Summary
Sepsis is a leading cause of death among inpatients at hospitals. However, with early detection, death rate can drop substantially. In this study, we present the top-performing algorithm for Sepsis II prediction in the DII National Data Science Challenge using the Cerner Health Facts data involving more than 100,000 adult patients. This large sample size allowed us to dissect the predictability by age-groups, race, genders, and care settings and up to 192 hr of sepsis onset. This large data collection also allowed us to conclude that the last six biometric records on average are informative to the prediction of sepsis. We identified biomarkers that are common across the treatment time and novel biomarkers that are uniquely presented for early prediction. The algorithms showed meaningful signals days ahead of sepsis onset, supporting the potential of reducing death rate by focusing on high-risk populations identified from heterogeneous data integration.
Subject areas: Medicine, disease, complication, artificial intelligence
Graphical Abstract

Highlights
-
•
We report a top-performing algorithm for sepsis prediction
-
•
We analyzed the timeliness and robustness for predicting sepsis
Medicine; disease; complication; artificial intelligence
Introduction
Sepsis is a life-threatening condition that occurs when the body's response to an infection causes tissue damage, organ failure, or death (Singer et al., 2016). In the U.S., nearly 1.7 million people develop sepsis and 270,000 people die from it each year (CDC, 2020a). Additionally, over one-third of people who die in U.S. hospitals have sepsis (CDC, 2020b). Internationally, an estimated 30 million people develop sepsis and 6 million people die from sepsis each year (Kumar et al., 2019), and an estimated 4.2 million newborns and children are affected (Demirer et al., 2019). Due to its high mortality rate, fast deterioration, and difficulty in treatment, sepsis has become a major public health concern around the world. It accounts for more than $23.66 billion (6.2%) of total US hospital costs in 2013 (Torio and Moore, 2016).
Death rate of sepsis can be significantly reduced by early diagnosis. It is estimated that for every hour sepsis goes undiagnosed, the death rate increases between 4 and 8% (Kumar et al., 2006; Seymour et al., 2017). Though the sepsis research community has been indefatigably working on establishing models predicting sepsis onset, three important questions remain. First, how far in advance can sepsis be detected and what is the corresponding accuracy? Second, how much data do we need to gather so that we can confidently predict a person will develop sepsis? Third, how does model performance differ among genders, races, age-groups, and care settings? Furthermore, it is yet to be discovered whether novel biomarkers can be revealed by the state-of-the-art machine learning techniques and whether a more rationally weighted strategy could be used to predict sepsis.
In this study, we address the above challenges by taking advantage of a large, heterogeneous cohort of 100,000 patients collected from a various set of care settings. Among the patients, 31,377 patients were septic, and others were matched non-septic individuals based on gender, age, admission type, and length of hospital stay. Previous large-scale studies include Barton et al. that used 3,679 patients with sepsis up to 48 hr of septic onset (Barton et al., 2019), Nemati et al. that used ∼5800 patients (Nemati et al., 2018), and Mao et al. that used 4107 patients (Mao et al., 2018). There were other studies that used a less number of patients (hundreds) with sepsis (Le et al., 2019; Schamoni et al., 2019). A recent community benchmark study used 2,921 patients with sepsis for the training set (Reyna et al., 2019). The other large-scale study is Komorowski et al. (47,220 septic patients), and it focused on predicting treatment outcome instead of detecting sepsis (Komorowski et al., 2018). The other large study is Delahanty et al. (2019) which included 54,661 patients with sepsis and examined gradient boosted trees in predicting sepsis (Delahanty et al., 2019). Overall, the size of this study is satisfactory to evaluate prediction modeling of sepsis, which also allowed us to examine the performance and predictive features up to hundreds of hours ahead of sepsis onset. The validity of this study is further supported by a community-wise benchmark study, DII National Data Science Challenge, which used this data set to benchmark performance, and the results presented here are based on the top-ranking algorithm. We dissected the model performance by genders, races, age-groups, and, most importantly, care settings. Then, we determined biomarkers (including early biomarkers) for predicting sepsis onset, as well as patterns related to forecasting time and segments of the data that are needed to build the model.
