Abstract
Since its emergence as a pneumonia-like outbreak in the Chinese city of Wuhan in late 2019, the novel coronavirus disease COVID-19 has spread widely to become a global pandemic. The first case of COVID-19 in India was reported on 30 January 2020 and since then it has affected more than ten million people and resulted in around 150,000 deaths in the country. Over time, the viral genome has accumulated mutations as it passes through its human hosts, a common evolutionary mechanism found in all microorganisms. This has implications for disease surveillance and management, vaccines and therapeutics, and the emergence of reinfections. Sequencing the viral genome can help monitor these changes and provides an extraordinary opportunity to understand the genetic epidemiology and evolution of the virus as well as tracking its spread in a population. Here we review the past year in the context of the phylogenetic analysis of variants isolated over the course of the pandemic in India and highlight the importance of continued sequencing-based surveillance in the country.
Keywords: SARS-CoV-2, COVID-19, genomics, variants, sequencing, clades
Introduction
The SARS-CoV-2 genome
COVID-19 is caused by the RNA virus SARS-CoV-2, a betacoronavirus with a nearly 30 kb positive-sense, single-strand RNA genome that encodes 29 proteins (Wu et al. 2020). These include structural proteins utilized by the virus to package its RNA as well as proteins for enabling its entry and propagation in the host by hijacking the host cellular machinery for viral replication. SARS-CoV-2 is an enveloped virus with a host-derived lipid membrane. The viral capsid assembly is mediated by several structural proteins encoded by the virus, the most important being the S (spike protein that forms a crown-like structure), M (a hydrophobic membrane protein), E (an integral membrane protein or envelope protein) and N (an abundant nucleocapsid protein that binds the RNA genome) proteins (figure 1). The Spike protein encoded by the S gene has a receptor-binding domain specifically evolved to bind to the human angiotensin-converting enzyme-2 (ACE2) receptor found on the surface of many human cells, including those of the nasal cavity, lungs, kidneys, intestines, brain, heart and blood vessels (Li et al. 2020a, b). Respiratory transmission is the primary route of infection via the nose and mouth when infected individuals in close contact with uninfected people spread the viral particles that bind to the epithelial cells of the new hosts and enter their body. A few studies suggest a correlation between the extent of ACE2 expression in individuals and the clinical outcome of SARS-CoV-2 infection, especially in elderly populations and those with comorbidities (Li et al. 2020a, b; Wang et al. 2020).
Figure 1.
The SARS-CoV-2 genome is ~30Kb and consists of genes encoding structural and non-structural proteins. The structural proteins are nucleocapsid (N), spike (S), membrane (M), and envelope (E) proteins. Each box indicates a gene. The numbers on the axis indicate genome coordinates.
So far in the pandemic, it has been the elderly, those with comorbidities (including hypertension, diabetes, asthma and chronic lung disorders), and immune-compromised systems that have been the most susceptible to the adverse effects of COVID-19 infection (Mueller et al. 2020; Moderbacher et al. 2020). The demographics of the most affected populations may however change due to adaptations of the viral genes, or depend on other host and environmental factors. Studies correlating the incidence and severity of COVID-19 with the host genetic make-up among Indian populations are still underway. Acquiring immunity to the virus via unchecked exposure can lead to unacceptable levels of mortality in susceptible populations, as seen repeatedly in countries across the globe (Azkur et al. 2020; Catanzaro et al. 2020). Over 86 million people have been afflicted with the disease, with nearly 1.9 million deaths reported so far throughout the world. Early diagnosis and treatment of COVID-19 are crucial, and the recent success in the development of vaccines is promising (Anderson et al. 2020; Polack et al. 2020; Voysey et al. 2020), but prevention measures will prove to be the most effective in mitigating the worldwide spread of the disease and decrease the scope for harmful mutants to evolve. A key aspect of prevention would include a focused approach towards surveillance and monitoring of the mutations in the virus, by constant and widespread analysis of its genome sequence.
Genome sequencing-based phylogenetic analysis
The first genome sequences of the novel betacoronavirus became available on the global public repository, Global Initiative on Sharing All Influenza Data (Elbe and Buckland-Merrett 2017) or GISAID (https://www.gisaid.org/) around 10 January 2020, named as the original virus from Wuhan (WIV 04-reference or hCoV-19/Wuhan/WIV04/2019). Since then the repository has amassed over 320,000 sequences from all over the globe. India was the 5th country in the world to sequence the viral genome (isolated from the first patients in Kerala) for inclusion in GISAID (ICMR 2020). Sequencing efforts across many labs in India have since led to the submission of more than 6000 SARS-CoV-2 viral genomes. Over time, viruses accumulate mutations that alter the genomic sequence, either due to random replication errors or via a defense mechanism of the host called RNA editing (Van Dorp et al. 2020a, b). The mutations are called synonymous when there is no change to the amino acid encoded and non-synonymous when the protein acquires a change due to the mutation. SARS-CoV-2 has acquired new mutations at the rate of ~2 changes per month so far. Thus, the viral sequences seen today differ from the Wuhan variant at around 20 points in their genomes.
Phylogenetic analysis of the GISAID sequences highlights multiple clusters of related genomes, called clades, grouped based on common mutations. The nomenclature of SARS-CoV-2 lineages is explained in table 1. As shown in figure 2, Clade O was the ancestral type which originated from Wuhan (Wu et al. 2020; Zhou et al. 2020). In January and early February, this diversified into Clades 19A and 19B (also known as L and S) (Tang et al. 2020). The L-type was more prevalent (~70%) in the early stages of the outbreak in Wuhan, even though the S-type was closer to the ancestral type, and then its frequency decreased over the next few months. A new clade, A2a or Clade G, the ancestor of clades 20A-C, was then identified in February, characterized by a specific non-synonymous mutation (D614G) in the Spike protein or gene S.
Table 1.
