Summary
Topologically associating domains (TADs) are fundamental units of three-dimensional (3D) nuclear organization. The regions bordering TADs—TAD boundaries—contribute to the regulation of gene expression by restricting interactions of cis-regulatory sequences to their target genes. TAD and TAD-boundary disruption have been implicated in rare-disease pathogenesis; however, we have a limited framework for integrating TADs and their variation across cell types into the interpretation of common-trait-associated variants. Here, we investigate an attribute of 3D genome architecture—the stability of TAD boundaries across cell types—and demonstrate its relevance to understanding how genetic variation in TADs contributes to complex disease. By synthesizing TAD maps across 37 diverse cell types with 41 genome-wide association studies (GWASs), we investigate the differences in disease association and evolutionary pressure on variation in TADs versus TAD boundaries. We demonstrate that genetic variation in TAD boundaries contributes more to complex-trait heritability, especially for immunologic, hematologic, and metabolic traits. We also show that TAD boundaries are more evolutionarily constrained than TADs. Next, stratifying boundaries by their stability across cell types, we find substantial variation. Compared to boundaries unique to a specific cell type, boundaries stable across cell types are further enriched for complex-trait heritability, evolutionary constraint, CTCF binding, and housekeeping genes. Thus, considering TAD boundary stability across cell types provides valuable context for understanding the genome’s functional landscape and enabling variant interpretation that takes 3D structure into account.
Keywords: Topologically associating domain, Hi-C, heritability, evolutionary conservation, TAD stability, genome 3D structure, genome topology, complex disease
Introduction
The three-dimensional (3D) conformation of the genome facilitates the regulation of gene expression.1, 2, 3, 4 Using chromosome-conformation-capture technologies (3C, 4C, 5C, Hi-C),5, 6, 7 recent studies have demonstrated that modulation of gene expression via 3D chromatin structure is important for many physiologic and pathologic cellular functions, including cell-type identity, cellular differentiation, and risk for multiple rare diseases and cancer.8, 9, 10, 11, 12, 13, 14 Nonetheless, many fundamental questions about the functions of and evolutionary constraints on 3D genome architecture remain. For example, how does genetic variation in different 3D contexts contribute to the risk of common complex disease? Furthermore, disease-causing regulatory variation is known to be tissue-specific; however, only recently has there been characterization of 3D-structure variation across multiple cell types and individuals.13,15,16 Understanding how different attributes of 3D genome architecture influence disease risk in a cell-type-specific manner is crucial for interpreting human variation and, ultimately, moving from disease associations to an understanding of disease mechanisms.17
3D genome organization can be characterized at different scales. Globally, chromosomes exist in discrete territories in the cell nucleus.7 On a sub-chromosomal scale, chromatin physically compartmentalizes into topologically associating domains (TADs). TADs are megabase-scale genomic regions that self-interact but rarely contact regions outside the domain (Figure 1A).7,18, 19, 20 They are formed and maintained through interactions between CTCF zinc-finger transcription factors and cohesin ring-shaped complexes, among other proteins both known and unknown.7,21 TADs are identified based on regions of enriched contact density in Hi-C maps (Figure 1A). TADs modulate gene regulation by limiting interactions of cis-regulatory sequences to target genes.7 The extent to which chromatin 3D topology affects gene expression is still debated.22 In extensively rearranged Drosophila balancer chromosomes, few genes had expression changes.23 In contrast, subtle chromatin interaction changes in induced pluripotent stem cells (iPSCs) from seven related individuals were associated with proportionally large differential gene expression.24 Thus, further cell-type-specific investigation into properties of TAD organization and disruption will need to clarify which parts of the genome are sensitive to changes in 3D structure and how these changes influence gene regulation and traits.
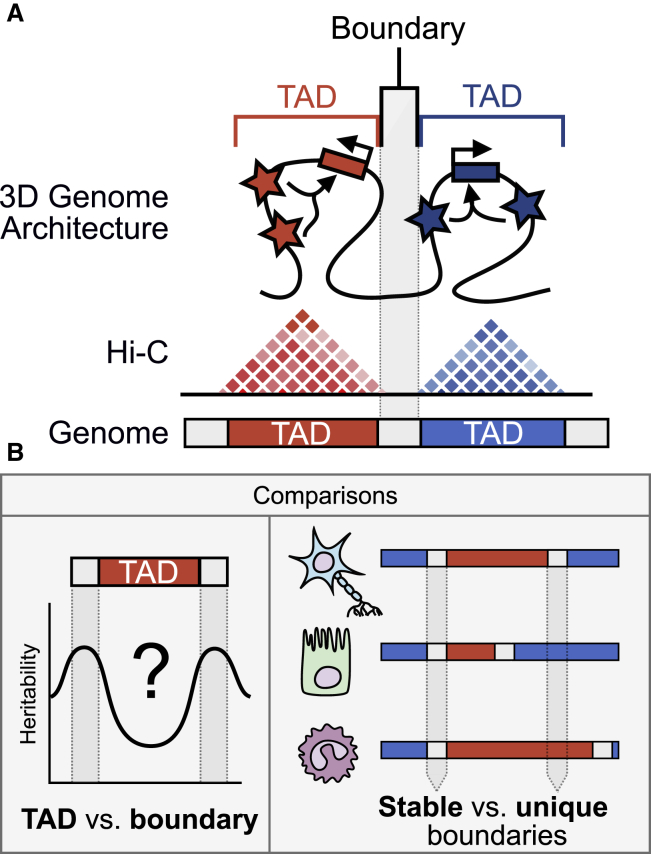
Figure 1.
Schematic depiction of our analyses of 3D chromatin TAD-boundary stability and function
(A) Chromatin is organized in 3D space into topologically associating domains (TADs), which are identified by Hi-C experiments. Regions within a TAD are much more likely to interact with one another than are regions outside of the TAD. Regions bordering TADs are TAD boundaries. Boxes with right-angled arrows represent genes, and stars represent gene regulatory elements, such as enhancers.
(B) This work addresses two main questions: (1) How are complex-trait heritability and evolutionary sequence conservation partitioned between TADs and TAD boundaries? (2) Do stable TAD boundaries (i.e., those observed across multiple tissues) have different contributions to trait heritability or sequence conservation than TAD boundaries unique to specific tissues?
At the highest level, TAD organization can be divided into two basic features: the TAD and the TAD boundary. TADs are the self-associating, loop-like domains that contain interacting cis-regulatory elements and target genes. TAD boundaries—regions in between TADs—are insulatory elements that restrict interactions of cis-regulatory sequences, such as enhancers, to target genes.7 Previous work suggests the functional importance of maintaining both the self-associating TADs and the insulatory boundaries. For example, in cross-species multiple sequence alignments, syntenic break enrichment at TAD boundaries suggests a long-term evolutionary preference for rearrangements that “shuffle” intact TADs, rather than “break” them.25,26 Additionally, 3D genome structure correlates with similar functional features, such as histone modifications and replication timing, across species.27 TADs also often contain clusters of co-regulated genes—e.g., cytochrome genes and olfactory receptors.7,19,28 Intra-TAD structural variation that deletes or duplicates enhancers has been implicated in polydactyly, B cell lymphoma, and aniridia.29 Together, these data suggest that the genome is under pressure to preserve TADs as functional units.
Other evidence suggests the greater importance of maintaining TAD boundaries. TAD boundaries are enriched for housekeeping genes and transcription start sites.7,18 Removing insulatory TAD boundaries leads to ectopic gene expression in cultured cells and in vivo. For example, TAD structure disruption at the EPHA4 locus leads to inappropriate rewiring of developmental genes implicated in limb-formation defects.7,29,30 In cancer, large structural alterations that disrupt TAD boundaries cause pathogenic gene expression in acute myeloid leukemia (AML) and medulloblastoma.31,32 Structural variation (SV) that disrupts TAD boundaries causes gain-of-function, loss-of-function, and misexpression in many forms of rare neurodevelopmental disease.29 Accordingly, TAD boundaries and CTCF sites have evidence of purifying selection on SVs.33,34 Finally, human haplotype breakpoints do not align with chromatin boundaries, which indicates that recombination might be deleterious at TAD boundaries.35 Collectively, these findings suggest that TAD boundaries are functionally important and constrained, especially on the scale of human evolution.
