Abstract
Molecular chaperone Hsp70 plays important roles in the pathology of amyloid diseases by inhibiting aberrant aggregation of proteins. However, the biophysical mechanism of the interaction of Hsp70 with the intrinsically disordered proteins (IDPs) is unclear. Here, we report that Hsp70 inhibits aggregation of islet amyloid polypeptide (IAPP) at substoichiometric concentrations under diverse solution conditions, including in the absence of ATP. The inhibitory effect is strongest if Hsp70 is added in the beginning of aggregation but progressively less if added later, indicating a role for Hsp70 in preventing nucleation of IAPP. However, ensemble measurement of the binding affinity suggests poor interactions between Hsp70 and IAPP. Therefore, we hypothesize that the interaction must involve a rare species (e.g., the oligomeric intermediates of IAPP). Size exclusion chromatography and field flow fractionation are then used to fractionate the constituent species. Multiangle light scattering and fluorescence correlation spectroscopy measurements indicate that the dominant fraction in size exclusion chromatography contains a few nanomolar Hsp70-IAPP complexes amid several μmoles of free Hsp70. Using single-particle two-color coincidence detection measurements, we detected a minor fraction that exhibits fluorescence bursts arising from heterogeneous oligomeric complexes of IAPP and Hsp70. Taken together, our results indicate that Hsp70 interacts poorly with the monomers but strongly with oligomers of IAPP. This is likely a generic feature of the interactions of Hsp70 chaperones with the amyloidogenic IDPs. Whereas high-affinity interactions with the oligomers prevent aberrant aggregation, poor interaction with the monomers averts interference with the physiological functions of the IDPs.
Significance
Molecular chaperone Hsp70 plays key role in delaying and inhibiting aberrant aggregation of proteins in amyloid diseases. Here, we investigate the effect of Hsp70 on the aggregation of islet amyloid polypeptide (IAPP), which is involved in the pathology of type 2 diabetes. Low substoichiometric amount of Hsp70 is found to dramatically delay aggregation of IAPP, even without the help of the cochaperones and ATP. Application of a combination of ensemble and single-molecule biophysical techniques unravels that Hsp70 interacts poorly with the monomers of IAPP but strongly with the oligomers, which are found to be rare and heterogeneous. These observations suggest a generic mechanism for how chaperones may inhibit amyloid formation without disturbing the physiological functions of the amyloidogenic intrinsically disordered proteins.
Introduction
Pathology of type 2 diabetes mellitus (T2DM) is characterized by dysfunction of pancreatic β-cells, progressive decline in insulin secretion, and loss of the β-cell mass. Etiology of β-cell damage is multifactorial (1); however, growing evidence suggests that amyloid aggregation of the islet amyloid polypeptide (IAPP) is a major cause of death of the β-cells, particularly at the late stage of T2DM (2,3). IAPP, a 37-residue peptide hormone, is coexpressed and cosecreted with insulin from the pancreatic β-cells. The triggers for aggregation of IAPP in the pancreas are not clear, but several different mechanisms, such as defects in IAPP biosynthesis, folding, trafficking, and/or processing in response to insulin resistance and inflammation, have been proposed to be responsible (4,5). In vitro, IAPP aggregates into soluble oligomers and insoluble fibrils at micromolar concentrations (6). Aggregates of IAPP, particularly the soluble oligomers, are considered to be the major toxic species affecting cellular integrity (7, 8, 9).
Heat shock protein Hsp70 is a central component of the cellular chaperone system that plays essential roles in protein homeostasis under normal and stress conditions by preventing protein misfolding and aggregation (10,11) and protecting the cells from the cytotoxicity caused by amyloid aggregation (12). Expression of Hsp70 is found to be lower in T2DM in the insulin sensitive tissues (13,14). Furthermore, overexpression of Hsp70 in obese mice delays or prevents the diabetes-associated symptoms such as inflammation and insulin resistance (15). Chaperone complementation studies, either by overexpression or by exogenous addition of Hsp70, were shown to minimize the cytotoxic effects of the amyloids in model organisms of T2DM and other amyloid diseases (16,17). Furthermore, Hsp70 is found codeposited with IAPP in the islet amyloids in T2DM (18). Therefore, elevating the levels of Hsp70 and also improving its specificity for IAPP are being proposed as therapeutic strategies in T2DM (16,19,20).
In vitro, Hsp70 along with the cochaperones Hsp40 and the nucleotide exchange factor (NEF) have been reported to inhibit aggregation of amyloid-β, α-synuclein, polyglutamine polypeptides, τ protein, and IAPP (21, 22, 23, 24, 25). Furthermore, the Hsp70 machinery can also cause disaggregation of the soluble oligomers and/or the insoluble fibrils in the presence of ATP (26). Hsp70 is hypothesized to act as a “foldase” in an ATP-dependent catalytic cycle that involves binding to the exposed hydrophobic patches of the “misfolded” or the “aggregation-competent” conformers of the globular proteins, refolding it, then releasing it in the “nonamyloidogenic” monomeric forms (12,27). However, it is unclear how Hsp70 interacts with the intrinsically disordered proteins (IDPs) and how this interaction is regulated. Biochemical studies have shown ATP-independent inhibition of amyloid aggregation by Hsp70 and several other chaperone proteins (28,29). For example, Chien et al. have reported that Hsp70 delays aggregation of IAPP at substoichiometric (1:200) concentrations, even in absence of ATP (25). Therefore, inhibitory action of Hsp70 does not necessarily require the catalytic activity, suggesting a “holdase” model of action of Hsp70. However, the substoichiometric nature of the interaction indicates that Hsp70 must interact with one or more on-pathway intermediates of IAPP. However, biophysical characterization of these intermediates are challenging because of its low abundance and transient and heterogeneous nature (30). Furthermore, Hsp70 is also known to exist as monomers, dimers, and higher oligomers (31, 32, 33, 34). Hence, little is known about the properties of the Hsp70/IAPP complexes. We think that investigations using single-molecule techniques can help to characterize these complexes, overcoming the limitations imposed by the heterogeneity and the transient nature of the interacting species.
Here, we report that Hsp70 chaperone proteins of diverse origins, such as the human Hsc70 (UniProt: P11142), Escherichia coli DnaK (UniProt: P0A6Y8), and Mycobacterium tuberculosis Hsp70 (MTB-Hsp70; UniProt: P9WMJ9), inhibit amyloid aggregation of human IAPP under diverse solution conditions via “holdase” activity. We have characterized the Hsp70/IAPP complexes using fluorescence-based single-molecule techniques. Because Hsc70 contains four native cysteine residues, site-specific labeling of this protein with fluorophores is difficult. Therefore, we have used fluorescently labeled DnaK, which contains only one cysteine residue, for characterization of Hsp70-IAPP complexes. Our results indicate that binding of Hsp70 to the prenucleation oligomers causes strong inhibition of the amyloid aggregation of IAPP.
Materials and Methods
All the chemicals, unless mentioned otherwise were procured from Sigma-Aldrich (St. Louis, MO). Tetramethylrhodamine (TMR)-maleimide and EDANS-maleimide were purchased from Thermo Fisher Scientific (Waltham, MA).
Chemical synthesis of IAPP
IAPP was chemically synthesized at 0.1-mmol scale using an automated peptide synthesizer (aapptec, Louisville, KY) following a protocol described earlier by Marek et al. (35). Crude IAPP was dissolved in 30 mM acetic acid. The disulfide bond between the cysteines at positions 2 and 7 was induced using hydrogen peroxide. The peptide was stored in the form of lyophilized powder at −20°C. The lyophilized powder was dissolved in 10 mM phosphate buffer (pH 7.4) containing 4 M guanidine hydrochloride (GdnCl). This solution was purified by size exclusion chromatography (SEC) using a Superdex Peptide column (10/300; GE Healthcare, Dornstadt, Germany) in 20 mM phosphate buffer (pH 7.4) containing 4 M GdnCl. Only the monomeric fraction was collected. Finally, a PD-10 Desalting Column (GE Healthcare, Dornstadt, Germany) was used to remove the GdnCl. The stock solution was aliquoted into 100-μL vials, flash frozen, and stored at −80°C. The mass and purity of the peptide was verified by mass spectrometry (Fig. S1). Labeling of IAPP with TMR was performed on the resin after deprotection of the N-terminus of IAPP using 20% piperidine. The resin was washed extensively with dimethylformamide followed by dichloromethane and methanol to remove free TMR. The peptide was then cleaved from the resin and purified using the procedure mentioned above.
Expression and purification of E. coli DnaK, DnaJ, MTB-Hsp70, and human Hsc70
Plasmids for E. coli DnaK, DnaJ, Mycobacterium tuberculosis Hsp70 and human Hsc70 are generous gifts from Dr. Shekhar Mande (National Centre for Cell Science, Pune, India), Dr. Soumit Sankar Mandal (Indian Institute of Science Education and Research, Tirupati, India), Prof. Lila Gierasch (University of Massachusetts, Amhearst, MA), and Prof. Lewis Kay (University of Toronto, Toronto, Canada), respectively. Expression and purification of the proteins was performed using the published protocols (36, 37, 38, 39). The DnaK protein was purified first with ion exchange chromatography using 0–1 M NaCl in a HiTrap column (GE Healthcare, Dornstadt, Germany). The primary fraction of DnaK was eluted in 150 mM NaCl. DnaJ, Hsc70, and MTB-Hsp70 were purified by Ni-NTA column chromatography (HisTrap; GE Healthcare) using standard protocol. All the Hsp proteins were finally purified by SEC using a Superdex 200 column (GE Healthcare, Dornstadt, Germany) in a phosphate-buffered saline (PBS) (pH 7.4) buffer containing 1 mM EDTA and 1 mM sodium azide. EDTA is added to enhance stability of the proteins, preventing degradation by the trace amounts of metalloproteases that might be present in the solutions. The fraction containing monomeric Hsp70 was collected, aliquoted into 50-μL vials, flash frozen, and stored at −80°C. The purity of the proteins was examined by SDS-PAGE (Fig. S2).
