Abstract
Disaccharide phosphorylases (DSPs) are carbohydrate-active enzymes with outstanding potential for the biocatalytic conversion of common table sugar into products with attractive properties. They are modular enzymes that form active homo-oligomers. From a mechanistic as well as a structural point of view, they are similar to glycoside hydrolases or glycosyltransferases. As the majority of DSPs show strict stereo- and regiospecificities, these enzymes were used to synthesize specific disaccharides. Currently, protein engineering of DSPs is pursued in different laboratories to broaden the donor and acceptor substrate specificities or improve the industrial particularity of naturally existing enzymes, to eventually generate a toolbox of new catalysts for glycoside synthesis. Herein we review the characteristics and classifications of reported DSPs and the glycoside products that they have been used to synthesize.
Keywords: Disaccharide phosphorylase, Classification, Structure and domains, Substrate specificity, Enzyme engineering
1. Introduction
Carbohydrates are known to play essential roles in a myriad of biological processes [1,2]. The functions depend on their constituent carbohydrates and linkage types [3,4]. The carbohydrate-active enzymes (CAZy) involved in the cleavage and formation of glycosidic linkages were classified in the CAZy database [[5], [6], [7]] and mainly categorized into three classes: glycoside hydrolases (GHs), glycosyltransferases (GTs), and glycoside phosphorylases (GPs) [8]. GHs are the most commonly used in the industry by far, being used to hydrolyze polysaccharides like starch and cellulose [9]. Besides, GHs can be used in the transglycosylation mode. Mutations in the active site of Agrobacterium sp. β-Glucosidase (AsβG) have allowed the efficient synthesis of oligosaccharides [10], and the AsβG mutant uses activated sugars (such as fluoro or p-nitrophenyl glycosides) to increase the reaction rate and product yield [11]. GTs have also been widely studied for the biological implication that they play an important role in the synthesis of carbohydrate chains in vivo [12]. Although utilization of GTs is currently limited by several problems such as instability, insolubility, and high costs of the activated donor substrates, no doubt overcoming these problems will represent major contributions to the expanding field of glycobiology and industrial application as a strong tool for the synthesis of oligosaccharides and glycoconjugates [13]. GPs (E.C. 2.4.1.-, usually named using a combination of “the name of the substrate” and “phosphorylase”) are a group of enzymes catalyzing reversible phosphorolysis of glycans into the corresponding sugar 1-phosphates and shortened glycan chains [14,15]. The reversibility of the reaction also enables the production of lengthened glycans from the sugar 1-phosphate donors and glycan acceptors of choice. Sugar 1-phosphate donors for GPs are relatively cheap and accessible compared with the nucleotide sugars required for GTs, therefore making GPs attractive as biocatalysts for oligosaccharides production [16]. The known GPs utilize disaccharides, oligosaccharides (maltodextrins, cellodextrins) or polysaccharides (starch, glycogen) as donor substrates. Here, we restrict our discussion mainly to disaccharide phosphorylases (DSPs) for two reasons [17]. First, the DSPs embrace enough structural data and essentially clear and thorough mechanistic explanations [18]. Second, applications of phosphorylase enzymes in glycoside synthesis were developed chiefly using DSPs, as disaccharides are the smallest type of oligosaccharides, and a large number of these compounds, have been reported to exhibit physiological activity [19]. These enzymes are a central theme of this review that is focused on their catalytic properties, structure-function relationships, considerations of enzymatic synthesis, and enzyme engineering.
2. Overview of disaccharide phosphorylases
The characterized DSPs comprise enzymes that can act on a wide range of glycosidic linkages (Table 1), with the exception for α-(1 → 6), β-(1 → 6) and β-(1 → 1) linkages, for which no specific DSPs have been characterized at the moment [20]. Most of the DSPs known so far use d-Glucose 1-phosphate (D-Glc 1-phosphate), including α- and β-form as a donor substrate, forming disaccharides; only a minority of DSPs use d-acetylglucosamine 1-phosphate (D-GlcNAc 1-phosphate), d-Galactose-1-phosphate (D-Gal-1-phosphate), or d-Mannose-1-phosphate (D-Man-1-phosphate) as their donor substrates (Table 1). Structural and mechanistic studies have led to the classification of known phosphorylases into the main group of glycoside hydrolase (GH)-like enzymes and a smaller group of enzymes related to glycosyltransferases (GT). According to whether retain or invert the anomeric configuration of the disaccharides substrate in the sugar 1-phosphate product, DSPs belonging to either GHs or GTs are further categorized into retaining or inverting classes [21]. Specifically, inverting DSPs with published crystal structures are found in GH families GH-65, GH-94, GH-112, and GH-130 [22]. Anomeric configuration of the glycosyl moiety of the donor is inverted, hence it must be attacked directly by inorganic phosphate from the opposite side relative to the glycosidic bond present in the substrate [23]. However, retaining DSPs are found in family GH-13 and GT-4 without changing the anomeric configuration between the substrate and the phosphorylated product [24].
Table 1.
Classification of disaccharide phosphorylases.
| Mechanism | Family | Linkage | Donor | Acceptor | Product |
|---|---|---|---|---|---|
| Retaining (IR) | GT4 | α,α-(1 → 1) | α-D-Glc-1-P | D-Glc | Trehalose |
| Retaining (DD) | GH13 | α1→β2 | α-D-Glc-1-P | D-Fru | Sucrose |
| α-(1 → 2) | α-D-Glc-1-P | Glycerate | Glucosylglycerate | ||
| α-(1 → 2) | α-D-Glc-1-P | Glycerol | Glucosylglycerol | ||
| Inverting (SD) | GH65 | α,α-(1 → 1) | β-D-Glc-1-P | D-Glc/D-Glc-6-P | Trehalose/trehalose-6-P |
| α-(1 → 2) | β-D-Glc-1-P | D-Glc | Kojibiose | ||
| α-(1 → 3) | β-D-Glc-1-P | D-Glc | Nigerose | ||
| α-(1 → 4) | β-D-Glc-1-P | D-Glc | Maltose | ||
| α-(1 → 2) | β-D-Glc-1-P | Glycerol | Glucosylglycerol | ||
| α-(1 → 3) | β-D-Glc-1-P | L-Rha | α-D-Glc-(1 → 3)-L-Rha | ||
| GH94 | β-(1 → 2) | α-D-Glc-1-P | D-Glc | Sophorose | |
| β-(1 → 3) | α-D-Glc-1-P | D-Glc | Laminaribiose | ||
| β-(1 → 4) | α-D-Glc-1-P | D-Glc | Cellobiose | ||
| β-(1 → 4) | α-D-GlcNAc-1-P | D-GlcNAc | Chitobiose | ||
| β-(1 → 4) | α-D-Glc-1-P | d-Gluconic acid | Cellobionic acid | ||
| GH112 | β-(1 → 3) | α-D-Gal-1-P | D-GalNAc/D-GlcNAc | Galacto-N-biose/lacto-N-biose | |
| β-(1 → 4) | α-D-Gal-1-P | L-Rha | β-D-Gal-(1 → 4)-L-Rha | ||
| GH130 | β-(1 → 4) | α-D-Man-1-P | D-Glc | β-D-Man-(1 → 4)-D-Glc | |
| β-(1 → 4) | α-D-Man-1-P | D-GlcNAc | β-D-Man-(1 → 4)-D-GlcNAc | ||
| β-(1 → 2) | α-D-Man-1-P | D-Man | Mannobiose |
DD = double displacement, SD = single displacement, IR = internal return.
