Summary
Focal segmental glomerulosclerosis (FSGS) is the main pathology underlying steroid-resistant nephrotic syndrome (SRNS) and a leading cause of chronic kidney disease. Monogenic forms of pediatric SRNS are predominantly caused by recessive mutations, while the contribution of de novo variants (DNVs) to this trait is poorly understood. Using exome sequencing (ES) in a proband with FSGS/SRNS, developmental delay, and epilepsy, we discovered a nonsense DNV in TRIM8, which encodes the E3 ubiquitin ligase tripartite motif containing 8. To establish whether TRIM8 variants represent a cause of FSGS, we aggregated exome/genome-sequencing data for 2,501 pediatric FSGS/SRNS-affected individuals and 48,556 control subjects, detecting eight heterozygous TRIM8 truncating variants in affected subjects but none in control subjects (p = 3.28 × 10−11). In all six cases with available parental DNA, we demonstrated de novo inheritance (p = 2.21 × 10−15). Reverse phenotyping revealed neurodevelopmental disease in all eight families. We next analyzed ES from 9,067 individuals with epilepsy, yielding three additional families with truncating TRIM8 variants. Clinical review revealed FSGS in all. All TRIM8 variants cause protein truncation clustering within the last exon between residues 390 and 487 of the 551 amino acid protein, indicating a correlation between this syndrome and loss of the TRIM8 C-terminal region. Wild-type TRIM8 overexpressed in immortalized human podocytes and neuronal cells localized to nuclear bodies, while constructs harboring patient-specific variants mislocalized diffusely to the nucleoplasm. Co-localization studies demonstrated that Gemini and Cajal bodies frequently abut a TRIM8 nuclear body. Truncating TRIM8 DNVs cause a neuro-renal syndrome via aberrant TRIM8 localization, implicating nuclear bodies in FSGS and developmental brain disease.
Keywords: TRIM8, genomics, monogenic, epilepsy, FSGS, SRNS, nuclear body
Main Text
Nephrotic syndrome (NS [MIM: PS256300]) is the second leading cause of chronic kidney disease in the first three decades of life.1 Children with NS manifest with edema and severe proteinuria, which arise from podocyte foot process effacement within the glomerular filtration barrier.2,3 Patients with steroid-resistant nephrotic syndrome (SRNS) invariably progress to end-stage renal disease (ESRD) and frequently have focal segmental glomerulosclerosis (FSGS [MIM: PS603278]) on renal biopsy.1,2,4,5 In 11%–45% of children with FSGS/SRNS, pathogenic variants can be identified in one of 59 known monogenic disease genes.6, 7, 8, 9, 10, 11, 12 While these Mendelian etiologies predominantly exhibit recessive modes of inheritance in 44/59 disease genes or dominant vertical inheritance with incomplete penetrance in much of the remainder,9 the contribution of de novo variants (DNVs) to NS is mostly unknown.
Monogenic NS genes are predominantly expressed in the glomerular podocyte.2,3,11,13 Their encoded proteins coalesce into molecular pathways, which are essential for podocyte development or homeostasis and impaired by patient mutations.2, 3, 4,11,13 Nucleoporins have been implicated in genetic forms of NS,14, 15, 16, 17, 18 thus suggesting that nuclear maintenance and homeostasis are key in the pathobiology of nephrotic syndrome. Nuclear bodies, which are defined sites within the nucleus for biochemical reactions and gene regulation, have not been linked to NS to date.19, 20, 21, 22
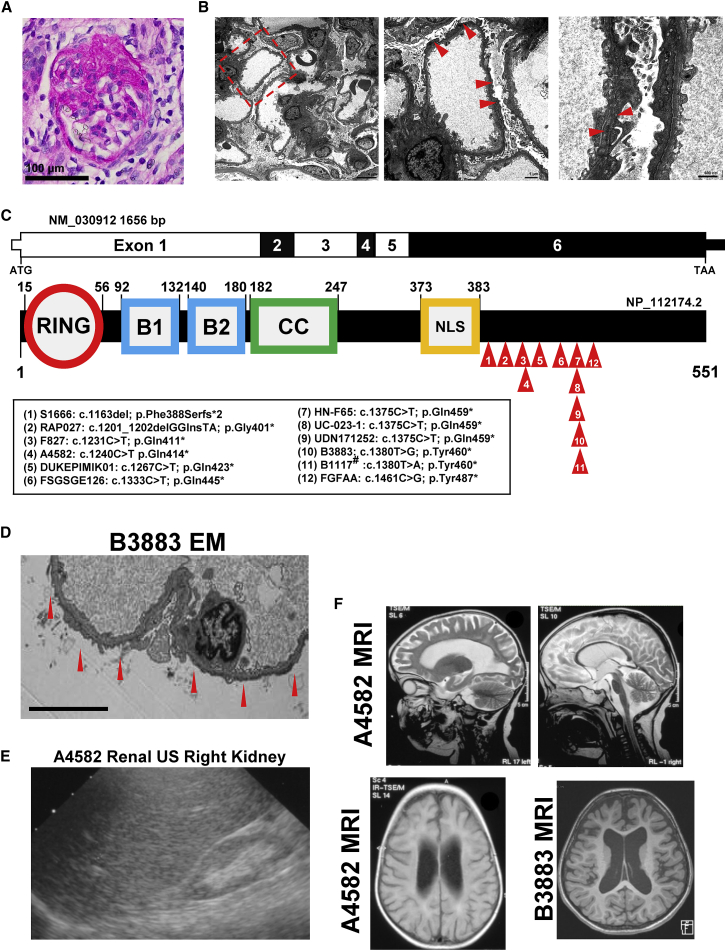
We started from the observation of an individual with a rarely observed clinical presentation. The index subject was a male child (case UC-023-1) of Asian ancestry with syndromic NS (Tables 1 and S1; Figures 1A–1C). He developed seizures at 2 years of life. At age 4 years, he developed NS that was resistant to steroid therapy. Renal biopsy revealed FSGS by light microscopy and electron microscopy (Figures 1A and 1B). Notably, electron microscopy showed extensive foot process effacement and distinctive alterations of the glomerular basement membrane (GBM), including thinning, lamellation, and blebbing (Figure 1B).
Table 1.