Various nomenclatures of SARS-CoV-2 clades*
| Nomenclature | Salient feature |
|---|---|
| GISAID Clade nomenclature (Global Initiative on Sharing All Influenza Data (GISAID) 2020) | The original strain sequenced from Wuhan is called the O strain. All subsequent clades are named based on specific amino acid mutations. For example, the earliest diverging clades, called L and S, were based on the amino acid observed at the 84th position in the gene ORF8. The clade with D614G mutation in Spike is called the G clade. Subclades of a clade are also named based on other signature amino acid mutations, such as GH, GR. |
| New Nextstrain Nomenclature (Year-Letter) (Hodcroft et al. 2020a, b; Hadfield et al. 2018) | Each clade name consists of the year when the clade emerged and a capital letter starting with A for each year. Clades are defined by signature mutations. New major clades are named once the frequency of a clade exceeds 20% in a representative global sample and that clade differs in at least two positions from its parent clade. The system is currently using the clade names 19A, 19B, 20A, 20B, and 20C. Clades/strains of immediate importance are named after the parental cluster, for example 20B/501Y.V1, or 20A.EU1 |
| Nomenclature by Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN) tool (Rambaut et al. 2020) | Proposed specifically in the context of rapid genomic data available for SARS-CoV-2, this hierarchical, dynamic nomenclature describes a lineage as a cluster of sequences seen in a geographically distinct region with evidence of ongoing transmission in that region. All lineages start with either ‘A’ or ‘B’, tracing back to the original two strains of SARS-CoV-2 sequenced from Wuhan. Further numbers are appended to the letters based on multiple sources of information, including phylogenetic information as well as a variety of metadata associated with that sequence. The finer scale of this nomenclature system can help tease apart outbreak investigations and as rates of international travel increases will facilitate tracking viral imports across the globe. As an example, the more transmissible strain first identified in the UK is given a specific lineage name of B.1.1.7, whereas that first identified in South Africa is called B.1.351. |
*Given the unprecedented scale of genomic data generated from the viral strains, there have been several nomenclature systems proposed to effectively identify and track them. All of them base their roots on the first two strains of SARS-CoV-2 from Wuhan. The most common naming conventions currently used are outlined.
Figure 2.
A simplified phylogenetic tree showing the divergence of clades from the ancestral root (Wuhan) and their corresponding clade-defining mutations. The boxes indicate clades. Boxes of the same color indicate derived clades sharing the same mutations as the parent. The clade nomenclature (19 and 20) as specified by Nextstrain is given within the box, while the GISAID clade is mentioned in parentheses. The text on the right shows the clade defining mutations of the respective clade and is in the order of protein, amino acid and position followed by the amino acid change. The month above the branches indicates the inferred month of emergence of specific clades. Currently, 20A, 20B and 20C are the globally dominant clades.
The D614G mutation replaced the 614th amino acid D (aspartic acid) with G (glycine) in the Receptor Binding Domain (RBD) of the Spike protein. Glycine being a less bulky amino acid than aspartic acid it is believed to contribute to a more flexible hinge region in the Spike that enables more efficient cutting for receptor binding (Korber et al. 2020; Turoňová et al. 2020). This offered the virus a selective advantage in infection and transmission, making it predominant all over the world (Zhang et al. 2020). At present, almost all new infections of COVID-19, in India as well as globally, are by viruses containing this mutation. Monitoring such mutations is critical in the context of vaccines and therapeutics developed globally. Importantly, the D614G mutation falls outside the region that is responsible for raising neutralizing antibodies and has not been a cause for concern in the context of vaccine efficacy and therapeutics (Li et al. 2020a, b).
Over the last 10 months, we have analyzed over five thousand SARS-CoV-2 genomes isolated from Indian patients of COVID-19 to build a phylogeny with 6888 mutation events (Singh et al. 2020). In the following sections, we review the rise and spread of different variants of the SARS-CoV-2 virus across India.
Spread of SARS-CoV-2 in India
Summary of early spread: Rise and decline of an India-specific variant
In late-March, a unique cluster of sequences was identified in India which could not be classified into any previously annotated global clades. This cluster, named the clade I/A3i, is characterized by a set of four mutations as described in our earlier work (Banu et al. 2020). Clade I/A3i potentially arose from a single outbreak and rapidly spread across the country and has a lower mutation rate compared to other clades. The evolution of the I/A3i clade is largely determined by changes in the Nucleocapsid (N) and Membrane (M) genes, in contrast with the predominant A2a clade, which is characterized by changes in the Spike (S) gene. When first characterized in late May, 42% of all genomes sequenced in India belonged to this clade. Members of the Clade I/A3i formed the predominant class of isolates from the states of Delhi, Telangana, Maharashtra, Karnataka, and Tamil Nadu and were the second largest in membership in Haryana, Madhya Pradesh, West Bengal, Odisha, Uttar Pradesh, and Bihar. Globally, around 300 genomes sampled from Singapore, Malaysia, Australia, United States, Canada, Taiwan, Japan, Thailand, Philippines, Oman, Guam, and Saudi Arabia belonged to this clade and a few of them had a sampling date earlier than the earliest sample of this cluster from India. Though originally a dominant clade, its representation has become non-existent in recent samples as predicted by its mutation profile (Banu et al. 2020). Currently, considering all the genomic data available from India, 547 genomes (10%) from 17 of the 20 states from which the genomes originated fall under the clade I/A3i.
Current SARS-CoV-2 variants in India
The A2a takeover from A3i and other minor variants in India is summarized in figure 3. The first instance of the A2a variant was identified from samples collected in early March with increasing prevalence in the following months. Currently, two subtypes of A2a are dominant across India, characterized by differing mutations in the N gene and other ORFs, in addition to the D614G spike mutation.
