Abstract
The objective of this study was to validate a fully automated total kidney volume measurement method for pre-clinical rodent trials that is fast, accurate, reproducible, and to provide these resources to the research community. Rodent studies that involve imaging are crucial for monitoring treatment efficacy in diseases such as polycystic kidney disease. Previous studies utilize manual or semi-automated segmentations, which are time consuming and potentially biased. To develop our automated system, a total of 150 axial magnetic resonance images (MRI) from a variety of mouse models were manually segmented and used to train/validate an automated algorithm. To test the longitudinal application of the model, four mutant and four wild-type mice were imaged sequentially over three to twelve weeks via MRI. Segmentations of the kidneys (excluding the renal pelvis) were generated by the automated method and two different readers, with one reader repeating the measurements. Similarity metrics and longitudinal analysis were calculated to assess the performance of the automated compared to the manual methods. The automated approach required no user input, besides a final visual quality control step. Similarity metrics of the automated method versus the manual segmentations were on par with inter- and intra-reader comparisons. Thus, our fully automated approach described here can be safely used in longitudinal, pre-clinical trials that involve the segmentation of rodent kidneys in T2-weighted MRIs.
Keywords: Deep learning, murine, polycystic kidney disease, segmentation, similarity metrics, total kidney volume
Graphical Abstract
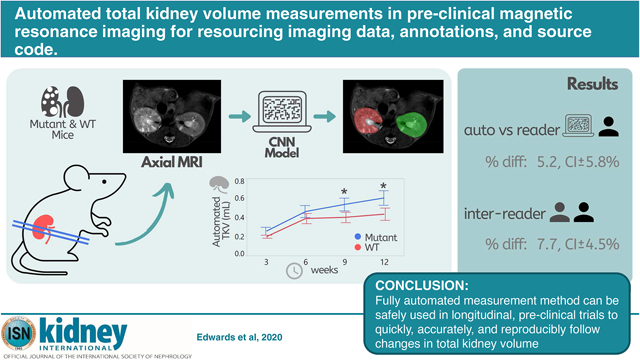
Translational Statement
This study developed a fully automated method for measuring total kidney volume for pre-clinical imaging in a mutant model of polycystic kidney disease as well as wild-type mice. This study also established both inter-reader and intra-reader variability for the measurement of total kidney volume in pre-clinical imaging. Similar studies and algorithmic approaches can be used to establish methods for clinical imaging data, and are needed for accurate disease prognosis and clinical decision making. We are providing the imaging data, annotations, and source code to the research community.
INTRODUCTION
The measurement of organ volume has been shown to correlate with clinical manifestations and morbidity of diseases such as total kidney volume (TKV) in autosomal dominant polycystic kidney disease (ADPKD),1, 2 and is used to ascertain the effectiveness of treatment interventions.3 Research, clinical trials, and increasingly clinical nephrology use these measurements to monitor disease progression in both animal models4 and patients,5 evaluate the effectiveness of therapies,6 and predict outcomes.7
Currently, pre-clinical studies are occurring at an unprecedented rate to look for new treatments to slow progression of PKD. A key advantage of MRI in animal models of PKD is the ability to use in vivo imaging, which allows for longitudinal volumetric studies that use the same animal.8 Numerous studies have previously been performed which involve manual,4, 9–12 semi-automated,13, 14 and registration-based automated segmentation of mouse kidneys.15, 16
Many of the methods deemed as automated still require user input. A majority of these pre-clinical studies utilize manual segmentation, which is time consuming, costly, and introduces observer bias. Therefore, our lab has assessed the variability in measurements of TKV and developed an automated analysis program to measure TKV in MR scans of murine models of the disease.
RESULTS
Intra- and Inter-observer Variability of Manual Kidney Segmentations
Figure 1 shows the results of the Bland-Altman analysis of TKV as measured manually by two different readers (inter-observer variance) and repeat measurements by Reader 2 (intra-observer variance). When Reader 1 was compared with Reader 2, there was a mean percentage difference of 7.7% and 95% confidence interval of ±4.5%. When Reader 2 performed repeat measurements of the same image, there was a mean percentage difference of −0.5% and 95% confidence interval of ±3.9%. Regression analysis indicated that there is high agreement in TKV among all methods, with an R-squared value of ≥0.99.
Figure 1.