In addition to the need for further characterization of the constraint on and functions of TADs versus TAD boundaries, there is also a gap in our understanding of the variability in TAD organization across cell types. TADs and TAD boundaries have been characterized as largely invariant across cell types18,19,36, 37, 38 and species.7,18,26,39,40 However, previous pairwise comparisons of five 3D maps suggest that 30%–50% of TADs differ across cell types.37,41 More comprehensive recent investigations have observed large differences in the percent of boundaries not shared across cell lines (20%–80%), which contrasts with previous claims of extensive TAD conservation.42,43 Boundaries shared across two cell types have evidence of stronger SV purifying selection than boundaries unique to a cell type, suggesting that shared boundaries are more intolerant of disruption.33 Additionally, stratifying boundaries by their strength (in a single cell type) facilitated discovery that greater CTCF binding confers stronger insulation and that super-enhancers are preferentially insulated by the strongest boundaries.44 Stratifying by hierarchical properties of TADs—TADs often have sub-TADs—demonstrated that boundaries flanking higher-level structures are enriched for CTCF, active epigenetic states, and higher gene expression.45
Despite these preliminary indications that the stability of components of the 3D architecture might influence functional constraint, there has been no comprehensive analysis comparing genomic features and disease associations between 3D structural elements stable across multiple cell types and those that are unique to single cell types. Quantifying stability across cell types is important for interpreting new variation within the context of the 3D genome given our knowledge that disease-associated regulatory variation is often tissue-specific.13,15,16
To investigate differences in TAD boundaries across cell types, we quantify boundary “stability” as the number of tissues that share a TAD boundary. If a TAD boundary is found in many tissues, it is “stable,” whereas if it is found in few tissues, it is “unique” (Figure 1B). Using this characterization, we address two main questions that aim to expand our framework for cell-type-aware interpretation of genetic variation and disease associations in the context of the 3D genome (Figure 1B):
-
1.
How do TADs and TAD boundaries differ in their contribution to complex-trait heritability and their evolutionary constraint?
-
2.
Are there functional and evolutionary differences in TAD boundaries that are stable across multiple cell types versus TAD boundaries that are unique to specific tissues?
Synthesizing 3D genome maps across 37 diverse cell types with multiple functional annotations and genome-wide association studies (GWASs), we show that TAD boundaries are more enriched for heritability of common complex traits and more evolutionarily conserved than TADs. Furthermore, genetic variation in TAD boundaries stable across multiple cell types contributes more to the heritability of immunologic, hematologic, and metabolic traits than variation in TAD boundaries unique to a single cell type. Finally, these cell-type-stable TAD boundaries are also more evolutionarily constrained and enriched for functional elements. Together, our work suggests that TAD boundary stability across cell types provides valuable context for understanding the genome’s functional landscape and enabling variant interpretation that accounts for genome 3D structure
Methods
We examine heritability and functional annotation enrichment across the 3D genome landscape in two ways: (1) across the genome in windows centered and scaled around each TAD and (2) in fixed-size TAD boundaries defined with varying resolution (40–200 kb) at the ends of each TAD. We then characterize the stability of TAD boundaries across diverse cellular contexts. By splitting boundaries into quartiles of stability—from those unique to a single tissue to those shared across many tissues—we test whether there is a relationship between boundary stability and annotation enrichment. The annotations considered include contribution to complex trait heritability enrichment, base-pair-level evolutionary constraint, CTCF binding, and genic content. We demonstrate the robustness of our results by using multiple definitions of TAD boundaries, TADs called by a variety of methods, and different measurements of the annotations investigated to replicate our experiments.
Defining TADs
TAD maps for 37 different cell types were obtained from the 3D genome browser (Table S1).46 All TAD maps were systematically predicted from Hi-C data with the hidden Markov model (HMM) pipeline from Dixon et al.18,36,46 The maps were defined with respect to the same 40 kb windows, except in the case of seven cell types (GM12878, HMEC, HUVEC, IMR90, K562, KBM7, and NHEK) that were defined with respect to 25 kb windows. For details about the length and number of TADs per map, see supplemental information.
Quantifying partitioned heritability with S-LDSC
We conducted partitioned heritability by using stratified-LD Score Regression v1.0.1 (S-LDSC) to test whether an annotation of interest (e.g., TADs or TAD boundaries) is enriched for heritability of a trait.47,48 We considered GWAS summary statistics from a previously described representative set of 41 diseases and complex traits (average n = 329,378, M = 1,155,239, h2SNP = 0.19, Table S2).49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59 Previous studies using these traits had GWAS replicates (genetic correlation > 0.9) for six traits (BMI, height, high cholesterol, type 2 diabetes, smoking status, years of education). For these, we considered only the GWAS with the largest sample size. All GWASs involved subjects of European ancestry only. We used 1000 Genomes for the LD reference panel60 and HapMap Project Phase 3 (HapMap 3)61 excluding the MHC region to estimate heritability enrichment and standardized effect size. Heritability was estimated from common variants with minor-allele frequency (MAF) > 0.05, and standard errors were computed by LDSC via a block-jackknife.
Heritability enrichment
S-LDSC estimates the heritability enrichment, defined as the proportion of heritability explained by single-nucleotide polymorphisms (SNPs) in the annotation divided by the proportion of SNPs in the annotation. The enrichment of annotation c is estimated as
where h2(c) is the heritability explained by common SNPs in annotation c, h2 is the heritability explained by the common SNPs over the whole genome, |c | is the number of common SNPs that lie in the annotation, and M is the number of common SNPs considered over the genome.48,50 To investigate trends across all traits, we computed the average heritability enrichment and a confidence interval. When compared to meta-analysis using a random-effects model conducted with Rmeta,48,50,62,63 the trends are consistent (Figure S1); therefore, we report results based on averaging to simplify interpretation and reduce over-representation of higher-powered GWAS traits.
Standardized effect size
In contrast to heritability enrichment, the standardized effect size (τ∗c) quantifies effects that are unique to the focal annotation compared to a set of other annotations.50,64 The estimate of τ∗c is conditioned on 86 diverse annotations from the baseline v. 2.1 model; these include coding, UTR, promoter and intronic regions, histone marks (H3K4me1, H3K4me3, H3K9ac, and H3K27ac), DNase I hypersensitivity sites (DHSs), chromHMM and Segway predictions, super-enhancers, FANTOM5 enhancers, GERP annotations, MAF bins, LD relation, and conservation annotations.48,64,65
Heritability enrichment across the TAD landscape
We partitioned the genome with respect to TAD annotations by using two different strategies. In the first, motivated by Krefting et al.,25 we considered TADs plus 50% of their total length flanking each side and subdivided these into 20 equal-sized partitions. Hence, the center 10 bins (6–15) are inside the TAD. Bins 1–5 are upstream of the TAD, and 16–20 are downstream of the TAD. In cases where a TAD is adjacent to another TAD, the ± 50% region flanking the TAD (bins 1–5 and 16–20) often partially extends into a neighboring TAD (Figure S2A). However, the ± 50% flanking region extends into the center of a neighboring TAD less than 20% of the time (Figure S2B). We ran S-LDSR on these 20 bins across TAD maps from 37 cell types to calculate heritability enrichment over 41 traits. We investigated the heritability enrichment (or depletion) trends averaged across all traits and cell types, by cell type, and by trait. Second, we analyzed heritability in fixed-size TAD boundary windows of 40, 100, and 200 kb (see subsection on TAD stability below).
For the analyses by cell type and by trait, we clustered the heritability landscapes to determine whether related cell types or related traits had similar patterns of heritability across the 3D genome. To do so, correlation distance was used as the distance metric with average linkage clustering. When clustering traits by their heritability landscape across the 3D genome, we identified two agglomerative clusters and termed these “boundary enriched” and “boundary depleted.”
Evaluating robustness on other TAD callers
To assess the influence of technical variation of TAD calling on our findings, we assessed the heritability patterns in human embryonic stem cells across TADs called by seven diverse methods (Armatus, Arrowhead, DomainCaller, HiCseg, TADbit, TADtree, and TopDom). The TADs were called and published by Dali et al.66 with Hi-C from Dixon et al.36
Sequence-level conservation across the TAD landscape
We considered PhastCons element overlap and score to quantify evolutionary constraint across the TAD landscape. Other researchers previously determined PhastCons elements by fitting a phylo-HMM across a group of 46 vertebrate genomes to predict conserved elements.67 We downloaded these conserved element loci from the UCSC table browser.68,69 Each element has a score describing its level of conservation (a transformed log-odds score between 0 and 1000). We intersected the PhastCons elements with regions of interest (e.g., TAD boundaries) across the TAD landscape. Across each region, we quantified the number of PhastCons base pairs (regardless of score) and the average PhastCons element score.
Evolutionary constraint in TADs versus boundary windows
To specifically measure the constraint in TAD boundaries versus TADs, we investigated base-pair-level conservation at 100 kb TAD boundaries (below) and matched randomly shuffled equally sized windows in TADs. For the windows in TADs, we shuffled the 100 kb boundaries for each of the 37 cell types three times and required them to fall inside TADs (n = 111). For both the TAD boundaries and TAD set, we calculated overlap with conserved (PhastCons) elements. To investigate whether conserved element overlap is influenced by the density of CTCF binding and exons, we repeated this analysis after subtracting bases (from both the boundaries and TAD windows) overlapping CTCF ChIP-seq peaks or exons.
Quantifying boundary overlap and stability
For each cell type, we defined a set of boundaries with regard to the same windows across the genome.