Results
Non-linear model capturing changes over time to predict sepsis
The data were obtained from the DII National Data Science Challenge, which originally came from the Cerner Health Facts database (Table S1). The total patient number is 106 million, and the hospitalization population is 297 thousands. Among the hospitalized patients, we selected patients who were hospitalized for at least 8 hr before sepsis onset, and consequently, anyone that was septic prior to admission was also excluded. Seven thousand seven hundred thirty two septic examples were excluded due to insufficient observation windows. Furthermore, we dropped patients who did not have medication, labs, or events records. Otherwise, missingness can be a confounding factor in studying predictive factors. The negative examples are sub-sampled by propensity score matched to the patients with sepsis based on age, gender, race, admission type, and length of stay. Because the data we will take to predict sepsis are up to 4 hr before sepsis onset, we also truncated the negative examples at their respective matched sepsis onset time of patients with sepsis—4 hr (Figure 1). According to the matched pairs, we truncated the vital data with the same length from admission to sepsis onset (and later to 4 hr, 5 hr, 6 hr, etc ahead of sepsis onset time). Otherwise, the length of hospital stay will bias the analytic results. This step allows a reasonable number of examples for training, as well as a matched population. The selected cases include a mixture of care settings (17,870 urgent care cases, 1,089 from trauma centers, 63,769 from emergency care centers, 20,888 from elective treatment, and 2,675 unknowns). Among them, 31,377 patients were septic.
Figure 1.
Overview of model construction for selecting patients, predicting sepsis onset, and feature importance analysis
We removed the hospitalized patients without sufficient observation time and selected negative examples according to matched age, gender, hospital stay time, and admission type. Data were truncated by the matched sepsis onset time—4 hr. Input data can be categorized into two sets: demographics and time series records. All demographic features are categorical, and thus we one-hot encoded them into multiple binary features. For time series data, the last eight points of records were included in the baseline model. A variety of additional features including slope, mean, maximal and minimal values for each feature, and first time observations were included as additional features. Diverse base learners were compared for their classification performance. We further tested how much time ahead the model can predict sepsis and the number of useful records for good model performance. Lastly, feature importance was analyzed using SHAP values (the SHAP figure is only for illustration purposes, modified from [https://github.com/slundberg/shap]), and grouped by time points and feature categories. See also Tables S1 and S2.
Other than typical sepsis diagnosis criteria, the data also required the inclusion of patients (septic or non-septic) to be those that must have a minimum of eight hours of hospitalization records. The rationale behind the choice of eight hours is that we attempt to construct a model that can be used in clinical settings to predict sepsis ahead of its onset. Features that are available for constructing models can be categorized into two types: demographic data including gender, race, admission type, admission source, care settings, and age-group (Table S1, Figure 1), and longitudinal data including event time and different types of biometrics, such as albumin, alkaline phosphatase, heart rate, blood pressure (Table S1, Figure 1). Outcome is indicated by a binary label of 1 for septic or 0 for non-septic. This data set followed the Sepsis-II definition (Gül et al., 2017; Obonyo et al., 2018; Patki, 2018) (see challenge webpage [https://sbmi.uth.edu/dii-challenge/usecase.htm]:). SIRS (systemic inflammatory response syndrome) was defined as meeting the 2/4 criteria within ±3 hr from the point the suspect infection (when the microbiology order was placed). Based on discussion with the Cerner clinical and statistical consultants, concerns were raised regarding retrospectively recalculating a Sepsis-III score or other scores without bias, due to inconsistency of old data with the new definition that leaves many samples in the database unqualified. The clinical and statistical consultant recommended that using Sepsis-II corresponding to the years of collection was more appropriate. We acknowledge that this presents a limitation of this study, in which Sepsis-III was not used. As the data of this long-spanning study were collected during the period of Sepsis-II, we focus on Sepsis-II in this analysis.
We one-hot encoded categorical, demographic information into 43 discrete features (Figure 1, Tables S1 and S2). For the longitudinal data, each time point, which we will term as “record” in the rest of this paper, is represented by a vector representing measurements of diverse biometrics (Figure 1). This results in 144 features for each record. Although the data are produced at a resolution of 30 min, many of the time points have no biometric measurement available (missing data). We started with a base model taking the last eight records, which were recorded a median of 65 hr before sepsis onset—4 hr, ranging from 0 (in case of only one record) to 700 hr. This gives us a total of 1195 features (144X8+43) for each patient. The features are listed in Table S3.