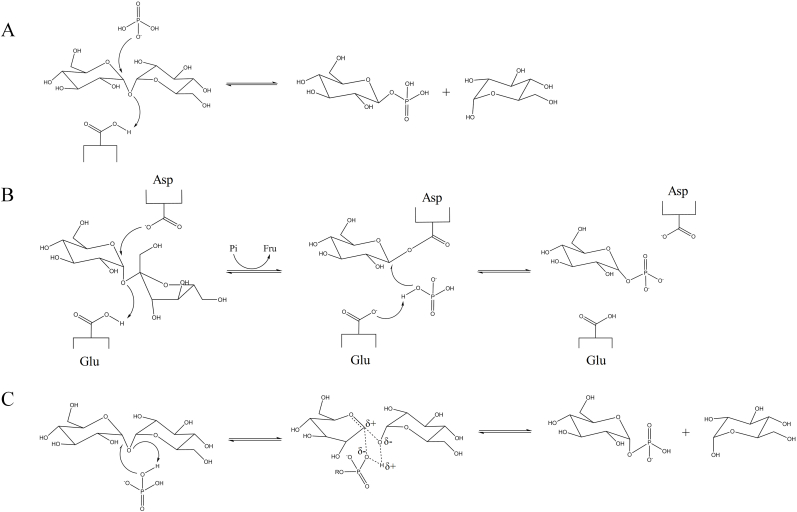
Although various DSPs reported, their reaction mechanisms can be boiled down to three types as Fig. 1 shows [25]. The majority of DSPs are inverting enzymes that follow a single displacement mechanism (Fig. 1A), catalyzing a direct nucleophilic attack of phosphate on the anomeric carbon. A single catalytic carboxylic residue (the catalytic acid) activates the breakdown of glycosidic bond by donating a proton [26]. In contrast, retaining DSPs, follow two different reaction mechanisms. Sucrose phosphorylases (SP), on the one hand, utilize a GH-like double displacement mechanism [27,28], illustrated in Fig. 1B. This mechanism involves one of the carboxylic residues (the catalytic nucleophile) attacks the anomeric carbon, resulting in a covalent glycosyl–enzyme intermediate that can be hydrolyzed in the next step [29,30]. The other residue (the catalytic acid/base) protonates the leaving group in the first step and subsequently deprotonates water in the second step. The double inversion of the anomeric configuration results in net retention [31,32]. The retaining trehalose phosphorylases (THP), on the other hand, are believed to follow a direct front-side nucleophilic displacement, often referred to a so-called internal return-like mechanism [33]. In this case, the glycosidic oxygen is protonated by a phosphate hydroxyl group, and the glycosidic bond is destabilized by a nucleophilic attack at C-1 atom with the oxygen atom donating a proton to the glycosidic oxygen [34]. As Fig. 1C shows, a ternary complex is formed in which the phosphate molecule deprotonates the leaving group while simultaneously attacking the anomeric center from the front side. It is important that both the proton donation and the nucleophilic attack at the C-1 atom take place from the same side, which leads to the retention of the anomeric configuration. The anticipated transition state is stabilized by a conserved amino acid, which is glutamine or asparagine [35].
Fig. 1.
The three reaction mechanisms employed by DSPs. (A) The single displacement mechanism of the inverting trehalose phosphorylase (EC 2.4.1.64), (B) the double displacement mechanism of the retaining sucrose phosphorylase (EC 2.4.1.7), and (C) the internal displacement mechanism of the retaining trehalose phosphorylase (EC 2.4.1.231).
3. Enzyme structure and recognition of the substrate
3.1. Architecture of disaccharide phosphorylases
Except for family GT4 where no structure is reported, the 3-dimensional structure has been solved for at least one phosphorylase of each family encompassing DSPs. The list of 3-D structures of DSPs is given in Table 2. There are marked contrasts between the diversity of 3-D folds observed for DSPs, and the functional assignment of catalytic amino acid residues furthermore proposed a structural classification into ‘clans’. Two major classes GH65 together with GH94 phosphorylases belong to clan GH-L, which have a common (α/α)6 protein fold. In addition, SPs belonging to GH13 are classified into clan GH-H sharing a common (β/α)8 barrel fold. However, since catalytic domains of GH112 galacto-N-biose/lacto-N-biose I phosphorylases (GLNBP) consist of a partially broken (β/α)8 barrel fold, they were classified into clan GH-A instead of GH-H. Moreover, 4-O-β-d-mannosyl-d-glucose phosphorylases (MGP) belonging to GH130 adopt the predominant structural fold 5-Blade β-propeller. Assignments of these DSPs into GH support evolutionary relationships between these enzyme classes in terms of both structure and catalytic function. Remarkably, (retaining) THPs belonging to GT4 adopt the predominant structural fold GT-B, composed of two distinct N-terminal and C-terminal Rossmann-like domains of six or seven parallel β-sheets linked to a-helices, connected by a linker region and an interdomain cleft.
Table 2.
Three-dimensional structures of disaccharide phosphorylases.
| Family | Fold | Clan | EC No. | Enzyme | Organism | PDB code | Ligand | Ref. |
|---|---|---|---|---|---|---|---|---|
| GT4 | GT-B | 2.4.1.231 | α,α-Trehalose phosphorylase | |||||
| GH13 | (β/α)8 | GH-H | 2.4.1.7 | Sucrose phosphorylase | Bifidobacterium adolescentis | 5M9X, 5MAN, 2GDV | Glycosylated resveratrol, sucrose, nigerose | [36] |
| 2.4.1.352 | Glucosylglycerate phosphorylase | |||||||
| 2.4.1.359 | 1,2-α-glucosylglycerol phosphorylase | |||||||
| GH65 | (α/α)6 | GH-L | 2.4.1.64 | α,α-trehalose phosphorylase | ||||
| 2.4.1.216 | Trehalose 6-phosphate phosphorylase | |||||||
| 2.4.1.230 | Kojibiose phosphorylase | Caldicellulosiruptor saccharolyticus | 3WIQ, 3WIR | Kojibiose, Glc | [37] | |||
| 2.4.1.279 | Nigerose phosphorylase | |||||||
| 2.4.1.8 | Maltose phosphorylase | Lactobacillus brevis | 1H54 | [38] | ||||
| 2.4.1.332 | Glucosylglycerol phosphorylase | Bacillus selenitireducens | 4KTP, 4KTR | Glc, isofagomine and glycerol | [39] | |||
| 2.4.1.282 | 3-O-α-d-glucosyl-l-rhamnose phosphorylase | |||||||
| GH94 | (α/α)6 | GH-L | 2.4.1.31 | Laminaribiose phosphorylase | Paenibacillus sp. | 6GH2, 6GH3,6GGY | G1P, Man1P, sulfate | [40] |
| 2.4.1.20 | Cellobiose phosphorylase | Cellulomonas uda | 3S4A, 3S4B, 3RSY | Cellobiose, Glc, sulfate and glycerol | [35,41,42] | |||
| Clostridium thermocellum | 3QDE | Phosphate | ||||||
| Cellvibrio gilvus | 3QFY, 2CQS, 3QFZ, 2CQT, 3QG0 | Sulfate, phosphate, isofagomine, 1-deoxynojirimycin | ||||||
| 2.4.1.280 | N,N′-diacetylchitobiose phosphorylase | Vibrio proteolyticus | 1V7W | GlcNAc | [43] | |||
| 2.4.1.321 | Cellobionic acid phosphorylase | Saccharophagus degradans | 4ZLF | Cellobionic acid | [44] | |||
| GH112 | (β/α)8 | GH-A | 2.4.1.211 | 1,3-β-Galactosyl-N-acetylhexosamine phosphorylase | Bifidobacterium longum | 2ZUT | GalNAc | [45,46] |
| 2.4.1.247 | β-d-galactosyl-(1 → 4)-l-rhamnose phosphorylase | |||||||
| GH130 | 5-Blade β-propeller | 2.4.1.281 | 4-O-β-d-mannosyl-d-glucose phosphorylase | Bacteroides fragilis | 3WAS | Man-Glc + PO4 | [47,48] | |
| Ruminococcus albus | 5AYC | Man-Glc + SO4 | ||||||
| 2.4.1.320 | 1,4-β-Mannosyl-N-acetylglucosamine phosphorylase | |||||||
| 2.4.1.339 | β-1,2-Mannobiose phosphorylase |
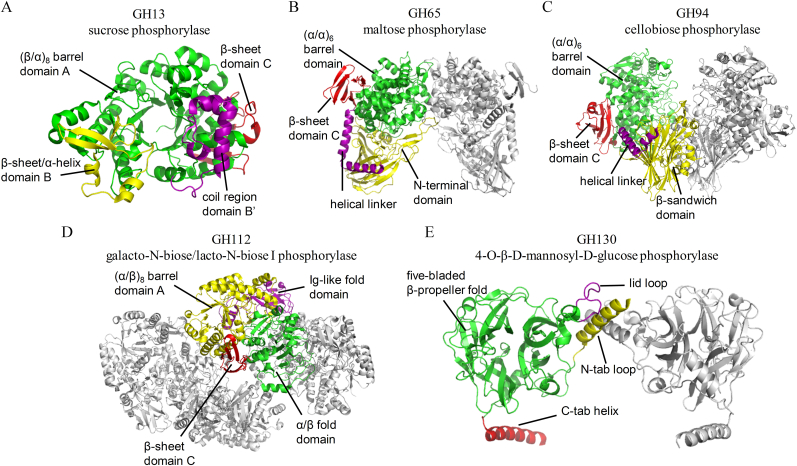
Consistent with their natural state in solution, most DSPs crystallize as dimers, although GH112 and GH130 members form higher homooligomeric complexes. All DSPs are composed of four domains that through direct and/or indirect contacts with the catalytic domain and other monomer tune enzyme donor and acceptor specificity and stabilize the oligomer architecture. The domain organization of the DSPs is summarized in Fig. 2. Specifically, Bifidobacterium adolescentis sucrose phosphorylase (BaSP) [36,49], belonging to GH13 family, consists of four domains. Among them, catalytic domain A ((β/α)8-barrel, green) is interrupted by two long loops displaying structural elements and therefore classified as domains B (composed of two β-sheets and two α-helices, yellow) and B’ (composed of a coil containing two α-helices, purple). C-Terminal domain C (red) forms a five-stranded antiparallel β-sheet (Fig. 2A). The majority of the dimer interactions are confined to the two domains B (yellow) although some interactions between the catalytic domains (green) were also observed [38]. For example, Lactobacillus brevis maltose phosphorylase (LbMP), belonging to GH65 family, includes the two monomers contacting each other essentially through loops close to the active site entrance. The four structural domains of the monomers are represented in different colors (Fig. 2B). The domain organization of LbMP is similar to other GH65 disaccharide phosphorylases, which include kojibiose phosphorylase from Caldicellulosiruptor saccharolyticus (CsKP) [37] and