the conservation level of each residue and further used to judge boundary regions.
Domain architecture of a protein is defined as the arrangement of its constituent domains [21]. Research on multi-domain proteins shows that some domain combinations are highly recurrent, while some combinations never appear. Such information can be used to enhance domain identification. Based on domain co-occurrence, CODD [22], dPUC [23] and DAMA [24], used different algorithms to predict domain architecture. Recently, dPUC2 [25] took into account order in which domains preferentially co-occur to improve domain architecture prediction, since it is observed that domains not only have combination preferences but also have order preferences in protein sequences.
2.1.2. Ab initio methods
Homology alignment-based methods can achieve high prediction accuracy when close templates can be identified. However, the prediction accuracy decreases sharply for targets lacking homologous templates. Ab initio methods can overcome this limitation to some extent.
Some ab initio methods use statistical approaches to predict boundary regions. For example, Domain Guess by Size [26] detects domain boundaries based on the distributions of chains and domain lengths. Domain boundary prediction can be seen as a binary classification problem for each residue in a target sequence. Each residue is labeled as being either a domain boundary residue or not. In view of the advantages of machine learning methods in the classification problem, many tools have been developed to detect domain boundaries using different machine learning algorithms.
In the early stage, ab initio methods usually use simple architectures of artificial neural networks and similar features, such as residue composition, predicted secondary structure, and solvent accessibility. These features focus on features that are associated with specific residues and short-range information. For example, CHOPnet [27] is a three-layer feed-forward artificial neural network that uses amino acid composition and predicts secondary structure and solvent accessibility to encode the residues. PPRODO [28] adopts a feed-forward back-propagation network with a single hidden layer and used a PSSM generated by PSI-BLAST [29] as an input. KemaDom [30] combines three SVM classifiers and each of them uses a subset of these features, including simple physiochemical information, amino acid entropy, secondary structure, the structures of five-residue segments, and solvent accessibility. DOMpro [31] uses recursive neural networks as an architecture and features like predicted secondary structure and solvent accessibility. These methods use a sliding window to choose a segment of a sequence as an input, predicting whether the residue located at the center of the side window is a domain boundary. In this stage, many methods ignore long-range information, and the overall accuracy of these methods is only approximately 25–40%.
To improve the accuracy of domain boundary prediction, some methods have tried to incorporate more features, such as an inter-domain linker index, physicochemical properties, and long-range interactions. These features can capture more long-range information about a domain. However, including all these features may cause the curse of dimensionality. And learning in a high-dimensional space will consume more computing resources and time and increase the risk of overfitting. For example, IGRN [32] uses enhanced general regression networks (EGRN) and PSSMs, secondary structure, solvent accessibility, and an inter-domain linker index as the input feature. To avoid the curse of dimensionality, it filters noise and less discriminative features using an auto-associative network. This auto-associative network includes an encoding unit, a bottleneck unit, and a decoding unit. Then the output of this auto-associative network is used as an input for a general regression neural network to predict whether a residue is in the domain boundary.
Due to the limitation of the amount of protein structure data available, only a few proteins have accurate structure-based domain annotations that can be used as a training data set. Small-sample data is a challenge for machine learning methods, which are data-driven. If a sample set is too small, machine learning methods may can’t learn meaningful information from the data. SVM is widely used at this stage because of its classification ability for high-dimensional small sample data. SVM maps the input data into a high-dimensional feature space and then finds a hyperplane that can separate two different classes in this space. Coupled with improved input features, SVM achieves better classification performance and strong generalization ability.
These methods include DomainDiscovery [33], which predicts domain boundaries by using SVM with PSSM, secondary structure, solvent accessibility, and an inter-domain linker index. DoBo [34] introduces evolutionary domain information that is included in homologous proteins into a protein domain boundary prediction. The domain architecture of homologous proteins can be used to reveal the potential domain boundary sites of a target protein sequence.
The following methods have been tried to consider the physicochemical properties of residues. DomSVR [35] uses a support vector regression to predict domain boundaries. Protein sequences are encoded by the physicochemical and biological properties, which are derived from the AAindex database [36]. Then, a principal component analysis (PCA) is used to choose the most important indices to encode protein sequences. DROP [37] encodes residues into a 3000-dimensional vector, where each element represents a different property, including PSSMs and over 2000 physicochemical properties. A random forest algorithm is used to select optimal features. Finally, an SVM is trained to predict domain boundaries.
In addition to improving feature extraction, there have been other attempts to improve the accuracy of domain boundary prediction by adopting innovative methods. PDP-CON [38] combines the results from six single domain boundary prediction classifiers by implementing an n-star quality consensus approach to yield a better prediction result. DomHR [39] is an indirect method that predicts domain boundaries based on a creative hinge region strategy. It defines a hinge region as an area centered on the boundary between domain regions and boundaries regions. The key step of DomHR [39] is in domain-hinge-boundary (DHB) feature generation.
