Summary
Human infertility is a multifactorial disease that affects 8%–12% of reproductive-aged couples worldwide. However, the genetic causes of human infertility are still poorly understood. Synaptonemal complex (SC) is a conserved tripartite structure that holds homologous chromosomes together and plays an indispensable role in the meiotic progression. Here, we identified three homozygous mutations in the SC coding gene C14orf39/SIX6OS1 in infertile individuals from different ethnic populations by whole-exome sequencing (WES). These mutations include a frameshift mutation (c.204_205del [p.His68Glnfs∗2]) from a consanguineous Pakistani family with two males suffering from non-obstructive azoospermia (NOA) and one female diagnosed with premature ovarian insufficiency (POI) as well as a nonsense mutation (c.958G>T [p.Glu320∗]) and a splicing mutation (c.1180−3C>G) in two unrelated Chinese men (individual P3907 and individual P6032, respectively) with meiotic arrest. Mutations in C14orf39 resulted in truncated proteins that retained SYCE1 binding but exhibited impaired polycomplex formation between C14ORF39 and SYCE1. Further cytological analyses of meiosis in germ cells revealed that the affected familial males with the C14orf39 frameshift mutation displayed complete asynapsis between homologous chromosomes, while the affected Chinese men carrying the nonsense or splicing mutation showed incomplete synapsis. The phenotypes of NOA and POI in affected individuals were well recapitulated by Six6os1 mutant mice carrying an analogous mutation. Collectively, our findings in humans and mice highlight the conserved role of C14ORF39/SIX6OS1 in SC assembly and indicate that the homozygous mutations in C14orf39/SIX6OS1 described here are responsible for infertility of these affected individuals, thus expanding our understanding of the genetic basis of human infertility.
Keywords: C14orf39/SIX6OS1, mutations, non-obstructive azoospermia, premature ovarian insufficiency, chromosome synapsis, synaptonemal complex
Introduction
Human infertility, defined as the failure to achieve a clinical pregnancy after 12 months or more of regular unprotected sexual intercourse,1 is estimated to affect 8%–12% of reproductive-aged couples worldwide.2 The etiology of human infertility is multifactorial, and genetic factors have been proposed to be involved in most cases.3 Although aberrations in the number and structure of chromosomes can explain a proportion of the affected individuals, the etiology and pathology for the large remaining number of infertile individuals, for whom single-gene variants could be the underlying causes, are still unclear.4 To date, several genes, such as STAG3 (MIM: 608489), BRCA2 (MIM: 600185), TEX11 (MIM:300311), HFM1 (MIM: 615684), and SHOC1 (MIM: 618038), have been associated with non-obstructive azoospermia (NOA) or premature ovarian insufficiency (POI).5, 6, 7, 8, 9, 10, 11 However, compared with the known genetic defects in more than 400 genes affecting mouse fertility,12 little is known about the genetic basis of human infertility.
Synaptonemal complex (SC), formed between paired homologous chromosomes during the prophase of meiosis I, is a conserved zipper-like structure that facilitates interhomolog crossover formation and successful production of gametes. The fully formed SC is composed of two parallel lateral elements (LEs) and a single central element (CE), which are bridged by numerous transverse filaments (TFs). So far, eight SC components have been identified in mammals, including two LE proteins, SYCP3 and SYCP2; one TF protein, SYCP1; and five CE proteins, SYCE1, SYCE2, SYCE3, TEX12, and SIX6OS1.13 Genetic disruption in any of them invariably causes mouse infertility,14, 15, 16, 17, 18, 19 except for Sycp3 and Sycp2 female mutant mice, which are subfertile.20,21 However, only mutations in SYCP3 (MIM: 604759), SYCE1 (MIM: 611486), and SYCP2 (MIM: 604105) have been associated with human infertility,22, 23, 24, 25 although the pathogenicity of these mutations was determined by in silico analysis or deduced from studies of knockout mice rather than by functional identification using animal models with the same mutation. Recently, humanized mutant mice were generated to evaluate the pathogenicity of the SYCE1 nonsense mutation (c.721C>T [p.Gln241*]) discovered in POI-affected individuals, which did not result in a truncated protein but induced nonsense-mediated mRNA degradation.25
C14orf39 (MIM: 617307) is the ortholog of murine Six6os1, which encodes a CE component of the SC and specifically interacts with SYCE1 via its α-helical domain.19 Knockout of Six6os1 in mice resulted in chromosomal asynapsis and meiotic arrest, which eventually led to azoospermia and ovarian failure.19 A heterozygous missense variant in C14orf39 (c.1570C>T [p.Leu524Phe]) was associated with higher recombination rates in human females, but not in males, in the Icelandic population.26 In contrast, another genome-wide association study conducted in the US population showed a strong association between C14orf39 and the recombination rate in males.27 Although these findings could be explained by ethnic differences, the relation between C14orf39 mutations and human fertility remains uncertain.
Here, we identified three pathogenic homozygous variants in C14orf39/SIX6OS1 (GenBank: NM_174978.3) from a consanguineous Pakistani infertile family with two males suffering from NOA and one female diagnosed with POI and two Chinese NOA-affected individuals. Functional analyses showed that the affected males displayed severe synaptic defects and meiotic arrest and, consequently, azoospermia. The pathogenicity of these mutations is validated by the Six6os1 mutant mouse model, which recapitulated the phenotypes of the NOA- and POI-affected individuals.
Material and methods
Clinical samples
Fifty consanguineous Pakistani families with at least two infertile siblings in each and 60 infertile Chinese men with meiotic arrest were collected. After we obtained informed consent, the individuals donated blood samples and testicular tissues for scientific research. Routine semen analyses were performed at least twice according to the WHO guidelines.28 The reproductive hormones in the serum of individuals were determined in the local laboratory. The karyotype analyses of all individuals and the ultrasound examination of the affected female’s reproductive system were performed in the local hospitals. This study was approved by the institutional ethics committee of the University of Science and Technology of China (USTC) with the approval number 2019-KY-168.
WES and variant filtration
WES was performed as we previously described.29 Variants of the Pakistani family were filtered according to the following criteria: (1) variants that conform to the autosomal recessive inheritance were included; (2) variants located in the linkage regions with the logarithm of the odds scores >0.58 were included;30 (3) variants with minor allele frequencies >0.05 in any of the public databases, including 1000 Genomes project, ESP6500, or gnomAD database, and variants that were homozygous in our in-house WES database generated from 578 fertile men (41 Pakistani, 254 Chinese, and 283 European) were excluded; (4) variants potentially affecting protein sequence (nonsense, missense, splice-site variants, and coding indels) were included; (5) variants within genes not expressed in the testis were excluded; (6) variants predicted to be deleterious by more than half of the software (provided by ANNOVAR) covering them were included;31 and (7) variants within genes dispensable for spermatogenesis in mice on the basis of SpermatogenesisOnline1.0 or literature search were excluded.32 The remaining variants were subsequently verified by Sanger sequencing in all the available family members. Primer sequences are shown in Table S2, and the flow chart of the variant filtering process is described in Figure S1.
Spermatocyte nuclear surface spreading and immunofluorescence staining
The surface spreads of spermatocytes were prepared according to previously described protocols.33,34 For immunofluorescence, slides were blocked with 3% nonfat milk in phosphate-buffered saline (PBS) for 30 min and then incubated with primary antibodies overnight at 37°C in a humidified chamber. After three washes in PBS containing 0.05% Triton X-100 (PBST), secondary antibodies were applied for 1 h at 37°C. Subsequently, the slides were washed three times in PBST and mounted with the Vectashield Medium (Vector Laboratories, H-1000). Images were captured with an Olympus BX61 microscope with a CCD camera (QImaging, QICAM Fast 1394) and processed with Image-Pro Plus software (Media Cybernetic). The antibodies are listed in Table S1.
