Abstract
Ligand-activated signaling through the type 1 insulin-like growth factor receptor (IGF1R) is implicated in many physiological processes ranging from normal human growth to cancer proliferation and metastasis. IGF1R has also emerged as a target for receptor-mediated transcytosis, a transport phenomenon that can be exploited to shuttle biotherapeutics across the blood–brain barrier (BBB). We employed differential hydrogen–deuterium exchange mass spectrometry (HDX-MS) and nuclear magnetic resonance (NMR) to characterize the interactions of the IGF1R ectodomain with a recently discovered BBB-crossing single-domain antibody (sdAb), VHH-IR5, in comparison with IGF-1 binding. HDX-MS confirmed that IGF-1 induced global conformational shifts in the L1/FnIII-1/-2 domains and α-CT helix of IGF1R. In contrast, the VHH-IR5 sdAb-mediated changes in conformational dynamics were limited to the α-CT helix and its immediate vicinity (L1 domain). High-resolution NMR spectroscopy titration data and linear peptide scanning demonstrated that VHH-IR5 has high-affinity binding interactions with a peptide sequence around the C-terminal region of the α-CT helix. Taken together, these results define a core linear epitope for VHH-IR5 within the α-CT helix, overlapping the IGF-1 binding site, and suggest a potential role for the α-CT helix in sdAb-mediated transcytosis.
Subject terms: Biochemistry, Drug discovery, Structural biology
Introduction
Human insulin like-growth factor receptor (IGF1R) is a member of a small family of transmembrane receptor tyrosine kinases (RTKs) that also includes the insulin receptor (IR) and a third member, the orphan insulin receptor-related receptor (IRR)1. Aside from IR, IGF1R is a secondary cell-signaling RTK involved in the regulation of glucose uptake and energy metabolism. IGF1R itself has important functions in fetal and prenatal growth and in cancer proliferation, by promoting the immortalization of transformed cells through binding to two insulin-like growth factors, IGF-1 and IGF-21–3. Implication of IGF1R in cancer has prompted the development of anti-cancer strategies using neutralizing antibodies and small-molecule inhibitors of the RTK4. Both IR and, more recently, IGF1R have emerged as therapeutic targets for brain-specific neurological disorders, especially for drug-delivery across the blood–brain barrier (BBB)5.
Many diseases of the central nervous system (CNS) have limited treatment options because of the BBB, whose relative impermeability severely impedes the physical delivery of blood-borne drugs to the CNS. The BBB represents a particular hurdle to CNS delivery of antibody-based therapeutics6 and efficient crossing of large macromolecules must engage endocytotic pathways through a process called receptor-mediated transcytosis (RMT)7,8. To date, transferrin receptor (TfR)9–11, low-density lipoprotein (LDL)12,13, and insulin receptor (IR)14,15 have been explored as RMT targets, with IR-targeted therapies in advanced clinical trials14. However, targeting these common receptors has substantial development risks related to species-dependent expression in brain endothelial cells16, organ selectivity required for improved safety profiles17, as well as potential side effects associated with interfering with their normal physiological functions18. Our recent work has focused on IGF1R, based on its elevated expression in brain endothelial cells relative to peripheral tissue19,20, and on its established role in the delivery of one of its endogenous ligands, IGF-1, across the BBB5,21.
IGF1R is a heavily glycosylated transmembrane protein with a complex dimeric (αβ)2 architecture and with the two-chain (α and β) ectodomains held together by tertiary structural interactions along with inter-chain and intra-chain disulfide bonds1,22. The modular architecture of IGF1R includes a leucine-rich region (L1), a cysteine-rich region (CR), the second leucine-rich region (L2) and the N-terminal part of a type-III fibronectin-like domain FnIII-2 in the α-chain and the C-terminal part of FnIII-2 leading to two other fibronectin-like domains FnIII-1/3 in the β-chain of the ectodomain1,22. The intact FnIII-2 domain in the IGF1R precursor also contains a long insertion domain (ID) connecting the α- and β- chains generated by proteolytic cleavage at a site within ID1. Extensive structural studies of the IGF1R protein family have revealed a novel mechanism of receptor auto-inhibition and ligand-triggered receptor activation22–24. In such a mechanism, IGF-1 appears to engage only one L1-CR-L2 segment of the dimeric IGF1R along with one of the all-important α-CT helices exposed at the extreme C-termini of the α-chains (hence α-CTs) and to release an auto-inhibiting structure of the ectodomain leading to spontaneous receptor auto-phosphorylation22–24. A significant structural feature of the IGF1R/IGF-1 interaction is the malleability of the receptor architecture and the disparate roles played by the two α-CT helices in IGF1R, one as part of the binding site for IGF-1 and the second remaining apparently free with partial contacts with the L1′ and the FnIII-2 regions of the dimeric receptor23,24. The differing environment of the two α-CT helices elicited by IGF-1 binding to IGF1R has been identified as the structural origin23,24 of negative cooperativity observed for the IGF-1/IGF1R ligand-receptor system25,26.
We recently discovered a unique class of single-domain antibodies (sdAbs) specific for the IGF1R ectodomain5,27, among which the single-domain antibody IGF1R-5 (referred here to as VHH-IR5), does not appear to interfere with normal kinase functions of IGF1R and its interactions with the endogenous ligand21. As such, VHH-IR5 not only has the potential to act as a BBB-crossing shuttle, but may help decipher the fundamental mechanisms of cell signaling through IGF1R and receptor transcytosis. In this work, we focus on elucidating the mode of action of VHH-IR5, with a specific goal of defining its binding epitope on IGF1R in relation to the natural IGF1R ligand IGF-1. Results from differential hydrogen–deuterium exchange mass spectrometry (HDX-MS) and nuclear magnetic resonance (NMR) spectroscopy, and binding studies with IGF1R fragments outline an apparently shared binding site between the sdAb VHH-IR5 and the natural ligand IGF-1 convergent on the α-CT helix motif, an epitope on IGF1R not targeted, to the best of our knowledge, by other antibodies reported to date28–31.
Results
Section I: comparison of IGF-1 and VHH-IR5 binding and functional activation of IGF1R
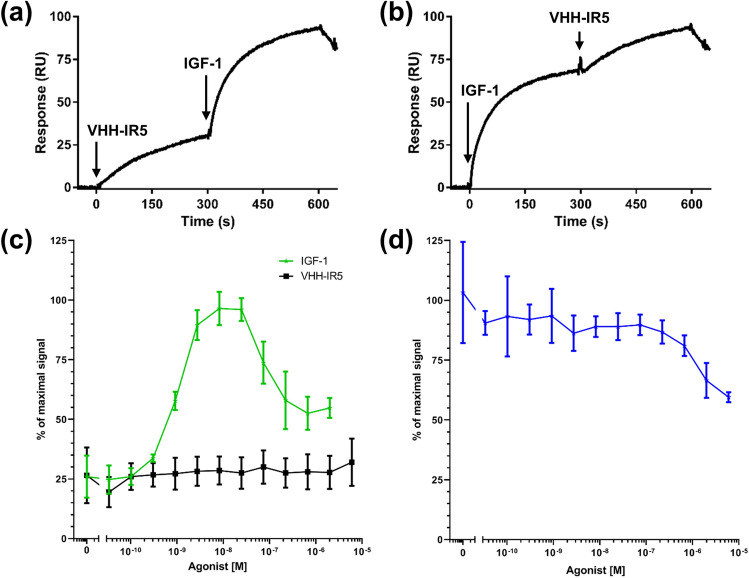
Previous work has suggested that VHH-IR5, an IGF1R-binding sdAb (of the llama origin27) with BBB crossing activity, does not interfere with the interaction between IGF1R and its native ligand, IGF-121,27. Figure 1a shows that saturation of surface-immobilized IGF1R ectodomain (eIGF1R) with VHH-IR5 does not preclude the subsequent binding of IGF-1. Reversing the order of pre-loading produced the same result (Fig. 1b). Qualitatively, pre-loading of either VHH-IR5 (Fig. 1a) or IGF-1 (Fig. 1b) also does not alter the shape of SPR sensograms of the other ligand (Fig. S2a,b). Thus, occupancy of eIGF1R by VHH-IR5 (or by IGF-1) has little effect on IGF-1 (or VHH-IR5) binding, outlining apparently independent and non-competitive binding sites on eIGF1R for these two ligands. Quantitatively, VHH-IR5 has a binding affinity of KD = 0.6 nM for eIGF1R along with a slow rate of dissociation, koff = 3.4 × 10–4 s−1, as seen by the characteristic plateau for the dissociation phase of the VHH-IR5 sdAb-eIGF1R complex (Fig. S2a). We measured an affinity of KD = 8.2 nM, and a faster dissociation rate constant (8.2 × 10–3 s−1) for IGF-1 binding to the eIGF1R ectodomain (Fig. S2b), in agreement with previous reported values (KD ~ 10–20 nM)29. VHH-IR5, therefore, has a higher affinity for eIGF1R as compared to IGF-1. In addition, VHH-IR5 is also specific for IGF1R as it does not bind the insulin receptor (Fig. S2c) in contrast to IGF-1 which interact with both (Fig. S2d).