With the development of deep learning technology and the increase in the number of protein data sets, artificial neural networks with multiple hidden layers can now be used to predict domain boundaries. Deep learning methods can generate data representations automatically from big datasets and generally achieve better prediction accuracy. For example, ConDo [40], which was developed by Hong et al., utilizes neural networks that were trained on long-range, coevolutionary features, in addition to conventional local window features, to detect domains. Some residues in domains are far away from others in a sequence but are actually close in a 3D structure and form hydrogen bonds or disulfide bonds. These long-range interactions are important for structural stability. RNN (including LSTM) is highly valued because of its ability to learn long-range information. DeepDom [41] and DNN-dom [42] are two methods developed recently that use RNNs to predict domain boundaries.
DeepDom [41] uses a stacked bidirectional Long Short Term Memory (LSTM). LSTM uses a cell state to remember information from the input data that has been processed so it can learn global information from protein sequences. DeepDom uses a sliding window to encode an input sequence into equal-length fragments, and each residue is encoded by a six-dimensional vector. The first five dimensions represent five physical–chemical properties, and the sixth encoding dimension is a padding indicator. The LSTM would predict the probability of the residue located at the center of the sliding window being a boundary, not being a boundary, or padding residue.
DNN-Dom [42] combines a convolutional neural network and bidirectional gate recurrent units (BGRUs) models to predict the domain boundary of a protein. Convolutional neural networks are utilized to extract multi-scale local contexts, and the outputs of CNNs are fed into BGRUs, which are used to learn the long-range interactions. The imbalance of positive samples with negative samples is a challenge for deep learning methods. To deal with the imbalance of samples, that is, as there are more non-boundary samples than boundary samples, DNN-Dom uses a balanced Random Forest where each tree in the RF is trained with balanced samples to predict the probability that the input sample is the boundary of a domain. It uses four kinds of input features representing each residue, including amino acid composition information, a protein position specific matrix, solvent accessibility, and protein secondary structure. These features contain local information, global information, and high-level latent information that can improve prediction performance. DNN-Dom has achieved great performance with the CASP datasets (from CASP 9 to CASP 12) and other independent protein domain datasets.
Most of the above sequence-based methods do not consider discontinuous domain predictions, while about 18% of proteins in the current PDB library have at least one discontinuous domain. ThreaDomEX [43] and FUpred [44] are two methods that pay special attention to discontinuous domain detection. ThraeDomEX can detect a discontinuous domain mainly by incorporating DomEx [45], which can assemble non-consecutive segments following multiple threading template alignments. FUpred detects domain boundaries based on contact map prediction, which is predicted by deep residual neural networks. When predicting a domain boundary, FUpred will generate an FUscore that maximizes the number of intra-domain contacts while minimizing the number of inter-domain contacts. Thus, it can identify discontinuous domains.
2.2. Structure-based methods
Structure-based methods are quite different from sequence-based methods, and structure-based methods need experimental or predicted protein structures for domain identification. For example, CATHEDRAL [46] compares target protein structure against a structure template library derived from the CATH [47] database to detect domains. DIAL [48] identifies domains by clustering substructures with similar structures. Table 2 lists most of the structure-based protein domain identification methods with a brief description and URL when available.
Table 2.
Structure-based protein domain identification methods.
| Category | Method | Description | Year | URL | Reference |
|---|---|---|---|---|---|
| Structure-based | DomainParser | Use flow network represent protein structure, and identify domain based on maximum-flow/minimum-cut theorem. | 2000 | http://compbio.ornl.gov/structure/domainparser/ | [49] |
| PDP | Identify the dividing site that makes the contact density of the two parts lower than a threshold as the domain boundary. | 2003 | http://123d.ncifcrf.gov/ | [50] | |
| DIAL | Identify the domain by clustering substructures on the basis of their spatial distances. | 2005 | http://www.ncbs.res.in/~faculty/mini/DIAL/home.html | [48] | |
| CATHEDRAL | Identifiy the domain by comparing target structure with structure templates in CATH. | 2007 | http://cathwww.biochem.ucl.ac.uk/cgi-bin/cath/CathedralServer.pl | [46] | |
| DDOMAIN | Identify the dividing site that makes the distance between the two parts exceed the threshold as the domain boundary. | 2007 | http://sparks.informatics.iupui.edu | [51] | |
| DHcL | Identify the domain by calculating the van der Waals model of protein. | 2008 | http://sitron.bccs.uib.no/dhcl/ | [52] | |
| Sword | Assign structural domains through the hierarchical merging of protein units. SWORD provides different domain assignments using different merge schemes. | 2017 | www.dsimb.inserm.fr/sword/ | [53] | |
| Predcitedstructure-based | SnapDRAGON | DRAGON generates 100 models, and then structure-based domain assignment is used to parse the models into domains. Finally, a result is derived from the consistency of the predicted boundaries. | 2002 | [55] | |
| RosettaDOM | RosettaDOM is a hybrid method that uses homology-based methods to predict domain boundaries when homologous templates can be found. When lacking templates, Rosetta is used to generate models, and final domain boundary predictions are derived from the models. | 2005 | [54] | ||
| OPUS-Dom | Generate a large ensemble of folded structure decoys by VECFOLD, and predicted domain boundaries are derived from the consistency of the domain boundary in the set of 3D models. | 2009 | [56] |
Since the above methods need templates with known domain information, some other methods that are template independent have been developed based on the structural characteristics of domains. DomainParser [49] is an efficient domain decomposition algorithm based on graph-theoretic. Residues were represented as nodes and residue-residue contacts were represented as edge. Capacity values were calculated for each edge depending on the strength of interaction. DomainParser divided the protein into two domains by finding the boundary that minimizes the edge capacity between the two sub-graph. PDP [50] and DDOMAIN [51] split proteins into domains depending on the assumption that there are more intra-domain residue contacts than inter-domain contacts. PDP splits proteins into two candidate domains. Then, contacts between candidate domains are normalized by domain sizes. Two segments are confirmed as domains if the contacts between these segments are less than half of the average contact density for the whole domain. Finally, contacts between all domains are checked, and two domains are combined into one if their normalized contacts are greater than a manually selected threshold. The final step allows PDP to find discontinuous domains. DDOMAIN uses normalized contacts similar to PDP. Unlike PDP, which only considers the number of contacts, DDOMAIN defines contact energy dependent on the number and distance of contacts. Moreover, DDOMAIN uses a threshold that is learned from a training data set to determine whether a protein is divided into two domains. Different from compactness-based approaches, DHcL [52] decomposes protein domains by calculating a van der Waals model of a protein.