Quantitative polymerase chain reaction
Total RNAs were extracted with RNAiso Plus reagents (TaKaRa, 9109), and the cDNAs were synthesized with the PrimeScript RT reagent kit (TaKaRa, RR047A) according to the manufacturer’s protocol. The quantitative polymerase chain reaction (qPCR) was performed with TransStart Top Green qPCR SuperMix (TransGen Biotech, AQ132) and a StepOne Real-Time PCR System (Applied Biosystems). We calculated relative mRNA levels by normalizing to ACTB (MIM: 102630). The relative mRNA levels in the control were regarded as 1, and the fold changes in the affected individuals were compared to this. Primer sequences are listed in Table S2.
Yeast two-hybrid assay
C14orf39 was cloned from human testis cDNA with the PrimeStar HS DNA Polymerase (TaKaRa, R010A) and then inserted into the pGADT7 vector and transformed into Y187 stain as prey. Mouse Syce1 was cloned from mouse testis cDNA, inserted into the pGBKT7 vector, and transformed into Y2HGold strain as bait. The mutagenesis was accomplished by overlap PCR. The construction of the plasmids was achieved by homologous recombination with the Exnase enzyme (Vazyme, C113). Yeast transformation was conducted according to the manual (PT1172-1) from Clontech Laboratories, and two-hybrid assays were performed as we previously described.33 We plated the mating products on SD/−Trp/−Leu (−LW) and SD/−Trp/−Leu/−His/−Ade (−LWHA) plates for 3–5 days to test the interactions. The pGBKT7 and pGADT7 vectors were used as negative control.
Polycomplex formation
Wild-type and mutated C14orf39 coding sequences were fused to the C terminus of EGFP, and the coding sequence of SYCE1 (GenBank: NM_001143765.1) was fused to the C terminus of mCherry. COS-7 cells (SCSP-508) were kindly provided by Stem Cell Bank (Chinese Academy of Sciences) and cultured in 24-well plates with a coverslip at the bottom and transfected with the vectors with Lipofectamine 3000 according to the manufacturer’s instructions. After 36 h of transfection, the cells were fixed in 4% polyformaldehyde for 10 min and the nuclei were stained with Hoechst 33342 (Invitrogen, H21492). After three washes in PBS, the coverslips were mounted on slides with Vectashield Medium. The images were captured with a Nikon Eclipse Ti2 inverted microscope with a C2 plus laser scanning confocal head under the control of NIS elements imaging software version 5.01.
Histological analysis
Testicular tissues were fixed in Bouin’s solution overnight and then embedded into the paraffin and sectioned at 5 μm thickness. The tissue slides were deparaffinized by xylene and rehydrated with gradient ethanol and then sequentially stained with hematoxylin and eosin. After dehydration and transparency, the tissue sections were sealed with neutral resin. Ovaries were fixed with 4% formaldehyde solution and only stained with hematoxylin. The images were captured via a Nikon ECLIPSE 80i microscope with a DR-Ri1 camera and processed with NIS-elements BR software. Immunofluorescence staining on the testicular sections was conducted as we previously described.35 The antibodies are listed in Table S1.
Generation of Six6os1 mutant mice
Six6os1 (GenBank: NM_029444.2) mutant mice carrying mutation analogous to the affected individuals was generated via CRISPR/Cas9 technology as we described previously.33 The sgRNA sequence and primers used for genotyping are listed in Table S2. The mice were maintained under specific-pathogen-free conditions in the laboratory animal center of USTC. All experiments involving animals were approved by the institutional animal ethics committee of USTC.
Statistical analysis
Statistical significance was assessed with an unpaired, two-tailed Student’s t test or Welch’s t test. All p values and statistical tests were specified in the text and figure legends. A p value of <0.05 was considered statistically significant.
Results
Identification of C14orf39 pathogenic variants
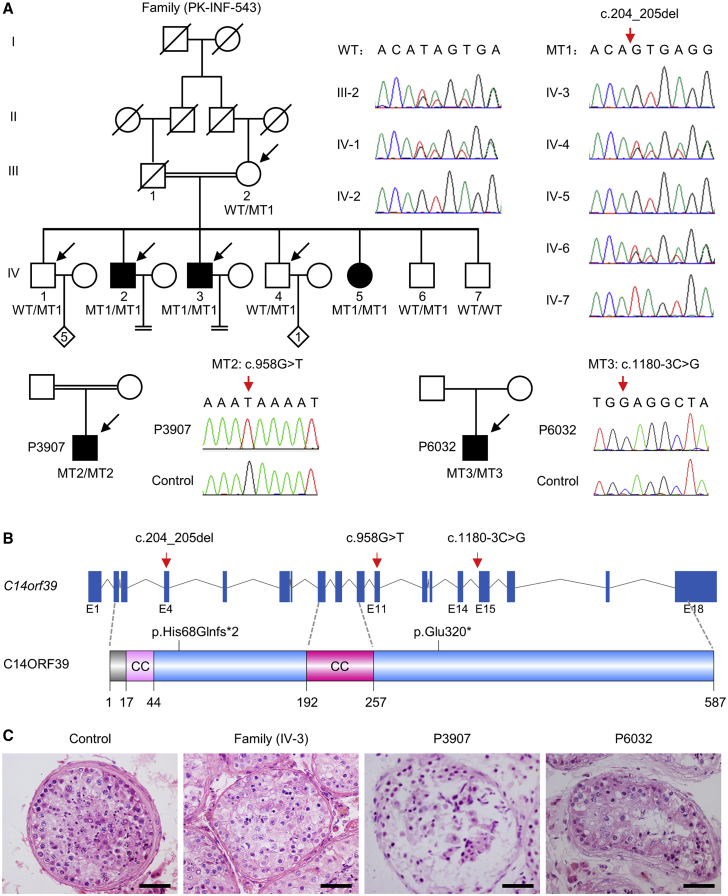
To reveal the genetic causes of human infertility, we have collected 50 consanguineous Pakistani families with at least two infertile siblings in each and 60 Chinese men with meiotic arrest. Among them, C14orf39 mutations were identified from one consanguineous Pakistani family and two Chinese men (Figure 1A). For the family PK-INF-543, we performed WES analyses of the two affected individuals with NOA (IV-2 and IV-3), the two fertile brothers (IV-1 and IV-4), and their mother (III-2) (Figure 1A). More than 400,000 variants per individual were detected from the WES data that covered at least 99% of the targeted sequence at 90× or greater depth. Considering that the individuals were born to a consanguineous marriage, variants following autosomal recessive inheritance were prioritized. These variants were filtered with a series of criteria, and two candidate pathogenic variants, a 1-bp insertion in DHRS4L2 (c.629_630insA [p.His211Alafs*69] [MIM: 615196] [GenBank: NM_198083.4]) and a 2-bp deletion in C14orf39 (c.204_205del [GenBank: NM_174978.3]) were identified (Figure S1). To determine the transmission patterns of these variants in this family, we performed targeted Sanger sequencing in all available family members. The DHRS4L2 variant was ruled out because of its heterozygosity in the diseased sister, IV-5 (Figure S2). Only the mutation in C14orf39 conformed to Mendelian segregation in all family members. This frameshift variant was confirmed at the mRNA level in testicular tissues of individual IV-3 (Figures 1A and S3). Besides, this C14orf39 variant (c.204_205del [p.His68Glnfs∗2]) is absent in the 1000 Genomes Project database and has low frequencies in the ESP6500 and gnomAD databases (Table S3). Given that this homozygous mutation is expected to cause 520 amino acid deletions in the full-length C14ORF39 protein consisting of 587 amino acids, and the meiotic arrest phenotype of Six6os1 knockout mice, we think that the mutation in C14orf39 (c.204_205del [p.His68Glnfs∗2]) is the most likely pathogenic variant in this family.
