Abstract
Direct analysis of whole blood on bloodstained textiles is achieved with thread spray mass spectrometry (MS). This capability satisfies investigators’ first priority in crime scene investigations, which is determining if a stain is blood. This thread spray method explores the use of evidentiary fabric threads for rapid determination of hemoglobin directly from whole blood within textiles without prior extraction steps. The multiplicity of information that can be derived from the thread spray MS method distinguishes it from the current presumptive Bluestar® method, by enabling the detection of hemoglobin (both α- and β-chains), the heme co-factor and lipids all from a single blood sample. Lipid composition was found to differ for blood samples originating from human, canine, and horse species. The robustness of the thread spray MS method as a forensic analytical platform was evaluated in three ways: 1) its successful applicability to samples previously tested by the Bluestar® presumptive method, offering a confirmatory test without prior sample pre-treatment, 2) successful detection of heme from previously washed fabrics, which demonstrated the unprecedented selectivity of the thread spray method, and 3) the ability to analyze samples stored under ambient conditions for up to 30-days. These results attest to the potential capabilities of the thread spray MS platform in forensic serology, and its application for direct analysis of evidentiary garments, which confer the advantages of rapid analysis and the reduction of the false positive and negative identification rates for blood on textiles.
Keywords: Analytical methods, Ambient ionization, Mass spectrometry, Hemoglobin, Bloodstain analysis
Graphical Abstract
INTRODUCTION
Forensic serology has become a vital first step in forensic investigations, particularly when crime scenes involve stains that may be of biological origin such as blood.1 Serological testing begins on site with a presumptive test to determine if the apparent stain is blood,2 and if the bloodstain is of human origin or not. For this reason, several presumptive tests have been developed to help identify the possible presence of blood. New alternative light source (ALS) methods, like the forensic Bluestar® kit, a luminol-based blood identifying agent, have gained popularity due to their rapid analysis time and their ability to not interfere with subsequent DNA analysis.3,4 However, ALS methods still suffer from low accuracy, increasing the risk of false positive results.5 A major challenge faced by crime scene investigators is determining the best approach for analyzing absorbent (fabrics, carpet, and bedding) and non-absorbent materials (painted wood, wallpaper, etc.). For example, analysis of absorbent material such as textiles can result in large false positives due to household detergents that contain alpha-amylase, which can lead to an unwanted chemiluminescent reaction. The wicking of blood in various textiles can also lead to difficulties in interpreting its appearance on fibers. As forensic laboratories become backlogged due to the extensive sample preparation and analysis time needed for DNA extraction,6–8 the need for more accurate and selective serological testing has become critical in forensic investigations.
In recent years, the introduction of ambient ionization techniques has enabled direct mass spectrometric (MS) analyses of multiple types of complex mixtures for forensic applications. In ambient ionization, the sample present on an untreated ambient surface is probed without prior sample preparations. The analysis process typically involves the interaction of the sample surface with high energy particles (e.g., charged liquid droplets,9–11 plasma,12 or laser13) to desorb and transfer the analyte(s) of interest to the mass spectrometer for characterization. This ambient MS methodology has been applied for (i) detection of explosives from conductive14 and non-conductive surfaces,15 (ii) rapid analysis of illicit drugs both in tissues and biofluids,16–18 (iii) finger printing,19 detection of forged documents,20 alongside other counterfeit products,21,22 and (iv) detection of spermicides23 and condom lubricants24 in a sexual assault case. In addition to the absence of sample preparation, the experimental setup for ambient ionization is simple, making it possible to be used for real-time field analyses, especially when coupled