Heterozygous truncating variants in TRIM8 in 12 families with nephrotic syndrome and neurologic disease
| Nucleotide changea | Amino acid changea | Exon (seg) | Sex | Ethnic origin | Renal disease onset (y) | ESRD onset (y) | Renal disease | DD onset (y) | Epilepsy onset (y) | Neuro disease | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nephrotic syndrome | |||||||||||
| S1666 | c.1163delT | p.Phe388Serfs∗2 | 6 (MW) | F | European | 2.2 | 3 | SRNS; Bx FSGS; Tx Age 7.5; Tx Rej | 2.2 | 4.5 | epilepsy, GDD, mild CC atrophy |
| F827 | c.1231C>T | p.Gln411∗ | 6 (DN) | M | Turkish | 4.5 | 4.8 | SRNS; Bx FSGS; Tx Age 5; No Rej | 1 | 2.5 | epilepsy, GDD, cerebral atrophy |
| A4582 | c.1240C>T | p.Gln414∗ | 6 (DN) | F | German | Birth | 1.1 | SRNS; Bx DMS; Tx Age 4.9; No Rej | <1 | 2 | epilepsy, GDD, cerebral atrophy, hypotonia |
| FSGSGE126 | c.1333C>T | p.Gln445∗ | 6 (DN) | F | Italian | 13.7 | 19.7 | SRNS; Bx FSGS | 1.5 | 1.5 | epilepsy, posterior fossa dilatation and cerebral atrophy, spastic dystonic quadriplegia |
| UC-023-1 (Index) | c.1375C>T | p.Gln459∗ | 6 (DN) | M | Korean | 4 | 5 | SRNS; Bx FSGS; Tx Age 7; No Rej | <1 | 2 | epilepsy, GDD, autism |
| HN-F65 | c.1375C>T | p.Gln459∗ | 6 (DN) | M | German | 7.9 | 9.8 (CKD3) | HTN; NRP; SR; Bx FSGS | 2.5 | 2.5 | epilepsy, GDD, secondary microcephaly |
| B1117b | c.1380T>A | p.Tyr460∗ | 6 (DN#) | M | Hispanic | 2.5 | >5 | SRNS; Bx FSGS | <1 | 2.5 | epilepsy, GDD |
| B3883 | c.1380T>G | p.Tyr460∗ | 6 (ND) | F | Middle Eastern | 6 | 8 (CKD2) | NRP; Bx FSGS | 0.5 | 1.5 | epilepsy, GDD, CCB atrophy, hypotonia |
| FG-FAc | c.1461C>G | p.Tyr487∗ | 6 (DN) | M | Irish/Hispanic | 6 | 14 | SRNS; Bx FSGS; Tx Age 14; No Rej | 1.5 | NA | mild GDD, Tourette’s syndrome-like symptoms, autism spectrum |
| Epilepsy | |||||||||||
| RAP027 | c.1201_1202delGGInsTA | p.Gly401∗ | 6 (DN) | F | European/South Asian | 3 | 5 | SRNS; Bx DMS | 1 | 3 | GDD, seizures, mild CC atrophy |
| DUKEPIMIK01 | c.1267C>T | p.Gln423∗ | 6 (DN) | M | African American | 11 | 12 | NS; Bx FSGS; Tx Age 12.5 | 0.5 | 2 | epilepsy, GDD |
| UDN171252 | c.1375C>T | p.Gln459∗ | 6 (DN) | F | European | 3 | 5 | NS; Bx FSGS | <1 | 2.5 | epilepsy, GDD, mesial temporal sclerosis |
Abbreviations: Bx, biopsy; CC, cerebral and cerebellar; CCB, cerebral, cerebellar and brainstem; CKD2, chronic kidney disease stage 2; CKD3, chronic kidney disease stage 3; DD, developmental delay; DMS, diffuse mesangial sclerosis; DN, de novo; ESRD, end-stage renal disease; F, female; FSGS, focal segmental glomerulosclerosis; GDD, global developmental delay; HTN, hypertension; M, male; MW, mother wild-type but paternal DNA not available; NA, not applicable; ND, no data; NRP, nephrotic-range proteinuria; NS, nephrotic syndrome; Rej, rejection; Seg, segregation; SR, steroid resistant; SRNS, steroid-resistant nephrotic syndrome; Tx, transplant; y, years.
Position change corresponds to TRIM8 transcript GenBank: NM_030912 and TRIM8 protein GenBank: NP_112174.2.
This patient was independently recruited and reported23 while the current manuscript was in preparation.
FG-FA corresponds to a pair of monozygotic twins (genetically confirmed) who share the same TRIM8 variant and a nearly identical phenotype.
Figure 1.
Truncating TRIM8 mutations identified in 12 families with nephrotic syndrome and neurologic disease
(A) Periodic acid Schiff (PAS) staining of paraffin sections of kidney cortex from a renal biopsy of the index case subject UC-023-1 was performed. A representative glomerulus shows a lesion of focal segmental glomerulosclerosis with segmental obliteration of capillary lumina by extracellular matrix accompanied by loss of overlying podocytes and broad adhesions to Bowman’s capsule (PAS, scale bar: 100 μm).
(B) Ultrastructural analysis of kidney tissue from a renal biopsy of the index case UC-023-1 was performed by transmission electron microscopy. A low-power view (left, 2,500×) of the glomerular capillaries shows patent lumina and unremarkable mesangium. There is nearly complete (>90%) effacement of the podocyte foot processes with microvillous transformation of the podocyte cytoplasm. A higher-power view (middle, 6,300×) shows a glomerular capillary with thinning and irregularities of the glomerular basement membrane as well as marked foot process effacement with microvillous change. There are no immune-type electron dense deposits. In the highest-power view (right, 16,000×), two adjacent glomerular capillaries show thinning (124–131 nm) of their glomerular basement membranes accompanied by textural irregularities with focal mild lamellation (arrowhead). Scale bars: 4 μm in left, 1 μm in middle, 600 nm in right.
(C) Coding exon (upper bar) and protein domain (lower bar) structures of TRIM8 are shown with arrows indicating position of heterozygous mutations identified in nine subjects. B1, B-box domain 1; B2, B-box domain 2; CC, coiled-coil domain; NLS, nuclear localization signal; RING, ring finger domain. #This patient was independently recruited and published, while the current manuscript was in preparation.
(D) Renal biopsy was performed in subject B3883 at age 8 years. Ultrastructural findings by electron microscopy (EM) at 3,000× include diffuse podocyte foot process effacement (red arrows) and thin glomerular basement membranes. Scale bar: 6 μm.
(E) Renal ultrasound (RUS) is shown for subject A4582 at 2.75 years, demonstrating a hyperechogenic and atrophied (4.68 cm) right kidney after progression to ESRD.
(F) Brain magnetic resonance imaging is shown for subject A4582 at 2.75 years and B3883 at 6 years of life. T2 transverse images of A4582 show deep sulci, atrophied gyri, and thin corpus callosum (top left) and atrophy of corpus callosum, pons and midbrain, while cerebellum preserved (top right). T2 flare axial image of A4582 (bottom left) shows atrophied white matter and hyperintense periventricular signal intensity. T1 axial image of B3883 shows increased ventricle size and sulcal spaces.