Figure 1.
Identification of homozygous variants in C14orf39 from NOA- and POI-affected individuals
(A) The pedigrees of the families. Double horizontal lines represent the consanguineous unions. Squares and circles denote male and female members, respectively. The number in the diamond indicates the number of offspring whose gender is unknown. Solid symbols indicate the affected members, and open symbols denote unaffected members. Slashes represent deceased members, and the equal sign indicates infertility. Members indicated by arrows were selected for whole-exome sequencing. Sanger sequencing chromatograms of C14orf39 are shown on the right side. The red arrows indicate the corresponding mutations.
(B) Schematic map of the mutation positions in C14orf39 at the genomic and protein levels. The gene composition is based on the Ensembl database (GRCh38, transcript ID: ENST00000321731.8). The blue solid squares represent exons, and the lines represent introns. Two putative coiled-coil (CC) domains of C14ORF39 were predicted by the web-based COILS program.
(C) Histological analyses of human testicular tissues by H&E staining. Scale bars indicate 50 μm.
We also identified two other C14orf39 homozygous mutations as the only candidate pathogenic variants in two Chinese men with meiotic arrest. Individual P3907 had a nonsense mutation (c.958G>T [p.Glu320∗]), and individual P6032 carried a splicing mutation (c.1180−3C>G). Both variants were validated by Sanger sequencing and have not been reported in any public databases (Figure 1A and Table S3). Therefore, three pathogenic variants in C14orf39 were identified in infertile individuals of different ethnic groups (Figure 1B), implying that C14ORF39 provides a genetic contribution to human fecundity.
Clinical characteristics of the affected individuals
On the basis of the WHO guidelines, at least two semen analyses were conducted in affected individuals IV-2 and IV-3 and showed that they had normal semen volume but no sperm (Table 1). To examine the spermatogenesis of individuals carrying homozygous C14orf39 mutations, we obtained testicular biopsies from three affected men (IV-3, P3097, and P6032) and a control man. H&E staining on the testicular sections revealed that many spermatogenic cells, including spermatogonia, spermatocytes, and spermatozoa, were observed in the seminiferous tubules of the control, whereas in the affected individuals, spermatogonia and spermatocytes were found, but post-meiotic cells were absent in all of their available seminiferous tubules (Figure 1C). These results indicated that the affected males suffered from non-obstructive azoospermia and their spermatogenesis was arrested at the spermatocyte stage. Notably, the unmarried sister (IV-5, 30 years old) in the family PK-INF-543 was considered as an individual with POI,36 characterized by irregular menstrual cycles, amenorrhea at the age of 24, high levels of follicle-stimulating hormone and luteinizing hormone, and low levels of anti-Müllerian hormone (Table 1 and Supplemental notes).
Table 1.
Clinical characteristics of the family members
| Ref values | IV-1 | IV-2 | IV-3 | IV-4 | IV-5 | IV-6 | IV-7 | III-1 | |
|---|---|---|---|---|---|---|---|---|---|
| Age (years)a | – | 43 | 40 | 37 | 33 | 30 | 28 | 24 | 58 |
| Gender | – | M | M | M | M | F | M | M | F |
| Height/weight (cm/kg) | – | 176/74 | 178/69 | 170/71 | 172/66 | 164/54 | 174/65 | 169/62 | 166/60 |
| Karyotype | – | 46,XY | 46,XY | 46,XY | 46,XY | 46,XX | 46,XY | 46,XY | 46,XX |
| Diagnosis of disease | – | – | NOA | NOA | – | POI | – | – | – |
| Semen analysisb | |||||||||
| Semen volume (mL) | >1.5 | 3.0 | 2.5 ± 0.87 | 2.0 ± 0.41 | – | – | – | – | – |
| Sperm count (millions/mL) | >15 | 42 | 0 | 0 | – | – | – | – | – |
| Hormone analysisc | |||||||||
| Testosterone (ng/dL) | 249–836 | – | 436.5 | 426.1 | – | – | – | – | – |
| FSH (mIU/mL) | M: 1.4–15.4 F: 1.4–9.9 |
– | 7.41 | 6.79 | – | 49.06 | – | – | – |
| LH (mIU/mL) | M: 1.2–7.8 F: 1.7–15 |
– | 4.83 | 3.95 | – | 47.69 | – | – | – |
| Progesterone (ng/mL) | M: 0.2–1.22 F: 0.1–1.40 |
– | 0.37 | 0.24 | – | 0.21 | – | – | – |
| Prolactin (ng/mL) | M: 3–14.7 | – | 6.42 | 9.86 | – | – | – | – | – |
| AMH (ng/mL) | F: 0.9–9.5 | – | – | – | – | 0.021 | – | – | – |
| Physical examinationd | |||||||||
| Ovaries (mL) | >3.5 | – | – | – | – | L: 1 mL; R: invisible | – | – | – |
| Uterus (cm∗cm) | >9∗8 | – | – | – | – | 5.5∗2.1 | – | – | – |
Ref, reference; NOA, non-obstructive azoospermia; POI, premature ovarian insufficiency; M, male; F, female; FSH, follicle-stimulating hormone; LH, luteinizing hormone; AMH, anti-Müllerian hormone; L, left; R, right.
Ages at the manuscript submission.
Reference values were published by WHO in 2010.
Reference values were suggested by the local clinical laboratory.
Physical examination was performed by a consultant gynecologist.
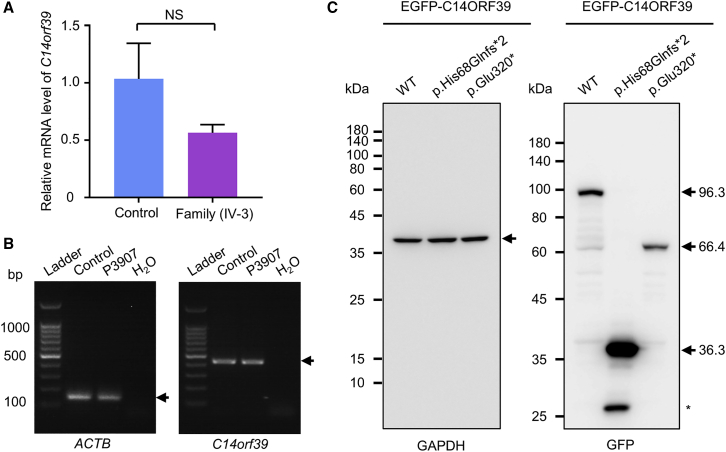
Effects of the identified mutations on C14orf39 expression
Considering that both the C14orf39 frameshift mutation (c.204_205del) and nonsense mutation (c.958G>T) introduce a premature stop codon, these mutations may result in mRNA degradation or truncated proteins. To test these possibilities, we first performed qPCR and reverse-transcription PCR (RT-PCR) to evaluate the relative transcriptional expression of C14orf39. The mRNA levels of C14orf39 in the testis of individual IV-3, carrying the homozygous frameshift mutation, were 56.4% of that in the control (p = 0.0527; Welch’s t test; Figure 2A), and individual P3907, harboring the homozygous nonsense mutation, showed comparable expression to the control group (Figure 2B). To clarify whether these mutations result in truncated proteins, we expressed wild-type and mutated C14ORF39 with an N-terminal EGFP fusion in COS-7 cells, followed by immunoblot examination. The mutant proteins were observed at the expected sizes (Figure 2C), indicating that these C14orf39 mutations led to the production of truncated proteins.