glucosylglycerol phosphorylase from Bacillus selenitireducens (BsGGP) [39]. Moreover, Paenibacillus sp. laminaribiose phosphorylase, belonging to GH94 family, contains two subunits per asymmetric unit, which are related by a non-crystallographic twofold axis [40]. Each PsLBP monomer consists of four domains as Fig. 2C shown. The domain organization in PsLBP is similar to that observed in other GH94 disaccharide phosphorylases, which include CBPs from Cellulomonas uda (CuCBP) [42], Cellvibrio gilvus (CgCBP) [35], Clostridium thermocellum (CtCBP) [41], ChBP from Vibrio proteolyticus (VpChBP) [43], and cellobionic acid phosphorylase from Saccharophagus degradans (SdCBAP) [44]. However, also have to point out that Bifidobacterium longum 1,3-β-galactosyl-N-acetylhexosamine phosphorylase (BlGLNBP) shows a ribbon diagram of the tetramer structure of ligand-free form [45]. The GLNBP monomer consists of four domains just like other DSPs (Fig. 2D). Remarkably, Bacteroides fragilis 4-O-β-d-mannosyl-d-glucose phosphorylase (BfMGP) appeared to form a homohexamer with 3222 (D3) symmetry, which consisted of three dimers related by the crystallographic 3-fold axis [48] (Fig. 2E).
Fig. 2.
Structures of disaccharide phosphorylases. A) GH13 sucrose phosphorylase from Bifidobacterium adolescentis (PDB ID: 5M9X), B) GH65 maltose phosphorylase from Lactobacillus brevis (PDB ID: 1H54), C) GH94 cellobiose phosphorylase from Cellvibrio gilvus (PDB ID: 2CQT), D) GH112 galacto-N-biose/lacto-N-biose I phosphorylase from Bifidobacterium longum (PDB ID: 2ZUT) and E) GH130 4-O-β-d-mannosyl-d-glucose phosphorylase from Bacteroides fragilis (PDB ID: 3WAS) are shown.
3.2. Substrate specificity of disaccharide phosphorylases
All of the DSPs except for SPs, which have been found in nature, show strict stereo- and regiospecificities, for phosphorolysis they prefer a single disaccharide in the forward direction, and in reverse reactions, the high specificity of the enzymes is reflected in a preference for both the donor and acceptor, in addition to the regioselectivity of the synthesized glycosidic bond [50,51]. The dual donor and acceptor specificity is given by the structure of the enzyme active site that recognizes the substrates via not only stacking interactions but also through a network of hydrogen bonds and van der Waals interactions [[52], [53], [54]]. A modification of any sugar hydroxyl group is allowed only at those positions which are not in direct or indirect contact with the enzyme. Otherwise, such a change of the hydroxyl group won't work unless the alteration is compensated by a new interaction of the ligand with the enzyme, which is a complicated and unclear condition in enzyme engineering [51]. It is reported that at least three hydroxyl groups of sugar residue bound are in contact with amino acids at the active site [35,38,41,43,44]. However, for SPs, the subsite −1 in the active pocket is strictly specific for glucosylated donors, while the subsite +1 shows relaxed specificity for acceptors. The structural microenvironment of subsite +1 can undergo drastic rearrangements during the catalytic process because of its unique double displacement mechanism. This conformational flexibility is probably responsible for the activity promiscuity of SPs [58].
4. Engineering of disaccharide phosphorylases and related enzymes
4.1. Thermostability
Carbohydrate conversions in the industry are preferably operated at elevated temperatures to prevent microbial contamination and to avoid excessive viscosity. Directed evolution has been performed extensively on DSPs to improve the industrial property. For example, SPs are promising biocatalysts for the production of a wide range of compounds, but their industrial applications have been hampered by the low thermostability of known representatives. The most thermostable SPs known to date is BaSP, with an optimal temperature of 58 °C [55,56]. Unfortunately, it quickly loses activity at the industrially relevant temperature of 60 °C. A combination of sequence-based and structure-based mutagenesis was applied to BaSP in pursuit of a variant with higher kinetic stability [57]. Based on B-factor calculation, three aspartate residues at positions 445–447 were chosen as the most flexible region of the entire protein. The enzyme's residual activity could be increased substantially by simultaneously random mutation. The structure-based rational design was then developed to introduce additional salt bridges and alleviate a potential electrostatic repulsion at the dimer interface. Combining all mutations, the half-life time of BaSP at 60 °C was increased dramatically from 24 h to 62 h [58]. Moreover, for CtCBP, site-directed mutagenesis based on structure-guided homology analysis and random mutagenesis was applied to improve thermal stability and temperature optimum of the enzyme. By comparison of the protein sequences and structures of CBP homologs, key amino acid residues responsible for enhanced stability were identified, donating a few variants accurately. Large libraries of random mutants at different mutagenesis frequencies were then constructed to further improve the thermostability of CtCBP. Eventually, the best mutant (CM3) with the halftime of inactivation at 70 °C extended from 8.3 to 24.6 min was achieved with optimal temperature increasing from 60 to 80 °C [59]. Also, GLNBP is the key enzyme in the enzymatic production of lacto-N-biose I [60], which is supposed to represent the Bifidus factor in human milk oligosaccharides [61]. For industrial use, the thermostability of BlGLNBP was improved by directed evolution in which five substitutions in the amino acid sequence were selected from a random mutagenesis library. The best mutant exhibited 20 °C higher thermostability than the wild type [46].
4.2. Alteration of substrate specificity
Any change of substrate specificity of DSPs is an extremely difficult task since these enzymes exhibit very high selectivity at both donor and acceptor site(s) [62]. A possible solution is to optimize the enzymes by protein engineering and, more specifically, via directed evolution [63,64]. As numerous X-ray crystal structures are available for DSPs, either in the presence of phosphate or sulfate, disaccharides, sugar 1-phosphate donors, and different sugar acceptors. These structures provide valuable resources about the active sites and catalytic mechanisms that can be used to guide the engineering of DSPs for noncognate and unnatural substrates [9]. There have been several trials with a certain degree of success in an alteration of donor or acceptor selectivity on CBPs [65], SPs, MPs [66] and THPs [53]. For example, structure-guided site-directed mutagenesis has been performed extensively on CuCBP, and T508I/N667A mutant was achieved with 7.5 times higher specific activity on lactose than the wild-type, although cellobiose phosphorolytic activity of the mutant was still predominant [67]. Besides, a single mutation (E649C) in CuCBP created an enzyme variant capable of using methyl β-glucoside, ethyl β-glucoside, and phenyl β-glucoside as acceptors [68]. Moreover, combining another mutation at this position with the other four mutations (T508I/N667A/N156D/N163D) further broadened the acceptor specificity of the mutant which was able to catalyze transglycosylation to methyl α-d-glucopyranoside [69]. Moreover, several mutations of Ruminococcus albus cellobiose phosphorylase (RaCBP) have been designed to modify the interactions of the +1 subsite with 2-hydroxyl group of the acceptor. As a result, the C485A, Y648F, and Y648V mutations significantly influenced the mutant specificity in terms of the glucosyl transfer to 2-deoxyglucose, mannose, and GlcNAc [70].