Figure 1.
sdAb-IGF1R binding and effects on IGF1R signalling. (a) SPR sensorgrams showing the pre-loading of the sdAb VHH-IR5 to surface-immobilized human eIGF1R followed by the binding of human IGF-1. Experiments were carried out in duplicate with SEC-purified eIGF1R, IGF-1 and VHH-IR5 (see Fig. S1 of the Supplementary Materials). (b) SPR sensorgrams showing the pre-loading of human IGF-1 to surfaced immobilized eIGF1R followed by the binding of VHH-IR5. (c) Cell-based assays showing IGF1R activation by IGF-1 (green) and and lack of activation by VHH-IR5 (black) alone. (d) Cell-based assays showing effects of VHH-IR5 on IGF1R activation in the presence of 3 nM IGF-1 (blue).
Next, we demonstrated in a cell-based assay that VHH-IR5 binding does not induce significant activation of IGF1R, in sharp contrast to the effects of IGF-1 (Fig. 1c). Very importantly, increasing concentrations of VHH-IR5 had little further effects on the activating level of IGF-1, even when present up to > 500-fold molar excess (Fig. 1d), demonstrating essentially mutual independence of these two ligands. This finding is in agreement with previous studies that showed VHH-IR5 did not induce phosphorylation of the downstream kinase AKT21, which is the expected outcome of IGF1R activation by the endogenous ligand IGF-122,32. Taken together, both biophysical binding (SPR) and cell-based assays indicate that VHH-IR5 and IGF-1 likely have independent binding sites on IGF1R and that VHH-IR5 binding is unlikely to alter the ligand-sensing capabilities of IGF1R on the cell-surface.
Section II: comparison of IGF-1 and VHH-IR5 binding to IGF1R using HDX-MS
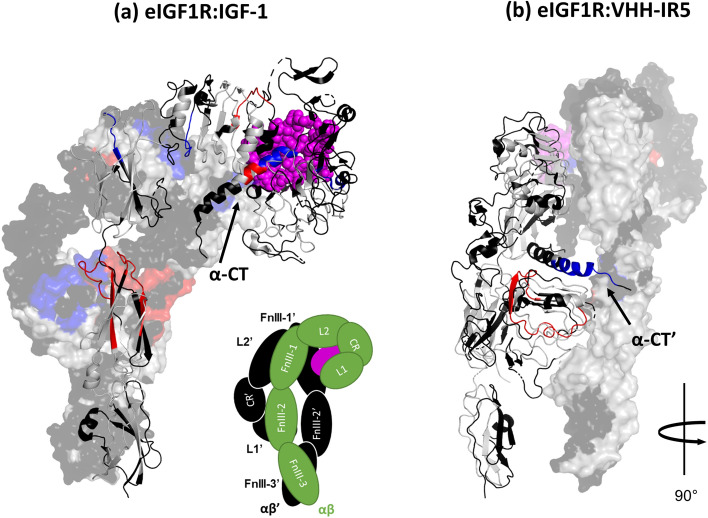
We next assessed the global structural consequences of IGF-1 and VHH-IR5 binding to the soluble eIGF1R using differential bottom-up HDX-MS profiles33. This method is extremely sensitive to conformational changes in proteins and has been employed for mapping antibody-epitope interactions34,35. The HDX-MS experiments were performed using the fully-glycosylated form of eIGF1R to preserve native eIGF1R conformations, similar to a strategy utilized for the insulin receptor (IR)36. Overall, 47% sequence coverage was achieved across 134 peptides (Table 1 and Table S1). Despite the resulting gaps in overall sequence coverage (Fig. S4), HDX-MS data still afforded key insights into the eIGF1R-binding characteristics of both protein ligands (Fig. 2). The HDX responses to the native ligand, IGF-1 (Fig. 2a) are similar to what has been reported previously, validating our experimental approach. For example, conformational stabilizations represented by a decrease in deuteration upon binding were observed at the contact site between the L domain and IGF-1 (residues I9-L16 and L57-L63 specifically), and at the dimer interface between the two L2 domains (L2/L2′, Y417-L424) and the two FnIII-1 domains (FnIII-1/FnIII-1′, R488-F493) (Fig. 2a, blue regions). There was increased deuteration or conformational destabilization for the L2 residues C323-L331, which may be related to the hinging motion of L1-CR away from the L2 domain observed in the full-length IGF1R23. An increase in deuteration within the FnIII-2 domain (I598-L611 and Y769-N789) likely results from the disruption of the L1/FnIII-2′ interaction associated with ligand recruitment. Bimodal exchange behaviour in a sequence segment spanning insertion domain residues Y628-D649, first identified by Houde et al.37, was also observed here in the uncomplexed, IGF-1-bound and VHH-IR5-bound states of eIGF1R. Inspection of the unlabelled isotopic profile (Fig. S5b) shows that the observed bimodality is not a result of overlapped profiles from unrelated peptides. However, further comparison of the exchange profiles between the three states was not possible due to inherent variability of the deuteration in this region (Fig. S5a).
Table 1.
Summary of HDX-MS experimental parameters53.
| Data set | Control | IGF-1 | VHH-IR5 |
|---|---|---|---|
| HDX reaction details | PBS, pD = 7.0, 25 °C | ||
| HDX time course (min) | 1, 3, 10, 60 | ||
| # of Peptides found across all samples | 133 | ||
| Sequence coverage | 47% | ||
| Average peptide length/redundancy | 11.7/1.7 | ||
| Replicates (biological or technical) | 3 (technical) | ||
| Repeatability (average standard deviation, %) | 1.6 | 1.5 | 1.4 |
| Significant differences in HDX | Two-state student T-Test performed at each time point (> 2 SD, p value 0.05) | ||
*All HDX-MS experiments were carried out with recombinant and SEC-purified eIGF1R (Fig. S3).
Figure 2.
A global survey of HDX-MS profiles of eIGF1R in response to IGF-1 and VHH-IR5 binding. (a) IGF-1 response profile projected on a structural model of the 1:1 eIGF1R:IGF-1 complex (PDB: 6PYH). The (αβ) ′ monomer, except for α-CT′, is rendered as a transparent surface, while the (αβ) monomer and α-CT′ are shown as a ribbon cartoon. The schematic in the inset shows the domain organization of IGF1R where the (αβ) monomer is colored in green, and (αβ)′ in black. (b) VHH-IR5 response profile projected on the same structural model as in (a) (PDB: 6PYH), except that the structure is rotated counterclockwise by 90°. Significant structural destabilizations are shown in red, stabilizations in blue, lack of significant changes in grey, and missing sequence coverage in black. Residues are colored based on differences in deuteration at a single time point (± 2 SD, p = 0.02). In both (a) and (b), IGF-1 molecules are shown as magenta spheres. Of note, residues of the rhesus eIGF1R used for HDX-MS data collection were mapped by BLAST onto the mouse eIGF1R before rendering the HDX-MS results onto the 3D structure of mouse eIGF1R (PDB: 6PYH).