Figure 2.
The effects of C14orf39 mutations on gene expression
(A) Quantitative PCR analyses of the relative C14orf39 expression in the testes from the control and individual IV-3. Data are from four repeated experiments and represent means ± SD. Statistical significance was determined using Welch’s t test. NS, not significant. ACTB was used as an internal control.
(B) RT-PCR analysis of blood samples from a man with obstructive azoospermia and individual P3907 carrying the C14orf39 nonsense mutation (c. 958G>T [p.Glu320∗]). ACTB served as a loading control. Arrows indicate targeted bands.
(C) Heterologous expression of C14ORF39 in COS-7 cells. N-terminal EGFP-fused wild-type (WT) and two mutated C14orf39 (c.204_205del [p.His68Glnfs∗2] and c.985G>T [p.Glu320∗]) plasmids were transfected into COS-7 cells. After 36 h of transfection, whole-cell lysates were prepared and separated by SDS-PAGE. C14ORF39 was detected by GFP antibody and GAPDH served as a loading control. Arrows indicate the targeted proteins, and the asterisk indicates non-specific band.
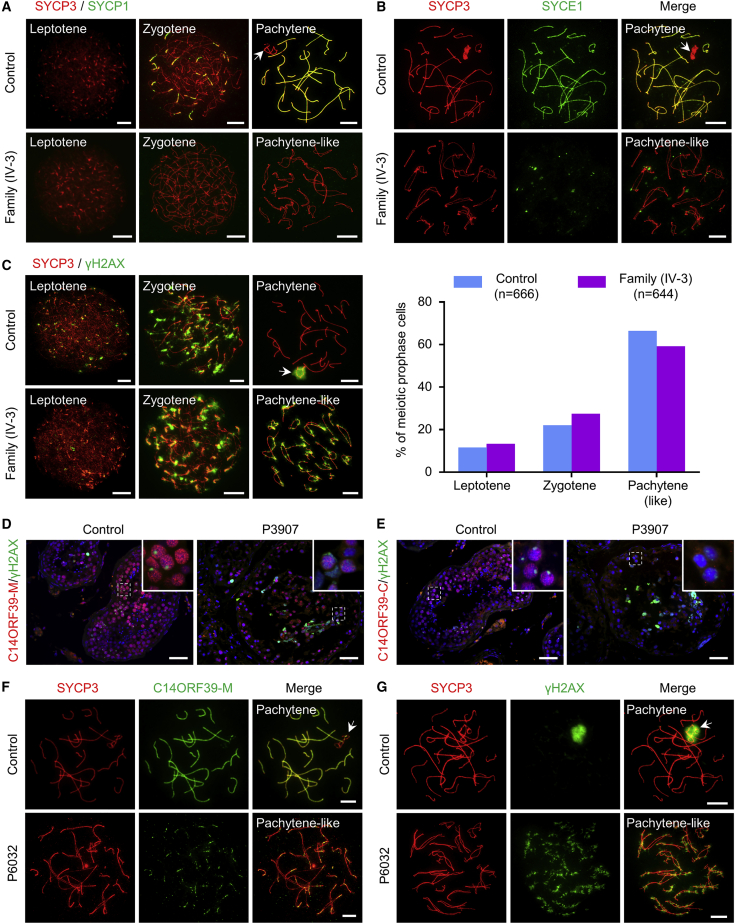
Synaptic defects and meiotic arrest were observed in individuals with homozygous C14orf39 mutations
To test whether these mutations in C14orf39 affected the assembly of SCs in vivo, we obtained testicular biopsies from affected males and prepared spermatocyte spreads for subsequent immunofluorescence staining. For individual IV-3 from the family PK-INF-543, we first stained the spermatocyte spreads for SYCP3 and SYCP1 to assess chromosome synapsis (Figure 3A). In control cells, SYCP1 signals first appeared as short stretches between the paired chromosomes at zygotene and extended to the full length of the chromosomal axes at pachytene. In spermatocytes of individual IV-3, however, the SYCP1 signal was undetectable on the chromosomal axes, even in the most advanced spermatocytes, when fully assembled lateral elements were extensively co-aligned in some regions. Consistent with this finding, the SYCE1 signals were also absent from the chromosomal axes of spermatocytes in individual IV-3 (Figure 3B). These results indicated that the C14orf39 frameshift mutation (c.204_205del [p.His68Glnfs∗2]) caused complete asynapsis of homologous chromosomes.
Figure 3.
The spermatocytes of individuals carrying homozygous C14orf39 mutations displayed severe synaptic defects and meiotic arrest
(A) Synapsis analyses of the spermatocytes from the control and individual IV-3 by immunofluorescence staining with antibodies against SYCP3 (red) and SYCP1 (green).
(B) Representative images of surface-spread spermatocytes from the control and individual IV-3 stained with antibodies against SYCP3 (red) and SYCE1 (green).
(C) Meiotic prophase I analysis. Representative images of surface-spread spermatocytes from the control and individual IV-3 stained with antibodies against SYCP3 (red) and γH2AX (green). Statistical results are shown on the right side. The “n” indicates the number of meiotic prophase cells examined.
(D and E) Immunofluorescence staining of testicular sections from the control and individual P3097 with antibodies against γH2AX (green) and C14ORF39-M (D, red) or C14ORF39-C (E, red). The nuclei were stained with Hoechst 33342 (blue). The insets show magnified images of the field in white dashed rectangles. Scale bars indicate 50 μm.
(F and G) Representative images of surface-spread spermatocytes from the control and individual P6032 stained with antibodies against SYCP3 (red) and C14ORF39-M (F, green) or γH2AX (G, green). The white arrows in (A), (B), and (F) indicate sex chromosomes, and the arrows in (C) and (G) indicate the sex body. Scale bars denote 10 μm in (A), (B), (C), (F), and (G).
Because severe synaptic defects always cause meiotic arrest in mammals,37 we thus examined the progression of meiotic prophase I by immunostaining the spermatocytes for SYCP3 and γH2AX, a marker of DNA breaks. Consistent with our previous report,38 for the control, we observed that 33.6% of spermatocytes were at leptotene and zygotene, characterized by small patches and bright focal domains of γH2AX signals, respectively, and 66.4% of spermatocytes at pachytene showed a typical sex body with intense γH2AX signals around the X and Y chromosomes. For individual IV-3, after analyzing 644 spermatocytes, we found that the leptotene and zygotene nuclei were similar to that of the control, however, the most advanced spermatocytes displayed shorter and thicker LEs but persistent γH2AX signals on autosomes and failed to form sex body (Figure 3C). These results indicated that meiosis was arrested at the pachytene-like stage in affected individual IV-3.
For individual P3907, we performed immunofluorescence staining on testicular sections by using antibodies recognizing the middle segment (C14ORF39-M) or C terminus of C14ORF39 (C14ORF39-C) (Figure S4). To examine the progression of meiotic prophase I, we used the γH2AX antibody concurrently. In control seminiferous tubules, C14ORF39 was located in the spermatocyte nuclei and displayed linear signals in the spermatocytes with a typical sex body, which is consistent with its function as a CE component of SCs. Interestingly, the signals of the C14ORF39-M antibody were observed in the spermatocyte nuclei of individual P3907 (Figure 3D), while the C-terminal antibody signals were absent (Figure 3E), which not only reaffirmed the existence of truncated C14ORF39 but also suggested that synapsis of homologous chromosomes in individual P3907 was incomplete. Notably, spermatocytes with a typical sex body were easily found in the control, but no such cells were found in any of the available seminiferous tubules of individual P3907 carrying the homozygous C14orf39 nonsense mutation (c.958G>T), similar to the observations in the affected individual (IV-3) from the family PK-INF-543.
