Abstract
The COVID-19 coronavirus has spread rapidly around the world and has caused global panic. Chest CT images play a major role in confirming positive COVID-19 patients. The computer aided diagnosis of COVID-19 from CT images based on artificial intelligence have been developed and deployed in some hospitals. But environmental influences and the movement of lung will affect the image quality, causing the lung parenchyma and pneumonia area unclear in CT images. Therefore, the performance of COVID-19’s artificial intelligence diagnostic algorithm is reduced. If chest CT images are reconstructed, the accuracy and performance of the aided diagnostic algorithm may be improved. In this paper, a new aided diagnostic algorithm for COVID-19 based on super-resolution reconstructed images and convolutional neural network is presented. Firstly, the SRGAN neural network is used to reconstruct super-resolution images from original chest CT images. Then COVID-19 images and Non-COVID-19 images are classified from super-resolution chest CT images by VGG16 neural network. Finally, the performance of this method is verified by the pubic COVID-CT dataset and compared with other aided diagnosis methods of COVID-19. The experimental results show that improving the data quality through SRGAN neural network can greatly improve the final classification accuracy when the data quality is low. This proves that this method can obtain high accuracy, sensitivity and specificity in the examined test image datasets and has similar performance to other state-of-the-art deep learning aided algorithms.
Keywords: COVID-19, Computer aided diagnosis, Chest CT images, Super-resolution images, Convolutional neural network
Introduction
COVID-19 pneumonia is a respiratory infectious disease, and the infection rate of the disease is very high [1–7]. On January 8, 2020, the pathogen of this outbreak was confirmed as a new coronavirus, namely Corona Virus Disease 2019, which has been named as COVID-19 by WHO. On January 30, the WHO announced that the new coronavirus-infected pneumonia epidemic would be listed as an “emergency public health event of international concern”. Up to now, the virus has appeared in various countries, and the number of infected people has greatly increased [8, 9]. To date (April 11th, 2020), there have been more than 1.72 million confirmed cases all around the world. This has led to a public health emergency of international concern, and put all health organizations on high alert.
At present, the diagnosis of new coronavirus infection pneumonia mainly depends on the new viral nucleic acid detection kit. Throat swab sampling has the risk of not being collected, and sometimes repeated sampling is required; It is not suitable for patients with early infection or patients with low throat virus titer, which is likely to cause false negatives; Sampling staff are easy to be exposed which is risky; Unskilled technicians can easily fail to extract nucleic acid and cause false negatives, or contaminate other people’s samples, causing false positives. X-ray images are a commonly used method for COVID-19 detection, but their sensitivity is often lower than 3D chest CT images, and it is easy to diagnose early or mild diseases as normal. A recent study showed that early admissions were found to have lower chest X-ray abnormalities than during hospitalization [10]. CT scanning can make up for the limitations of RT-PCR analysis and X-rays. It is the most powerful and effective method for detecting COVID-19 pneumonia and the severity of pneumonia, so it is of great significance for the diagnosis of pneumonia [11]. Especially for some patients who have false negative RT-PCR results or false positive nucleic acid detection, the results of CT imaging play a vital role.
At present, in many countries, there is a shortage of medical staff in response to the new coronary pneumonia problem. The number of radiologists is far less than patients, which may lead to inefficient treatment. The overwhelming demand for pneumonia diagnosis caused by COVID-19 is inspiring researchers to develop efficient and intelligent diagnostic methods that can respond quickly. AI has played an important role in the imaging detection of new coronary pneumonia, which can significantly improve the diagnosis speed, diagnosis accuracy and precision [12, 13].
There have been many methods of using artificial intelligence to diagnose COVID-19. F. Shi and D.G. Shen reviewed the rapid responses in the community of medical imaging (empowered by AI) toward COVID-19 [14]. Maghdid et al. used simple convolutional neural networks (CNN) and a modified pre-trained model AlexNet to experiment on X-ray and CT scan image data sets. The two neural network models achieved accuracy of 98% and 94.1% [15]. Narin et al. used three kinds of convolutional neural network models (ResNet50, InceptionV3 and InceptionResNetV2), among which ResNet50 had the best classification effect, while the accuracy of InceptionV3 and InceptionResNetV2 were 97% and 87% respectively [16]. Hemdan et al. designed COVID-Net with seven different models. The results showed that VGG19 and DenseNet have good and similar performance, and the f1-scores for identifying normal and COVID-19 are 0.89 and 0.91 respectively [17]. Ghoshal et al. proposed a COVID-19 screening model based on X-rays. The detection of COVID-19 and non-COVID-19 reached 96.00% and 70.65% respectively [18]. Khali et al. compared some recent deep convolutional neural network architectures, and finally found that the fine-tuned versions of Resnet50, MobileNet_V2, and Inception_Resnet_V2 have relatively satisfactory performance, and can achieve an accuracy rate of more than 96% [19]. Abbas et al. classified X-ray images based on their previously developed CNN (known as decomposition, transfer and synthesis, or DeTrac), and achieved an accuracy of 95.12% [20]. Apostolopoulos et al. adopted the Mobile_Net network and found that CNN can extract important features found only in specific X-ray images by training from scratch. Based on these features, the accuracy of COVID diagnosis has reached nearly 99% [21]. Gozes et al. proposed a deep learning model combining the existing AI model and clinical understanding. The classification results of coronavirus and non-coronavirus reached an AUC value of 0.996 [22]. Xu et al. established an early screening deep learning model to distinguish COVID-19 from influenza A viral pneumonia and healthy cases, and the final overall accuracy was 86.7% [23]. Wang et al. used human-machine collaboration to design a customized network architecture