Abstract
Cell adhesion proteins of the Cadm (SynCAM/Necl) family regulate myelination and the organization of myelinated axons. In the peripheral nervous system (PNS), intercellular contact between Schwann cells and their underlying axons is believed to be mediated by binding of glial Cadm4 to axonal Cadm3 or Cadm2. Nevertheless, given that distinct neurons express different combinations of the Cadm proteins, the identity of the functional axonal ligand for Cadm4 remains to be determined. Here, we took a genetic approach to compare the phenotype of Cadm4 null mice, which exhibit abnormal distribution of Caspr and Kv1 potassium channels, with mice lacking different combinations of Cadm1–Cadm3 genes. We show that in contrast to mice lacking the single Cadm1, Cadm2, or Cadm3 genes, genetic ablation of all three phenocopies the abnormalities detected in the absence of Cadm4. Similar defects were observed in double mutant mice lacking Cadm3 and Cadm2 (i.e., Cadm3−/−/Cadm2−/−) or Cadm3 and Cadm1 (i.e., Cadm3−/−/Cadm1−/−), but not in mice lacking Cadm1 and Cadm2 (i.e., Cadm1−/−/Cadm2−/−). Furthermore, axonal organization abnormalities were also detected in Cadm3 null mice that were heterozygous for the two other axonal Cadms. Our results identify Cadm3 as the main axonal ligand for glial Cadm4, and reveal that its absence could be compensated by the combined action of Cadm2 and Cadm1.
SIGNIFICANCE STATEMENT Myelination by Schwann cells enables fast conduction of action potentials along motor and sensory axons. In these nerves, Schwann cell-axon contact is mediated by cell adhesion molecules of the Cadm family. Cadm4 in Schwann cells regulates axonal ensheathment and myelin wrapping, as well as the organization of the axonal membrane, but the identity of its axonal ligands is not clear. Here, we reveal that Cadm mediated axon-glia interactions depend on a hierarchical adhesion code that involves multiple family members. Our results provide important insights into the molecular mechanisms of axon-glia communication, and the function of Cadm proteins in PNS myelin.
Keywords: axon-glia interaction, cell adhesion, myelin, node of Ranvier, Schwann cells, SynCAM
Introduction
Myelinating Schwann cells in the peripheral nervous system (PNS) and oligodendrocytes in the CNS are engaged in a continuous dialogue with the axons they ensheath. Axon-glial contact not only affects myelin membrane wrapping, but also controls the precise placement of ion channels along the axolemma, and the generation of specific axoglial contact sites that are required for the formation of a functional myelin units (Stadelmann et al., 2019; Wilson et al., 2020). Such interactions are mediated by distinct cell adhesion systems that are present at the axon-glial interface at the nodes of Ranvier, the paranodal junction (PNJ), and along the internodes below the compact myelin (Rasband and Peles, 2015). Recent studies revealed that accurate myelination in the CNS involves the coordinate action of both paranodal and internodal adhesion systems (Djannatian et al., 2019; Elazar et al., 2019a).
Intercellular adhesion mediated by members of the Cadm family, also known as Nectin like (Necls) and synaptic adhesion molecules (SynCAMs), regulates different developmental aspects of the nervous system, including axon guidance (Niederkofler et al., 2010; Frei et al., 2014), synapse formation and plasticity (Biederer et al., 2002; Robbins et al., 2010; Perez de Arce et al., 2015; Ribic et al., 2019), and myelination (Maurel et al., 2007; Spiegel et al., 2007; Park et al., 2008; Golan et al., 2013; Chen et al., 2016; Elazar et al., 2019a,b). Not surprisingly, variations in Cadm genes are associated with diverse neurologic and mental health-related conditions (Day et al., 2016; Pasman et al., 2018; Xu et al., 2018; Dohrn and Saporta, 2020). The Cadm proteins belong to a small group of the immunoglobulin superfamily cell adhesion molecules that contains four different members (Cadm1–Cadm4; Mandai et al., 2015). They are transmembrane glycoproteins that contain three immunoglobulin-like (Ig) domains in their extracellular region and a short intracellular domain which mediates their interactions with protein 4.1 and PDZ-domain proteins (Zhou et al., 2005; Hoy et al., 2009; Einheber et al., 2013; Rademacher et al., 2016; Meng et al., 2019). Cadms mediate Ca2+-independent cell-adhesion by binding homophilically, as well as heterophilically to other members of the family (Kakunaga et al., 2005; Fogel et al., 2007; Spiegel et al., 2007; Liu et al., 2019). Cadm1 (Necl2), Cadm 2 (Necl3), and Cadm3 (Necl1; Kakunaga et al., 2005; Fogel et al., 2011), but not Cadm4 (Necl4; Liu et al., 2019), also form homophilic and heteromeric cis dimers on the cell membrane. In addition, the interaction between Cadm family members is modulated by differential glycosylation (Fogel et al., 2007, 2010; Galuska et al., 2010). In the PNS, Cadm proteins are differentially expressed in Schwann cells and axons, i.e., Cadm1, Cadm2, and Cadm3 are mostly found in neurons, whereas Cadm4 and to a much lesser extent Cadm1, are present in myelinating Schwann cells (Maurel et al., 2007; Spiegel et al., 2007). Cadm4 is present at the internodal Schwann cell-axon interface, directly apposing Cadm2 and Cadm3 in the axolemma. The extracellular domain of Cadm4 preferentially binds to Cadm2-expressing and Cadm3-expressing cells, as well as to cultured sensory neurons, but not to Schwann cells (Spiegel et al., 2007), suggesting that in the PNS, axon-glia interaction is primarily mediated by binding of glial Cadm4 to axonal Cadm2 and Cadm3 (Maurel et al., 2007; Spiegel et al., 2007). Similarly, in the CNS, genetic deletion of Cadm2 and Cadm3 completely abolished the binding of the extracellular domain of Cadm4 to spinal cord neurons (Elazar et al., 2019b). However, since both sensory (Zheng et al., 2019) and motor (Maciel et al., 2018) neurons express different combinations of Cadm genes, the identity of the functional axonal ligand for Cadm4 is presently unclear. Given that loss-of-function phenotypes of receptors and of their ligands are often similar, we compared between Cadm4 null mice and mice lacking each of the other single Cadm genes, as well as mice lacking different allelic Cadm combinations. This genetic analysis identified Cadm3 as the main axonal ligand for Cadm4 in the PNS, and revealed a compensatory contribution of the two other axonal Cadms.
Materials and Methods
Animal handling and generation of knock-out mice
All experiments were performed in compliance with the relevant laws and institutional guidelines and were approved by the Weizmann's Institutional Animal Care and Use Committee. Mice were kept in Weizmann's animal facility, in aseptic SFP conditions until used for experiments. Knock-out mice for Cadm1, Cadm2, Cadm3, and Cadm4 were generated as described previously (Fujita et al., 2005, 2006; Golan et al., 2013). Cadm1KO, Cadm2KO, and Cadm3 KO mice were crossed to generate triple KO (Cadm123 KO) mice. Genotyping was done by PCR of genomic DNA using the following primers: Cadm1WT allele, GATGTGTGCTGACTTAGGAACGGTC and GAGTGATTAACAACGTGCAGGCAAT; targeted, GATGTGTGCTGACTTAGGAACGGTC and TGCGAGGCCAGAGGCCACTTGTGTAGC; Cadm2WT allele, AGGCACAAGTTGCCAGCCA and ATTCCACCCCGTATTTCCA; targeted, AGGCACAAGTTGCCAGCCA and CAAGTTAGCACGTGGCCACC; Cadm3WT allele, GCCCTGACTGAAACAGCGAC and CAGGGAGCCTGTTGGCTTTG; targeted, GCCCTGACTGAAACAGCGAC and CTTTGCTCCACACCCGAGTG; Cadm4WT allele, CGCAGTTCTGATCCAGCATC and ATCAGAGGACAGCTCATGAG; targeted, CGCAGTTCTGATCCAGCATC and GGCAGGAGGGTTGCCATGA.