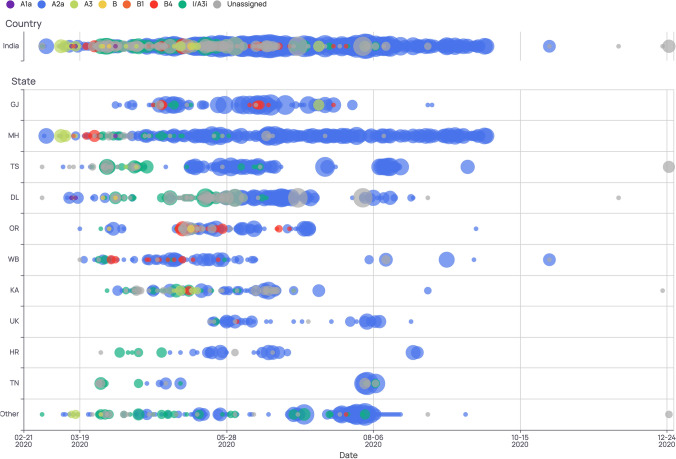
Figure 3.
Timeline of clade distribution in India (top) and across different states (state abbreviations are indicated on the left). Clades are differentiated by colors as per the legend on the top while size of the bubble indicates their prevalence. As seen in the country track on the top, A3i clade (green) was prevalent during the months of March-May and was eventually overtaken by the A2a clade (blue).
The variant landscape is mostly concordant across states in India. However, there appear to be instances of high representation of specific variants in selected states. These include the ORF3a mutation L46F from Telangana and the Spike mutation L54F seen in Gujarat (Singh et al. 2020; Hassan et al. 2020). Another Spike mutation N440K was first identified in late June in the state of Andhra Pradesh and has been present in ~6% of the samples collected from India since then (Jolly et al. 2020). The top three genes where most mutations have been identified are ORF1a (2333), ORF1b (1278) and S (714). Figure 4 shows the frequency of mutations identified in Indian samples across all the viral genes and table 2 summarizes the mutations in Indian variants identified over the last 10 months of the pandemic. The clade analysis and variant information can be explored interactively at our website https://data.ccmb.res.in/gear19/. Most variants appear overrepresented in states that currently sequence and submit the most samples to GISAID. More sequencing from underrepresented areas needs to be carried out for a clearer picture of country-wide trends of the viral spread.
Figure 4.
Plot showing the number of mutations identified across genes in the SARS-CoV-2 genome, from Indian samples. ORF1a, ORF1b and S show higher frequency of mutations compared to the rest of the genes.
Table 2.
Most prevalent SARS-CoV-2 mutations in India
| Variant | Protein_Position | Protein_Mutation | Prevalence | Percentage |
|---|---|---|---|---|
| A23403G | S:614 | D614G | 4430 | 82.29 |
| C14408T | ORF1b:314 | P314L | 4375 | 81.12 |
| G28881A | N:203 | R203K | 2096 | 38.87 |
| G28883C | N:204 | G204R | 2086 | 38.68 |
| G28882A | N:203 | R203K | 2086 | 38.68 |
| G25563T | ORF3a:57 | Q57H | 1256 | 23.29 |
| C5700A | ORF1a:1812 | A1812D | 1209 | 22.42 |
| C28854T | N:194 | S194L | 1096 | 20.32 |
| G11083T | ORF1a:3606 | L3606F | 761 | 14.11 |
| C13730T | ORF1b:88 | A88V | 655 | 12.15 |
| C28311T | N:13 | P13L | 650 | 12.05 |
| C6312A | ORF1a:2016 | T2016K | 593 | 11 |
| C8917T | ORF1a:2884 | F2884F | 447 | 8.29 |
| C6573T | ORF1a:2103 | S2103F | 301 | 5.58 |
| G9389A | ORF1a:3042 | D3042N | 300 | 5.56 |
| C25528T | ORF3a:46 | L46F | 295 | 5.47 |
| T1947C | ORF1a:561 | V561A | 234 | 4.34 |
| C9693T | ORF1a:3143 | A3143V | 215 | 3.99 |
| C3267T | ORF1a:1001 | T1001I | 192 | 3.56 |
| G26173T | ORF3a:261 | E261* | 190 | 3.52 |
| C21034T | ORF1b:2523 | L2523F | 187 | 3.47 |
| G28183T | ORF8:97 | S97I | 187 | 3.47 |
| T28277C | N:2 | S2P | 163 | 3.02 |
| C1218T | ORF1a:318 | S318L | 121 | 2.24 |
| G21724T | S:54 | L54F | 115 | 2.13 |
| G28878A | N:202 | S202N | 104 | 1.93 |
| A4372G | ORF1a:1369 | G1369G | 103 | 1.91 |
| T28144C | ORF8:84 | L84S | 103 | 1.91 |
| G29474T | N:401 | D401Y | 93 | 1.72 |
| A21551T | ORF1b:2695 | N2695L | 87 | 1.61 |
| A21550C | ORF1b:2695 | N2695L | 87 | 1.61 |
| C10815T | ORF1a:3517 | S3517F | 84 | 1.56 |
| C6310A | ORF1a:2015 | S2015R | 80 | 1.48 |
| A2292C | ORF1a:676 | Q676P | 74 | 1.37 |
| C18568T | ORF1b:1701 | L1701F | 73 | 1.35 |
| C16726T | ORF1b:1087 | H1087Y | 71 | 1.32 |
| C21575T | S:5 | L5F | 70 | 1.3 |
| G23593T | S:677 | Q677H | 70 | 1.3 |
| G11417T | ORF1a:3718 | V3718F | 67 | 1.24 |
| G1820A | ORF1a:519 | G519S | 63 | 1.17 |
| C20384T | ORF1b:2306 | A2306V | 61 | 1.13 |
| G3871T | ORF1a:1202 | K1202N | 60 | 1.11 |
| C19862T | ORF1b:2132 | A2132V | 58 | 1.08 |
| G8371T | ORF1a:2702 | Q2702H | 58 | 1.08 |
| C26447T | E:68 | S68F | 58 | 1.08 |
| T25556G | ORF3a:55 | V55G | 58 | 1.08 |
| G21974T | S:138 | D138Y | 56 | 1.04 |
| C23604A | S:681 | P681H | 53 | 0.98 |
| G28899T | N:209 | R209I | 53 | 0.98 |
| G28209C | ORF8:106 | E106Q | 52 | 0.96 |
| C26060T | ORF3a:223 | T223I | 52 | 0.96 |
| T8022G | ORF1a:2586 | V2586G | 52 | 0.96 |
| G28221T | ORF8:110 | E110* | 47 | 0.87 |
| C11195T | ORF1a:3644 | L3644F | 47 | 0.87 |
| C19154T | ORF1b:1896 | T1896I | 46 | 0.85 |
| C6027T | ORF1a:1921 | P1921L | 44 | 0.82 |
| T22882G | S:440 | N440K | 43 | 0.8 |
| C22227T | S:222 | A222V | 5 | 0.09 |
| G23012A | S:484 | E484K | 3 | 0.06 |
| A23063T | S:501 | N501Y | 2 | 0.04 |
| G22992A | S:477 | S477N | 1 | 0.02 |
Summary of the top 61 non-synonymous Indian variants of SARS-CoV-2 (arranged by prevalence) listing the genomic mutation and the corresponding amino acid change in the associated viral protein (out of 6888 total variants; https://data.ccmb.res.in/gear19/). The position of the change in the genome (column 1) and on the protein sequence (column 2) is indicated. Prevalence trends in terms of frequency and proportion in the total sequences from the Indian samples are also provided. The last few variants are relatively new and hence have a low prevalence among the samples sequenced in India so far, however, they are strong candidates for increased viral transmission and/or immune escape.
