Summary
Whereas large-scale statistical analyses can robustly identify disease-gene relationships, they do not accurately capture genotype-phenotype correlations or disease mechanisms. We use multiple lines of independent evidence to show that different variant types in a single gene, SATB1, cause clinically overlapping but distinct neurodevelopmental disorders. Clinical evaluation of 42 individuals carrying SATB1 variants identified overt genotype-phenotype relationships, associated with different pathophysiological mechanisms, established by functional assays. Missense variants in the CUT1 and CUT2 DNA-binding domains result in stronger chromatin binding, increased transcriptional repression, and a severe phenotype. In contrast, variants predicted to result in haploinsufficiency are associated with a milder clinical presentation. A similarly mild phenotype is observed for individuals with premature protein truncating variants that escape nonsense-mediated decay, which are transcriptionally active but mislocalized in the cell. Our results suggest that in-depth mutation-specific genotype-phenotype studies are essential to capture full disease complexity and to explain phenotypic variability.
Keywords: SATB1, de novo variants, neurodevelopmental disorders, intellectual disability, seizures, teeth abnormalities, HPO-based analysis, cell-based functional assays
Main text
SATB1 (MIM: 602075) encodes a dimeric/tetrameric transcription factor1 with crucial roles in development and maturation of T cells.2, 3, 4 Recently, a potential contribution of SATB1 to brain development was suggested by statistically significant enrichment of de novo variants in two large neurodevelopmental disorder (NDD) cohorts,5,6 although its functions in the central nervous system are poorly characterized.
Through international collaborations7, 8, 9 conforming to local ethical guidelines and the declaration of Helsinki, we identified 42 individuals with a rare (likely) pathogenic variant in SATB1 (GenBank: NM_001131010.4), a gene under constraint against loss-of-function and missense variation (pLoF: o/e = 0.15 [0.08–0.29]; missense: o/e = 0.46 [0.41–0.52]; gnomAD v2.1.1).10 Twenty-eight of the SATB1 variants occurred de novo, three were inherited from an affected parent, and five resulted from (suspected) parental mosaicism (Figure S1). Reduced penetrance is suggested by two variants inherited from unaffected parents (identified in individuals 2 and 12; Table S1A), consistent with recent predictions of incomplete penetrance being more prevalent in novel NDD syndromes.6 Inheritance status of the final four could not be established (Table S1A). Of note, two individuals also carried a (likely) pathogenic variant affecting other known disease genes, including NF1 (MIM: 162200; individual 27) and FOXP2 (MIM: 602081; individual 42) which contributed to (individual 27) or explained (individual 42) the observed phenotype (Table S1A).
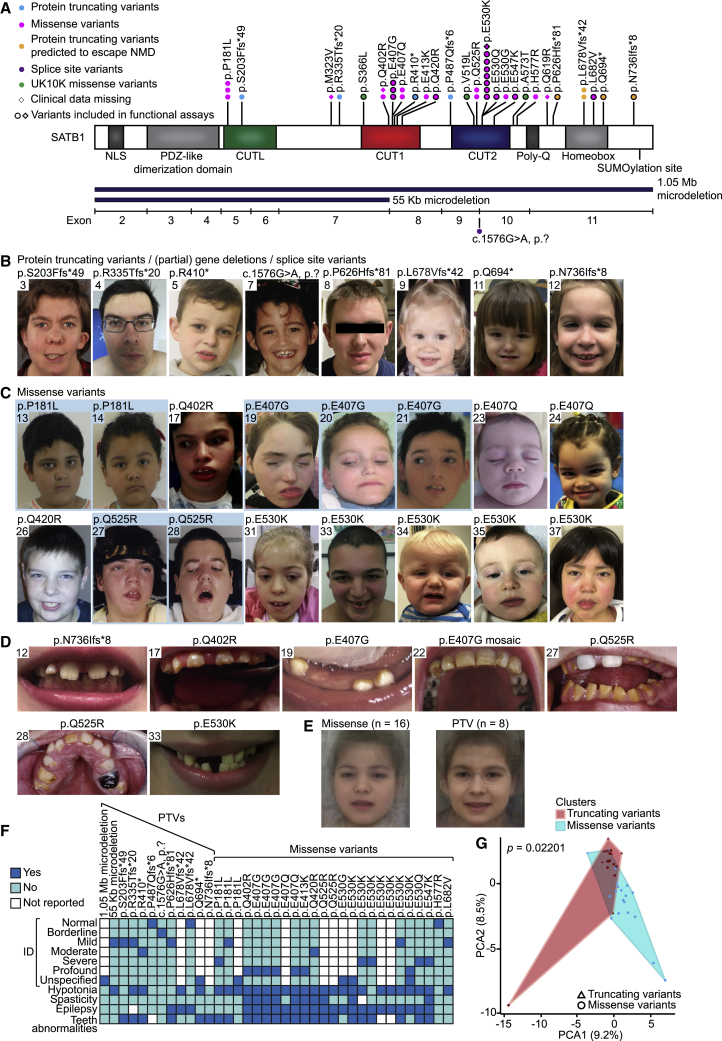
Thirty individuals carried 15 unique SATB1 missense variants, including three recurrent variants (Figure 1A), significantly clustering in the highly homologous DNA-binding domains CUT1 and CUT2 (p = 1.00e−7; Figures 2A and S2).11,12 Ten individuals harbored premature protein truncating variants (PTVs; two nonsense, seven frameshift, one splice site; Tables S1A and S2), and two individuals had a (partial) gene deletion (Figure S3). For 38 affected individuals and 1 mosaic parent, clinical information was available. Overall, we observed a broad phenotypic spectrum, characterized by neurodevelopmental delay (35/36, 97%), intellectual disability (ID) (28/31, 90%), muscle tone abnormalities (abnormal tone 28/37, 76%; hypotonia 28/37, 76%; spasticity 10/36, 28%), epilepsy (22/36, 61%), behavioral problems (24/34, 71%), facial dysmorphisms (24/36, 67%; Figures 1B, 1C, and S4A), and dental abnormalities (24/34, 71%) (Figures 1D and S4B; Tables 1 and S1). Individuals with missense variants were globally more severely