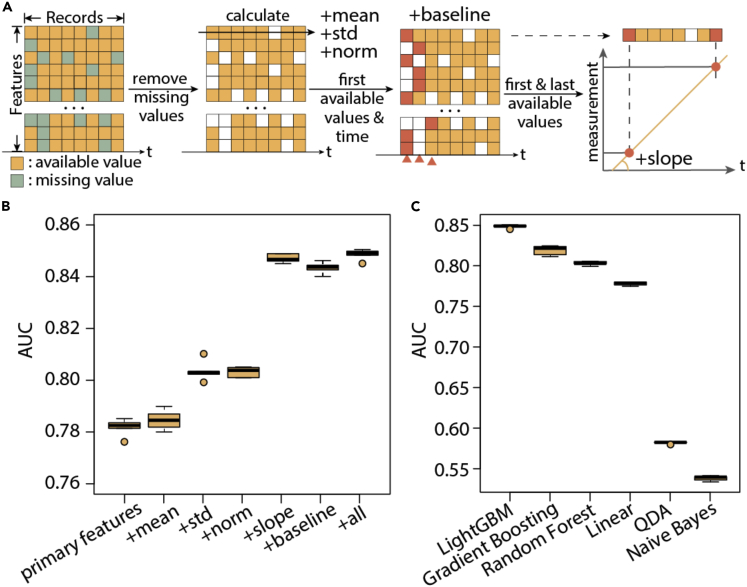
Apart from the above basic predictive features, we included the following clinically relevant features (Figures 1 and 2A, Table S2). First, we recorded the first available measurement for each feature, as well as their corresponding time of records relevant to sepsis onset. These features were included to approximate the baseline biometric values since these values could be quite different for every patient. Second, we calculated the slope of each biometric by dividing between the difference of the first and last available measurement by the time difference. These features provide information of the changes over time. Third, the mean and standard deviation of non-missing entries are recorded as additional features. This set of additional features profile the whole time series of a patient. Fourth, using the mean and standard deviation of each feature, we normalized observed features and created a new set of features, of which we took the last eight records. When the training set and test set are drawn from the same population, the fourth set of features may not add additional value. We included this set of features to account for batch effect and potentially improve the robustness of the model when delivered to new cohorts. Through five-fold cross-validation, we found the above features progressively improved the model performance from area under the receiver operating characteristic curve (AUC) = 0.78178 to AUC = 0.84854 (specificity = 0.77 at 0.8 recall and specificity = 0.827 at 0.7 recall, Figure 2B, p < 1e-5). The partial AUC above 0.8 recall (pAUC80) is 0.128, compared to a random baseline of 0.02 (Figures 2 and 3); the partial AUC above 0.7 recall (pAUC70) is 0.181, compared to a random baseline of 0.045. The most useful additional features are slope and baseline observations, reflecting the importance of capturing changes of biometrics in predicting sepsis.
Figure 2.
Post-processing of features can improve model performance.
(A) Illustration of primary features and additional features: missing values are removed from calculating the mean, standard deviation, first available values, and time for each longitudinal feature.
(B) Performance improvement by adding derived features separately to the base model.
(C) Performance using different base learners on the features. In the boxplots in (B) and (C), center lines refer to median performance, bounds of box refer to the first quartile and the third quartile of the data, whiskers refer to min and max of the data (except outliers), and spheres refer to outliers which are defined as data points below Q1 - 1.5IQR or beyond Q3 + 1.5IQR.
Figure 3.
Evaluation by specificity and pAUC
Specificity and pAUC at 80% recall for different (A) base learners, (B) feature sets, (C) age-groups, (D) admission types, (E) gender, (F) race. See also Figure S8.
We further explored a range of base learners and compared their performance in learning information from the above features (Figures 2C and 3A). Overall, we found LightGBM showed the best performance (AUC = 0.84854), followed by gradient boosting trees (0.81904, p < 1e-5) and random forest (0.80312). The other base learners did not show as competitive performance as the above: linear regression (0.7779), Naive Bayes (0.53812), quadratic discriminant analysis (0.58206). LightGBM further demonstrated advantage in training speed (Figure S1). Thus, in the following analysis, we focused on the property of the LightGBM models built with all features described above. Of note, none of the linear or posterior inference models demonstrated satisfactory performance, which corroborates that the relationships between the features are largely non-linear and thus simplistic cutoffs on biometrics to predict sepsis onset are not ideal. This effect prompted us to further categorize and investigate the predictive features in later sections.
Determination of how much time ahead and the number of observations is needed for sepsis prediction in different clinical settings
One critical, unanswered question is how far in advance sepsis can be detected or whether we can prioritize at-risk populations. This data set provides a valuable resource to examine this question for a large time range. To determine this duration, we progressively cut the data toward the beginning of the records from four hours from sepsis onset to 192 hr (8 days, Figure 4A), at which we still have a reasonable number of patients with sepsis (2579 patients with sepsis and 8679 total patients, Figure S2). Certainly, for days of records, we may no longer directly predict sepsis but more focus on prioritization of at-risk population. Of note, the control examples are cut off correspondingly from the end by record hours. As expected, the performance of the model drops as we include less proximate information of the records from 0.84854 (4 hr ahead) to 0.70818 (192 hr ahead). At 12 hr ahead, the performance was 0.8078, at 24 hr ahead, the performance was 0.78836, and at 48 hr ahead, the performance was 0.77628 (Figure 4B). These performances indicate that meaningful models can be established for identifying sepsis at least 1-2 days ahead of onset.