The first positive case in India was reported from Kerala in January of a patient who had traveled from Wuhan, and the state provides a unique opportunity to study viral diversity (Yadav et al. 2020). A study conducted on 200 samples identified 4 novel genetic variants and 89 variants that were exclusive to Kerala and not present in other parts of the country (Radhakrishnan et al. 2020). This work is currently being scaled up by the local state government to gain insights into the transmission and needs to be performed across all Indian states.
As of December 2020, a new country-wide consortium named INSACOG has been established to identify new and circulating variants by genome sequencing across multiple states (MoHFW 2020). One of the main goals of this consortium is to sequence 5% of all COVID-19 positive cases in the country.
New global variants and causes for concern
The mutation landscape of SARS-CoV-2 has been under constant global scrutiny to understand the effect of these changes on the infectivity and antigenicity of the virus. While most mutations are of little to no consequence, sometimes the virus acquires a mutation that gives it an advantage over other strains. The Spike protein is used by the virus to enter human cells via the ACE2 receptor. Thus, Spike mutations can potentially facilitate better affinity or binding and enable easier entry into the host cell, as seen in the case of the D614G mutation described in the preceding section. The receptor-binding domain (RBD) in the spike protein is the most variable part of the coronavirus genome (Zhou et al. 2020). Mutations can putatively also render the virus resistant to neutralization by host antibodies and thus need to be identified and monitored for the efficacy of antibody therapeutics. Figure 5 shows the position of some of the key Spike mutations that can alter its biology in terms of transmission, infectivity and enabling immune evasion.
Figure 5.
Representation of the spike gene indicating key mutations that are a cause for concern and require monitoring in India. The various sub-domains of the spike gene are shown as colored boxes and defined in the legend. The position and color of each line indicates the location of the particular mutation, defined above the gene.
Some of the spike mutations recently identified that are of concern include the N439K, N440K, Q493K and E484K, which are prone to immune escape (Andreano et al. 2020; Thomson et al. 2020; Weisblum et al. 2020). Of these, the N440K variant has been found in ~42% of the samples from Andhra Pradesh and E484K in 3 samples from Maharashtra (Jolly and Scaria 2020; Singh et al. 2020). Most of the other mutations are absent in currently sequenced samples from Indian isolates and need to be actively monitored. Table 3 highlights the key Spike mutations of global concern which are a priority for surveillance in the Indian landscape.
Table 3.
S gene mutations of concern that require monitoring via genome sequencing
| Mutation | Cause for concern |
|---|---|
| △69H-70V | Immune escape, diagnostic failure in assays targeting S gene, identified as part of lineage of UK Variant of Concern (VOC) 202012/01 (B.1.1.7 or 501Y.V1), part of Cluster 5 mink set |
| A222V | Fast growing lineage in Europe |
| N439K | Enhanced binding affinity to hACE2 receptor and can likely evade neutralizing antibodies |
| N440K | High frequency in Andhra Pradesh |
| Y453F | Enhanced binding affinity to hACE2 receptor and can likely evade neutralizing antibodies, part of Cluster 5 mink set |
| N501Y | Enhanced binding affinity to hACE2 receptor, possible role in increased transmission, identified as part of lineage of UK Variant of Concern (VOC) 202012/01, identified as part of lineage of South African 501Y.V2 (B.1.351), and as part of lineage 501Y.V2 (B.1.351) in South Africa, and 501Y.V3 (P.1) in Brazil |
| D614G | Enhanced binding affinity to hACE2 receptor, increased transmission, current predominantly prevalent strain |
| P681H | Immediately adjacent to the furin cleavage site, identified as part of lineage of UK Variant of Concern (VOC) 202012/01, identified as part of lineage in Nigeria (B.1.1.207) |
| E484K | Reduced susceptibility to neutralization by antibodies, identified as part of lineage of South African 501Y.V2 (B.1.351), identified as part of lineage in Brazil (B.1.1.28), and as part of the B.1.1.28 lineage in Brazil (501Y.V3 or P.1) |
Indian and global mutations identified in the S-gene that cause an alteration in the spike protein and may be detrimental to the human population in terms of viral transmission, infectivity and immune escape.
European lineages identified in Denmark and Spain
SARS-CoV-2 was recently introduced into minks from humans and since then has adapted to the mink host. A unique strain called Cluster 5 was identified in both hosts which encompasses three amino-acid changes (I692V, M1229I and Y453F) and two deletions (del 69–70) in the spike protein (Oude Munnink et al. 2020; Van Dorp et al. 2020a, b). This variant was last seen in September across genomes. Recent surveillance studies by European consortia have identified several other strains of SARS-CoV-2 that show increased transmission. Sequencing and analysis efforts by Spain have identified the EU1 and EU2 strains, which harbor two mutations in their Spike proteins (A222V and S477N respectively) (Hodcroft et al. 2020a, b). These strains were associated with the surge of cases in various European countries during the summer. In Indian isolates, five samples with A222V and one sample with S477N mutations have been identified till date. Additional studies are required to understand their potential implications in terms of diagnostics, therapeutics and vaccines under development.