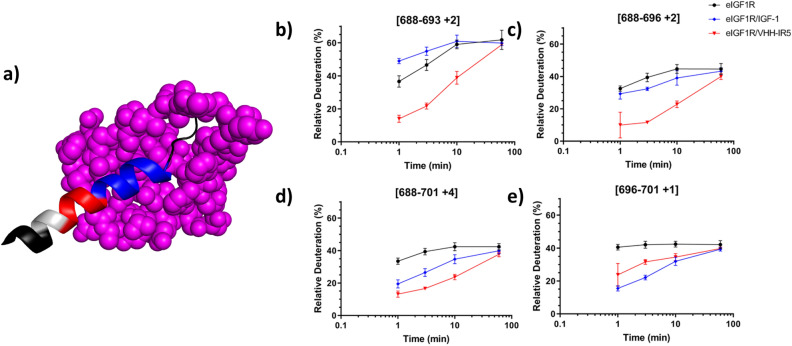
Interestingly, many of the dramatic changes in deuteration elicited by IGF-1 were almost entirely absent in response to VHH-IR5 binding (Fig. 2b), suggesting that VHH-IR5 does not induce the same global structural changes in IGF1R. However, both IGF-1 and VHH-IR5 appear to engage the α-CT structural motif located at the C-terminal end of the eIGF1R α-chain, although key differences were observed in the deuteration pattern of the α-CT helix (Fig. 3a) and its immediate surroundings. First, the HDX-MS profile for the IGF-1/eIGF1R interaction (Fig. 3a) closely tracks engagement of the α-CT helix and the associated conformational malleability38. Specifically, an increase in deuteration was observed with residues Y688-E693, which suggests that this sequence segment is conformationally more dynamic relative to apo-eIGF1R, despite elongation of the α-CT helix in the IGF-1 bound state23. This is followed by progressively stronger reduction in deuteration as peptide coverage shifts towards the C-terminus of the α-CT motif, implying the strongest structural stabilization for residues N694-F701 (Fig. 3b–e). In contrast, binding of VHH-IR5 induced stabilization across residues in the N-terminal region of the α-CT helix (ID residues K690-F701), with the strongest effect localized at residues K690-F692 (Fig. 3b–e). Additionally, VHH-IR5 triggered structural destabilization within the L1 domain (A61-L87) of eIGF1R (Fig. 2b), which is in close contact with the α-CT-helix in IGF1R23, 24.
Figure 3.
Detailed analysis of the HDX-MS profiles in the IGF1R α-CT region. (a) HDX profile of the α-CT’ bound to IGF-1 (magenta) (based on PDB: 6PYH). Residues 684-706 of mouse IGF1R α-CT’ (residues 683-705 in rhesus/human eIGF1R) are shown as a ribbon cartoon. Significant structure destabilization is shown in red and stabilization in blue, while lack of significant changes in grey and missing coverage in black. (b–e) HDX-MS kinetics of peptides covering the α-CT helix. Data collected in quadruplicate, and error bars represent 1× SD. Deuteration was normalized to theoretical maximum uptake of 45%. Free eIGF1R is shown as black circles, eIGF1R/IGF-1 as blue diamonds, and eIGF1R/VHH-IR5 as red triangles.
Section III: epitope mapping of VHH-IR5 using IGF1R insertion domain (ID) fragments containing the α-CT helix
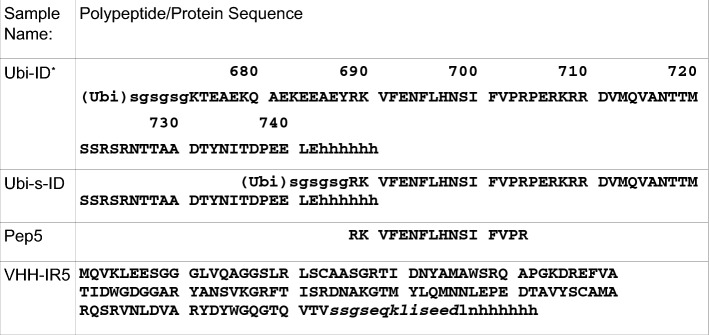
In order to further define the role of the α-CT helix motif in ligand engagement, we employed NMR to examine the binding behaviour of VHH-IR5 with IGF1R fragments derived from the insertion domain (Table 2). Two fragments, IGF1R674-742 and IGF1R689-742, in the form, respectively, of Ubi-ID and Ubi-s-ID fusion proteins (Table 2) retain the covalent junction between the extreme C-terminus of the α-chain and the N-terminus of the β-chain in IGF1R that is present in pro-IGF1R1. Interestingly, both Ubi-ID and Ubi-s-ID, the latter lacking the N-terminal part of the long α-CT helix, have specific binding with VHH-IR5, as shown by localized perturbations in the 1H-15N SOFAST-HMQC map of 15N-labelled Ubi-ID and Ubi-s-ID by unlabelled VHH-IR5 (Fig. S6). Ubiquitin moieties present at the N-terminus of Ubi-ID and Ubi-s-ID served as an internal control for the interpretation of NMR signal perturbations. The ubiquitin protein is not expected to bind VHH-IR5 and indeed the ubiquitin moiety in the fusion protein showed minimal HMQC signal perturbations (Figs. 4 and S6).
Table 2.
Polypeptide and protein constructs used in NMR and related binding studies.
*(Ubi) denotes the ubiquitin moiety, which has an amino-acid sequence of MQIFVKTLTG-KTITLEVEPS-DTIENVKAKI-QDKEGIPPDQ-QRLIFAGKQL- EDGRTLSDYN-IQKESTLHLV-LRLRGG. Ubi-ID is, therefore, a 157-residue fusion protein and Ubi-s-ID, a 142-residue fusion protein. For Ubi-ID, Ubi-s-ID and Pep5, upper-case letters designate the amino-acid sequence of the uncleaved single-chain human IGF1R precursor1. In lower-case letters are the linker sequence, sgsgsg (for Ubi-ID and Ubi-s-ID), and the c-Myc tag ssgseqkliseed (in VHH-IR5) and a His6 tag added for protein expression and purification. All expressed proteins contain an artificial Met residue at the N-terminus added by the E. Coli host during protein expression.
Figure 4.
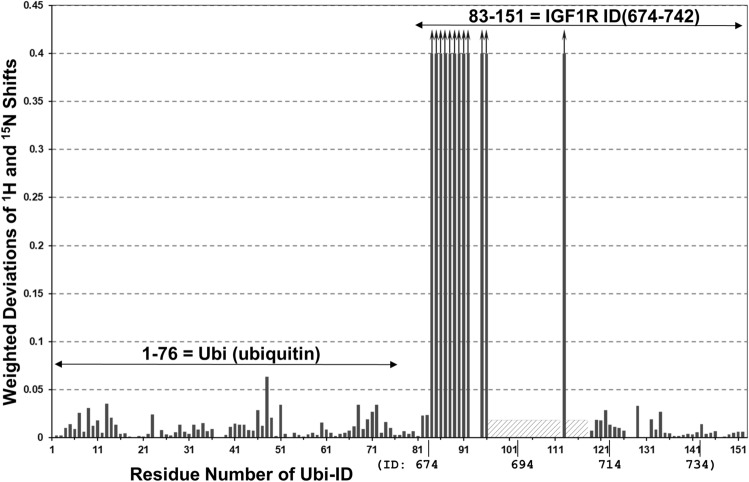
NMR signal perturbations of the 15N-labelled Ubi-ID protein by VHH-IR5. Weighted deviations are calculated as the square root of the weighted frequency shifts along both the 1H and 15N dimensions of the HSQC spectra of the free Ubi-ID as compared to a 1:1 complex of Ubi-ID with unlabelled VHH-IR5 (Fig. S3a) and plotted according to residue-specific assignments achieved for Ubi-ID (Fig. S4). The ubiquitin moiety from 1 to 76 exhibits relatively small differences between the free protein and its complex with VHH-IR5. Some residues of the IGF1R ID 694-742 moiety show pronounced perturbations, especially for T675-E687 (or residues 84-96 of Ubi-ID). Bars with an arrow indicate those residues whose HSQC signals disappeared in the complex of 15N-labelled Ubi-ID with VHH-IR5. Hatched boxes indicate that no HSQC signals were found for this region of Ubi-ID (R689-R709), except for R704. HSQC signals of the ID segment re-emerge from residue 119 to 151 (R710-E742), with essentially no responses to VHH-IR5 binding.
The observed NMR HMQC perturbations for the longer ID fragment were localized in the N-terminal region, i.e., residues T675-K683, A686 and E687 (Fig. 4); in other words, further upstream to the α-CT helix characterized by HDX-MS (Fig. 3). Surprisingly, the HSQC map (as compared to SOFAST-HMQC, vida infra) of Ubi-ID appears to lack assignable signals for the entire C-terminal region of the α-CT helix, i.e., residues R689-V702 and E706-E709, including residues E706-R709 within an extended segment after the α-CT helix (Fig. S7). These signals are missing at both near-neutral pH 6.8 and pH 5.5 (Fig. S7), despite that this latter lower pH condition normally favors optimal NMR signal intensities. The more sensitive proton-15N SOFAST-HMQC experiment39 revealed a few additional crosspeaks of very low intensities (Fig. S8a,b), suggesting that the largely invisible region of Ubi-ID (Figs. 4 and S7) must be a consequence of severe line broadening of the NH and/or 15N NMR signals of the IGF1R689-709 segment. As well, the SOFAST-HMQC spectrum of Ubi-ID became sharper at an elevated temperature (308 vs. 298 K, Fig. S8c), especially in the spectral region where new SOFAST-HMQC signals (i.e. the observable α-CT residues) are located (Fig. S6). These same SOFAST-HMQC signals also experience more significant changes in NH/15N chemical shifts upon temperature elevation (Fig. S8c). Taken together, these NMR lineshape behaviors strongly implicate the α-CT residues as being involved in conformational dynamics such as frame shifts of the α-CT helix (Fig. 3) or other yet-to-be-clarified structural transitions.