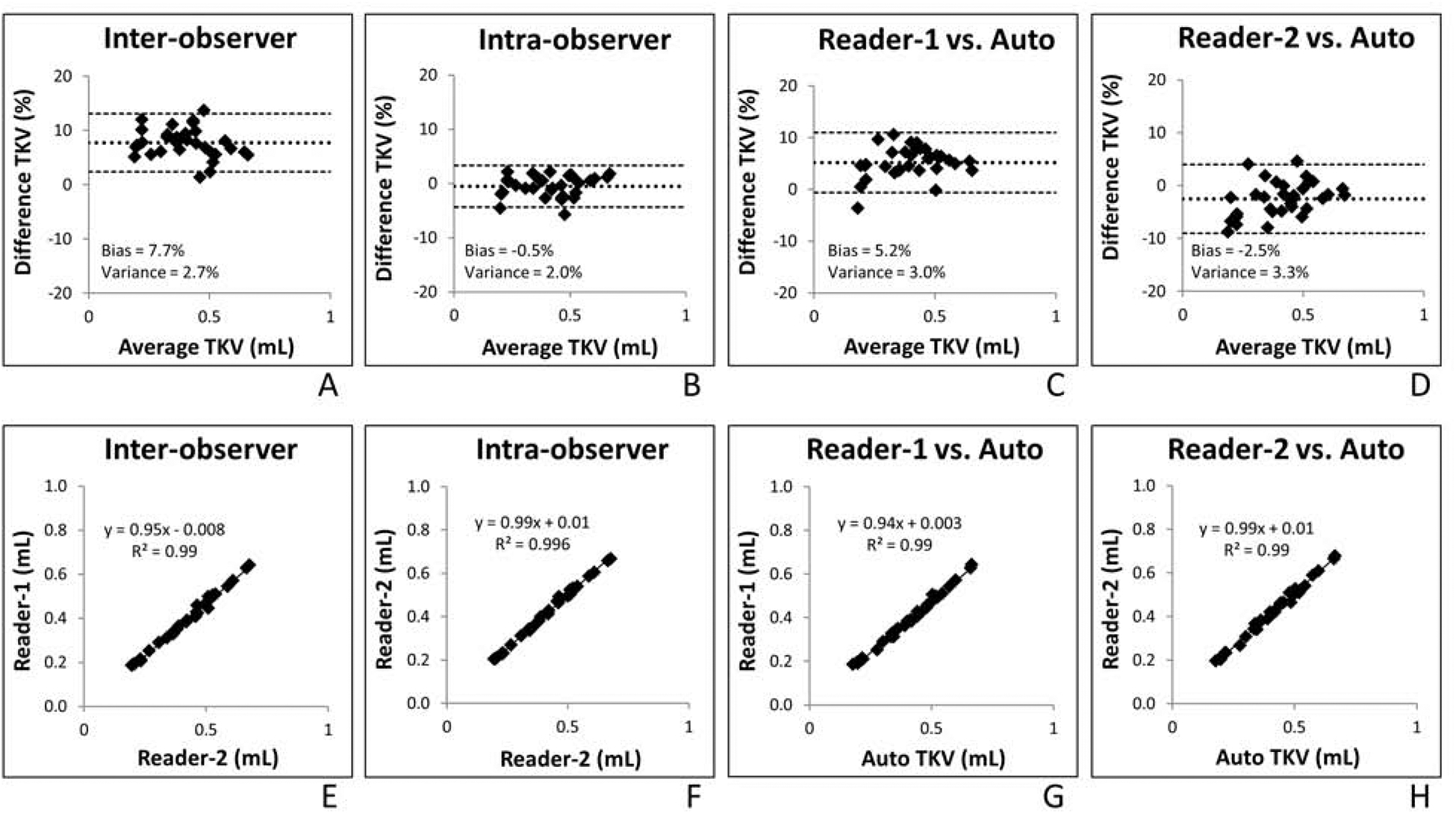
Bland-Altman and regression analysis of inter- (A,E) and intra-observer (B,F) total kidney volume (TKV) measurements (measured in milliliters) in addition to the automated method compared with reader one (C,G) and reader two (D,H). Bland-Altman plots show mean difference (solid line) and 95% confidence interval (dotted lines). Regression analysis shows correlation between the compared methods.
Validation of Automated Segmentation Algorithm
The automated method compared with each reader in relation to percentage difference of TKV was similar to that of inter- and intra-observer variance as suggested by the Bland-Altman plots in Figure 1. When Reader 1 was compared with the automated method, there was a mean percentage difference of 5.2% and 95% confidence interval of ±5.8%. When Reader 2 was compared with the automated method, there was a mean percentage difference of −2.5% and 95% confidence interval of ±6.5%.