Lineage B.1.351 identified in S. Africa
The last month of the year 2020 began with worrying news regarding new variants of SARS-CoV-2 that show increased transmissibility, first identified in the UK and S. Africa. Recent reports from South Africa mention concerns regarding lineage B.1.351 which has a mutation in the RBD of the Spike protein (N501Y) which may be associated with faster transmission and possible adverse illness in young and healthy individuals (Tegally et al. 2020). Characterized by another non-synonymous Spike mutation, the variant replaces asparagine (N) with tyrosine (Y) in the RBD and increases viral affinity to the ACE2 receptor on the host cells. This might explain the dominant spread of 501Y.V2 in the region over the last couple of months, though further studies are needed to understand its epidemiology.
Lineage B.1.1.7 identified in the UK
Even more concerning has been the latest report by the COVID-19 Genomics UK Consortium (COG-UK), detailing the variant VUI-202012/01 (lineage B.1.1.7) that is associated with fast-growing outbreaks across London, Kent, and the other UK counties (COG-UK 2020; Volz et al. 2020). A four-fold increase in cases in a span of just 10 weeks prompted immediate monitoring and investigation of the new variant, followed by global measures to limit its spread. Lineage B.1.1.7 is believed to be 70% more transmissible than other strains and has mutated at a much faster rate than other variants (European Centre for Disease Prevention and Control 2020). Sequencing of this strain has identified a cluster of about 23 mutations (Public Health England 2020), 17 of which are non-synonymous including N501Y and P681H as well as the two deletions (69-70 del and 144 del) in Spike protein, associated with a capacity to escape previous immune responses (Kemp et al. 2020). Viral fusion with host cells is facilitated by the cleavage of S into S1 and S2 sub-regions (via host enzyme furin) and the P681H mutation adjacent to the cleavage site is thus also a cause for concern (figure 5). So far, 54 Indian isolates have P681H mutation and it is present in ~4% of isolates from Maharashtra. Over 12% of samples analyzed by the COG-UK currently belong to this lineage. Despite large-scale restrictions on global travel being implemented from the latter half of December 2020, the VUI-202012/01 variant has already spread to a few other European countries from the UK, while the South African variant 501Y.V2 has now been identified in the UK (European Centre for Disease Prevention and Control 2020). Variants with efficient transmission can thus spread very rapidly all over the globe without appropriate care and surveillance.
The VUI-202012/01 has recently also been identified in the Indian population, with instances of infected travelers from the UK testing positive for the B.1.1.7 lineage fast approaching 100 cases. So far there is no evidence of community transmission of the variant in India but this can only be confirmed once a sufficient number of positive samples have been sequenced across the country. If this strain indeed possesses a transmission advantage it is likely to overtake the D614G and drive the pandemic in 2021, unless strict measures are adopted for global containment. Understanding the genomic epidemiology of the virus in India will be crucial for anticipating variant emergence, tracing transmission networks, discerning selective pressure and evaluating disease severity of outbreaks.
Lineage P.1 identified in Brazil
A variant circulating in Manaus, Brazil was identified from international travellers in Japan in viral genomes sampled from mid-late December 2020 to early January 2021. The new lineage P.1 (descendant of B.1.1.28) has 17 amino acid mutations including those of concern such as E484K, K417T, and N501Y in spike gene. This coincided with a resurgence of infections in Manaus and a rapid increase in the number of COVID-19 hospitalizations in January 2021 despite high seroprevalence; 76% of the population had already been shown to have antibodies to the virus in October 2020 (Sabino et al. 2021). A new sublineage P.2 (that independently acquired the spike E484K mutation associated with immune evasion) has now been detected in many locations in Brazil including in Manaus. Three cases of reinfection have recently been identified in Brazil, one of which belongs to P.1 and two others belong to P.2 lineage (Resende et al. 2021; Naveca et al. 2021; Vasques Nonaka et al. 2021). Such variants containing multiple mutations that can drive higher transmission and/or immune escape are strong contenders for enabling reinfections and lowering vaccine efficacy globally.
Genomic epidemiology: implications for origin and surveillance
Zoonotic transmission and origin of the virus
Genomic studies can aid in the identification of the origin of the virus and possible sources of transmission to humans. Zoonoses are infectious diseases transmitted from animals to humans and can evolve to become efficiently transmissible human-to-human infections such as malaria, SARS, HIV, pandemic influenzas and, most recently, COVID-19. Transmission of the causative pathogens from animals to humans and livestock can be via many routes such as infected meat (wet markets, wildlife trade, contaminated feed), direct contact with the pathogen (carcasses, fresh meat), contamination of water and produce, as well as air-borne and vector-borne (mosquitoes, ticks, rodents) routes. The current pandemic caused by the SARS-CoV-2 virus is believed to have originated from a wildlife food market in China's Wuhan city towards the end of 2019 (Wu et al. 2020; Zhou et al. 2020). Current evidence points to its origin from a bat-borne virus and the global pandemic represents the first time that the virus has been transmitted into humans (Andersen et al. 2020; MacKenzie and Smith 2020; Zhou et al. 2020). Consequently, this is a novel pathogen for the human immune system and many individuals are susceptible to its devastating systemic effects. Even prior to this pandemic, some of the earlier known coronaviruses have been transmitted to humans via intermediate hosts such as civet cats (Severe Acute Respiratory Syndrome (SARS) in 2002) and dromedary camels (Middle East Respiratory Syndrome (MERS) in 2012) although their spread has not been as prevalent globally (Song et al. 2005; Hemida et al. 2014). In the last century, the world has seen at least six major outbreaks of novel coronaviruses causing a range of diseases from a mild cold to infections with high mortality. COVID-19 has been the most devastating pandemic in this century, but it was preceded by the recent outbreaks of SARS and MERS, as well as the H5N1 bird flu, Ebola, HIV, Lyme disease, Rift Valley fever, Lassa fever and Nipah virus infections.