The lone cysteine residue (at position 15) of DnaK was used for labeling with the fluorophores, such as EDANS-maleimide, TMR-maleimide and Atto425-maleimide using standard maleimide labeling protocol (40).
Measurement of kinetics of aggregation of IAPP
A 100-μM stock solution of IAPP was diluted to 20 μM into PBS (pH 7.4) containing 1 mM EDTA, 2 mM sodium azide, and 4 μM Thioflavin T (ThT) into a round-bottom glass test tube (OD = 10 mm) placed inside a temperature-controlled cell holder in the spectrofluorometer (PTI QuantaMaster; Horiba, Kyoto, Japan). The solution was stirred continuously using a Teflon coated stirrer (3 mm × 6 mm) bead at ∼300 rpm at 37°C. Fluorescence of ThT (λex = 438 nm and λem = 480 nm) was monitored continuously. The duration of the “lag” phase is measured by noting the point at which the tangent to the early growth phase of the curve intersects the time axis. To characterize the effects of Hsp70 on various stages of aggregation, 160 nM Hsc70 or 200 nM DnaK was added in the beginning (t = 0 h), early “lag” (t = 1 h), late in the “lag” (t ∼ 2 h), and in the growth phase (t ∼ 2.4 h) of aggregation of 20 μM IAPP. Aggregation of TMR-IAPP (2 μM) was monitored using λem = 600 nm with λex = 520 nm (41). The kinetics of interaction between EDANS-DnaK and TMR-IAPP was monitored using intermolecular Förster resonance energy transfer (FRET). Fluorescence of EDANS was monitored using λex = 350 nm and λem = 480 nm. FRET efficiency was calculated using the following: FRET efficiency E = 1 − FDA/FD, where FDA is donor emission obtained in presence of the acceptor and FD is donor fluorescence in the absence of acceptor (40). FD was obtained from the fluorescence of EDANS-DnaK monitored in presence of 2 μM unlabeled IAPP.
For the measurement of binding using fluorescence anisotropy, 400 nM TMR-IAPP was incubated with different concentrations of DnaK (0–5 μM) in PBS buffer (pH 7.4) containing 1 mM EDTA and 2 mM sodium azide. The instrument “g” factor is calculated using free TMR-IAPP.
Atomic force microscopy imaging
A 10 μL of IAPP/DnaK (20 μM/0.5 μM) solution was collected from the end of the experiment involving the kinetics of aggregation described above. The solution was adsorbed onto a freshly cleaved mica sheet for ∼2 min. The surface of the mica was then washed gently with Milli-Q water and dried overnight. The sample was imaged using atomic force microscopy (AFM; AFMWorkshop, Signal Hill, CA) in the tapping mode.
Fluorescence correlation spectroscopy measurements
The fluorescence correlation spectroscopy (FCS) experiments were performed using a homebuilt cuvette-FCS setup described elsewhere (42). We used two excitation lasers: a solid-state laser with λ = 450 nm (Thorlabs, Newton, NJ) and a He-Ne laser with λ = 543 nm (Newport, Irvine CA). For measurement of binding of TMR-IAPP and DnaK, TMR-IAPP (400 nM) solution was incubated with 0–5 μM DnaK in PBS buffer (pH 7.4) in presence of 1 mM EDTA and 1 mM NaN3 at room temperature (RT). The autocorrelation data were analyzed using a single-component diffusion model. The visibly distorted autocorrelation curves occurring because of the appearance of photon bursts are removed from the analysis. The diffusion time obtained was used to calculate the hydrodynamic radii (Rh) (43). Calibration of the FCS observation volume was performed using free rhodamine B (42).
Burst analysis
Photon count traces obtained from the FCS measurements of 100 nM TMR-DnaK in presence or absence of 20 μM IAPP were recorded with a bin time of 100 μs. First, the photon counting histograms (PCH) are calculated from raw photon count data collected over 300 s. Number of photons (ΔN) belonging to the bursts observed in the TMR-DnaK plus IAPP sample is estimated by comparing the PCH (i.e., ΔN = Ʃn × [P2(n) − P1(n)]). Here, P1(n) and P2(n) are number of events recording “n” photons in the bin time from the TMR-DnaK and the TMR-DnaK + IAPP samples, respectively. Because mean photon count rates are nearly same in both the samples, P2(n) − P1(n) is negative for small “n” but positive for larger “n.” Because we are interested in the contributions from the bursts the range of “n” in the above, summation is chosen such that P2(n) > P1(n). The fraction of photons corresponding to bursts is estimated as ΔN/N, where N = Ʃn × P2(n) for all “n.” Quantitative analysis of the photon bursts, particularly in presence of a larger concentration of the monomer background, is quite complex (44). Hence, we performed an approximate estimation. First, the PCH data were fitted using a Gaussian to determine the baseline and the standard deviation (SD), although the appropriate function to fit the PCH data is quite complex (45). Then the threshold for burst detection is set at 3 × SD above the baseline. The basis for choice of such a threshold value is that it yields nearly “zero” bursts per minute in the control sample (i.e., 100 nM TMR-DnaK in absence of IAPP (Fig. S3)). The number of bursts, total photon counts (n) per burst, and the temporal size (τ) of the bursts are then calculated from the raw photon count data using Mathematica. The brightness of each burst is calculated as ε = n/τ.
Two-color coincidence detection
Atto425-labeled DnaK (2 μM) and TMR-IAPP (4 μM) were incubated at RT overnight, fractionated in SEC using a Superdex 200 column, and PBS buffer at pH 7.4. The fraction corresponding to 17 mL, after 6× dilution, is used in two-color coincidence detection (TCCD). The experiments are performed in our cuvette-FCS setup equipped with two detection channels (viz, channel 1 (λ = 510 ± 80 nm and the channel 2 (λ = 605 ± 70 nm)). First, the sample is excited using 450-nm laser, and photons are collected with a bin time of 0.5 ms. Photon bursts are detected using a threshold count, which is 5 × SD above the baseline. The FRET efficiency (ε) for each burst is estimated using ε = F′ch2/(Fch1 + Fch2), where Fch1 and Fch2 correspond to total number of photons detected in channels 1 and 2, respectively (46). F′ch2 = Fch2 − α × Fch1, where α is the leak of the fluorescence of free Atto425 in the channel 2. In our setup, α = 0.22. Contribution from direct excitation of TMR by the 450-nm laser is ignored, here, because it is found to be less than 5%. It may be noted that r = 0 and 1 correspond to noncoincident events, respectively, due to ch1 and ch2 only events. Apparent number of monomers of atto425-DnaK per burst is estimated from the ratio of the brightness of the burst and the brightness of a monomer. Brightness of a burst is estimated as ε = (Fch1 + Fch2)/τ, where τ is the temporal width of the burst. Brightness of the monomer (viz, counts per molecule per second is estimated from FCS measurement (42). For estimation of apparent number of TMR-IAPP molecules per burst and the stoichiometry of the TMR-IAPP and atto425-DnaK molecules in each burst, both 450 and 543 nm laser are used for excitation. Channel 1 is used for identification and estimation of the intensity of the atto425 bursts. Leak corrected coincident bursts in channel 2 are used for estimation of brightness of TMR bursts.
Field flow fractionation- multiangle light scattering and SEC-multiangle light scattering measurements
The field flow fractionation (FFF) and the multiangle light scattering (MALS) instruments are from Postnova Analytics (Landes am Lech, Germany). The FFF was used to separate the DnaK/IAPP complexes from the free IAPP. The MALS is connected in line with the FFF for the measurements of the molecular weight (MW) and radius of gyration (Rg) of the proteins. IAPP (20 μM) was incubated with 10 μM DnaK in PBS (pH 7.4) buffer at RT. A 100-μL aliquot of this mixture was injected in the FFF system. We have used a 5-kDa cellulose membrane, 0.20 mL/min focus flow for 6 min, 0.25 mL/min forward flow, and 3 mL/min cross flow in the FFF. The dn/dc measurements were performed using a refractive index (RI) detector (Postnova Analytics). The SEC-MALS measurements were performed in the same setup. In this case, instead of using the FFF membrane assembly, a Superdex 200 column was used for fractionation of the DnaK/IAPP complexes. The flow rate used in SEC is 0.5 mL/min.
Results
Inhibition of amyloid aggregation of IAPP by “holdase” activity of Hsp70 proteins
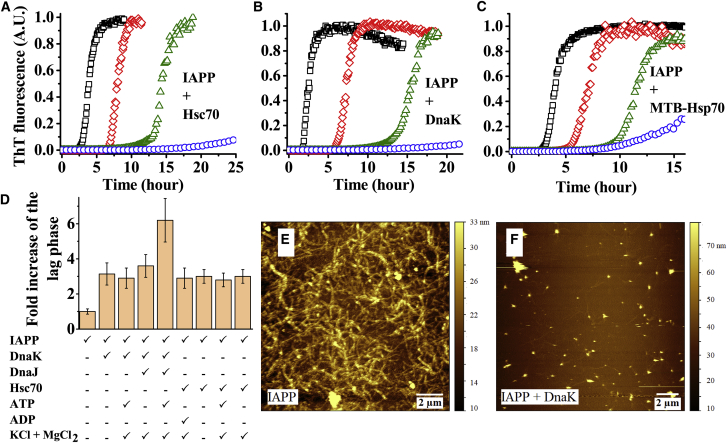
First, we examine whether Hsp70 inhibits aggregation of IAPP in absence of any ATP-dependent activity. Fig. 1 A shows that the time course of aggregation of IAPP (20 μM) is characterized by a lag (t = 0–2.5 h), a growth (t = 2.5–5 h), and a saturation phase (t > 5 h), consistent with the nucleation-dependent aggregation of amyloid proteins (41). Addition of substoichiometric concentrations of Hsc70 with molar ratio as low as 1:400 can be seen to delay the kinetics of aggregation dramatically. For example, the duration of the lag phase is extended by more than 3-fold in presence of Hsc70. Furthermore, the rate of increase of fluorescence (i.e., the rate of aggregation in the growth phase) is also slowed down considerably in presence of higher concentration of Hsc70 (e.g., at 1: 100 molar ratio). We then investigated whether the Hsp70 from other organisms, such as E. coli DnaK and MTB-Hsp70 also could affect the aggregation of IAPP. Fig. 1, B and C show that both DnaK and MTB-Hsp70 inhibit aggregation of IAPP strongly, and the inhibitory activity requires only substoichiometric concentrations of the chaperone proteins. It may be noted that sequence identity between the Hsp70 proteins used here is ∼50% (47). Therefore, despite large differences in the primary sequences, Hsp70 across the different species possibly share common mechanisms in protection against amyloid aggregation. This is consistent with the highly conserved nature of the overall structural organization of the variants of Hsp70 (47).