To identify the genetic cause of NS and, potentially, his epileptic encephalopathy, we performed parent-proband trio-based exome sequencing (ES). Evaluation of ES variant data under both dominant and recessive models did not reveal a causative variant in any gene from a curated list of 625 genes known to be associated to Mendelian kidney diseases,24 which include 59 monogenic NS disease genes9 and 20 glomerulonephritis disease genes.9 This suggested that the syndromic phenotype of this individual was likely caused by a variant in a gene not previously implicated in known Mendelian forms of kidney disease. Under a dominant de novo Mendelian genetic hypothesis, we identified a heterozygous nonsense variant (c.1375C>T [p.Gln459∗]) in TRIM8 (MIM: 606125) in subject UC-023-1, while his unaffected parents were wild type at this position (Table 1; Figures 1C and S1). The results were confirmed using CLIA-certified genome sequencing (GS), which also excluded pathogenic structural variants. TRIM8 encodes a 551 amino acid tripartite E3 ubiquitin ligase, which regulates NF-κB, interferon, and STAT signaling pathways in cancer and the immune system25, 26, 27, 28, 29, 30 (Figure 1C). The variant was deemed deleterious and likely to be causal because (1) the variant is predicted to cause protein truncation, because the resulting mRNA transcript is likely to escape nonsense-mediated decay as it resides in the last exon; (2) it is exceedingly rare and, in fact, absent from the Genome Aggregation Database (gnomAD) population, which contains ES or GS data from 141,456 adults who are not known to have severe pediatric disease;31,32 and (3) the TRIM8 locus exhibits high loss-of-function (LoF) intolerance (gnomAD probability for loss-of-function intolerance [pLI] 0.99; loss-of-function observed/expected upper bound fraction [LOEUF] 0.26), meaning LoF alleles are not frequently observed relative to gene size and expected mutation rate.31
To determine whether TRIM8 DNVs are a monogenic cause of NS and establish whether NS and developmental delay/epilepsy are allelic in these cases, we aggregated and analyzed ES or GS from a cohort of 60,114 subjects, including 2,501 SRNS/FSGS-affected individuals, 48,556 control subjects, and 9,057 individuals with a broad diagnosis of epilepsy. First, we queried for truncating TRIM8 variants in ES/GS data from 2,501 pediatric nephrosis subjects (defined as SRNS or nephrotic-range proteinuria with biopsy-proven FSGS) and from 48,556 control subjects (Table 2). The control subjects were children or adults without NS or related kidney disease, as determined by a clinical nephrologist and/or available clinical data.33 We identified heterozygous truncating TRIM8 variants in eight unrelated probands but no variants in control subjects, demonstrating statistically significant overrepresentation in affected individuals (two-sided Fisher’s exact p = 3.28 × 10−11; OR = inf, CI 33.2-inf) and, thereby, establishing TRIM8 truncating variants as a monogenic cause of NS (Tables 1 and 2; Figures 1C and S1). Accordingly, all TRIM8 variants were absent from the gnomAD (N = 141,456 individuals). Furthermore, variant data from all cases did not reveal pathogenic variants in known NS disease genes. Importantly, segregation analysis confirmed de novo inheritance of TRIM8 variants in all six families with available parental DNA (p = 2.21 × 10−15) (Table 1, Figure S1). Post hoc review of the clinical data demonstrated that all eight NS-affected individuals also had presented with developmental delay and seven of eight developed epilepsy (Tables 1, S1), strongly indicating that the renal and neurodevelopmental phenotypes are not coincidental but indeed caused by pathogenic variants in the same gene.
Table 2.
Heterozygous truncating TRIM8 variants discovered in 2,501 FSGS/SRNS-affected individuals and 48,556 control subjects
| Cohort | TRIM8+NS subjects (n) | Total pediatric NS subjects (n) | TRIM8+control subjects (n) | Total control subjects (n) | OR (95% CI) | p value |
|---|---|---|---|---|---|---|
| CUIMC | 1 | 369 | 0 | 19,533 | Inf (1.36 - Inf) | 1.85 × 10−2 |
| BCH | 4 | 1,382 | 0 | 660 | Inf (0.31 - Inf) | 3.16 × 10−1 |
| BIDMC | 1 | 220 | 0 | 363 | Inf (0.04 – inf) | 3.77 × 10−1 |
| TUMG | 1 | 89 | 0 | 15,000 | Inf (4.37 - Inf) | 5.83 × 10−3 |
| NephroS | 1 | 441 | 0 | 13,000a | Inf (0.77 - Inf) | 3.21 × 10−2 |
| Total | 8 | 2,501 | 0 | 48,556 | Inf (33.20 - Inf) | 3.28 × 10−11 |
Abbreviations: BCH, Boston Children’s Hospital; CI, confidence interval; BIDMC, Beth Israel Deaconess Medical Center; CUIMC, Columbia University Irving Medical Center; Inf, infinity; N, number; NephroS, National Study of Nephrotic Syndrome; NS, nephrotic syndrome; TUMG, Technical University Munich in Germany; WES, whole exome sequencing; WGS, whole genome sequencing.
Control subjects from WGS of UK Biobank participants.33
To confirm this hypothesis, we analyzed ES data for TRIM8 truncating variants from 9,057 individuals with a broad diagnosis of epilepsy, aggregated from multiple investigators and consortia and hosted at the genetic warehouse at the Institute for Genomic Medicine (IGM) at Columbia University (see supplemental material and methods). We identified three additional subjects with heterozygous de novo truncating TRIM8 variants and with complete clinical data available to interrogate for kidney disease (Tables 1 and 2; Figures 1B and S1). Strikingly, clinical chart review and reverse phenotyping revealed that nephrotic syndrome arose in all three individuals after the diagnosis of epilepsy/developmental delay (Tables 1 and S1). Altogether, these findings establish that de novo truncating TRIM8 variants are the genetic basis of a neuro-renal syndrome in children characterized by early-onset developmental delay with epileptic encephalopathy and FSGS and distinctive glomerular basement membrane alterations.
Phenotypic evaluation of all 12 individuals revealed a common natural history in individuals with TRIM8 variants. They all exhibited early-onset developmental delay (onset <1 to 2.5 years) and 11/12 developed epilepsy (onset <1 to 4.5 years) (Tables 1 and S1). All individuals developed NS with variable age of onset (onset birth to 13.7 years) but generally delayed compared to the onset of neurologic disease (Tables 1 and S1). Only one monozygotic twin pair did not develop epilepsy (FG-FA) and had a relatively mild developmental brain phenotype. Brain atrophy was observed in six of eight individuals with available imaging (Tables 1 and S1; Figure 1F). SRNS developed in 8 of 12 subjects (Tables 1 and S1). FSGS was observed in 10 of 12 biopsies, whereas diffuse mesangial sclerosis was noted in RAP027 and in A4582 (Tables 1 and S1; Figures 1D and 1E). Five subjects underwent renal transplantation; follow-up studies showed that none of them developed clinical or pathological FSGS recurrence with up to 24 years of follow up, and one individual (S1666) developed rejection at 8 years (Tables 1 and S1).