Genome sequencing has enabled the retrospective dating of the first known cases of COVID-19 as appearing in December 2019 in Wuhan and many of the early cases in the Chinese city were epidemiologically pinned to a local wet market. There have been some reports suggesting an earlier origin of the human to human transmission in locations other than China but these need to be examined further and verified by sequencing the viral samples to establish their lineage of origin (Apolone et al. 2020; Basavaraju et al. 2020). The zoonotic origin of the SARS-CoV-2 pandemic is still unknown and the reservoir host species unproven. The viral genome sequence clusters with SARS-CoV and has been placed within the SARS- related coronaviruses (SARSr-CoVs) found in bats, in the subgenus Sarbecovirus of the genus Betacoronavirus. With 96.2% genomic sequence similarity to a horseshoe bat coronavirus (RaTG13; Zhou et al 2020), and a high degree of relatedness with other bat coronaviruses, SARS-CoV-2 is believed to have originated from a bat reservoir although information regarding the intermediate host, if any, remains sparse (Andersen et al., 2020). Typically, pathogens that cause outbreaks in humans, including the current COVID-19 pandemic, evolve in 'reservoir' hosts such as rodents, bats and small mammals (Shereen et al. 2020). Over time they become endemic within their populations, causing relatively no harm to these animals. A few key mutations then allow them to infect humans directly, or via 'intermediate hosts' that are closely related mammals, such as livestock. This pandemic is an urgent example of the increasing danger from zoonotic transmissions as humans come into greater conflict with their environment.
The inter-species transmission from humans to minks and back described in the previous section is another example of the creation of host reservoirs that offer conditions for viral evolution and adaptation. Tracking mutations across these genomes can aid in understanding viral diversity and transmission after such events of zoonotic crossover. In the light of SARS-CoV-2 infection eventually becoming endemic to the human population, it will be important to have mechanisms in place to monitor disease crossover to non-human species in contact with human habitation, including pets, livestock and wildlife.
Vaccine efficacy and immune evasion mutations
SARS-CoV-2 can theoretically evolve to evade immunity when brought under the stress of therapeutic or preventive interventions. A prevalent mutation of the Spike receptor binding motif (RBM) - N439K - has enhanced binding affinity to the hACE2 receptor, and can likely evade neutralizing antibodies since it is a part of the epitope recognized by these antibodies (Thomson et al. 2020). Another mutation in the RBD region, E484K has been described in lineages in South Africa (501Y.V2 (B.1.351)) and Brazil (descended from the B.1.1.28 lineage) and is shown to reduce the neutralization potency of some human sera by >10-fold. It is of concern that this mutation can impact binding and can escape even a potent polyclonal serum targeting multiple neutralizing epitopes (Greaney et al. 2020; Andreano et al. 2020; Weisblum et al. 2020).
The currently approved vaccines raise a host immune response against multiple epitopes of the viral proteins, decreasing the chances of a few mutations facilitating efficient vaccine escape and there is hope that immune evasion will therefore be controlled before such variants spiral out of control. Currently, none of the variants of SARS-CoV-2 appear to have higher virulence or contribute to greater disease severity.
However, such mutations that maintain virulence and viral fitness need to be identified and monitored to inform the future of Covid-19 vaccines and therapeutics, so that combinations of antibodies based on distinct epitopes can be designed for laboratory analysis of escape prevalence (Weisblum et al. 2020). A recent study has characterized the novel N501Y and other Spike mutations for the potential of infection as well as vaccine-based immune evasion (Shang and Axelsen 2020). Even as the vaccination process has now been initiated globally, the current vaccines are also being evaluated for their potential against the new viral mutations as they arise.
Surveillance and detection
An earlier study based on in silico analysis of 2086 whole-genome sequences from India documented extensive deletion of amino acid residues in the C-terminal region of the envelope glycoprotein in some SARS-CoV-2 genomes (Kumar et al. 2021). These amino acid deletions map to the C-terminal region of E protein which is just beyond the reverse primer binding site used in the detection of positive cases; thus, E gene-based RT-qPCR could still detect these isolates. However, a handful of genomes from the State of Odisha had deletion even in the primer binding site. This opens yet another front for genomic sequencing and surveillance to ensure accurate testing in the months to come.
The variant VUI-202012/01 in the UK includes a deletion in the Spike gene (69–70del) that does not amplify in RT-PCR tests using the S-gene primers while the other two primer pairs can be used to detect the viral presence. This can be exploited for a first pass identification of an outbreak involving this variant using the routine RT-PCR based testing methods and further confirmed by genome sequencing. A typical whole-genome sequencing strategy involves tiled primers-based amplification of the entire viral genome, as described by the ARTIC network (DNA pipelines R&D; Farr et al. 2020). The amplified product is then sequenced on high throughput platforms such as Illumina (short reads) or Oxford Nanopore (long reads). The entire procedure starting from viral RNA to sequencing takes 3–4 days at an average cost of INR 7000 and is invaluable for (i) discovering and tracking new mutations that appear from local transmissions and (ii) monitoring the import of harmful variants from elsewhere.
Conclusion
As the COVID-19 pandemic enters its second year, it is crucial to keep a lookout for new and emergent strains and localized disease outbreaks. The evolution of SARS-CoV-2 can render it more infectious via adaptive mutations that increase affinity or enhance binding to host cells, while escape mutations that can help it evade the immune response have serious implications for vaccines and therapeutics and can adversely impact the severity and mortality of the disease. As multiple vaccines are rolled out in the year ahead, the virus will be subjected to new selection pressures and evolution modes. India has so far not been sequencing SARS-CoV-2 isolates to full capacity, having deposited only about 6,400 genomes of the over 10.4 million recorded cases (0.06%). Exploiting advances in genomic epidemiology by monitoring and increasing sequencing efforts following local spikes will go a long way in staying on top of mutations of concern while their biology and effects are studied in greater detail.