Despite their great potential for the synthesis of high added value sugars, developing economical production processes with SPs remains challenging [32]. The BaSP wild-type enzyme preferentially forms maltose and generates the rare disaccharide kojibiose as a side product. Accordingly, through mutation, several hits were achieved that notably improved the selectivity and activity towards kojibiose synthesis, unveiling that the double mutant L341I-Q345S exhibits a selectivity of 95% with only a modest loss of activity [63]. Besides, the +1 subsite of SPs was also modified in an attempt to promote activity on bulky aromatics like catechin, epicatechin and resveratrol, and Gln345 was mutated to Phe (Q345F) of BaSP to establish π-π-stacking-mediated coordination of the acceptor. The domain shift brings about an enlarged and multifunctional active site for polyphenol glucosylation [36]. Meanwhile, for another SP from Thermoanaerobacterium thermosaccharolyticum (TtSP), an active site loop was predicted to hinder the binding of bulky aromatics, and Arg134 was mutated to Ala (R134A) to unbolt the loop of active sites [71]. In a different study, the Q345F variant was further engineered to improve the activity and selectivity of SP towards nigerose formation without the need for a cosolvent. A mutant with four hits (R135Y-D342G-Y344Q-Q345F) was designed that forms nigerose with greater selectivity and a 68-fold improved catalytic efficiency in aqueous solution [72].
Lactobacillus acidophilus maltose phosphorylase (LaMP) catalyzes both phosphorolysis of maltose and formation of maltose by reverse phosphorolysis with β-glucose 1-phosphate and glucose as donor and acceptor, respectively. Substitution of LaMP His413–Glu421, His413–Ile418 and His413–Glu415 from loop 3, by corresponding segments from Ser426–Ala431 in THP and Thr419–Phe427 in KP from Thermoanaerobacter brockii ATCC35047, thus conferred LaMP with phosphorolytic activity towards trehalose and kojibiose, respectively [73].
Thermoanaerobacter brockii trehalose phosphorylase (TbTHP) catalyzes the reversible phosphorolysis of trehalose to glucose-1-phosphate and glucose. Through semirational and random mutagenesis, enzyme variant R448S is achieved as a new biocatalyst with improved affinity for galactose as an acceptor for the industrial production of lactotrehalose (α-d-glucopyranosyl-(1,1)-α-D-gal-actopyranoside) [74]. Moreover, wild-type inverting trehalose phosphorylase from Caldanaerobacter subterraneus (CsTHP) exhibiting activity on galactose as acceptor was used in the first reaction for linking glucose to galactose by α-1,1-α-bond and an optimized CsTHP variant (L649G/A693Q/W371Y) has been created for the production of β-Gal-1P from lactotrehalose and inorganic phosphate through iterative saturation mutagenesis (ISM) [75].
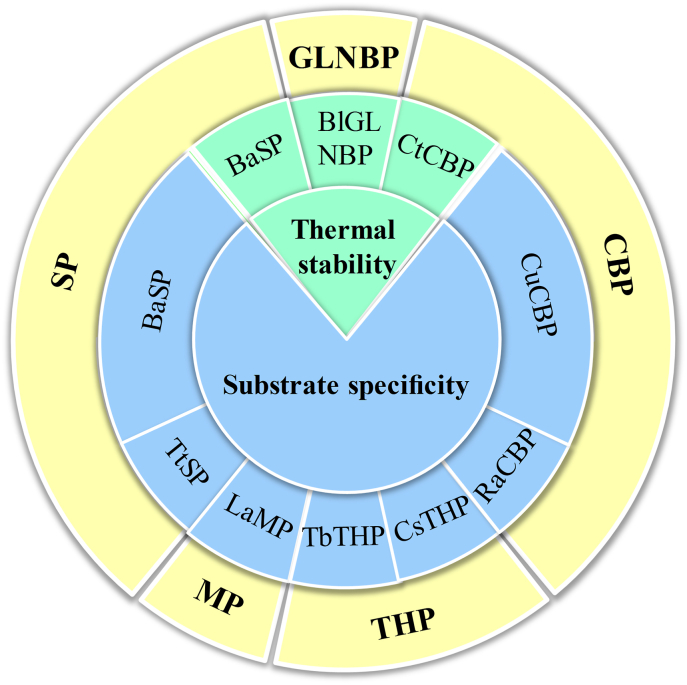
These findings suggest the potential for improving thermal stability and changing the substrate specificity of DSPs using gene mutagenesis. In summary, relevant pieces of literature are sort out, showing the distribution of the effort on the enzyme engineering of DSPs (Fig. 3). For DSP engineering, the amount of attempts to alter substrate specificity on DSP are more than those to improve thermal stability, even though altering substrate specificity is more difficult and challenging. The most studied DSPs in the aspect of enzyme engineering are SPs and CBPs, probably relating to their extensive application in producing value-added products.
Fig. 3.
The distribution of efforts made on DSPs engineering. SP, sucrose phosphorylase; CBP, cellobiose phosphorylase; GLNBP, galacto-N-biose/lacto-N-biose I phosphorylase; MP, maltose phosphorylase; THP, trehalose phosphorylase; BaSP, Bifidobacterium adolescentis sucrose phosphorylase; TtSP, Thermoanaerobacterium thermosaccharolyticum sucrose phosphorylase; BlGLNBP, Bifidobacterium longum galacto-N-biose/lacto-N-biose I phosphorylase; CtCBP, cellobiose phosphorylase; CuCBP, Cellulomonas uda cellobiose phosphorylase; RaCBP, Ruminococcus albus cellobiose phosphorylase; CsTHP, Caldanaerobacter subterraneus trehalose phosphorylase; TbTHP, Thermoanaerobacter brockii trehalose phosphorylase; LaMP, Lactobacillus acidophilus maltose phosphorylase.
5. Biotechnological use of disaccharide phosphorylases
While DSP synthetic reactions can be used for disaccharides synthesis, DSP phosphorolysis reactions can be exploited for the degradation of glyco-oligomers, leading to the production of useful sugar-1-phosphates. The reversibility of phosphorolysis can be exploited for the utilization of abundantly available natural sugar as a starting material [76,77]. Due to the industrial interests in DSPs as a result of their cheap and readily available substrates, in the last decades, a big effort has been put into the discovery of new DSP activities, as well as engineering DSPs to overcome the limitations associated with their strict specificity in terms of substrates or linkage [8,19,78]; efforts to improve their thermostability and catalytic efficiency with non-natural substrates have also been reported, since these properties govern their potential for use in the production of novel disaccharides and small glyco-conjugated compounds with potentially useful pharmacological properties [63,79]. DSPs can be applied either as sole catalysts and/or in combination with other phosphorylases or other carbohydrate-active enzymes to produce small glycoconjugated molecules and rare disaccharides.
A single DSP can be used as a sole catalyst for the production of a disaccharide of interest. Morimoto and co-workers have shown that SP from Leuconostoc mesenteroides (LmSP) exhibits activity towards eight different ketohexose acceptors while using α-D-Glc-1-phosphate as a donor, allowing the production of eight corresponding rare or absent d-glucosyl-ketohexoses in nature [80]. Kraus and co-workers reported several variants of BaSP, which switch the regioselectivity of the transfer reaction from α-(1,2) to α-(1,3), thus enabling the efficient synthesis and isolation of nigerose [72]. Luley-Goedl et al. constructed a biocatalytic process for the synthesis of glucosylglycerol as an industrial fine chemical in which SP catalyzes regioselective glucosylation of glycerol using sucrose as the donor substrate [81]. Moreover, an efficient and scalable kojibiose production process was established from sucrose and glucose catalyzed by SP with a yield of 74% [63]. Besides, a non-reducing disaccharide α-d-galactosyl α-D-glucoside was synthesized by THP using trehalose as a glucosyl donor and d-galactose as an acceptor [82]. SP can also catalyze the 2-O-a-glucosylation of l-ascorbic acid from sucrose with high efficiency and perfect site-selectivity [83]. Besides, CgCBP was used to prepare 1,5-anhydro-4-O-β-D glucopyranosyl-d-fructose from 1,5-anhydro-d-fructose and αfD-glucose 1-phosphate [84].