These specific phenotypes suggested a strong correlation with patient genotypes. All 12 TRIM8 variants cause truncation between amino acids 390 and 487, which is C-terminal to a nuclear localization signal (NLS) (Table 1, Figure 1C). Because all variants reside within the last exon, the mRNA transcripts are predicted to escape nonsense-mediated decay.34,35 In addition, we interrogated TRIM8 LoF variants in gnomAD and identified 7 heterozygous truncating variants out of 141,456 subjects. These gnomAD variants were identified in subjects ranging from ages 30 to 75 years and are spatially distinct from TRIM8 subject variants, causing truncation prior to the NLS or farther C-terminal than the cluster of variants we identified (amino acids 390–487) (Figure S3). This supports a cluster effect for TRIM8 variants, in which earlier or later TRIM8 truncating events are less likely to cause the full neuro-renal syndrome and suggests that TRIM8 subject variants do not cause disease through haploinsufficiency.
Furthermore, we observed the same recurrent variant (c.1375C>T [p.Gln459∗]) in three independent subjects, who were confirmed to be unrelated by genome-wide analysis for hidden relatedness and principal component analysis for ancestry. Two additional affected individuals harbor p.Tyr460∗ variants, arising from two different nucleotide changes at the same position (Table 1). Recurrent de novo truncating variants affecting the same nucleotide are exceedingly rare events. The observation of three of such variants (in the case of c.1375C>T [p.Gln459∗]) would have a probability below 0.94 × 10−18, even if we assume we had conducted ES in trios from all 60,114 subjects studied here. This further supports a hotspot effect for TRIM8 DNVs causing the clinical syndrome of FSGS and developmental brain disease.
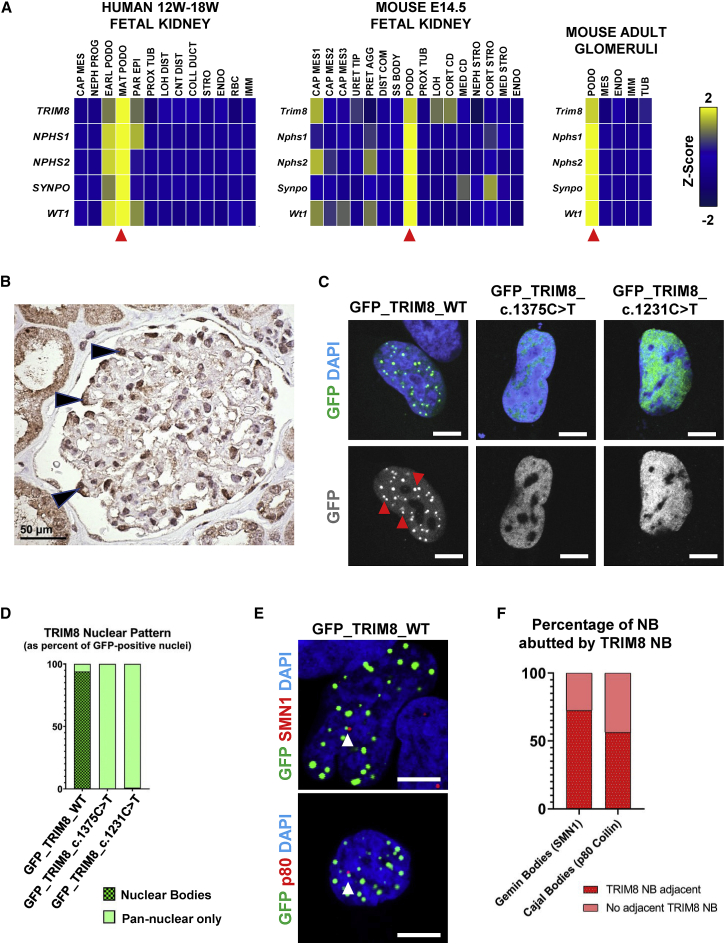
To assess the biological role of TRIM8 in renal disease, we interrogated published single-cell RNA sequencing (scRNA-seq) datasets for its endogenous expression in mammalian kidneys. Consistent with prior literature,27 we observed that TRIM8/Trim8 mRNA is predominantly expressed in podocytes in scRNA-seq data from two independent human fetal kidney studies,36,37 mouse fetal kidney,38 and adult mouse glomeruli39 (Figures 2A, S4A, and S4B). By immunohistochemistry, we determined that TRIM8 protein localized to the renal cortex and, particularly, glomerular podocytes of adult human kidneys (Figure 2B). Of note, we also observed that TRIM8 staining localized to discrete nuclear foci, especially within proximal tubular epithelium (Figure S4C). Overall, these findings indicate that TRIM8 is expressed in podocytes, suggesting its variants may cause a primary podocytopathy.
Figure 2.
TRIM8 is expressed in podocytes of mammalian kidneys and localizes to nuclear bodies
(A) TRIM8 mRNA (z-score) was predominantly expressed by podocytes (red arrows) from single-cell mRNA sequencing data.26,28,29 In human fetal kidneys from 12 to 19 weeks gestation (left), TRIM8 expression was highest in the mature podocyte cluster, marked by expression of NPHS1, NPHS2, SYNPO, and WT1, relative to other developmental cell-type and nephron segment clusters. Trim8 expression in E14.5 mouse kidneys (middle) was, similarly, highest in the podocyte cluster (Nphs1, Nphs2, Synpo, Wt1) with lower expression in one cap mesenchyme cluster and two tubular segment clusters. In adult mouse glomeruli (right), Trim8 mRNA was predominantly expressed in podocytes relative to other clusters. CAP MES, cap mesenchyme; CNT DIST, distal connecting tubule; COLL DUCT, collecting duct; CORT CD, cortical collecting duct; CORT STRO, cortical stroma; DIST COM, distal comma shaped body; EARL PODO, early podocyte; ENDO, endothelium; IMM, immune cells; LOH, Loop of Henle; LOH DIST, distal Loop of Henle; MAT PODO, mature podocyte; MED CD, medullary collecting duct; MED STRO, medullary stroma; NEPH PROG, nephron progenitor; NEPH STRO, nephrogenic stroma; PAR EPI, parietal epithelial cell; PODO, podocyte; PRET AGG, pretubular aggregate; PROX TUB; proximal tubule; RBC, red blood cells; SS Body, mid S-shaped body; STRO, stroma; TUB, tubule; URET TIP, ureteric tip.
(B) Immunohistochemistry of adult human kidney tissue demonstrates immunoreactivity for TRIM8 within the nuclei, as well as the cytoplasm, of individual glomerular podocytes (arrowheads). There is also staining of the adjacent tubular epithelial cells. Scale bar: 50 μm.