COVID-Net for X-ray image classification, which achieved a test accuracy of 92.4% [24]. Zhang et al. developed a new deep anomaly detection model with a sensitivity of 96% for COVID-19 and a specificity of 70.65% for non-COVID-19 [18]. Chowdhury et al. proposed to use SqueezeNet to classify patients with normal chest X-ray images and patients with pneumonia, and compared with three other different deep CNN networks, confirming that SqueezeNet’s performance is high, reaching an accuracy rate of 98.3% [25]. Wang et al. used a CNN-based convolutional neural network model, and the results showed that the overall accuracy was 82.9%, the sensitivity was 84%, and the specificity was 80.5% [26]. Zheng et al. used the method of combining the U-Net segmentation model with the CNN classification model. Firstly, the pre-trained U-Net model was used to segment the lung region, and then the segmented lung region was sent to the classification network. The specificity and AUC values are 90.7%, 91.1% and 0.959 respectively [27]. Jin et al. developed an AI system for diagnosing COVID-19 based on convolutional neural networks and showed a sensitivity of 94.06% and a specificity of 95.47% respectively in the external test queue [28]. Chen et al. firstly extracted the smallest square containing the effective area on the CT image, and then used the Unet ++ model for training and prediction. Finally, the accuracy, sensitivity and specificity were 95.2%, 100% and 93.6%, respectively [29]. Jin et al. designed a combined model called “3D Unet ++-ResNet-50”. The final classification result reached 97.4% sensitivity and 92.2% specificity [30]. Tang et al. used a random forest model (RF) model to train CT images of COVID-19 patients and non-patients. The test results obtained were 87.9% accuracy, 93.3% sensitivity, and 74.5% specificity [31]. Li et al. established a COVNet deep learning model for extracting visual features from volumetric chest CT examination to classify and identify the three groups of cases of COVID-19, CAP, and non-pneumonia, and finally a classification model with 90% overall sensitivity and 96% specificity is obtained [32]. Ying et al. trained and recognized the ResNet-50 deep learning network. The experimental results show that the model can accurately identify COVID-19 patients with a recall of 93% and an accuracy of 86% [33]. Shi et al. trained CT images of 1658 COVID-19 patients and 1027 CAP patients and proposed an infection Size Aware Random Forest method (iSARF). The experimental results showed that the method had a sensitivity of 90.7% and a specificity of 83.3% with an accuracy of 87.9% [34]. The above studies show that the use of AI can assist in the detection and diagnosis of COVID-19, which can significantly improve the speed and accuracy of COVID-19 diagnosis.
During the acquisition process of CT images, some volume effects, noise effects, offset field effects, and motion effects are likely to occur due to environmental influences, resulting in blurry and uneven images, which may have ring-shaped, bar-shaped or motion artifacts. According to the research of Gagne, the movement of the lungs during the CT image acquisition process will affect the contour area of the target, causing a decline in image quality [35]. From the perspective of the quality of hardware facilities or the requirements of patients and doctors, it is very difficult to collect high-quality CT images, and high-dose radiation can easily cause cancerous diseases and genetic damage [36–39]. In recent years, researchers have conducted a lot of research work on image super-resolution reconstruction, and convolutional neural networks have gradually been applied to the field of image super-resolution [40, 41]. Ledig designed a new loss function combining content loss and adversarial loss based on the SRGAN proposed by the Generative Adversarial Network (GAN), and reconstructed a 4x magnification factor to obtain a more realistic photo [42]. Li used it in textile images to restore rich textures [43]. Xiaoran combined SRGAN and DCNN to achieve super-resolution reconstructed synthetic aperture radar (SAR) images, so as to achieve the purpose of accurately extracting the target area to achieve automatic signal testing [44]. Sood used SRGAN, SRCNN, SRResNet, and Sparse Representation models for magnetic resonance (MR) resolution improvement respectively. Finally, according to the average opinion score (MOS); it is found that the processing result of the SRGAN method is visually closer to the original HR image [45].
If the chest CT images of the suspected patients are reconstructed by super-resolution methods, they can overcome the limitations of the imaging equipment by human operation and imaging environment, and obtain clearer and higher-contrast images, which may help to improve the accuracy of the AI algorithm for COVID-19 diagnosis. In this study, we propose a method that combines the use of the SRGAN model to improve the resolution of chest CT images and the use of image classification model based on VGG16 deep convolutional neural network, which is used to predict and screen chest CT images. We used this method to test the original image dataset of different quality in the public COVID-CT-Dataset [46]. The experimental results show that the method in this paper significantly improves the diagnostic accuracy of COVID-19, which is of great significance for dealing with the problem of insufficient image data quality. This work provides a new idea for the AI algorithm applied to the auxiliary detection of COVID-19 pneumonia.
The rest of the paper is organized as follows
Section II introduces the basic principles and steps of this method. Firstly, the principle and structure of SRGAN and the effect of image enhancement on the COVID-CT-Dataset are studied. Then, the structure of VGG and its application in chest CT image classification are introduced. Section III introduces the classification experiments, results and discussion of COVID-19 and non-COVID-19 using the method proposed in this paper. Section IV summarizes this work.
Methods
This method comprises two main components
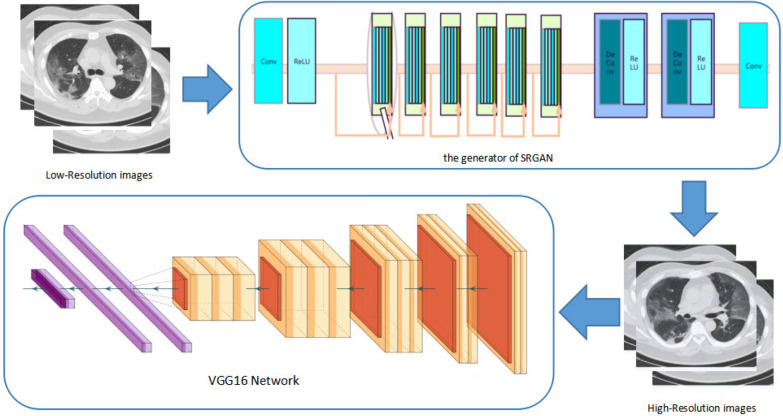
Improving the resolution of chest CT images and classifying CT images. The component of improving resolution of chest CT images uses the SRGAN neural network to reconstruct super-resolution CT images. The component of classifying COVID-19 CT images uses the VGG16 neural network to classify the COVID-19 images and non-COVID-19 images. The flow chart of this method is shown in Fig. 1. The components are described in detail in the following subsections.