Figure 4.
Performance comparison in different time and population settings
(A) We first tested how many hours ahead we can predict sepsis onset by progressively cutting off data according to record time.
(B) Model performance by cutting the records according to how many hours ahead of sepsis onset to be included. The performance drops as we attempt to make predictions earlier, i.e., more hours ahead of sepsis.
(C) Illustration of selecting last n records for model input.
(D) Performance improves as more records ahead of sepsis were included.
(E) Model performance by patients from different admission types.
(F) Model performance by different age-groups.
(G) Model performance by different genders.
(H) Model performance by different races. In the boxplots in (B) to (H), center lines refer to median performance, bounds of box refer to the first quartile and the third quartile of the data, whiskers refer to min and max of the data (except outliers), and spheres refer to outliers which are defined as data points below Q1 - 1.5IQR or beyond Q3 + 1.5IQR.
See also Figures S2, S3, and S8.
We next examined the minimal number of longitudinal records that are needed to predict sepsis well. The rationale behind this analysis is to infer the amount of information we lost if we limit our view to the number of records we look at, as clinicians might focus on the 1-2 most recent records. We thus started with taking one, two, three, and progressively increasing the number of biometric records we include in the model (Figure 4C). We found that until the six records, the model still makes meaningful additions in performance. We focused on examining the models without additional features so that the effect of derived features from the entire time course will not affect this evaluation. Using the last record only, the model had an average AUC of 0.73382; using the last two records, the average AUC was 0.74764 (p < 1e-5, compared to using the last one); using the last three records, the average AUC was 0.75974 (p = 1e-5 compared to using the last two); using the last four records, the average AUC was 0.76654 (p = 0.0001, compared to using the last three); using the last five records, the average AUC was 0.7707 (p = 0.0281, compared to using the last four); and using the last six records, the average AUC was 0.77964 (p < 1e-5, compared to using the last five), and using the last seven records, the average AUC was 0.77762, which is slightly worse than last six records (p = 0.1797, Figure 3D). Certainly, all these are significantly worse (p < 1e-5) than taking into account additional features (AUC: 0.84854), which capture the dynamics and overall profiles of patients.
Sepsis prediction model performance is robust across care settings, age-groups, genders, and races
Previous meta-analysis has shown that diagnostic tests for sepsis gathered from different admission types and care settings are drastically different in their accuracy, ranging 0.68–0.99 in ICUs, 0.96–0.98 in hospitals, and 0.87–0.97 in emergency departments (Fleuren et al., 2020). It is therefore natural to hypothesize that prediction models may also differ in their accuracy by the data collection source. We thus separated the test set by the data source and evaluated the performance by admission types. Records resulted from elective procedures showed the best performance of AUC = 0.8528 (specificity = 0.780 at 0.8 recall, pAUC80 = 0.132), followed by urgent care (AUC = 0.849, specificity = 0.771 at 0.8 recall, pAUC80 = 0.129) and emergency admission (AUC = 0.8473, specificity = 0.754 at 0.8 recall, pAUC80 = 0.127); records from trauma center (AUC = 0.8343, pAUC80 = 0.129) showed slightly weaker performance (Figures 3D, 4E, and S8)
We went on to evaluate the performance separated by different age-groups and overall found minimal difference. Performance is lower for the young age group, possibly due to smaller sample size, < 20 (AUC = 0.833, specificity = 0.731 at 0.8 recall, pAUC80 = 0.124), 20∼30 (AUC = 0.836, specificity = 0.749 at 0.8 recall, pAUC80 = 0.119), 30∼40 (AUC = 0.840, specificity = 0.752 at 0.8 recall, pAUC80 = 0.124) and generally performance increases as the patient population gets older, 40∼50 (AUC = 0.847, specificity = 0.780 at 0.8 recall, pAUC80 = 0.123), 50∼60 (AUC = 0.849, specificity = 0.763 at 0.8 recall, pAUC80 = 0.129), and 60∼70 (AUC = 0.844, specificity = 0.762 at 0.8 recall, pAUC80 = 0.126), 70∼80 (0.847, specificity = 0.777 at 0.8 recall, pAUC80 = 0.128), >80 (0.851, specificity = 0.776 at 0.8 recall, pAUC80 = 0.131) (Figures 4F, 3C, and S8). There was no difference between females (AUC = 0.850, specificity = 0.776 at 0.8 recall, pAUC80 = 0.128) and males (AUC = 0.846, p = 0.0506, specificity = 0.764 at 0.8 recall, pAUC80 = 0.127) (Figures 4G, 3E, and S8). The model performs slightly worse for Asian (AUC = 0.836, 2% of total population, p = 0.0263 and p = 0.1129, respectively, specificity = 0.755 at 0.8 recall, pAUC80 = 0.117) than African American (AUC = 0.852, specificity = 0.788 at 0.8 recall, pAUC80 = 0.130, 18% of total population) and Caucasians (0.844, specificity = 0.763 at 0.8 recall, pAUC80 = 0.127, 73% of total population, ), possibly related to a much smaller population size for Asian (Figures 4H, 3F, and S8).