to portable mass spectrometers.25,26 Further simplification in instrumentation was recently achieved through the development of substrate-based ambient ionization methods that do not require nebulizer gases and active pumps for the generation of gas-phase ions. These include paper spray,27 thread spray,16,28 membrane spray,29 wood spray,30 and blade spray,31 to name a few. Despite recent advances in the field of substrate-based ambient ionization applications, the direct analysis of fresh blood is still challenging. When analyzing small illicit drugs in whole blood samples, blood must be dried (e.g., 10 μL of blood can take >2 h to dry) to minimize matrix effects. To reduce drying time, Espy et. al. used coagulants to clot the blood immediately upon application providing the ability to analyze blood samples without a need for long wait times.32 Ren et. al. showed that the detection of hemoglobin, (Hb), protein directly from fresh blood was not possible when the ordinary hydrophilic paper substrate was used in paper spray MS.33 Instead, silanized hydrophobic paper was used to decrease protein/paper interactions. Later, Zhang et. al. used Whatman filter paper grade 42, which has much smaller pore size (2.5 μm, compared to the 11 μm pore size for the typical filter paper grade 1 substrate), to achieve Hb detection from diluted human blood detritus (i.e., blood without plasma).34
In this work, we attempt to establish a forensic confirmatory MS-based test for the analysis of whole (fresh) blood samples through the classification of human, canine, and horse blood. The use of thread spray is ideal for whole blood analysis via identification and characterization of Hb for various reasons: (i) matrix effects are inherently minimized by using a single thread instead of the entire whole blood spot. The reduction of analyte volume in turn decreases the amount of blood introduced to the mass spectrometer during the spray process; (ii) high sensitivity – previous studies have confirmed ultra-sensitivity with thread spray MS for tissue-like residues collected by pushing the thread substrate through the tissue once.16 This indicates the feasibility of analyzing smaller/diluted sample volumes by thread spray; (iii) minimally destructive – single thread substrates can be removed from any fabric without complete destruction; and (iv) wide availability - thread is naturally available from a variety domestic materials including carpets, car seat covers, and clothing. We demonstrate not only the direct detection of Hb to determine the presence of blood, but we also show that the lipid composition detected in the positive-ion mode could serve as a means to discriminate between blood of human origin versus canine or horse origins. In addition, we show that samples previously subjected to the commercial Bluestar® presumptive blood test can be directly analyzed and confirmed by our developed thread spray MS-based platform without the need for additional sample pre-treatment.
EXPERIMENTAL SECTION
Thread Spray Ionization Experimental Setup.
The experimental workflow for direct analysis of bloodstained fabrics by thread spray ionization is as illustrated in Figure 1. Blood (10 – 15 μL) was deposited on a piece of fabric (100% cotton) and allowed to soak for 5–10 minutes. A single thread (~0.3 mm OD, 35 mm L) was pulled from the fabric on which the blood sample had been deposited, and inserted into a glass capillary (0.8 mm ID, 1.63 mm OD, and 30 mm long)16,28. On average, a 10 μL blood sample produced a spot size of about 43.5 mm2, and the corresponding pulled thread was found to contain <10% of the total deposited blood (see Figure S1). The assembly of the cellulose thread and glass capillary was positioned 5 mm in front of the mass spectrometer inlet. To initiate thread spray ionization, a 10 – 20 μL aliquot of spray solvent was added to the glass capillary to extract analytes present within the blood sample. The spray solvent utilized was MeOH/H2O (1:1, v/v pH 3) unless otherwise stated. The wetted thread was electrically charged by applying a direct current (DC) high voltage (3 – 5 kV), which resulted in the release of tiny charged droplets that carry the extracted analyte to the mass spectrometer for characterization.
Figure 1.
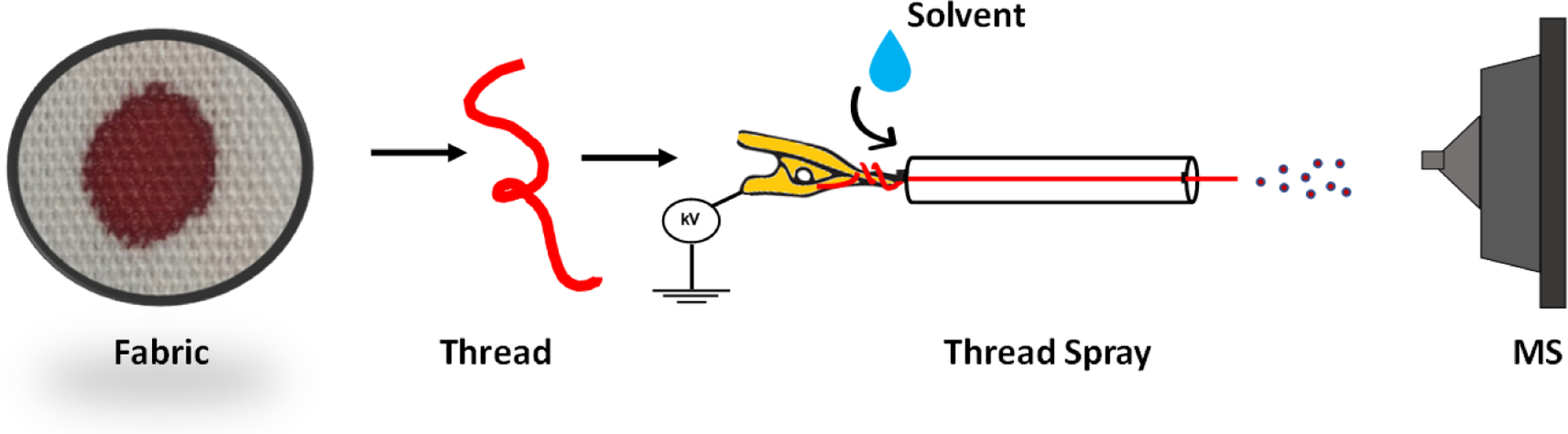
Experimental setup for thread spray mass spectrometry for the analysis of fabrics pre-coated with whole blood sample.