affected than those with PTVs: 57% of individuals with a missense variant had severe/profound ID whereas this level of ID was not observed for any individuals with PTVs. Furthermore, hypotonia, spasticity, and (severe) epilepsy were more common in individuals with missense variants than in those with PTVs (92% versus 42%, 42% versus 0%, 80% versus 18%, respectively) (Figure 1F, Tables 1 and S1A). To objectively quantify these observations, we divided our cohort into two variant-specific clusters (missense versus PTVs) and assessed the two groups using a Partitioning Around Medoids clustering algorithm13 on 100 features derived from standardized clinical data (Human Phenotype Ontology [HPO]; Figure S5A and Data S1).14 A total of 38 individuals were subjected to this analysis, of which 27 were classified correctly as either belonging to the PTV or missense variant group (p = 0.022), confirming the existence of at least two separate clinical entities (Figures 1G and S5B). Moreover, computational averaging of facial photographs15 revealed differences between the average facial gestalt for individuals with missense variants when compared to individuals with PTVs or deletions (Figures 1B–1E and S4, Table S1B).
Figure 1.
Clinical evaluation of SATB1 variants in neurodevelopmental disorders
(A) Schematic representation of SATB1 (GenBank: NM_001131010.4/NP_001124482.1), including functional domains, with truncating variants labeled in cyan, truncating variants predicted to escape NMD in orange, splice site variants in purple, missense variants in magenta, and UK10K rare control missense variants in green. Deletions are shown in dark blue below the protein schematic, above a diagram showing the exon boundaries. We obtained clinical data for all individuals depicted by a circle.
(B and C) Facial photographs of individuals with (partial) gene deletions and truncations (B) and of individuals with missense variants (C). All depicted individuals show facial dysmorphisms and although overlapping features are seen, no consistent facial phenotype can be observed for the group as a whole. Overlapping facial dysmorphisms include facial asymmetry, high forehead, prominent ears, straight and/or full eyebrows, puffy eyelids, downslant of palpebral fissures, low nasal bridge, full nasal tip and full nasal alae, full lips with absent cupid’s bow, prominent cupid’s bow, or thin upper lip vermilion (Table S1B). Individuals with missense variants are more alike than individuals in the truncating cohorts, and we observed recognizable overlap between several individuals in the missense cohort (individuals 17, 27, 31, 37, the siblings 19, 20, and 21, and to a lesser extent individuals 24 and 35). A recognizable facial overlap between individuals with (partial) gene deletions and truncations could not be observed. Related individuals are marked with a blue box.
(D) Photographs of teeth abnormalities observed in individuals with SATB1 variants. Dental abnormalities are seen for all variant types and include widely spaced teeth, dental fragility, missing teeth, disorganized teeth positioning, and enamel discoloration (Table S1B).
(E) Computational average of facial photographs of 16 individuals with a missense variant (left) and 8 individuals with PTVs or (partial) gene deletions (right).
(F) Mosaic plot presenting a selection of clinical features.
(G) The Partitioning Around Medoids analysis of clustered HPO-standardized clinical data from 38 individuals with truncating (triangle) and missense (circle) variants shows a significant distinction between the clusters of individuals with missense variants (blue) and individuals with PTVs (red). Applying Bonferroni correction, a p value smaller than 0.025 was considered significant.
For analyses displayed in (F) and (G), individuals with absence of any clinical data and/or low-level mosaicism for the SATB1 variant were omitted (for details, see supplemental materials and methods).
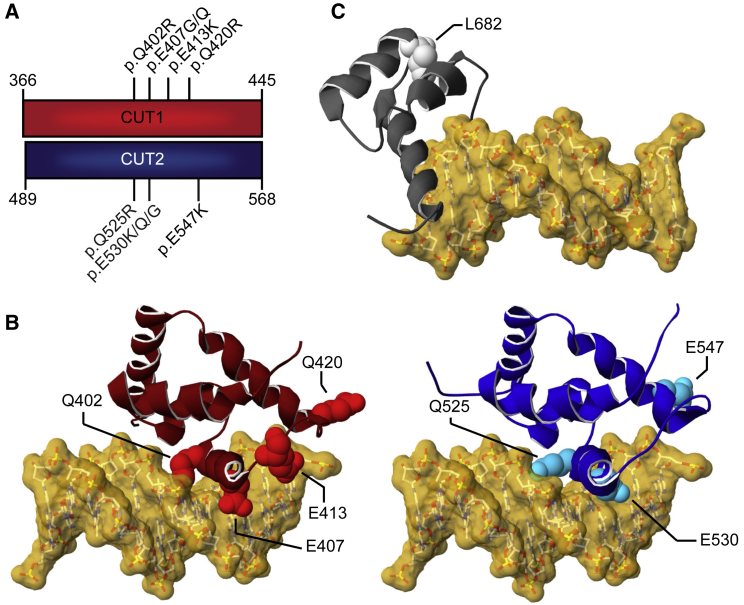
Figure 2.