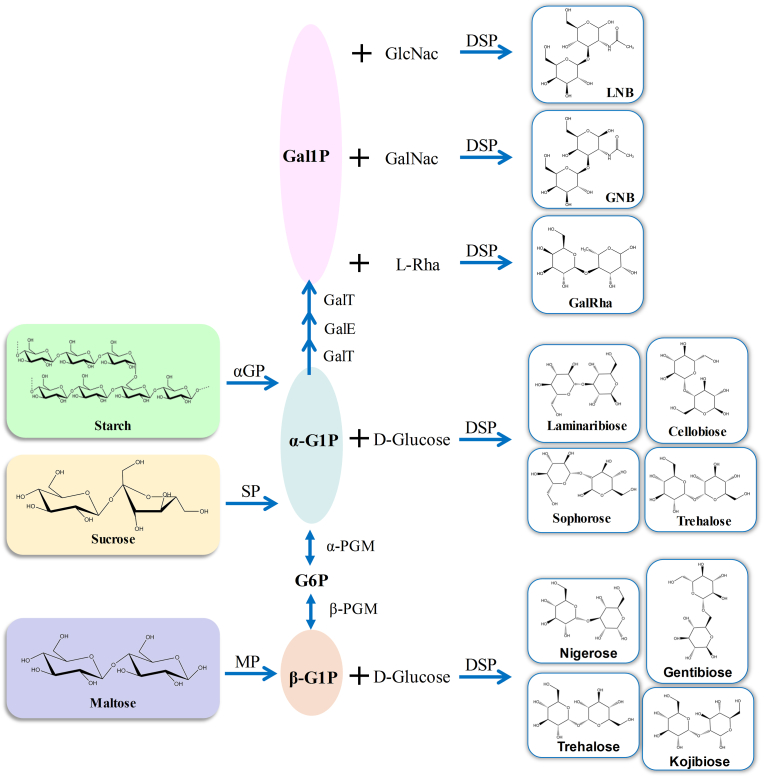
DSPs can also be used in conjunction with other biocatalysts to expand the range of possible products through a “one-pot” enzymatic approach [21,85]. The concept of one-pot phosphorolysis/reverse reactions that circumvent the use of costly sugar 1-phosphate has been often applied to large-scale preparations of oligosaccharides as functional biomaterial from sucrose, maltose, and starch (Fig. 4). A common strategy is the use of SPs as in situ generators of α-D-Glc-1-phosphate, which can then be used as the substrates for the downstream reactions catalyzed by other phosphorylases and/or other biocatalysts. Recent examples consist of the one-pot enzymatic approaches from sucrose and the corresponding acceptor included synthesis of d-galactosyl-β-1,3-N-acetyl-d-hexosamine: lacto-N-biose I (Galβ1→3GlcNAc, LNB) [60] and galacto-N-biose (Galβ1→3GalNAc, GNB) [86], d-Galactosyl-1→4-l-rhamnose (GalRha) [87], laminaribiose [88], kojibiose [63], cellobiose [89] and so on. Maltose can be donated to produce β-D-Glc-1-phosphate, and then coupled with corresponding DSPs to catalyze glucose producing trehalose [90]. Starch can also generate α-D-Glc-1-phosphate catalyzed by α-glucan phosphorylase, and be used as a donor to transform sugar acceptor producing various disaccharides, like trehalose [91], laminaribiose [77], cellobiose [76], nigerose [92].
Fig. 4.
One-pot enzymatic approaches to produce various disaccharides catalyzed by corresponding DSPs. αGP, α-glucan phosphorylase; α-G1P, α-glucose 1-phosphate; G6P, glucose 6-phosphate; β-G1P, β-glucose 1-phosphate; α-PGM, α-phosphoglucomutase; β-PGM, β-phosphoglucomutase; GalT, UDP-glucose—hexose-1-phosphate uridylyltransferase; GalE, UDP-glucose 4-epimerase; Gal1P, galactose 1-phosphate; GlcNac, N-acetyl-glucosamine; GalNac, N-acetyl-galactosamine; L-Rha; l-Rhamnose.
6. Concluding remarks
DSPs are a rather small group of unique enzymes, knowledge and quantity of which is still increasing. All known DSPs are classified in one GT families and five GH families so far. With exception of GT4 family, the three-dimensional structures of at least one phosphorylase in each family are solved. These lay a solid foundation for further studying the structure-function relationship and engineering enzymes to adapt to an industrial condition in substrate selectivity and thermostability aspects. The high selectivity of DSPs in both phosphorolysis and synthesis directions was a serious limitation for their wider biotechnological application. Broadening or changing the substrate specificity of DSPs by protein engineering would lead to a simple, high-yield, and economic synthesis of various functional disaccharides. Moreover, the weak stability of some DSPs (SP, CBP, and GLNBP) would weaken their industrial possibility. Improving the thermostability of DSPs by structure-guided mutation and random mutagenesis have scored some achievement to facilitate the pace of the industrial application of DSPs. Thanks to the results of various process and protein engineering efforts, numerous compounds with the industrial appeal can now be obtained with the help of DSPs.
Despite the significant achievements in the art, there are still many questions to answer, from a view of both basic and applied research. One of them is whether DSPs can be used to produce other functional disaccharides, such as lactose (β-d-galacto-pyranosyl-1,4-d-glucose), melibiose (α-d-galactopyranosyl-1,6-d-glucose), and isomaltose (α-d-glucopyranosyl-1,6-d-glucose), from low-cost substrates. Furthermore, the discovery and characterization of a few enzymes with novel natural specificities will open up new directions for the development of useful phosphorylase-mediated biocatalytic processes, such as β-glucan phosphorylase or another phosphorylase to produce β-G1P from starch or sucrose. Another interesting question is completely unknown why the phosphorolytic processing of carbohydrates is absent from pentoses present in plant hemicelluloses, like d-xylose or l-arabinose. But more importantly, there is not a general rule or some specific sites can get from the above-mentioned attempts of enzyme engineering, to pave the way for modification of other DSPs.