Mass Spectrometry.
Mass spectra were acquired on a Thermo Fisher Scientific Velos Pro LTQ linear ion trap mass spectrometer (San Jose, CA, U.S.A.). The tip of the thread was held and positioned coaxial to the MS inlet via a copper alligator clip, which was connected to an external high-voltage supply. The thread spray ionization method generated ions without gas assistance, and thus, a close interface distance (5 mm) between the tip and the MS inlet was used to optimize signal intensity. The MS parameters used were as follows: 200 °C capillary temperature, 3 microscans, and 60% S-lens voltage. Thermo Fisher Scientific Xcalibur 2.2 SP1 software was applied for MS data collection and processing. Tandem MS with collision-induced dissociation (CID) was utilized for analyte identification. An isolation window of 1.5 Th (m/z units) and a normalized collision energy of 30 – 35% (manufacturer’s unit) were selected for CID experiment.
Scanning Electron Microscopy (SEM) and Energy-Dispersive X-ray Spectroscopy (EDS).
To investigate the surface interactions of the thread surface/fibers containing blood, scanning electron microscopy (SEM) was performed. Sample preparation for SEM and EDS consists of drying blood samples fixed in 2.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4, for a minimum of 72 hours. Samples were then rinsed twice every 5 minutes with 1X PBS and then twice every 5 minutes in nanopure water. Samples were then allowed to dry overnight in ambient air. Upon SEM analysis, samples were sputter-coated with Au for approximately 1 minute. Electron micrographs were recorded using a Quanta 200 Scanning Electron Microscope. EDS samples were prepared similarly to the SEM protocol with a thinner coat of Au (~ 30 seconds). The EDS detector on the Quanta 200 was utilized for surface analysis of cotton fibers containing blood that had previously been analyzed by thread spray. Pure cotton fibers were also analyzed as a control. A beam energy of 20 kV was used. The iron content, as well as other minerals introduced onto the thread surface from processing, were monitored for all samples. The corresponding signals for iron were normalized to oxygen and compared for all samples.
Chemicals and Reagents.
Methanol (99.9%, HPLC grade), acetic acid, cytochrome c, lysozyme, and myoglobin were purchased from Sigma-Aldrich (St. Louis, MO). All protein samples were prepared in 100% 18.2 MΩ water from a Milli-Q water purification system (Millipore, Billerica, MA). 100% Cotton was purchased from JoAnn Fabrics, Columbus, OH) while Thoroughbred II TrafficMaster (synthetic polyester) carpet was purchased from Home Depot (Columbus, Ohio). Kimble 51 expansion borosilicate glass melting point capillaries (O.D. 1.5 mm) were purchased from Kimble Chase (Rockwood, TN). Forensic Bluestar® was purchased from Arrowhead Forensics (Lenexa, KS). Human blood was donated by healthy volunteers with IRB exemption. Remel™ horse defibrinated blood was purchased from Thermo Scientific (Waltham, MA), and beagle whole blood was purchased from BIOIVT (Hicksville, NY).
RESULTS AND DISCUSSION
Spray Solvent Optimization
The use of a thread spray MS method for direct detection of capsaicinoids in pepper spray28 and analysis of illicit drugs in dried blood16 has previously been demonstrated in our group. In the present study, we focused on detecting hemoglobin and lipids from untreated bloodstain samples. We first optimized the process using three standard proteins: myoglobin (MW 16, 700 Da), lysozyme (MW 14, 307 Da), and cytochrome c (MW 12, 384 Da). Figure 2A depicts the positive-ion mass spectrum of cytochrome c prepared in pure water. Narrow charge state distribution (CSD) of +6 to +9 was observed indicating a folded conformation of the protein. A similar CSD was observed when cytochrome c was analyzed from aqueous ammonium acetate solution (100 μM), although the signal intensity reduced slightly in this case (Figure S2a). Likewise, lysozyme was analyzed via thread spray MS using pure water and 100 μM ammonium acetate solutions. Both analyses yielded mass spectra with a charge state distribution of +8 to +10 confirming lysozyme’s native structure (Figures 2B and S2B). The intensity-weighted average charge state (qave; equation 1)34 can also be used to compare the spread in protein CSD induced by different spray conditions to quantify unfolding events.
| Eqn 1 |
where N is the number of detected charge states, qi is the charge of the ith charge state and wi is the intensity of the ith charge state.