Although protein domain is an important concept and has been used in many fields in the biological sciences for many years, there is still no authoritative definition of what a domain is. The variety of definitions of a domain reflects different perspectives and the different problems being tackled. As a result, many methods have been developed to detect domains, while some of them annotate the same protein in different ways. Therefore, some proteins will be decomposed into different domains using different tools. Considering that a protein may be divided into different but equally valid domain, SWORD [53] was developed to generate multiple alternative domain architectures for a target protein. It defines protein units (PUs), a structural descriptor between secondary structures and domains. PUs will gradually merge into large fragments, and different merge schemes will enable SWORD to provide several different domain assignments.
Furthermore, there are some methods used predicted protein models to detect domains, such as RosettaDom [54], SnapDRAGON [55], and OPUS-Dom [56]. In general, these methods predict a large number of model structures of target sequences using ab initio methods such as Rosetta, DRAGON [57], [58], [59], and VECFOLD. Then, a structure-based domain assignment tool such as Taylor [60] is used to detect domain boundaries for each model generated by ab initio methods. Finally, the predicted domain boundaries of a target sequence are obtained by counting the domain boundaries of these 3D models. These methods often give reliable results but usually need significant computational resources.
3. Protein domain libraries
The rapid increase in protein sequence and structure data has generated a pressing need to classify them systematically. Proteins are generally classified into different groups based on their sequence or structural similarities. Then the functional properties of a newly identified protein can be inferred from a well-characterized protein in the same group to which it is predicted to belong. Here, we focus on the classification of protein domains. Consistent with the methods to identify domains, there are also two kinds of domain databases, namely sequence-based and structure-based.
3.1. Sequence-based domain databases
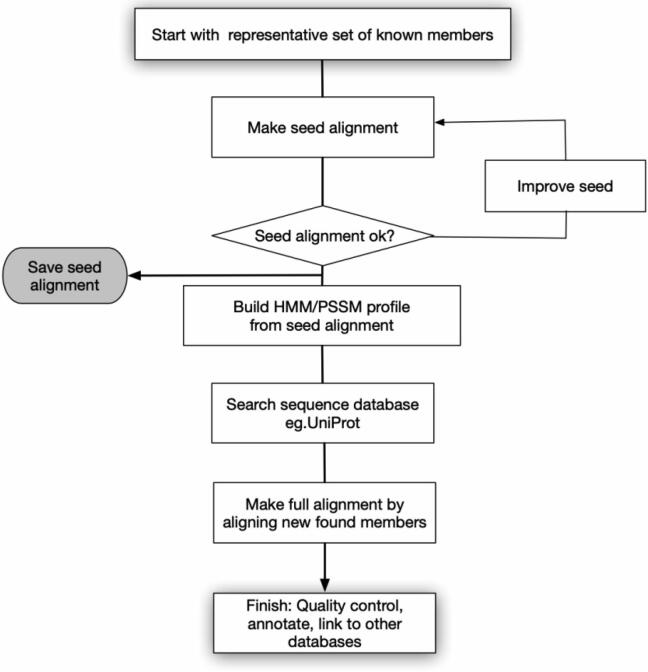
Despite great progress having been made in the field of ab initio methods of protein domain boundary prediction, most of them are not suitable for large-scale sequence analysis. Therefore, most automatic domain clustering methods are homology-based, followed by varying levels of expert-driven validation. A general flow chart of the construction of sequence-based domain family databases is shown in Fig. 2. It usually starts with a representative set of sequences belonging to a family that are selected as a ‘seed’. Then a multiple sequence alignment of these seeds produces conservative patterns, and profiles are generated based on it. The profiles produced from the previous step are used to search against a protein sequence database (e.g., UniProt) to find all sequences belonging to this family and then generate profiles based on all family members. Three widely used sequence-based domain databases are introduced below.
Fig. 2.
Diagram of homology alignment-based methods to construct a domain database.