Intriguingly, as many as 10 new SOFAST-HMQC peaks appeared from within the IGF1R689-742 fragment of the shorter Ubi-s-ID construct, which can be traced to the sequence segment without significant HMQC signals in Ubi-ID (Fig. S6). All these re-appeared SOFAST-HMQC signals exhibited specific perturbations in the presence of VHH-IR5. The binding interactions were then examined in complementary experiments using 15N-labelled VHH-IR5 titrated with unlabelled Ubi-ID and Ubi-s-ID. Figure 5a and b show that 15N-labelled VHH-IR5 responds specifically to binding of Ubi-ID and Ubi-s-ID, in a similar fashion as the 15N-labelled Ubi-ID and Ubi-s-ID responding to unlabelled VHH-IR5 (Fig. S6). Despite lacking the N-terminal residues K674-Y688 of the long α-CT helix, binding of the Ubi-s-ID construct perturbs essentially the same HSQC signals of VHH-IR5 as binding to Ubi-ID (Fig. 5a,b). Very importantly, the same set of perturbed residues in VHH-IR5 include some of CDR2, e.g. T51 and I52 and many in the framework region, e.g. V13, L21, A25, E47, F48, Q83, M84, A93 and V121 (Fig. 5a,b), based on the NMR assignments of free VHH-IR5 (Fig. S9).
Figure 5.
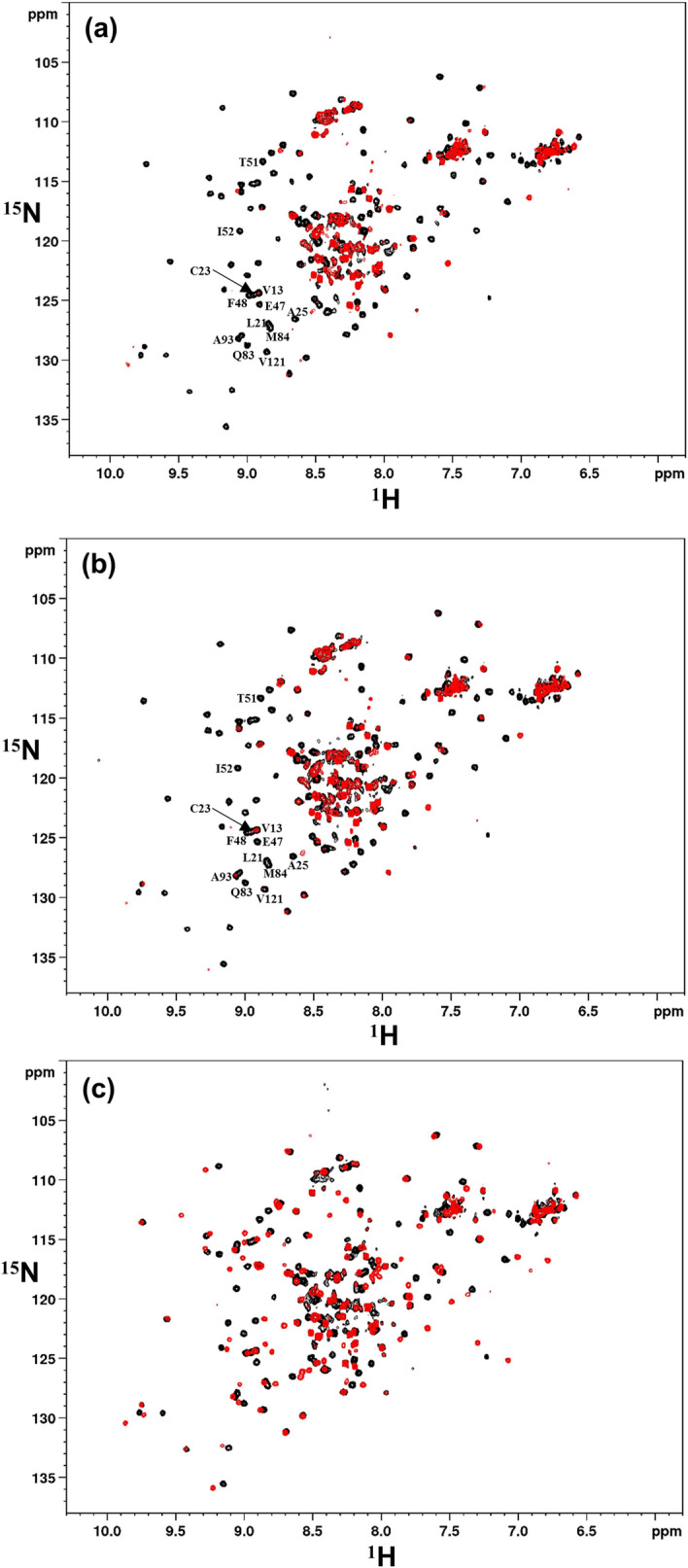
Comparative responses of VHH-IR5 to IGF1R ID fragments. HSQC spectra of 15N-labelled VHH-IR5 identify effects of interactions with three unlabelled IGF1R ID fragments. In black are the HSQC spectra of free VHH-IR5 collected immediately before additions of Ubi-ID, Ubi-s-ID and Pep5 while superimposed in red are the HSQC spectra of the sdAb complexes at ~ 1:1 molar ratio. (a) spectral comparisons showing the effects of Ubi-ID binding. (b) spectra showing the effects of Ubi-s-ID binding. Note that Ubi-ID (a) and Ubi-s-ID (b) binding induce almost the same perturbations, i.e. disappearances of many HSQC signals of 15N-labelled VHH-IR5, especially those of residues T51 and I52 at the beginning of the CDR2 loop and many other residues in the framework region. All perturbed residues were labelled using the resonance assignments of 15N-labelled VHH-IR5 (Fig. S5). (c) Widespread spectral displacements of 15N-labelled VHH-IR5 were produced by binding of the peptide fragment Pep5.
To further explore the interactions of VHH-IR5 with the α-CT helix, a short peptide derived from the ID region of IGF1R similar to the fragment VFENFLHNSIFVPRPE (α-CT691-706) was produced (Pep5, Table 2). Previously, this portion of the α-CT helix was utilized for reconstituting a hybrid IR-IGF1R micro-receptor40. More importantly, Pep5 contains all of the amino acids in the α-CT region of IGF1R that were seen to have modified HDX-MS patterns upon VHH-IR5 and/or IGF-1 binding. The synthetic Pep5 (unlabelled) was used here to titrate into 15N-labelled VHH-IR5 for selective observations of responses in 15N-labelled VHH-IR5. Instead of broadening and the resulting disappearance of many HSQC signals of free VHH-IR5, Pep5 binding to VHH-IR5 led to the appearance of new HSQC signals (i.e., those from the Pep5-sdAb complex) accompanying the disappearance of the HSQC signals of the free VHH-IR5 (Fig. 5c). Such widespread differences between free and ligand-bound proteins are characteristic of slow off-rates and indicative of high-affinity binding interactions in contrast to weaker molecular interactions giving rise to NMR signal broadening (Fig. 5a,b) and disappearance41. A smaller peptide fragment of the IGF1R ID such as Pep5 (Table 2), or the related α-CT691-706 peptide40, very likely has a high binding affinity to VHH-IR5, suggesting that it may represent the linear epitope of this sdAb.