CRediT authorship contribution statement
Shangshang Sun: Conceptualization, Visualization, Writing - original draft. Chun You: Conceptualization, Writing - review & editing.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Key Research Program of the Chinese Academy of Sciences (Grant No. ZDRW-ZS-2016-3), the National Natural Science Foundation of China (Grant No. 21778073) and Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (Grant No. TSBICIP-KJGG-003).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Lairson L.L., Withers S.G. Mechanistic analogies amongst carbohydrate modifying enzymes. Chem Commun. 2004;20:2243–2248. doi: 10.1039/b406490a. [DOI] [PubMed] [Google Scholar]
- 2.Field R.A. Glycobiology: challenging reaction equilibria. Nat Chem Biol. 2011;7:658–659. doi: 10.1038/nchembio.668. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen M.M., Pedersen C.M. Catalytic glycosylations in oligosaccharide synthesis. Chem Rev. 2018;118:8285–8358. doi: 10.1021/acs.chemrev.8b00144. [DOI] [PubMed] [Google Scholar]
- 4.Dimakos V., Taylor M.S. Site-Selective Functionalization of hydroxyl groups in carbohydrate derivatives. Chem Rev. 2018;118(23):11457–11517. doi: 10.1021/acs.chemrev.8b00442. [DOI] [PubMed] [Google Scholar]
- 5.Cantarel B.L., Coutinho P.M., Rancurel C., Bernard T., Lombard V., Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mika L.T., Cséfalvay E. Németh Á. Catalytic conversion of carbohydrates to initial platform chemicals: chemistry and sustainability. Chem Rev. 2018;118(2):505–613. doi: 10.1021/acs.chemrev.7b00395. [DOI] [PubMed] [Google Scholar]
- 7.Panza M., Pistorio S.G., Stine K.J., Demchenko A.V. Automated chemical oligosaccharide synthesis: novel approach to traditional challenges. Chem Rev. 2018;118(17):8105–8150. doi: 10.1021/acs.chemrev.8b00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakai H., Kitaoka M., Svensson B., Ohtsubo K. Recent development of phosphorylases possessing large potential for oligosaccharide synthesis. Curr Opin Chem Biol. 2013;17(2):301–309. doi: 10.1016/j.cbpa.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Davies G.J., Gloster T.M., Henrissat B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr Opin Struct Biol. 2005;15(6):637–645. doi: 10.1016/j.sbi.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Mackenzie L.F., Wang Q., Warren R.A.J., Withers S.G. Glycosynthases: mutant glycosidases for oligosaccharide synthesis. J Am Chem Soc. 1998;120(22):5583–5584. doi: 10.1021/ja980833d. [DOI] [Google Scholar]
- 11.Wen L., Edmunds G., Gibbons C., Zhang J., Gadi M.R., Zhu H., Fang J., Liu X., Kong Y., Wang P.G. Toward automated enzymatic synthesis of oligosaccharides. Chem Rev. 2018;118(17):8151–8187. doi: 10.1021/acs.chemrev.8b00066. [DOI] [PubMed] [Google Scholar]
- 12.Williams G.J., Thorson J.S. Natural product glycosyltransferases: properties and applications. Adv Enzymol Relat Area Mol Biol. 2009;76:55–119. doi: 10.1002/9780470392881.ch2. [DOI] [PubMed] [Google Scholar]
- 13.Lairson L.L., Henrissat B., Davies G.J., Withers S.G. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 14.Kitaoka M. Phosphorylases in the production of oligosaccharides. ACS Symp Ser. 2007;972:195–206. doi: 10.1021/bk-2007-0972.ch014. [DOI] [Google Scholar]
- 15.Browner M.F., Fletterick R.J. Phosphorylase: a biological transducer. Trends Biochem Sci. 1992;17(2):66–71. doi: 10.1016/0968-0004(92)90504-3. [DOI] [PubMed] [Google Scholar]
- 16.Krasnova L., Wong C.H. Oligosaccharide synthesis and translational innovation. J Am Chem Soc. 2019;141(9):3735–3754. doi: 10.1021/jacs.8b11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luley-Goedl C., Nidetzky B. Carbohydrate synthesis by disaccharide phosphorylases: reactions, catalytic mechanisms and application in the glycosciences. Biotechnol J. 2010;5(12):1324–1338. doi: 10.1002/biot.201000217. [DOI] [PubMed] [Google Scholar]
- 18.Puchart V. Glycoside phosphorylases: structure, catalytic properties and biotechnological potential. Biotechnol Adv. 2015;33(2):261–276. doi: 10.1016/j.biotechadv.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Beerens K., De Winter K., Van de Walle D., Grootaert C., Kamiloglu S., Miclotte L., Van de Wiele T., Van Camp J., Dewettinck K., Desmet T. Biocatalytic synthesis of the rare sugar kojibiose: process scale-up and application testing. J Agric Food Chem. 2017;65(29):6030–6041. doi: 10.1021/acs.jafc.7b02258. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima M., Tanaka N., Furukawa N., Nihira T., Kodutsumi Y., Takahashi Y., Sugimoto N., Miyanaga A., Fushinobu S., Taguchi H., Nakai H. Mechanistic insight into the substrate specificity of 1,2-β-oligoglucan phosphorylase from Lachnoclostridium phytofermentans. Sci Rep. 2017;42671:7. doi: 10.1038/srep42671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pergolizzi G., Kuhaudomlarp S., Kalita E., Field R.A. Glycan phosphorylases in multi-enzyme synthetic processes. Protein Pept Lett. 2017;24(8):696–709. doi: 10.2174/0929866524666170811125109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitaoka M. Diversity of phosphorylases in glycoside hydrolase families. Appl Microbiol Biotechnol. 2015;99(20):8377–8390. doi: 10.1007/s00253-015-6927-0. [DOI] [PubMed] [Google Scholar]
- 23.Nishimoto M., Kitaoka M. Identification of the putative proton donor residue of lacto-N-biose phosphorylase (EC 2.4.1.211) Biosci Biotechnol Biochem. 2007;71(6):1587–1591. doi: 10.1271/bbb.70064. [DOI] [PubMed] [Google Scholar]
- 24.Goedl C., Nidetzky B. Sucrose phosphorylase harbouring a redesigned, glycosyltransferase-like active site exhibits retaining glucosyl transfer in the absence of a covalent intermediate. Chembiochem. 2009;10(14):2333–2337. doi: 10.1002/cbic.200900429. [DOI] [PubMed] [Google Scholar]
- 25.Desmet T., Soetaert W. Broadening the synthetic potential of disaccharide phosphorylases through enzyme engineering. Process Biochem. 2012;47(1):11–17. doi: 10.1016/j.procbio.2011.10.039. [DOI] [Google Scholar]
- 26.Desmet T., Soetaert W. Enzymatic glycosyl transfer: mechanisms and applications. Biocatal Biotransform. 2011;29:1–18. doi: 10.3109/10242422.2010.548557. [DOI] [Google Scholar]
- 27.Schwarz A., Brecker L., Nidetzky B. Acid-base catalysis in Leuconostoc mesenteroides sucrose phosphorylase probed by site-directed mutagenesis and detailed kinetic comparison of wild-type and Glu237-->Gln mutant enzymes. Biochem J. 2007;403(3):441–449. doi: 10.1042/BJ20070042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirza O., Skov L.K., Sprogøe D., van den Broek L.A., Beldman G., Kastrup J.S., Gajhede M. Structural rearrangements of sucrose phosphorylase from Bifidobacterium adolescentis during sucrose conversion. J Biol Chem. 2006;281(46):35576–35584. doi: 10.1074/jbc.M605611200. [DOI] [PubMed] [Google Scholar]
- 29.Doudoroff M., Barker H.A., Hassid W.Z. Studies with bacterial sucrose phosphorylase; the mechanism of action of sucrose phosphorylase as a glucose-transferring enzyme (transglucosidase) J Biol Chem. 1947;168(2):725–732. [PubMed] [Google Scholar]
- 30.Voet J.G., Abeles R.H. The mechanism of action of sucrose phosphorylase. Isolation and properties of a β-linked covalent glucose-enzyme complex. J Biol Chem. 1970;245:1020–1031. [PubMed] [Google Scholar]
- 31.Mieyal J.J., Abeles R.H. 17 disaccharide phosphorylases. Enzymes. 1972;7:515–532. doi: 10.1016/S1874-6047(08)60461-8. [DOI] [Google Scholar]
- 32.Wiesbauer J., Goedl C., Schwarz A., Brecker L., Nidetzky B. Substitution of the catalytic acid–base Glu237 by Gln suppresses hydrolysis during glucosylation of phenolic acceptors catalyzed by Leuconostoc mesenteroides sucrose phosphorylase. J Mol Catal B Enzym. 2010;65:24–29. doi: 10.1016/j.molcatb.2009.12.007. [DOI] [Google Scholar]
- 33.Goedl C., Schwarz A., Mueller M., Brecker L., Nidetzky B. Mechanistic differences among retaining disaccharide phosphorylases: insights from kinetic analysis of active site mutants of sucrose phosphorylase and alpha,alpha-trehalose phosphorylase. Carbohydr Res. 2008;343(12):2032–2040. doi: 10.1016/j.carres.2008.01.029. [DOI] [PubMed] [Google Scholar]