Distinction between Wild-Type and Mutant Mice
The mean and SD TKVs were plotted at each time point for each method and separated by genotype (mutant vs wild-type). As seen in Figure 2, the mean TKV is always smaller in the wild-type at each time point compared to the mutant mice. All three methods (automated, Reader 1, and Reader 2) demonstrate a significant separation of mouse type at nine and twelve weeks of age.
Figure 2.
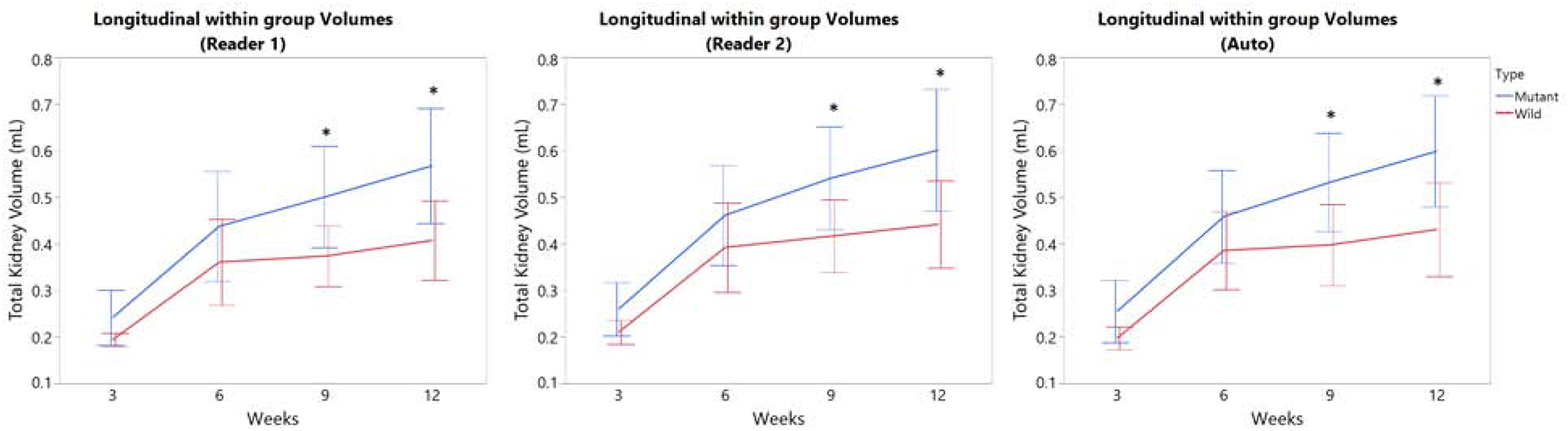
Total kidney volume (TKV) of wild type (WT) and mutant mice was plotted over time (3 – 12 weeks of age) for the automated method (left), Reader 1 (middle), and Reader 2 (right). All three methods demonstrate a significant separation of mouse type at nine weeks of age (*P < 0.05; error bars indicate SD).
DISCUSSION
The analysis of kidney volume in PKD is one of the most important metrics currently used for characterizing disease status. Prior to our work, there was no alternative to manually tracing kidneys in model systems of PKD. Because of the time taken to trace these structures, as well as the time required to train a person to perform these measurements, and the potential for inter-operator variability, in this study we developed and validated a fully automated segmentation method for TKV. The automated segmentations are computed in a matter of minutes (depending on computing power), whereas manual segmentations take 20–40 minutes. Unlike manual or even semi-automated segmentation methods, this automated method will produce the same exact results each time it is applied to the same image.
Pre-clinical trials are often comprised of both a control group and treatment group(s), therefore it is essential that the automated method is sensitive enough to detect volume differences between groups appropriately.17 Figure 2 shows that manual segmentations and the automated segmentation both show a significant separation in the wild-type and mutant groups at nine weeks of age. While the overall agreement was excellent, visual comparisons suggested minor disagreement on whether to include or exclude the renal pelvis on a small subset of slices. Although it is common practice to exclude the renal pelvis, variability might decrease if readers are instructed to always include this structure.
The automated method presented in this study has not yet been applied to external images. It is important to note that signal intensities vary across sites, scanners, and MRI acquisitions. It is likely that a larger training dataset with more diverse cases from different MRI machines could achieve a more robust model due to the nature of deep learning algorithms. Automated algorithms such as these often need to be retrained on outside datasets due to the variations in MRI signals. We therefore provide the imaging data, annotations, and source code to the research community in order for other groups to either use the same model, or develop their own.