Fig. 1.
Workflow of proposed method for classifying the COVID-19 status in CT images
Improving the resolution of chest CT images
The design basis of SRGAN is Generative Adversarial Network (GAN). This deep neural network generates images by using both the generator and the discriminator. The generator keeps trying to generate images from small arrays, and then the discriminator is used to judge whether the generated images are real or fake [47]. The GAN structure is shown in Fig. 2.
Fig. 2.
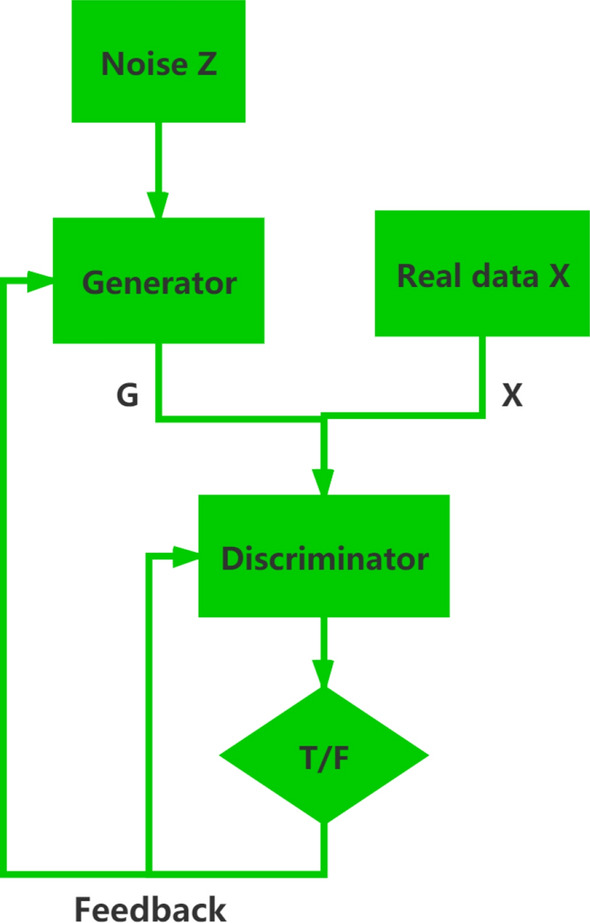
The basic structure of GAN
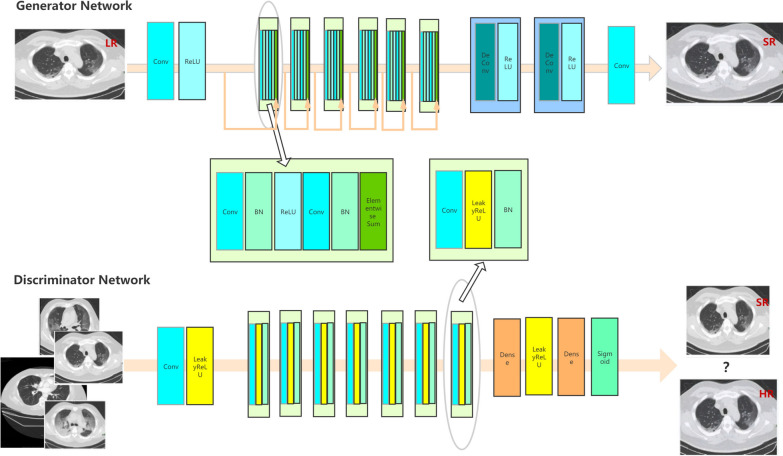
Structure of SRGAN
Super Resolution Generative Adversarial Network is a network designed for super-resolution reconstruction based on Generative Adversarial Network (GAN). Like GAN, SRGAN network structure is also divided into two parts, namely SRGAN generating network and SRGAN discriminating network. As shown in Fig. 3. The residual network structure in which “shortcut connection” is added to the generated network is to prevent problems such as degradation or gradient diffusion caused by the increase of the network layer. The basic structure of the residual block is shown in Fig. 4. The “shortcut connection” can skip multiple layers of training and perform identity mapping to deepen the network layer number so that the network can be optimized faster. Each residual block consists of two convolutional layers plus batch normalization and ReLU activation layer. After the low-resolution image enters the SRGAN network, it firstly enters the convolutional layers for feature extraction, then performs nonlinear activation via the ReLU activation function, and then enters the residual blocks for training. After coming out of the residual blocks, it enters the convolutional layers, and then enlarges the image feature maps by upsampling. The method used for the upsampling operation is scaling convolution, using the nearest neighbor interpolation to enlarge the picture, and finally enters the convolutional layer to perform the convolution operation to output a high-resolution picture.
Fig. 3.
The architecture of SRGAN model
Fig. 4.
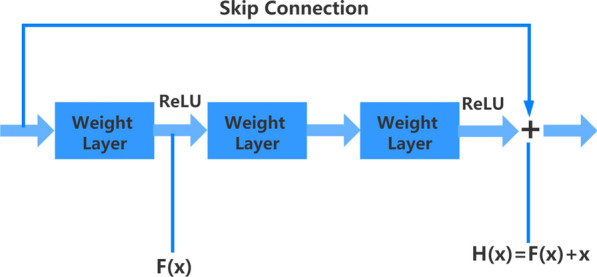
The basic structure of the residual block
Loss Function of SRGAN
The loss function of SRGAN is one of its main innovations, which makes it different from other resolution algorithm and high-frequency details are preserved. For the preservation of detailed information, SRGAN is achieved through perceptual loss. Its loss function is the weighted sum of content loss and adversarial loss. Specifically:
| 1 |
Among them is content loss, is adversarial loss, and is the total loss.