Game theory-based feature analysis reveals important players in predicting sepsis onset
The above described model integrates discrete demographic and clinical features. This allows us to investigate the important factors predicting sepsis. A technical challenge in finding independent contribution is addressed by a recent advance in game theory application: an improved SHapley Additive exPlanation (SHAP) analysis, which substantially improved the speed of calculation and made the Shapley values feasible to obtain for large-scale feature analysis (Lundberg et al., 2018; Shapley, 1988). Mimicking the process of finding out the contribution of players in a football game, the SHAP analysis assigns the independent contribution of each of the features considering the existence of other features. This feature contribution analysis can be carried out for the prediction of an individual patient (Figure S3 for examples) or summarized for each feature. Compared to direct correlation analysis, SHAP analysis is more capable of addressing confounding features as a consequence of shared patterns with another important feature.
We implemented SHAP analysis with LightGBM and identified the most important features for predicting sepsis. The top features are the most recent records of heart rate and respiration rate, followed by temperature and end-tidal CO2, i.e., parameters related to cardiovascular and respiratory functions (Figures 5A and S4, Table S3). This is not a circulation in feature analysis, as we are restricting the data input far before the actual sepsis diagnosis; especially, as will be shown later, these factors remain to be important when we are looking ahead much time. This indicates that the model is capable of capturing and integrating early signs of sepsis before a diagnosis using the SIRS criteria can be made. Being in the coronary care unit was identified as an important risk factor among the demographic information (Figures 5B, S4, and S5, Table S3).
Figure 5.
SHAP analysis identifies important features for predicting sepsis onset
(A) Top 10 features for predicting sepsis onset.
(B) Ten most important demographic features for predicting sepsis onset.
(C) Ten most important biometrics features for predicting sepsis onset.
(D) Difference in the importance of records collected at different proximity to sepsis onset—4 hr. The smaller the record number, the closer the record is to sepsis onset.
See also Figures S4 and S5, Tables S3 and S4.
Because SHAP values are directly additive, we can combine the features in several ways to either eliminate the time factor in feature analysis. Specifically, we combined the SHAP values of longitudinal data by the type of measurement (Table S4, Figure S6). The top features are a group that related to heart rate (HR_monitored (4.4e-3), HR (2.9e-3), HR_Apical (9.4e-4), peripheral pulse (2.3e-4), pulse (1.7e-4)), a group related to respiration function (respiration rate (2.7e-3), end-tidal CO2 (2.8e-4), oxygen saturation/SPO2 (2.2e-4), partial pressure of CO2/PCO2 (1.4e-4), partial pressure of oxygen/PaO2 (1.1e-4), tidal volume (1.3e-4)), a group related to body temperature (temperature (6.6e-4), oral temperature (3.6e-4)), important biometrics (white blood cell (2.0e-4), Braden scale (1.9e-4), Glasgow coma score (1.7e-4)) (Figure 5C). Again, the majority of the parameters are closely related to respiratory function and heart rate and lastly body temperature, which by itself is a sepsis diagnosis criterion.
We next combined the absolute shap values of longitudinal data by the proximity to sepsis onset—4 hr, in order to study the relative contribution of all features related to a specific record. As expected, the most recent record is most predictive of sepsis, showing an average SHAP value of 3.6e-4, and it declines gradually as the records go further away from the onset of sepsis (Figure 5D). Records from fourth to eighth records ahead of sepsis are of almost the same SHAP values, indicating that these biometrics represent certain baseline characteristics that put the patients at risk. Yet, at least the second (1.7e-4) and third (1.1e-4) records should be seriously considered, as the SHAP values are at a similar scale for the most recent records. This result is consistent with the progressive cut experiment we carried out in the previous sections and supports that longitudinal analysis of biometric data is necessary to establish accurate models for sepsis prediction.