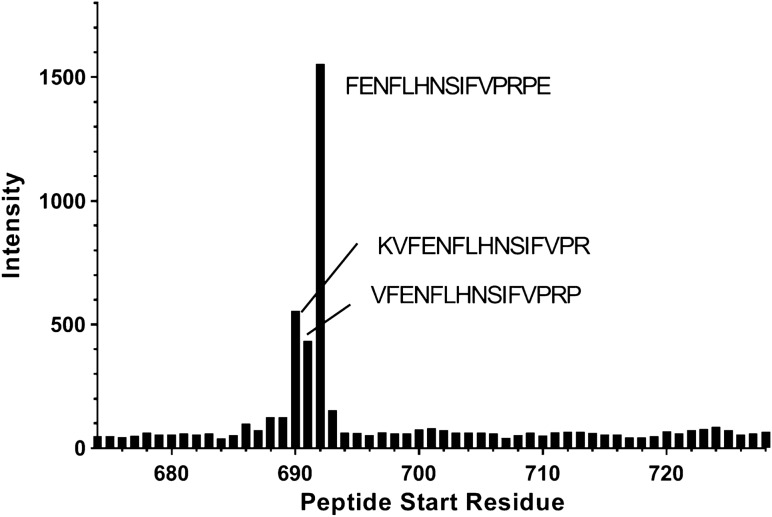
To confirm our results with Pep5, the epitope of VHH-IR5 was further examined using a library of 15-residue peptides that covers the same 674-742 region of IGF1R contained in Ubi-ID (Table 2). Interestingly, the first peptide KTEAEKQAEKEEAEY showed no binding at all to VHH-IR5 (Fig. 6 and Table S2) despite containing all the residues of Ubi-ID with large HSQC signal perturbations (Fig. 4). Binding interactions only become observable for peptides KVFENFLHNSIFVPR, VFENFLHNSIFVPRP, and FENFLHNSIFVPRPE covering IGF1R residues K690-E706, which mirror the strong NMR signal perturbation for residue R704 (Fig. 4). There was essentially no difference between Ubi-ID and Ubi-s-ID for their binding to 15N-labelled VHH-IR5 (Fig. 5) since both carry the same sequence K690-E706 of IGF1R (Table 2). NMR signal perturbations in the N-terminal region of the α-CT helix therefore reflects conformational changes induced by VHH-IR5 binding to the epitope segment as represented by Pep5, which, for yet unknown mechanisms, has severely broadened NMR signals as part of the longer ID fragments in Ubi-ID (Fig. S8). This latter experimental NMR observations may be related to HDX-MS results (Fig. 3), outlining significant structural stabilizations of this important region of IGF1R in response to VHH-IR5 binding.
Figure 6.
VHH-IR5-binding profile of 15-residue peptide fragments covering IGF1R 674-742. The horizontal axis corresponds to the first residue in each peptide and the vertical axis is the ELISA readout (Table S2) in the PepScan assay52. A high level of binding of peptide 692-FENFLHNSIVPRPE-706 (containing R704) should be noted in light of the large NMR signal perturbations of the same Arg residue.
Discussion
In the search for sdAbs targeting IGF1R, VHH-IR5 has demonstrated the characteristics required for drug delivery across the BBB21: efficient BBB transmigration, no detectable impact on the functional activation of IGF1R and no interference with IGF1R/IGF-1 interaction (Fig. 1). The uniqueness of this BBB-crossing sdAb is further highlighted by the nature of the binding epitope of VHH-IR5, revealed here using an array of orthogonal and complementary methodologies. In summary, HDX-MS and NMR results converged on an epitope sequence consisting of residues FENFLHNSIFVPR located near the extreme C-terminus of the IGF1R α-chain (the α-CT). This region of the IGF1R ectodomain demonstrated significant decreases in deuteration in HDX-MS profiles (Fig. 3), equating a structural/conformational stabilization upon binding either driven by allostery or as a direct consequence of binding contacts or both. Of particular interest is the finding that IGF1R fragments containing FENFLHNSIFVPR all retain binding to VHH-IR5 as shown by use of NMR spectroscopy (Figs. 4 and 5). The nature of the VHH-IR5 epitope is further highlighted by two shorter fragments of IGF1R having increased binding affinities for VHH-IR5: RKVFENFLHNSIFVPR (Pep5, Table 2 and Fig. 5) and FENFLHNSIFVPRPE (Fig. 6). These peptide segments terminate, 6 and 4 residues, respectively, before the α-chain C-terminus of IGF1R (Table 2), which is exposed by enzymatic cleavage during IGF1R maturation (Fig. S6)1. Taken together, the sdAb VHH-IR5 has a prototypical linear epitope42, which is available on the surface of the IGF1R structure, allowing binding of VHH-IR5 to native IGF1R on the cell surface.
At first glance, the identified VHH-IR5 epitope directly overlaps with part of the IGF-1 binding site, residues F692-N694, L696-N698, and I700-F701 identified previously37,38,43 and seen here again using HDX-MS (Fig. 3). VHH-IR5 binding therefore necessitates availability of this sequence motif in both apo-IGF1R and in the IGF-1/IGF1R complex since these two ligands do not appear to interfere with each other for IGF1R binding (Fig. 1). As a disulfide-linked homodimer, IGF1R contains two potential binding sites and IGF-1 has been shown to engage only one of these sites on full-length IGF1R at biologically relevant concentrations, as a consequence of negative cooperativity23,25,26. Upon binding to IGF1R, IGF-1, along with the L1 domain of one monomer, engages the α-CT' helix of the other monomer, and induces a dramatic structural rearrangement along with the enlongation of the helical structure of the free α-CT toward the N-terminal region (residues 670-681)23. For non-competitive binding to be possible, the second α-CT site must remain competent for binding VHH-IR5 and vice versa for IGF-1. Alternatively, both binding sites can be equally available for independent ligation of IGF-1 and VHH-IR5 as shown by SPR evidence of simultaneous IGF-1 and VHH-IR5 interaction with eIGF1R (Fig. 1a). Interestingly, despite dramatic structural rearrangement, the unligated α-CT of the 1:1 IGF-1/IGF1R complex indeed maintains a surface-exposed configuration23, leaving this second α-CT available for VHH-IR5 complexation as we observed here for the IGF1R ectodomain (Fig. 1).
The precise mechanism for how VHH-IR5 interacts with the rest of IGF1R without activating downstream signalling events remains largely unknown. However, there are clearly identifiable differences between structural changes of the IGF1R ectodomain in response to IGF-1 and VHH-IR5 binding. The HDX-MS responses in FnIII-1, -2, and L2 of eIGF1R to IGF-1 binding closely mirror the recently-determined symmetric conformation of the IGF1R ectodomain in complex with two IGF-1 molecules37,38. Such responses are notably absent upon binding to VHH-IR5. There is, instead, a decrease in deuteration at the L1 domain of the eIGF1R-sdAb complex (Fig. 2b), which suggests structural relaxation of this L1 region subsequent to VHH-IR5 complexation with the α-CT. This same L1 region interacts with the α-CT helix in an apo-form of eIGF1R38. In the case of VHH-IR5, its main binding effect is in stabilizing the entire helix and limiting its conformational sampling. Taken together, while the IGF-1 binding elicits more global conformational rearrangements in eIGF1R, VHH-IR5 appears to exhibit a localized binding response through a predominantly linear epitope at the α-CT. VHH-IR5 binding may also be negatively cooperative in the sense that it can only bind a single α-CT site on IGF1R. Since VHH-IR5 alone does not activate IGF1R, VHH-IR5 must induce conformational changes in IGF1R that are different from the large structural shifts induced by IGF-1, such that associations of the IGF1R transmembrane domains and the subsequent receptor auto-phosphorylation cannot take place. Such a distinct mode of receptor engagement by VHH-IR5 may underline the structural basis of the transcytosis capacity of this unique sdAb without a dramatic impact on the normal functions of IGF1R. Future structural investigations are needed, particularly with more complete HDX-MS sequence coverages, for a detailed understanding of the novel sdAb-IGF1R interaction.
Limitations
This work is focused on assigning the epitope of an IGF1R-specific sdAb VHH-IR5 and comparing the molecular interactions with those observed for the natural ligand IGF-1. Consequently, the ectodomain of IGF1R was selected for the binding studies and for the comparison. There are therefore limitations in the data that prevent detailed assessments of the global architecture of the sdAb-IGF1R complex. Indeed, our HDX-MS data accurately reflect structural shifts in the bound α-CT motif reported by Li et al. for a full-length IGF1R23. Our HDX-MS findings suggests a limited degree of asymmetry upon IGF-1 binding to the IGF1R ectodomain, which is not in conflict with asymmetric and simultaneous binding of VHH-IR5 and IGF1 in the full length receptor. It is also plausible that the low deuteration content may mask the underlying asymmetry in the HDX profiles44. We are unable to explore this question in greater depth within the scope of this current study.