Immunofluorescent staining
Teased sciatic nerves were prepared and immunolabeled as previously described (Eshed-Eisenbach et al., 2020). Briefly, sciatic nerves were dissected and immersed in 4% paraformaldehyde for 20 min at room temperature (RT). Teasing of sciatic nerves was done by desheathing followed by teasing on SuperFrost Plus slides (Menzel-Gläser, Thermo Scientific), air dried overnight, and then kept frozen at −20°C till used. Teased peripheral nerves were all stained according to the following methodology. When required, samples were postfixed for 7 min using cold methanol (−20°C) following by consecutive PBS washes. Blocking and permeabilization were done by incubation for 1 h in 5% normal goat serum and 0.5% Triton X-100, in PBS at RT. Primary antibodies were diluted in 5% normal goat serum and 0.1% Triton X-100, in PBS and incubated overnight at 4°C. Secondary antibodies were incubated for 40 min at RT in 5% normal goat serum and 0.1% Triton X-100, in PBS. Samples were mounted with Fluoromount-G (SouthernBiotech). Images were taken using Nikon eclipse 90i microscope or 3DHistech Panoramic Midi II scanner.
Antibodies
The following primary antibodies were used: rat anti-neurofilament (NF-H; MAB5448, Millipore Bioscience Research Reagents), chicken anti-NF-H (AB_2313552 AVES labs), rabbit anti-Caspr (Peles et al., 1997), rabbit anti- Kv1.2 (Ogawa et al., 2008), mouse anti- Kv1.2 (K14/16; NeuroMab), rat anti-NrCAM (Lustig et al., 2001). Fluorophore-coupled antibodies included the following: 488-coupled anti-rabbit was purchased from Invitrogen; Cy3-coupled anti-rabbit, Cy3-coupled anti-mouse, and Cy5-coupled anti-rat were obtained from Jackson ImmunoResearch.
Electronic microscopy
Mice were scarified and sciatic nerves were exposed and fixed by continuous dripping of fresh fixative containing 4% PFA, 2.5% glutaraldehyde in 0.1 m cacodylate buffer (pH 7.4). Nerves were then excised and incubated overnight in the same fixative at RT and stored at 4°C. Nerves were processed as previously described (Elazar et al., 2019a). Samples were examined using a FEI Tecnai T12 transmission electron microscope equipped with a XF416 TVIP camera. In parallel, the resin embedded sections were stained with 1% toluidine blue and then analyzed using a 3DHistech Panoramic Midi II scanner.
Quantitation and statistical analysis
Samples from at least three animals were used per genotype in all quantitation performed on teased sciatic nerves. Statistical significance was determined using Student's two-tailed t test. Graphs were prepared using GraphPad Prism software. Images were taken using Nikon eclipse 90i microscope or 3DHistech Panoramic Midi II scanner.
Results
Loss of Cadm4, but not other Cadms, disrupts axonal organization
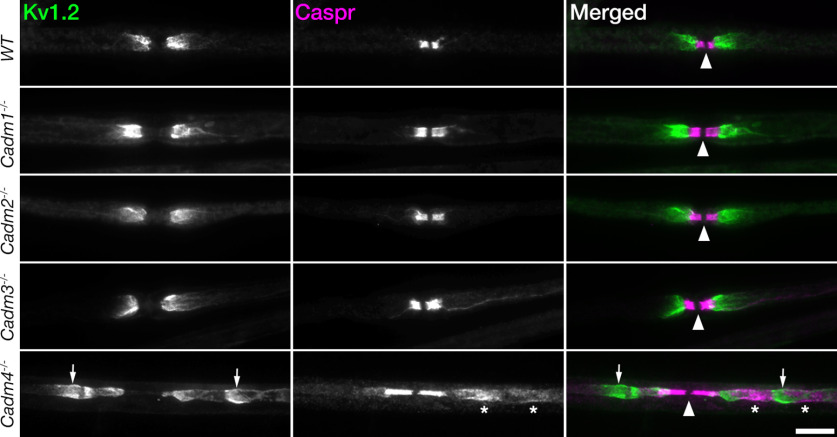
We previously found that genetic deletion of Cadm4 results in pronounced disorganization of myelinated axons in the PNS, as reflected by the disruption of Caspr-labeled paranodal junctions, and clustering of Kv1.2 potassium channels away from the juxtaparanodal region (JXP) (Fujita et al., 2005, 2006; Golan et al., 2013). To examine whether one of the neuronal Cadms (i.e., Cadm1–Cadm3) serves as an axonal ligand for glial Cadm4, we compared the distribution of Caspr and Kv1.2 in sciatic nerve of Cadm4 null mice, to mice lacking either Cadm1, Cadm2, or Cadm3 (Fig. 1). Similar to wild-type mice, all the latter three single Cadm mutants exhibited normal accumulation of Caspr and Kv1.2 channels at the paranodal junction and the juxtaparanodal region, respectively. In contrast, and as expected, Cadm4 null mice displayed prominent disruption of the axonal organization (Fig. 1).
Figure 1.
Genetic deletion of Cadm4, but not of other family members, results in axonal organization abnormalities. Immunolabeling of teased sciatic nerve fibers isolated from 2.5-month-old wild-type mice (WT), or mice lacking Cadm1 (Cadm1−/−), Cadm2 (Cadm2−/−), Cadm3 (Cadm3−/−), and Cadm4 (Cadm4−/−), using antibodies to Kv1.2 potassium channels to label the juxtaparanodal region and Caspr to label the paranodal junction. Images of the merged channels are shown on the right panels. Arrowhead mark the nodes of Ranvier. Note the abnormal labeling of Caspr (asterisks) and Kv1.2 (arrows) away from the paranodal junction and the juxtaparanodal region, respectively. Scale bar: 10 μm.