Figure 1.
Effect of Hsp70 on aggregation of IAPP. (A–C) Kinetics of aggregation of 20 μM IAPP in presence of substoichiometric concentrations of human Hsc70 (A), E. coli DnaK (B), and MTB-Hsp70 (C) is shown. The squares, diamonds, triangles, and circles represent molar ratios of 0:1, 1:400, 1:200, and 1:100 of Hsp70/IAPP, respectively. (D) Fold increase of the “lag” phase of aggregation of 20 μM IAPP by 80 nM Hsp70 under various solution conditions is shown. Concentration of DnaJ used is 80 nM and of ATP and ADP is 2 mM is shown. The solutions are prepared in 10 mM phosphate buffer at pH 7.4, containing 150 mM NaCl, 1 mM EDTA, and 2 mM sodium azide. The experiments are performed at 37°C with continuous stirring. In the ATP/ADP-dependent experiments, NaCl and EDTA are substituted with 100 mM KCl and 5 mM MgCl2. (E and F) AFM images of the 20 μM IAPP incubated for 48 h in absence or presence of 0.5 μM DnaK are shown. All the experiments have been performed at least twice. To see this figure in color, go online.
To probe whether the inhibitory effect of Hsp70 requires specific solution conditions, we performed the kinetic experiments in various solution conditions, such as in absence or presence of NaCl, KCl, MgCl2, ATP, and ADP. Fig. 1 D shows that the “lag phase” of the aggregation of IAPP is greatly extended in presence of low substoichiometric concentrations of Hsp70 under all of the solution conditions we tested. The kinetic data are shown in Fig. S4. Notably, the inhibitory effects of Hsp70 are not enhanced significantly in presence of ATP. This is not surprising because inherent ATPase activity of Hsp70 is known to be poor (11). However, the inhibitory effect of DnaK is enhanced considerably when ATP and the cochaperone DnaJ are present together, consistent with the previously reported results (25,48). Furthermore, the inhibitory effect of Hsc70 is observed to be stronger if NaCl is substituted with KCl. The strong effects of Hsp70 under the diverse conditions led us to test whether the observed effects are specific to Hsp70. Hence, we examined the effect of an unrelated protein of similar size (viz, bovine serum albumin). Fig. S4 B shows that bovine serum albumin has no significant effect on the kinetics of aggregation of IAPP.
Some literature reports suggest that Hsp70 prevents amyloid formation but it promotes amorphous aggregation (28). We have performed AFM measurements on the aggregates of IAPP prepared upon incubation with or without DnaK. Clearly, large quantities of amyloid fibrils are observed in the sample containing IAPP alone (Fig. 1 E). However, very little aggregation is observed in presence of DnaK (Fig. 1 F). Hence, Hsp70 inhibits fibril formation. Furthermore, it does not seem to promote nonfibrillar aggregation of IAPP.
Taken together, the data presented above indicate that Hsp70 is a strong inhibitor of amyloid aggregation of IAPP under diverse solution conditions. Inhibition of the aggregation in the absence of ATP indicates that it is not mediated by the catalytic cycle of the “foldase” activity of Hsp70, rather it is mediated by the “holdase” activity (25). Unlike the “foldase” activity, which requires the appropriate cochaperones, specific solution conditions, and higher concentrations of the chaperone (49,50), the “holdase” activity of Hsp70 appears to be quite robust and highly efficient. The strong effects of Hsp70 observed at low substoichiometric concentrations cannot be explained by its interactions with the monomeric IAPP (21,25). Therefore, Hsp70 possibly interacts with one or more on-pathway intermediates of IAPP. The intermediates could be the prenucleation oligomers or the postnucleation fibrillar species.
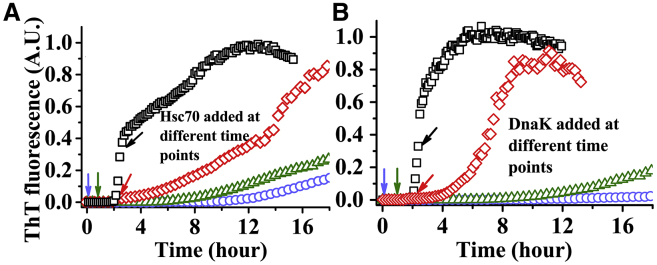
Inhibitory effect of Hsp70 is the strongest at the prenucleation phase
We then examined whether the inhibitory role of Hsp70 is mediated via its interactions with the prenucleation oligomers or with the postnucleation fibrillar species or both. Fig. 2 A shows that the time course of aggregation of a 20 μM IAPP is delayed differently depending on whether the Hsc70 (160 nM) is added in the beginning (i.e., at t = 0), in the early part of the lag phase (at t = 1 h), at the end of the lag phase (at t ∼ 2 h) or in the growth phase (at t ∼ 2.4 h). The inhibitory effect is clearly the highest if Hsc70 is added at t = 0. The effect is progressively less if added later. Fig. 2 B shows that DnaK (200 nM) also exhibits similar effects as Hsc70. Strongest inhibition by the chaperones when it is introduced at t = 0 suggests that the primary mechanism of the inhibitory action of Hsp70 possibly involves prevention of the nucleation via its interactions with the prenucleation species of IAPP. It may, however, be noted that the addition of the chaperones, particularly Hsc70 in the growth phase, affects the rate of increase of the fluorescence of ThT significantly. Hence, Hsc70 could also interact with the fibrils and slow down its growth. Interaction and the effect of Hsp70, particularly in presence of the cochaperones and ATP, on the disaggregation of the amyloid fibrils have been demonstrated earlier (26). However, interaction of Hsp70 with the oligomeric intermediates remains poorly characterized.
Figure 2.
Effect of Hsp70 on different stages of IAPP aggregation. 160 nM Hsc70 (A) or 200 nM DnaK (B) is added to a 20-μM IAPP solution at different stages of aggregation (viz, at t = 0 (circles)), in the middle of “lag”’ phase (t = 1 h) (triangles), in the late lag phase (t ∼ 2 h) (diamonds), and in the growth phase (t ∼ 2.4 h) (squares). Arrows indicate the time points when Hsp70 was added. Clearly, the effect of Hsp70 is highest if added at t = 0. Solutions are prepared in PBS buffer containing 1 mM EDTA and 2 mM sodium azide at pH 7.4 and incubated at 37°C with continuous stirring. These experiments have been performed twice. To see this figure in color, go online.
To probe the interactions between Hsp70 and IAPP further, we consider using fluorescence-based methodologies, including single-molecule techniques. Although Hsc70 is the most relevant chaperone in this study, site-specific labeling of Hsc70 with fluorophores is difficult because of the presence of four cysteine residues in its primary sequence. However, DnaK contains only one cysteine residue (at position 15), which could be used for fluorescence labeling. Because DnaK exhibits similarly strong and robust inhibition of amyloid aggregation of IAPP, we decided to use fluorescently labeled DnaK to investigate the interactions of Hsp70 with IAPP. Hence, hereafter most of the experiments have been performed using wild type or fluorescently labeled DnaK. However, we have also used Hsc70 whenever possible to examine the validity of the crucial observations obtained using DnaK.
Binding of DnaK with the prefibrillar species of IAPP monitored using intermolecular FRET
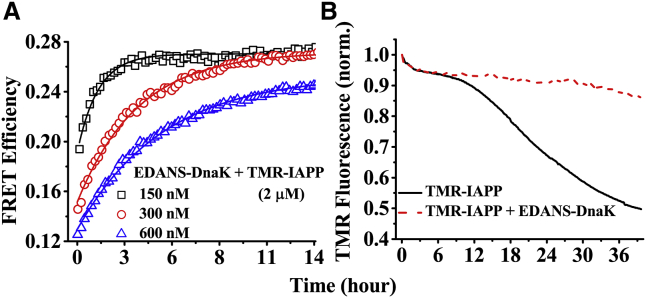
To monitor the intermolecular interactions, we used FRET between EDANS-DnaK and TMR-IAPP. The kinetics of intermolecular FRET exhibits two distinct phases, a fast phase (t <mixing time of the samples, ∼5 s) and a slow phase (t ∼ 1 h) (Fig. 3 A). The fast phase most likely indicates binding with the preformed oligomers and/or with the monomers of IAPP. The extremely slow kinetics of the second phase is consistent with binding of DnaK with the oligomers or protofibrils of IAPP that form slowly with time. Therefore, formation of the multimers is likely the rate-limiting process in this phase.
Figure 3.
Kinetics of interaction between EDANS-DnaK and TMR-IAPP. (A) Kinetics of intermolecular FRET between EDANS-DnaK (150–600 nM) and TMR-IAPP (2 μM) monitored using donor (EDANS) fluorescence is shown. (B) Kinetics of aggregation of TMR-IAPP monitored by fluorescence of TMR in presence and absence of 300 nM EDANS-DnaK is shown. Comparison of the kinetics between (A and B) indicates that the interaction occurs in the early phase of aggregation. The solutions are prepared in PBS buffer containing 1 mM EDTA and 1 mM sodium azide at pH 7.4 and incubated at 37°C with continuous stirring. These experiments have been performed twice. To see this figure in color, go online.