Figure 2.

Optimization of thread spray using the analysis of 100 μM aqueous protein solutions deposited on 100 % cotton. Positive-ion mode thread spray mass spectra for (A) cytochrome c (B) lysozyme and (C) cytochrome c at pH 3.0 and (D) myoglobin at pH 3.0. The spray voltage was 4 kV for all thread spray optimization experiments.
The average charge state (qave) for aqueous solutions of cytochrome c and lysozyme were calculated to be 7.3 and 8.3, respectively, again confirming folded states for both proteins. The relative ion signal derived from the neutral aqueous solutions was higher for lysozyme than cytochrome c, which resulted in a more resolved spectrum. This result is likely due to the fact that lysozyme is slightly more basic (pI 11.35)36 than cytochrome c, (pI 10.0)37 which can lead to higher ionization efficiency. Ion signal for cytochrome c was dramatically increased when the pH of the analyte solution was decreased from 7 to 3 (Figure 2C) using acetic acid. A broad charge state distribution of +12 to +22 (qave 15.8) was observed under this acidified thread spray condition. As shown in Figure 2D, abundant ion signal was also detected for thread spray MS analysis of acidified aqueous of myoglobin, yielding a maximum charge state of +22 (qave 14.6). All the multiply charged species were detected in apo form (i.e., heme cofactor lost) indicating that the presence of acid caused myoglobin to unfold during the thread spray MS experiment. These results clearly show that large protein analytes can be detected from hydrophilic cellulose threads and that the process is much more effective under denaturing conditions.
Direct hemoglobin and from untreated whole blood
A number of bloodstain presumptive tests rely on the peroxidase activity of Hb.38 These presumptive blood tests can be beneficial due to their simplicity but they suffer from a lack of specificity, reacting with common household cleaners such as bleach.39 In some scenarios, a test result only needs to determine if the sample in question is blood or not, and in some cases, it is useful to know the specific species involved (e.g., human versus animal origins). We envisioned that specific molecular information derived from MS will provide more discriminatory power when compared to other chemical tests. We opted to detect the hemoglobin protein, heme co-factor, and/or lipid profiles from blood-contaminated fabrics using the thread spray ambient ionization platform. To facilitate hemoglobin detection from untreated blood, the denaturation power of the spray solvent was increased via acidification using acetic acid. Figure 3A was recorded at the onset of the thread spray process, which showed the elution of hemoglobin from thread. Two conformations of the Hb can be observed in Figure 3A, the first centered at m/z 2100 indicating a more folded hemoglobin and the second conformation is found centered at a lower m/z region signifying a more unfolded protein. The presence of hemoglobin was further confirmed by identifying both the α-chain (MW 15154.4 ± 0.2 Da) and β-chain (MW 15953 ± 0.5 Da) monomeric peaks of hemoglobin. After about 1.5 min of continuous spray, the increased contact of the sample with the acidified solution containing high organic content caused hemoglobin to unfold to expose the non-covalently bound iron containing heme co-factor. This resulted in the detection of the diagnostic heme group in high abundance at m/z 616 (Figure 3B). The reduction of Hb signal (relative to heme) after 1.5 min of thread spray is not surprising given that the presence of high content of methanol in the spray solvent can cause protein precipitation, which subsequently hinders its detection.40 This is a desirable phenomenon because after we obtain data on Hb (at the start of the spray), the precipitated proteins allow the detection of smaller organic analytes such as heme and lipids. In particular, lipid analysis from biological samples often use MeOH/H2O solvent systems explaining the emergence of other ions in the m/z region of 700–900, which we attribute to lipids belonging to the phosphatidylcholine (PC) class, commonly present in blood.33
Figure 3.