Triple mutant mice lacking Cadm1–3 exhibit diverse axonal organization defects
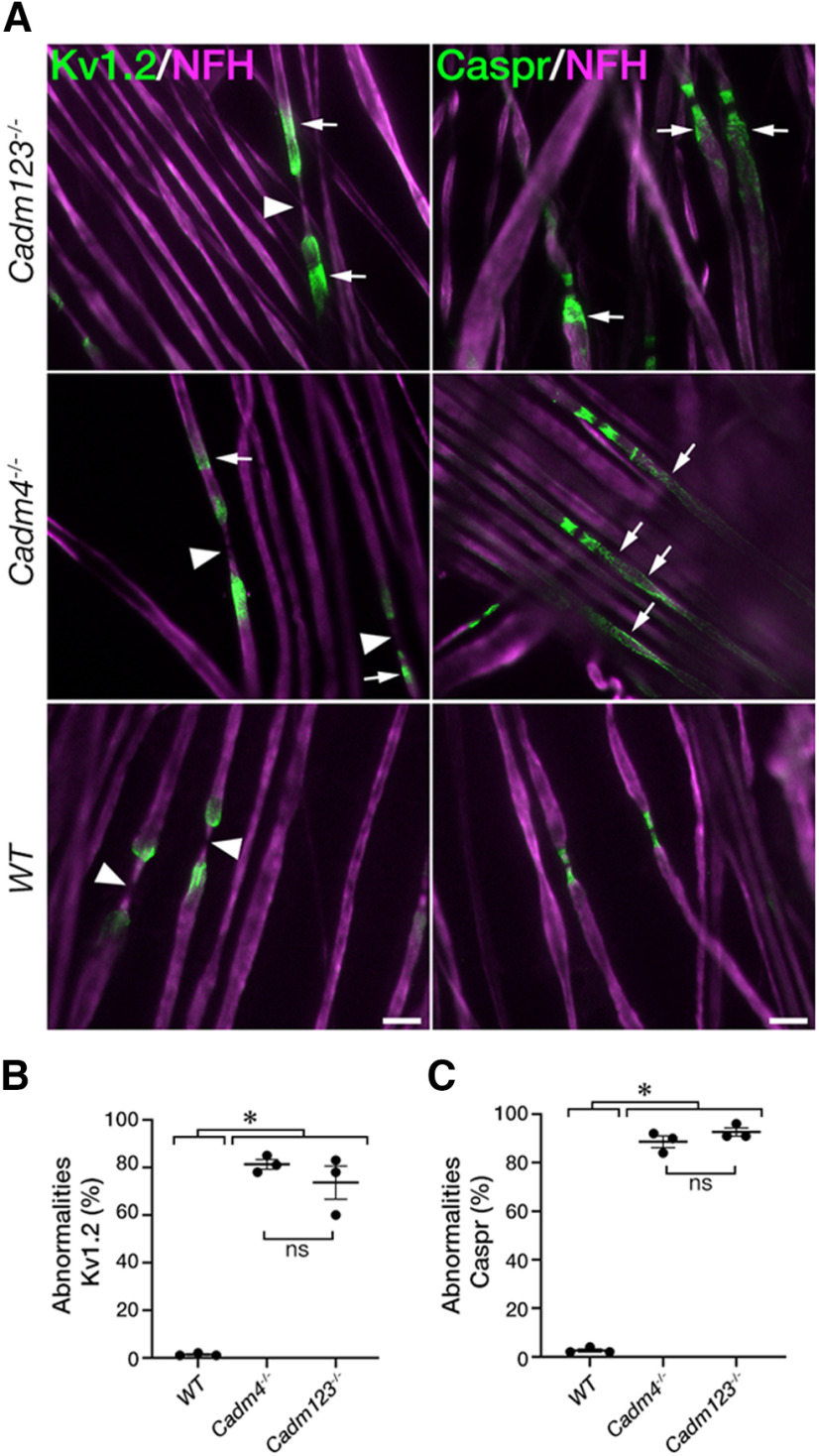
The observation that Cadm1−/−, Cadm2−/−, or Cadm3−/− null mice do not display similar abnormalities as detected in the absence of Cadm4, suggests that the axonal Cadms compensate for each other, or alternatively, that other non-Cadm proteins serve as a ligand for glial Cadm4. To further examine these possibilities, we generated mutant mice lacking all three axonal Cadm proteins (Cadm1-3; Cadm1−/−/Cadm2−/−/Cadm3−/−). Similar to the single nulls (Fujita et al., 2005, 2006; Golan et al., 2013), homozygous Cadm1−/−/Cadm2−/−/Cadm3−/− mice display no apparent neurologic abnormalities and exhibit PNS myelin that was morphologically indistinguishable from wild-type animals (Fig. 2A). Nevertheless, immunolabeling of teased sciatic nerves isolated from these mice revealed clear abnormalities in the distribution of Caspr and Kv1.2 channels compared with each of the single mutants (Fig. 2B). While in the single mutants Kv1.2 potassium channels were restricted to the juxtaparanodal region and the mesaxon, in Cadm1−/−/Cadm2−/−/Cadm3−/− mice they were located away from this site and often showed elongated, diffuse or irregular appearance (Fig. 2B, arrows). The triple mutant also displayed aberrant distribution of Caspr away from the paranodal junction and almost complete absence of this protein from the inner mesaxon, i.e., the site where the inner most Schwann cell membrane is attached to the axolemma. As depicted in Figure 2C, Caspr abnormalities could be divided into three major subgroups, including aberrant extension beyond the paranodal junction in what appeared as unwinding paranodal loops, abnormal accumulation in patches away from the paranodal junction, and diffuse Caspr immunoreactivity along the internodes. The abnormalities observed for Kv1.2 channels included the presence of gaps withing the juxtaparanodal region, the appearance of irregular membrane clusters, and their focal accumulation along the internodes (Fig. 2C), which all resemble the abnormalities reported for mice lacking Cadm4 in Schwann cells (Fujita et al., 2005, 2006; Golan et al., 2013). Quantitative analysis showed that 63 ± 0.07% of nodal environs of Cadm1−/−/Cadm2−/−/Cadm3−/− mice displayed abnormal distribution of Kv1.2 channels compared with 2 ± 0.004% in Cadm1−/−/, 3 ± 0.002% in Cadm2−/−, and 9 ± 0.02% in Cadm3−/− (Fig. 2D). Abnormal Caspr immunoreactivity was detected in 70 ± 0.02% of the sites in Cadm1−/−/Cadm2−/−/Cadm3−/− mice compared with 7 ± 0.01% in Cadm1−/−/, 5 ± 0.02% in Cadm2−/−, and 17 ± 0.05% in Cadm3−/− (Fig. 2E).
Figure 2.
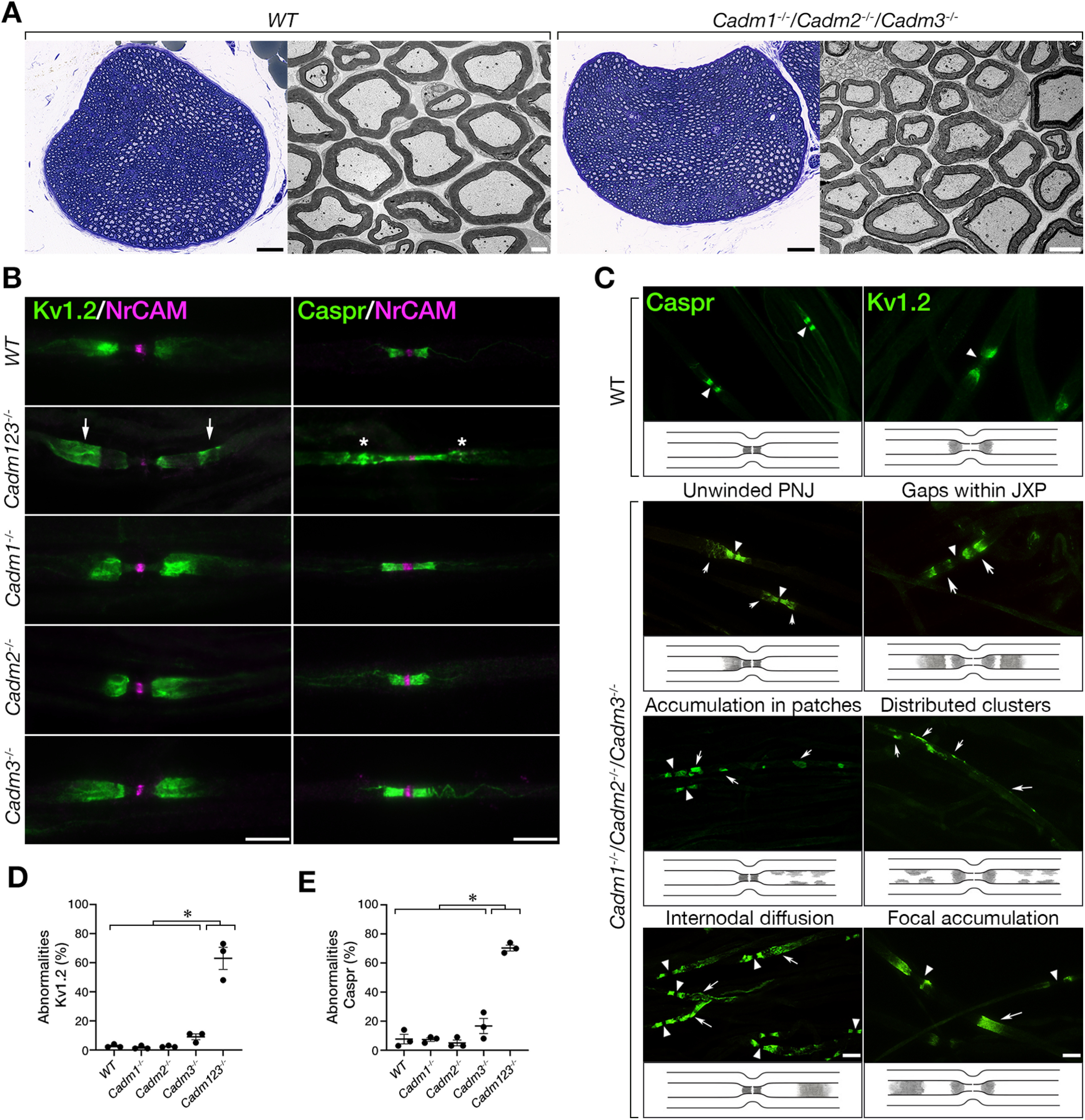
Mice lacking all three axonal Cadms exhibit abnormal axonal organization. A, Cross sections of sciatic nerves isolated from wild-type (WT) and the triple Cadm1−/−/Cadm2−/−/Cadm3−/− null mice reveal normal myelin morphology. Toluidine blue (left panels) and electron microscopy (right panels) images are shown for each genotype. B, Immunolabeling of teased sciatic nerve fibers isolated from 2.5-month-old WT mice (WT), or mice lacking Cadm1–Cadm3 (Cadm123−/−) using antibodies to Kv1.2 and NrCAM or Caspr and NrCAM as indicated. NrCAM was used to mark the location of the nodes of Ranvier. The single Cadm1 (Cadm1−/−), Cadm2 (Cadm2−/−), Cadm3 (Cadm3−/−) null mice were used as additional controls. Arrows and asterisks mark abnormal distribution of Kv1.2 and Caspr, respectively. C, Summary of the abnormalities detected. Teased sciatic nerve fibers from the indicated genotypes were labeled using antibodies to Caspr or Kv1.2. Schematic illustrations describing the distribution of Caspr and Kv1.2 are shown below each panel. Caspr abnormalities include unwinding of the paranodal mesaxonal line, accumulation of Caspr in patches away from the paranodes, and its diffusion along the internodes. Kv1.2 abnormalities include the presence of gaps within the juxtaparanodal area, and the appearance of intermittent clusters as well as focal accumulation of these channels throughout the internodes. Arrowheads and arrows mark the nodes and the various abnormalities, respectively. D, E, Quantitative analysis of the percentage of Kv1.2 (D) and Caspr (E) abnormalities in the different genotypes. Graphs represent mean ± SEM of 300 sites corresponding to the paranodal junction and the juxtaparanodal region per each genotype (n = 3); *p < 0.0001. Scale bars: 10 μm.