METHODS
Training/Validation Data
The model was trained on 100 cases, and validated on 50 cases. The 150 cases consisted of mice with varied disease severity and at a range of ages. The test set is a completely held out set and is what we evaluated in this paper.
Testing Study Cohort
This study was reviewed and approved by the Mayo Clinic Institutional Animal Care and Use Committee (IACUC). The cohort consisted of Wild Type C57Bl6 × 129s6Svev/Tac (n=4; 2F/2M) and C57Bl6 × 129s6Svev/Tac mutants Pkd1 RC/RC model mice (n=4; 2F/2M). The mutant mice mirror the human manifestation of PKD1 both genetically and phenotypically.18 One of the mutant mice died mid-experiment and was replaced with another mutant mouse of the same age at week 9.
Image Acquisitions
Imaging was performed using an Avance DRX 700WB (Bruker BioSpin, Billerica, Massachusetts, USA) spectrometer. Complete coverage of the kidneys was obtained by an axial TurboRARE T2-weighted acquisition reconstructed with 0.1mm in plane resolution, and 1mm slice thickness (matrix size = 256 × 256 × Z, with Z chosen large enough to cover the full extent of the kidneys). Total scan time ranged from 5–10 minutes. In the testing study cohort, each mouse was imaged at 4 different time points (3, 6, 9, and 12 weeks of age). The time points for each mouse were performed within two days of each other to ensure consistent imaging parameters and to limit environmental variations.
Image Analysis
Regions of interest (ROI) were traced on each scan using an imaging software package (Analyze, version 12.0, Biomedical Imaging Resource, Mayo Clinic). Each reader was instructed to exclude the renal pelvis when the renal pelvis is not enclosed by the kidney capsule within the image slice. Manual segmentation took 20–40 minutes depending on the case. The TKVs were calculated by first summing the number of voxels contained within the segmentation on each slice, then multiplying the number of voxels by the voxel volume obtained from the image header. For the testing data, two double-blinded readers (1 and 2), both experienced in manual MRI segmentation, performed the kidney segmentations on all scans. For intra-reader analysis, Reader 2 repeated the measurements at two different time points (> 3 months apart).
Automated method
The neural network model was adapted from our previous model for measurement of total kidney volume from coronal T2-weighted MR images from clinical scans.19. The source code, images, and annotations are made publically available at: https://github.com/TLKline/AutoTKV_MouseMRI
Statistical analysis
The axial T2-weighted MR images acquired per mouse (n=8) at each time point (3, 6, 9, and 12 weeks) were used for the statistical analysis. A total of 32 images allowed for the comparison of different images over a wide range of ages and differences in phenotypes. For validating the fully automated method, comparison statistics were used to evaluate the ability of each method to separate wild-type and mutant groups. This was achieved by plotting the TKV according to time point and separating by mouse type. TKV measurements and growth rates for each method were also assessed using Bland-Altman and linear regression plots.
Acknowledgements
This work was supported by the Mayo Clinic Robert M. and Billie Kelley Pirnie Translational PKD Center and the NIDDK [grant numbers P30DK090728, K01DK110136]. The authors thank Ms. Lynnae M. Henry for her administrative support in preparing and formatting this manuscript.
Abbreviations:
- ADPKD
Autosomal dominant polycystic kidney disease
- CT
Computed tomography
- EMA
European medicines agency
- FA
flip angle
- FDA
US Food and Drug Administration
- htTKV
height-adjusted total kidney volume
- IACUC
institutional animal care and use committee
- MRI
magnetic resonance imaging
- TKV
total kidney volume
- PKD
polycystic kidney disease
- Res
resolution (μm/pxl)
- RF
radiofrequency
- ROI
Region Of Interest
- TE
echo time
- TR
repetition time
- US
ultrasound
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE STATEMENT
None of the authors have financial interests to disclose and the authors declare no conflicts of interest.