100 kb boundaries
We defined 100 kb boundaries (results shown in main text) as regions 100 kb upstream of the TAD start and 100 kb downstream of the TAD end. For example, if a TAD was at chr1: 2,000,000–3,000,000, we would define its TAD boundaries to be at chr1:1,900,000–2,000,000 (boundary around the start) and chr1: 3,000,000–3,100,000 (boundary around the end). To quantify stability, we examined each 100 kb window across the genome. We removed boundaries that had any overlap with genomic gaps (centromeric/telomeric repeats from UCSC table browser).68,69 If there was a TAD boundary in the window for any of the cell types, we counted how many cell types (out of 37) shared the boundary. If only one cell type had a boundary at that location, it was considered a “unique” boundary, whereas if it was observed in many cell types, it was considered “stable.” These boundaries were divided into quartiles of cell-type-stability.
40 kb and 200 kb bookend boundaries
To test whether our results were robust to different resolutions of boundary definitions, we defined 40 kb and 200 kb bookend boundaries (see results in Supplemental Information). 40 kb boundaries are 40 kb windows surrounding (±20 kb) TAD start and stop sites. For example, if a TAD was located at chr1: 2,000,000–3,000,000, we would define its TAD boundaries to be at chr1: 1,980,000–2,020,000 and chr1: 2,980,000-3,020,000. 200 kb bookend boundaries are 200 kb upstream of the TAD start and 200 kb downstream of the TAD end. For example, if a TAD was at chr1: 2,000,000–3,000,000, we would define its TAD boundaries to be at chr1: 1,800,000–2,000,000 and chr1: 3,000,000–3,200,000. We removed boundaries that had any overlap with genomic gaps.68,69 Both sets of boundaries were divided into quartiles of cell-type-stability.
Boundaries distant from genomic gap or blacklist regions
To investigate whether boundaries near genome assembly gaps or repetitive sequences affect the relationship between annotation enrichment and stability quartile, we defined a very conservative set of 100 kb TAD boundaries by excluding those within 5 Mb of a genomic gap (UCSC table browser68,69) or blacklist region (Amemiya et al.70).
Germ-layer-informed boundary-stability measure
Of the 37 cell types considered, some are more closely related than others, therefore we grouped 34 of them by germ-layer origin (endoderm [n = 12], mesoderm [n = 13], ectoderm [n = 9]; Table S1). Germ layers for each of the cell types were defined via ENCODE documentation of common cell types.71,72 Embryonic stem cell, mesendoderm, and trophoblast were omitted because they have no single germ-layer classification. We defined a measurement of stability on the basis of whether each 100 kb boundary (above) was found in cells from one, two, or all three germ layers.
Quantifying TAD boundary similarity across cell types
To quantify TAD boundary similarity between two cell types, we calculate the Jaccard similarity coefficient by counting the number of shared boundaries (intersection) and dividing by the total boundaries over both tissues (union). For the TAD boundary similarity heatmaps, we clustered the cell types by using complete linkage (i.e., farthest neighbor) with the Jaccard distance (1-stability).
Heritability and annotation enrichment by TAD boundary stability
Complex-trait heritability
S-LDSC was conducted on each quartile of stability for all 41 traits. Partitions for each quartile include TAD boundaries of that stability (see above). We computed a linear regression on log-scaled enrichment values by regressing log10(heritability enrichment) on quartile of stability. by regressing log10(heritability enrichment) on quartile of stability.
Evolutionary constraint
Evolutionary constraint was quantified by PhastCons67 as described above. The PhastCons elements were intersected with the TAD boundaries, partitioned by stability. The two overlap quantifications are the number of PhastCons base pairs per boundary regardless of score (base pairs per boundary) and the average PhastCons element score per boundary (average score of elements in the boundary).
CTCF enrichment
CTCF binding sites were determined through ChIP-seq analyses from ENCODE.71,72 We downloaded all CTCF ChIP-seq data with the following criteria: experiment, released, ChIP-seq, human (hg19), all tissues, adult, BED NarrowPeak file format. We excluded any experiments with biosample treatments. Across all files, the CTCF peaks were concatenated, sorted, and merged into a single file; thus, overlapping peaks were merged into a single larger peak. We quantified the number of CTCF ChIP-seq peaks per TAD boundary (peaks per boundary) and the number of CTCF peak base pairs overlapping each boundary (base pairs per boundary).
Genes and protein-coding genes
RefSeq genes were downloaded from the UCSC table browser68,69,73 and filtered to include coordinates of only one transcript per gene (the longest) and only autosomal and sex chromosome genes. From the simplified list of RefSeq genes, a subset of protein-coding genes was also created (these were identified on the basis of RefSeq accession numbers starting with NM). The simplified RefSeq gene list contains 27,090 genes. The simplified protein-coding RefSeq gene list contains 19,225 genes. We quantified the number of genes or protein-coding genes per TAD boundary stratified by boundary stability.
Housekeeping genes
Housekeeping genes (N = 3804) are from Eisenberg & Levanon (2013).74 We retrieved the coordinates by intersecting with the RefSeq genes (above), resulting in coordinates for 3681 genes (coordinates for a small number of genes were not found in the RefSeq list).68,69,73 We quantified the number of housekeeping genes or protein-coding genes per TAD boundary stratified by boundary stability.
Defining GWAS phenotypic classes
To determine whether similar traits had similar heritability patterns across the 3D genome, we defined eight different phenotypic classes (Table S2): cardiopulmonary (n = 4), dermatologic (n = 7), hematologic (n = 5), immunologic (n = 4), metabolic (n = 7), neuropsychiatric (n = 8), reproductive (n = 4), and skeletal (n = 2). Our clusters originated from domains in the GWAS Atlas;75 however, the categories were modified to place more emphasis on disease pathophysiology instead of organ system (e.g., Crohn disease and Rheumatoid Arthritis were moved from the gastrointestinal and connective-tissue categories, respectively, to an immunologic category). Similar categories were also combined (e.g., metabolic and endocrine, cardiovascular and respiratory).
Data analysis and figure generation
All analyses were conducted with the hg19 genome build. Intersections of genomic regions were computed with the pybedtools wrapper for BedTools.76,77 Data and statistical analyses were conducted in Python 3.5.4 (Anaconda distribution) and R 3.6.1. Figure generation was aided by Matplotlib, Seaborn, and Inkscape.78, 79, 80 This work was conducted in part with the resources of the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University, Nashville, TN.
Results
Estimating complex-trait heritability across the 3D genome landscape
Disruption of 3D genome architecture plays a role in rare disease and cancer; however, the contribution of common variation in different 3D contexts to common phenotypes is unknown. To investigate complex-trait heritability patterns across the 3D genome landscape, we use 37 TAD maps from the 3D Genome Browser (Table S1).46 The cellular contexts include primary tissues, stem cells, and cancer cell lines;36, 37, 38,71,72,81,82 for simplicity, we will refer to these as “cell types.” All TAD maps were systematically predicted from Hi-C data with the HMM pipeline from Dixon et al.18 at either 40 kb or 25 kb resolution (Supplemental Information).46
We estimated common-trait heritability enrichment among common variants within these 3D genome annotations by using stratified-LD score regression (S-LDSC).47,48 S-LDSC is a method of partitioning heritability across the genome by using GWAS summary statistics and LD patterns to test whether variants in an annotation of interest (e.g., TADs or TAD boundaries) are enriched for heritability of a trait in comparison to the rest of the genome. We considered GWAS summary statistics from a previously described representative set of 41 diseases and complex traits.49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59
To investigate patterns of heritability across the 3D genome landscape, we used two strategies for defining genomic partitions. In the first, we analyzed TADs plus 50% of their length on each side. Motivated by the approach to partitioning TADs from Krefting et al.,25 we subdivided these regions into 20 equally sized partitions. Bins 1–5 and 16–20 “bookend” the TAD, whereas the center bins 6–15 are inside the TAD (see Methods). In addition to characterizing heritability patterns in bins across the TAD landscape, we also explicitly defined TAD boundary windows as fixed-size (40 kb, 100 kb, or 200 kb) regions bookending TADs. We conducted S-LDSC across the 37 cell types for the 41 traits to estimate the enrichment (or depletion) of heritability for each trait across the 20 partitions over the TAD landscape and the 100 kb TAD boundaries.
TAD boundaries are enriched for complex-trait heritability and evolutionary sequence conservation
Regions flanking TADs are enriched for complex-trait heritability; whereas partitions in TADs are marginally depleted for heritability overall (1.07× enrichment in flanking regions versus 0.99× enrichment in TADs, p = 1 × 10−193) (Figure 2A). We also observed enrichment in regions flanking TADs when when we used the 100 kb TAD boundary definition (1.07× background, p = 0.001, Figure S3). The results are consistent whether averaged across traits or meta-analyzed with a random-effects model48,50,62 (r2 = 0.85, p = 7 × 10−9, Figure S1); therefore, further analyses of heritability across traits will use averaging for simplicity and interpretability. There is also a spike of heritability enrichment in the center of TADs; we explore this further in a subsequent section.