Pfam [8] is one of the most comprehensive domain family databases. It is manually curated using a seed alignment first, and then a profile HMM is built based on the seed alignment. A profile HMM is queried against a sequence database called pfamseq, which is derived from the UniProt Knowledgebase (UniProtKB) [61] Reference Proteomes to find other members in the same domain family. Pfam consists of two types of subsets: high-quality Pfam-A families that are generated by manually checking seed alignments and HMMs, and less reliable Pfam-B families, which are produced automatically by applying the ADDA algorithm [62].
Similar to Pfam, SMART [63], [64] (Simple Modula Architecture Research Tool) uses HMMs to search and annotate protein sequences that belong to the same family. It is synchronized with UniProt [65], Ensembl [66], and STRING [67]. SMART holds manually curated HMMs, and structure information is encompassed to select seed alignment. To improve its ability to find homologous sequences, especially remote homologous sequences, SMART uses three iterative homologue search methods—HMMer [10], MoST [68], and WiseTools [69]. Sequences considered to be homologues are added to a multiple alignment, which is used to construct profiles and HMMs. This tool also offers a ‘genomic’ mode that annotates proteins from complete sequenced genomes.
PROSITE [70], [71] provides protein information about domains, families, and functional sites. It identifies protein families and protein domains using generalized profiles and uses patterns to identify short sequence motifs, which often have an important impact on structure or function. Patterns are regular expressions that can be used to identify highly conserved structures and motifs. These areas typically include 10 to 20 amino acids and have important functions such as active sites or binding sites. PROSITE includes a collection of rules called ProRules [72] to define protein annotations and the conditions under which they apply. It uses patterns and profiles to search against UniProtKB [61] and annotate protein databases via ProRule. Combining profiles and patterns with ProRule, PROSITE can annotate proteins more accurately.
3.2. Structure-based domain databases
Sequence-based protein domain database depend on sequence alignments to identify domains that belong to the same family. In the Twilight Zone [73] of sequence similarity (<30% sequence identity), the reliability of sequence comparisons decreases quickly. Structure-based protein domain identification can break through this restriction, although the number of proteins with known structures is much less than that of known sequences. Structure-based domain databases usually classify proteins on hierarchical levels. Some levels of hierarchy include Class, Architecture, Fold/topology, Superfamily, and Family. Two popular structure-based protein domain databases are SCOP [74] and CATH [47]. The basic principle of these two databases is finding conserved substructures that are repeated in different proteins through structure alignment.
SCOP [74] (Structural Classification of Proteins) mainly annotates domains and constructs domain families by manual inspection. It organizes domains and discrete units into families and superfamilies based on structural features and evolutionary relationships, and superfamilies are further organized into folds and classes. Similar structures and sequences means that a protein has an evolutionary relationship and similar functions. Comparing the structures of proteins and organizing them into different levels can help researchers explore proteins with unknown function.
Like SCOP [74], CATH [47], [75] classifies proteins in four main levels: class (C), architecture (A), topology (T), and homologous superfamily (H). CATH combines automatic procedures with manual curation to identify protein domain structures and clusters them. It uses a number of sensitive structure-comparison and sequence comparison tools (including SSAP [76], HMMER3 hmmer.org, PRC [77]) to assist the manual curation of these remote evolutionary relationships.
3.3. Integrated domain databases
Since a variety of domain family databases are available now, and each source database has its own biological focus, it may be difficult to choose which database to use or how to meaningfully combine the results from different sources. InterPro [78] and Genome3D [79] were designed as comprehensive databases to combine data from other databases.
InterPro [78] integrates 14 protein family classification databases and maps these family resources to the primary sequences of UniProt [9]; as of September 2020, it has annotated 79.1% of the protein sequences of UniProt. It does not generate annotations itself but rather integrates information from other member databases. Member databases generate representative signatures for each group of homologous proteins. Then, InterPro manually inspects these signatures to ensure accuracy. The new signatures passing quality control are added to InterPro to be used to identify and annotate protein sequences.
Genome3D [79] also integrates domain family annotations from different databases like InterPro. It not only collects information from SCOP [74] and CATH [47], but also uses five domain prediction methods (Gene3D [80], SUPERFAMILY [81], FUGUE [82], Phyre [83], and pDomTHREADER [84]) to identify domains. Gene3D and SUPERFAMILY construct HMMs to describe the sequence features of SCOP or CATH superfamilies and use these HMMs to identify domains in new sequences. Other methods can detect more distant homologues belonging to the SCOP (FUGUR, Phyre) or CATH (FUGUE, pDomTHREADER) superfamilies. Since none of these methods is guaranteed to provide a correct answer, Genome3D displays prediction results from all these methods so that users can identify which result is more likely to be correct.
3.4. Basic statistics of what is in available domain databases
Based on the above sequence-based and structure-based domain databases, we have counted how many domain families have been annotated for the three kingdoms of life: eukaryote, bacteria and archaea, which is shown in Table 3. From Table 3, it is easy to see that eukaryotes have the most domain families, followed by bacteria and archaea nearly among all the counted databases. The number and percentage shared by the three domains of life have also been counted. Around 4.4%-16.2% domain families have been shared by the three domains of life according to the different counted databases. Furthermore, we also counted the protein domain average length of the three kingdoms of life in CATH and SCOP from which we can get the downloaded domain length. Table 4 shows that the domain average length of all the three kingdoms in SCOP is longer than that of CATH, which is mainly attributable to the different protocols used by the two databases. Meanwhile, the average length of domains of bacterial seems longer than that of eukaryota and archaea in both CATH and SCOP, especially in the SCOP database.