3D protein modeling of SATB1 missense variants in DNA-binding domains
(A) Schematic representation of the aligned CUT1 and CUT2 DNA-binding domains. CUT1 and CUT2 domains have a high sequence identity (40%) and similarity (78%). Note that the recurrent p.Gln402Arg, p.Glu407Gly/p.Glu407Gln, and p.Gln525Arg, p.Glu530Gly/p.Glu530Lys/p.Glu530Gln variants affect equivalent positions within the respective CUT1 and CUT2 domains, while p.Gln420Arg in CUT1 and p.Glu547Lys in CUT2 affect cognate regions.
(B) 3D model of the SATB1 CUT1 domain (left; PDB: 2O4A) and CUT2 domain (right; based on PDB: 2CSF) in interaction with DNA (yellow). Mutated residues are highlighted in red for CUT1 and cyan for CUT2, along the ribbon visualization of the corresponding domains in burgundy and dark blue, respectively.
(C) 3D-homology model of the SATB1 homeobox domain (based on PDB: 1WI3 and 2D5V) in interaction with DNA (yellow). The mutated residue is shown in light gray along the ribbon visualization of the corresponding domain in dark gray.
(B and C) For more detailed descriptions of the different missense variants in our cohort, see Supplemental data.
Table 1.
Summary of clinical characteristics associated with (de novo) SATB1 variants
|
All individuals |
Individuals with PTVs and (partial) gene deletions |
Individuals with missense variants |
||||
|---|---|---|---|---|---|---|
| % | Present/total assessed | % | Present/total assessed | % | Present/total assessed | |
| Neurologic | ||||||
| Intellectual disability | 90 | 28/31 | 80 | 8/10 | 95 | 20/21 |
| Normal | 10 | 3/31 | 20 | 2/10 | 5 | 1/21 |
| Borderline | 0 | 0/31 | 0 | 0/10 | 0 | 0/21 |
| Mild | 26 | 8/31 | 60 | 6/10 | 10 | 2/21 |
| Moderate | 10 | 3/31 | 10 | 1/10 | 10 | 2/21 |
| Severe | 19 | 6/31 | 0 | 0/10 | 29 | 6/21 |
| Profound | 19 | 6/31 | 0 | 0/10 | 29 | 6/21 |
| Unspecified | 16 | 5/31 | 10 | 1/10 | 19 | 4/21 |
| Developmental delay | 97 | 35/36 | 100 | 12/12 | 96 | 23/24 |
| Motor delay | 92 | 34/37 | 92 | 11/12 | 92 | 23/25 |
| Speech delay | 89 | 32/36 | 83 | 10/12 | 92 | 22/24 |
| Dysarthria | 30 | 6/20 | 9 | 1/11 | 56 | 5/9 |
| Epilepsy | 61 | 22/36 | 18 | 2/11 | 80 | 20/25 |
| EEG abnormalities | 79 | 19/24 | 29 | 2/7 | 100 | 17/17 |
| Hypotonia | 76 | 28/37 | 42 | 5/12 | 92 | 23/25 |
| Spasticity | 28 | 10/36 | 0 | 0/12 | 42 | 10/24 |
| Ataxia | 22 | 6/27 | 17 | 2/12 | 27 | 4/15 |
| Behavioral disturbances | 71 | 24/34 | 58 | 7/12 | 77 | 17/22 |
| Sleep disturbances | 41 | 12/29 | 27 | 3/11 | 50 | 9/18 |
| Abnormal brain imaging | 55 | 17/31 | 43 | 3/7 | 58 | 14/24 |
| Regression | 17 | 6/35 | 8 | 1/12 | 22 | 5/23 |
| Growth | ||||||
| Abnormalities during pregnancy | 24 | 8/33 | 27 | 3/11 | 23 | 5/22 |
| Abnormalities during delivery | 32 | 10/31 | 55 | 6/11 | 20 | 4/20 |
| Abnormal term of delivery | 6 | 2/31 | 10 | 1/10 | 5 | 1/21 |
| Preterm (<37 weeks) | 6 | 2/31 | 10 | 1/10 | 5 | 1/21 |
| Postterm (>42 weeks) | 0 | 0/31 | 0 | 0/10 | 0 | 0/21 |
| Abnormal weight at birth | 16 | 5/32 | 22 | 2/9 | 13 | 3/23 |
| Small for gestational age (<p10) | 9 | 3/32 | 11 | 1/9 | 9 | 2/23 |
| Large for gestational age (>p90) | 6 | 2/32 | 11 | 1/9 | 4 | 1/23 |