REFERENCES
- 1.Grantham JJ, Torres VE, Chapman AB, et al. Volume progression in polycystic kidney disease. N Engl J Med 2006; 354: 2122–2130. [DOI] [PubMed] [Google Scholar]
- 2.Torres VE, Chapman AB, Devuyst O, et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med 2012; 367: 2407–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caroli A, Perico N, Perna A, et al. Effect of longacting somatostatin analogue on kidney and cyst growth in autosomal dominant polycystic kidney disease (ALADIN): a randomised, placebo-controlled, multicentre trial. Lancet 2013; 382: 1485–1495. [DOI] [PubMed] [Google Scholar]
- 4.Wallace DP, Hou YP, Huang ZL, et al. Tracking kidney volume in mice with polycystic kidney disease by magnetic resonance imaging. Kidney Int 2008; 73: 778–781. [DOI] [PubMed] [Google Scholar]
- 5.Grantham JJ, Torres VE. The importance of total kidney volume in evaluating progression of polycystic kidney disease. Nat Rev Nephrol 2016; 12: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Higashihara E, Torres VE, Chapman AB, et al. Tolvaptan in autosomal dominant polycystic kidney disease: three years’ experience. Clin J Am Soc Nephrol 2011; 6: 2499–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Irazabal MV, Rangel LJ, Bergstralh EJ, et al. Imaging classification of autosomal dominant polycystic kidney disease: a simple model for selecting patients for clinical trials. J Am Soc Nephrol 2015; 26: 160–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irazabal MV, Mishra PK, Torres VE, et al. Use of Ultra-high Field MRI in Small Rodent Models of Polycystic Kidney Disease for In Vivo Phenotyping and Drug Monitoring. J Vis Exp 2015: e52757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erokwu BO, Anderson CE, Flask CA, et al. Quantitative magnetic resonance imaging assessments of autosomal recessive polycystic kidney disease progression and response to therapy in an animal model. Pediatr Res 2018; 83: 1067–1074. [DOI] [PubMed] [Google Scholar]
- 10.Doctor RB, Serkova NJ, Hasebroock KM, et al. Distinct patterns of kidney and liver cyst growth in pkd2(WS25/-) mice. Nephrol Dial Transplant 2010; 25: 3496–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke M, Baessler B, Vechtel J, et al. Magnetic resonance T2 mapping and diffusion-weighted imaging for early detection of cystogenesis and response to therapy in a mouse model of polycystic kidney disease. Kidney Int 2017; 92: 1544–1554. [DOI] [PubMed] [Google Scholar]
- 12.Zhou X, Bao H, Takakura A, et al. Polycystic kidney disease evaluation by magnetic resonance imaging in ischemia-reperfusion injured PKD1 knockout mouse model: comparison of T2- weighted FSE and true-FISP. Invest Radiol 2010; 45: 24–28. [DOI] [PubMed] [Google Scholar]
- 13.Fei B, Flask C, Wang H, et al. Image Segmentation, Registration and Visualization of Serial MR Images for Therapeutic Assessment of Polycystic Kidney Disease in Transgenic Mice. Conf Proc IEEE Eng Med Biol Soc 2005; 1: 467–469. [DOI] [PubMed] [Google Scholar]
- 14.Almajdub M, Magnier L, Juillard L, et al. Kidney volume quantification using contrast-enhanced in vivo X-ray micro-CT in mice. Contrast Media Mol Imaging 2008; 3: 120–126. [DOI] [PubMed] [Google Scholar]
- 15.Hadjidemetriou S, Reichardt W, Hennig J, et al. Volumetric analysis of MRI data monitoring the treatment of polycystic kidney disease in a mouse model. MAGMA 2011; 24: 109–119. [DOI] [PubMed] [Google Scholar]
- 16.Gleason SS, Sari-Sarraf H, Abidi MA, et al. A new deformable model for analysis of X-ray CT images in preclinical studies of mice for polycystic kidney disease. IEEE Trans Med Imaging 2002; 21: 1302–1309. [DOI] [PubMed] [Google Scholar]
- 17.Reichardt W, Romaker D, Becker A, et al. Monitoring kidney and renal cyst volumes applying MR approaches on a rapamycin treated mouse model of ADPKD. MAGMA 2009; 22: 143–149. [DOI] [PubMed] [Google Scholar]
- 18.Hopp K, Ward CJ, Hommerding CJ, et al. Functional polycystin-1 dosage governs autosomal dominant polycystic kidney disease severity. J Clin Invest 2012; 122: 4257–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kline TL, Korfiatis P, Edwards ME, et al. Performance of an Artificial Multi-observer Deep Neural Network for Fully Automated Segmentation of Polycystic Kidneys. J Digit Imaging 2017; 30: 442–448. [DOI] [PMC free article] [PubMed] [Google Scholar]