(C) A human podocyte cell line was transfected with N-terminal GFP-tagged wild-type TRIM8 or TRIM8 mutant constructs based on NS patient variants c.1375C>T and c.1231C>T. Cells were imaged by confocal microscopy. Representative images of GFP-tagged protein and DAPI localization are shown, revealing that wild-type TRIM8 localizes to nuclear bodies (NBs) (red arrows) while patient mutants exhibit pan-nuclear staining overlapping with DAPI signal. Scale bars: 7 μm.
(D) The presence of TRIM8 NBs in (C) is determined from the sum of three independent experiments, as the percentage of 100 GFP-positive cells with NB localization versus those with pan-nuclear staining. Wild-type TRIM8 localized to NBs in 94% of GFP-positive cells (94/100). In contrast, mutated TRIM8 localized diffusely to the nucleoplasm in 100% (100/100) and 99% (99/100) of cells for the c.1375C>T and c.1231C>T constructs, respectively.
(E) A human podocyte cell line was transfected with N-terminal GFP-tagged wild-type TRIM8. IF and confocal microscopy was performed for endogenous Gemini body marker SMN1 (top) or Cajal body marker p80 Coilin (p80, bottom), demonstrating these bodies frequently abut a GFP-TRIM8 NB (white arrow). Representative images are shown. Scale bars: 7 μm.
(F) The percentage of endogenous SMN1-positive Gemini bodies or p80 Coilin-positive Cajal bodies that are abutted by a GFP-TRIM8 nuclear body, as in (E), is determined from the sum of three independent experiments. 72.5% of Gemini bodies (58/80 NB in 38 transfected cells) and 56.4% of Cajal bodies (57/101 NB in 21 transfected cells) abutted a TRIM8 NB.
To assess the molecular consequence of TRIM8 case-associated variants on its protein localization, we performed transient overexpression of N-terminally GFP-tagged cDNA constructs modeling the wild-type allele and two independent TRIM8 variants (p.Gln411∗ and p.Gln459∗) in an immortalized human podocyte cell line. These two variants are representative of our described allelic series, which truncate the C terminus of the protein. This region is not well conserved across human TRIM paralogs in contrast to the N-terminal region, which contains the well-conserved tripartite domains (Figure S5). Immunofluorescence (IF) and confocal microscopy showed that wild-type TRIM8 localized to discrete nuclear bodies (NBs) in 94% of transfected nuclei (Figures 2C, 2D, and S6), consistent with the literature.27 Expression of MYC-tagged wild-type TRIM8 demonstrated similar localization to nuclear bodies (Figure S8A). In contrast, TRIM8 bearing the case-associated variants p.Gln411∗ or p.Gln459∗ mis-localized diffusely to the nucleoplasm in transfected cells (Figures 2C, 2D, and S6). This pattern of localization of GFP-tagged wild-type and case-associated variant TRIM8 constructs was confirmed in immortalized mouse podocytes and the neural BE(2)-M17 cell line (Figures S8B and S8C). To determine the identity of TRIM8 NBs, we co-stained for established markers of Gemin bodies, Cajal bodies, PML bodies, cleavage bodies, or nuclear speckles but did not observe co-localization with wild-type TRIM8. However, we found that 72.5% of endogenous SMN1-positive Gemini bodies and 56.4% of p80 Coilin-positive Cajal bodies abutted a TRIM8 NB by confocal microscopy (Figures 2E and 2F). These findings suggest that TRIM8 can aggregate into NB that are likely disrupted by variants that truncate in a critical region from amino acid 390–487.
It still remained unclear whether TRIM8 variants cause disease by haploinsufficiency, dominant-negative, gain-of-function, or toxic effects. To explore this question indirectly, we simultaneously targeted TRIM8 orthologs in zebrafish (79% and 78% similarity to trim8a [GenBank: NP_001265783.1] and trim8b [GenBank: NP_001135847.1], respectively) via CRISPR-Cas9 single-guide RNAs (sgRNAs) that target N-terminal regions likely to induce nonsense-mediated decay (exon 5 and 3, respectively; Figure S7A). After determining high targeting efficiency using heteroduplex analysis in F0 mosaic mutants (∼100%; Figures S7B–S7E), we simultaneously disrupted both genes and assessed (1) gross morphology indicative of renal function impairment, which include pericardial and periorbital edema and (2) glomerular filtration defects using dextran-FITC conjugates according to our established procedures.40 Double F0 mosaic mutants showed no significant lethality and no gross morphological anomalies (n = 30–45 larvae/experiment, repeated twice) and did not display any detectable increase in proteinuria when compared to gRNA alone injected larval batches (n = 21–25 larvae/condition, repeated; Figures S7F and S7G). These findings argue against the possibility that TRIM8 variants cause epilepsy and nephrotic syndrome through loss-of-function mechanisms (haploinsufficiency or dominant-negative effects) and support our genetic and cell-based studies that implicate gain-of-function or toxic effects.
In summary, beginning with the observation of a rare clinical presentation, we show that de novo truncating variants in TRIM8 cause a neuro-renal syndrome through the analysis of genomic sequencing of 60,114 subjects from world-wide cohorts (Table 2). We detected a genotype-phenotype correlation between C-terminally truncating TRIM8 variants and syndromic nephrotic syndrome, in which variants clustered within a restricted region of the TRIM8 C terminus (Figure 1C, Table 1). At the cellular level, we found that case-associated variants disrupt TRIM8 localization to nuclear bodies, which might associate proximally to Gemini and Cajal bodies. Our combined in vitro data suggest nuclear sub-compartments as candidates in NS pathogenesis.
TRIM8 variants cause a unique subgroup of syndromic nephrotic syndrome. All subjects were diagnosed with renal disease after (10/12) or at the time of (2/12) the development of neurologic disease. Previous case reports have described truncating DNVs in TRIM8 in a total of eight children with developmental delay and epilepsy, but only three of these subjects were described to have nephrotic syndrome (Figure S2),23,41, 42, 43 which left it unsolved whether the renal disease was associated to TRIM8 variants or just coincidental. The early ascertainment of these cases is also consistent with our observation that renal disease presents later in the course of these patients and may not have been apparent yet at the time of these reports. This underscores the importance of establishing a molecular genetic diagnosis in TRIM8 affected subjects with neurologic manifestations and should prompt clinicians to screen immediately for features of nephrosis.