| Abnormal head circumference at birth | 7 | 1/14 | 17 | 1/6 | 0 | 0/8 |
| Microcephaly (<p3) | 0 | 0/14 | 0 | 0/6 | 0 | 0/8 |
| Macrocephaly (>p97) | 7 | 1/14 | 17 | 1/6 | 0 | 0/8 |
| Abnormal height | 21 | 6/29 | 9 | 1/11 | 28 | 5/18 |
| Short stature (<p3) | 14 | 4/29 | 0 | 0/11 | 22 | 4/18 |
| Tall stature (>p97) | 7 | 2/29 | 9 | 1/11 | 6 | 1/18 |
| Abnormal head circumference | 23 | 7/31 | 11 | 1/9 | 27 | 6/22 |
| Microcephaly (<p3) | 23 | 7/31 | 11 | 1/9 | 27 | 6/22 |
| Macrocephaly (>p97) | 0 | 0/31 | 0 | 0/9 | 0 | 0/22 |
| Abnormal weight | 48 | 13/27 | 11 | 1/9 | 67 | 12/18 |
| Underweight (<p3) | 22 | 6/27 | 11 | 1/9 | 28 | 5/18 |
| Overweight (>p97) | 26 | 7/27 | 0 | 0/9 | 39 | 7/18 |
| Other phenotypic features | ||||||
| Facial dysmorphisms | 67 | 24/36 | 64 | 7/11 | 68 | 17/25 |
| Dental/oral abnormalities | 71 | 24/34 | 55 | 6/11 | 78 | 18/23 |
| Drooling/dysphagia | 38 | 12/32 | 25 | 3/12 | 45 | 9/20 |
| Hearing abnormalities | 7 | 2/30 | 18 | 2/11 | 0 | 0/19 |
| Vision abnormalities | 55 | 17/31 | 73 | 8/11 | 45 | 9/20 |
| Cardiac abnormalities | 19 | 6/32 | 27 | 3/11 | 14 | 3/21 |
| Skeleton/limb abnormalities | 38 | 13/34 | 18 | 2/11 | 48 | 11/23 |
| Hypermobility of joints | 30 | 8/27 | 30 | 3/10 | 29 | 5/17 |
| Gastrointestinal abnormalities | 53 | 17/32 | 27 | 3/11 | 67 | 14/21 |
| Urogenital abnormalities | 17 | 5/30 | 0 | 0/11 | 26 | 5/19 |
| Endocrine/metabolic abnormalities | 30 | 9/30 | 0 | 0/11 | 47 | 9/19 |
| Immunological abnormalities | 32 | 8/25 | 25 | 2/8 | 35 | 6/17 |
| Skin/hair/nail abnormalities | 24 | 8/34 | 9 | 1/11 | 30 | 7/23 |
| Neoplasms in medical history | 0 | 0/34 | 0 | 0/11 | 0 | 0/23 |
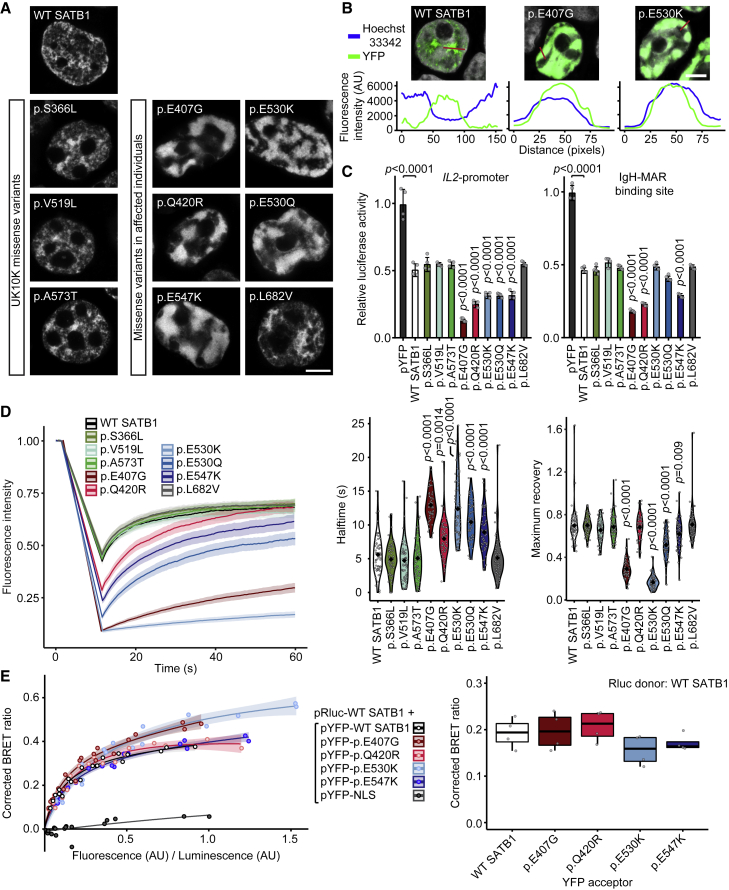
We performed functional analyses assessing consequences of different types of SATB1 variants for cellular localization, transcriptional activity, overall chromatin binding, and dimerization capacity. Based on protein modeling (Figure 2, descriptions of 3D protein modeling in supplemental information), we selected five missense variants (observed in 14 individuals) in CUT1 and CUT2 affecting residues that interact with, or are close to, the DNA backbone (mosaic variant c.1220A>G [p.Glu407Gly] and de novo variants c.1259A>G [p.Gln420Arg], c.1588G>A [p.Glu530Lys], c.1588G>C [p.Glu530Gln], c.1639G>A [p.Glu547Lys]), as well as the only homeobox domain variant (c.2044C>G [p.Leu682Val], de novo). As controls, we selected three rare missense variants from the UK10K consortium, identified in healthy individuals with a normal IQ: c.1097C>T (p.Ser366Leu) (gnomAD allele frequency 6.61e−4), c.1555G>C (p.Val519Leu) (8.67e−6), and c.1717G>A (p.Ala573Thr) (1.17e−4) (Figure 1A, Table S3).16 When overexpressed as YFP-fusion proteins in HEK293T/17 cells, wild-type SATB1 localized to the nucleus in a granular pattern, with an intensity profile inverse to the DNA-binding dye Hoechst 33342 (Figures 3A and 3B). In contrast to wildtype and UK10K control missense variants, the p.Glu407Gly, p.Gln420Arg, p.Glu530Lys/p.Glu530Gln, and p.Glu547Lys variants displayed a cage-like clustered nuclear pattern, strongly co-localizing with the DNA (Figures 3A, 3B, and S6).