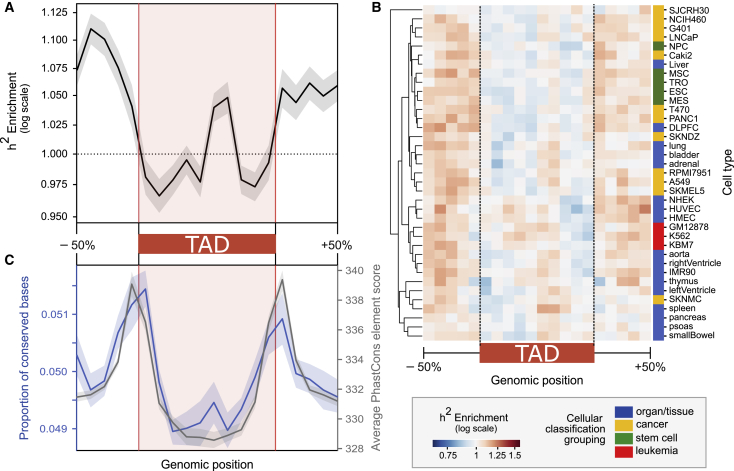
Figure 2.
Regions flanking TADs are enriched for heritability of diverse common complex traits and evolutionary sequence conservation
(A) Contribution to trait heritability (h2) is enriched across variation in TAD-flanking regions and in the center of TADs when averaged across 41 common complex phenotypes and TAD maps from 37 cell types (p = 1 × 10−193). Enrichment was computed within 20 equally sized bins centered on each TAD ± 50% of its length.
(B) Heritability patterns are consistent across the 3D genome landscape for 37 cell types.
(C) Regions flanking TADs have increased sequence-level constraint. They have a higher proportion of conserved bases (overlap with PhastCons elements; p = 5 × 10−11) (left blue axis) and a higher average conservation score across those overlapping PhastCons elements (right gray axis; p = 3 × 10−29).
Error bands signify 99% confidence intervals. Trends are similar for fixed-size 100 kb TAD boundaries bookending TADs; TAD boundaries are enriched for heritability (p = 0.001, Figure S3) and conservation (p = 3 × 10−29, Figure S4A).
The complex-trait heritability enrichment flanking TADs is also consistent across cell types (Figure 2B). The heritability enrichment values are significant but relatively small in magnitude. This is expected in light of the large genomic regions considered by this analysis—only a small fraction of the base pairs in a boundary are likely to be functionally relevant.
To assess functionality via a complementary approach, we compared between-species sequence-level conservation for TADs and boundaries. Regions flanking TADs are more evolutionarily conserved than sequences in TADs (Figure 2C). We quantified evolutionary conservation in terms of the proportion of base pairs in a region in a conserved element identified by PhastCons elements and by the average PhastCons element score across the region. On average, 5.02% of regions flanking TADs are overlapped by PhastCons elements, versus 4.97% of TADs (p = 5 × 10−11, Figure 2C). Furthermore, across these PhastCons elements, regions flanking TADs have average higher conservation scores than TADs (334 versus 331, p = 3 × 10−29, Figure 2C). The 100 kb TAD boundary set corroborates these results; 5.21% of bases in TAD boundaries are conserved versus 4.91% in intra-TAD 100 kb windows (p = 3x10−29, Figure S4A). This supports previous findings underscoring the importance of maintaining TAD boundaries.
The heritability enrichment and conservation at TAD boundaries are most likely due to their known overlap with functional elements such as CTCF binding sites and genes. Many such elements are enriched for heritability and conservation themselves.48 To assess whether the heritability enrichment flanking TADs is greater than expected given the known functional elements overlapping TAD boundaries, we calculated standardized enrichment effect sizes (τ∗).50,64 This statistic quantifies heritability unique to the focal annotation by conditioning on a broad set of 86 gene regulatory, evolutionary, gene, allele frequency, and LD-based annotations (baseline v2.1).48,50,64,65 TAD boundaries did not show more heritability than expected on the basis of their enrichment for the 86 other annotations (Figure S5). Similarly, to assess whether the greater evolutionary conservation flanking TADs is the result of the known enrichment in functional elements, we evaluated the conservation of bases in 100 kb boundaries and matched intra-TAD windows that do not overlap CTCF ChIP-seq peaks or exons. Filtering the base pairs that overlap CTCF peaks, we found that TAD boundaries still overlap more PhastCons elements and have a higher average PhastCons element score than windows in TADs (Figure S4). When removing all exonic base pairs, we found that TAD boundaries have less overlap with PhastCons elements than do windows in TADs. However, the conserved non-exonic regions of TAD boundaries have higher conservation scores than conserved non-exonic regions in TADs (Figure S4). Thus, existing annotations probably capture most of the relevant functional elements (e.g., CTCF, genes, and other regulatory element-binding sites) that determine and maintain boundary function.
TAD boundaries vary in stability across cellular contexts
The heritability enrichment patterns we observed are similar across cell types, and TADs have been characterized as largely invariant across cell types.18,19,36, 37, 38 However, previous work suggests distinct functional properties among TAD boundaries with different insulatory strengths, hierarchical structures, and cell types.33,44,45 Thus, we hypothesized that the stability of TAD boundaries across cell types would be informative about their functional roles and conservation. To characterize the stability of TAD boundaries across diverse cellular contexts, we focused on the 100 kb bookended TAD boundaries (described above), since these can be directly compared across the 37 cell types. The maps for each cell type are defined with respect to the same 100 kb windows across the genome, so we identify shared, or “stable,” boundaries on the basis of these 100 kb windows (Figure 3A). Our results are robust to different definitions of TAD boundaries, including 40 kb windows surrounding (±20 kb) TAD start and stop sites (“40 kb boundaries”) and 200 kb windows flanking the TAD start and stop sites (“200 kb bookend boundaries”) (see Figure S6 and Methods).
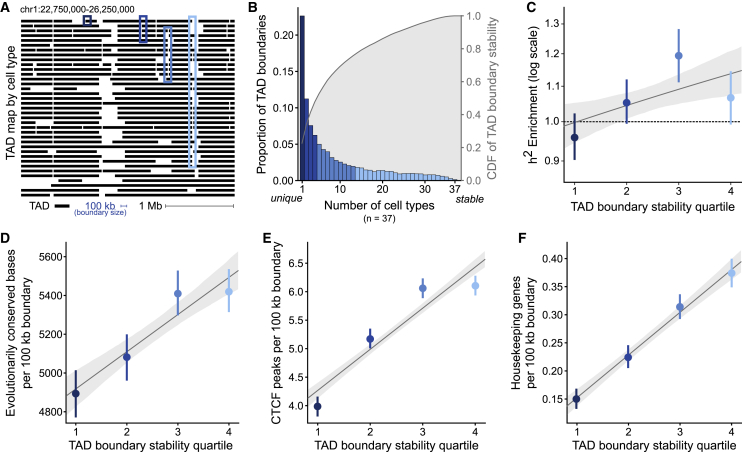
Figure 3.
Stable TAD boundaries are enriched for complex-trait heritability, evolutionary conservation, and functional elements
(A) Example TAD maps from 37 cell types (rows) for a 3.5 Mb window from human chromosome 1 (hg19). Each black line represents the genomic extent of a TAD. Example boundaries of different stability quartiles are outlined in blue (quartile 1 [most cell-type unique] in the darkest blue and quartile 4 [most cell-type stable] in light blue).
(B) Histogram of TAD boundaries by the number of cell types they are observed in (this quantifies their “stability,” colored by quartiles). The right axis and gray distribution represent the empirical cumulative distribution function (CDF) of boundary stability shown in the histogram.
(C–F) Across TAD-boundary stability quartiles, there is a correlation between increased cell-type stability and increased (C) complex-trait heritability enrichment (p = 0.006), (D) conserved bases (overlap with PhastCons elements, p = 6 × 10−13), (E) CTCF binding (overlap with ChIP-seq peaks, p = 1 × 10−83), and (F) housekeeping genes (p = 8 × 10−58). All error bars signify 95% confidence intervals. These trends hold at different boundary definitions (40 kb and 200 kb), for germ-layer informed measures of cell type stability, and for other measurements of conservation, CTCF binding, and gene overlap (Figures S9–S12).
Using the cross-cell-type TAD boundary intersection, we found that boundaries vary substantially across cell types. Less than 10% of TAD boundaries are shared in 25+ of the 37 cell types, and 22.6% of TAD boundaries are unique to a single cell type (Figure 3B). With the more granular 40 kb boundaries, 33.9% of boundaries are unique to one tissue (Figure S6A). Even with the permissive 200 kb resolution boundaries, 18.3% of boundaries are unique to a single tissue (Figure S6B). To quantify boundary stability for further analyses, we bin boundaries into their cell-type stability quartile: boundaries present in only one context of 37 (cell-type unique) are in the first quartile of stability, boundaries in 2–4 cell types are in the second quartile, boundaries in 5–13 cell types are in the third quartile, and boundaries in 14 or more of the 37 contexts are the fourth quartile of cell type stability (Figure 3B, examples in Figure 3A).