For individual P6032, the identified C14orf39 splicing mutation (c.1180−3C>G) was also expected to produce a truncated protein that was longer than that in individual P3907. To determine the localization of the truncated protein and to evaluate the synapsis of homologous chromosomes, we incubated the spermatocytes with SYCP3 and C14ORF39-M antibodies. The signals of C14ORF39-M antibody were observed as short stretches at the paired regions of homologous chromosomes (Figure 3F), suggesting that the truncated protein produced by the splicing mutation of C14orf39 localized to the chromosomal axes. It bears mentioning that we never observed the C14ORF39-M signals that extended along the entire length of the autosomal axes in the spermatocytes of individual P6032, indicating incomplete synapsis in this man. Furthermore, γH2AX staining results showed that no spermatocytes were able to form a typical sex body (Figure 3G), suggesting meiotic arrest at the pachytene-like stage in this individual.
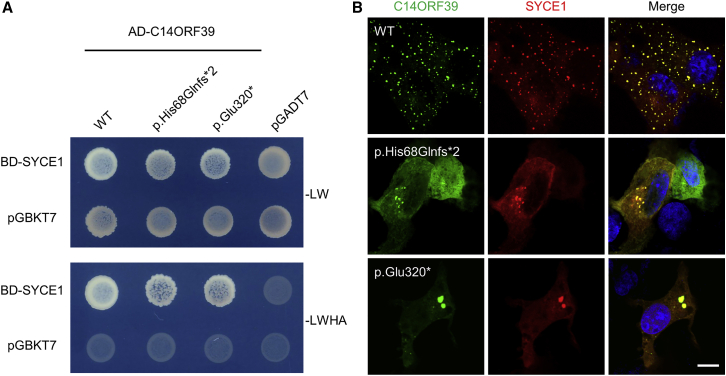
Mutant C14ORF39 retained SYCE1 binding but decreased polycomplex formation
It has been reported that C14ORF39/SIX6OS1 specifically interacts with SYCE1.19 We thus wondered whether these mutations disrupt the interaction of C14ORF39 with SYCE1. For this purpose, we performed yeast two-hybrid (Y2H) assays to detect the protein-protein interaction (Figure 4A). Since human C14ORF39 and SYCE1 were both auto-activated in the Y2H system when used as bait, we instead used human C14ORF39 as prey and mouse SYCE1 as bait. Diploid yeast cells co-expressing Gal4-AD-fused wild-type or mutated C14ORF39 (p.His68Glnfs∗2 and p.Glu320∗) and Gal4-BD-fused SYCE1 grew well on the selective medium plates, suggesting that these mutations in C14orf39 did not disrupt the interaction between C14ORF39 and SYCE1.
Figure 4.
Mutant C14ORF39 retained SYCE1 binding but diminished polycomplex formation
(A) Yeast two-hybrid assays for pairwise protein interaction. WT and mutated C14ORF39 (p.His68Glnfs∗2 and p.Glu320∗) were used as prey, and mouse SYCE1 was used as bait. The pGBKT7 and pGADT7 vectors were used as negative controls. The pictures were taken after 4 days of growth. −LW, medium lacking Leu and Trp amino acids; −LWHA, medium lacking Leu, Trp, His, and Ade.
(B) Polycomplex formation assays. EGFP-fused WT and mutated C14ORF39 (green) were co-expressed with mCherry-fused SYCE1 (red) in COS-7 cells. The nuclei were stained with Hoechst 33342 (blue). The experiments were repeated three times, and the representative images are shown. Scale bars indicate 10 μm.
Because C14ORF39/SIX6OS1 can form punctate aggregates (named as polycomplex) when co-expressed with SYCE1,19 we next examined whether these mutations affected polycomplex formation. As expected, large abundant punctate aggregates were evident in the cytoplasm of COS-7 cells that co-expressed wild-type C14ORF39 and SYCE1. Notably, the two C14ORF39 mutants (p.His68Glnfs∗2 and p.Glu320∗) also formed aggregates when co-expressed with SYCE1, but the number of punctate aggregates was greatly reduced (Figure 4B). These results indicated that the C14orf39 mutations impaired polycomplex formation between C14ORF39 and SYCE1, which partly explains the synaptic defects observed in affected individuals.
Six6os1 mutant mice recapitulated the NOA and POI phenotypes of individuals with homozygous C14orf39 mutations
To confirm that the identified C14orf39 mutations indeed cause human infertility, we characterized the phenotypes of Six6os1 mutant mice. The C14orf39 frameshift mutation (c.204_205del) identified in affected individuals from the consanguineous family is located at the N terminus, and the phenotypes of both male and female individuals were consistent with reported observations in Six6os1 knockout mice.19 Unlike the phenotype caused by the homozygous frameshift mutation (c.204_205del), the other two C14orf39 mutations (c.958G>T and c.1180−3C>G) led to incomplete synapsis in the spermatocytes of the two affected Chinese men, possibly because of the presence of longer truncated proteins. Thus, we generated a mouse model that retained the N terminus of Six6os1 in order to mimic that in our Chinese individuals. These mutant mice harbored a 4-bp deletion (c.907_910del [p.Asn303Serfs*9]) that did not affect Six6os1 mRNA abundance but did lead to a truncated protein lacking 272 C-terminal amino acids, termed Six6os1ΔC/ΔC mice (Figures S4 and S5).
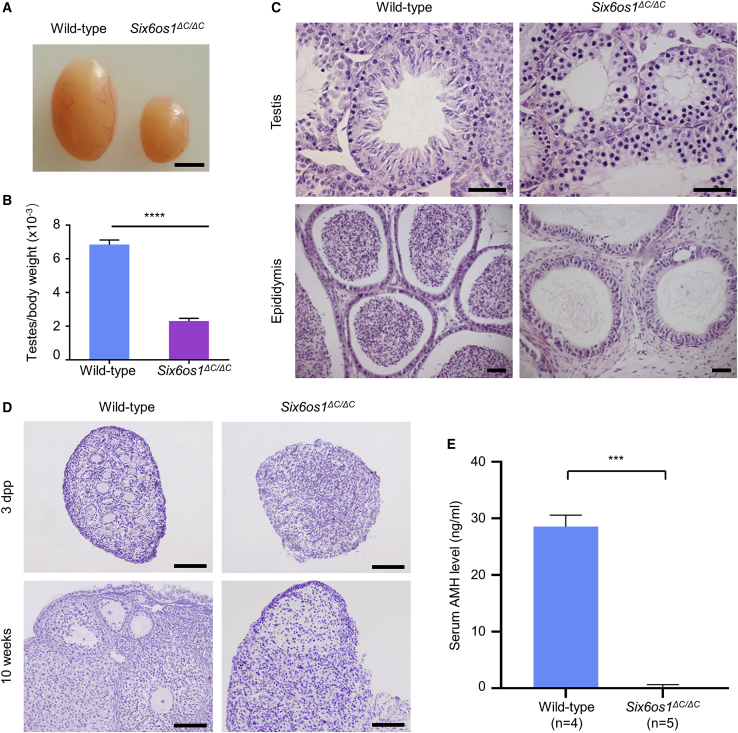
Two-month-old Six6os1ΔC/ΔC males showed a reduced testis size and a significant decrease (p < 0.0001; unpaired Student’s t test; 66.3%) in the testis to body weight ratio compared to their wild-type littermates (Figures 5A and 5B). Histological analyses of testicular sections revealed no post-meiotic germ cells in the seminiferous tubules of the Six6os1ΔC/ΔC mice. In agreement with these results, no sperm were found in the epididymis of the Six6os1 mutant mice (Figure 5C), suggesting a complete meiotic arrest.