Identification of predictive features for sepsis onset depending on how much time ahead we make the predictions
To investigate whether the predictive features will change as we change the numbers of hours ahead of sepsis as the input data, we separately calculated the feature importance at each time point from 4 hrs ahead of sepsis onset till 192 hrs ahead of sepsis onset. At each hour cutoff, we carried out 5-fold cross-validation and correspondingly generated five sets of SHAP values for each feature. We then took the average of the five SHAP values as the importance for each feature. The SHAP values are additive, while the contributions of standard deviation, mean, slope, and others may be either negative or positive (depending on the correlation direction). Thus, we next mapped the processed features back to their original features and took the sum of the absolute value for each group to represent the original feature importance. For example, the time series, the mean, the standard deviation, the normalized values of HR will be grouped into one single feature importance for HR. This process was repeated for the models of 4 hrs ahead till 192 hrs ahead (Figure 6A). For each time point cutoff, we took the top 5 features, and this gave us a total of 7 features across all time points: HR, monitored HR (HR_Monitored), intensive care setting, respiration rate, hematocrit (HCT), Caucasian, and diastolic blood pressure (DBP).
Figure 6.
Change of feature importance with numbers of hours ahead of sepsis onset
(A) Calculation of grouped feature importance for each hour cutoff. Different colors of SHAP values represent different groups of features.
(B) Dynamics of feature importance according to the numbers of hours ahead of sepsis onset.
We found that although the majority of these top features appear to be important throughout the time course (e.g., HR, respiration rate), others showed dynamics in their importance in predicting sepsis. For example, HCT only becomes an important predictive feature when the time cutoff is above 136 hr. DBP has a stable increase in its importance as we require further time ahead to make predictions. The importance of intensive care setting quickly drops as the time cutoff approaches sepsis onset (around 48 hr) (Figure 6B). These observations support dynamic changes of predictive features for sepsis onset, from predicting which population is at risk to which individuals' biometrics reflect potential sepsis onset. As in clinical settings, it is impossible to know ahead of time how far away a patient is to sepsis onset; this result suggests the importance to consider both early features and late features mentioned above in analysis.
Discussion
In this study, we presented the top-performing algorithm in the DII National Data Science Challenge for sepsis prediction, involving the largest sepsis detection study to date (with over 30,000 patients with sepsis). This data set gave us the unique opportunity to dissect the model performance, depending on a variety of care settings, genders, races, and age-group factors. We found being Asian is the only factor that negatively affects model performance, while performances of different care settings do not differ statistically. This result is rather surprising considering that physicians' diagnosis for sepsis was reported to differ in accuracy in different care settings (Fleuren et al., 2020).
Furthermore, this large-scale data collection allowed us to estimate the number of useful records to make reliable predictions. We found the records are useful until the six records prior to sepsis onset—4 hr. This has important clinical decision implications, highlighting the importance to examine longitudinal changes of a patient in determining his/her risk in developing sepsis. Indeed, we found that the slope of biometrics was one of the top contributing processed features. Additionally, we step wisely estimated the predictability by time ahead of onset by progressively removing the records prior to onset and thus provided a reference to confidence of the predictions. We found even back to 24-48 hr ahead, the performance of the model remains strong, and up to eight days, we can still prioritize at-risk population, which supports the notion of early detection of sepsis to reduce death rate.
Respiratory function, HR, and body temperature as expected are the three major features for early sepsis prediction throughout the time course, even much earlier than a formal sepsis diagnosis. Previous studies have pointed to the importance of HR variability for prioritizing patients at high risk of sepsis (Aboab et al., 2008; de Castilho et al., 2018; Tang et al., 2009). We found that a fast HR is a strong indicator of sepsis. Similarly, fast respiration rate is indicative of sepsis onset. For very early time points, variation of HCT appeared as a strong, novel biomarker for sepsis onset prediction.
We would like to point out several limitations and future directions of the study. First, one potential limitation is the usage of Sepsis II criteria in this study. The primary reasons we chose Sepsis II instead of Sepsis III are the relative amount of data available and the clinical usage of this model. Sepsis III requires organ failure (SOFA criterion). Validation of the performance using similarly collected Sepsis III data is an important next step. Second, although the data set is large and the data source is heterogeneous in this study, it will be informative to further validate the models using independent cohorts. Third, while this study focused on cross-validation evaluation, new insights might be brought in if perspective data can be collected and used for validation.