Figure 3.
SATB1 missense variants stabilize DNA binding and show increased transcriptional repression
(A) Direct fluorescence super-resolution imaging of nuclei of HEK293T/17 cells expressing YFP-SATB1 fusion proteins. Scale bar = 5 μm.
(B) Intensity profiles of YFP-tagged SATB1 and variants, and the DNA binding dye Hoechst 33342. The graphs represent the fluorescence intensity values of the position of the red lines drawn in the micrographs on the top (SATB1 proteins in green, Hoechst 33342 in white, scale bar = 5 μm). For each condition a representative image and corresponding intensity profile plot is shown.
(C) Luciferase reporter assays using reporter constructs containing the IL2-promoter region and the IgH matrix associated region (MAR) binding site. UK10K control variants are shaded in green, CUT1 domain variants in red, CUT2 domain variants in blue, and the homeobox variant in gray. Values are expressed relative to the control (pYFP; black) and represent the mean ± SEM (n = 4, p values compared to wildtype SATB1 [WT; white], one-way ANOVA and post hoc Bonferroni test).
(D) FRAP experiments to assess the dynamics of SATB1 chromatin binding in live cells. Left, mean recovery curves ± 95% C.I. recorded in HEK293T/17 cells expressing YFP-SATB1 fusion proteins. Right, violin plots with median of the halftime (central panel) and maximum recovery values (right panel) based on single-term exponential curve fitting of individual recordings (n = 60 nuclei from three independent experiments, p values compared to WT SATB1, one-way ANOVA and post hoc Bonferroni test). Color code as in (C).
(E) BRET assays for SATB1 dimerization in live cells. Left, mean BRET saturation curves ± 95% C.I. fitted using a non-linear regression equation assuming a single binding site (y = BRETmax ∗x / (BRET50 / x); GraphPad). The corrected BRET ratio is plotted against the ratio of fluorescence/luminescence (AU) to correct for expression level differences between conditions. Right, corrected BRET ratio values at mean BRET50 level of WT SATB1, based on curve fitting of individual experiments (n = 4, one-way ANOVA and post hoc Bonferroni test, no significant differences). Color code as in (C).
When compared to WT YFP-SATB1 or UK10K variants, most variants identified in affected individuals show a nuclear cage-like localization (A), stronger co-localization with the DNA-binding dye Hoechst 33342 (B), increased transcriptional repression (C), reduced protein mobility (D), and unchanged capacity of interaction with WT SATB1 (E).
To assess the effects of SATB1 missense variants on transrepressive activity, we used a luciferase reporter system with two previously established downstream targets of SATB1, the IL2-promoter and IgH-MAR (matrix associated region).17, 18, 19 All five functionally assessed CUT1 and CUT2 missense variants demonstrated increased transcriptional repression of the IL2-promoter, while the UK10K control variants did not differ from wild type (Figure 3C). In assays using IgH-MAR, increased repression was seen for both CUT1 variants and for one of the CUT2 variants (Figure 3C). The latter can be explained by previous reports that the CUT1 domain is essential for binding to MARs, whereas the CUT2 domain is dispensable.20,21 Taken together, these data suggest that etiological SATB1 missense variants in CUT1 and CUT2 lead to stronger binding of the transcription factor to its targets.
To study whether SATB1 missense variants affect the dynamics of chromatin binding more globally, we employed fluorescent recovery after photobleaching (FRAP) assays. Consistent with the luciferase reporter assays, all CUT1 and CUT2 missense variants, but not the UK10K control variants, affected protein mobility in the nucleus. The CUT2 variant p.Gln420Arg demonstrated an increased halftime, but showed a maximum recovery similar to wild type, while the other CUT1 and CUT2 variants demonstrated both increased halftimes and reduced maximum recovery. These results suggest stabilization of SATB1 chromatin binding for all tested CUT1 and CUT2 variants (Figure 3D).
In contrast to the CUT1 and CUT2 missense variants, the homeobox variant p.Leu682Val did not show functional differences from wild type (Figures 3A–3D and S6), suggesting that, although it is absent from gnomAD, and the position is highly intolerant to variation and evolutionarily conserved (Figures S2, S7A, and S7B), this variant is unlikely to be pathogenic. This conclusion is further supported by the presence of a valine residue at the equivalent position in multiple homologous homeobox domains (Figure S7C). Additionally, the mild phenotypic features in this individual (individual 42) can be explained by the fact that the individual carries an out-of-frame de novo intragenic duplication of FOXP2, a gene known to cause NDD through haploinsufficiency.22
We went on to assess the impact of the CUT1 and CUT2 missense variants (p.Glu407Gly, p.Gln420Arg, p.Glu530Lys, p.Glu547Lys) on protein interaction capacities using bioluminescence resonance energy transfer (BRET). All tested variants retained the ability to interact with wildtype SATB1 (Figure 3E), with the potential to yield dominant-negative dimers/tetramers in vivo and to disturb normal activity of the wild-type protein.