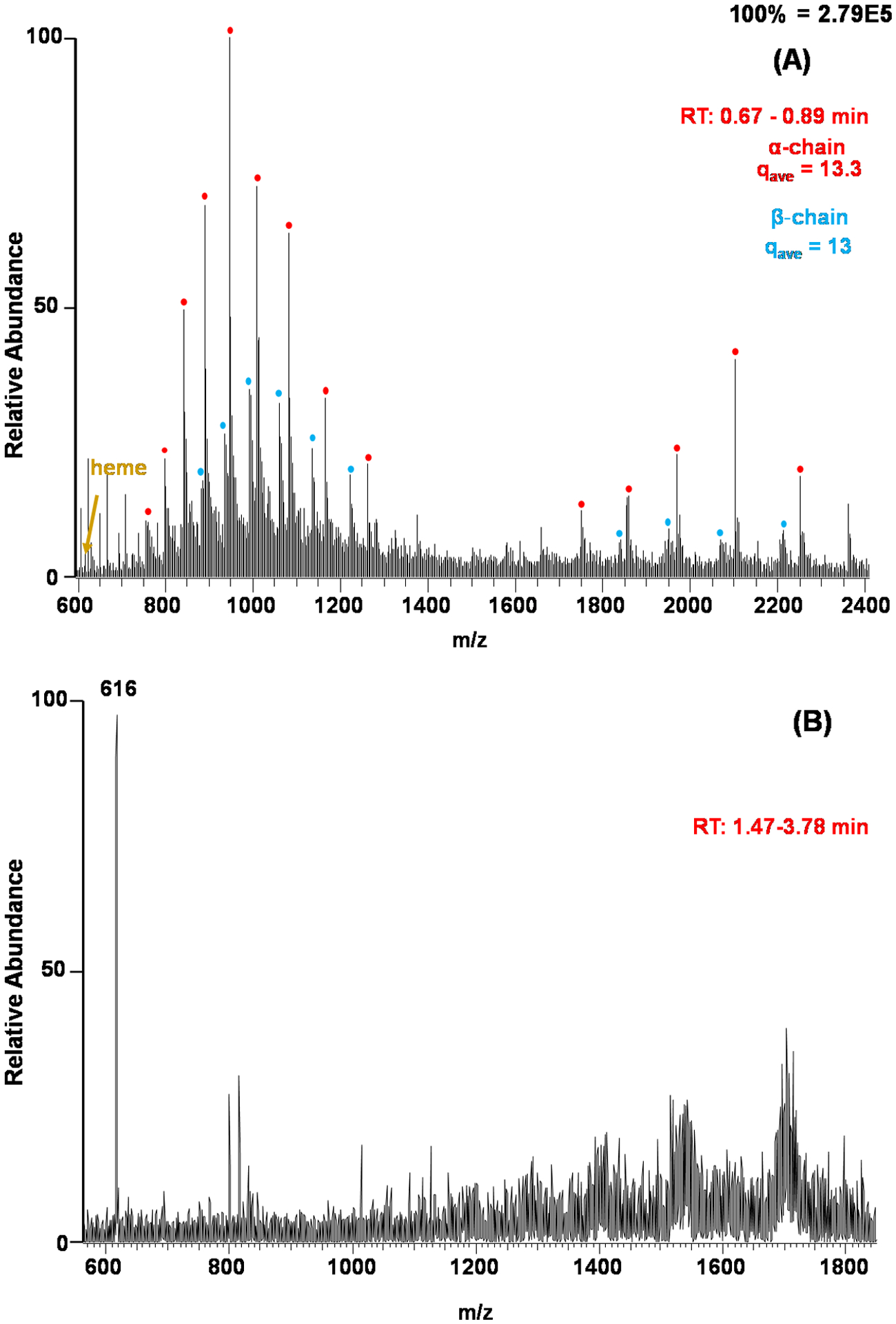
Full mass spectra of hemoglobin found in fresh blood on 100% cotton thread fibers recorded (A) at the onset of the spray within 0.67 – 0.89 min and (B) after 1.47 min of spray time. The spray solvent was a MeOH/H2O, 1:1, solution with 5% acetic acid (pH 3). Hemoglobin α-chain = 15154.4 ± 1 Da and β-chain = 15871.5 ± 1 Da were detected in several charge states.