While 44 of 59 known NS disease genes exhibit recessive modes of inheritance9 and the remainder 15 mostly cause dominantly inherited forms of FSGS with age of onset later in adolescence or adulthood, the contribution of de novo variants to pediatric NS is poorly understood. This may reflect ascertainment bias due to the genetic approaches employed previously to discover these etiologies, mostly in consanguineous families. Our approach of combining trio-ES/GS with large cohort analysis to validate TRIM8 DNVs offers a useful framework to be applied for SRNS-affected individuals lacking a molecular diagnosis to potentially discover causative de novo variants and/or support a rare genetic finding. This is especially critical as de novo variants are increasing in prevalence in society in association with advanced paternal age.44, 45, 46
TRIM8 disease-associated variants are restricted to the last exon (Figures 1C and S2).23,41, 42, 43 Combining our findings with previous reports showed that four individuals from distinct ethnic backgrounds share the identical variant at position c.1375 and two share nonsense variants at position c.1380 (Figure S2).23,41 This observation suggests that this region is a recurrent germline variant hotspot (Figure S2, Table 1), as has been demonstrated for other rare dominant monogenic causes of syndromic epilepsy47 and nephrotic syndrome.48
The mechanisms by which TRIM8 variants cause disease are likely complex. While LoF intolerance metrics for TRIM8 support haploinsufficiency, the presence of adult gnomAD subjects with earlier truncating events (Figure S3) do not support this genetic mechanism for this severe neuro-renal syndrome caused by C-terminal TRIM8 variants. Moreover, these metrics are not always definitive, as there are other examples of de novo variants causing disease through a gain-of-function mechanism despite strong LoF intolerance at the locus.49 Alternatively, TRIM8 variants may cause dominant-negative loss-of-function of the wild-type allele, given that TRIM8 can dimerize through its coiled-coil domain27 and that we observe that C-terminal case-associated variants cause TRIM8 mis-localization (Figures 2C, 2D, and S6). However, the absence of a phenotype in trim8a/trim8b knockout models supports the hypothesis that human variants cause disease through gain-of-function or toxic effects, as do the absence of reported neurologic and renal phenotypes in other TRIM8 loss-of-function zebrafish30 and mouse29,50 models. Still, caution is warranted in translating such observations from zebrafish to human because there are established compensatory mechanisms for truncating variants in Danio rerio that may confound these results.51 Further studies employing knock-in models and mammalian organisms are warranted to delineate the precise mechanism of human TRIM8 variants.
Based on our findings (Figures 2C–2F, S4C, and S6), TRIM8 may play a critical role in nuclear bodies that are abrogated by disease-associated variants. The precise identity of these NB is not known, although Gemini and Cajal bodies can associate proximally to them (Figures 2E and 2F) and have broad functions in snRNP biogenesis, histone mRNA processing, and telomere maintenance.19, 20, 21, 22 Furthermore, our findings suggest that the C-terminal TRIM8 variants may impair the normal condensation of TRIM8 protein into liquid droplets, although it remains unclear how the C-terminal region regulates liquid-liquid phase separation,52, 53, 54 thus posing intriguing questions and opening a new avenue of research in renal diseases. It will be critical to understand the cellular role of endogenous TRIM8 in these NBs and how its functions are impacted by disease-associated variants introduced in podocytes.
In conclusion, we provide insight into the genetic and molecular etiology of a neuro-renal syndrome caused by a cluster of de novo variants in TRIM8, which reveal a yet unexplored mechanism of cellular pathogenesis in SRNS, FSGS, and developmental brain disease.
Data and Code Availability
The datasets supporting the current study have not been deposited in a public repository due to restriction by patient consent but are available from the corresponding author on request.
Declaration of Interests
F.H. is a co-founder of Goldfinch Biopharma Inc. The other authors declare that they have no competing financial interests. No part of this manuscript has been previously published.
Acknowledgments
F.H. is the William E. Harmon Professor of Pediatrics. This research is supported by grants from the NIH to F.H. (DK-076683-13) and to S.S.-C., F.H., and M.R.P. (RC2-DK122397); from the DoD to S.S.-C. (GRANT12019690); and by the NCATS to S.S.-C. (UL1-TR001873). A.J.M. was supported by grants from the NIH (T32DK-007726, 5K12HD052896-13), ASN Lipps Research Program PKD Foundation Fellowship and Foundation for Kidney Research (FP01025169), and by Boston Children’s Hospital Manton Center for Orphan Disease Research. K.K. was funded by an International Research Support Initiative Program fellowship from the Higher Education Commission of Pakistan. Sequencing by the Broad Institute of MIT and Harvard Center for Mendelian Genomics (Broad CMG) was funded by the NHGRI, the NEI, and the NHLBI grant UM1-HG008900 and, partly, NHGRI (R01-HG009141). CUIMC cohort sequencing was partly supported by AstraZeneca Centre for Genomics Research, Precision Medicine and Genomics, IMED Biotech Unit, Cambridge, United Kingdom. The Yale CMG (UM1-HG006504) is funded by the NHGRI. The GSP Coordinating Center (U24-HG008956) provided logistical and general study coordination. Research reported in this manuscript was supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH (U01-HG007672 and U01-HG007942). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Sequencing for studies by M.A.S. was funded by the NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London, London, UK. M.A.S is supported by a Medical Research Council, Precision Medicine award.
Published: January 27, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ajhg.2021.01.008.
Contributor Information
Friedhelm Hildebrandt, Email: friedhelm.hildebrandt@childrens.harvard.edu.
Simone Sanna-Cherchi, Email: ss2517@cumc.columbia.edu.