Methods
Surface plasmon resonance (SPR)
The binding of IGF-1 and VHH-IR5 to immobilized IGF1R ectodomain (eIGF1R) was determined using BIACORE 3000 (GE Healthcare). Approximately 3000 Resonance Units (RU) of recombinant human eIGF1R (R&D Systems, Cat# 391-GR-050) were immobilized on a sensor chip CM5 after the quality of the eIGF1R was confirmed by size-exclusion chromatography (SEC, see Fig. S1a). Immobilization was carried out at 10 µg/mL in 10 mM acetate at pH 4.0 using the amine coupling kit (GE Healthcare). The remaining reactive sites were blocked with 1 M ethanolamine at pH 8.5. An ethanolamine blocked surface was used as a reference. Binding studies were carried out at 25 °C in 10 mM HEPES, pH 7.4 containing 150 mM NaCl, 3 mM EDTA and 0.005% surfactant P20 (Polyoxyethylenesorbitan, GE Healthcare). Various concentrations of SEC-purified IGF-1 and VHH-IR5 (Fig. S1b,c) were flowed over the immobilized eIGF1R and reference surfaces at 20 uL/min. The concentrations employed for VHH-IR5 were 0.75, 1, 2.5, 5, 5, 7.5 and 10 nM, while for IGF-1 binding, they were 10, 25, 50, 100 and 250 nM. The SPR co-injections (saturation of the surface with injection of analyte 1, followed by injection of a mixture of analyte 1 + analyte 2) utilized VHH-IR5 at 10 nM and IGF-1 at 250 nM. Surfaces were regenerated, after binding of VHH-IR5 with 10 mM glycine at pH 2.0 with a contact time of 24 s. No regeneration was used for IGF-1 binding experiments. Data were analysed with BIAevaluation 4.1 (GE Healthcare). The data was fit to a 1:1 binding model without further attempts at quantifying the binding stoichiometry (Text S1).
Cell-based IGF1R functional assay
The receptor functional assay was carried out using the PathHunter eXpress IGF1R kit (DiscoverX), which uses engineered HEK293 cells expressing a (Pro-Link or PK-) tagged IGF-1 Receptor and an Enzyme Acceptor (EA-) tagged SH2 domain. Upon receptor activation, EA-SH2 binds to the phosphorylated PK-IGF1R and reconstitutes an active β-galactosidase enzyme which hydrolyzes a substrate to generate a chemiluminescent readout.
Cells were thawed and plated in a 384-well white wall clear bottom plate (Greiner, North Carolina, USA) at 20,000 cells/well in the cell plating 17 reagent media. After a 24-h incubation at 37 °C in an atmosphere with 5% CO2, cells were treated with IGF-1 (R&D, Minneapolis, USA) (with concentrations varying from 0.03—2000 nM), VHH-IR5 (0.03–6000 nM) or 3 nM IGF-1 + VHH-IR5 (0.03–6000 uM) for 180 min at RT. The chemiluminescent substrate was added and cells were further incubated at RT for 60 min. The resulting luminescence was measured using the CLARIOstar plate reader (BMG LABTECH, Ortenberg, Germany).
Expression and purification of the IGF1R ectodomain
Codon-optimized cDNAs (Genscript) for the rhesus IGF1R ectodomain (eIGF1R, residues 1-902) with a C-terminal His-tag was subcloned into pTTTM109 expression vector. A pool of CHO cells (CHOBRI55E1-JN, proprietory to The National Research Council of Canada) was initiated and the CHOBRI TM55E1 pool was selected with methionine sulfoximine for approximately two weeks. A fed batch production was then performed in optiFlasks. The eIGF1R protein was purified by immobilized metal ion affinity chromatography followed by size-exclusion chromatography in 1× PBS (HiPrep 26/60Superdex 200) (Fig. S3a). Eluting fractions were monitored by SDS-PAGE (Fig. S3b), pooled, concentrated to 10.17 mg/mL and stored at − 80 °C.
Peptides and expression of isotope-labelled proteins for NMR Spectroscopy
Fusion proteins of IGF1R ID fragments, Ubi-ID and Ubi-s-ID, were constructed by adding the sequence of human ubiquitin (Ubi) to the N-terminus of the two IGF1R fragments (Table 2). The DNA sequences were codon-optimized for E. coli, synthesized and inserted into the pD441-SR expression vector (Atum). The plasmids were electroporated into a modified BL21 E. coli host (F–ompT lon hsdSB,rB-,mB-)gal dcm [malB+]K-12(λS) rhaB elaD) and selected with 50 µg/mL kanamycin. The expression plasmid for VHH-IR5 was constructed similarly using the normal BL21 E. Coli and contained a c-Myc tag and a His6 tag linked to the C-terminus of the sdAb using a SSGS spacer (Table 2). Further details for VHH-IR5 production are provided in Text S2 (Supplementary Materials).
Protein expression was induced in the presence of kanamycin by addition of 1 mM IPTG at 24 °C overnight. Uniform 15N- and 13C/15N-labeled proteins were obtained by growing the cells in defined media containing 2 g/L d-glucose (U-13C6-99%, Cambridge Isotope Laboratories, Inc) and 1 g/L ammonium sulfate (15N2-99%, Cambridge Isotope Laboratories, Inc) as the sole carbon and nitrogen source, respectively. Cell pellets were re-suspended in ~ 150 mL lysis buffer (50 mM sodium phosphate, 300 mM NaCl, pH 8), sonicated on ice for 2 min, and clarified at 10,000 rpm at 4 °C for 30 min. The clarified lysate was mixed with 3 mL Ni–NTA resin by gently vortexing, washed with 100 mL lysis buffer + 20 mM imidazole, and eluted with lysis buffer + 250 mM imidazole. Eluted protein was dialyzed into 50 mM sodium phosphate, 50 mM NaCl at either pH 6.8 or pH 5.5. The peptide Pep5 (Table 2) was synthesized chemically using the solid-phase method (CanPeptide Inc., Pointe-Claire, Quebec, Canada) and purified by use of HPLC. Identities of the purified peptide Pep5 and Ubi-fusion proteins were confirmed by mass spectrometry: Pep5—theoretical FW of 2003.3 versus experimental mass of 2003.2; Ubi-ID—theoretical FW of 17,937.03 versus experimental intact mass of 17,938; Ubi-s-ID—theoretical FW of 16,172.17 versus experimental intact mass of 16,173. The integrity of the bacterially-expressed VHH-IR5 (Text S2) was verified by 1H-15N NMR HSQC and by sequence-specific residue assignments using 3D NMR spectra of 15N/13C-labelled VHH-IR5 protein.
Bottom up HDX-MS
HDX-MS was performed by first mixing 20 μM eIGF1R with 25 μM ligand (IGF-1 or VHH-IR5) in PBS at a 1:1 volume ratio. Labelling was initiated by adding 3 μL of 90% D2O (0.1× PBS, pD 6.9) to 3 μL of an IGF1R-complex at 19 °C. The labelling reaction was quenched by the addition of a 54 μL ice-cold solution of 250 mM TCEP, 2 M Guanidine, 100 mM glycine–HCl at pH 2.0. Quenched and deuterium-labelled samples were immediately injected into a 20 μL sample loop (at 20 pmol eIGF1R/25 pmol ligand). Samples were non-specifically digested online with a Poroszyme Immobilized Pepsin Cartridge (2.1 × 30 mm, Thermo Scientific) at room temperature for 1.5 min at 75 μL/min, and desalted at 400 μL/min with a C18 PepMap100 trap at 1 °C (1 × 5 mm, 5 µm, Thermo Scientific) in mobile phase A (0.23% Formic Acid in water, pH 2.55). Peptides were eluted from a BioBasic-18 (50 × 0.32 mm, 5 µm, Thermo Scientific) column with a separation gradient spanning 10–35% mobile phase B (0.23% Formic Acid in Acetonitrile) over 20 min. Samples were injected into Q-TOF Premier mass spectrometer (Waters) in quadruplicate technical replicates, and deuteration assignment and statistical analysis of individual timepoints (2 × SD, p = 0.02) were achieved using MS Studio45. Unlabelled eIGF1R was injected and analyzed by data-dependent acquisition, and peptides were identified with Mascot. Further experimental details can be found in Table 1. Sequence alignment of the rhesus eIGF1R used for HDX-MS on to the mouse eIGF1R was performed with BLAST46 prior to displaying the HDX-MS data on the 3D structure (PDB: 6PYH) of mouse eIGF1R (Figs. 2 and 3). The sequences of rhesus and human eIGF1R are highly homologous and completely in frame with each other.
NMR spectroscopy
All NMR experiments were carried out at a sample temperature of 298 K on Bruker Avance-III 600 MHz and 800 MHz NMR spectrometers equipped with a 5 mm PATXI sample probe and Z-field gradient accessories. Assignments of the 1H-15 N correlations (i.e. HSQC or HMQC) spectra for Ubi-ID (Table 2) were achieved using 3D HNCA, HN(CO)CA, CBCA(CO)NH, HNCACB, HN(CA)CO, and HNCO spectra47 collected at 600 MHz. The NMR sample was prepared from a 0.5 mM uniformly 15N-/13C-labeled Ubi-ID dissolved in a buffer that was 50 mM in sodium phosphate and 50 mM in NaCl at pH 5.5 followed by the addition of 10% D2O. The SOFAST-HMQC39 pulse sequence with 0.3 s recycle delay was used to record 2D 1H-15N correlation spectra of Ubi-ID and Ubi-s-ID (Table 2) titrated with varying concentrations of the unlabelled sdAb VHH-IR5.