Table 3.
Statistics on the number of domain families annotated for the three kingdoms of life in different domain databases.
| Database | Source |
Shared by the three kingdoms | ||
|---|---|---|---|---|
| Eukaryota | Bacteria | Archaea | ||
| Pfam | 8437 | 5857 | 1735 | 890 (6.2%) |
| SMART | 1120 | 428 | 241 | 166 (11.4%) |
| PROSITE | 2185 | 1262 | 641 | 500 (16.2%) |
| CATH | 2972 | 3665 | 918 | 399 (5.9%) |
| SCOP | 3166 | 2626 | 749 | 264 (4.4%) |
Table 4.
The domain average length of the three kingdoms in CATH and SCOP.
| Database | Mean length | Std | Eukaryota |
Bacteria |
Archaea |
|||
|---|---|---|---|---|---|---|---|---|
| Mean | Std | Mean | Std | Mean | Std | |||
| CATH | 150.4 | 90.9 | 147.9 | 90.8 | 154.8 | 91.8 | 137.2 | 81.0 |
| SCOP | 196.8 | 129.7 | 179.5 | 128.9 | 215.4 | 129.5 | 189.5 | 115.1 |
4. Discussion and conclusion
The exact identification of protein domains and their boundaries is one of the most important problems in the study of protein structure and function. Therefore, a number of domain prediction methods and databases have been developed, which can be divided into two categories: sequence-based and structure-based.
With known three-dimensional structures, accuracy is often not the problem. The problem that needs be considered is the ambiguity in a domain definition. To the best of our knowledge, Sword [53], developed recently, is the only method which has tried to address this problem by producing multiple alternative decompositions of a protein. Therefore, more innovative multipartitioning algorithms are needed to tackle this problem.
The difficulty of obtaining protein experimental structure limits the application scope of structure-based protein domain identification methods. Sequence-based methods have been developed based on the assumption that domain family members share some common sequence features. When there are close templates, such methods can achieve high prediction accuracy. However, this prediction accuracy decreases sharply when homologous templates are unavailable. Therefore, a number of approaches independent of templates have been developed, and most of them are based on machine learning. Despite extensive research, predicting domain boundaries from sequence data alone is still a challenging problem. The prediction accuracy of most of these methods is not high enough to be applied in large-scale sequence annotation. Another problem is that sequence-based methods generally do not consider discontinuous domains prediction. With the development of machine learning algorithms and the improvement of contact map prediction, there will be great progress in protein domain prediction accuracy and discontinuous domain detection.
Coupled with the development of domain identification methods, a variety of protein domain databases have been constructed to classify protein sequence and structure. A newly identified protein can be classified into a corresponding family through searching the available protein domain family databases. InterPro [78], which is an integrated domain family resource, has annotated 79.1% protein sequences in UniProt [9]. It is hoped that with the improvement of the protein domain detection methods, the domain annotation ratio of protein sequences will increase.
CRediT authorship contribution statement
Yan Wang: Conceptualization, Writing - review & editing, Formal analysis, Funding acquisition. Hang Zhang: Conceptualization, Writing - review & editing, Data curation. Haolin Zhong: Writing - review & editing. Zhidong Xue: Conceptualization, Supervision, Writing - review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by National Natural Science Foundation of China under Grant 61772217, and Fundamental Research Funds for the Central Universities under Grant 2016YXMS104 and 2017KFYXJJ225.