The detection of lipids was interesting, so we further optimized the thread spray MS method to differentiate between animal and human blood. Although all mammalian blood contains hemoglobin, lipid profiles between humans and most animals are slightly different due to minor differences in blood composition.41–43 For example, an extensive lipid profile of nine animal species including dog, cat, pig, horse, bovine, hamster, mouse, rat and non-human primate along with human showed three separated groups of similarities: i) dog and cat, ii) pig, non-human primate and rat, and iii) mouse, hamster and human.41 We elected to analyze blood samples from two domestic animals (canine and horse) and to compare their lipid profiles to that of human blood. We anticipated this experiment to add an extra layer of confidence by enabling confirmatory forensic analysis where animal blood and human blood are concerned. Typical positive-ion mode mass spectra recorded from direct analysis of human, canine, and horse blood samples are provided in Figure 4. The lipid profile from human blood showed three different classes of lipids: phosphatidylcholine [PC], phosphatidylethanolamine [PE], and sphingomyelin [SM], which were confirmed in MS/MS experiments (Figure S3). Principal component analysis (PCA) of the spectra recorded showed a clear distinction between human blood samples from that of horse and canine blood (Figure S4). The lipid composition detected by thread spray MS is consistent with several studies performed on healthy human biofluids and tissues using paper spray.44,45
Figure 4.
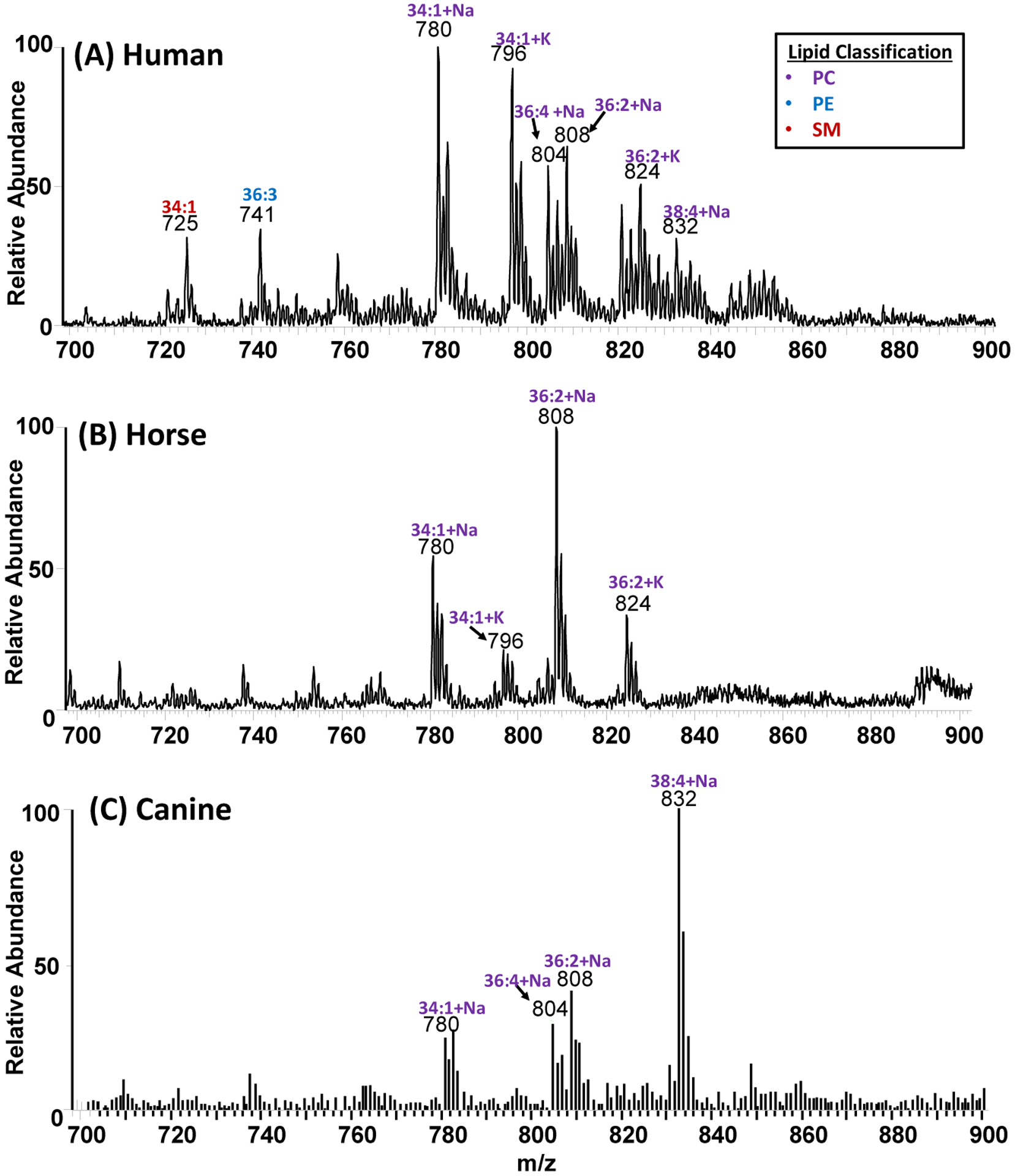
Lipid profiles of phosphatidylcholine (PC), phosphatidylethanolamine (PE), and sphingomyelin (SM) lipids recorded using positive-ion mode thread spray MS for A) human B) horse C) canine. Blood samples (5 μL) were deposited on cotton thread and analyzed directly without pre-treatment
The thread spray ambient ionization method has shown high efficiency toward ionization of different analyte types (i.e., proteins, heme, and lipids). This is an advantage over paper spray, which works well for small organic compounds. For thread spray, the natural sharp tips presented by tiny sub-fibers facilitate analyte extraction for direct analysis. These sub-fibers induce weaker binding of the protein to the thread substrate due to their roughness and shorter diffusion distance as a result of their complete immersion in the spray solvent, allowing efficient transport to the mass spectrometer. The sub-fibers in a single thread are unidirectional, which facilitates close packing and hence less free pores within the thread substrate (Figure S5a). When whole blood is applied to a single cotton thread, it largely remained at the thread surface (due to blood coagulation), but some blood was found at the interstitial spaces between the sub-fibers (Figure S5b) as well. These observations imply that blood residues present on thread are easily accessible and can be extracted more readily by the optimized spray solvent compared with the highly porous paper substrate used in paper spray.