NMR experiments for VHH-IR5 (Table 2) resonance assignments were carried out at 800 MHz with a 0.3 mM uniformly 15N-/13C-labeled VHH-IR5 dissolved in the NMR buffer (see above) at pH 6.8 followed by the addition of 10% D2O. Multi-dimensional heteronuclear NMR data, i.e. HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH, HN(CA)CO, and HNCO were collected using pulse sequences from the Bruker pulse sequence library. HN(CA)CO and HNCO experiments were carried out with a non-uniform (sparse) sampling (NUS) schedule generated using the jittered rejection sampling algorithm48. These NUS schedules were individually optimized for the 13C and 15N dimensions, separately, each using a 50% of sparsity with denser sampling at short evolution times, resulting in, for example, a total of 32 sampled data points from a uniform (Nyquist) grid of 64. Water suppression was achieved using gradient selection for all 3D NMR experiments. The 3D HN(CA)CO and HNCO spectra were processed from time-domain data matrices after filling the NUS data along with extrapolation in each of the indirect dimensions using an in-house implementation of the Convex-Accelerated Maximum Entropy Algorithm (CAMERA)49. The filled 3D NUS-NMR data matrices were processed using the same processing parameters as for complete data matrices, but leaving out linear prediction, by use of NMRPipe50. All other 3D NMR spectra were processed using NMR Pipe with linear prediction for the two evolution dimensions. Analysis of 3D NMR spectra and resonance assignments were achieved using nmrView51.
Fragment scanning of the IGF1R Insertion Domain
A total of 55 overlapping peptides of 15 residues in length were derived from the 674-742 sequence segment of human IGF1R using the PepScan procedure42,52. Binding activities of the surface-immobilized peptides were quantified through ELISA using an anti-His antibody specific to a His-tag present in VHH-IR5 (Table 2). Concentration of VHH-IR5 was 10 μg/mL in all loading solutions.
Supplementary Information
Acknowledgements
We thank Mélanie Duchesne, Brian Cass, Hongtao Qi and Eric Grazzini of the Protein Production Team at the Montreal Royalmount Laboratories, NRC for the production and purification of rhesus eIGF1R. We also thank anonymous reviewers for their insightful critiques, which facilitated greatly the integration of our new experimental findings into the fast-emerging and complex structural biology of the IR/IGF1R protein family. This work was supported by the Targeting the Brain for Better Biotherapeutics (TBBB) Program at the Human Health Therapeutics Research Centre of the National Research Council of Canada (NRCC Publication No. NRC-HHT_53514A).
Abbreviations
- HDX
Hydrogen–deuterium exchange
- HDX-MS
Hydrogen–deuterium exchange mass spectrometry
- NMR
Nuclear magnetic resonance
- HSQC
Heteronuclear single quantum coherence
- HMQC
Heteronuclear multiple quantum coherence
- SOFAST-HMQC
Band-selective optimized flip-angle short transient HMQC
- HNCA, HN(CO)CA, CBCA(CO)NH, HNCACB, HN(CA)CO, and HNCO
A set of heteronuclear 3D NMR experiments for backbone resonance assignments of 15N- and 13C-labelled proteins
- NUS
Non-uniform (sparse) sampling
- SPR
Surface plasmon resonance
- RU
Resonance unit
- PDB
Protein data bank
- SD
Standard deviation
- CDR
Complementarity-determining region
- IGF1R
Type-1 insulin-like growth factor receptor
- eIGF1R
The ectodomain of IGF1R
- IGF-1,2
Type-1/2 insulin-like growth factor (or insulin-like growth factor 1, 2)
- BBB
Blood brain barrier
- sdAb
Single-domain antibody
- α-CT
The C-terminus of the α-subunit of IGF1R
- L1 and L2
Leucine-rich domains 1 or 2
- L1-CR
L1 constant region
- FnIII-1,2,3
Fibronectin type III domains 1,2,3 of IGF1R
- RTK
Receptor tyrosine kinase
- IR
Insulin receptor
- IRR
Insulin receptor–related receptor
- CNS
Central nervous system
- RMT
Receptor mediated transcytosis
- TfR
Transferrin
- LDL
Low-density lipoprotein
- SH2
Src homology 2 domain
- EA
Enzyme acceptor
- ID
Insertion domain
- HEK293
Human embryonic kidney cell line 293
- CHO
Chinese hamster ovary cells
- RT
Room temperature
- PBS
Phosphate Buffered Saline
- SDS-PAGE
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- TCEP
Tris(2-carboxyethyl)phosphine
- EDTA
Ethylenediaminetetraacetic acid
- HPLC
High-performance liquid chromatography
Author contributions
J.S., J.J.H. and J.K. conceived and designed HDX-MS experiments. J.S. performed HDX-MS experiments, interpreted the data and wrote the manuscript. F.N. designed IGF1R fragment constructs, interpreted NMR data and wrote the manuscript. M.A. and L.M. designed specialized plasmids for expressing intact ubiquitin fusion proteins in E. Coli. P.W. produced isotope-labelled proteins, P.X. collected NMR data and both analyzed the NMR spectra. K.K. and G.H. designed SPR experiments and H.v.F. collected and analyzed the data. D.S. designed receptor activation experiments and E.B. performed cell-based assays and analyzed the data. K.K. and D.S. designed IGF1R peptide (PepScan) experiments. J.S. and F.N. contributed equally to manuscript drafting and all authors reviewed and edited the manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-83198-w.
References
- 1.Adams TE, Epa VC, Garrett TP, Ward CW. Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol. Life Sci. 2000;57:1050–1093. doi: 10.1007/PL00000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pollak MN, Schernhammer ES, Hankinson SE. Insulin-like growth factors and neoplasia. Nat. Rev. Cancer. 2004;4:505–518. doi: 10.1038/nrc1387. [DOI] [PubMed] [Google Scholar]
- 3.Chitnis MM, Yuen JS, Protheroe AS, Pollak M, Macaulay VM. The type 1 insulin-like growth factor receptor pathway. Clin. Cancer Res. 2008;14:6364–6370. doi: 10.1158/1078-0432.CCR-07-4879. [DOI] [PubMed] [Google Scholar]
- 4.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: an update. Nat. Rev. Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 5.Stanimirovic DB, Sandhu JK, Costain WJ. Emerging technologies for delivery of biotherapeutics and gene therapy across the blood-brain barrier. BioDrugs. 2018;32:547–559. doi: 10.1007/s40259-018-0309-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abbott NJ, Patabendige AA, Dolman DE, Yusof SR, Begley DJ. Structure and function of the blood–brain barrier. Neurobiol. Dis. 2010;37:13–25. doi: 10.1016/j.nbd.2009.07.030. [DOI] [PubMed] [Google Scholar]
- 7.Deeken JF, Löscher W. The blood–brain barrier and cancer: transporters, treatment, and Trojan horses. Clin. Cancer Res. 2007;13:1663–1674. doi: 10.1158/1078-0432.CCR-06-2854. [DOI] [PubMed] [Google Scholar]
- 8.Pulgar VM. Transcytosis to cross the blood brain barrier, new advancements and challenges. Front. Neurosci. 2018;12:1019. doi: 10.3389/fnins.2018.01019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niewoehner J, et al. Increased brain penetration and potency of a therapeutic antibody using a monovalent molecular shuttle. Neuron. 2014;81:49–60. doi: 10.1016/j.neuron.2013.10.061. [DOI] [PubMed] [Google Scholar]
- 10.Haqqani AS, et al. Intracellular sorting and transcytosis of the rat transferrin receptor antibody OX26 across the blood–brain barrier in vitro is dependent on its binding affinity. J. Neurochem. 2018;146:735–752. doi: 10.1111/jnc.14482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boado RJ, Lu JZ, Hui EK, Pardridge WM. Reduction in brain heparan sulfate with systemic administration of an IgG Trojan horse-sulfamidase fusion protein in the mucopolysaccharidosis type IIIA mouse. Mol. Pharm. 2018;15:602–608. doi: 10.1021/acs.molpharmaceut.7b00958. [DOI] [PubMed] [Google Scholar]