References
- 1.Dawson N., Sillitoe I., Marsden R.L., Orengo C.A. Bioinformatics. Springer; 2017. The classification of protein domains; pp. 137–164. [DOI] [PubMed] [Google Scholar]
- 2.Peng W., Wang J., Cai J., Chen L., Li M., Wu F.X. Improving protein function prediction using domain and protein complexes in PPI networks. BMC Syst Biol. 2014;8:35. doi: 10.1186/1752-0509-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z., Zhao C., Wang Y., Sun Z., Wang N. PANDA: protein function prediction using domain architecture and affinity propagation. Sci Rep. 2018;8(1):3484. doi: 10.1038/s41598-018-21849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piovesan D., Giollo M., Leonardi E., Ferrari C., Tosatto S.C. INGA: protein function prediction combining interaction networks, domain assignments and sequence similarity. Nucleic Acids Res. 2015;43(W1):W134–140. doi: 10.1093/nar/gkv523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y. I-TASSER, Fully automated protein structure prediction in CASP8. Proteins Struct Funct Bioinf. 2009;77(S9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Rost B. CHOP: parsing proteins into structural domains. Nucleic Acids Res. 2004;32(suppl_2):W569–W571. doi: 10.1093/nar/gkh481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47(D1):D464–D474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sara E.G., Jaina M., Alex B., Eddy S.R., Aurélien L., Potter S.C. The Pfam protein families database in 2019. Nucleic Acids Res. 2018;D1:D1. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bairoch A., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28(1):45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eddy S.R., Mitchison G., Durbin R. Maximum discrimination hidden Markov models of sequence consensus. J Comput Biol. 1995;2(1):9–23. doi: 10.1089/cmb.1995.2.9. [DOI] [PubMed] [Google Scholar]
- 11.Remmert M., Biegert A., Hauser A., Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 12.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 13.Bernardes J., Zaverucha G., Vaquero C., Carbone A. Improvement in protein domain identification is reached by breaking consensus, with the agreement of many profiles and domain co-occurrence. PLoS Comput Biol. 2016;12(7) doi: 10.1371/journal.pcbi.1005038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ugarte A., Vicedomini R., Bernardes J., Carbone A. A multi-source domain annotation pipeline for quantitative metagenomic and metatranscriptomic functional profiling. Microbiome. 2018;6(1):149. doi: 10.1186/s40168-018-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryson K., McGuffin L.J., Marsden R.L., Ward J.J., Sodhi J.S., Jones D.T. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33(suppl_2):W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gewehr J.E., Zimmer R. SSEP-Domain: protein domain prediction by alignment of secondary structure elements and profiles. Bioinformatics. 2006;22(2):181–187. doi: 10.1093/bioinformatics/bti751. [DOI] [PubMed] [Google Scholar]
- 17.von Öhsen N., Sommer I., Zimmer R. Biocomputing 2003. World Scientific; 2002. Profile-profile alignment: a powerful tool for protein structure prediction; pp. 252–263. [PubMed] [Google Scholar]
- 18.Xue Z., Xu D., Wang Y., Zhang Y. ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics. 2013;29(13):i247–i256. doi: 10.1093/bioinformatics/btt209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins Struct Funct Bioinf. 2007;69(S8):108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 20.Wu S., Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35(10):3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu L., Tanwar D.K., Penha E.D.S., Wolf Y.I., Koonin E.V., Basu M.K. Grammar of protein domain architectures. PNAS. 2019;116(9):3636–3645. doi: 10.1073/pnas.1814684116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Terrapon N., Gascuel O., Maréchal E., Bréehélin L. Detection of new protein domains using co-occurrence: application to Plasmodium falciparum. Bioinformatics. 2009;25(23):3077–3083. doi: 10.1093/bioinformatics/btp560. [DOI] [PubMed] [Google Scholar]
- 23.Ochoa A., Llinás M., Singh M. Using context to improve protein domain identification. BMC Bioinf. 2011;12:90. doi: 10.1186/1471-2105-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernardes J.S., Vieira F.R., Zaverucha G., Carbone A. A multi-objective optimization approach accurately resolves protein domain architectures. Bioinformatics. 2016;32(3):345–353. doi: 10.1093/bioinformatics/btv582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ochoa A., Singh M. Domain prediction with probabilistic directional context. Bioinformatics. 2017;33(16):2471–2478. doi: 10.1093/bioinformatics/btx221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheelan S.J., Marchler-Bauer A., Bryant S.H. Domain size distributions can predict domain boundaries. Bioinformatics. 2000;16(7):613–618. doi: 10.1093/bioinformatics/16.7.613. [DOI] [PubMed] [Google Scholar]
- 27.Liu J., Rost B. Sequence-based prediction of protein domains. Nucleic Acids Res. 2004;32(12):3522–3530. doi: 10.1093/nar/gkh684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sim J., Kim S.Y., Lee J. PPRODO: prediction of protein domain boundaries using neural networks. Proteins Struct Funct Bioinf. 2005;59(3):627–632. doi: 10.1002/prot.20442. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L., Wang W., Ling S., Jia C., Wang F. KemaDom: a web server for domain prediction using kernel machine with local context. Nucleic Acids Res. 2006;34(suppl_2):W158–W163. doi: 10.1093/nar/gkl331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng J., Sweredoski M.J., Baldi P. DOMpro: protein domain prediction using profiles, secondary structure, relative solvent accessibility, and recursive neural networks. Data Min Knowl Disc. 2006;13(1):1–10. [Google Scholar]