Forensic perspective and comparison with Bluestar® blood test
The Bluestar® presumptive blood tests exhibit a chemiluminescence signal when reacted with iron in hemoglobin, thereby indicating the possible presence of blood. However, the intensity and duration of illumination are time sensitive and may give rise to false positives. In addition, it is widely known that during the textile manufacturing process, the textile (e.g., cotton, polyester, and nylon) is often exposed to varying amounts of metals such as iron, (Fe).46 A possible method to confirm that the iron content on the fabric is due to the presence of hemoglobin is through energy dispersive X-ray spectroscopy (EDS).47 We performed EDS microanalysis on thread fibers in order to characterize the elemental composition of blank untreated cotton compared to cotton containing bloodstain. Indeed, the results (Table S1 and Figure S6) showed a marked increase in Fe content in the presence of blood. However, the Fe signal decreased significantly when a week-old blood sample was analyzed indicating limited confirmatory abilities for EDS on stored samples.
The main advantage of this thread spray MS method is its multiplicity. As already explained, the doublet signal derived from α- and β-chains is a strong indicator for the presence of hemoglobin. The heme co-factor can also be detected with the Fe intact, further confirming the presence of blood. In collisionally activated dissociation (MS/MS) experiments, the heme group at m/z 616 dissociates to give a fragment ion at m/z 557 (Figure 5A). Using similar MS/MS experiments, we were able to successfully re-analyze heme from a previously analyzed cotton thread (Figure S7b). The thread spray MS approach is also applicable to other commonly encountered materials like cotton/polyester and synthetic polyester blend carpet (Figure S7). This confirms the compatibility of thread spray with various textiles. The diameter of thread substrates removed from these textiles may be different, but electrospray occurs from sub-fibers28 indicating that the actual physical dimensions of the threads within distinct fabrics will not limit analyte detection. The main factor to consider when utilizing different thread types is the solvent composition used during the extraction and analysis processes. Different types of analyte-thread intermolecular interactions are easily overcome through solvent optimization.28 From this observation, we anticipate that this thread spray method can be extended beyond the fabrics tested here (i.e., 100% cotton, cotton/polyester and synthetic polyester blends) to include other commonly encountered natural and man-made fabrics.
Figure 5.