Studying the virus under a genomic lens has played a pivotal role in tackling key challenges in pandemic management so far. Other issues beyond the scope of this article include the role of mutations in reinfections and disease severity. The extent to which genomic surveillance can help answer these questions and control outbreaks is only limited by the availability of data and will be crucial to controlling the pandemic in the future.
Acknowledgements
We thank the COVID-19 volunteers and coronavirus genome sequencing team at CSIR-Centre for Cellular and Molecular Biology. Dr Karthikeyan Vasudevan of CSIR-CCMB is gratefully acknowledged for inputs on zoonotic transmission. This work was supported by the Council of Scientific and Industrial Research, India.
Footnotes
This article is part of the Topical Collection: COVID-19: Disease Biology & Intervention.
Corresponding editor: BJ Rao
Contributor Information
Divya Tej Sowpati, Email: tej@ccmb.res.in.
Rakesh K. Mishra, Email: mishra@ccmb.res.in
References
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EJ, Rouphael NG, Widge AT, Jackson LA, Roberts PC, Makhene M, Chappell JD, Denison MR, et al. Safety and immunogenicity of SARS-CoV-2 mRNA-1273 vaccine in older adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E, Piccini G, Licastro D, Casalino L, Johnson NV, Paciello I, Dal Monego S, Pantano E et al. 2020 SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv10.1101/2020.12.28.424451 [DOI] [PMC free article] [PubMed]
- Apolone G, Montomoli E, Manenti A, Boeri M, Sabia F, Hyseni I, Mazzini L, Martinuzzi D, et al. Unexpected detection of SARS-CoV-2 antibodies in the pre-pandemic period in Italy. Tumori J. 2020 doi: 10.1177/0300891620974755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azkur AK, Akdis M, Azkur D, Sokolowska M, van de Veen W, Brüggen MC, O’Mahony L, Gao Y, Nadeau K, Akdis CA. Immune response to SARS-CoV-2 and mechanisms of immunopathological changes in COVID-19. Allergy Eur. J. Allergy Clin. Immunol. 2020;75:1564–1581. doi: 10.1111/all.14364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banu S, Jolly B, Mukherjee P, Singh P, Khan S, Zaveri L, Shambhavi S, Gaur N et al. 2020 A distinct phylogenetic cluster of indian severe acute respiratory syndrome Coronavirus 2 isolates. Open Forum Infect. Dis.7 ofaa434 [DOI] [PMC free article] [PubMed]
- Basavaraju SV, Patton ME, Grimm K, Rasheed MA, Lester S, Mills L, Stumpf M, Freeman B, et al. Serologic testing of US blood donations to identify SARS-CoV-2-reactive antibodies. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzaro M, Fagiani F, Racchi M, Corsini E, Govoni S, Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:1–10. doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COG-UK 2020 COG-UK update on SARS-CoV-2 Spike mutations of special interest-Report 1. https://www.cogconsortium.uk/wp-content/uploads/2020/12/Report-1_COG-UK_20-December-2020_SARS-CoV-2-Mutations_final_updated2.pdf
- DNA Pipelines R&D, Farr B, Rajan D, et al. 2020. COVID-19 ARTIC v3 Illumina library construction and sequencing protocol. protocols.io. 10.17504/protocols.io.bibtkann
- Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob. Challenges. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control 2020 Rapid increase of a SARS-CoV-2 variant with multiple spike protein mutations observed in the United Kingdom. https://www.ecdc.europa.eu/sites/default/files/documents/SARS-CoV-2-variant-multiple-spike-protein-mutations-United-Kingdom.pdf
- European Centre for Disease Prevention and Control 2020 Risk related to spread of new SARS-CoV-2 variants of concern in the EU/EEA2 https://www.ecdc.europa.eu/en/publications-data/threat-assessment-brief-rapid-increase-sars-cov-2-variant-united-kingdom
- Global Initiative on Sharing All Influenza Data (GISAID) 2020 Clade and lineage nomenclature aids in genomic epidemiology studies of active hCoV-19 viruses. https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/
- Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY and Bloom JD 2020 Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv 10.1101/2020.12.31.425021 [DOI] [PMC free article] [PubMed]
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan SS, Choudhury PP, Roy B, Jana SS. Missense mutations in SARS-CoV2 genomes from Indian patients. Genomics. 2020;112:4622–4627. doi: 10.1016/j.ygeno.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida MG, Chu DK, Poon LL, Perera RA, Alhammadi MA, Ng HY, Siu LY, Guan Y, Alnaeem A, Peiris M, et al. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg. Infect. Dis. 2014;20:1231. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodcroft EB, Hadfield J, Neher RA and Bedford T 2020 Year-letter genetic clade naming for SARS-CoV-2 on Nextstrain.org. https://nextstrain.org/blog/2020–06–02-SARSCoV2-clade-naming
- Hodcroft EB, Zuber M, Nadeau S, Crawford KHD, Bloom JD, Veesler D, Vaughan TG, Comas I et al. 2020 Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020 . medRxiv10.1101/2020.10.25.20219063
- ICMR 2020 India is the 5th country globally to isolate the COVID-19 virus strain. https://www.icmr.gov.in/pdf/press_realease_files/Press_Release_ICMR_13March2020.pdf
- Jolly B, Rophina M, Shamnath A, Imran M, Bhoyar RC, Divakar MK, Rani PR, Ranjan G et al. 2020 Genetic epidemiology of variants associated with immune escape from global SARS-CoV-2 genomes. bioRxiv10.1101/2020.12.24.424332
- Jolly B and Scaria V 2020 Phylovis - Genomic epidemiology of novel coronavirus in India. http://clingen.igib.res.in/genepi/phylovis/
- Kemp SA, Collier DA, Datir R, Gayed S, Jahun A, Hosmillo M, Ferreira IA, Rees-Spear C et al. 2020 Neutralising antibodies drive Spike mediated SARS-CoV-2 evasion. medRxiv10.1101/2020.12.05.20241927