Previous studies have reported a range of performance, which are reexamined in this study. First, there is a strong variation in reported accuracies, ranging from ∼0.65 to 0.9 (e.g., reported results in (Barton et al., 2019; Gwadry-Sridhar et al., 2011; Liu et al., 2019; Mao et al., 2018; Michelson et al., 2019; Nemati et al., 2018; Taylor et al., 2016)). It is unclear if these differences come from algorithms, populations, the time frame of the data, or care settings. Using the top-performing algorithm in a benchmark study, we dissected each of the above factors using a very large data set and now give an estimation of the expected performance by care settings, genders, age-groups, races, numbers of hours ahead, and number of records. We conclude that the time frame of the data appears as a major influential factor, while predictive features may change along the time course. This information can serve as an important reference for future studies and applications.
Limitations of studies
We would like to point out several limitations and future directions of the study. First, one potential limitation is the usage of Sepsis II criteria in this study. The primary reasons we chose Sepsis II instead of Sepsis III are the relative amount of data available and the clinical usage of this model. Sepsis III requires organ failure (SOFA criterion). Validation of the performance using similarly collected Sepsis III data is an important next step. Second, although the data set is large and the data source is heterogeneous in this study, it will be informative to further validate the models using independent cohorts. Third, while this study focused on cross-validation evaluation, new insights might be brought in if perspective data can be collected and used for validation.
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yuanfang Guan (gyuanfan@umich.edu).
Material availability
This study did not generate new unique reagents.
Data and code availability
Code is available at [https://github.com/GuanLab/sepsis].
Methods
All methods can be found in the accompanying Transparent methods supplemental file.
Acknowledgments
This work is supported by R35-GM133346, NSF #1452656. We thank Christopher Carpenter for English editing.
Author contributions
Challenge data preparation: X.J. and L.C. Implementation of challenge model: Y.G. and X.C. Post-challenge analysis and manuscript writing: Y.G. Figures: X.W. and D.Y. All authors edited and approved the submission.
Declaration of interests
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Published: February 19, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102106.
Supplemental information
References
- Aboab J., Polito A., Orlikowski D., Sharshar T., Castel M., Annane D. Hydrocortisone effects on cardiovascular variability in septic shock: a spectral analysis approach. Crit. Care Med. 2008;36:1481–1486. doi: 10.1097/CCM.0b013e31816f48f2. [DOI] [PubMed] [Google Scholar]
- Barton C., Chettipally U., Zhou Y., Jiang Z., Lynn-Palevsky A., Le S., Calvert J., Das R. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput. Biol. Med. 2019;109:79–84. doi: 10.1016/j.compbiomed.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Castilho F.M., Ribeiro A.L.P., Nobre V., Barros G., de Sousa M.R. Heart rate variability as predictor of mortality in sepsis: a systematic review. PLoS One. 2018;13:e0203487. doi: 10.1371/journal.pone.0203487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Clinical information. 2020. https://www.cdc.gov/sepsis/clinicaltools/index.html
- CDC . Centers for Disease Control and Prevention; 2020. CDC data & statistics.https://www.cdc.gov/datastatistics/index.html [Google Scholar]
- Delahanty R.J., Alvarez J., Flynn L.M., Sherwin R.L., Jones S.S. Development and evaluation of a machine learning model for the early identification of patients at risk for sepsis. Ann. Emerg. Med. 2019;73:334–344. doi: 10.1016/j.annemergmed.2018.11.036. [DOI] [PubMed] [Google Scholar]
- Demirer R.M., Murat Demirer R., Demirer O. 2019 Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT) 2019. Early prediction of sepsis from clinical data using artificial intelligence. [DOI] [Google Scholar]
- Fleuren L.M., Klausch T.L.T., Zwager C.L., Schoonmade L.J., Guo T., Roggeveen L.F., Swart E.L., Girbes A.R.J., Thoral P., Ercole A. Machine learning for the prediction of sepsis: a systematic review and meta-analysis of diagnostic test accuracy. Intensive Care Med. 2020;46:383–400. doi: 10.1007/s00134-019-05872-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gül F., Arslantaş M.K., Cinel İ., Kumar A. Changing definitions of sepsis. Turk. J. Anaesthesiol. Reanim. 2017;45:129–138. doi: 10.5152/TJAR.2017.93753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwadry-Sridhar F., Hamou A., Lewden B., Martin C., Bauer M. Predicting sepsis: a comparison of analytical approaches. Lecture notes of the institute for computer sciences, social informatics and telecommunications engineering. 2011. [DOI]