To examine the time evolution of the aggregation status of TMR-IAPP, we monitored the fluorescence of TMR under the same experimental conditions. Because fluorescence of TMR undergoes self-quenching upon oligomerization, this assay, introduced by Garai and Frieden, is suitable for monitoring both fibrillar and nonfibrillar aggregation (41). The kinetics of fluorescence of TMR-IAPP exhibits three distinct phases (Fig. 3 B). These different phases are interpreted as the oligomerization phase (t < 2 h), the “lag” phase (t ∼ 10 h), and the growth phase, based on its similarities with the previously reported kinetics of aggregation of TMR-labeled amyloid-β peptides (41). In presence of DnaK, the “lag” phase is extended beyond 30 h. The data presented in Fig. 3 A indicate that the intermolecular FRET reaches saturation in ∼10 h. Thus, intermolecular FRET, which indicates interaction, occurs primarily during the oligomerization and the “lag” phase of aggregation. Therefore, binding of DnaK with the prefibrillar species of IAPP is possibly responsible for the delaying the kinetics of aggregation.
Ensemble and single-molecule measurements of the binding of DnaK with TMR-IAPP
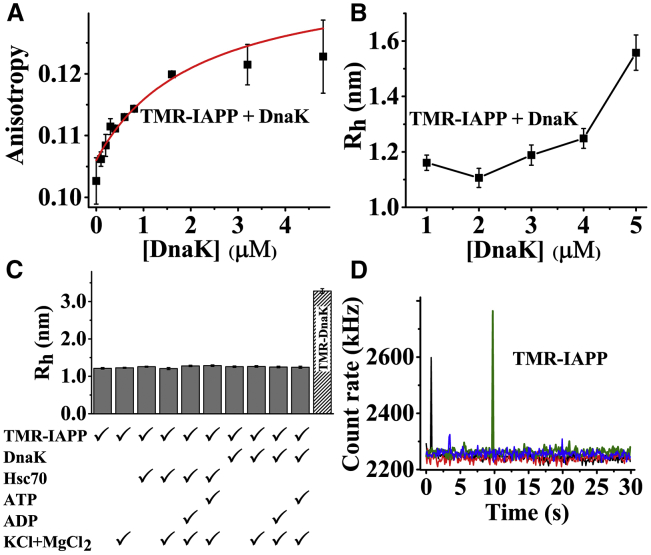
To determine the binding constant of the interactions, we measured fluorescence anisotropy of TMR-IAPP in the presence of unlabeled DnaK. Because IAPP is a small peptide, anisotropy of TMR-IAPP (MW = 4.4 kDa) is expected to increase considerably upon binding to DnaK (MW = 69 kDa). Fig. 4 A shows that anisotropy of 400 nM TMR-IAPP increases in presence of 0–5 μM DnaK in a concentration-dependent manner. Although the increase in anisotropy of TMR-IAPP indicates binding to DnaK, the increase is quite small (∼20%). The small increase may indicate poor binding, which is contradictory with the observations that even a very low concentration of Hsp70 is shown to alter the aggregation of IAPP strongly (see Fig. 1). It could also arise if the Hsp70 binds to a rare species of TMR-IAPP or if the TMR moiety rotates freely in the IAPP/Hsp70 complexes.
Figure 4.
Characterization of the IAPP-DnaK interactions by ensemble and single-molecule techniques. (A) Ensemble fluorescence anisotropy and (B) hydrodynamic radii (Rh) from FCS measurements of 400 nM TMR-IAPP in presence of 0–5 μM DnaK are shown. The symbols represent data. The solid line in (A) is guide to the eye. (C) Rh of 110 nM TMR-IAPP in presence of 1 μM of DnaK or Hsc70 under various solution conditions is shown. (D) Presence of oligomeric TMR-IAPP is reflected in the fluorescence bursts in the photon count traces obtained from four identical FCS measurements on the 400 nM TMR-IAPP sample is shown. The solutions are prepared in 10 mM phosphate buffer (pH 7.4) containing 150 mM NaCl, 1 mM EDTA, and 2 mM azide and incubated for O/N at RT. In some of the samples, as mentioned in (C), NaCl and EDTA have been substituted by 100 mM KCl and 5 mM MgCl2. The experiments have been performed four times. To see this figure in color, go online.
To examine the interactions further, we use a single-molecule technique (viz, FCS) to measure the hydrodynamic radius (Rh) of TMR-IAPP in presence of DnaK. Because Rh is proportional to MW1/3, it is expected to increase by ∼2.6 times if IAPP binds to monomeric DnaK. Fig. 4 B shows that Rh of TMR-IAPP (400 nM) increases in presence of 0–5 μM DnaK in a concentration-dependent manner. Surprisingly, the shape of the Rh versus [DnaK] plot is considerably different from that was obtained from the anisotropy measurements presented in Fig. 4 A. Furthermore, the increase of Rh, even in presence of 5 μM DnaK, is only ∼30%, which is significantly smaller than the expected 160% increase if all the monomeric IAPP are bound to DnaK. Therefore, most of the IAPP in the solution must be free. We have also examined the interactions between DnaK (and Hsc70) and TMR-IAPP under various solution conditions, such as in presence or absence NaCl, KCl, MgCl2, ATP, and ADP. The autocorrelation data obtained under these solution conditions are shown in Fig. S5. Fig. 4 C shows that Rh of TMR-IAPP remains unaltered and almost equal to that of the free IAPP under all the conditions tested here, indicating no significant interaction between DnaK (or Hsc70) and the predominant species of IAPP. Hence, the chaperones most likely bind primarily to the oligomers rather than the monomers of IAPP. The small increase of anisotropy or Rh is possibly indicative of the presence of a relatively small population of the oligomers of IAPP. The presence of the oligomers may be observed from the photon count traces obtained from the FCS measurements (Fig. 4 D). Clearly, several of the photon count traces exhibit presence of fluorescence bursts, indicating the presence of oligomers of IAPP. Consequently, large variability is observed among the autocorrelation curves as well (Fig. S6). We note, here, that the presence of the larger photon bursts distorts the autocorrelation data (Fig. S6). The autocorrelation data that are visibly distorted are excluded from the analysis in determination of Rh. Hence, the nature of the binding curves obtained from the ensemble anisotropy measurements and FCS measurements differ considerably (see Fig. 4, A and B). Furthermore, appearances of the photon bursts indicate that fluorescence of the oligomers of TMR-IAPP is not fully quenched, although the extent of quenching is not known.
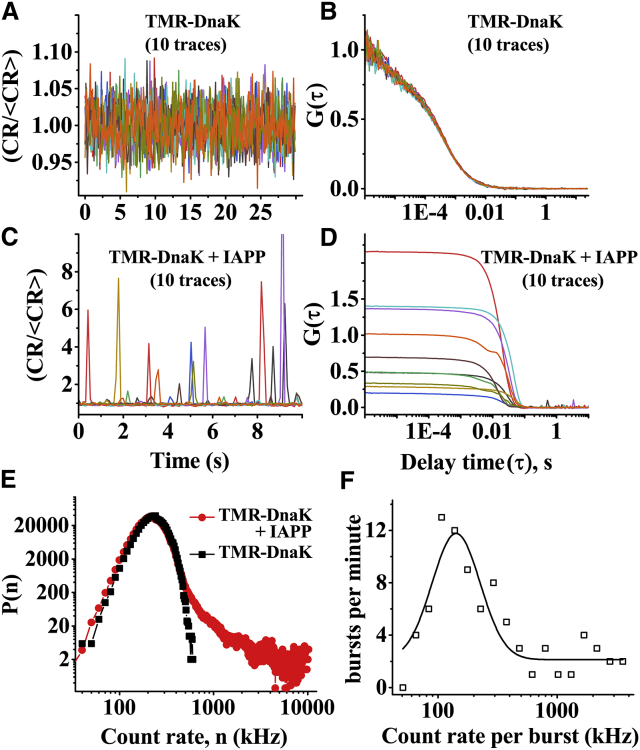
Characterization of the complexes using photon burst analysis
It may be argued that the above-mentioned heterogeneity is a characteristic of TMR-IAPP. To verify, we performed FCS measurements of TMR-DnaK (100 nM) in the presence of unlabeled IAPP (20 μM). Many fluorescence bursts are observed in the photon count traces of TMR-DnaK in presence (Fig. 5 C) but not in the absence of IAPP (Fig. 5 A). The autocorrelation traces obtained in presence of IAPP are also highly heterogeneous, as may be expected (Fig. 5 D). The observed photon bursts are indicative of clustering the TMR-DnaK molecules, possibly on the oligomers or the protofilaments of IAPP. Quantification of the oligomers in the midst of a vast majority of the free monomers is difficult (46). However, comparison of the PCH of TMR-DnaK in presence and absence of IAPP shows that ∼8% of the total photons are associated with the photon bursts. Hence, ∼8% of the 100 nM TMR-DnaK must be associated with the complexes with IAPP oligomers. We then counted the number of the bursts per minute using a photon count threshold of mean + 3 × SD. We observed ∼16 photon bursts per minute. Furthermore, the probability of detecting a photon burst in any time bin (equal to the number of the bins associated with the bursts/total number of bins in 1 min) is found to be ∼0.01. In our FCS setup, the average number of molecules (<N>) observed in the confocal volume from 1 nM solution is ∼1.1 (43). Hence, the number concentration of the oligomeric complexes giving rise to the photon bursts must be in the low picomolar range (51). The size of the photon bursts exhibits a broad distribution (Fig. 5 F). The peak of the distribution corresponds to ∼130 kHz. Counts per molecule of the monomeric TMR-DnaK obtained from the FCS measurement of the purified TMR-DnaK solution is ∼8 kHz. Hence, the average brightness of the complexes is ∼16 times higher than monomeric TMR-DnaK. Therefore, oligomers of IAPP interact with high affinity with multiple molecules of DnaK. We note, here, that the approach outlined above is suitable only for the large bursts because the small complexes would get masked in the shot noise of the photon counts. Hence, this approach is not suitable for characterization of the small complexes.
Figure 5.