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BK, Rohit A, Prithvisagar KS, Rai P, Karunasagar I, Karunasagar I. Deletion in the C-terminal region of the envelope glycoprotein in some of the Indian SARS-CoV-2 genome. Virus Res. 2021;291:198222. doi: 10.1016/j.virusres.2020.198222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect. Dis. Poverty. 2020;9:1–7. doi: 10.1186/s40249-019-0617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Wu J, Nie J, Zhang L, Hao H, Liu S, Zhao C, Zhang Q, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie JS, Smith DW. COVID-19: a novel zoonotic disease caused by a coronavirus from China: what we know and what we don’t. Microbiol. Aust. 2020;41:45–50. doi: 10.1071/MA20013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moderbacher CR, Ramirez SI, Dan JM, Grifoni A, Hastie KM, Weiskopf D, Belanger S, Abbott RK, et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;184:996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MoHFW 2020 Genomic Surveillance for SARS-CoV-2 In India - Indian SARS-CoV-2 Genomics Consortium (INSACOG) 1–18. https://www.mohfw.gov.in/pdf/IndianSARSCoV2PDFGenomicsConsortiumGuidanceDocument.pdf
- Mueller AL, McNamara MS, Sinclair DA. Why does COVID-19 disproportionately affect older people? Aging. 2020;12:9959–9981. doi: 10.18632/aging.103344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveca F, Nascimento V, Souza V, Corado A, Nascimento F, Silva G, Costa A, Duarte D et al. 2020 Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585. Accessed 09 Feb 2021
- Oude Munnink BB, Sikkema RS, Nieuwenhuijse DF, Molenaar RJ, Munger E, Molenkamp R, Van Der Spek A, Tolsma P, et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2020 doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Public Health England 2020 Investigation of novel SARS-COV-2 variant. Variant of Concern 202012/01. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/947048/Technical_Briefing_VOC_SH_NJL2_SH2.pdf
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan C, Divakar MK, Jain A, Viswanathan P, Bhoyar RC, Jolly B, Imran M, Sharma D et al. 2020 Initial insights into the genetic epidemiology of SARS-CoV-2 isolates from Kerala suggest local spread from limited introductions. bioRxiv10.1101/2020.09.09.289892 [DOI] [PMC free article] [PubMed]
- Rambaut A, Holmes EC, Hill V, OTooleMcCroneRuisdu PlessisPybus AJCLO. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende PC, Bezerra JF, de Vasconcelos RHT, Arantes I, Appolinario L, Mendonça AC, Paixao AC, Rodrigues ACD et al. 2021 Spike E484K mutation in the first SARS-CoV-2 reinfection case confirmed in Brazil, 2020. https://virological.org/t/spike-e484k-mutation-in-the-first-sars-cov-2-reinfection-case-confirmed-in-brazil-2020/584. Accessed 09 Feb 2021
- Sabino EC, Buss LF, Carvalho MP, Prete CA, Crispim MA, Fraiji NA, Pereira RH, Parag KV et al. 2021 Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. The Lancet397 452–455 10.1016/S0140-6736(21)00183-5 [DOI] [PMC free article] [PubMed]
- Shang E and Axelsen PH 2020 The potential for SARS-CoV-2 to evade both natural and vaccine-induced immunity. bioRxiv10.1101/2020.12.13.422567
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Avvaru AK, Banu S, Sharma D and Sowpati DT 2020 Genome evolution analysis resource for COVID-19 (GEAR-19). https://data.ccmb.res.in/gear19/
- Song HD, Tu CC, Zhang GW, Wang SY, Zheng K, Lei LC, Chen QX, Gao YW, et al. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA. 2005;102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Wu C, Li X, Song Y, Yao X, Wu X, Duan Y, Zhang H, et al. On the origin and continuing evolution of SARS-CoV-2. Natl. Sci. Rev. 2020;7:1012–1023. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S et al. 2020 Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. bioRxiv10.1101/2020.12.21.20248640
- Thomson EC, Rosen LE, Shepherd JG, Spreafico R, da Silva Filipe A, Wojcechowskyj JA, Davis C, Piccoli L et al. 2020 The circulating SARS-CoV-2 spike variant N439K maintains fitness while evading antibody-mediated immunity. bioRxiv10.1101/2020.11.04.355842 [DOI] [PMC free article] [PubMed]
- Turoňová B, Sikora M, Schürmann C, Hagen WJ, Welsch S, Blanc FE, von Bülow S, Gecht M, et al. In situ structural analysis of SARS-CoV-2 spike reveals flexibility mediated by three hinges. Science. 2020;370:203–208. doi: 10.1126/science.abd5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dorp L, Richard D, Tan CC, Shaw LP, Acman M, Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nature. 2020;11:5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dorp L, Tan CC, Lam SD, Richard D, Owen C, Berchtold D, Orengo C and Balloux F 2020 Recurrent mutations in SARS-CoV-2 genomes isolated from mink point to rapid host-adaptation. bioRxiv10.1101/2020.11.16.384743
- Vasques Nonaka CK, Miranda Franco M, Gräf T, Almeida Mendes AV, Santana de Aguiar R, Giovanetti M and Solano de Freitas Souza B 2021 Genomic Evidence of a Sars-Cov-2 Reinfection Case With E484K Spike Mutation in Brazil Preprints 10.20944/preprints202101.0132.v1
- Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ et al. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: Insights from linking epidemiological and genetic data. https://www.imperial.ac.uk/mrc-global-infectious-disease-analysis/covid-19/report-42-sars-cov-2-variant/
- Voysey M, Clemens SA, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2020 doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, Muecksch F, Rutkowska M, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav PD, Potdar VA, Choudhary ML, Nyayanit DA, Agrawal M, Jadhav SM, Majumdar TD, Shete-Aich A, et al. Full-genome sequences of the first two SARS-CoV-2 viruses from India. Indian J. Med. Res. 2020;151:200–209. doi: 10.4103/ijmr.IJMR_663_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Jackson CB, Mou H, Ojha A, Peng H, Quinlan BD, Rangarajan ES, Pan A, et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat. Commun. 2020 doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]