- 32.Yoo P.D., Sikder A.R., Zhou B.B., Zomaya A.Y. BMC bioinformatics. Springer; 2008. Improved general regression network for protein domain boundary prediction; p. S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sikder AR, Zomaya AY. Improving the performance of DomainDiscovery of protein domain boundary assignment using inter-domain linker index. In: BMC bioinformatics: 2006. BioMed Central: 1-9. [DOI] [PMC free article] [PubMed]
- 34.Eickholt J., Deng X., Cheng J. DoBo: Protein domain boundary prediction by integrating evolutionary signals and machine learning. BMC Bioinf. 2011;12(1):43. doi: 10.1186/1471-2105-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen P., Liu C., Burge L., Li J., Mohammad M., Southerland W. DomSVR: domain boundary prediction with support vector regression from sequence information alone. Amino Acids. 2010;39(3):713–726. doi: 10.1007/s00726-010-0506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawashima S., Pokarowski P., Pokarowska M., Kolinski A., Katayama T., Kanehisa M. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2007;36(suppl_1):D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ebina T., Toh H., Kuroda Y. DROP: an SVM domain linker predictor trained with optimal features selected by random forest. Bioinformatics. 2011;27(4):487–494. doi: 10.1093/bioinformatics/btq700. [DOI] [PubMed] [Google Scholar]
- 38.Chatterjee P., Basu S., Zubek J., Kundu M., Nasipuri M., Plewczynski D. PDP-CON: prediction of domain/linker residues in protein sequences using a consensus approach. J Mol Model. 2016;22(4):72. doi: 10.1007/s00894-016-2933-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X.-y., Lu L.-j., Song Q., Yang Q.-q., Li D.-p., Sun J.-m., Li T.-h., Cong P.-s. DomHR: accurately identifying domain boundaries in proteins using a hinge region strategy. PLoS ONE. 2013;8(4) doi: 10.1371/journal.pone.0060559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong S.H., Joo K., Lee J. ConDo: protein domain boundary prediction using coevolutionary information. Bioinformatics. 2019;35(14):2411–2417. doi: 10.1093/bioinformatics/bty973. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y, Wang D, Xu D: DeepDom: Predicting protein domain boundary from sequence alone using stacked bidirectional LSTM. In: Pacific Symposium on Biocomputing Pacific Symposium on Biocomputing: 2019. World Scientific: 66-75 [PMC free article] [PubMed]
- 42.Shi Q., Chen W., Huang S., Jin F., Dong Y., Wang Y. DNN-Dom: predicting protein domain boundary from sequence alone by deep neural network. Bioinformatics. 2019;35(24):5128–5136. doi: 10.1093/bioinformatics/btz464. [DOI] [PubMed] [Google Scholar]
- 43.Wang Y., Wang J., Li R., Shi Q., Xue Z., Zhang Y. ThreaDomEx: a unified platform for predicting continuous and discontinuous protein domains by multiple-threading and segment assembly. Nucleic Acids Res. 2017;45(W1):W400–W407. doi: 10.1093/nar/gkx410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng W., Zhou X., Wuyun Q., Pearce R., Li Y., Zhang Y. FUpred: detecting protein domains through deep-learning based contact map prediction. Bioinformatics. 2020 doi: 10.1093/bioinformatics/btaa217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue Z., Jang R., Govindarajoo B., Huang Y., Wang Y. Extending protein domain boundary predictors to detect discontinuous domains. PLoS ONE. 2015;10(10) doi: 10.1371/journal.pone.0141541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Redfern O.C., Harrison A., Dallman T., Pearl F.M., Orengo C.A. CATHEDRAL: a fast and effective algorithm to predict folds and domain boundaries from multidomain protein structures. PLoS Comput Biol. 2007;3(11) doi: 10.1371/journal.pcbi.0030232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sillitoe I., Dawson N., Lewis T.E., Das S., Lees J.G., Ashford P. CATH: expanding the horizons of structure-based functional annotations for genome sequences. Nucleic Acids Res. 2019;47(D1):D280–D284. doi: 10.1093/nar/gky1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pugalenthi G., Archunan G., Sowdhamini R. DIAL: a web-based server for the automatic identification of structural domains in proteins. Nucleic Acids Res. 2005;33(suppl_2):W130–W132. doi: 10.1093/nar/gki427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y., Xu D., Gabow H.N. Protein domain decomposition using a graph-theoretic approach. Bioinformatics. 2000;16(12):1091–1104. doi: 10.1093/bioinformatics/16.12.1091. [DOI] [PubMed] [Google Scholar]
- 50.Alexandrov N., Shindyalov I. PDP: protein domain parser. Bioinformatics. 2003;19(3):429–430. doi: 10.1093/bioinformatics/btg006. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H., Xue B., Zhou Y. DDOMAIN: dividing structures into domains using a normalized domain–domain interaction profile. Protein Sci. 2007;16(5):947–955. doi: 10.1110/ps.062597307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koczyk G., Berezovsky I.N. Domain Hierarchy and closed Loops (DHcL): a server for exploring hierarchy of protein domain structure. Nucleic Acids Res. 2008;36(suppl_2):W239–W245. doi: 10.1093/nar/gkn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Postic G., Ghouzam Y., Chebrek R., Gelly J.-C. An ambiguity principle for assigning protein structural domains. Sci Adv. 2017;3(1) doi: 10.1126/sciadv.1600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D.E., Chivian D., Malmström L., Baker D. Automated prediction of domain boundaries in CASP6 targets using Ginzu and RosettaDOM. Proteins Struct Funct Bioinf. 2005;61(S7):193–200. doi: 10.1002/prot.20737. [DOI] [PubMed] [Google Scholar]
- 55.George R.A., Heringa J. SnapDRAGON: a method to delineate protein structural domains from sequence data. J Mol Biol. 2002;316(3):839–851. doi: 10.1006/jmbi.2001.5387. [DOI] [PubMed] [Google Scholar]