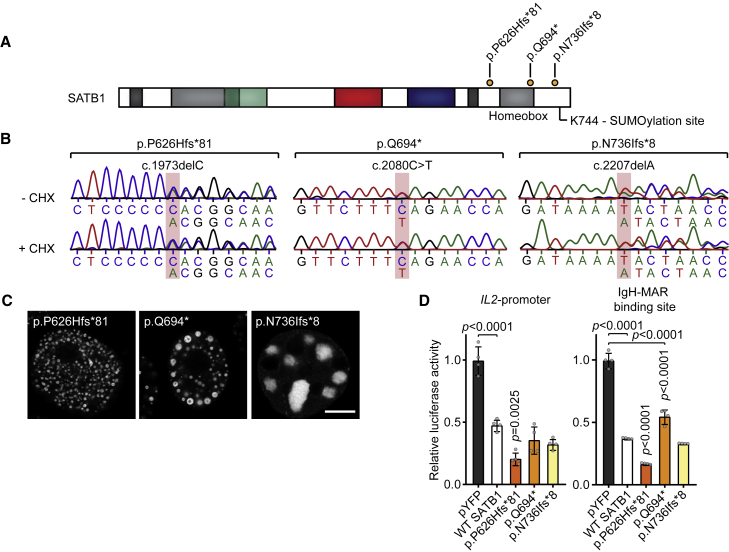
The identification of SATB1 deletions suggests that haploinsufficiency is a second underlying disease mechanism. This is supported by the constraint of SATB1 against loss-of-function variation and the identification of PTV carriers that are clinically distinct from individuals with missense variants. PTVs are found throughout the locus and several are predicted to undergo NMD by in silico models of NMD efficacy (Table S4).23 In contrast to these predictions, we found that one of the PTVs, c.1228C>T (p.Arg410∗), escapes NMD (Figures S8A and S8B). However, the p.Arg410∗ variant would lack critical functional domains (CUT1, CUT2, homeobox) and indeed showed reduced transcriptional activity in luciferase reporter assays when compared to wild-type protein (Figure S8), consistent with the haploinsufficiency model.
Four unique PTVs that we identified were located within the final exon of SATB1 (Figure 1A) and predicted to escape NMD (Table S4). Following experimental validation of NMD escape (Figures 4A and 4B), three such variants (c.1877delC [p.Pro626Hisfs∗81], c.2080C>T [p.Gln694∗], and c.2207delA [p.Asn736Ilefs∗8]) were assessed with the same functional assays that we used for missense variants. When overexpressed as YFP-fusion proteins, the tested variants showed altered subcellular localization, forming nuclear puncta or (nuclear) aggregates, different from patterns observed for missense variants (Figures 4C, S9A, and S9B). In luciferase reporter assays, the p.Pro626Hisfs∗81 variant showed increased repression of both the IL2-promoter and IgH-MAR, whereas p.Gln694∗ only showed reduced repression of IgH-MAR (Figure 4D). The p.Asn736Ilefs∗8 variant showed repression comparable to that of wild-type protein for both targets (Figure 4D). In further pursuit of pathophysiological mechanisms, we tested protein stability and SUMOylation, as the previously described Lys744 SUMOylation site is missing in all assessed NMD-escaping truncated proteins (Figure 4A).24 Our observations suggest the existence of multiple SATB1 SUMOylation sites (Figure S10) and no effect of NMD-escaping variants on SUMOylation of the encoded proteins (Figure S10) nor any changes in protein stability (Figure S9C). Although functional assays with NMD-escaping PTVs hint toward additional disease mechanisms, HPO-based phenotypic analysis or qualitative evaluation could not confirm a third distinct clinical entity (p = 0.932; Figures S5 and S11, Table S5).
Figure 4.
SATB1 protein-truncating variants in the last exon escape NMD
(A) Schematic overview of the SATB1 protein, with truncating variants predicted to escape NMD that are included in functional assays labeled in orange. A potential SUMOylation site at position Lys744 is highlighted.
(B) Sanger sequencing traces of proband-derived EBV-immortalized lymphoblastoid cell lines treated with or without cycloheximide (CHX) to test for NMD. The mutated nucleotides are shaded in red. Transcripts from both alleles are present in both conditions showing that these variants escape NMD.
(C) Direct fluorescence super-resolution imaging of nuclei of HEK293T/17 cells expressing SATB1 truncating variants fused with a YFP tag. Scale bar = 5 μm. Compared to WT YFP-SATB1 (Figure 3), NMD-escaping variants show altered localization forming nuclear puncta or aggregates.
(D) Luciferase reporter assays using reporter constructs containing the IL2-promoter and the IgH matrix associated region (MAR) binding site. Values are expressed relative to the control (pYFP; black) and represent the mean ± SEM (n = 4, p values compared to WT SATB1 [white], one-way ANOVA and post hoc Bonferroni test). All NMD-escaping variants are transcriptionally active and show repression of the IL2-promoter and IgH-MAR binding site.