Figure 5.
Six6os1ΔC/ΔC mice recapitulated the NOA and POI phenotypes of individuals with homozygous C14orf39 mutations
(A) Testis morphology of 2-month-old wild-type and Six6os1ΔC/ΔC mice. Scale bar indicates 2 mm.
(B) Testis to body weight ratio. The data are obtained from three mice for each genotype and represent means ± SD. The significance was determined via unpaired Student’s t test. ∗∗∗∗p < 0.0001.
(C) Histological sections of testes and epididymides from wild-type and Six6os1ΔC/ΔC mice. Scale bars denote 50 μm.
(D) Histological sections of ovaries from wild-type and Six6os1ΔC/ΔC female mice at the indicated ages were stained with hematoxylin. Scale bars indicate 50 μm.
(E) The serum AMH levels of 3- to 4-month-old wild-type and Six6os1ΔC/ΔC female mice. Data are expressed as means ± SD, and n shows the number of mice analyzed. Welch’s t test was used to determine statistical significance. ∗∗∗p < 0.001. AMH, anti-Müllerian hormone.
Given that the sister (IV-5) carrying a homozygous C14orf39 mutation was diagnosed with POI, we thus analyzed the ovaries of 3-days-postpartum (dpp) and 10-week-old female mice histopathologically. We easily detected numerous follicles of different developmental stages in the ovarian sections of wild-type mice. However, no follicles were found in the female Six6os1 mutant mice at each age examined (Figure 5D). Furthermore, the serum AMH levels of 3- to 4-month-old Six6os1ΔC/ΔC female mice averaged 0.22 ng/mL, significantly lower than that in wild-type mice, which showed an average serum AMH level of 28.57 ng/mL (p < 0.001; Welch’s t test; Figure 5E). These results suggested that Six6os1ΔC/ΔC female mice mimicked the POI phenotype of the affected sister.
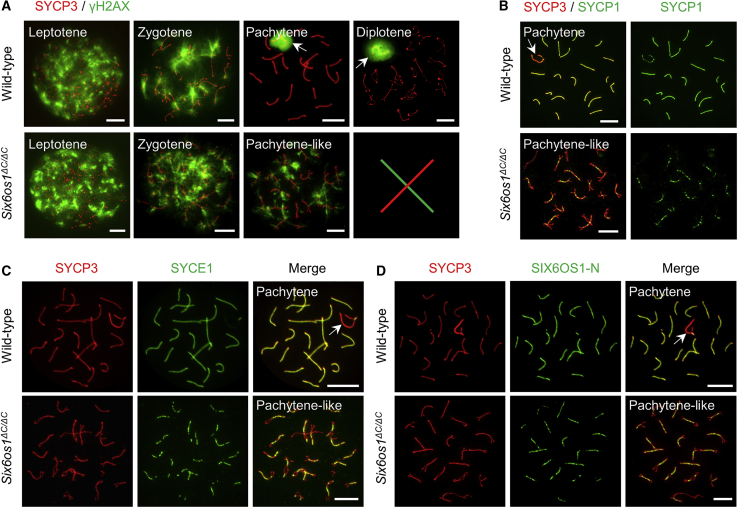
To identify the exact substage at which meiotic arrest occurred in Six6os1ΔC/ΔC male mice, we examined the progression of meiotic prophase I, as we did for the affected individuals. In wild-type spermatocytes, γH2AX signals that were diffused in the nuclei at leptotene gradually diminished as synapsis ensued during zygotene and finally disappeared from autosomes. The γH2AX signals ultimately accumulated on sex chromosomes to form the sex body from the pachytene to diplotene stage. However, in Six6os1 mutant spermatocytes, γH2AX signals appeared and diminished normally but were persistent on the chromosomes and failed to form the sex body, thereby indicating a meiotic arrest, as observed in the affected men (Figure 6A). Next, we investigated the chromosomal synapsis by staining the spermatocytes with antibodies against SYCP3 and SYCP1. In wild-type pachytene spermatocytes, SYCP1 showed a continuous linear signal along the entire length of the autosomal axes. However, only short stretches of SYCP1 signals were observable in the most advanced spermatocytes of Six6os1ΔC/ΔC mice (Figure 6B). Similar results were observed for another synapsis marker, SYCE1 (Figure 6C), further indicating that the homologous chromosomes underwent incomplete synapsis in the mutant mice. Considering the essential role of SIX6OS1 in synapsis initiation, we inferred that the truncated SIX6OS1 should be located on the chromosomal axes. To test this, we stained the surface-spread spermatocytes with antibodies against SYCP3 and the N terminus of SIX6OS1 (SIX6OS1-N). In agreement with our presumption, the SIX6OS1 threads were detected in the synapsed regions of homologous chromosomes in Six6os1 mutant spermatocytes (Figure 6D). Taken together, these results showed that Six6os1ΔC/ΔC male mice recapitulated the NOA phenotype found in the affected men.
Figure 6.
Six6os1ΔC/ΔC male mice displayed meiotic arrest and incomplete synapsis between homologous chromosomes
(A) The progression of meiotic prophase I was analyzed by immunostaining surface-spread spermatocytes from wild-type and Six6os1ΔC/ΔC mice with antibodies against SYCP3 (red) and γH2AX (green). Representative images of leptotene, zygotene, pachytene, and diplotene spermatocytes from wild-type mice are shown. The meiosis of Six6os1ΔC/ΔC mice was arrested at the pachytene-like stage.
(B and C) Immunofluorescence staining of surface-spread spermatocytes from wild-type and Six6os1ΔC/ΔC mice with antibodies against SYCP3 (red) and SYCP1 (B, green) or SYCE1 (C, green).
(D) Representative images of surface-spread spermatocytes from wild-type and Six6os1ΔC/ΔC mice stained with antibodies against SYCP3 (red) and SIX6OS1-N (green). The white arrows in (A) indicate the sex body, and the arrows in (B), (C), and (D) indicate sex chromosomes. Scale bars denote 10 μm.
Discussion
SC is essential for crossover formation between homologous chromosomes and the successful production of gametes in mammals. The deficiency of SC components in mice consistently results in infertility or subfertility.14, 15, 16, 17, 18, 19 Here, for the first time, we identified three homozygous variants in C14orf39 from both male and female infertile individuals. All of these C14orf39 mutations led to severe synaptic defects and meiotic arrest in affected individuals, which was strongly supported by the observations in our Six6os1 (the murine ortholog of human C14orf39) mutant mice that recapitulated the phenotypes of our NOA- and POI-affected individuals, as well as in previously reported Six6os1 knockout mice.19 Our results thus confirm an important and conserved role for C14ORF39/SIX6OS1 in chromosomal synapsis and demonstrate that mutations in C14orf39 can result in human infertility.
The N terminus of C14ORF39/SIX6OS1 contains two SYCE1 binding regions: one located at the amino acid region from residues 1–67 and the other from 68–267. The integrity of the two SIX6OS1-SYCE1 binding interfaces is essential for SC assembly in vivo.39 The C14orf39 frameshift mutation (c.204_205del [p.His68Glnfs∗2]) retains the first but loses the second binding interface and thus adversely affects protein function. Consistent with this finding, the mutant protein can still interact with SYCE1, but its ability to form aggregates with SYCE1 is diminished. More importantly, individuals with this frameshift mutation showed complete asynapsis of homologous chromosomes, confirming the pathogenicity of this mutation. Additionally, our in vitro and in vivo studies showed that both the C14orf39 nonsense mutation (c.958G>T [p.Glu320∗]) and splicing mutation (c.1180−3C>G) resulted in the generation of truncated proteins. Interestingly, although the two binding interfaces are retained, the mutant C14ORF39 protein (p.Glu320∗) also displayed an impaired polycomplex formation between C14ORF39 and SYCE1 in COS-7 cells. Furthermore, the individuals carrying these two homozygous C14orf39 mutations also exhibited synaptic defects and meiotic arrest, thereby providing in vivo evidence for the pathogenicity of these two mutations.