As for content loss, it is used in the G network. Before SRGAN, the loss function was generally calculated by the mean square error between and . Although this calculation method can get a high signal-to-noise ratio, it also loses a lot of high-frequency details. Therefore, SRGAN creatively defines the VGG loss based on the ReLU activation layer of the pre-trained 19-layer VGG network, to find the Euclidean distance of the image and reference image feature representation. A feature map of a certain layer is extracted on the VGG after training, and the real image is compared with the generated feature map. The content loss is calculated as:
| 2 |
And for Adversarial loss, SRGAN adds the adversarial portion of the GAN to the perceptual loss. Through deceiving the network, the generative network could better generate images. The mathematical formula is:
| 3 |
Here is the training image, represents the generative network. And means the probability that the discriminator regards the generated fake image as real. The larger is the better the discriminator could identify the real or fake images.
Applying SRGAN to Improve the Resolution of Chest CT Images
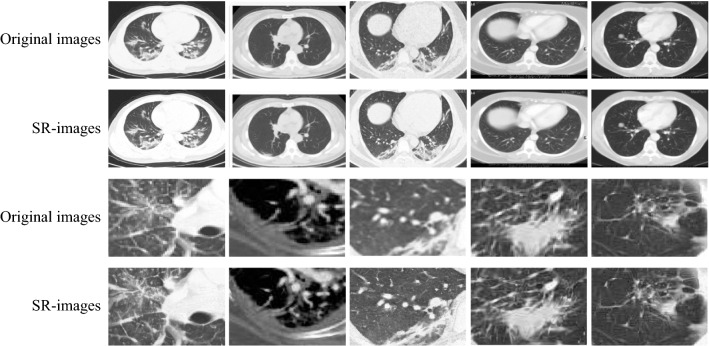
After the model training is finished, input the CT images in our hands as the low-resolution CT images into the model to improve the quality. Thus the texture can be clearer, the feature expression ability of the images is enhanced, and the accuracy of COVID-19 recognition will be higher. The effect before and after SRGAN processing is shown in Fig. 5.
Fig. 5.
Comparison of original images and Super-Resolution images
Classifying COVID-19 CT images
In recent years, a large number of researchers have used deep convolutional neural network models to extract and classify images.The image texture feature and spatial information can be automatically learned by convolution operation on the image through convolutional layers. Various biomedical applications have adopted CNN models [48–50]. The visual geometry group network (VGGNet) is a classic deep convolutional neural network (DCNN) jointly developed by the Oxford University visual geometry group and Google DeepMind researchers [51]. This network is based on the CNN model and is a neural network with multi-layer operation. It is widely used because of its simple structure and good performance. Through repeated stacking of small convolutional kernels and the use of maximum pooling layers, 16-layer VGG16 and 19-layer VGG19 were successfully constructed.
As we all know, the effectiveness of CNN training is affected by the size of the training set. If the training set is relatively small, overfitting is likely to occur. A number of previous studies have shown that in a new task or domain, the representation of the CNN learned in the previous task can be transferred or expanded to a certain extent, that is, transfer learning. And the transfer results in the fields of information retrieval [52], object localization [53] and biomedical image analysis [54] are often superior to other conventional methods. This article is based on VGGNet. VGG has been pre-trained on a large labeled natural image dataset like ImageNet, so it can significantly reduce training time and computational load.
The structure of VGG16 is shown in Figure 6 6, the input image of is received by the conv1 layer, and then propagated through a set of convolutional layers which have a receptive field of . The convolution stride is 1 pixel. Following this is the process of downsampling, in which five max-pooling layers with stride equals to 2 are taken. After going through a set of conv layers, there are three fully connected layers with the channel size as 4096, 4096 and 1000 respectively. Each neuron in fc layer accepts the activations input from the previous layer neuron.
Fig. 6.
Comparative results of different resolution images
1000 is the number of existing categories in the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC). After training more than one million images, VGG-16 can classify images into 1000 categories. Because the features of the pre-trained VGG layer contain information such as image color and edges, they are finally specific to the class attributes. Therefore, we only made some adjustments at the end.
Specifically, after VGG16, the average pooling layer AveragePooling2D layer, dropout layer and fully connected layer are added. The size of the Fc layer is set to the number of new data categories, namely COVID-19 and normal.
Convolutional layer
For image feature extraction, the convolution operation is the key point. The features are extracted through the process of convolving the previous layers with the feature maps of the output. And at the same time, the convolutional kernels are updated continuously. This is the training process, which can be expressed by mathematical formula as:
| 4 |
Here (x,y) is a pixel, is the i-th feature map of the l-th layer, represents the convolutional kernel that connects the i-th input feature map and the j-th output feature map on the l-th layer, F denotes the size of convolutional kernel, represents the j-th bias of the l-th layer, and means the 2-D convolutional operation. For each convolutional layer, the nonlinear activation function is connected to increase the nonlinear characteristic of the network and thus make the model have stronger classification expression ability. This could be expressed as:
| 5 |
Here denotes the nonlinear activation function of ReLU.
Pooling layer
Pooling is divided into maximum and average pooling, which is designed to reduce the number of the training parameters. The pooled window size we usually use is , and after the process of pooling, the four pixel values will be calculated to one result. For maximum pooling and average pooling, the returned results are the maximum and average of the four values respectively. Here in the VGG neural network, we use the maximum pooling as:
| 6 |
Here P represents the size of pooling window.
Activation function
After the features of the image are extracted through a set of convolutional and pooling layers, the softmax activation layer is at last to complete the task of classification. This is expressed as:
| 7 |
Here represents the predicted label of the i-th class, is the input of the softmax layer, denotes the weighted sum of the i-th node of the output of the last fc layer, K represents the number of class, and L is the number of the layer. The output is the posterior probability of each sample, in which the maximum is the predicted class.
Loss function
Some rules are required to update the network parameters after the forward propagation, which can be expressed by loss functions, like the MSE or cross-entropy loss function. Through the process of minimizing the loss value, the network is optimized. And this is just the process of training. The cross-entropy loss function could better reflect the similarity of the training samples and the model distribution. This is expressed as:
| 8 |
Here W and b are weight and bias sets of all the layers in the neural network respectively, and is the real label of the i-th class.