Although there is high variability in the landscape of TAD boundaries across different cell types, we found that biologically similar cell types have more similar TAD boundary maps. For example, cell type classes (e.g., organ or tissue, stem cell, and cancer) generally cluster together. The two neuroblastoma cell lines cluster together, as do left ventricle, right ventricle, aorta, and skeletal muscle (Figure S7B). This trend of biologically similar clusters also held at the 40 kb and 200 kb boundary resolution (Figures S7A and S7C). Previous studies have found contrasting results about the level and patterns of similarity across cell types (Supplemental Information), but our similarity quantifications between cell types agree with some previous estimates.13,37,42
In summary, although TADs and TAD boundaries have been characterized as largely invariant across cell types, we demonstrate that there is substantial variability between cell types.18,19,36, 37, 38 We also find that biologically related cell types have more similar TAD maps, providing preliminary evidence for the cell-type specificity of the 3D genome and providing further rationale for investigating differences in TAD maps between cell types.
Stable TAD boundaries are enriched for complex-trait heritability, evolutionary constraint, and functional elements
When stratifying the 100 kb boundaries by their cell-type stability we found a positive relationship between cell-type-stability and trait-heritability enrichment (r2 = 0.045, p = 0.006, Figure 3C). The most stable boundaries (fourth quartile, darkest blue) have 1.07× enrichment of trait heritability, as opposed to 0.96× enrichment in unique boundaries (first quartile). This positive relationship between heritability and boundary stability holds at both the 40 kb and 200 kb resolution (Figures S8A and S8D).
We also explored the relationship between TAD boundary stability and other evolutionary and functional attributes. Although TAD boundaries, when compared to TADs, are enriched for CTCF binding,18,44 evidence of evolutionary constraint (Figure 2C,33,35) and housekeeping genes are enriched at TAD boundaries7,18 (compared to TADs), it is unknown how these features relate to boundary stability across cell types.
We found that TAD boundary stability is positively correlated with increased evolutionary sequence constraint (Figure 3D, p = 3 × 10−13); compared to cell-type-unique TAD boundaries, boundaries in the highest quartile of stability have an additional 527 base pairs of overlap with PhastCons elements (5,420 versus 4,893 per 100 kb boundary). This extends previous observations that investigated two cell types to show that shared boundaries have evidence of stronger purifying selection on structural variants than boundaries present in only one of the cell types.33 On the basis of on our result, we conclude that stable boundaries are more intolerant of disruption, not only on the scale of structural variants, but also at the base-pair level.
TAD boundary stability is also correlated with increased CTCF binding (Figure 3E, p = 1 × 10−83). Boundaries in the highest quartile of stability have 1.5× more CTCF sites on average than TAD boundaries unique to one cell type (6.1 versus 4.0). This aligns with previous findings that boundary insulatory strength (in a single cell type) is positively associated with CTCF binding;18,44 however, it expands this finding to stability across cell types.
Finally, we found that TAD boundary stability is correlated with increased overlap with genes (1.56×, Figures S9A–S9C, p = 1 × 10−74), protein-coding genes (1.65×, Figures S9D-F, p = 7x10−90), and housekeeping genes (2.50×, Figures 3F, S9G-I, p = 8 × 10−58). Boundaries in the highest quartile of stability overlap 2.5× more housekeeping genes than do cell-type-unique TAD boundaries (0.37 versus 0.15 per 100 kb boundary). The relationship between stable TAD boundaries and housekeeping-gene enrichment might result from many factors, including strong enhancer-promoter interactions, specific transcription-factor binding, or chromatin insulation caused by highly active sites of transcription.12
Motivated by the observation that closely related cell types have more similar boundary maps (Figure S7) and given the non-uniform sampling of cell types considered here, we defined an additional measure of boundary stability based on cellular development. We determined the germ layer of origin (endoderm, mesoderm, ectoderm) for each of the 37 cell types and stratified boundaries on the basis of their presence across cells of different origins. Consistent with our results based on the raw count of cell types, boundaries observed in cell types from all three germ layers are enriched for trait heritability, conserved bases, CTCF binding, and housekeeping genes in comparison to boundaries unique to one germ layer (Figure S12). This shows that the greater contribution to complex trait heritability for more stable boundaries is probably robust to the sample of cell types considered.
Although our measure of TAD boundary stability correlates highly with these functional annotations, we note a slight drop-off in enrichment at the fourth quartile (compared to the third quartile), especially for trait heritability, conservation, and CTCF binding (Figures 3C–3E). We identify two factors—one technical and one biological—contributing to this. First, TADs must necessarily start and stop at the edges of chromosomes, centromeres, and gap regions; these regions will be identified as highly stable TAD boundaries independent of their functional importance and constraint. When boundaries within 5 Mb of genomic gaps68,69 or blacklist regions are removed,70 the enrichment drop-off is diminished (Figure S13). Second, the 37 cellular contexts considered are not uniformly sampled; some are more closely related than others. Thus, a boundary present in a well-sampled set of cell types might appear more stable than a boundary present in less densely sampled cell types. The germ-layer-based definition of stability has lower resolution but is less subject to sampling biases. We do not observe a decrease in the enrichment for heritability or other functional annotations among the most stable set when we use the germ-layer stability scores (Figure S12). Thus, it will be important in future work to incorporate more detailed understanding of the developmental relationships of the considered cell types into comparisons of TAD maps.
In summary, TAD boundaries stable across multiple cell types are enriched for complex-trait heritability, evolutionary constraint, CTCF binding, and housekeeping genes. These trends hold at different boundary definitions (40 kb and 200 kb), for germ-layer-informed measures of cell type stability, and for other measurements of conservation, CTCF binding, and gene overlap (Figures S9–S12).
The heritability landscape across the 3D genome varies across phenotypes
The previous analyses have shown that trait heritability is generally enriched at TAD boundaries and further enriched in boundaries stable across cell types. Given preliminary evidence that different traits have unique enrichment profiles among different functional annotations,48 we hypothesized that variation in TAD boundaries might influence certain traits more than others. To investigate trait-specific heritability across the TAD landscape, we computed heritability enrichment profiles across the 3D genome partitions by trait and hierarchically clustered them (Figure 4A). We observed two distinct trait clusters (Figure 4A).
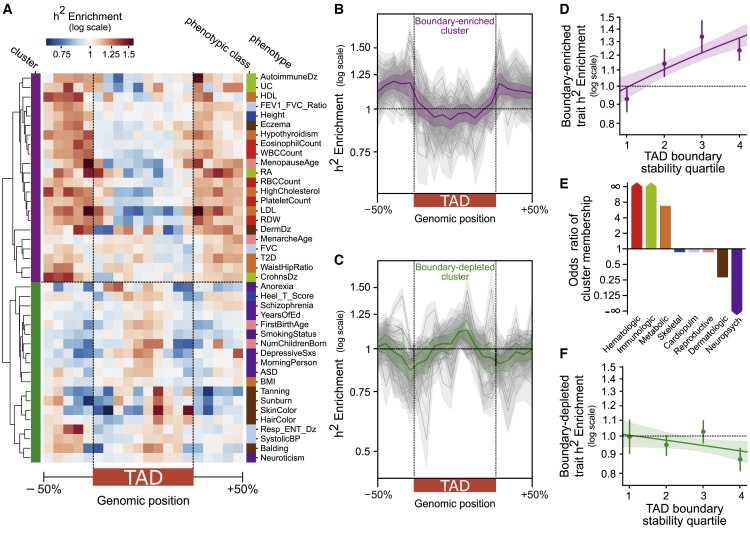
Figure 4.
The heritability landscape across the 3D genome varies across phenotypes
(A) Trait heritability patterns across the 3D genome organize into two clusters. Some traits are strongly enriched for complex-trait heritability at TAD boundaries (“boundary-enriched” cluster, purple), whereas others are weakly depleted at TAD boundaries and enriched centrally within the TAD (“boundary-depleted” cluster, green).
(B) Heritability enrichment landscape over TADs for traits in the boundary-enriched cluster (n = 22). The gray lines represent the heritability pattern for each trait in the cluster; the purple line is the average over all the traits.
(C) Heritability enrichment landscape over TADs for traits in the boundary-depleted cluster (n = 19). The green line is the average over all the traits.
(D) The positive correlation between boundary stability and trait heritability (Figure 3C) is driven by the subset of traits in the boundary-enriched cluster (r2 = 0.23, p = 2 × 10−6).
(E) Odds of cluster membership across phenotype categories. The boundary-enriched cluster is predominantly hematologic, immunologic, and metabolic traits. The boundary-depleted cluster is predominantly neuropsychiatric traits.
(F) There is a weak negative correlation between boundary stability and trait heritability for traits in the boundary-depleted cluster (r2 = 0.04, p = 0.09).
Error bars signify 99% confidence intervals in (B) and (C) and 95% confidence intervals in (D) and (F).
One cluster of traits (“boundary-enriched” cluster) is strongly enriched for complex-trait heritability at regions flanking TADs (Figure 4B) and in the 100 kb TAD boundaries (Figure S3). Across TAD maps in 37 cell types, these traits have on average 1.16× heritability enrichment at 100 kb TAD boundaries in comparison to genomic background (p = 1 × 10−7, Figure S3). The other cluster of traits (“boundary-depleted” cluster) shows a weak inverted pattern in comparison to the boundary-enriched cluster; there is marginal heritability depletion at TAD boundaries (0.97× enrichment, p = 0.06, Figure S3) and a spike of heritability enrichment within the TAD center (Figure 4C).