- 56.Wu Y., Dousis A.D., Chen M., Li J., Ma J. OPUS-Dom: applying the folding-based method VECFOLD to determine protein domain boundaries. J Mol Biol. 2009;385(4):1314–1329. doi: 10.1016/j.jmb.2008.10.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aszoadi A., Taylor W. Folding polypeptide α-carbon backbones by distance geometry methods. Biopolymers: Original Res Biomol. 1994;34(4):489–505. [Google Scholar]
- 58.Aszodi A., Gradwell M., Taylor W. Global fold determination from a small number of distance restraints. J Mol Biol. 1995;251(2):308–326. doi: 10.1006/jmbi.1995.0436. [DOI] [PubMed] [Google Scholar]
- 59.Aszódi A., Taylor W.R. Hierarchic inertial projection: a fast distance matrix embedding algorithm. Comput Chem (Oxford) 1997;21(1):13–23. [Google Scholar]
- 60.Taylor W.R. Protein structural domain identification. Protein Eng. 1999;12(3):203–216. doi: 10.1093/protein/12.3.203. [DOI] [PubMed] [Google Scholar]
- 61.Apweiler R., Bairoch A., Wu C.H., Barker W.C., Boeckmann B., Ferro S. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32(suppl_1):D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heger A., Holm L. Exhaustive enumeration of protein domain families. J Mol Biol. 2003;328(3):749–767. doi: 10.1016/s0022-2836(03)00269-9. [DOI] [PubMed] [Google Scholar]
- 63.Schultz J., Milpetz F., Bork P., Ponting C.P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci. 1998;95(11):5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Letunic I., Doerks T., Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43(D1):D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Consortium U. Activities at the universal protein resource (UniProt) Nucleic Acids Res. 2014;42(D1):D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Flicek P., Amode M.R., Barrell D., Beal K., Billis K., Brent S. Ensembl 2014. Nucleic Acids Res. 2014;42(D1):D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., Von Mering C. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucl Acids Res. 2012;41(D1):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tatusov R.L., Altschul S.F., Koonin E.V. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci. 1994;91(25):12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Birney E., Thompson J.D., Gibson T.J. PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res. 1996;24(14):2730–2739. doi: 10.1093/nar/24.14.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sigrist C.J., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M. PROSITE: a documented database using patterns and profiles as motif descriptors. Briefings Bioinf. 2002;3(3):265–274. doi: 10.1093/bib/3.3.265. [DOI] [PubMed] [Google Scholar]
- 71.Sigrist C.J., De Castro E., Cerutti L., Cuche B.A., Hulo N., Bridge A. New and continuing developments at PROSITE. Nucleic Acids Res. 2012;41(D1):D344–D347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sigrist C.J., De Castro E., Langendijk-Genevaux P.S., Le Saux V., Bairoch A., Hulo N. ProRule: a new database containing functional and structural information on PROSITE profiles. Bioinformatics. 2005;21(21):4060–4066. doi: 10.1093/bioinformatics/bti614. [DOI] [PubMed] [Google Scholar]
- 73.Feng D.-F., Doolittle R.F. [21] progressive alignment of amino acid sequences and construction of phylogenetic trees from them. Methods Enzymol. 1996;266:368–382. doi: 10.1016/s0076-6879(96)66023-6. [DOI] [PubMed] [Google Scholar]
- 74.Andreeva A., Kulesha E., Gough J., Murzin A.G. The SCOP database in 2020: expanded classification of representative family and superfamily domains of known protein structures. Nucleic Acids Res. 2020;48(D1):D376–D382. doi: 10.1093/nar/gkz1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Orengo C.A., Michie A.D., Jones S., Jones D.T., Swindells M.B., Thornton J.M. CATH–a hierarchic classification of protein domain structures. Structure. 1997;5(8):1093–1109. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 76.Orengo C.A., Taylor W.R. SSAP: sequential structure alignment program for protein structure comparison. Methods Enzymol. 1996;266:617–635. doi: 10.1016/s0076-6879(96)66038-8. [DOI] [PubMed] [Google Scholar]
- 77.Brandt B.W., Heringa J. webPRC: the Profile Comparer for alignment-based searching of public domain databases. Nucleic Acids Res. 2009;37(Web Server issue):W48–W52. doi: 10.1093/nar/gkp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitchell A., Chang H.-Y., Daugherty L., Fraser M., Hunter S., Lopez R. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43(D1):D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis T.E., Sillitoe I., Andreeva A., Blundell T.L., Buchan D.W., Chothia C. Genome3D: a UK collaborative project to annotate genomic sequences with predicted 3D structures based on SCOP and CATH domains. Nucleic Acids Res. 2012;41(D1):D499–D507. doi: 10.1093/nar/gks1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dawson N.L., Lewis T.E., Das S., Lees J.G., Lee D., Ashford P. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017;45(D1):D289–D295. doi: 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gough J., Chothia C. SUPERFAMILY, HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucleic Acids Res. 2002;30(1):268–272. doi: 10.1093/nar/30.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi J., Blundell T.L., Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310(1):243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 83.Bennett-Lovsey R.M., Herbert A.D., Sternberg M.J., Kelley L.A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins Struct Funct Bioinf. 2008;70(3):611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- 84.Lobley A., Sadowski M.I., Jones D.T. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25(14):1761–1767. doi: 10.1093/bioinformatics/btp302. [DOI] [PubMed] [Google Scholar]