Previous study has shown that homologous chromosomes are unable to synapse in Six6os1 knockout mice.19 In contrast, the homologous chromosomes undergo partial synapsis in some closely co-aligned regions in our Six6os1ΔC/ΔC mice. These phenotypic differences are also observed between our Pakistani and Chinese affected individuals, indicating that the N terminus of C14ORF39/SIX6OS1 is sufficient to interact with SYCE1 and forms a complex with SYCE3 and SYCP1,17,40 thereby initiating SC assembly. Furthermore, the incomplete synapsis in Six6os1ΔC/ΔC mice is reminiscent of Syce2 and Tex12 knockout mice,16,18 suggesting that the C terminus of C14ORF39/SIX6OS1 plays an indispensable role in facilitating SC extension along the entire chromosomal axes.
It is worth noting that these three homozygous mutations in C14orf39 are identified from different ethnic populations. The first C14orf39 frameshift mutation was found in one of 50 Pakistani consanguineous families with at least two infertile siblings in each, which accounts for 2% of familial infertility. The other two C14orf39 mutations (one nonsense mutation and one splicing mutation) were found in two of 60 infertile Chinese men with meiotic arrest. In addition, a missense variant (p.Leu524Phe) in C14orf39 has been associated with higher recombination rates in human females in the Icelandic population.26 These results indicate that mutations in C14orf39 are not rare and are closely linked with human infertility, and individuals with NOA or POI should undertake mutation screening of C14orf39.
To date, several mutations in SC components have been reported to be associated with human infertility.22, 23, 24, 25 Notably, the mutations in C14orf39 we identified and the previously reported SYCE1 mutations are all homozygous and segregate with the disease via an autosomal recessive inheritance. However, SYCP3 and SYCP2 heterozygous mutations are also reported in azoospermic men or women with recurrent pregnancy loss and probably display a dominant-negative effect.24,41 These cases suggest that in human infertility, mutations occur frequently in the SC-coding genes and screening for mutations in these genes should be considered for adoption in the clinical practice of human-assisted reproduction.
In conclusion, our present data demonstrate that homozygous mutations in C14orf39 cause severe synaptic defects and meiotic arrest, which consequently leads to NOA and POI in humans. These findings highlight the essential and conserved role of C14ORF39/SIX6OS1 in SC assembly and will facilitate genetic diagnoses for clinical infertility while also enabling development of possible treatments for infertile individuals in the future.
Data and code availability
The accession numbers for the C14orf39 variants reported in this paper are ClinVar: SCV001468897, SCV001468898, and SCV001468899. The WES datasets supporting the current study have not been deposited in a public repository because of privacy and ethical restrictions but are available from the corresponding authors on request. The relevant NCBI accession numbers are GenBank: NM_174978.3 (for the reference sequence of human C14orf39), NP_777638.3 (for the reference sequence of human C14ORF39 protein), and NM_029444.2 (for the reference sequence of mouse Six6os1).
Declaration of interests
The authors declare no competing interests.
Acknowledgments
We are very grateful to all individuals for their informed consent to donate their DNA and tissue biopsies for scientific research and to the physicians who take care of them. This work was supported by the National Natural Science Foundation of China (31630050, 31890780, 32061143006, 31871514, and 82071709), the National Key Research and Developmental Program of China (2016YFC1000600, 2018YFC1003700, and 2018YFC1003900), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB19000000), and the Fundamental Research Funds for the Central Universities (YD2070002006 and WK9100000004).
Published: January 27, 2021
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ajhg.2021.01.010.
Contributor Information
Yuanwei Zhang, Email: zyuanwei@ustc.edu.cn.
Qinghua Shi, Email: qshi@ustc.edu.cn.
Web resources
1000 Genomes Project, https://www.internationalgenome.org/home
ESP6500, https://evs.gs.washington.edu/EVS/
OMIM, https://omim.org/
SpermatogenesisOnline 1.0, https://mcg.ustc.edu.cn/bsc/spermgenes/
Supplemental information
References
- 1.Zegers-Hochschild F., Adamson G.D., de Mouzon J., Ishihara O., Mansour R., Nygren K., Sullivan E., Vanderpoel S., International Committee for Monitoring Assisted Reproductive Technology. World Health Organization International Committee for Monitoring Assisted Reproductive Technology (ICMART) and the World Health Organization (WHO) revised glossary of ART terminology, 2009. Fertil. Steril. 2009;92:1520–1524. doi: 10.1016/j.fertnstert.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Vander Borght M., Wyns C. Fertility and infertility: Definition and epidemiology. Clin. Biochem. 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Krausz C., Cioppi F., Riera-Escamilla A. Testing for genetic contributions to infertility: potential clinical impact. Expert Rev. Mol. Diagn. 2018;18:331–346. doi: 10.1080/14737159.2018.1453358. [DOI] [PubMed] [Google Scholar]
- 4.Zorrilla M., Yatsenko A.N. The Genetics of Infertility: Current Status of the Field. Curr. Genet. Med. Rep. 2013;1:247–260. doi: 10.1007/s40142-013-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caburet S., Arboleda V.A., Llano E., Overbeek P.A., Barbero J.L., Oka K., Harrison W., Vaiman D., Ben-Neriah Z., García-Tuñón I. Mutant cohesin in premature ovarian failure. N. Engl. J. Med. 2014;370:943–949. doi: 10.1056/NEJMoa1309635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinberg-Shukron A., Rachmiel M., Renbaum P., Gulsuner S., Walsh T., Lobel O., Dreifuss A., Ben-Moshe A., Zeligson S., Segel R. Essential Role of BRCA2 in Ovarian Development and Function. N. Engl. J. Med. 2018;379:1042–1049. doi: 10.1056/NEJMoa1800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin Y., Zhang F., Chen Z.J. BRCA2 in Ovarian Development and Function. N. Engl. J. Med. 2019;380:1086. doi: 10.1056/NEJMc1813800. [DOI] [PubMed] [Google Scholar]
- 8.Yatsenko A.N., Georgiadis A.P., Röpke A., Berman A.J., Jaffe T., Olszewska M., Westernströer B., Sanfilippo J., Kurpisz M., Rajkovic A. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 2015;372:2097–2107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang F., Silber S., Leu N.A., Oates R.D., Marszalek J.D., Skaletsky H., Brown L.G., Rozen S., Page D.C., Wang P.J. TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol. Med. 2015;7:1198–1210. doi: 10.15252/emmm.201404967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J., Zhang W., Jiang H., Wu B.L., Primary Ovarian Insufficiency Collaboration Mutations in HFM1 in recessive primary ovarian insufficiency. N. Engl. J. Med. 2014;370:972–974. doi: 10.1056/NEJMc1310150. [DOI] [PubMed] [Google Scholar]