The traits in the boundary-enriched cluster are predominantly hematologic (e.g., counts of white and red blood cells), immunologic (e.g., rheumatoid arthritis, Crohn disease), and metabolic traits (e.g., type 2 diabetes, lipid counts) (Figure 4E). The traits in the boundary-depleted cluster are mostly neuropsychiatric (e.g., schizophrenia, years of education, Autism spectrum disorder) and dermatologic (e.g., skin color, balding) (Figure 4E). This stratification of complex diseases into phenotypic classes does not perfectly reflect the traits’ pathophysiology. For example, some dermatologic traits fall into the boundary-enriched cluster. However, these dermatologic traits, such as eczema, also have a substantial immunologic and hematologic basis, which is a hallmark of other traits in the boundary-enriched cluster. Additionally, body mass index (BMI) clustered with the psychiatric-predominant boundary-depleted cluster instead of with other metabolic traits in the boundary-enriched cluster. This is interesting in light of previous findings that BMI heritability is enriched in central nervous system (CNS)-specific annotations rather than metabolic-tissue (liver, adrenal, pancreas) annotations.48 Skeletal, cardiopulmonary, and reproductive traits do not consistently segregate into one of the clusters (Figure 4E). This is most likely because of the small sample size and heterogeneity of traits in these phenotypic classes.
The relationship between heritability enrichment in TAD boundaries and the trait clusters is not confounded by GWAS trait sample size (n), number of SNPs (M), or the traits’ SNP-based heritability (h2SNP) (Figure S14). Despite using a diverse set of cell types, we recognize that the heritability pattern differences between traits could be affected by the representation of investigated cell types. However, given that the pattern of heritability enrichment is consistent across all cell types (Figure 2B), we are confident that no single cluster of cell types is driving the differences in heritability patterns between traits. Furthermore, these patterns are maintained even when we call TADs by a variety of computational methods (Armatus, Arrowhead, DomainCaller, HiCseg, TADbit, TADtree, TopDom), suggesting that the finding of immunologic and hematologic heritability enrichment at TAD boundaries is robust to technical variation (Figure S15).
Although analysis across all traits revealed a positive relationship between boundary cell-type-stability and heritability enrichment (Figure 3C), we found that this trend is driven by traits in the boundary-enriched cluster: they have further heritability enrichment in cell-type-stable boundaries (r2 = 0.23, p = 2 × 10−6, Figure 4D). The most stable boundaries (fourth quartile) have 1.23× enrichment of trait heritability as compared to 0.93× enrichment in unique boundaries (first quartile). In contrast, traits in the boundary-depleted cluster have a non-significant negative relationship between stability and heritability (r2 = 0.04, p = 0.09, Figure 4F). These trends also hold when the germ-layer-informed measurement of boundary stability is used (Figures S12C and S12D). Thus, boundary stability might be more relevant when interpreting variation associated with hematologic, immunologic, and metabolic traits.
Discussion
Although we are beginning to understand the role of 3D genome disruption in rare disease and cancer, we have a limited framework for integrating maps of 3D genome structure into the study of genome evolution and the interpretation of common disease-associated variation. Here, we show that TAD boundaries, in comparison to TADs, are enriched for common complex-trait heritability. Additionally, in exploring TAD boundaries stable across cell types, we find they are further enriched for heritability of hematologic, immunologic, and metabolic traits, as well as evolutionary constraint, CTCF binding, and housekeeping genes. These findings demonstrate a relationship between 3D genome structure and the genetic architecture of common complex disease and reveal differences in the evolutionary pressures acting on different components of the 3D genome.
Previous work has predominantly characterized the importance and evolutionary constraint of different components of the 3D genome from the perspective of SV and rearrangement events. We address the relationship between genome 3D structure across cell types at the level of common single nucleotide variation. We consider evolutionary constraint within humans (∼100,000 ya) and constraint across diverse vertebrate species (∼13-450 mya).
At the scale of common human variation, we show that TAD boundaries are enriched for common variants that account for the heritability of common complex traits. This relationship between 3D genome structure and common disease-associated variation aligns with the finding of Whalen et al.35 that human haplotype breakpoints—which are associated with increased variation as a result of the mutagenic properties of recombination—are depleted at chromatin boundaries. Together, these findings suggest that TADs and TAD boundaries differ in their tolerance to genetic variation.
Over vertebrate evolution, we show that TAD boundaries have more sequence-level constraint than TADs. This provides a complementary perspective to that of Krefting et al.,25 who found that human TAD boundaries are enriched for syntenic breaks when they compared humans to 12 other vertebrate species, and they thus concluded that intact TADs are shuffled over evolutionary time. While shuffling a TAD may “move” its genomic location, preserving the TAD unit also requires maintaining at least part of its boundary. Our work suggests that even though TADs are shuffled, the boundary-defining sequences are under more constraint than the sequences within the TAD. This is further supported by the high concordance of TAD boundaries within syntenic blocks across different species and by depletion of SVs at TAD boundaries in humans and primates.7,18,26,33,39,40
Slight variation in 3D structure can cause large changes in gene expression.22,24 For example, CTCF helps maintain and form TAD boundaries; consequently, altering CTCF binding often leads to functional gene expression changes, e.g., oncogenic gene expression in gliomas.28 We hypothesize that altering gene regulation though common-variant disruption of transcription-factor motifs, such as CTCF, that are important in 3D structure organization contributes to the enrichment for complex-disease heritability. However, variation at TAD boundaries most likely also modifies genes or regulatory elements, such as enhancers, that are known to be enriched at boundaries without disrupting the TAD architecture. A deeper mechanistic understanding of TAD formation will be critical to further understanding how TAD-boundary disruption contributes to both rare and common disease at potentially nucleotide-level and cell-type resolution.
Our finding of divergent patterns of TAD boundary heritability enrichment for different traits (enrichment for hematologic, immunologic, and metabolic traits versus depletion for psychiatric and dermatologic traits) suggests that the 3D genome architecture might play differing roles in the genetic architecture of different traits. As a preliminary test of this hypothesis, we evaluated the relationship between boundary stability and intra-TAD heritability enrichment. We find that, for traits with heritability depletion at boundaries (psychiatric, dermatologic traits), TADs with stable boundaries have greater intra-TAD heritability enrichment (Figure S16). Thus, for these traits, we speculate that stable boundaries might function to insulate important intra-TAD functional elements (e.g., enhancers or genes). This idea is consistent with previous work showing that super-enhancers are insulated by the strongest boundaries (in a single cell type).44 However, for the boundary-enriched traits (hematologic, immunologic, metabolic), we hypothesize that essential functional elements are enriched at the stable boundaries (rather than inside the TAD). This is supported by previous work that detected a positive association between genome-wide binding of CTCF, a transcription factor intimately involved in TAD boundary formation, and eczema, an immunologic trait that we identified as part of the boundary-enriched trait cluster.83 Thus, it will be important to further explore how TAD boundaries (or other functional elements at TAD boundaries) might play different regulatory roles in different traits and diseases. This will be especially interesting to consider from an evolutionary perspective in light of evidence that certain subtypes of TADs, depending on the regulatory role of genes they contain, are under different selective pressures.84
Finally, we identify substantial variation among 3D maps across cell types. Whereas TAD stability across cell types is greater than expected by chance, our findings expand the number and diversity of compared cell types and identify a large proportion of boundaries unique to single cell types (see Supplemental Information). Furthermore, using our measurement of cell-type stability to stratify TAD boundaries identifies meaningful biological differences: stable boundaries are enriched for common-trait heritability, evolutionary constraint, and functional elements. Although we identify this enrichment for stable boundaries, we anticipate that cell-type-specific TAD boundaries often have functional significance relevant to their context; however, we are underpowered to detect trait-heritability enrichment in cell-type-specific TAD boundaries.
Several limitations should be considered when interpreting our results. First, they are based on available Hi-C data and existing methods for calling TADs. The Hi-C data were generated by different groups, so there could be batch- or protocol-specific effects. However, previous work suggests that biological differences dominate lab-of-origin effects in comparisons of structural similarity.42 Furthermore, we showed that the conclusions are robust to the computational method used (Figure S15) and that our stability results are not contingent on the specific set of cell types considered (Figure S12). Nonetheless, higher-resolution Hi-C across diverse cell types in multiple replicates is needed. Second, there is no standard for defining TAD boundaries. We use two complementary approaches and show our conclusions are robust. The first approach considers heritability across the 3D structural landscape by partitioning TADs and their flanking regions into 20 equal-size bins and enable comparison with previous work.25 The second defines fixed-size boundaries at multiple resolutions: 40, 100, and 200 kb. Continued efforts to integrate data from multiple TAD-calling algorithms to more precisely define TAD boundaries, especially given their hierarchical nature, will further refine our observations.45,85 Despite the complexities inherent in identifying TAD boundaries, our findings replicate with all our boundary definitions and with different TAD calling pipelines.