Cadm1−/−/Cadm2−/−/Cadm3−/− triple nulls phenocopy mice lacking Cadm4
We next compared between Cadm1−/−/Cadm2−/−/Cadm3−/− and Cadm4−/− null mice. Immunolabeling of teased sciatic nerves isolated from adult mice using antibodies to Caspr and Kv1.2 channels revealed the presence of remarkably similar abnormalities in both genotypes, but not in wild-type mice (Fig. 3A). Quantitative analysis showed no significant difference in appearance of these proteins in the two genotypes (Fig. 3B,C). Cadm1−/−/Cadm2−/−/Cadm3−/− mice displayed abnormal distribution of Kv1.2 channels in 74 ± 0.07% and 81 ± 0.02% in Cadm1−/−/Cadm2−/−/Cadm3−/− and Cadm4−/− mice, respectively (Fig. 3B). Likewise, we noted abnormal distribution of Caspr in 93 ± 0.01% of the paranodes compared with 89 ± 0.02% in Cadm4−/− null mice (Fig. 3C). These results demonstrate that deletion of all three neuronal Cadm proteins phenocopies mice lacking Cadm4, indicating that Cadm1–Cadm3 serve as the exclusive axonal ligand for glial Cadm4.
Figure 3.
Cadm1-3 null mice phenocopy mice lacking Cadm4. A, Double immunolabeling of teased sciatic nerve fibers isolated from 2.5-month-old mice lacking Cadm1–Cadm3 (Cadm123−/−), Cadm4 (Cadm4−/−), or wild-type (WT) mice using antibodies to neurofilament (NFH) and either Kv1.2 or Caspr as indicated. Arrowheads and arrows mark the nodes and abnormal distribution of Kv1.2 and Caspr, respectively. B, C, Quantification of Kv1.2 channels (B) and Caspr (C) abnormalities in the different genotypes, presented as a percentage of total number of nodes. Graphs represent mean ± SEM of 300 sites corresponding to the paranodal junction and the juxtaparanodal region per each genotype (n = 3); *p < 0.0001. ns, not significant. Scale bars: 10 μm.
Cadm3 is the main functional axonal ligand for glial Cadm4
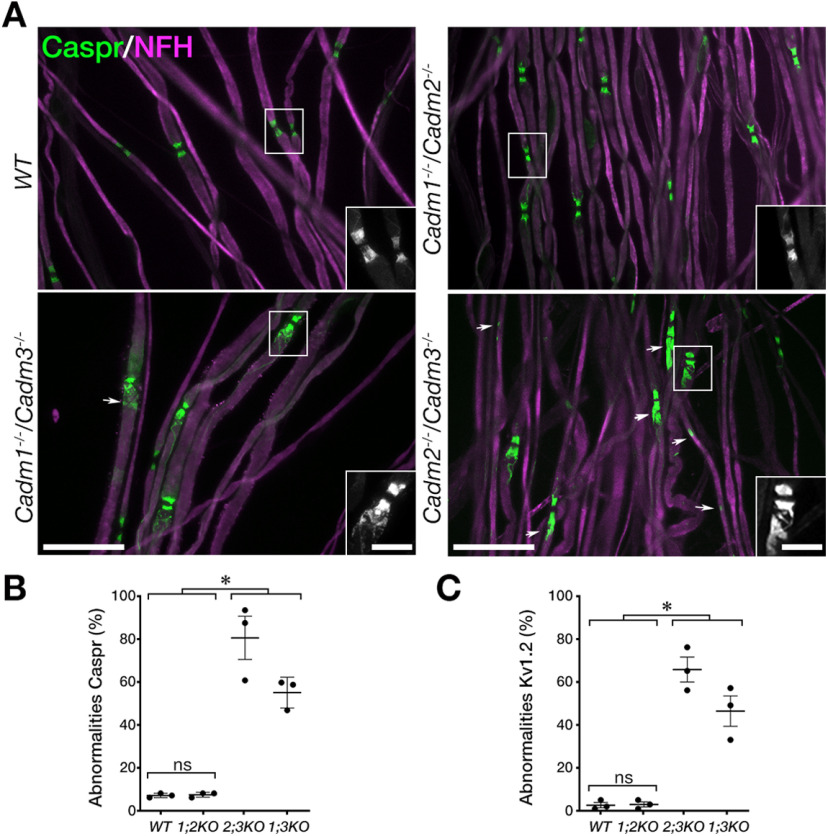
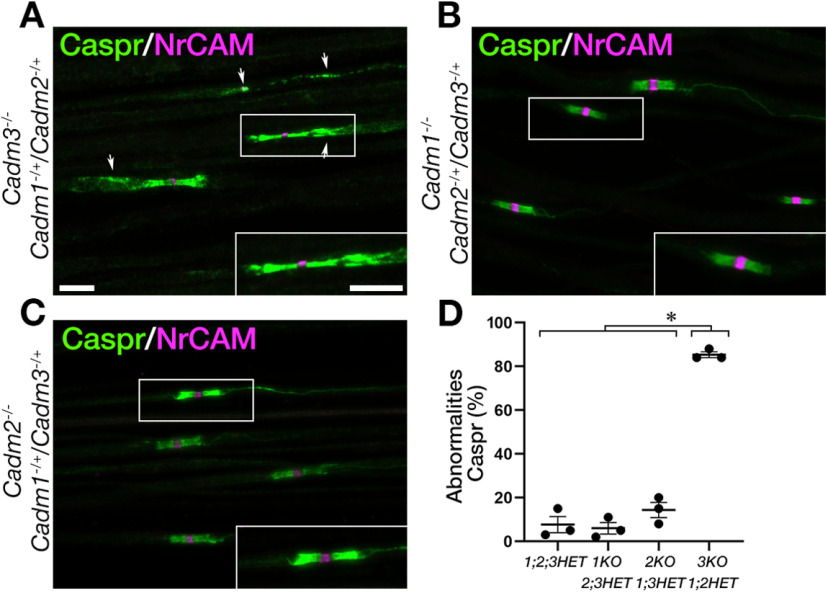
The observation that Cadm1−/−/Cadm2−/−/Cadm3−/− but not the single genes mutants phenocopies Cadm4−/− null mice, supports the notion that axonal Cadms compensate for each other. To further corroborate the existence of such a functional redundancy, we examine the distribution of Caspr in mice lacking different combinations of Cadm1–Cadm3 genes (Fig. 4A). We found that double mutant mice lacking Cadm3 and Cadm2 (i.e., Cadm3−/−/Cadm2−/−) or Cadm3 and Cadm1 (i.e., Cadm3−/−/Cadm1−/−) display abnormal distribution of Caspr at the paranodes corresponding to unusual “opening” of the paranodal loops, as well as intermittent accumulation of Caspr along the internodes. In contrast, no such abnormalities were detected in double mutant mice lacking both Cadm1 and Cadm2 (i.e., Cadm1−/−/Cadm2−/−), indicating that Cadm3 is a primary axonal ligand for glial Cadm4. Quantification of the results revealed that abnormal distribution of Caspr was detected at 55 ± 0.04% and 81 ± 0.1% of the sites in Cadm3−/−/Cadm1−/− and Cadm3−/−/Cadm2−/− mice, respectively, which was similar to mice lacking all three neuronal Cadms (71 ± 0.02%; Fig. 4B). In contrast, we detected Caspr abnormalities in only 7 ± 0.01% of the sites in Cadm1−/−/Cadm2−/− mice, and 7 ± 0.01% in wild-type animals. Comparable results were obtained for Kv1.2 as well (WT 3 ± 0.01%, Cadm1−/−/Cadm2−/− 3 ± 0.01%, Cadm3−/−/Cadm2−/− 66 ± 0.1%, Cadm3−/−/Cadm1−/− 46 ± 0.1%, Cadm1−/−/ Cadm2−/− Cadm3−/− 77 ± 0.04%; Fig. 4C). These experiments identify Cadm3 as the main axonal ligand for Cadm4 in the