The network finally converges and stable network will be obtained. As for this experiment, we put all the lung CT images into the VGG16 network and then to obtain their class attributes.
Experiment results and disscussion
Experiment CT images dataset
The data we used for this experiment were all obtained from the dataset published online by COVID-CT-Dataset [46]. Zhao collected a total of 760 preprints from medRxiv1 and bioRxiv2, which were released from January 19 to March 25. A total of 470 CT images were acquired, of which 275 were COVID-19 images and the other 195 were normal CT images.
Experiments
In this work, a new collaborative learning method for classifying COVID-19 and non-COVID-19 images is presented based on the SRGAN and VGG16 models. As mentioned above, we firstly send the original lung CT image data set to the pre-trained SRGAN network. After a 4x resolution improvement, we can get high-resolution images. As shown earlier in the article, it also looks clearer. Then we uniformly adjust the high-resolution images to the size of , and send them to the deep neural network designed for image classification based on the VGG16 classic model. Finally, the classification results were obtained and we compared them with some previous studies.
VGGNet should uniformly adjust the image size before training and processing images. For the data set used in this experiment, there are large differences in the quality and resolution between images. In order to more clearly understand the impact of image resolution on the classification results, we adjusted the CT image data set to the size of , , and before and after being processed by the SRGAN model. In this experiment, we use Keras framework with TensorFlow as backend.
Evaluation index
According to the results obtained in each experiment, we used precision, sensitivity, accuracy, specificity and F1-score as evaluation indicators. Since the goal of this experiment is to diagnose whether a person has COVID-19 through CT image data, each of these four indicators has important practical significance.
For the precision indicator, it refers to the proportion of true pneumonia in all cases where the prediction result is positive COVID-19. In other words, if the precision value is too small, there will be many healthy people who are misdiagnosed as COVID-19 patients. This will not only cause a waste of medical supplies, but also make people who have contacted this person feel panic. Sensitivity, also known as recall, refers to the proportion of patients with COVID-19 who are actually diagnosed. In response to this highly infectious virus, each patient who has not been diagnosed will cause great harm to people around him or even a wider range of people. Accuracy is the proportion that the diagnosis result is correct. For specificity, it refers to the proportion of people diagnosed with non-COVID-19 who actually do not have COVID-19.The higher the above indicators, the better the classification result, which means that the performance of our diagnostic method is better.
Comparison and discussion
At present, a large number of researchers have invested a lot of energy in the problem of using deep learning to classify COVID-19. For each experiment, the data set used is different, and some different results have been obtained. Because medical data itself involves privacy issues, it is not easy to obtain large quantities and high-quality data.
The effect of different resolutions of images
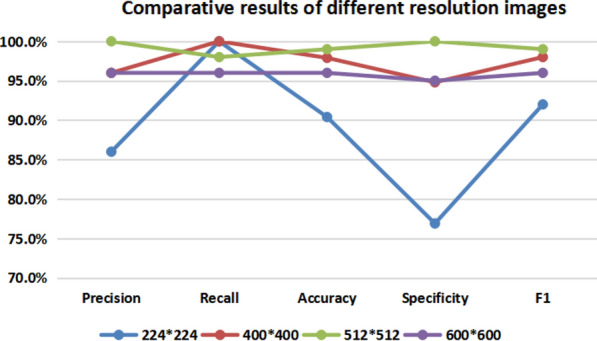
For the effect of the resolution mentioned above on the classification results, we separately adjusted the CT images to , , and . After the images of these sizes are trained and predicted based on the VGG16 classification model, the results obtained are as Table 1.
Table 1.
The classification results of different resolutions of images
| Results | 224 × 224 (%) | 400 × 400(%) | 512 × 512(%) | 600 × 600(%) |
|---|---|---|---|---|
| Precision | 86.0 | 96.0 | 100.0 | 96.0 |
| Recall | 100.0 | 100.0 | 98.0 | 96.0 |
| Accuracy | 90.4 | 97.9 | 99.0 | 96.0 |
| Specificity | 76.9 | 94.8 | 100.0 | 95.0 |
| F1 | 92.0 | 98.0 | 99.0 | 96.0 |
As can be seen from the Table 1 and Fig. 6, within a certain range, the resolution of the images can be improved to obtain more excellent results. When the resolution reaches about , the numerical values of the experimental results tend to be saturated. At this time, if the resolution is being improved, the performance of the classification begins to show a downward trend. This proves that the size of used in our experiment is basically the most suitable for the classification application of COVID-19.
Compare with the results of the other method under the same dataset
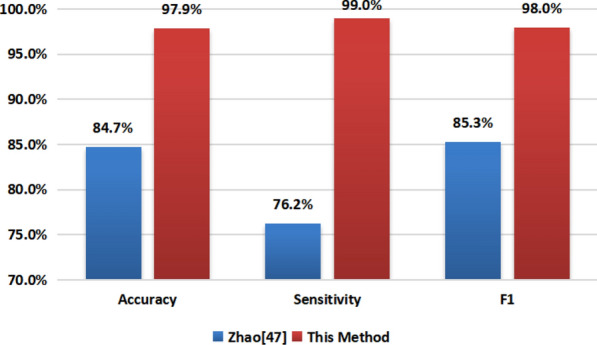
For the results of this experiment, we firstly compared with Zhao[47] using the same data set. For the same 275 COVID-19 and 195 healthy CT images, Zhao directly used a CNN classification model based on the convolutional neural network model. The method we used was to improve the resolution through SRGAN before using the VGG16-based Model to classify. The results are shown in Table 2.
Table 2.