- 34.Klimacek M., Sigg A., Nidetzky B. On the donor substrate dependence of group-transfer reactions by hydrolytic enzymes: insight from kinetic analysis of sucrose phosphorylase-catalyzed transglycosylation. Biotechnol Bioeng. 2020;117(10):2933–2943. doi: 10.1002/bit.27471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hidaka M., Kitaoka M., Hayashi K., Wakagi T., Shoun H., Fushinobu S. Structural dissection of the reaction mechanism of cellobiose phosphorylase. Biochem J. 2006;398(1):37–43. doi: 10.1042/BJ20060274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraus M., Grimm C., Seibel J. Switching enzyme specificity from phosphate to resveratrol glucosylation. Chem Commun. 2017;53(90):12181–12184. doi: 10.1039/c7cc05993k. [DOI] [PubMed] [Google Scholar]
- 37.Okada S., Yamamoto T., Watanabe H., Nishimoto T., Chaen H., Fukuda S., Wakagi T., Fushinobu S. Structural and mutational analysis of substrate recognition in kojibiose phosphorylase. FEBS J. 2014;281(3):778–786. doi: 10.1111/febs.12622. [DOI] [PubMed] [Google Scholar]
- 38.Egloff M.P., Uppenberg J., Haalck L., van Tilbeurgh H. Crystal structure of maltose phosphorylase from Lactobacillus brevis: unexpected evolutionary relationship with glucoamylases. Structure. 2001;9(8):689–697. doi: 10.1016/s0969-2126(01)00626-8. [DOI] [PubMed] [Google Scholar]
- 39.Touhara K.K., Nihira T., Kitaoka M., Nakai H., Fushinobu S. Structural basis for reversible phosphorolysis and hydrolysis reactions of 2-O-α-glucosylglycerol phosphorylase. J Biol Chem. 2014;289(26):18067–18075. doi: 10.1074/jbc.M114.573212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhaudomlarp S., Walpole S., Stevenson C.E.M., Nepogodiev S.A., Lawson D.M., Angulo J., Field R.A. Unravelling the specificity of laminaribiose phosphorylase from Paenibacillus sp. YM-1 towards donor substrates glucose/mannose 1-phosphate by using X-ray crystallography and saturation transfer difference NMR spectroscopy. Chembiochem. 2019;20(2):181–192. doi: 10.1002/cbic.201800260. [DOI] [PubMed] [Google Scholar]
- 41.Bianchetti C.M., Elsen N.L., Fox B.G., Phillips G.N., Jr. Structure of cellobiose phosphorylase from Clostridium thermocellum in complex with phosphate. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2011;67(Pt 11):1345–1349. doi: 10.1107/S1744309111032660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Hoorebeke A., Stout J., Kyndt J., De Groeve M., Dix I., Desmet T., Soetaert W., Van Beeumen J., Savvides S.N. Crystallization and X-ray diffraction studies of cellobiose phosphorylase from Cellulomonas uda. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66(Pt 3):346–351. doi: 10.1107/S1744309110002642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hidaka M., Honda Y., Kitaoka M., Nirasawa S., Hayashi K., Wakagi T., Shoun H., Fushinobu S. Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold. Structure. 2004;12(6):937–947. doi: 10.1016/j.str.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Nam Y.W., Nihira T., Arakawa T., Saito Y., Kitaoka M., Nakai H., Fushinobu S. Crystal structure and substrate recognition of cellobionic acid phosphorylase, which plays a key role in oxidative cellulose degradation by microbes. J Biol Chem. 2015;290(30):18281–18292. doi: 10.1074/jbc.M115.664664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hidaka M., Nishimoto M., Kitaoka M., Wakagi T., Shoun H., Fushinobu S. The crystal structure of galacto-N-biose/lacto-N-biose I phosphorylase: a large deformation of a TIM barrel scaffold. J Biol Chem. 2009;284(11):7273–7283. doi: 10.1074/jbc.M808525200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koyama Y., Hidaka M., Nishimoto M., Kitaoka M. Directed evolution to enhance thermostability of galacto-N-biose/lacto-N-biose I phosphorylase. Protein Eng Des Sel. 2013;26(11):755–761. doi: 10.1093/protein/gzt049. [DOI] [PubMed] [Google Scholar]
- 47.Nakae S., Ito S., Higa M., Senoura T., Wasaki J., Hijikata A., Shionyu M., Ito S., Shirai T. Structure of novel enzyme in mannan biodegradation process 4-O-β-D-mannosyl-D-glucose phosphorylase MGP. J Mol Biol. 2013;425(22):4468–4478. doi: 10.1016/j.jmb.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 48.Ye Y., Saburi W., Odaka R., Kato K., Sakurai N., Komoda K., Nishimoto M., Kitaoka M., Mori H., Yao M. Structural insights into the difference in substrate recognition of two mannoside phosphorylases from two GH130 subfamilies. FEBS Lett. 2016;590(6):828–837. doi: 10.1002/1873-3468.12105. [DOI] [PubMed] [Google Scholar]
- 49.Sprogøe D., van den Broek L.A., Mirza O., Kastrup J.S., Voragen A.G., Gajhede M., Skov L.K. Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis. Biochemistry. 2004;43(5):1156–1162. doi: 10.1021/bi0356395. [DOI] [PubMed] [Google Scholar]
- 50.Franceus J., Desmet T. A GH13 glycoside phosphorylase with unknown substrate specificity from Corallococcus coralloides. Amylase. 2019;3(1):32–40. doi: 10.1515/amylase-2019-0003. [DOI] [Google Scholar]
- 51.Watson K.A., McCleverty C., Geremia S., Cottaz S., Driguez H., Johnson L.N. Phosphorylase recognition and phosphorolysis of its oligosaccharide substrate: answers to a long outstanding question. EMBO J. 1999;18(17):4619–4632. doi: 10.1093/emboj/18.17.4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lairson L.L., Watts A.G., Wakarchuk W.W., Withers S.G. Using substrate engineering to harness enzymatic promiscuity and expand biological catalysis. Nat Chem Biol. 2006;2(12):724–728. doi: 10.1038/nchembio828. [DOI] [PubMed] [Google Scholar]
- 53.Goedl C., Nidetzky B. The phosphate site of trehalose phosphorylase from Schizophyllum commune probed by site-directed mutagenesis and chemical rescue studies. FEBS J. 2008;275(5):903–913. doi: 10.1111/j.1742-4658.2007.06254.x. [DOI] [PubMed] [Google Scholar]
- 54.Schwarz A., Nidetzky B. Asp-196-->Ala mutant of Leuconostoc mesenteroides sucrose phosphorylase exhibits altered stereochemical course and kinetic mechanism of glucosyl transfer to and from phosphate. FEBS Lett. 2006;580(16):3905–3910. doi: 10.1016/j.febslet.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 55.Cerdobbel A., Desmet T., De Winter K., Maertens J., Soetaert W. Increasing the thermostability of sucrose phosphorylase by multipoint covalent immobilization. J Biotechnol. 2010;150(1):125–130. doi: 10.1016/j.jbiotec.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 56.Verhaeghe T., Aerts D., Diricks M., Soetaert W., Desmet T. The quest for a thermostable sucrose phosphorylase reveals sucrose 6'-phosphate phosphorylase as a novel specificity. Appl Microbiol Biotechnol. 2014;98(16):7027–7037. doi: 10.1007/s00253-014-5621-y. [DOI] [PubMed] [Google Scholar]
- 57.Cerdobbel A., De Winter K., Aerts D., Kuipers R., Joosten H.J., Soetaert W., Desmet T. Increasing the thermostability of sucrose phosphorylase by a combination of sequence- and structure-based mutagenesis. Protein Eng Des Sel. 2011;24(11):829–834. doi: 10.1093/protein/gzr042. [DOI] [PubMed] [Google Scholar]
- 58.Franceus J., Desmet T. Sucrose phosphorylase and related enzymes in glycoside hydrolase family 13: discovery, application and engineering. Int J Mol Sci. 2020;2526(7):21. doi: 10.3390/ijms21072526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye X., Zhang C., Zhang Y.H. Engineering a large protein by combined rational and random approaches: stabilizing the Clostridium thermocellum cellobiose phosphorylase. Mol Biosyst. 2012;8(6):1815–1823. doi: 10.1039/c2mb05492b. [DOI] [PubMed] [Google Scholar]
- 60.Nishimoto M., Kitaoka M. Practical preparation of lacto-N-biose I, a candidate for the bifidus factor in human milk. Biosci Biotechnol Biochem. 2007;71(8):2101–2104. doi: 10.1271/bbb.70320. [DOI] [PubMed] [Google Scholar]
- 61.Bode L. Human milk oligosaccharides: prebiotics and beyond. Nutr Rev. 2009;67(Suppl 2):S183–S191. doi: 10.1111/j.1753-4887.2009.00239.x. [DOI] [PubMed] [Google Scholar]
- 62.Palm D., Goerl R., Burger K.J. Evolution of catalytic and regulatory sites in phosphorylases. Nature. 1985;313(6002):500–502. doi: 10.1038/313500a0. [DOI] [PubMed] [Google Scholar]