PNS, and revealed a compensatory contribution of the two other axonal Cadms. They also suggest that Cadm-mediate axon-glia interactions involve multiple family members. To further explore this point, we asked whether deletion of Cadm3 along with a 50% reduction in the expression of the other two genes, will result in a phenotype. As depicted in Figure 5A,D, Cadm3−/− null mice that were heterozygous for Cadm1 and Cadm2 (Cadm1−/+/Cadm2−/+Cadm3−/−), exhibit abnormal localization of Caspr. As expected, no such abnormalities were detected in Cadm1−/− or Cadm2−/− null mice that were heterozygous for the other two neuronal Cadm genes (Fig. 5B–D). Quantitative analysis revealed that 85 ± 0.01% of nodal environs of Cadm3−/−/Cadm1−/+/Cadm2−/+ mice displayed abnormal distribution of Caspr compared with 6 ± 0.03% in Cadm1−/−/Cadm2−/+/Cadm3−/+, and 14 ± 0.04% in Cadm2−/−/Cadm1−/+/Cadm3−/+ (Fig. 5D). Taken together, our results demonstrate that Cadm3 is the main axonal ligand for glial Cadm4 and that its absence can be compensated by the combined action of Cadm1 and Cadm2.
Figure 4.
Cadm3 is the main functional axonal ligand for Schwann cell Cadm4. A, Immunolabeling of teased sciatic nerve fibers isolated from 2.5-month-old wild-type (WT) animals or mice lacking either Cadm2 and Cadm3 (Cadm2−/−/Cadm3−/−), Cadm1 and Cadm2 (Cadm1−/−/Cadm2−/−), or Cadm1 and Cadm3 (Cadm1−/−/Cadm3−/−) using antibodies to Caspr and neurofilament (NFH). Insets represent higher magnification images of the boxed areas. Arrows mark abnormal distribution of Caspr. B, C, Quantification of Caspr (B) and Kv1.2 (C) abnormalities in the different genotypes, presented as a percentage of total number of nodes. Graphs represent mean ± SEM of 300 sites corresponding to the paranodal junction and the juxtaparanodal region per each genotype (n = 3); *p < 0.0001. Scale bars: 10 μm.
Figure 5.
Cadm-mediated axoglial interaction involves multiple members of the family. A–C, Caspr immunolabeling of teased sciatic nerve fibers isolated from 2.5-month-old mice lacking Cadm3 and one allele of Cadm1 and Cadm2 (Cadm3−/−/Cadm1−/+/Cadm2−/+; A), mice lacking Cadm1 and one allele of Cadm2 and Cadm3 (Cadm1−/−/Cadm2−/+/Cadm3−/+; B), or mice lacking Cadm2 and one allele of Cadm1 and Cadm3 (Cadm2−/−/Cadm1−/+/Cadm3−/+; C). Arrows mark abnormal distribution of Caspr. Insets shows higher magnification of the boxed area in each panel. D, Quantification of Caspr abnormalities in the different genotypes, presented as a percentage of total number of nodes. Graph represents mean ± SEM of 300 from three animals per each genotype (n = 3); *p < 0.0001. Scale bars: 10 μm.
Discussion
Given that loss-of-function of receptors and their cognate ligands often results in similar phenotypes, we took a comparative genetic approach to identify the axonal Cadm ligand for Schwann cell Cadm4. We reasoned that this approach would be useful since members of this family are co-expressed in sensory and motor neurons (Maciel et al., 2018; North et al., 2019; Sharma et al., 2020) and are engaged in homophilic and multiple heterophilic interactions (Fogel et al., 2007; Maurel et al., 2007; Spiegel et al., 2007). Our results show that in peripheral myelinated neurons, Cadm3 is the chief axonal ligand for glial Cadm4. They further reveal a compensatory contribution of Cadm1 and Cadm2. Such redundancy may explain the discrepancy between results obtained from single gene deletion experiments, in which myelination is normal or only mildly affected (Park et al., 2008; Golan et al., 2013; Zhu et al., 2013), to those affecting/involving a few members of the family such as gain of function experiments (Elazar et al., 2019b), the use of dominant negative proteins (Hughes and Appel, 2019), ligand-receptor interference by soluble Cadm's extracellular domains (Spiegel et al., 2007) and or deletion of Cadm4 together with other axoglial adhesion systems (Djannatian et al., 2019; Elazar et al., 2019a).