The comparison of experimental results with the same dataset
| Results | Zhao [46] | This method |
|---|---|---|
| Accuracy | 84.7% | 97.9% |
| Sensitivity | 76.2% | 99.0% |
| F1 | 85.3% | 98.0% |
| Method | CNN | SRGAN+VGG |
As can be seen from the Table 2 and Fig 7, for the accuracy index, the classification results of Zhao achieved an accuracy rate of 84.7%, while in our experiment, the accuracy was 97.9%. Our results relatively increased by 13.2 percentage points. This is a big improvement. When it comes to the sensitivity index, the result of Zhao et al. is 76.2%, and our corresponding value is 99%. Our sensitivity here is a very high value, which means that if our method is used to diagnose COVID-19, almost no possible patient will be missed, which is of great significance for the diagnosis of highly infectious COVID-19 extraordinary. From the perspective of math, F1 is a weighted sum of accuracy and sensitivity, which represents a comprehensive performance of the classification results in these two aspects. As can be seen from the results, relative to the 85.3% result obtained by Zhao et al., Our value has reached 98%, which has achieved a 4.7% increase. Based on the above, the method we used in this experiment for this data set has far exceeded the research in [46].
Fig. 7.
The comparative results of our method and Zhao [46]
Compare our results with other state-of-the-art deep learning aided methods
In response to the use of different data sets, we have found the results of ten predecessors this time. Those are the experimental results of others who have previously classified positive and negtive COVID-19. Then we compared our experimental results with them, which are as shown in Table 3 and Fig 8.
Table 3.
The comparative experimental results of different methods
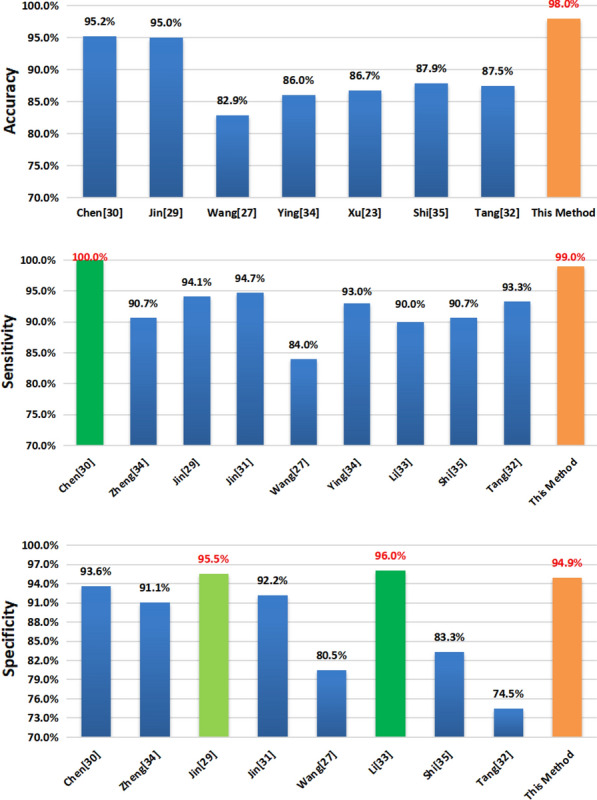
| Results | Chen [29] | Zheng [33] | Jin [28] | Jin [30] | Wang [26] | Ying [33] | Xu [23] | Li [32] | Shi [34] | Tang [31] | This Method |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | 95.2% | NA | 95.0% | NA | 82.9% | 86.0% | 86.7% | NA | 87.9% | 87.5% | 98.0% |
| Sensitivity | 100.0% | 90.7% | 94.1% | 94.7% | 84.0% | 93.0% | NA | 90.0% | 90.7% | 93.3% | 99.0% |
| Specificity | 93.6% | 91.1% | 95.5% | 92.2% | 80.5% | NA | NA | 96.0% | 83.3% | 74.5% | 94.9% |
| Method | Unet++ | U-Net +CNN | CNN | Unet++ +CNN | CNN | ResNet-50 | CNN | ResNet-50 | RF | RF | SRGAN +VGG16 |
Fig. 8.
Comparative accuracy, sensitivity and specificity results of different methods
Apart from the fact that our data set itself is not dominant, we can see that based on the method proposed in this paper combining the SRGAN and VGG16 models, Our final results reached 98%, 99%, and 94.9% in accuracy, sensitivity, and specificity respectively. All indicators show that this method is relatively suitable.
For experiments and research conducted by different organizations, the models, methods, and analysis indicators used for the classification results are different. In order to be able to clearly see our experimental results, we made three histogram observations for accuracy, sensitivity and specificity. The value of the last orange bar in each histogram is corresponding to our experiment results.
As for the indicator of accuracy, before our experiment, Chen et al. used Unet ++ to train and classify the data, and obtained a good result of 95.2%, which is the highest value among the results we listed. The method used by Jin et al. is to firstly use U-Net to extract regions of interest and then use the CNN model for classification, which also achieved a good result of 95%. However, the final accuracy of the method used in our experiment reached 98%, which was 2.8 percentage points higher than the previous highest result.
Regarding sensitivity, the experimental results of Chen et al. reached a good result of 100%, which is unsurpassable, but the final result of our method also reached 99%, ranking second, which is relatively a good result.
For specificity, the classification method based on CNN network model used by Jin et al. and the ResNet-50 method used by Li et al. both have achieved good results, which are 95.5% and 96.0% respectively. The results of our experiment did not reach the height of the two in this index, but achieved 94.9%.
In order to compare all the indicator results of all studies together, we made a result radar chart to represent all the values, as shown in Fig. 9. It is obvious shown that the farther the line is from the center of the circle, the better the performance of the corresponding index. The chart shows that the accuracy, sensitivity and specificity lines corresponding to our results are all close to the periphery of the circle, which means that all performance results are high. And this also shows that our method has higher or similar performance than the other state-of-the-art methods.
Fig. 9.