Our study demonstrates that while statistical analyses5,6 can provide the first step toward identification of NDDs, a mutation-specific functional follow-up is required to gain insight into the underlying mechanisms and to understand phenotypic differences within cohorts (Table S6). Multiple mechanisms and/or more complex genotype-phenotype correlations are increasingly appreciated in newly described NDDs, such as those associated with RAC1, POL2RA, KMT2E, and PPP2CA.25, 26, 27, 28 Interestingly, although less often explored, such mechanistic complexity might also underlie well-known (clinically recognizable) NDDs. For instance, a CUT1 missense variant in SATB2, a paralog of SATB1 that causes Glass syndrome through haploinsufficiency (MIM: 612313),29 affects protein localization and nuclear mobility in a similar manner to the corresponding SATB1 missense variants (Figures S12 and S13).30 Taken together, these observations suggest that mutation-specific mechanisms await discovery both for newly described and well-established clinical syndromes.
In summary, we demonstrate that at least two different previously uncharacterized NDDs are caused by distinct classes of rare (de novo) variation at a single locus. We combined clinical investigation, in silico models, and cellular assays to characterize the phenotypic consequences and functional impacts of a large number of variants, uncovering distinct pathophysiological mechanisms of the SATB1-associated NDDs. This level of combined analyses is recommended for known and yet undiscovered NDDs to fully understand disease etiology.
Declaration of Interests
K.M., T.B.P., and T.S.-S. are employees of GeneDx, Inc. K.R. is employee of Ambrygen Genetics.
Acknowledgments
We are extremely grateful to all families participating in this study. In addition, we wish to thank the members of the Genome Technology Center and Cell culture facility, Department of Human Genetics, Radboud university medical center, Nijmegen, for data processing and cell culture of proband-derived cell lines. This work was financially supported by Aspasia grants of the Dutch Research Council (015.014.036 to T.K. and 015.014.066 to L.E.L.M.V.), Netherlands Organization for Health Research and Development (91718310 to T.K.), the Max Planck Society (J.d.H., S.E.F.), Oxford Brookes University, the Leverhulme Trust, and the British Academy (D.F.N.), and grants from the Swiss National Science Foundation (31003A_182632 to A.R.), Lithuanian-Swiss cooperation program to reduce economic and social disparities within the enlarged European Union (A.R., V. Kučinskas) and the Jérôme Lejeune Foundation (A.R.). We wish to acknowledge ALSPAC, the UK10K consortium, the 100,000 Genomes Project, “TRANSLATE NAMSE,” and Genomic Answers for Kids program (see supplemental acknowledgments). In addition, the collaborations in this study were facilitated by ERN ITHACA, one of the 24 European Reference Networks (ERNs) approved by the ERN Board of Member States, co-funded by European Commission. The aims of this study contribute to the Solve-RD project (E.d.B., H.G.B., S.B., A.-S.D.-P., L.F., C.G., A.J., T.K., A.V., L.E.L.M.V.), which has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No 779257.
Published: January 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.01.007.
Web Resources
OMIM, https://www.omim.org/
RCSB Protein Data Bank, http://www.rcsb.org/pdb/home/home.do
Supplemental information
Format (.JSON file)
References
- 1.Wang Z., Yang X., Chu X., Zhang J., Zhou H., Shen Y., Long J. The structural basis for the oligomerization of the N-terminal domain of SATB1. Nucleic Acids Res. 2012;40:4193–4202. doi: 10.1093/nar/gkr1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alvarez J.D., Yasui D.H., Niida H., Joh T., Loh D.Y., Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- 3.Cai S., Lee C.C., Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 4.Kitagawa Y., Ohkura N., Kidani Y., Vandenbon A., Hirota K., Kawakami R., Yasuda K., Motooka D., Nakamura S., Kondo M. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 2017;18:173–183. doi: 10.1038/ni.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Satterstrom F.K., Kosmicki J.A., Wang J., Breen M.S., De Rubeis S., An J.Y., Peng M., Collins R., Grove J., Klei L., Autism Sequencing Consortium. iPSYCH-Broad Consortium Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180:568–584.e23. doi: 10.1016/j.cell.2019.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplanis J., Samocha K.E., Wiel L., Zhang Z., Arvai K.J., Eberhardt R.Y., Gallone G., Lelieveld S.H., Martin H.C., McRae J.F., Deciphering Developmental Disorders Study Evidence for 28 genetic disorders discovered by combining healthcare and research data. Nature. 2020;586:757–762. doi: 10.1038/s41586-020-2832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson R., Johnston L., Taruscio D., Monaco L., Béroud C., Gut I.G., Hansson M.G., ’t Hoen P.B., Patrinos G.P., Dawkins H. RD-Connect: an integrated platform connecting databases, registries, biobanks and clinical bioinformatics for rare disease research. J. Gen. Intern. Med. 2014;29(Suppl 3):S780–S787. doi: 10.1007/s11606-014-2908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lelieveld S.H., Reijnders M.R., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 12.Lelieveld S.H., Wiel L., Venselaar H., Pfundt R., Vriend G., Veltman J.A., Brunner H.G., Vissers L.E.L.M., Gilissen C. Spatial Clustering of de Novo Missense Mutations Identifies Candidate Neurodevelopmental Disorder-Associated Genes. Am. J. Hum. Genet. 2017;101:478–484. doi: 10.1016/j.ajhg.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman, L. R.P.J. (1987). Clustering by means of medoids https://wis.kuleuven.be/stat/robust/papers/publications-1987/kaufmanrousseeuw-clusteringbymedoids-l1norm-1987.pdf.