- Komorowski M., Celi L.A., Badawi O., Gordon A.C., Aldo Faisal A. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat. Med. 2018 doi: 10.1038/s41591-018-0213-5. [DOI] [PubMed] [Google Scholar]
- Kumar A., Roberts D., Wood K.E., Light B., Parrillo J.E., Sharma S., Suppes R., Feinstein D., Zanotti S., Taiberg L. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med. 2006;34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- Kumar S., Tripathy S., Jyoti A., Singh S.G. Recent advances in biosensors for diagnosis and detection of sepsis: a comprehensive review. Biosens. Bioelectron. 2019;124-125:205–215. doi: 10.1016/j.bios.2018.10.034. [DOI] [PubMed] [Google Scholar]
- Le S., Hoffman J., Barton C., Fitzgerald J.C., Allen A., Pellegrini E., Calvert J., Das R. Pediatric severe sepsis prediction using machine learning. Front. Pediatr. 2019;7:413. doi: 10.3389/fped.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Greenstein J.L., Granite S.J., Fackler J.C., Bembea M.M., Sarma S.V., Winslow R.L. Data-driven discovery of a novel sepsis pre-shock state predicts impending septic shock in the ICU. Sci. Rep. 2019;9:6145. doi: 10.1038/s41598-019-42637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg S.M., Nair B., Vavilala M.S., Horibe M., Eisses M.J., Adams T., Liston D.E., Low D.K.-W., Newman S.-F., Kim J., Lee S.-I. Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat. Biomed. Eng. 2018;2:749–760. doi: 10.1038/s41551-018-0304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Q., Jay M., Hoffman J.L., Calvert J., Barton C., Shimabukuro D., Shieh L., Chettipally U., Fletcher G., Kerem Y. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open. 2018;8:e017833. doi: 10.1136/bmjopen-2017-017833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelson A., Yu S., Gupta A., Lai A.M., Kollef M.H., Payne P.R.O. A Machine Learning Approach to Sepsis Prediction in Non-intensive Care Unit Patients. D104. CRITICAL CARE: A FINE BALANCE - SEPSIS DEFINITIONS, OUTCOMES and EPIDEMIOLOGY. 2019. [DOI]
- Nemati S., Holder A., Razmi F., Stanley M.D., Clifford G.D., Buchman T.G. An interpretable machine learning model for accurate prediction of sepsis in the ICU. Crit. Care Med. 2018;46:547–553. doi: 10.1097/CCM.0000000000002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obonyo N.G., Schlapbach L.J., Fraser J.F. Sepsis: changing definitions, unchanging treatment. Front. Pediatr. 2018;6:425. doi: 10.3389/fped.2018.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patki V. Sepsis definitions - changing perspectives. J. Pediatr. Crit. Care. 2018 doi: 10.21304/2018.0504.00407. [DOI] [Google Scholar]
- Reyna M., Shashikumar S.P., Moody B., Gu P., Sharma A., Nemati S., Clifford G. Early Prediction of Sepsis from Clinical Data: The PhysioNet/Computing in Cardiology Challenge 2019. 2019 Computing in Cardiology Conference (CinC) 2019. [DOI]
- Schamoni S., Lindner H.A., Schneider-Lindner V., Thiel M., Riezler S. Leveraging implicit expert knowledge for non-circular machine learning in sepsis prediction. Artif. Intell. Med. 2019;100:101725. doi: 10.1016/j.artmed.2019.101725. [DOI] [PubMed] [Google Scholar]
- Seymour C.W., Gesten F., Prescott H.C., Friedrich M.E., Iwashyna T.J., Phillips G.S., Lemeshow S., Osborn T., Terry K.M., Levy M.M. Time to treatment and mortality during mandated emergency care for sepsis. N. Engl. J. Med. 2017;376:2235–2244. doi: 10.1056/NEJMoa1703058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapley L.S. A value for n-person games. The Shapley Value. 1988. [DOI]
- Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.-D., Coopersmith C.M. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang C.H.H., Chan G.S.H., Middleton P.M., Savkin A.V., Lovell N.H. Spectral analysis of heart period and pulse transit time derived from electrocardiogram and photoplethysmogram in sepsis patients. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2009;2009:1781–1784. doi: 10.1109/IEMBS.2009.5334005. [DOI] [PubMed] [Google Scholar]
- Taylor R.A., Pare J.R., Venkatesh A.K., Mowafi H., Melnick E.R., Fleischman W., Hall M.K. Prediction of in-hospital mortality in emergency department patients with sepsis: a local big data-driven, machine learning approach. Acad. Emerg. Med. 2016;23:269–278. doi: 10.1111/acem.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torio C.M., Moore B.J. Healthcare Cost and Utilization Project (HCUP) Statistical Briefs. Agency for Healthcare Research and Quality (US); 2016. National Inpatient Hospital Costs: The Most Expensive Conditions by Payer, 2013: Statistical Brief #204. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code is available at [https://github.com/GuanLab/sepsis].