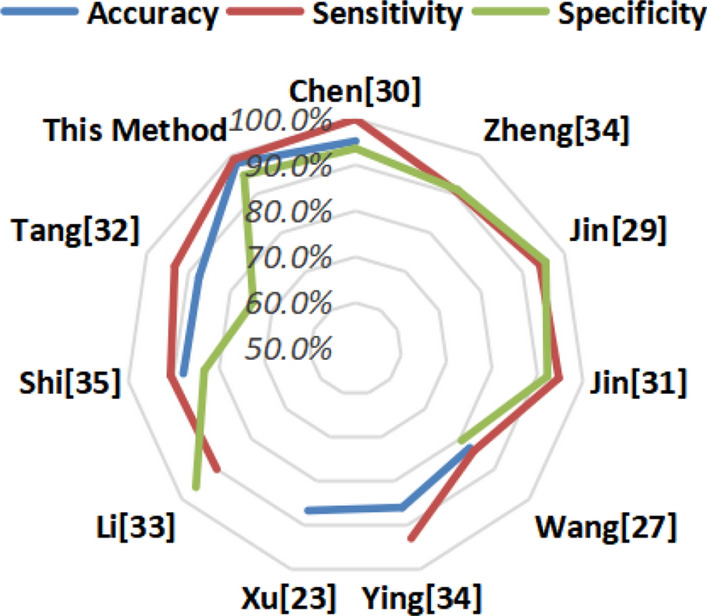
Radar chart of comparative experimental results of different methods
Conclusion
The reconstructed super-resolution chest CT images are clearer and higher-contrast than the original CT images to improve the accuracy of the AI algorithm for COVID-19 diagnosis. In this study, we propose a new aided diagnostic algorithm for COVID-19, which comprises two main components:the component of improving the resolution of chest CT images and the component of classifying CT images. The SRGAN network is used to improve the resolution of chest CT images and VGG16 convolutional neural network is used to classify the COVID-19 and non-COVID-19 from super-resolution CT images. We conducted this experiment on the currently public CT image dataset COVID-CT-Dataset. The overall accuracy rate reached 97.87%. We have compared and analyzed the results obtained in this experiment with the existing experimental results that can be found. The results show that our method is higher or similar than most of the current results in terms of sensitivity, accuracy, specificity and F1-sore. This work provides a new idea for the AI algorithm applied to the auxiliary detection of COVID-19 pneumonia. For the next step, we will continue to improve the method to achieve better accuracy.
Acknowledgements
The authors would like to thank the editor and reviewers for their valuable advices that have helped to improve the paper quality. This work is supported by the Fundamental Research Funds for the Central Universities (N181602014), National Key Research and Development Program of China (2018YFC1314501), National Natural Science Foundation of China (61971118).
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Wenjun Tan, Email: tanwenjun@cse.neu.edu.cn.
Pan Liu, Email: 2974586095@qq.com.
Xiaoshuo Li, Email: 414908645@qq.com.
Yao Liu, Email: 2228109254@qq.com.
Qinghua Zhou, Email: 475859322@qq.com.
Chao Chen, Email: cccovb@hotmail.com.
Zhaoxuan Gong, Email: gzxgzxppy@163.com.
Xiaoxia Yin, Email: xiaoxia.yin@gzhu.edu.cn.
Yanchun Zhang, Email: Yanchun.Zhang@vu.edu.au.
References
- 1.Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China of novel coronavirus-infected pneumonia. N Engl J Med. 2020. 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed]
- 2.Munster VJ, Koopmans M, van Doremalen N, et al. A novel coronavirus emerging in China-key questions for impact assessment. N Engl J Med. 2020 doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 3.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. The Lancet. 2020. 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed]
- 4.2019 to 2020: A data-driven analysis in the early phase of the outbreak. bioRxiv,2020: 2020.2001.2023. 916395.
- 5.Liu T, Hu J, Kang M, et al. Transmission dynamics of 2019 novel coronavirus (2019-nCoV). bioRxiv, 2020: 2020.2001. 2025.919787.
- 6.Wang C, Horby PW, Hayden FG, et al. A novel coronavirus outbreak of global health concern. Lancet. 2020. 10.1016/s0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed]
- 7.Read JM, Bridgen JR, Cummings DA, et al. Novel coronavirus 2019-nCoV–early estimation of epidemiological parameters and epidemic predictions. medRxiv. 2020. doi.org/10.1101/2020.01.23.20018549.
- 8.Holshue M, DeBolt C, First LS. Novel coronavirus in the United States. N Engl J Med. 2019;2020:31. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Wu L, Zhang J, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study, medRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 10.Wong HYF, Lam HYS, Fong AH-T, et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2020;201160. [DOI] [PMC free article] [PubMed]
- 11.Ye Z, Zhang Y, Wang Y, Chest CT, manifestations of new coronavirus disease, et al. (COVID-19): A pictorial review. Eur Radiol. 2019;2020:1–9. [DOI] [PMC free article] [PubMed]
- 12.Torti E, Musci M, Guareschi F, et al. Deep recurrent neural networks for edge monitoring of personal risk and warning situations. Scientific Programming, 2019, Scientific Programming, 01 January 2019, Vol.2019.
- 13.Bullock Joseph LA, Pham Katherine Hoffmann, Lam Cynthia, et al. Mapping the landscape of artificial intelligence applications against COVID-19, 2020, arXiv:2003.11336.
- 14.Shi F, Wang J, Shi J, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19. https://arxiv.org/ftp/arxiv/papers/2004/2004.02731.pdf [DOI] [PubMed]
- 15.Maghdid H, Asaad A, Ghafoor K, et al. Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. 2020, arXiv:2004.00038.
- 16.Narin A, Kaya C, Pamuk Z, et al. Automatic detection of Coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. 2020, arXiv:2003.10849. [DOI] [PMC free article] [PubMed]
- 17.Hemdan E, Shouman M, Karar M, et al. COVIDX-net: a framework of deep learning classifiers to diagnose COVID-19 in X-ray images. 2020, arXiv:2003.11055.
- 18.Zhang J, Xie Y, Li Y, et al. COVID-19 screening on chest X-ray images using deep learning based anomaly detection. 2020, arXiv:2003.12338.
- 19.Asnaoui K, Chawki Y, Idri A, et al. Automated methods for detection and classification pneumonia based on X-ray images using deep learning. 2020, arXiv:2003.14363.