Web Resources
CHOPCHOP, https://chopchop.cbu.uib.no
Ensembl Genome Browser, http://www.ensembl.org/index.html
gnomAD Browser, https://gnomad.broadinstitute.org/
Kidney Interactive Transcriptomics, https://humphreyslab.com/SingleCell/
OMIM, https://www.omim.org/
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental information
References
- 1.Harmon W. 2008. NAPRTCS 2008 Annual Report.https://www.naprtcs.org/system/files/2008_Annual_CKD_Report.pdf [Google Scholar]
- 2.Wiggins R.-C. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71:1205–1214. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 3.Somlo S., Mundel P. Getting a foothold in nephrotic syndrome. Nat. Genet. 2000;24:333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 4.Wharram B.L., Goyal M., Wiggins J.E., Sanden S.K., Hussain S., Filipiak W.E., Saunders T.L., Dysko R.C., Kohno K., Holzman L.B., Wiggins R.C. Podocyte depletion causes glomerulosclerosis: diphtheria toxin-induced podocyte depletion in rats expressing human diphtheria toxin receptor transgene. J. Am. Soc. Nephrol. 2005;16:2941–2952. doi: 10.1681/ASN.2005010055. [DOI] [PubMed] [Google Scholar]
- 5.Trautmann A., Schnaidt S., Lipska-Ziętkiewicz B.S., Bodria M., Ozaltin F., Emma F., Anarat A., Melk A., Azocar M., Oh J., PodoNet Consortium Long-Term Outcome of Steroid-Resistant Nephrotic Syndrome in Children. J. Am. Soc. Nephrol. 2017;28:3055–3065. doi: 10.1681/ASN.2016101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sadowski C.E., Lovric S., Ashraf S., Pabst W.L., Gee H.Y., Kohl S., Engelmann S., Vega-Warner V., Fang H., Halbritter J., SRNS Study Group A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J. Am. Soc. Nephrol. 2015;26:1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warejko J.K., Tan W., Daga A., Schapiro D., Lawson J.A., Shril S., Lovric S., Ashraf S., Rao J., Hermle T. Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome. Clin. J. Am. Soc. Nephrol. 2018;13:53–62. doi: 10.2215/CJN.04120417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W., Lovric S., Ashraf S., Rao J., Schapiro D., Airik M., Shril S., Gee H.Y., Baum M., Daouk G. Analysis of 24 genes reveals a monogenic cause in 11.1% of cases with steroid-resistant nephrotic syndrome at a single center. Pediatr. Nephrol. 2018;33:305–314. doi: 10.1007/s00467-017-3801-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connaughton D.M., Kennedy C., Shril S., Mann N., Murray S.L., Williams P.A., Conlon E., Nakayama M., van der Ven A.T., Ityel H. Monogenic causes of chronic kidney disease in adults. Kidney Int. 2019;95:914–928. doi: 10.1016/j.kint.2018.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tryggvason K., Patrakka J., Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N. Engl. J. Med. 2006;354:1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 11.Lovric S., Ashraf S., Tan W., Hildebrandt F. Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol. Dial. Transplant. 2016;31:1802–1813. doi: 10.1093/ndt/gfv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vivante A., Hildebrandt F. Exploring the genetic basis of early-onset chronic kidney disease. Nat. Rev. Nephrol. 2016;12:133–146. doi: 10.1038/nrneph.2015.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J., Shrestha R., Qiu C., Kondo A., Huang S., Werth M., Li M., Barasch J., Suszták K. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science. 2018;360:758–763. doi: 10.1126/science.aar2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun D.A., Lovric S., Schapiro D., Schneider R., Marquez J., Asif M., Hussain M.S., Daga A., Widmeier E., Rao J. Mutations in multiple components of the nuclear pore complex cause nephrotic syndrome. J. Clin. Invest. 2018;128:4313–4328. doi: 10.1172/JCI98688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Braun D.A., Sadowski C.E., Kohl S., Lovric S., Astrinidis S.A., Pabst W.L., Gee H.Y., Ashraf S., Lawson J.A., Shril S. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat. Genet. 2016;48:457–465. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyake N., Tsukaguchi H., Koshimizu E., Shono A., Matsunaga S., Shiina M., Mimura Y., Imamura S., Hirose T., Okudela K. Biallelic Mutations in Nuclear Pore Complex Subunit NUP107 Cause Early-Childhood-Onset Steroid-Resistant Nephrotic Syndrome. Am. J. Hum. Genet. 2015;97:555–566. doi: 10.1016/j.ajhg.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosti R.O., Sotak B.N., Bielas S.L., Bhat G., Silhavy J.L., Aslanger A.D., Altunoglu U., Bilge I., Tasdemir M., Yzaguirrem A.D. Homozygous mutation in NUP107 leads to microcephaly with steroid-resistant nephrotic condition similar to Galloway-Mowat syndrome. J. Med. Genet. 2017;54:399–403. doi: 10.1136/jmedgenet-2016-104237. [DOI] [PubMed] [Google Scholar]
- 18.Zhao F., Zhu J.Y., Richman A., Fu Y., Huang W., Chen N., Pan X., Yi C., Ding X., Wang S. Mutations in NUP160 Are Implicated in Steroid-Resistant Nephrotic Syndrome. J. Am. Soc. Nephrol. 2019;30:840–853. doi: 10.1681/ASN.2018080786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Staněk D., Fox A.H. Nuclear bodies: news insights into structure and function. Curr. Opin. Cell Biol. 2017;46:94–101. doi: 10.1016/j.ceb.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Zimber A., Nguyen Q.-D., Gespach C. Nuclear bodies and compartments: functional roles and cellular signalling in health and disease. Cell. Signal. 2004;16:1085–1104. doi: 10.1016/j.cellsig.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 21.Mao Y.S., Zhang B., Spector D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011;27:295–306. doi: 10.1016/j.tig.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer I.A., Dundr M. Nuclear bodies: Built to boost. J. Cell Biol. 2016;213:509–511. doi: 10.1083/jcb.201605049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warren M., Takeda M., Partikian A., Opas L., Fine R., Yano S. Association of a de novo nonsense mutation of the TRIM8 gene with childhood-onset focal segmental glomerulosclerosis. Pediatr. Nephrol. 2020;35:1129–1132. doi: 10.1007/s00467-020-04525-3. [DOI] [PubMed] [Google Scholar]
- 24.Groopman E.E., Marasa M., Cameron-Christie S., Petrovski S., Aggarwal V.S., Milo-Rasouly H., Li Y., Zhang J., Nestor J., Krithivasan P. Diagnostic Utility of Exome Sequencing for Kidney Disease. N. Engl. J. Med. 2019;380:142–151. doi: 10.1056/NEJMoa1806891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q., Yan J., Mao A.P., Li C., Ran Y., Shu H.B., Wang Y.Y. Tripartite motif 8 (TRIM8) modulates TNFα- and IL-1β-triggered NF-κB activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. USA. 2011;108:19341–19346. doi: 10.1073/pnas.1110946108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okumura F., Matsunaga Y., Katayama Y., Nakayama K.I., Hatakeyama S. TRIM8 modulates STAT3 activity through negative regulation of PIAS3. J. Cell Sci. 2010;123:2238–2245. doi: 10.1242/jcs.068981. [DOI] [PubMed] [Google Scholar]