Here, we introduce a method for quantifying the stability of a TAD boundary across cell types and demonstrate enrichment of complex-trait heritability, sequence-level constraint, and CTCF binding among stable TAD boundaries. Our work suggests the utility of incorporating 3D structural data across multiple cell types to aid context-specific non-coding variant interpretation. Starting from this foundation, much further work is needed to elucidate the molecular mechanisms, evolutionary history, and cell-type-specificity of TAD-structure disruption. Furthermore, although we have focused on properties of TAD boundaries stable across cell types, it will also be valuable to identify differences in TAD boundary stability across species and find human-specific structures across diverse cell types.27 Finally, as high-resolution Hi-C becomes more prevalent from diverse tissues and individuals, we anticipate that computational prediction of personalized cell-type-specific TAD structure86,87 will facilitate understanding of how specific genetic variants are likely to affect 3D genome structure, gene regulation, and disease risk.
Data and Code Availability
The datasets we generated are available in the TAD-stability-heritability GitHub repository [https://github.com/emcarthur/TAD-stability-heritability] and at Zenodo: https://doi.org/10.5281/zenodo.360155988 and include all results of our boundary calling (40 kb, 100 kb bookend, and 200 kb bookend) and all partitioned heritability analysis output (by cell type and trait). The repository also contains a Jupyter notebook with code for analysis, statistics, and figure generation.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors would like to thank Katherine S. Pollard, Emily Hodges, Geoff Fudenberg, Sarah Fong, Mary Lauren Benton, and other members of the Capra Lab for helpful discussions and manuscript comments. They would like to thank Margaux L.A. Hujoel and Steven Gazal for their help with LDSC implementation and heritability result interpretation. This work was conducted in part with the resources of the Advanced Computing Center for Research and Education at Vanderbilt University, Nashville, TN. This work was supported by the National Institutes of Health (NIH) General Medical Sciences award R35GM127087 to J.A.C., NIH National Human Genome Research Institute award F30HG011200 to E.M., and T32GM007347. The funding bodies had no role in the design of the study; collection, analysis, or interpretation of data; or in writing the manuscript. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Published: February 4, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ajhg.2021.01.001.
Web Resources
CTCF ChIP-seq peaks, https://www.encodeproject.org/
housekeeping genes, https://www.tau.ac.il/∼elieis/HKG/HK_genes.txt
information regarding the cell types from ENCODE, http://genome.ucsc.edu/ENCODE/cellTypes.html
GWAS traits formatted for LDSC from the Alkes Price lab, https://data.broadinstitute.org/alkesgroup/LDSCORE/independent_sumstats/
GitHub, https://github.com/emcarthur/TAD-stability-heritability
PhastCons elements, RefSeq Genes, and genome gaps, https://genome.ucsc.edu/cgi-bin/hgTables
TAD maps from 3D Genome Browser, http://3dgenome.fsm.northwestern.edu/publications.html
Supplemental information
References
- 1.Cavalli G., Misteli T. Functional implications of genome topology. Nat. Struct. Mol. Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cremer T., Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 3.Duggal G., Wang H., Kingsford C. Higher-order chromatin domains link eQTLs with the expression of far-away genes. Nucleic Acids Res. 2014;42:87–96. doi: 10.1093/nar/gkt857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Dily F., Baù D., Pohl A., Vicent G.P., Serra F., Soronellas D., Castellano G., Wright R.H.G., Ballare C., Filion G. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014;28:2151–2162. doi: 10.1101/gad.241422.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J., Rippe K., Dekker M., Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 6.Lieberman-Aiden E., van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B.R., Sabo P.J., Dorschner M.O. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon J.R., Gorkin D.U., Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol. Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fudenberg G., Getz G., Meyerson M., Mirny L.A. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat. Biotechnol. 2011;29:1109–1113. doi: 10.1038/nbt.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hnisz D., Weintraub A.S., Day D.S., Valton A.L., Bak R.O., Li C.H., Goldmann J., Lajoie B.R., Fan Z.P., Sigova A.A. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meaburn K.J., Gudla P.R., Khan S., Lockett S.J., Misteli T. Disease-specific gene repositioning in breast cancer. J. Cell Biol. 2009;187:801–812. doi: 10.1083/jcb.200909127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb. Perspect. Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonev B., Mendelson Cohen N., Szabo Q., Fritsch L., Papadopoulos G.L., Lubling Y., Xu X., Lv X., Hugnot J.-P., Tanay A., Cavalli G. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauerwald N., Kingsford C. Quantifying the similarity of topological domains across normal and cancer human cell types. Bioinformatics. 2018;34:i475–i483. doi: 10.1093/bioinformatics/bty265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bruijn S.E., Fiorentino A., Ottaviani D., Fanucchi S., Melo U.S., Corral-Serrano J.C., Mulders T., Georgiou M., Rivolta C., Pontikos N. Structural Variants Create New Topological-Associated Domains and Ectopic Retinal Enhancer-Gene Contact in Dominant Retinitis Pigmentosa. Am. J. Hum. Genet. 2020;107:802–814. doi: 10.1016/j.ajhg.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu J., Hu M., Li C. Integrative analyses of multi-tissue Hi-C and eQTL data demonstrate close spatial proximity between eQTLs and their target genes. BMC Genet. 2018;20:43. doi: 10.1186/s12863-019-0744-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguet F., Ardlie K.G., Cummings B.B., Gelfand E.T., Getz G., Hadley K., Handsaker R.E., Huang K.H., Kashin S., Karczewski K.J. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards S.L., Beesley J., French J.D., Dunning A.M. Beyond GWASs: illuminating the dark road from association to function. Am. J. Hum. Genet. 2013;93:779–797. doi: 10.1016/j.ajhg.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dixon J.R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J.S., Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nora E.P., Lajoie B.R., Schulz E.G., Giorgetti L., Okamoto I., Servant N., Piolot T., Van Berkum N.L., Meisig J., Sedat J. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sexton T., Yaffe E., Kenigsberg E., Bantignies F., Leblanc B., Hoichman M., Parrinello H., Tanay A., Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Rowley M.J., Corces V.G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018;19:789–800. doi: 10.1038/s41576-018-0060-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao J., Hafner A., Boettiger A.N. How subtle changes in 3D structure can create large changes in transcription. bioRxiv. 2020 doi: 10.7554/eLife.64320. 10.22.351395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghavi-Helm Y., Jankowski A., Meiers S., Viales R.R., Korbel J.O., Furlong E.E.M. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat. Genet. 2019;51:1272–1282. doi: 10.1038/s41588-019-0462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwald W.W., Li H., Benaglio P., Jakubosky D., Matsui H., Schmitt A., Selvaraj S., D’Antonio M., D’Antonio-Chronowska A., Smith E.N., Frazer K.A. Subtle changes in chromatin loop contact propensity are associated with differential gene regulation and expression. Nat. Commun. 2019;10:1054. doi: 10.1038/s41467-019-08940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krefting J., Andrade-Navarro M.A., Ibn-Salem J. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol. 2018;16:87. doi: 10.1186/s12915-018-0556-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vietri Rudan M., Barrington C., Henderson S., Ernst C., Odom D.T., Tanay A., Hadjur S. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10:1297–1309. doi: 10.1016/j.celrep.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang Y., Zhang Y., Ren B., Dixon J.R., Ma J. Comparing 3D Genome Organization in Multiple Species Using Phylo-HMRF. Cell Syst. 2019;8:494–505.e14. doi: 10.1016/j.cels.2019.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flavahan W.A., Drier Y., Liau B.B., Gillespie S.M., Venteicher A.S., Stemmer-Rachamimov A.O., Suvà M.L., Bernstein B.E. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spielmann M., Lupiáñez D.G., Mundlos S. Structural variation in the 3D genome. Nat. Rev. Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 30.Lupiáñez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015;161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gröschel S., Sanders M.A., Hoogenboezem R., de Wit E., Bouwman B.A.M., Erpelinck C., van der Velden V.H.J., Havermans M., Avellino R., van Lom K. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157:369–381. doi: 10.1016/j.cell.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 32.Northcott P.A., Lee C., Zichner T., Stütz A.M., Erkek S., Kawauchi D., Shih D.J.H., Hovestadt V., Zapatka M., Sturm D. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fudenberg G., Pollard K.S. Chromatin features constrain structural variation across evolutionary timescales. Proc. Natl. Acad. Sci. USA. 2019;116:2175–2180. doi: 10.1073/pnas.1808631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han L., Zhao X., Benton M.L., Perumal T., Collins R.L., Hoffman G.E., Johnson J.S., Sloofman L., Wang H.Z., Stone M.R., CommonMind Consortium Functional annotation of rare structural variation in the human brain. Nat. Commun. 2020;11:2990. doi: 10.1038/s41467-020-16736-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whalen S., Pollard K.S. Most chromatin interactions are not in linkage disequilibrium. Genome Res. 2019;29:334–343. doi: 10.1101/gr.238022.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon J.R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J.E., Lee A.Y., Ye Z., Kim A., Rajagopal N., Xie W. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518:331–336. doi: 10.1038/nature14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S., Aiden E.L. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitt A.D., Hu M., Jung I., Xu Z., Qiu Y., Tan C.L., Li Y., Lin S., Lin Y., Barr C.L., Ren B. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep. 2016;17:2042–2059. doi: 10.1016/j.celrep.2016.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dekker J. Two ways to fold the genome during the cell cycle: insights obtained with chromosome conformation capture. Epigenetics Chromatin. 2014;7:25. doi: 10.1186/1756-8935-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dekker J., Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS Lett. 2015;589(20 Pt A):2877–2884. doi: 10.1016/j.febslet.2015.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krijger P.H.L., de Laat W. Regulation of disease-associated gene expression in the 3D genome. Nat. Rev. Mol. Cell Biol. 2016;17:771–782. doi: 10.1038/nrm.2016.138. [DOI] [PubMed] [Google Scholar]