- 12.Wagner S, et al. Uptake mechanism of ApoE-modified nanoparticles on brain capillary endothelial cells as a blood–brain barrier model. PLoS ONE. 2012;7:e32568. doi: 10.1371/journal.pone.0032568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Sullivan CC, et al. ANG1005 for breast cancer brain metastases: correlation between. Breast Cancer Res. Treat. 2016;160:51–59. doi: 10.1007/s10549-016-3972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giugliani R, et al. Neurocognitive and somatic stabilization in pediatric patients with severe mucopolysaccharidosis Type I after 52 weeks of intravenous brain-penetrating insulin receptor antibody-iduronidase fusion protein (valanafusp alpha): an open label phase 1–2 trial. Orphanet. J. Rare Dis. 2018;13:110. doi: 10.1186/s13023-018-0849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boado RJ, Lu JZ, Hui EK, Pardridge WM. Insulin receptor antibody-sulfamidase fusion protein penetrates the primate blood–brain barrier and reduces glycosoaminoglycans in Sanfilippo type A cells. Mol. Pharm. 2014;11:2928–2934. doi: 10.1021/mp500258p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aday S, Cecchelli R, Hallier-Vanuxeem D, Dehouck MP, Ferreira L. Stem cell-based human blood–brain barrier models for drug discovery and delivery. Trends Biotechnol. 2016;34:382–393. doi: 10.1016/j.tibtech.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Couch JA, et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood–brain barrier. Sci. Transl. Med. 2013;5:1–12. doi: 10.1126/scitranslmed.3005338. [DOI] [PubMed] [Google Scholar]
- 18.Ohshima-Hosoyama S, et al. A monoclonal antibody-GDNF fusion protein is not neuroprotective and is associated with proliferative pancreatic lesions in parkinsonian monkeys. PLoS ONE. 2012;7:e39036. doi: 10.1371/journal.pone.0039036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bondy C, Werner H, Roberts CT, LeRoith D. Cellular pattern of type-I insulin-like growth factor receptor gene expression during maturation of the rat brain: comparison with insulin-like growth factors I and II. Neuroscience. 1992;46:909–923. doi: 10.1016/0306-4522(92)90193-6. [DOI] [PubMed] [Google Scholar]
- 20.Werner H, LeRoith D. Insulin and insulin-like growth factor receptors in the brain: physiological and pathological aspects. Eur. Neuropsychopharmacol. 2014;24:1947–1953. doi: 10.1016/j.euroneuro.2014.01.020. [DOI] [PubMed] [Google Scholar]
- 21.Ribecco-Lutkiewicz M, et al. A novel human induced pluripotent stem cell blood–brain barrier model: applicability to study antibody-triggered receptor-mediated transcytosis. Sci. Rep. 2018;8:1873. doi: 10.1038/s41598-018-19522-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kavran, J.M. et al. How IGF-1 activates its receptor. Elife3 (2014). [DOI] [PMC free article] [PubMed]
- 23.Li J, Choi E, Yu H, Bai XC. Structural basis of the activation of type 1 insulin-like growth factor receptor. Nat. Commun. 2019;10:4567. doi: 10.1038/s41467-019-12564-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Visualization of ligand-bound ectodomain assembly in the full-length human IGF-1 receptor by Cryo-em single-particle analysis. Structure. 2020;28:555–561. doi: 10.1016/j.str.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Surinya KH, et al. An investigation of the ligand binding properties and negative cooperativity of soluble insulin-like growth factor receptors. J. Biol. Chem. 2008;283:5355–5363. doi: 10.1074/jbc.M707054200. [DOI] [PubMed] [Google Scholar]
- 26.Christoffersen CT, et al. Negative cooperativity in the insulin-like growth factor-I receptor and a chimeric IGF-I/insulin receptor. Endocrinology. 1994;135:472–475. doi: 10.1210/endo.135.1.8013387. [DOI] [PubMed] [Google Scholar]
- 27.Stanimirovic, D., Kemmerich, K., Haqqani, A., Sulea, T., Arbabi-Ghahroudi, M., Massie, B., and Gilbert, R. Insulin-like growth factor 1 receptor -specific antibodies and uses thereof (United States, 2017, US20170015748A1).
- 28.Calzone FJ, et al. Epitope-specific mechanisms of IGF1R inhibition by ganitumab. PLoS ONE. 2013;8:e55135. doi: 10.1371/journal.pone.0055135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doern A, et al. Characterization of inhibitory anti-insulin-like growth factor receptor antibodies with different epitope specificity and ligand-blocking properties: implications for mechanism of action in vivo. J. Biol. Chem. 2009;284:10254–10267. doi: 10.1074/jbc.M809709200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soos MA, et al. A panel of monoclonal antibodies for the type I insulin-like growth factor receptor. Epitope mapping, effects on ligand binding, and biological activity. J. Biol. Chem. 1992;267:12955–12963. doi: 10.1016/S0021-9258(18)42367-8. [DOI] [PubMed] [Google Scholar]
- 31.Keyhanfar M, Booker GW, Whittaker J, Wallace JC, Forbes BE. Precise mapping of an IGF-I-binding site on the IGF-1R. Biochem. J. 2007;401:269–277. doi: 10.1042/BJ20060890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly GM, Buckley DA, Kiely PA, Adams DR, O'Connor R. Serine phosphorylation of the insulin-like growth factor I (IGF-1) receptor C-terminal tail restrains kinase activity and cell growth. J. Biol. Chem. 2012;287:28180–28194. doi: 10.1074/jbc.M112.385757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown KA, Wilson DJ. Bottom-up hydrogen deuterium exchange mass spectrometry: data analysis and interpretation. Analyst. 2017;142:2874–2886. doi: 10.1039/C7AN00662D. [DOI] [PubMed] [Google Scholar]
- 34.Zhu S, et al. Hydrogen-deuterium exchange epitope mapping reveals distinct neutralizing mechanisms for two monoclonal antibodies against diphtheria toxin. Biochemistry. 2019;58:646–656. doi: 10.1021/acs.biochem.8b01123. [DOI] [PubMed] [Google Scholar]
- 35.Puchades C, et al. Epitope mapping of diverse influenza Hemagglutinin drug candidates using HDX-MS. Sci Rep. 2019;9:4735. doi: 10.1038/s41598-019-41179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qiang G, et al. Identification of a small molecular insulin receptor agonist with potent antidiabetes activity. Diabetes. 2014;63:1394–1409. doi: 10.2337/db13-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Houde D, Demarest SJ. Fine details of IGF-1R activation, inhibition, and asymmetry determined by associated hydrogen/deuterium-exchange and peptide mass mapping. Structure. 2011;19:890–900. doi: 10.1016/j.str.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Xu Y, et al. How ligand binds to the type 1 insulin-like growth factor receptor. Nat. Commun. 2018;9:821. doi: 10.1038/s41467-018-03219-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schanda P, Kupce E, Brutscher B. SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 40.Menting JG, et al. Structural congruency of ligand binding to the insulin and insulin/type 1 insulin-like growth factor hybrid receptors. Structure. 2015;23:1271–1282. doi: 10.1016/j.str.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 41.Ni F. Recent developments in transferred NOE methods. Prog. Nucl. Magn. Reson. Spectrosc. 1994;26:517–606. doi: 10.1016/0079-6565(94)90000-0. [DOI] [Google Scholar]
- 42.Nilvebrant J, Rockberg J. An introduction to epitope mapping. Methods Mol. Biol. 2018;1785:1–10. doi: 10.1007/978-1-4939-7841-0_1. [DOI] [PubMed] [Google Scholar]
- 43.Whittaker J, et al. Alanine scanning mutagenesis of a type 1 insulin-like growth factor receptor ligand binding site. J. Biol. Chem. 2001;276:43980–43986. doi: 10.1074/jbc.M102863200. [DOI] [PubMed] [Google Scholar]
- 44.Slysz GW, Percy AJ, Schriemer DC. Restraining expansion of the peak envelope in H/D exchange-MS and its application in detecting perturbations of protein structure/dynamics. Anal. Chem. 2008;80:7004–7011. doi: 10.1021/ac800897q. [DOI] [PubMed] [Google Scholar]
- 45.Rey M, et al. Mass spec studio for integrative structural biology. Structure. 2014;22:1538–1548. doi: 10.1016/j.str.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, et al. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 47.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Cambridge: Academic Press; 1996. [Google Scholar]
- 48.Worley B. Subrandom methods for multidimensional nonuniform sampling. J. Magn. Reson. 2016;269:128–137. doi: 10.1016/j.jmr.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worley B. Convex accelerated maximum entropy reconstruction. J. Magn. Reson. 2016;265:90–98. doi: 10.1016/j.jmr.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 52.Geysen HM, Rodda SJ, Mason TJ, Tribbick G, Schoofs PG. Strategies for epitope analysis using peptide synthesis. J. Immunol. Methods. 1987;102:259–274. doi: 10.1016/0022-1759(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 53.Masson GR, et al. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat. Methods. 2019;16:595–602. doi: 10.1038/s41592-019-0459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.