- 63.Verhaeghe T., De Winter K., Berland M., De Vreese R., D'hooghe M., Offmann B., Desmet T. Converting bulk sugars into prebiotics: semi-rational design of a transglucosylase with controlled selectivity. Chem Commun (Camb) 2016;52(18):3687–3689. doi: 10.1039/c5cc09940d. [DOI] [PubMed] [Google Scholar]
- 64.De Winter K., Soetaert W., Desmet T. An imprinted cross-linked enzyme aggregate (iCLEA) of sucrose phosphorylase: combining improved stability with altered specificity. Int J Mol Sci. 2012;13(9):11333–11342. doi: 10.3390/ijms130911333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Groeve M.R., Desmet T., Soetaert W. Engineering of cellobiose phosphorylase for glycoside synthesis. J Biotechnol. 2011;156(4):253–260. doi: 10.1016/j.jbiotec.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 66.Tsumuraya Y., Brewer C.F., Hehre E.J. Substrate-induced activation of maltose phosphorylase: interaction with the anomeric hydroxyl group of alpha-maltose and alpha-D-glucose controls the enzyme's glucosyltransferase activity. Arch Biochem Biophys. 1990;281(1):58–65. doi: 10.1016/0003-9861(90)90412-r. [DOI] [PubMed] [Google Scholar]
- 67.De Groeve M.R., De Baere M., Hoflack L., Desmet T., Vandamme E.J., Soetaert W. Creating lactose phosphorylase enzymes by directed evolution of cellobiose phosphorylase. Protein Eng Des Sel. 2009;22(7):393–399. doi: 10.1093/protein/gzp017. [DOI] [PubMed] [Google Scholar]
- 68.De Groeve M.R., Tran G.H., Van Hoorebeke A., Stout J., Desmet T., Savvides S.N., Soetaert W. Development and application of a screening assay for glycoside phosphorylases. Anal Biochem. 2010;401(1):162–167. doi: 10.1016/j.ab.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 69.De Groeve M.R., Remmery L., Van Hoorebeke A., Stout J., Desmet T., Savvides S.N., Soetaert W. Construction of cellobiose phosphorylase variants with broadened acceptor specificity towards anomerically substituted glucosides. Biotechnol Bioeng. 2010;107(3):413–420. doi: 10.1002/bit.22818. [DOI] [PubMed] [Google Scholar]
- 70.Hamura K., Saburi W., Matsui H., Mori H. Modulation of acceptor specificity of Ruminococcus albus cellobiose phosphorylase through site-directed mutagenesis. Carbohydr Res. 2013;379:21–25. doi: 10.1016/j.carres.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 71.Dirks-Hofmeister M.E., Verhaeghe T., De Winter Karel, Desmet T. Creating space for large acceptors: rational biocatalyst design for resveratrol glycosylation in an aqueous system. Angew Chem Int Ed Engl. 2015;54(32):9289–9292. doi: 10.1002/ange.201503605. [DOI] [PubMed] [Google Scholar]
- 72.Kraus M., Görl J., Timm M., Seibel J. Synthesis of the rare disaccharide nigerose by structure-based design of a phosphorylase mutant with altered regioselectivity. Chem Commun (Camb) 2016;52(25):4625–4627. doi: 10.1039/c6cc00934d. [DOI] [PubMed] [Google Scholar]
- 73.Nakai H., Petersen B.O., Westphal Y., Dilokpimol A., Abou Hachem M., Duus J.Ø., Schols H.A., Svensson B. Rational engineering of Lactobacillus acidophilus NCFM maltose phosphorylase into either trehalose or kojibiose dual specificity phosphorylase. Protein Eng Des Sel. 2010;23(10):781–787. doi: 10.1093/protein/gzq055. [DOI] [PubMed] [Google Scholar]
- 74.Van der Borght J., Soetaert W., Desmet T. Engineering the acceptor specificity of trehalose phosphorylase for the production of trehalose analogs. Biotechnol Prog. 2012;28(5):1257–1262. doi: 10.1002/btpr.1609. [DOI] [PubMed] [Google Scholar]
- 75.Chen C., Van der Borght J., De Vreese R., D'hooghe M., Soetaert W., Desmet T. Engineering the specificity of trehalose phosphorylase as a general strategy for the production of glycosyl phosphates. Chem Commun (Camb) 2014;50(58):7834–7836. doi: 10.1039/c4cc02202e. [DOI] [PubMed] [Google Scholar]
- 76.Suzuki M., Kaneda K., Nakai Y., Kitaoka M., Taniguchi H. Synthesis of cellobiose from starch by the successive actions of two phosphorylases. N Biotechnol. 2009;26(3–4):137–142. doi: 10.1016/j.nbt.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 77.Sun S., Wei X., You C. The construction of an in vitro synthetic enzymatic biosystem that facilitates laminaribiose biosynthesis from maltodextrin and glucose. Biotechnol J. 2019;14(4):e1800493. doi: 10.1002/biot.201800493. [DOI] [PubMed] [Google Scholar]
- 78.Kraus M., Grimm C., Seibel J. Reversibility of a point mutation induced domain shift: expanding the conformational space of a sucrose phosphorylase. Sci Rep. 2018;10490(1):8. doi: 10.1038/s41598-018-28802-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Neill E.C. University of East Anglia; 2013. An exploration of phosphorylases for the synthesis of carbohydrate polymers. [Google Scholar]
- 80.Morimoto K., Yoshihara A., Furumoto T., Takata G. Production and application of a rare disaccharide using sucrose phosphorylase from Leuconostoc mesenteroides. J Biosci Bioeng. 2015;119(6):652–656. doi: 10.1016/j.jbiosc.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 81.Luley-Goedl C., Sawangwan T., Mueller M., Schwarz A., Nidetzky B. Biocatalytic process for production of α-glucosylglycerol using sucrose phosphorylase. Food Technol Biotechnol. 2010;48(3):276–283. [Google Scholar]
- 82.Chaen H., Nakada T., Mukai N., Nishimoto T., Tsujisaka Y. Efficient enzymatic synthesis of disaccharide, α-D-Galactosyl α-D-Glucoside, by trehalose phosphorylase from Thermoanaerobacter brockii. J Appl Glycosci. 2001;48(2):135–137. doi: 10.5458/jag.48.135. [DOI] [Google Scholar]
- 83.Gudiminchi R.K., Nidetzky B. Walking a fine line with sucrose phosphorylase: efficient single-step biocatalytic production of L-ascorbic acid 2-glucoside from sucrose. Chembiochem. 2017;18(14):1387–1390. doi: 10.1002/cbic.201700215. [DOI] [PubMed] [Google Scholar]
- 84.Kajiki T., Yoshinaga K., Komba S., Kitaoka M. Enzymatic synthesis of 1,5-Anhydro-4-O-β-D-glucopyranosyl-D-fructose using cellobiose phosphorylase and its spontaneous decomposition via β-Elimination. J Appl Glycosci. 2017;64:91–97. doi: 10.5458/jag.jag.JAG-2017_010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fessner W.D. Systems Biocatalysis: development and engineering of cell-free "artificial metabolisms" for preparative multi-enzymatic synthesis. N Biotechnol. 2015;32(6):658–664. doi: 10.1016/j.nbt.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 86.Ichikawa M., Schnaar R.L., Ichikawa Y. Application of sucrose phosphorylase reaction in one-pot enzymatic galactosylation - scavenger of phosphate and generation of glucose 1-phosphate in-situ. Tetrahedron Lett. 1995;36:8731–8732. [Google Scholar]
- 87.Nakajima M., Nishimoto M., Kitaoka M. Practical preparation of D-galactosyl-β1→4-L-rhamnose employing the combined action of phosphorylases. Biosci Biotechnol Biochem. 2010;74(8):1652–1655. doi: 10.1271/bbb.100263. [DOI] [PubMed] [Google Scholar]
- 88.Kitaoka M., Sasaki T., Taniguchi H. Conversion of sucrose into laminaribiose using sucrose phosphorylase, xylose isomerase and laminaribiose phosphorylase. Denpun Kagaku. 1993;4:40. doi: 10.5458/jag1972.40.311. [DOI] [Google Scholar]
- 89.Zhong C., Wei P., Zhang Y.H.P. A kinetic model of one-pot rapid biotransformation of cellobiose from sucrose catalyzed by three thermophilic enzymes. Chem Eng Sci. 2017;161:159–166. doi: 10.1016/j.ces.2016.11.047. [DOI] [Google Scholar]
- 90.Yoshida M., Nakamura N., Horikoshi K. Production of trehalose by a dual enzyme system of immobilized maltose phosphorylase and trehalose phosphorylase. Enzym Microb Technol. 1998;5 doi: 10.1016/S0141–0229(97)00132-4. [DOI] [Google Scholar]
- 91.Yoshida M., Nakamura N., Shizuoka, Horikoshi K., Saitama Production of trehalose from starch by maltose phosphorylase and trehalose phosphorylase from a strain of Piesiomonas. Starch Staerke. 1997;6 doi: 10.1002/star.19970490106. [DOI] [Google Scholar]
- 92.Nihira T., Miyajima F., Chiku K., Nishimoto M., Kitaoka M., Ohtsubo K., Nakai H. One pot enzymatic production of nigerose from common sugar resources employing nigerose phosphorylase. J Appl Glycosci. 2014;61(3):75–80. doi: 10.5458/jag.jag.JAG-2013_012. [DOI] [Google Scholar]