Characterization of the oligomeric IAPP/TMR-DnaK complexes from photon bursts analysis. Photon count traces (A and C) and autocorrelation data (B and D) from 10 identical measurements from 100 nM TMR-DnaK in absence of IAPP (A and B) or in presence of 20 μM IAPP (C and D) are shown. Clearly, both photon counts and autocorrelation data are highly heterogeneous in presence of IAPP. (E) comparison of photon counting histograms (PCH) indicates that ∼8% of TMR-DnaK is partitioned into the bursts. (F) Histogram of the brightness of the bursts is shown. The solid line is a fit assuming a Gaussian distribution. Mean brightness of the bursts (130 kHz) is ∼16 times brighter than the monomeric TMR-DnaK. The solutions are prepared in PBS buffer containing 1 mM EDTA and 1 mM sodium azide at pH 7.4 and incubated at RT. The experiments have been performed twice. To see this figure in color, go online.
Characterization of the complexes of DnaK/IAPP by MALS and FCS after fractionation by SEC and FFF
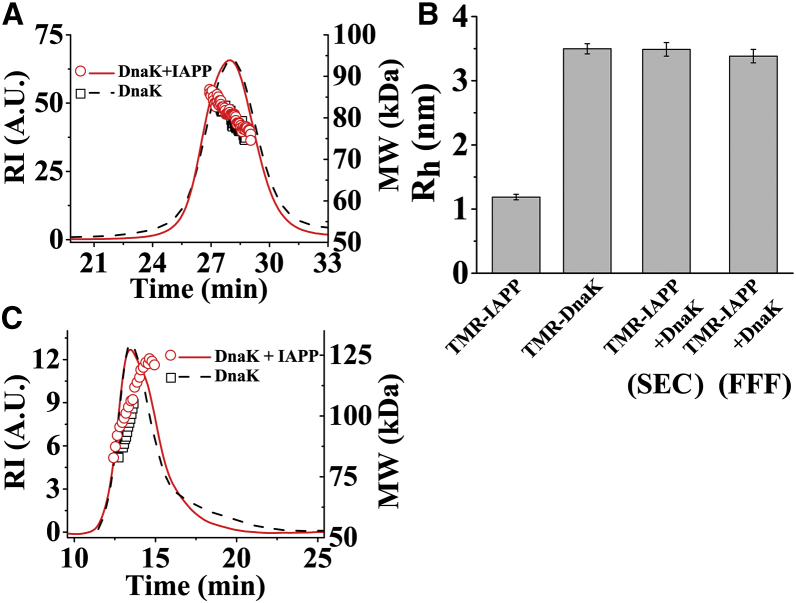
The FCS measurements presented above indicate that the major problem in characterization of the DnaK/IAPP interaction is heterogeneity of the complexes. Therefore, we use SEC to resolve the various species in the heterogeneous mixture of free IAPP, free DnaK, and the IAPP/DnaK complexes. The SEC is connected to a MALS instrument for determination of the MW and the size of the DnaK/IAPP complexes. Fig. 6 A shows the time course of the concentration of the proteins measured by RI of the solution. It may be seen that the RI (i.e., the concentration) profile shows a dominant peak at ∼28 min. The same figure also shows the MW obtained from analysis of the MALS data. It may be seen that the MW of the particles corresponding to this peak is ∼75–85 kDa, indicating that DnaK exists predominantly as monomers. To verify whether this peak consists of complexes of DnaK with oligomers of IAPP, we compared the data obtained using DnaK alone. However, the profiles of the RI and the MW of the samples containing DnaK/IAPP and DnaK alone appear to be similar. Fig. S7 shows that similar observations are made using the solutions of Hsc70/IAPP and Hsc70 alone. We then performed mass spectrometry measurements on the peak fraction to obtain an estimate of the relative concentrations of DnaK and IAPP in the DnaK/IAPP sample. Fig. S8 shows that these fractions contain DnaK, but no IAPP was detected. Therefore, concentration of the DnaK/IAPP complexes must be small. Thus, the major population observed in the RI and the MALS profiles must originate from the primarily free DnaK.
Figure 6.
Characterization of IAPP-DnaK complexes after fractionation by size exclusion chromatography (SEC) or field flow fractionation (FFF). (A) SEC-MALS and (C) FFF-MALS of 10 μM DnaK in absence and presence of 20 μM IAPP are shown. The solid lines indicate refractive index (RI), which is a measure of concentration, and empty symbols represent molecular weight (MW) measured by MALS. (B) Rh measured using FCS of TMR-IAPP, TMR-DnaK, and TMR-IAPP + DnaK after purification using SEC or FFF is shown. Clearly, Rh of TMR-IAPP + DnaK is similar to TMR-DnaK, indicating binding of TMR-IAPP to DnaK. The solutions are prepared in PBS buffer containing 1 mM EDTA and 1 mM sodium azide at pH 7.4 and incubated at RT for O/N. These experiments have been performed four times. To see this figure in color, go online.
To characterize the DnaK/IAPP complexes specifically, we then used FCS. In these experiments, we used TMR-IAPP instead of IAPP. Because the DnaK used here is not fluorescently labeled, the unbound DnaK would not contribute to the signal in FCS. First, SEC is used to fractionate the DnaK/TMR-IAPP complexes from the population of the free TMR-IAPP. The fraction exhibiting peak intensity is collected for FCS measurements. Fig. 6 B shows that Rh of the TMR-IAPP/DnaK complexes measured using FCS is ∼3.3 nm, which is almost same as that of the monomeric DnaK but quite larger than that of the free IAPP (Rh ∼ 1.2 nm). Therefore, FCS measurements indicate binding of monomeric DnaK to monomers or small oligomers of TMR-IAPP. Because Rh is proportional to (MW)1/3, the FCS measurements do not have the resolution to distinguish if these complexes contain monomers or small oligomers of IAPP. The brightness of these complexes is found to be ∼1.2 times the brightness of the free TMR-IAPP. Thus, the fluorescence of the oligomers of TMR-IAPP must be considerably quenched (43). The concentration of the TMR-IAPP/DnaK complexes detected in FCS is ∼45 nM (Table S1). We note, here, that the concentrations of DnaK and IAPP used in these experiments are several μM. Therefore, concentration of the complexes detected in FCS is very low, suggesting that most of the DnaK or the TMR-IAPP in the solution is free.
A well-recognized problem in characterization of noncovalent protein-protein complexes by SEC-MALS is that the complexes may dissociate because of interactions of the proteins with the matrix of the SEC column. To circumvent this problem, we attempt to resolve the various species of the IAPP/DnaK complexes by using FFF, which uses flow field to separate the molecules based on hydrodynamic size (52). Fig. 6 C compares the time courses of the RI and the MW obtained from the mixture of DnaK and IAPP and the DnaK alone. It may be seen that both the samples show a peak at ∼13 min in RI. The MW of the peak lies within 80–120 kDa, consistent with the SEC-MALS measurements. Once again, we performed FCS measurements on the TMR-IAPP/DnaK samples fractionated using FFF. Analysis of the FCS data presented in Fig. 6 B shows that Rh of the complexes is almost the same as that of the monomeric DnaK. Once again, the concentration of the complexes is very low (∼15 nM) (Table S1). Therefore, FFF-MALS/FCS data are consistent with the SEC-MALS/FCS data. The FCS autocorrelation data and the fits are shown in Fig. S9. Taken together, the FCS data indicate that the Rh of the complexes is the same as the complex of monomeric DnaK binding to monomers or small oligomers of IAPP. However, the population of the complexes is small.
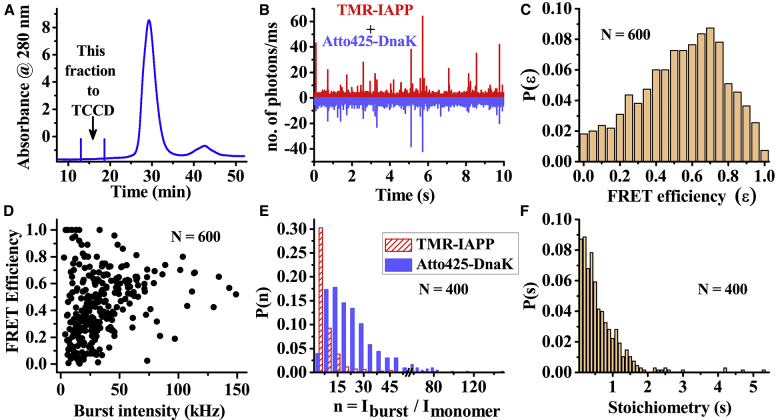
Single-particle TCCD of the oligomeric complexes
We then use TCCD to detect and characterize the oligomeric complexes using Atto425-labeled DnaK and TMR-IAPP. TCCD measurements are performed using our cuvette-FCS setup (42), which is equipped with two detection channels (viz, blue and red channels for the fluorescence of Atto-425 and TMR, respectively). SEC is then used to separate the complexes from the predominantly free monomeric population of DnaK and IAPP. As expected, the SEC profile exhibits a dominant peak at ∼28 min (Fig. 7 A). However, in this experiment, we focus our attention on a fraction eluted at ∼18 min, which corresponds to the elution time of larger species than the monomeric DnaK. The SEC profile exhibits nearly zero absorbance (at 280 nm). No protein band is observed in this fraction in the SDS-PAGE either (see Fig. S10). Hence, concentrations of DnaK and IAPP eluted in this fraction must be very low. However, single-molecule fluorescence measurements by TCCD exhibit large photon bursts in both the detection channels (Fig. 7 B). Extraction of the bursts from both the channels using a threshold counts of baseline + 5 × SD shows that more than 75% of the bursts are coincident. Thus, majority of the photon bursts arise from the complexes of Atto425-Hsp70 and TMR-IAPP. The histogram of FRET efficiency (ε) of the coincident bursts is found to be quite broad, ranging from 0 to 1, with a peak at ∼0.7 (Fig. 7 C). The spread in the values of ε is particularly wide for the smaller bursts (Fig. 7 D). This could arise because of the relatively higher photon shot noise for smaller bursts. However, considerable broadness of the distribution of ε for larger bursts is indicative of the heterogeneity of the complexes, possibly due to the variabilities in the stoichiometry of Hsp70 and IAPP molecules and/or due to the differences in the intermolecular arrangements. Although fluorescence of Atto425-Hsp70 does not exhibit self-quenching (data not shown), fluorescence of TMR-IAPP experiences considerable quenching upon oligomerization (see Fig. 3 B). Hence, it is not possible to estimate the absolute size of the oligomers from the intensity of the fluorescence bursts. Furthermore, the intensity also depends on the trajectory of the particles inside the confocal observation volume (45). Therefore, we attempt to obtain an approximate estimate of the number of the Atto425-DnaK and a lower-bound estimate of the TMR-IAPP molecules in the complexes from the brightness of the bursts (46). Fig. 7 E shows that the distribution of the number of Atto425-DnaK molecules per complex is broad, ranging from 2 to 100 mer, with a peak at around 15 mer. The distribution of TMR-IAPP monomers in the complexes is found to be relatively narrow, ranging from 2 to 30 mer, with a peak at 5 mer. However, the number of TMR-IAPP molecules per complex is most likely considerably underestimated because of the quenching of the fluorescence of TMR in the oligomeric forms. Stoichiometry of TMR-IAPP and Atto425-DnaK estimated from the ratio of the intensities in the red and the blue channels is found to vary from ∼0.1 to 2.0 with the average being 0.6 (Fig. 7 F). However, because of the presence of FRET between the fluorophore pair and self-quenching of TMR, the estimate of stoichiometry also should be considered as approximate.