The distribution of ion channels along myelinating axons strongly depends on Schwann cells and represents a sensitive indicator of myelin abnormalities (Arroyo et al., 2004). The precise positioning of these channels at the axolemma is regulated by their association with axoglial CAMs and cytoskeleton-linker proteins (Horresh et al., 2010; Einheber et al., 2013; Amor et al., 2017; Brivio et al., 2017). It was previously shown that genetic deletion of Cadm4 in Schwann cells results in abnormal organization of the axonal membrane, reflected by aberrant localization of Caspr and Kv1.2 channels along the internodes (Golan et al., 2013). A similar phenotype was also detected in mice lacking protein 4.1G, which binds Cadm4 and regulates its expression at the axon–glial interface (Ivanovic et al., 2012). The observation that similar axonal organization defects are obtained by deletion of axonal Cadms compared with glial Cadm4, indicates that the function of Cadm4 in shaping the axonal membrane is mediated by binding to Cadm3–Cadm1. We propose that similar to the role Cadms are playing in CNS synapse (Fogel et al., 2007; Perez de Arce et al., 2015), binding of glial Cadm4 to the axonal Cadms forms an adhesion scaffold that contributes to the organization of the axonal membrane by recruiting intracellular linker proteins and cytoskeletal components. In support of this view, binding of Cadm3 to Cadm4 recruits the polarity protein Par3 to the axoglial interface (Meng et al., 2019). In addition, deletion of protein 4.1B, which binds members of the Cadm proteins (Hoy et al., 2009; Yang et al., 2011), results in reduced axonal expression of Cadm3, Cadm1, and αII spectrin in myelinated DRG axons (Einheber et al., 2013).
Axoglial adhesion mediated by Cadm4 regulated myelination in both CNS and PNS (Maurel et al., 2007; Spiegel et al., 2007; Golan et al., 2013; Elazar et al., 2019a,b). Here, we found that Cadm3 is the main axonal ligand for glial Cadm4. This conclusion is supported by previous observations showing that (1) Cadm3 is localized at the axoglial interface in myelinated peripheral nerves (Kakunaga et al., 2005; Maurel et al., 2007; Spiegel et al., 2007), and is found together with Cadm4 in axogliasome-enriched fraction of sciatic nerves (Eichel et al., 2020); (2) the extracellular domain of Cadm4 binds to neurons expressing Cadm3 and Cadm2 and not to Schwann cells or oligodendrocytes (Maurel et al., 2007; Pellissier et al., 2007; Spiegel et al., 2007; Elazar et al., 2019b); (3) Cadm4 binding is abolished in cultured spinal cord neurons isolated from double Cadm2−/−/Cadm3−/− null mice (Elazar et al., 2019b); and (4) Cadm3 showed preferential binding to Cadm4 and Cadm1 (Fogel et al., 2007; Liu et al., 2019). Further support comes from experiments showing that while increased expression of Cadm4 lacking its intracellular domain (Cadm4dCT) in oligodendrocytes inhibits myelination, this effect could be rescued when Cadm4dCT OPCs were co-cultured with Cadm2−/−/Cadm3−/− neurons (Elazar et al., 2019b).
In contrast to genetic ablation of Cadm3 alone, deletion of Cadm3 together with either Cadm2 or Cadm1 resulted in an axonal organization defect, suggesting that Cadm1 and Cadm2 cooperate with each other generating a complex that could functionally substitute for Cadm3. Furthermore, the observation that deletion of Cadm3 together with only one allele of both Cadm1 and Cadm2 resulted in a phenotype, indicating that the function of the axonal Cadms is dose dependent. The involvement of Cadm1 was surprising as previous studies demonstrated that Cadm4 binds Cadm3 and Cadm2 but not Cadm1 (Maurel et al., 2007; Pellissier et al., 2007; Spiegel et al., 2007). Its contribution could point to a role for Cadm1 in regulating the binding of Cadm2 and Cadm3 to Cadm4 through the formation of cis-heterodimers. In support of this idea, it was previously demonstrated that Cadm1-Cadm2 form cis-heterodimers (Shingai et al., 2003) that modulate trans interactions (Frei et al., 2014). Such a mode of interaction may also explain why co-deletion of Cadm3 and Cadm1 resulted in a somewhat milder phenotype than the one obtained in mice lacking Cadm3 and Cadm2 (Fig. 4C, note the trend although it is not statistically significant). Our findings also have clinical implication as mutations in the extracellular domain of human CADM3 gene that replace a tyrosine with a cysteine residue (Y172C) results in axonal Charcot-Marie-Tooth neuropathy (Dohrn and Saporta, 2020). Notably, while the Y172C mutation did not seem to directly affect Cadm4 binding, it lies within the second Ig domain, an area that could change some of the regulatory cis-mediated trans interactions of the protein (Liu et al., 2019).
The redundant function of Cadm1–Cadm3 in myelinated axons described here is also relevant for the roles Cadm (SynCAM) proteins play during other neurodevelopmental processes such as the establishment of CNS synapses and axon guidance (Frei and Stoeckli, 2017). Different combinations of Cadm1-4 were found in synaptic sites by biochemical fractionation as well as proteomic identification of the synaptic cleft proteins (Fogel et al., 2007; Loh et al., 2016; Cijsouw et al., 2018). Notably, the role of Cadm proteins in synapse formation of hippocampal neurons was masked by overlapping functions of these gene family members, and was only revealed by multiple gene deletions (Fowler et al., 2017) Binding and coimmunoprecipitation experiments suggested that the binding between Cadm1 and Cadm2, as well as between Cadm3 and Cadm4 provide a synaptogenic adhesive code (Fogel et al., 2007). Such a code would be affected by the presence of other members of the Cadm family that are present at the presynaptic or postsynaptic membranes, as revealed by recent proteomic approaches (Cijsouw et al., 2018). Whether synaptogenic and myelinogenic Cadm-dependent adhesion code share molecular similarities will be an interesting topic to explore in future studies.
Footnotes
This work was supported by the National Institutes of Health Grant R01NS097428, the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, and the Israel Science Foundation. E.P. is the Incumbent of the Hanna Hertz Professorial Chair for Multiple Sclerosis and Neuroscience.
The authors declare no competing financial interests.