- 14.Köhler S., Carmody L., Vasilevsky N., Jacobsen J.O.B., Danis D., Gourdine J.P., Gargano M., Harris N.L., Matentzoglu N., McMurry J.A. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47(D1):D1018–D1027. doi: 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reijnders M.R.F., Miller K.A., Alvi M., Goos J.A.C., Lees M.M., de Burca A., Henderson A., Kraus A., Mikat B., de Vries B.B.A., Deciphering Developmental Disorders Study De Novo and Inherited Loss-of-Function Variants in TLK2: Clinical and Genotype-Phenotype Evaluation of a Distinct Neurodevelopmental Disorder. Am. J. Hum. Genet. 2018;102:1195–1203. doi: 10.1016/j.ajhg.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M., Lawson D., UK10K Consortium The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pavan Kumar P., Purbey P.K., Sinha C.K., Notani D., Limaye A., Jayani R.S., Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol. Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 18.Kumar P.P., Purbey P.K., Ravi D.S., Mitra D., Galande S. Displacement of SATB1-bound histone deacetylase 1 corepressor by the human immunodeficiency virus type 1 transactivator induces expression of interleukin-2 and its receptor in T cells. Mol. Cell. Biol. 2005;25:1620–1633. doi: 10.1128/MCB.25.5.1620-1633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siebenlist U., Durand D.B., Bressler P., Holbrook N.J., Norris C.A., Kamoun M., Kant J.A., Crabtree G.R. Promoter region of interleukin-2 gene undergoes chromatin structure changes and confers inducibility on chloramphenicol acetyltransferase gene during activation of T cells. Mol. Cell. Biol. 1986;6:3042–3049. doi: 10.1128/mcb.6.9.3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh R.P., Shi Q., Yang L., Reddick M.P., Nikitina T., Zhurkin V.B., Fordyce P., Stasevich T.J., Chang H.Y., Greenleaf W.J., Liphardt J.T. Satb1 integrates DNA binding site geometry and torsional stress to differentially target nucleosome-dense regions. Nat. Commun. 2019;10:3221. doi: 10.1038/s41467-019-11118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson L.A., Dickinson C.D., Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J. Biol. Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 22.MacDermot K.D., Bonora E., Sykes N., Coupe A.M., Lai C.S., Vernes S.C., Vargha-Khadem F., McKenzie F., Smith R.L., Monaco A.P., Fisher S.E. Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. Am. J. Hum. Genet. 2005;76:1074–1080. doi: 10.1086/430841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindeboom R.G.H., Vermeulen M., Lehner B., Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan J.A., Sun Y., Song J., Chen Y., Krontiris T.G., Durrin L.K. SUMO conjugation to the matrix attachment region-binding protein, special AT-rich sequence-binding protein-1 (SATB1), targets SATB1 to promyelocytic nuclear bodies where it undergoes caspase cleavage. J. Biol. Chem. 2008;283:18124–18134. doi: 10.1074/jbc.M800512200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haijes H.A., Koster M.J.E., Rehmann H., Li D., Hakonarson H., Cappuccio G., Hancarova M., Lehalle D., Reardon W., Schaefer G.B. De Novo Heterozygous POLR2A Variants Cause a Neurodevelopmental Syndrome with Profound Infantile-Onset Hypotonia. Am. J. Hum. Genet. 2019;105:283–301. doi: 10.1016/j.ajhg.2019.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell-Luria A.H., Pais L.S., Faundes V., Wood J.C., Sveden A., Luria V., Abou Jamra R., Accogli A., Amburgey K., Anderlid B.M., Deciphering Developmental Disorders (DDD) Study Heterozygous Variants in KMT2E Cause a Spectrum of Neurodevelopmental Disorders and Epilepsy. Am. J. Hum. Genet. 2019;104:1210–1222. doi: 10.1016/j.ajhg.2019.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynhout S., Jansen S., Haesen D., van Belle S., de Munnik S.A., Bongers E.M.H.F., Schieving J.H., Marcelis C., Amiel J., Rio M. De Novo Mutations Affecting the Catalytic Cα Subunit of PP2A, PPP2CA, Cause Syndromic Intellectual Disability Resembling Other PP2A-Related Neurodevelopmental Disorders. Am. J. Hum. Genet. 2019;104:139–156. doi: 10.1016/j.ajhg.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reijnders M.R.F., Ansor N.M., Kousi M., Yue W.W., Tan P.L., Clarkson K., Clayton-Smith J., Corning K., Jones J.R., Lam W.W.K., Deciphering Developmental Disorders Study RAC1 Missense Mutations in Developmental Disorders with Diverse Phenotypes. Am. J. Hum. Genet. 2017;101:466–477. doi: 10.1016/j.ajhg.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarate Y.A., Bosanko K.A., Caffrey A.R., Bernstein J.A., Martin D.M., Williams M.S., Berry-Kravis E.M., Mark P.R., Manning M.A., Bhambhani V. Mutation update for the SATB2 gene. Hum. Mutat. 2019;40:1013–1029. doi: 10.1002/humu.23771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J.S., Yoo Y., Lim B.C., Kim K.J., Choi M., Chae J.H. SATB2-associated syndrome presenting with Rett-like phenotypes. Clin. Genet. 2016;89:728–732. doi: 10.1111/cge.12698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Format (.JSON file)