Figure 7.
Characterization of TMR-IAPP/Atto425-DnaK complexes using TCCD. (A) SEC profile measured by absorbance at 280 nm of a solution containing 4 μM TMR-IAPP and 2 μM atto425-DnaK is shown. The fraction eluted at 17 min is examined using TCCD. (B) Photon counts/ms obtained from atto425-DnaK (lower panel) and TMR-IAPP (upper panel) using excitation with the blue (λ = 450 nm) laser are shown. (C) Histogram of the FRET efficiency (ε = ITMR/[Iatto425 + ITMR]) of the coincident bursts is shown. (D) Dependence of ε on the total burst intensity is shown. (E and F) Histograms of the apparent number of monomers (E) and approximate stoichiometry (F) of TMR-IAPP and atto425-DnaK molecules in the complexes are shown. Estimates in (F) are obtained using the coincident bursts with both (λ = 450 and 543 nm) lasers on. To see this figure in color, go online.
Discussion
Amyloid aggregation of proteins is involved in the pathology of multiple human diseases, such as Alzheimer’s, Parkinson’s disease, and T2DM. Extensive studies over the past three decades indicate that protein monomers first self-assemble into soluble low-MW oligomers; the oligomers then possibly act as sites for nucleation to form small fibrillar aggregates that grow to larger fibrils. To counteract the numerous pathways that lead to aberrant aggregation of proteins, cells utilize molecular chaperones as a defense mechanism (10,12,53,54). Hsp70 chaperone machinery is the central component of the chaperone systems, providing protection against aberrant aggregation of cellular proteins under stress conditions. The commonly depicted mechanism of action is that Hsp70 machinery, which involves the cochaperones, such as Hsp40 and the NEF, bind to the exposed hydrophobic patches of “misfolded” proteins or the fully unfolded polypeptide chains and then release it in the correctly “folded” form using an ATP-dependent catalytic cycle (55). In vitro, Hsp70/Hsp40/NEF machinery has been shown to inhibit aggregation and promote disaggregation of the aggregates of amyloid-β, α-synuclein, and polyglutamine polypeptides in presence of ATP (21,25,28). In vivo, overexpression or exogeneous addition of Hsp70 has been shown to inhibit amyloid aggregation and rescue cytotoxicity of the amyloids in cell cultures and model organisms (16,20,56).
Whereas the “foldase” activity of Hsp70 is mediated by the ATP-dependent catalytic cycle, recent in vitro studies have shown that Hsp70 can also inhibit amyloid aggregation in the absence of ATP via “holdase” activity (12,25). A recent study has demonstrated the protective role of the secretory Hsp70 in blocking the amyloid-β-induced neurotoxicity in Drosophila (56). Holdase activity of Hsp70 has also been shown to block formation of the toxic oligomers of α-synuclein (57). Our results demonstrate that Hsp70 strongly inhibits aggregation of IAPP under diverse solution conditions via “holdase” activity (see Figs. 1, 2, and 3). Hence, the “holdase” activity of Hsp70 is highly robust, unlike the “foldase” activity that requires specific solution conditions and specific cochaperones (26,50). Another striking feature of the “holdase” activity is that it requires very low substoichiometric concentration of Hsp70. Inhibition of amyloid formation by “holdase” action possibly occurs because of binding of Hsp70 to the polypeptides leading to blocking of the sites of growth. However, the extent of the inhibition would depend on the molecular mechanism of the interactions. For example, if Hsp70 binds predominantly to the monomers, then in absence of ATP-driven catalytic activity, stoichiometric amount of Hsp70 is required to observe significant effects. Our results demonstrate that Hsp70 can exert strong effects, even at low substoichiometric concentrations. Thus, it must interact with the intermediates/oligomers rather than the monomers of IAPP (25). The results presented in Fig. 2 indicate that the effects of Hsc70 or DnaK is the strongest when it is administered at t = 0, indicating its interaction with the prefibrillar oligomers of IAPP. This is further supported by the intermolecular FRET measurements, which show that DnaK interacts with the species of IAPP that form in the “lag” phase of aggregation (see Fig. 3). Therefore, we hypothesize that the Hsp70 inhibits nucleation via interactions with the oligomers of IAPP. Similar effects have been observed in the case of Hsp70 and DnaJB6 on the aggregation of other amyloid proteins (30,58).
Low concentration and heterogeneity of the complexes led us to employ single-molecule fluorescence-based techniques to investigate the nature of these complexes. Little is known so far about the biophysical properties of the Hsp70/IAPP complexes. In fact, ensemble anisotropy measurements and FCS measurement of TMR-IAPP in presence of Hsp70 indicate poor binding (see Fig. 4), contrary to the observed striking effect of Hsp70 on aggregation of IAPP. These apparently contradictory observations suggest that Hsp70 must interact with a rare species (e.g., the “intermediates” or the oligomers of IAPP). To detect and characterize the complexes, we have employed several strategies. The fluorescence bursts of the TMR-DnaK formed in presence of IAPP suggest that the oligomers of IAPP bind to multiple molecules of DnaK (see Fig. 5). We find that the concentration of the bursts (i.e., concentration of the complexes) is in the picomolar range. Determination of the size of the oligomeric complexes is difficult, but average brightness of the bursts suggests ∼10–100 molecules of DnaK per oligomer. An inherent limitation of the burst analysis in presence of a high background fluorescence, which arises from the large population of the free TMR-DnaK, is that detection of the small oligomers is difficult. Furthermore, analysis of the FCS autocorrelation data becomes unreliable in presence of the photon bursts. Therefore, we used SEC and FFF to separate the large oligomeric complexes. FCS and MALS measurements on the major (SEC or FFF) fractions indicate that the size of the DnaK/IAPP complexes is ∼90 kDa, possibly containing one molecule of DnaK and a few molecules of IAPP. However, concentration of the complexes is very low, ∼15–45 nM, indicating that majority of the DnaK and the IAPP in the mixture must be free. FCS measurements of TMR-IAPP in presence of considerably high molar excess of DnaK or Hsc70 performed under various solution conditions also support that the majority of the IAPP in the mixture is free (see Fig. 4). Taken together, our results suggest that binding of Hsp70 with the monomers of IAPP is poor, whereas affinity for binding to the oligomers is high.
Although the concentration of the oligomers is very low, the single-molecule techniques have been successful to detect these species (30,44,46,50,59). Klenerman and co-workers used TCCD measurements after dilution of the μM fluorescently labeled protein stock solutions to picomolar concentrations (46). Because, compared with the free protein molecules, the population of the complexes is very small, a microfluidic device was used to improve the rate of sampling of the complexes. Here, we have used SEC to improve the rate of sampling of the Hsp70/IAPP complexes by eliminating the population of the free monomers. The SEC-TCCD measurements clearly exhibit the oligomeric and heterogeneous nature of the complexes consisting of atto425-DnaK and TMR-IAPP (see Fig. 7). Analysis of the individual fluorescence bursts reveal that the distributions of FRET efficiency, and the apparent number and stoichiometry of the TMR-IAPP and Atto425-Hsp70 molecules in the complexes are quite broad. Broadness of the distributions reflects the heterogeneous nature of the complexes. However, MW of the complexes estimated from the average number of IAPP and Hsp70 molecules present per complex is ∼1 MDa and the stoichiometry is ∼0.6. Owing to the self-quenching of the fluorescence of TMR, the number of the IAPP molecules in the complexes is likely underestimated.