References
- Amor V, Zhang C, Vainshtein A, Zhang A, Zollinger DR, Eshed-Eisenbach Y, Brophy PJ, Rasband MN, Peles E (2017) The paranodal cytoskeleton clusters Na(+) channels at nodes of Ranvier. Elife 6:e21392 10.7554/eLife.21392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo EJ, Sirkowski EE, Chitale R, Scherer SS (2004) Acute demyelination disrupts the molecular organization of peripheral nervous system nodes. J Comp Neurol 479:424–434. 10.1002/cne.20321 [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297:1525–1531. 10.1126/science.1072356 [DOI] [PubMed] [Google Scholar]
- Brivio V, Faivre-Sarrailh C, Peles E, Sherman DL, Brophy PJ (2017) Assembly of CNS nodes of Ranvier in myelinated nerves is promoted by the axon cytoskeleton. Curr Biol 27:1068–1073. 10.1016/j.cub.2017.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Kim H, Jagot-Lacoussiere L, Maurel P (2016) Cadm3 (Necl-1) interferes with the activation of the PI3 kinase/Akt signaling cascade and inhibits Schwann cell myelination in vitro. Glia 64:2247–2262. 10.1002/glia.23072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cijsouw T, Ramsey AM, Lam TT, Carbone BE, Blanpied TA, Biederer T (2018) Mapping the proteome of the synaptic cleft through proximity labeling reveals new cleft proteins. Proteomes 6:48 10.3390/proteomes6040048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day FR, Helgason H, Chasman DI, Rose LM, Loh PR, Scott RA, Helgason A, Kong A, Masson G, Magnusson OT, Gudbjartsson D, Thorsteinsdottir U, Buring JE, Ridker PM, Sulem P, Stefansson K, Ong KK, Perry JRB (2016) Physical and neurobehavioral determinants of reproductive onset and success. Nat Genet 48:617–623. 10.1038/ng.3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djannatian M, Timmler S, Arends M, Luckner M, Weil MT, Alexopoulos I, Snaidero N, Schmid B, Misgeld T, Möbius W, Schifferer M, Peles E, Simons M (2019) Two adhesive systems cooperatively regulate axon ensheathment and myelin growth in the CNS. Nat Commun 10:4794. 10.1038/s41467-019-12789-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrn MF, Saporta M (2020) Hereditary motor neuropathies. Curr Opin Neurol 33:568–574. [DOI] [PubMed] [Google Scholar]
- Eichel MA, Gargareta VI, D'Este E, Fledrich R, Kungl T, Buscham TJ, Lüders KA, Miracle C, Jung RB, Distler U, Kusch K, Möbius W, Hülsmann S, Tenzer S, Nave KA, Werner HB (2020) CMTM6 expressed on the adaxonal Schwann cell surface restricts axonal diameters in peripheral nerves. Nat Commun 11:4514. 10.1038/s41467-020-18172-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einheber S, Meng X, Rubin M, Lam I, Mohandas N, An X, Shrager P, Kissil J, Maurel P, Salzer JL (2013) The 4.1B cytoskeletal protein regulates the domain organization and sheath thickness of myelinated axons. Glia 61:240–253. 10.1002/glia.22430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar N, Vainshtein A, Rechav K, Tsoory M, Eshed-Eisenbach Y, Peles E (2019a) Coordinated internodal and paranodal adhesion controls accurate myelination by oligodendrocytes. J Cell Biol 218:2887–2895. 10.1083/jcb.201906099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elazar N, Vainshtein A, Golan N, Vijayaragavan B, Schaeren-Wiemers N, Eshed-Eisenbach Y, Peles E (2019b) Axoglial adhesion by Cadm4 regulates CNS myelination. Neuron 101:224–231.e5. 10.1016/j.neuron.2018.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed-Eisenbach Y, Devaux J, Vainshtein A, Golani O, Lee SJ, Feinberg K, Sukhanov N, Greenspan DS, Susuki K, Rasband MN, Peles E (2020) Precise spatiotemporal control of nodal Na(+) channel clustering by bone morphogenetic protein-1/tolloid-like proteinases. Neuron 106:806–815.e6. 10.1016/j.neuron.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T (2007) SynCAMs organize synapses through heterophilic adhesion. J Neurosci 27:12516–12530. 10.1523/JNEUROSCI.2739-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel AI, Li Y, Giza J, Wang Q, Lam TT, Modis Y, Biederer T (2010) N-glycosylation at the SynCAM (synaptic cell adhesion molecule) immunoglobulin interface modulates synaptic adhesion. J Biol Chem 285:34864–34874. 10.1074/jbc.M110.120865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel AI, Stagi M, Perez de Arce K, Biederer T (2011) Lateral assembly of the immunoglobulin protein SynCAM 1 controls its adhesive function and instructs synapse formation. EMBO J 30:4728–4738. 10.1038/emboj.2011.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DK, Peters JH, Williams C, Washbourne P (2017) Redundant postsynaptic functions of SynCAMs 1-3 during synapse formation. Front Mol Neurosci 10:24. 10.3389/fnmol.2017.00024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei JA, Stoeckli ET (2017) SynCAMs - From axon guidance to neurodevelopmental disorders. Mol Cell Neurosci 81:41–48. 10.1016/j.mcn.2016.08.012 [DOI] [PubMed] [Google Scholar]
- Frei JA, Andermatt I, Gesemann M, Stoeckli ET (2014) The SynCAM synaptic cell adhesion molecules are involved in sensory axon pathfinding by regulating axon-axon contacts. J Cell Sci 127:5288–5302. 10.1242/jcs.157032 [DOI] [PubMed] [Google Scholar]
- Fujita E, Urase K, Soyama A, Kouroku Y, Momoi T (2005) Distribution of RA175/TSLC1/SynCAM, a member of the immunoglobulin superfamily, in the developing nervous system. Brain Res Dev Brain Res 154:199–209. 10.1016/j.devbrainres.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Ozeki S, Tanabe Y, Toyama Y, Maekawa M, Kojima N, Senoo H, Toshimori K, Momoi T (2006) Oligo-astheno-teratozoospermia in mice lacking RA175/TSLC1/SynCAM/IGSF4A, a cell adhesion molecule in the immunoglobulin superfamily. Mol Cell Biol 26:718–726. 10.1128/MCB.26.2.718-726.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galuska SP, Rollenhagen M, Kaup M, Eggers K, Oltmann-Norden I, Schiff M, Hartmann M, Weinhold B, Hildebrandt H, Geyer R, Mühlenhoff M, Geyer H (2010) Synaptic cell adhesion molecule SynCAM 1 is a target for polysialylation in postnatal mouse brain. Proc Natl Acad Sci USA 107:10250–10255. 10.1073/pnas.0912103107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan N, Kartvelishvily E, Spiegel I, Salomon D, Sabanay H, Rechav K, Vainshtein A, Frechter S, Maik-Rachline G, Eshed-Eisenbach Y, Momoi T, Peles E (2013) Genetic deletion of Cadm4 results in myelin abnormalities resembling Charcot-Marie-Tooth neuropathy. J Neurosci 33:10950–10961. 10.1523/JNEUROSCI.0571-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horresh I, Bar V, Kissil JL, Peles E (2010) Organization of myelinated axons by Caspr and Caspr2 requires the cytoskeletal adapter protein 4.1B. J Neurosci 30:2480–2489. 10.1523/JNEUROSCI.5225-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy JL, Constable JR, Vicini S, Fu Z, Washbourne P (2009) SynCAM1 recruits NMDA receptors via protein 4.1B. Mol Cell Neurosci 42:466–483. 10.1016/j.mcn.2009.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AN, Appel B (2019) Oligodendrocytes express synaptic proteins that modulate myelin sheath formation. Nat Commun 10:4125. 10.1038/s41467-019-12059-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovic A, Horresh I, Golan N, Spiegel I, Sabanay H, Frechter S, Ohno S, Terada N, Möbius W, Rosenbluth J, Brose N, Peles E (2012) The cytoskeletal adapter protein 4.1G organizes the internodes in peripheral myelinated nerves. J Cell Biol 196:337–344. 10.1083/jcb.201111127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakunaga S, Ikeda W, Itoh S, Deguchi-Tawarada M, Ohtsuka T, Mizoguchi A, Takai Y (2005) Nectin-like molecule-1/TSLL1/SynCAM3: a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule localizing at non-junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J Cell Sci 118:1267–1277. 10.1242/jcs.01656 [DOI] [PubMed] [Google Scholar]