- 20.Abbas A, Abdelsamea M, Gaber M. Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. 2020, arXiv:2003.13815. [DOI] [PMC free article] [PubMed]
- 21.Apostolopoulos I, Aznaouridis S, Tzani M (2020). Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. 2020, arXiv:2004.00338. [DOI] [PMC free article] [PubMed]
- 22.Gozes O, Frid-Adar M, Greenspan H, et al. Rapid AI Development Cycle for the Coronavirus (COVID-19) Pandemic: Initial Results for Automated Detection & Patient Monitoring using Deep Learning CT Image Analysis. 2020, arXiv:2003.05037.
- 23.Xu XJ, Jiang XG, Ma CL, et al. Deep learning system to screen Coronavirus disease 2019 pneumonia. arXiv:2002.09334
- 24.Wang L, Wong A. COVID-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. 2020, arXiv:2003.09871. [DOI] [PMC free article] [PubMed]
- 25.Chowdhury M, Rahman T, Khandakar A, et al. Can AI help in screening viral and COVID-19 pneumonia? 2020, arXiv:2003.13145.
- 26.Wang S, Kang B, Ma J, et al. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19). MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 27.Zheng C, Deng X, Fu Q, et al. Deep learning-based detection for COVID-19 from chest CT using weak label. MedRxiv, 2020.
- 28.Jin C, Cheny W, Cao Y, et al. Development and evaluation of an AI system for COVID-19 diagnosis. MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 29.Chen J, Wu L, Zhang J, et al. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 30.Jin S, Wang B, Xu H, et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 31.Tang Z, Zhao W, Xie X, et al. Severity assessment of coronavirus disease 2019 (COVID-19) using quantitative features from chest CT images. 2020, arXiv:2003.11988.
- 32.Li L, Qin L, Xu Z, et al. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020;200905.
- 33.Song Y, Zheng S, Li L, et al. Deep learning enables accurate diagnosis of novel Coronavirus (COVID-19) with CT images. MedRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 34.Shi F, Xia L, Shan F, et al. Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification. 2020, arXiv:2003.09860. [DOI] [PubMed]
- 35.Gaone I, Robinson D. 239 The impact of lung tumor motion during CT image acquisition upon target delineation. Radiother. Oncol. 2005;76:S114. doi: 10.1016/S0167-8140(05)81216-2. [DOI] [Google Scholar]
- 36.Ng C, Chandler A, Wei W, et al. Effect of duration of scan acquisition on CT perfusion parameter values in primary and metastatic tumors in the lung. Euro J Radiol. 2013;82(10):1811–1818. doi: 10.1016/j.ejrad.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 37.Li G, Luo S, You C, et al. A novel calibration method incorporating nonlinear optimization and ball-bearing markers for cone-beam CT with a parameterized trajectory. Med Phys. 2019;46(1):152–164. doi: 10.1002/mp.13278. [DOI] [PubMed] [Google Scholar]
- 38.Brenner DJ, Hall EJ. Computed tomography: an increasing source of radiation exposure. Eng J Med. 2007;357(22):2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 39.de González AB, Darby S. Risk of cancer from diagnostic X-rays: estimates for the U.K. and 14 other countries. Lancet. 2004;363(9406):345–351. doi: 10.1016/S0140-6736(04)15433-0. [DOI] [PubMed] [Google Scholar]
- 40.Wang G, Kalra M, Orton CG. Machine learning will transform radiology significantly within the next 5 years. Med Phys. 2017;44(6):2041–2044. doi: 10.1002/mp.12204. [DOI] [PubMed] [Google Scholar]
- 41.Wang G, Ye JC, Mueller K, et al. Image reconstruction is a new frontier of machine learning. IEEE Trans Med Image. 2018;37(6):1289–1296. doi: 10.1109/TMI.2018.2833635. [DOI] [PubMed] [Google Scholar]
- 42.Ledig C, Theis L L, Huszár F, et al. Photo-realistic single image super-resolution using a generative adversarial network, Proceedings of the IEEE conference on computer vision and pattern recognition. 2017: 4681–4690.
- 43.Li J, Wu L, Wang S, et al. Super resolution image reconstruction of textile based on SRGAN. In: 2019 IEEE International Conference on Smart Internet of Things (SmartIoT), Tianjin 2019, pp. 436–39.
- 44.Shi X, Zhou F, Yang S, et al. Automatic target recognition for synthetic aperture radar images based on super-resolution generative adversarial network and deep convolutional neural network. Remote Sens. 2019;11(2):135. doi: 10.3390/rs11020135. [DOI] [Google Scholar]
- 45.Rewa S, Binit T, Karthik C, et al. An application of generative adversarial networks for super resolution medical imaging. 2019;326–31.
- 46.Zhao J, Zhang Y, He X, et al. COVID-CT-Dataset: a CT scan dataset about COVID-19. 2020, arXiv:2003.13865.
- 47.Goodfellow I, NIPS 2016 tutorial: generative adversarial networks. 2016, arXiv:1701.00160.
- 48.Acharya U, Oh L, Hagiwara Y, et al. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput. Biol. Med. 2018;100:270–278. doi: 10.1016/j.compbiomed.2017.09.017. [DOI] [PubMed] [Google Scholar]
- 49.Hoo-Chang S, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. doi: 10.1109/TMI.2016.2528162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bar Y, Diamant I, Wolf L, Lieberman S, Konen E, Greenspan H. Chest pathology detection using deep learning with non-medical training. In: ISBI, 2015, pp. 294–297.
- 51.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. 2014, arXiv:1409.1556.
- 52.Chigorin A, Babenko A, Slesarev A, et al. Neural codes for image retrieval. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2014, pp. 584–599.
- 53.Darrell T, Girshick R, Donahue J, et al. Rich feature hierarchies for accurate object detection and semantic segmentation. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2014, pp. 580–587.
- 54.Zhang L, Gurudu S, Gotway M, et al. Fine-tuning convolutional neural networks for biomedical image analysis: actively and incrementally. In: Proceedings of the IEEE conference on computer vision and pattern recognition, 2017, pp. 4761–4772. [DOI] [PMC free article] [PubMed]