The mechanism of the interaction of Hsp70 with substrate proteins has been a matter of intense studies (38,55,60, 61, 62). Hsp70 is known to be highly promiscuous because of its interactions with a multitude of substrates with diverse polypeptide sequences (47). However, most of our knowledge about how Hsp70 binds to its substrates is based on the studies using model peptides from experiments using peptide libraries and phage display (63,64). The octapeptide substrate for Hsp70 generally contains a six-residue hydrophobic middle patch flanked by positively charged residues at both ends. Many polypeptide sequences contain such patches. Therefore, Hsp70 binds to fully unfolded polypeptides or the exposed hydrophobic patches in the globular proteins. For example, Hsp70 machinery can refold denatured luciferase in vitro (65). Therefore, binding of Hsp70 with globular proteins is considered to be regulated by misfolding or unfolding of the protein. However, little is known about how Hsp70 binds to the IDPs because the putative binding motifs in IDPs are possibly exposed all the time. Therefore, how its interaction with the IDPs is regulated is unclear. The molecular chaperone binding site prediction program Limbo (66) predicts residues 13–19 heptapeptide (ANFLVHS) of IAPP as a probable binding site for DnaK. Cryoelectron microscopy reveals that the IAPP monomers in the fibrils exhibit an S-fold structure comprising triple β-sheets, involving residues 14–20, 26–32, and 35–37 (67,68). Therefore, the peptide comprising residues 13–19 is an important part of the fibril. Hence, binding of Hsp70 to this peptide can potentially inhibit nucleation and growth of the IAPP fibrils. Our results indicate that DnaK binds strongly to the oligomers but weakly to the monomers. Therefore, we think that the high affinity of the interaction with the oligomers might arise from multivalent binding. The multivalent binding might involve oligomeric forms of IAPP and the multiple binding sites on Hsp70, similar to what has been proposed earlier for the interaction between apolipoprotein E and the oligomers of amyloid-β (40). Although the polypeptide substrates are believed to bind to the canonical substrate binding cleft in the substrate binding domain, the surface of the Hsp70 proteins is capable of interacting with a multitude of cochaperone proteins (47). In fact, diversity of the functions of Hsp70 are hypothesized to originate from its ability to interact with a diverse array of cochaperones (69). Furthermore, a second binding site that is important for the activity of the Hsp70 has been reported recently (70). A recent study suggests involvement of a noncanonical site of Hsp70 in blocking the oligomerization of α-synuclein (57). Hence, multivalent binding might involve the alternative binding sites and/or other protein-protein interaction sites on the surface of Hsp70. Alternatively, compared with the monomers, the oligomeric forms of IAPP might provide a larger binding surface, leading to oligomerization-dependent increase of affinity. Because the IDPs do not possess defined structure, high-affinity binding of Hsp70 to the oligomeric but not the monomeric forms of IAPP is possibly required to regulate the interaction, ensuring that the physiological functions of the monomeric IAPP are not affected. Furthermore, FCS measurements reported in Figs. 5 and 7 suggest that oligomers of IAPP bind to multiple molecules of Hsp70. Recruitment of multiple Hsp70 molecules to the protein rhodanase has been hypothesized to promote unfolding of the protein via an “entropic pulling” action (59). A similar mechanism of action may be involved in the disassembly of the oligomers or the fibrils of the amyloid proteins by the Hsp70 machinery in presence of ATP (26).
Thus, our data support that Hsp70 alone can inhibit aggregation of IAPP by the “holding” model (21,25) via binding to the oligomers of IAPP. This is possibly the first step in the catalytic cycle of the chaperone machinery (21,25). Alternatively, Hsp70 can direct the bound complexes to the degradation pathways (71). Several other heat shock proteins, such as Hsp90 and Hsp60, also exhibit similar protection against amyloid aggregation (21,72). We find that Hsp70 binds weakly with the monomers but strongly with the oligomeric forms of IAPP, showing affinity in the picomolar range. Thus, it is possible that Hsp70 could detect single particles of aberrant amyloid oligomers and nullify its toxic effects in vivo. This might be a common feature, which is important for the regulation of the interactions between the constitutively expressed molecular chaperones and the functionally important but amyloidogenic IDPs. Application of a combination of ensemble and single-molecule biophysical techniques, including SEC-TCCD, enables us to identify and characterize the rare and highly heterogeneous oligomers of IAPP that interact with Hsp70.
Author Contributions
K.G. conceptualized research. N.C., T.B.S., B.S., S.D., S.C., and M.M. performed research. K.G., T.B.S., and B.S. performed analysis. K.G. and N.C. wrote the study.
Acknowledgments
We acknowledge funding from the Department of Atomic Energy, India. This work is partly supported by an Early Career Research Award received by K.G. from the Science and Engineering Research Board, India.
Editor: James Shorter.
Footnotes
Neeraja Chilukoti and Timir Baran Sil contributed equally to this work.
Neeraja Chilukoti’s present address is Centre for Cellular & Molecular Biology, Habsiguda, Hyderabad, India.
Bankanidhi Sahoo’s present address is Dr. Reddy’s Laboratories, Bachupally, Hyderabad, India.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2020.12.019.
Supporting Material
References
- 1.Donath M.Y., Ehses J.A., Reinecke M. Mechanisms of β-cell death in type 2 diabetes. Diabetes. 2005;54(Suppl 2):S108–S113. doi: 10.2337/diabetes.54.suppl_2.s108. [DOI] [PubMed] [Google Scholar]
- 2.Höppener J.W., Oosterwijk C., Lips C.J. Extensive islet amyloid formation is induced by development of Type II diabetes mellitus and contributes to its progression: pathogenesis of diabetes in a mouse model. Diabetologia. 1999;42:427–434. doi: 10.1007/s001250051175. [DOI] [PubMed] [Google Scholar]
- 3.Hayden M.R., Tyagi S.C., Nicolls M.R. Type 2 diabetes mellitus as a conformational disease. JOP. 2005;6:287–302. [PubMed] [Google Scholar]
- 4.Jeong H.R., An S.S. Causative factors for formation of toxic islet amyloid polypeptide oligomer in type 2 diabetes mellitus. Clin. Interv. Aging. 2015;10:1873–1879. doi: 10.2147/CIA.S95297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonemoto I.T., Kroon G.J., Kelly J.W. Amylin proprotein processing generates progressively more amyloidogenic peptides that initially sample the helical state. Biochemistry. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meier J.J., Kayed R., Butler P.C. Inhibition of human IAPP fibril formation does not prevent β-cell death: evidence for distinct actions of oligomers and fibrils of human IAPP. Am. J. Physiol. Endocrinol. Metab. 2006;291:E1317–E1324. doi: 10.1152/ajpendo.00082.2006. [DOI] [PubMed] [Google Scholar]
- 7.Lorenzo A., Razzaboni B., Yankner B.A. Pancreatic islet cell toxicity of amylin associated with type-2 diabetes mellitus. Nature. 1994;368:756–760. doi: 10.1038/368756a0. [DOI] [PubMed] [Google Scholar]
- 8.Janson J., Ashley R.H., Butler P.C. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 9.Lin C.Y., Gurlo T., Butler P.C. Toxic human islet amyloid polypeptide (h-IAPP) oligomers are intracellular, and vaccination to induce anti-toxic oligomer antibodies does not prevent h-IAPP-induced beta-cell apoptosis in h-IAPP transgenic mice. Diabetes. 2007;56:1324–1332. doi: 10.2337/db06-1579. [DOI] [PubMed] [Google Scholar]
- 10.Balch W.E., Morimoto R.I., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 11.Hartl F.U., Hayer-Hartl M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009;16:574–581. doi: 10.1038/nsmb.1591. [DOI] [PubMed] [Google Scholar]
- 12.Mannini B., Chiti F. Chaperones as suppressors of protein misfolded oligomer toxicity. Front. Mol. Neurosci. 2017;10:98. doi: 10.3389/fnmol.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atalay M., Oksala N., Roy S. Heat shock proteins in diabetes and wound healing. Curr. Protein Pept. Sci. 2009;10:85–95. doi: 10.2174/138920309787315202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krause M., Heck T.G., Homem de Bittencourt P.I., Jr. The chaperone balance hypothesis: the importance of the extracellular to intracellular HSP70 ratio to inflammation-driven type 2 diabetes, the effect of exercise, and the implications for clinical management. Mediators Inflamm. 2015;2015:249205. doi: 10.1155/2015/249205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung J., Nguyen A.K., Febbraio M.A. HSP72 protects against obesity-induced insulin resistance. Proc. Natl. Acad. Sci. USA. 2008;105:1739–1744. doi: 10.1073/pnas.0705799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosas P.C., Nagaraja G.M., Asea A.A. Hsp72 (HSPA1A) prevents human islet amyloid polypeptide aggregation and toxicity: a new approach for type 2 diabetes treatment. PLoS One. 2016;11:e0149409. doi: 10.1371/journal.pone.0149409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klucken J., Shin Y., McLean P.J. Hsp70 reduces alpha-synuclein aggregation and toxicity. J. Biol. Chem. 2004;279:25497–25502. doi: 10.1074/jbc.M400255200. [DOI] [PubMed] [Google Scholar]
- 18.Press M., Jung T., Höhn A. Protein aggregates and proteostasis in aging: amylin and β-cell function. Mech. Ageing Dev. 2019;177:46–54. doi: 10.1016/j.mad.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Kavanagh K., Flynn D.M., Wagner J.D. Restoring HSP70 deficiencies improves glucose tolerance in diabetic monkeys. Am. J. Physiol. Endocrinol. Metab. 2011;300:E894–E901. doi: 10.1152/ajpendo.00699.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bongiovanni M.N., Aprile F.A., Vendruscolo M. A rationally designed Hsp70 variant rescues the aggregation-associated toxicity of human IAPP in cultured pancreatic islet β-cells. Int. J. Mol. Sci. 2018;19:1443. doi: 10.3390/ijms19051443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans C.G., Wisén S., Gestwicki J.E. Heat shock proteins 70 and 90 inhibit early stages of amyloid beta-(1-42) aggregation in vitro. J. Biol. Chem. 2006;281:33182–33191. doi: 10.1074/jbc.M606192200. [DOI] [PubMed] [Google Scholar]
- 22.Luk K.C., Mills I.P., Lee V.M. Interactions between Hsp70 and the hydrophobic core of alpha-synuclein inhibit fibril assembly. Biochemistry. 2008;47:12614–12625. doi: 10.1021/bi801475r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wacker J.L., Zareie M.H., Muchowski P.J. Hsp70 and Hsp40 attenuate formation of spherical and annular polyglutamine oligomers by partitioning monomer. Nat. Struct. Mol. Biol. 2004;11:1215–1222. doi: 10.1038/nsmb860. [DOI] [PubMed] [Google Scholar]