- 11.Yao C., Yang C., Zhao L., Li P., Tian R., Chen H. Bi-allelic SHOC1 loss-of-function mutations cause meiotic arrest and non-obstructive azoospermia. J. Med. Genet. 2020 doi: 10.1136/jmedgenet-2020-107042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matzuk M.M., Lamb D.J. The biology of infertility: research advances and clinical challenges. Nat. Med. 2008;14:1197–1213. doi: 10.1038/nm.f.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J., Colaiácovo M.P. Zipping and Unzipping: Protein Modifications Regulating Synaptonemal Complex Dynamics. Trends Genet. 2018;34:232–245. doi: 10.1016/j.tig.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries F.A., de Boer E., van den Bosch M., Baarends W.M., Ooms M., Yuan L., Liu J.G., van Zeeland A.A., Heyting C., Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19:1376–1389. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolcun-Filas E., Hall E., Speed R., Taggart M., Grey C., de Massy B., Benavente R., Cooke H.J. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 2009;5:e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolcun-Filas E., Costa Y., Speed R., Taggart M., Benavente R., De Rooij D.G., Cooke H.J. SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J. Cell Biol. 2007;176:741–747. doi: 10.1083/jcb.200610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schramm S., Fraune J., Naumann R., Hernandez-Hernandez A., Höög C., Cooke H.J., Alsheimer M., Benavente R. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet. 2011;7:e1002088. doi: 10.1371/journal.pgen.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamer G., Wang H., Bolcun-Filas E., Cooke H.J., Benavente R., Höög C. Progression of meiotic recombination requires structural maturation of the central element of the synaptonemal complex. J. Cell Sci. 2008;121:2445–2451. doi: 10.1242/jcs.033233. [DOI] [PubMed] [Google Scholar]
- 19.Gómez-H L., Felipe-Medina N., Sánchez-Martín M., Davies O.R., Ramos I., García-Tuñón I., de Rooij D.G., Dereli I., Tóth A., Barbero J.L. C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat. Commun. 2016;7:13298. doi: 10.1038/ncomms13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan L., Liu J.G., Zhao J., Brundell E., Daneholt B., Höög C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
- 21.Yang F., De La Fuente R., Leu N.A., Baumann C., McLaughlin K.J., Wang P.J. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J. Cell Biol. 2006;173:497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisinger A., Benavente R. Mutations in Genes Coding for Synaptonemal Complex Proteins and Their Impact on Human Fertility. Cytogenet. Genome Res. 2016;150:77–85. doi: 10.1159/000453344. [DOI] [PubMed] [Google Scholar]
- 23.Pashaei M., Rahimi Bidgoli M.M., Zare-Abdollahi D., Najmabadi H., Haji-Seyed-Javadi R., Fatehi F., Alavi A. The second mutation of SYCE1 gene associated with autosomal recessive nonobstructive azoospermia. J. Assist. Reprod. Genet. 2020;37:451–458. doi: 10.1007/s10815-019-01660-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schilit S.L.P., Menon S., Friedrich C., Kammin T., Wilch E., Hanscom C., Jiang S., Kliesch S., Talkowski M.E., Tüttelmann F. SYCP2 Translocation-Mediated Dysregulation and Frameshift Variants Cause Human Male Infertility. Am. J. Hum. Genet. 2020;106:41–57. doi: 10.1016/j.ajhg.2019.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hernández-López D., Geisinger A., Trovero M.F., Santiñaque F.F., Brauer M., Folle G.A., Benavente R., Rodríguez-Casuriaga R. Familial primary ovarian insufficiency associated with an SYCE1 point mutation: defective meiosis elucidated in humanized mice. Mol. Hum. Reprod. 2020;26:485–497. doi: 10.1093/molehr/gaaa032. [DOI] [PubMed] [Google Scholar]
- 26.Kong A., Thorleifsson G., Frigge M.L., Masson G., Gudbjartsson D.F., Villemoes R., Magnusdottir E., Olafsdottir S.B., Thorsteinsdottir U., Stefansson K. Common and low-frequency variants associated with genome-wide recombination rate. Nat. Genet. 2014;46:11–16. doi: 10.1038/ng.2833. [DOI] [PubMed] [Google Scholar]
- 27.Begum F., Chowdhury R., Cheung V.G., Sherman S.L., Feingold E. Genome-Wide Association Study of Meiotic Recombination Phenotypes. G3 (Bethesda) 2016;6:3995–4007. doi: 10.1534/g3.116.035766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization . World Health Organization; New York: 2010. WHO laboratory manual for the examination and processing of human semen. [Google Scholar]
- 29.Yin H., Ma H., Hussain S., Zhang H., Xie X., Jiang L., Jiang X., Iqbal F., Bukhari I., Jiang H. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet. Med. 2019;21:62–70. doi: 10.1038/s41436-018-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nyholt D.R. All LODs are not created equal. Am. J. Hum. Genet. 2000;67:282–288. doi: 10.1086/303029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Zhong L., Xu B., Yang Y., Ban R., Zhu J., Cooke H.J., Hao Q., Shi Q. SpermatogenesisOnline 1.0: a resource for spermatogenesis based on manual literature curation and genome-wide data mining. Nucleic Acids Res. 2013;41:D1055–D1062. doi: 10.1093/nar/gks1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiao Y., Fan S., Jabeen N., Zhang H., Khan R., Murtaza G. A TOP6BL mutation abolishes meiotic DNA double-strand break formation and causes human infertility. Sci. Bull. 2020;65:2120–2129. doi: 10.1016/j.scib.2020.08.026. [DOI] [PubMed] [Google Scholar]
- 34.Peters A.H., Plug A.W., van Vugt M.J., de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- 35.Jiang X., Yin S., Fan S., Bao J., Jiao Y., Ali A., Iqbal F., Xu J., Zhang Y., Shi Q. Npat-dependent programmed Sertoli cell proliferation is indispensable for testis cord development and germ cell mitotic arrest. FASEB J. 2019;33:9075–9086. doi: 10.1096/fj.201802289RR. [DOI] [PubMed] [Google Scholar]
- 36.Qin Y., Jiao X., Simpson J.L., Chen Z.J. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum. Reprod. Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian V.V., Hochwagen A. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 2014;6:a016675. doi: 10.1101/cshperspect.a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Leng M., Ma T., Yu D., Shi H., Shi Q. Cryopreservation has no effect on meiotic recombination and synapsis in testicular tissues. Fertil. Steril. 2009;91(4, Suppl):1404–1407. doi: 10.1016/j.fertnstert.2008.04.074. [DOI] [PubMed] [Google Scholar]
- 39.Sanchez-Saez F., Gomez H.L., Dunne O.M., Gallego-Paramo C., Felipe-Medina N., Sanchez-Martin M., Llano E., Pendas A.M., Davies O.R. Meiotic chromosome synapsis depends on multivalent SYCE1-SIX6OS1 interactions that are disrupted in cases of human infertility. Sci. Adv. 2020;6:eabb1660. doi: 10.1126/sciadv.abb1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J., Gu Y., Feng J., Zhou W., Yang X., Shen Y. Structural insight into the central element assembly of the synaptonemal complex. Sci. Rep. 2014;4:7059. doi: 10.1038/srep07059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolor H., Mori T., Nishiyama S., Ito Y., Hosoba E., Inagaki H., Kogo H., Ohye T., Tsutsumi M., Kato T. Mutations of the SYCP3 gene in women with recurrent pregnancy loss. Am. J. Hum. Genet. 2009;84:14–20. doi: 10.1016/j.ajhg.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the C14orf39 variants reported in this paper are ClinVar: SCV001468897, SCV001468898, and SCV001468899. The WES datasets supporting the current study have not been deposited in a public repository because of privacy and ethical restrictions but are available from the corresponding authors on request. The relevant NCBI accession numbers are GenBank: NM_174978.3 (for the reference sequence of human C14orf39), NP_777638.3 (for the reference sequence of human C14ORF39 protein), and NM_029444.2 (for the reference sequence of mouse Six6os1).