- Liu X, An T, Li D, Fan Z, Xiang P, Li C, Ju W, Li J, Hu G, Qin B, Yin B, Wojdyla JA, Wang M, Yuan J, Qiang B, Shu P, Cui S, Peng X (2019) Structure of the heterophilic interaction between the nectin-like 4 and nectin-like 1 molecules. Proc Natl Acad Sci USA 116:2068–2077. 10.1073/pnas.1810969116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KH, Stawski PS, Draycott AS, Udeshi ND, Lehrman EK, Wilton DK, Svinkina T, Deerinck TJ, Ellisman MH, Stevens B, Carr SA, Ting AY (2016) Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell 166:1295–1307.e21. 10.1016/j.cell.2016.07.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig M, Erskine L, Mason CA, Grumet M, Sakurai T (2001) Nr-CAM expression in the developing mouse nervous system: ventral midline structures, specific fiber tracts, and neuropilar regions. J Comp Neurol 434:13–28. 10.1002/cne.1161 [DOI] [PubMed] [Google Scholar]
- Maciel R, Bis DM, Rebelo AP, Saghira C, Züchner S, Saporta MA (2018) The human motor neuron axonal transcriptome is enriched for transcripts related to mitochondrial function and microtubule-based axonal transport. Exp Neurol 307:155–163. 10.1016/j.expneurol.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K, Rikitake Y, Mori M, Takai Y (2015) Nectins and nectin-like molecules in development and disease. Curr Top Dev Biol 112:197–231. 10.1016/bs.ctdb.2014.11.019 [DOI] [PubMed] [Google Scholar]
- Maurel P, Einheber S, Galinska J, Thaker P, Lam I, Rubin MB, Scherer SS, Murakami Y, Gutmann DH, Salzer JL (2007) Nectin-like proteins mediate axon Schwann cell interactions along the internode and are essential for myelination. J Cell Biol 178:861–874. 10.1083/jcb.200705132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Maurel P, Lam I, Heffernan C, Stiffler MA, McBeath G, Salzer JL (2019) Necl-4/Cadm4 recruits Par-3 to the Schwann cell adaxonal membrane. Glia 67:884–895. 10.1002/glia.23578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederkofler V, Baeriswyl T, Ott R, Stoeckli ET (2010) Nectin-like molecules/SynCAMs are required for post-crossing commissural axon guidance. Development 137:427–435. 10.1242/dev.042515 [DOI] [PubMed] [Google Scholar]
- North RY, Li Y, Ray P, Rhines LD, Tatsui CE, Rao G, Johansson CA, Zhang H, Kim YH, Zhang B, Dussor G, Kim TH, Price TJ, Dougherty PM (2019) Electrophysiological and transcriptomic correlates of neuropathic pain in human dorsal root ganglion neurons. Brain 142:1215–1226. 10.1093/brain/awz063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Horresh I, Trimmer JS, Bredt DS, Peles E, Rasband MN (2008) Postsynaptic density-93 clusters Kv1 channels at axon initial segments independently of Caspr2. J Neurosci 28:5731–5739. 10.1523/JNEUROSCI.4431-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Liu B, Chen T, Li H, Hu X, Gao J, Zhu Y, Zhu Q, Qiang B, Yuan J, Peng X, Qiu M (2008) Disruption of Nectin-like 1 cell adhesion molecule leads to delayed axonal myelination in the CNS. J Neurosci 28:12815–12819. 10.1523/JNEUROSCI.2665-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, Abdellaoui A, Nivard MG, Baselmans BML, Ong JS, Ip HF, van der Zee MD, Bartels M, Day FR, Fontanillas P, Elson SL, de Wit H, Davis LK, MacKillop J, Derringer JL, et al. (2018) GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci 21:1161–1170. 10.1038/s41593-018-0206-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peles E, Nativ M, Lustig M, Grumet M, Schilling J, Martinez R, Plowman GD, Schlessinger J (1997) Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J 16:978–988. 10.1093/emboj/16.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellissier F, Gerber A, Bauer C, Ballivet M, Ossipow V (2007) The adhesion molecule Necl-3/SynCAM-2 localizes to myelinated axons, binds to oligodendrocytes and promotes cell adhesion. BMC Neurosci 8:90. 10.1186/1471-2202-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez de Arce K, Schrod N, Metzbower SWR, Allgeyer E, Kong GKW, Tang AH, Krupp AJ, Stein V, Liu X, Bewersdorf J, Blanpied TA, Lucić V, Biederer T (2015) Topographic mapping of the synaptic cleft into adhesive nanodomains. Neuron 88:1165–1172. 10.1016/j.neuron.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher N, Schmerl B, Lardong JA, Wahl MC, Shoichet SA (2016) MPP2 is a postsynaptic MAGUK scaffold protein that links SynCAM1 cell adhesion molecules to core components of the postsynaptic density. Sci Rep 6:35283. 10.1038/srep35283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband MN, Peles E (2015) The nodes of Ranvier: molecular assembly and maintenance. Cold Spring Harb Perspect Biol 8:a020495. 10.1101/cshperspect.a020495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribic A, Crair MC, Biederer T (2019) Synapse-selective control of cortical maturation and plasticity by parvalbumin-autonomous action of SynCAM 1. Cell Rep 26:381–393.e6. 10.1016/j.celrep.2018.12.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins EM, Krupp AJ, Perez de Arce K, Ghosh AK, Fogel AI, Boucard A, Südhof TC, Stein V, Biederer T (2010) SynCAM 1 adhesion dynamically regulates synapse number and impacts plasticity and learning. Neuron 68:894–906. 10.1016/j.neuron.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, Ginty DD (2020) The emergence of transcriptional identity in somatosensory neurons. Nature 577:392–398. 10.1038/s41586-019-1900-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai T, Ikeda W, Kakunaga S, Morimoto K, Takekuni K, Itoh S, Satoh K, Takeuchi M, Imai T, Monden M, Takai Y (2003) Implications of nectin-like molecule-2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell-cell adhesion and transmembrane protein localization in epithelial cells. J Biol Chem 278:35421–35427. 10.1074/jbc.M305387200 [DOI] [PubMed] [Google Scholar]
- Spiegel I, Adamsky K, Eshed Y, Milo R, Sabanay H, Sarig-Nadir O, Horresh I, Scherer SS, Rasband MN, Peles E (2007) A central role for Necl4 (SynCAM4) in Schwann cell-axon interaction and myelination. Nat Neurosci 10:861–869. 10.1038/nn1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmann C, Timmler S, Barrantes-Freer A, Simons M (2019) Myelin in the central nervous system: structure, function, and pathology. Physiol Rev 99:1381–1431. 10.1152/physrev.00031.2018 [DOI] [PubMed] [Google Scholar]
- Wilson ER, Della-Flora Nunes G, Weaver MR, Frick LR, Feltri ML (2020) Schwann cell interactions during the development of the peripheral nervous system. Dev Neurobiol. Advance online publication. Retrieved April 12, 2020. doi: 10.1002/dneu.22744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Li L, Ren HT, Yin B, Yuan JG, Peng XZ, Qiang BQ, Cui LY (2018) Mutation of the cellular adhesion molecule NECL2 is associated with neuromyelitis optica spectrum disorder. J Neurol Sci 388:133–138. 10.1016/j.jns.2017.10.023 [DOI] [PubMed] [Google Scholar]
- Yang S, Weng H, Chen L, Guo X, Parra M, Conboy J, Debnath G, Lambert AJ, Peters LL, Baines AJ, Mohandas N, An X (2011) Lack of protein 4.1G causes altered expression and localization of the cell adhesion molecule nectin-like 4 in testis and can cause male infertility. Mol Cell Biol 31:2276–2286. 10.1128/MCB.01105-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, Ginty DD (2019) Deep sequencing of somatosensory neurons reveals molecular determinants of intrinsic physiological properties. Neuron 103:598–616.e7. 10.1016/j.neuron.2019.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Du G, Hu X, Yu S, Liu Y, Xu Y, Huang X, Liu J, Yin B, Fan M, Peng X, Qiang B, Yuan J (2005) Nectin-like molecule 1 is a protein 4.1N associated protein and recruits protein 4.1N from cytoplasm to the plasma membrane. Biochim Biophys Acta 1669:142–154. 10.1016/j.bbamem.2005.01.013 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Li H, Li K, Zhao X, An T, Hu X, Park J, Huang H, Bin Y, Qiang B, Yuan J, Peng X, Qiu M (2013) Necl-4/SynCAM-4 is expressed in myelinating oligodendrocytes but not required for axonal myelination. PLoS One 8:e64264. 10.1371/journal.pone.0064264 [DOI] [PMC free article] [PubMed] [Google Scholar]