- 24.Baughman H.E.R., Clouser A.F., Nath A. HspB1 and Hsc70 chaperones engage distinct tau species and have different inhibitory effects on amyloid formation. J. Biol. Chem. 2018;293:2687–2700. doi: 10.1074/jbc.M117.803411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chien V., Aitken J.F., Loomes K.M. The chaperone proteins HSP70, HSP40/DnaJ and GRP78/BiP suppress misfolding and formation of β-sheet-containing aggregates by human amylin: a potential role for defective chaperone biology in type 2 diabetes. Biochem. J. 2010;432:113–121. doi: 10.1042/BJ20100434. [DOI] [PubMed] [Google Scholar]
- 26.Gao X., Carroni M., Bukau B. Human Hsp70 disaggregase reverses Parkinson’s-linked α-synuclein amyloid fibrils. Mol. Cell. 2015;59:781–793. doi: 10.1016/j.molcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Repalli J., Meruelo D. Screening strategies to identify HSP70 modulators to treat Alzheimer’s disease. Drug Des. Devel. Ther. 2015;9:321–331. doi: 10.2147/DDDT.S72165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muchowski P.J., Schaffar G., Hartl F.U. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. USA. 2000;97:7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakono M., Kidani T. ATP-independent inhibition of amyloid beta fibrillation by the endoplasmic reticulum resident molecular chaperone GRP78. Biochem. Biophys. Res. Commun. 2017;493:500–503. doi: 10.1016/j.bbrc.2017.08.162. [DOI] [PubMed] [Google Scholar]
- 30.Kundel F., De S., Klenerman D. Hsp70 inhibits the nucleation and elongation of Tau and sequesters Tau aggregates with high affinity. ACS Chem. Biol. 2018;13:636–646. doi: 10.1021/acschembio.7b01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schönfeld H.J., Schmidt D., Bukau B. The DnaK chaperone system of Escherichia coli: quaternary structures and interactions of the DnaK and GrpE components. J. Biol. Chem. 1995;270:2183–2189. doi: 10.1074/jbc.270.5.2183. [DOI] [PubMed] [Google Scholar]
- 32.Aprile F.A., Dhulesia A., Cremades N. Hsp70 oligomerization is mediated by an interaction between the interdomain linker and the substrate-binding domain. PLoS One. 2013;8:e67961. doi: 10.1371/journal.pone.0067961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgner N., Schmidt C., Robinson C.V. Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Rep. 2015;11:759–769. doi: 10.1016/j.celrep.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu S., Hong L., Perrett S. Kinetics of the conformational cycle of Hsp70 reveals the importance of the dynamic and heterogeneous nature of Hsp70 for its function. Proc. Natl. Acad. Sci. USA. 2020;117:7814–7823. doi: 10.1073/pnas.1914376117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marek P., Woys A.M., Raleigh D.P. Efficient microwave-assisted synthesis of human islet amyloid polypeptide designed to facilitate the specific incorporation of labeled amino acids. Org. Lett. 2010;12:4848–4851. doi: 10.1021/ol101981b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montgomery D.L., Morimoto R.I., Gierasch L.M. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- 37.Swain J.F., Schulz E.G., Gierasch L.M. Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J. Biol. Chem. 2006;281:1605–1611. doi: 10.1074/jbc.M509356200. [DOI] [PubMed] [Google Scholar]
- 38.Rosenzweig R., Sekhar A., Kay L.E. Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. eLife. 2017;6:e28030. doi: 10.7554/eLife.28030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morán Luengo T., Kityk R., Rüdiger S.G.D. Hsp90 breaks the deadlock of the Hsp70 chaperone system. Mol. Cell. 2018;70:545–552.e9. doi: 10.1016/j.molcel.2018.03.028. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh S., Sil T.B., Garai K. High-affinity multivalent interactions between apolipoprotein E and the oligomers of amyloid-β. FEBS J. 2019;286:4737–4753. doi: 10.1111/febs.14988. [DOI] [PubMed] [Google Scholar]
- 41.Garai K., Frieden C. Quantitative analysis of the time course of Aβ oligomerization and subsequent growth steps using tetramethylrhodamine-labeled Aβ. Proc. Natl. Acad. Sci. USA. 2013;110:3321–3326. doi: 10.1073/pnas.1222478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sahoo B., Sil T.B., Garai K. A fluorescence correlation spectrometer for measurements in cuvettes. Biophys. J. 2018;115:455–466. doi: 10.1016/j.bpj.2018.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sil T.B., Sahoo B., Garai K. Quantitative characterization of metastability and heterogeneity of amyloid aggregates. Biophys. J. 2018;114:800–811. doi: 10.1016/j.bpj.2017.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puchalla J., Krantz K., Rye H. Burst analysis spectroscopy: a versatile single-particle approach for studying distributions of protein aggregates and fluorescent assemblies. Proc. Natl. Acad. Sci. USA. 2008;105:14400–14405. doi: 10.1073/pnas.0805969105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Müller J.D., Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiten D.R., Cox D., Wilson M.R. Single-molecule characterization of the interactions between extracellular chaperones and toxic α-synuclein oligomers. Cell Rep. 2018;23:3492–3500. doi: 10.1016/j.celrep.2018.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zuiderweg E.R.P., Hightower L.E., Gestwicki J.E. The remarkable multivalency of the Hsp70 chaperones. Cell Stress Chaperones. 2017;22:173–189. doi: 10.1007/s12192-017-0776-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lotz G.P., Legleiter J., Muchowski P.J. Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J. Biol. Chem. 2010;285:38183–38193. doi: 10.1074/jbc.M110.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feifel B., Sandmeier E., Christen P. Potassium ions and the molecular-chaperone activity of DnaK. Eur. J. Biochem. 1996;237:318–321. doi: 10.1111/j.1432-1033.1996.0318n.x. [DOI] [PubMed] [Google Scholar]
- 50.Nunes J.M., Mayer-Hartl M., Müller D.J. Action of the Hsp70 chaperone system observed with single proteins. Nat. Commun. 2015;6:6307. doi: 10.1038/ncomms7307. [DOI] [PubMed] [Google Scholar]
- 51.Scales N., Swain P.S. Resolving fluorescent species by their brightness and diffusion using correlated photon-counting histograms. PLoS One. 2019;14:e0226063. doi: 10.1371/journal.pone.0226063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hubin E., Verghese P.B., Broersen K. Apolipoprotein E associated with reconstituted high-density lipoprotein-like particles is protected from aggregation. FEBS Lett. 2019;593:1144–1153. doi: 10.1002/1873-3468.13428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richter K., Haslbeck M., Buchner J. The heat shock response: life on the verge of death. Mol. Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 54.Mannini B., Cascella R., Chiti F. Molecular mechanisms used by chaperones to reduce the toxicity of aberrant protein oligomers. Proc. Natl. Acad. Sci. USA. 2012;109:12479–12484. doi: 10.1073/pnas.1117799109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clerico E.M., Tilitsky J.M., Gierasch L.M. How hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J. Mol. Biol. 2015;427:1575–1588. doi: 10.1016/j.jmb.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fernandez-Funez P., Sanchez-Garcia J., Rincon-Limas D.E. Holdase activity of secreted Hsp70 masks amyloid-β42 neurotoxicity in Drosophila. Proc. Natl. Acad. Sci. USA. 2016;113:E5212–E5221. doi: 10.1073/pnas.1608045113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tao J., Berthet A., McConlogue L. Hsp70 chaperone blocks α-synuclein oligomer formation via a novel engagement mechanism. bioRxiv. 2020 doi: 10.1101/2020.05.15.098699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Österlund N., Lundquist M., Emanuelsson C. Amyloid-β oligomers are captured by the DNAJB6 chaperone: direct detection of interactions that can prevent primary nucleation. J. Biol. Chem. 2020;295:8135–8144. doi: 10.1074/jbc.RA120.013459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kellner R., Hofmann H., Schuler B. Single-molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc. Natl. Acad. Sci. USA. 2014;111:13355–13360. doi: 10.1073/pnas.1407086111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Horvath I., Blockhuys S., Wittung-Stafshede P. Interaction between copper chaperone Atox1 and Parkinson’s disease protein α-synuclein includes metal-binding sites and occurs in living cells. ACS Chem. Neurosci. 2019;10:4659–4668. doi: 10.1021/acschemneuro.9b00476. [DOI] [PubMed] [Google Scholar]
- 61.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One. 2011;6:e26319. doi: 10.1371/journal.pone.0026319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zietkiewicz S., Lewandowska A., Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J. Biol. Chem. 2006;281:7022–7029. doi: 10.1074/jbc.M507893200. [DOI] [PubMed] [Google Scholar]
- 63.Fourie A.M., Sambrook J.F., Gething M.J. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J. Biol. Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- 64.Flynn G.C., Chappell T.G., Rothman J.E. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- 65.Schröder H., Langer T., Bukau B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Van Durme J., Maurer-Stroh S., Schymkowitz J. Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput. Biol. 2009;5:e1000475. doi: 10.1371/journal.pcbi.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Röder C., Kupreichyk T., Schröder G.F. Cryo-EM structure of islet amyloid polypeptide fibrils reveals similarities with amyloid-β fibrils. Nat. Struct. Mol. Biol. 2020;27:660–667. doi: 10.1038/s41594-020-0442-4. [DOI] [PubMed] [Google Scholar]
- 68.Weirich F., Gremer L., Heise H. Structural characterization of fibrils from recombinant human islet amyloid polypeptide by solid-state NMR: the central FGAILS segment is part of the β-sheet core. PLoS One. 2016;11:e0161243. doi: 10.1371/journal.pone.0161243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kampinga H.H., Craig E.A. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H., Zhu H., Liu Q. An unexpected second binding site for polypeptide substrates is essential for Hsp70 chaperone activity. J. Biol. Chem. 2020;295:584–596. doi: 10.1074/jbc.RA119.009686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kettern N., Dreiseidler M., Höhfeld J. Chaperone-assisted degradation: multiple paths to destruction. Biol. Chem. 2010;391:481–489. doi: 10.1515/BC.2010.058. [DOI] [PubMed] [Google Scholar]
- 72.Mangione M.R., Vilasi S., San Biagio P.L. Hsp60, amateur chaperone in amyloid-beta fibrillogenesis. Biochim. Biophys. Acta. 2016;1860:2474–2483. doi: 10.1016/j.bbagen.2016.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.