- 27.Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomar D., Sripada L., Prajapati P., Singh R., Singh A.K., Singh R. Nucleo-cytoplasmic trafficking of TRIM8, a novel oncogene, is involved in positive regulation of TNF induced NF-κB pathway. PLoS ONE. 2012;7:e48662. doi: 10.1371/journal.pone.0048662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye W., Hu M.M., Lei C.Q., Zhou Q., Lin H., Sun M.S., Shu H.B. TRIM8 Negatively Regulates TLR3/4-Mediated Innate Immune Response by Blocking TRIF-TBK1 Interaction. J. Immunol. 2017;199:1856–1864. doi: 10.4049/jimmunol.1601647. [DOI] [PubMed] [Google Scholar]
- 30.Maarifi G., Smith N., Maillet S., Moncorgé O., Chamontin C., Edouard J., Sohm F., Blanchet F.P., Herbeuval J.P., Lutfalla G. TRIM8 is required for virus-induced IFN response in human plasmacytoid dendritic cells. Sci. Adv. 2019;5:x3511. doi: 10.1126/sciadv.aax3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1101/531210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turro E., Astle W.J., Megy K., Gräf S., Greene D., Shamardina O., Allen H.L., Sanchis-Juan A., Frontini M., Thys C., NIHR BioResource for the 100,000 Genomes Project Whole-genome sequencing of patients with rare diseases in a national health system. Nature. 2020;583:96–102. doi: 10.1038/s41586-020-2434-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J., Sun X., Qian Y., Maquat L.E. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang J., Sun X., Qian Y., LaDuca J.P., Maquat L.E. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol. Cell. Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menon R., Otto E.A., Kokoruda A., Zhou J., Zhang Z., Yoon E., Chen Y.C., Troyanskaya O., Spence J.R., Kretzler M., Cebrián C. Single-cell analysis of progenitor cell dynamics and lineage specification in the human fetal kidney. Development. 2018;145:dev164038. doi: 10.1242/dev.164038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lindström N.O., De Sena Brandine G., Tran T., Ransick A., Suh G., Guo J., Kim A.D., Parvez R.K., Ruffins S.W., Rutledge E.A. Progressive Recruitment of Mesenchymal Progenitors Reveals a Time-Dependent Process of Cell Fate Acquisition in Mouse and Human Nephrogenesis. Dev. Cell. 2018;45:651–660.e4. doi: 10.1016/j.devcel.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magella B., Adam M., Potter A.S., Venkatasubramanian M., Chetal K., Hay S.B., Salomonis N., Potter S.S. Cross-platform single cell analysis of kidney development shows stromal cells express Gdnf. Dev. Biol. 2018;434:36–47. doi: 10.1016/j.ydbio.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karaiskos N., Rahmatollahi M., Boltengagen A., Liu H., Hoehne M., Rinschen M., Schermer B., Benzing T., Rajewsky N., Kocks C. A Single-Cell Transcriptome Atlas of the Mouse Glomerulus. J. Am. Soc. Nephrol. 2018;29:2060–2068. doi: 10.1681/ASN.2018030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson B.R., Howell D.N., Soldano K., Garrett M.E., Katsanis N., Telen M.J., Davis E.E., Ashley-Koch A.E. In vivo Modeling Implicates APOL1 in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress. PLoS Genet. 2015;11:e1005349. doi: 10.1371/journal.pgen.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Assoum M., Lines M.A., Elpeleg O., Darmency V., Whiting S., Edvardson S., Devinsky O., Heinzen E., Hernan R.R., Antignac C. Further delineation of the clinical spectrum of de novo TRIM8 truncating mutations. Am. J. Med. Genet. A. 2018;176:2470–2478. doi: 10.1002/ajmg.a.40357. [DOI] [PubMed] [Google Scholar]
- 42.Sakai Y., Fukai R., Matsushita Y., Miyake N., Saitsu H., Akamine S., Torio M., Sasazuki M., Ishizaki Y., Sanefuji M. De Novo Truncating Mutation of TRIM8 Causes Early-Onset Epileptic Encephalopathy. Ann. Hum. Genet. 2016;80:235–240. doi: 10.1111/ahg.12157. [DOI] [PubMed] [Google Scholar]
- 43.McClatchey M.A., du Toit Z.D., Vaughan R., Whatley S.D., Martins S., Hegde S., Naude J.T.W., Thomas D.H., Griffiths D.F., Clarke A.J., Fry A.E., Genomics England Research Consortium Focal segmental glomerulosclerosis and mild intellectual disability in a patient with a novel de novo truncating TRIM8 mutation. Eur. J. Med. Genet. 2020;63:103972. doi: 10.1016/j.ejmg.2020.103972. [DOI] [PubMed] [Google Scholar]
- 44.Khandwala Y.S., Zhang C.A., Lu Y., Eisenberg M.L. The age of fathers in the USA is rising: an analysis of 168 867 480 births from 1972 to 2015. Hum. Reprod. 2017;32:2110–2116. doi: 10.1093/humrep/dex267. [DOI] [PubMed] [Google Scholar]
- 45.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., Gudjonsson S.A., Sigurdsson A., Jonasdottir A., Jonasdottir A. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayo J.A., Lu Y., Stevenson D.K., Shaw G.M., Eisenberg M.L. Parental age and stillbirth: a population-based cohort of nearly 10 million California deliveries from 1991 to 2011. Ann. Epidemiol. 2019;31:32–37.e2. doi: 10.1016/j.annepidem.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Schoch K., Meng L., Szelinger S., Bearden D.R., Stray-Pedersen A., Busk O.L., Stong N., Liston E., Cohn R.D., Scaglia F., UCLA Clinical Genomics Center. Undiagnosed Diseases Network A Recurrent De Novo Variant in NACC1 Causes a Syndrome Characterized by Infantile Epilepsy, Cataracts, and Profound Developmental Delay. Am. J. Hum. Genet. 2017;100:343–351. doi: 10.1016/j.ajhg.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barbaux S., Niaudet P., Gubler M.C., Grünfeld J.P., Jaubert F., Kuttenn F., Fékété C.N., Souleyreau-Therville N., Thibaud E., Fellous M., McElreavey K. Donor splice-site mutations in WT1 are responsible for Frasier syndrome. Nat. Genet. 1997;17:467–470. doi: 10.1038/ng1297-467. [DOI] [PubMed] [Google Scholar]
- 49.Miyake N., Takahashi H., Nakamura K., Isidor B., Hiraki Y., Koshimizu E., Shiina M., Sasaki K., Suzuki H., Abe R. Gain-of-Function MN1 Truncation Variants Cause a Recognizable Syndrome with Craniofacial and Brain Abnormalities. Am. J. Hum. Genet. 2020;106:13–25. doi: 10.1016/j.ajhg.2019.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koscielny G., Yaikhom G., Iyer V., Meehan T.F., Morgan H., Atienza-Herrero J., Blake A., Chen C.-K., Easty R., Di Fenza A. The International Mouse Phenotyping Consortium Web Portal, a unified point of access for knockout mice and related phenotyping data. Nucleic Acids Res. 2014;42:D802–D809. doi: 10.1093/nar/gkt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma Z., Zhu P., Shi H., Guo L., Zhang Q., Chen Y., Chen S., Zhang Z., Peng J., Chen J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019;568:259–263. doi: 10.1038/s41586-019-1057-y. [DOI] [PubMed] [Google Scholar]
- 52.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 53.Alberti S., Gladfelter A., Mittag T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strom A.R., Brangwynne C.P. The liquid nucleome - phase transitions in the nucleus at a glance. J. Cell Sci. 2019;132:jcs235093. doi: 10.1242/jcs.235093. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the current study have not been deposited in a public repository due to restriction by patient consent but are available from the corresponding author on request.