- 42.Sauerwald N., Singhal A., Kingsford C. Analysis of the structural variability of topologically associated domains as revealed by Hi-C. NAR Genom. Bioinform. 2020;2 doi: 10.1093/nargab/lqz008. lqz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eres I.E., Gilad Y. 2020. A TAD Skeptic: Is 3D Genome Topology Conserved? Trends Genet. Published online November 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong Y., Lazaris C., Sakellaropoulos T., Lozano A., Kambadur P., Ntziachristos P., Aifantis I., Tsirigos A. Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nat. Commun. 2018;9:542. doi: 10.1038/s41467-018-03017-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.An L., Yang T., Yang J., Nuebler J., Xiang G., Hardison R.C., Li Q., Zhang Y. OnTAD: hierarchical domain structure reveals the divergence of activity among TADs and boundaries. Genome Biol. 2019;20:282. doi: 10.1186/s13059-019-1893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y., Song F., Zhang B., Zhang L., Xu J., Kuang D., Li D., Choudhary M.N.K., Li Y., Hu M. The 3D Genome Browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. Genome Biol. 2018;19:151. doi: 10.1186/s13059-018-1519-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulik-Sullivan B.K., Loh P.R., Finucane H.K., Ripke S., Yang J., Schizophrenia Working Group of the Psychiatric Genomics Consortium. Patterson N., Daly M.J., Price A.L., Neale B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hormozdiari F., Gazal S., van de Geijn B., Finucane H.K., Ju C.J.T., Loh P.R., Schoech A., Reshef Y., Liu X., O’Connor L. Leveraging molecular quantitative trait loci to understand the genetic architecture of diseases and complex traits. Nat. Genet. 2018;50:1041–1047. doi: 10.1038/s41588-018-0148-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hujoel M.L.A., Gazal S., Hormozdiari F., van de Geijn B., Price A.L. Disease Heritability Enrichment of Regulatory Elements Is Concentrated in Elements with Ancient Sequence Age and Conserved Function across Species. Am. J. Hum. Genet. 2019;104:611–624. doi: 10.1016/j.ajhg.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boraska V., Franklin C.S., Floyd J.A.B., Thornton L.M., Huckins L.M., Southam L., Rayner N.W., Tachmazidou I., Klump K.L., Treasure J., Wellcome Trust Case Control Consortium 3 A genome-wide association study of anorexia nervosa. Mol. Psychiatry. 2014;19:1085–1094. doi: 10.1038/mp.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smoller J.W., Kendler K., Craddock N., Lee P.H., Neale B.M., Nurnberger J.N., Ripke S., Santangelo S., Sullivan P.S., Neale B.N., Cross-Disorder Group of the Psychiatric Genomics Consortium Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013;381:1371–1379. doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., International IBD Genetics Consortium (IIBDGC) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okbay A., Baselmans B.M.L., De Neve J.E., Turley P., Nivard M.G., Fontana M.A., Meddens S.F.W., Linnér R.K., Rietveld C.A., Derringer J. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 2016;48:624–633. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barban N., Jansen R., de Vlaming R., Vaez A., Mandemakers J.J., Tropf F.C., Shen X., Wilson J.F., Chasman D.I., Nolte I.M., BIOS Consortium. LifeLines Cohort Study Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 2016;48:1462–1472. doi: 10.1038/ng.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., RACI consortium. GARNET consortium Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H., Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.International HapMap 3 Consortium. Altshuler D.M., Gibbs R.A., Peltonen L., Altshuler D.M., Gibbs R.A., Peltonen L., Dermitzakis E., Schaffner S.F., Yu F. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hormozdiari F., van de Geijn B., Nasser J., Weissbrod O., Gazal S., Ju J.-T., Louise Anna C., Hujoel M., Engreitz J., Hormozdiari F., Price A.L. Functional disease architectures reveal unique biological role of transposable elements. Nat. Commun. 2018;10:4054. doi: 10.1038/s41467-019-11957-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lumley T. 2018. Rmeta: Meta-Analysis. R package version 3.0. [Google Scholar]
- 64.Gazal S., Finucane H.K., Furlotte N.A., Loh P.R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gazal S., Loh P.R., Finucane H.K., Ganna A., Schoech A., Sunyaev S., Price A.L. Functional architecture of low-frequency variants highlights strength of negative selection across coding and non-coding annotations. Nat. Genet. 2018;50:1600–1607. doi: 10.1038/s41588-018-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dali R., Blanchette M. A critical assessment of topologically associating domain prediction tools. Nucleic Acids Res. 2017;45:2994–3005. doi: 10.1093/nar/gkx145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019;47(D1):D853–D858. doi: 10.1093/nar/gky1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Amemiya H.M., Kundaje A., Boyle A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019;9:9354. doi: 10.1038/s41598-019-45839-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Davis C.A., Hitz B.C., Sloan C.A., Chan E.T., Davidson J.M., Gabdank I., Hilton J.A., Jain K., Baymuradov U.K., Narayanan A.K. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 75.Watanabe K., Stringer S., Frei O., Umićević Mirkov M., de Leeuw C., Polderman T.J.C., van der Sluis S., Andreassen O.A., Neale B.M., Posthuma D. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 76.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dale R.K., Pedersen B.S., Quinlan A.R. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waskom M., Botvinnik O., O’Kane D., Hobson P., Ostblom J., Lukauskas S., Gemperline D.C., Augspurger T., Halchenko Y., Cole J.B. 2018. mwaskom/seaborn: v0.9.0. 10.5281/zenodo.1313201. [Google Scholar]
- 79.Hunter J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007;9:90–95. [Google Scholar]
- 80.InkscapeProject 2018. https://inkscape.org
- 81.Leung D., Jung I., Rajagopal N., Schmitt A., Selvaraj S., Lee A.Y., Yen C.A., Lin S., Lin Y., Qiu Y. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–354. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lajoie B.R., Dekker J., Kaplan N. The Hitchhiker’s guide to Hi-C analysis: practical guidelines. Methods. 2015;72:65–75. doi: 10.1016/j.ymeth.2014.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reshef Y.A., Finucane H.K., Kelley D.R., Gusev A., Kotliar D., Ulirsch J.C., Hormozdiari F., Nasser J., O’Connor L., van de Geijn B. Detecting genome-wide directional effects of transcription factor binding on polygenic disease risk. Nat. Genet. 2018;50:1483–1493. doi: 10.1038/s41588-018-0196-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Torosin N.S., Anand A., Golla T.R., Cao W., Ellison C.E. 3D genome evolution and reorganization in the Drosophila melanogaster species group. PLoS Genet. 2020;16:e1009229. doi: 10.1371/journal.pgen.1009229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zufferey M., Tavernari D., Oricchio E., Ciriello G. Comparison of computational methods for the identification of topologically associating domains. Genome Biol. 2018;19:217. doi: 10.1186/s13059-018-1596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fudenberg G., Kelley D.R., Pollard K.S. Predicting 3D genome folding from DNA sequence with Akita. Nat. Methods. 2020;17:1111–1117. doi: 10.1038/s41592-020-0958-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schwessinger R., Gosden M., Downes D., Brown R.C., Oudelaar A.M., Telenius J., Teh Y.W., Lunter G., Hughes J.R. DeepC: predicting 3D genome folding using megabase-scale transfer learning. Nat. Methods. 2020;17:1118–1124. doi: 10.1038/s41592-020-0960-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McArthur, E., and Capra, J.A. emcarthur/TAD-stability-heritability. Zenodo. 10.5281/zenodo.3601559. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets we generated are available in the TAD-stability-heritability GitHub repository [https://github.com/emcarthur/TAD-stability-heritability] and at Zenodo: https://doi.org/10.5281/zenodo.360155988 and include all results of our boundary calling (40 kb, 100 kb bookend, and 200 kb bookend) and all partitioned heritability analysis output (by cell type and trait). The repository also contains a Jupyter notebook with code for analysis, statistics, and figure generation.