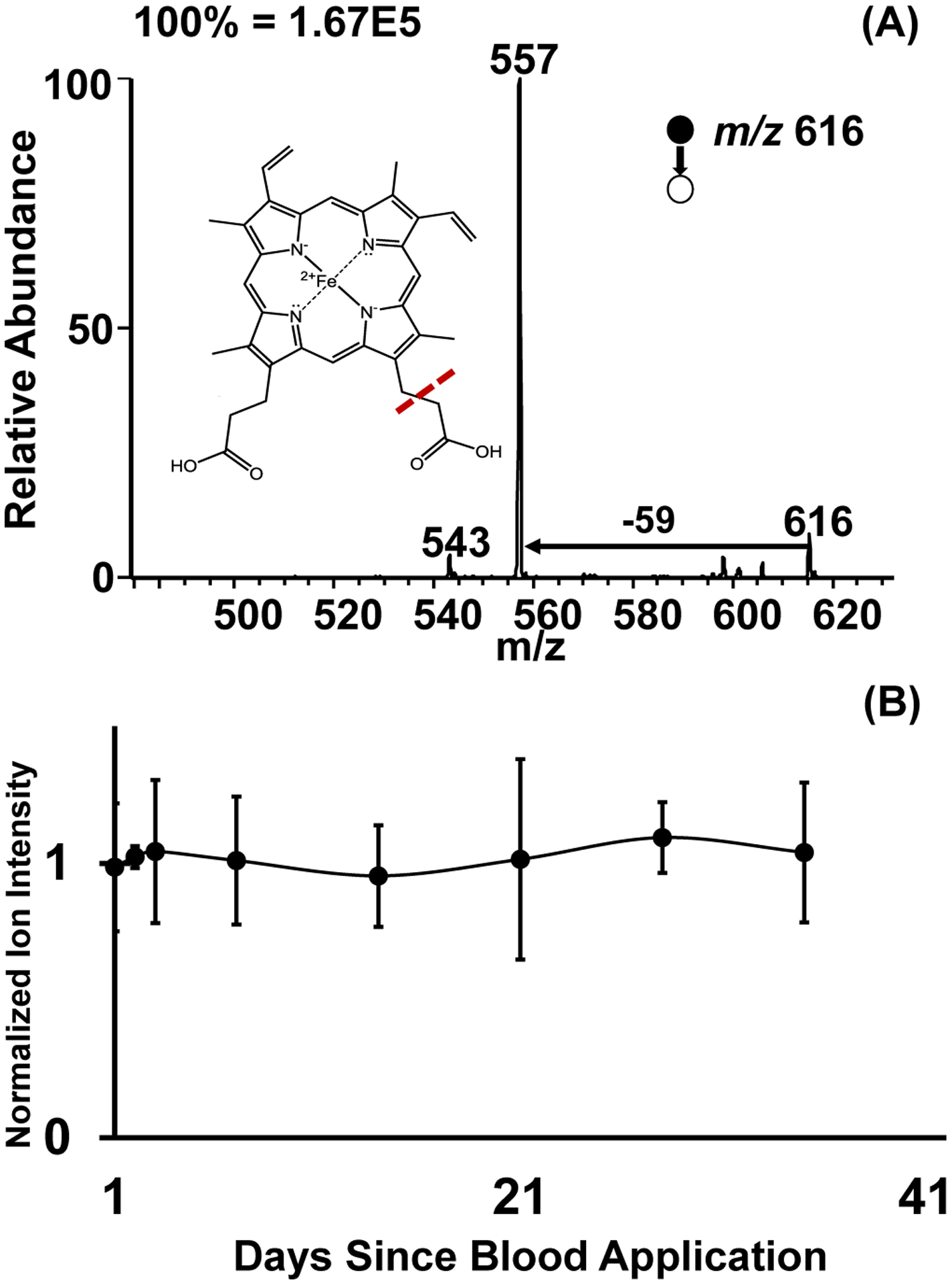
(A) Thread spray MS/MS fragmentation of the heme group (m/z 616) in positive mode and (B) Thread spray whole blood stability analysis utilizing a fragment peak (m/z 557) as an indicator of blood on cotton fabric normalized with fresh whole blood.
Most importantly, we investigated the possibility of detecting heme from old blood samples. Here, we monitored dried blood present on 100% cotton thread fibers stored under ambient conditions for 35 days. After each week, the stored threads were analyzed by the thread spray MS. Ion signal from heme in the stored samples were normalized against signal from fresh blood applied on 100% cotton thread. We used methanol containing 5% acetic acid as the spray solvent. The results from this storage experiment are summarized in Figure 5b, which shows that a stable signal from heme can be recorded in MS/MS analysis mode over a one-month period. This shows an advantage over EDS analysis, which showed markedly reduced Fe signal over a seven-day period, due to its inability to generate data from the top few microns of SEM specimens and limiting analysis of heterogeneous materials like thread. The analysis of heme provides a useful alternative to detect the presence of blood since studies have shown the instability of proteins in whole blood when stored at room temperature in the cellulose matrix,48 due potentially due to temperature and humidity effects. These results establish heme as a stable analyte at room temperature.
Therefore, using the heme signal, the potential of employing the thread spray approach in conjunction with the Bluestar® presumptive blood test was investigated. Two scenarios were considered: in the first, blood was applied to a fabric and first analyzed with the Bluestar® followed by the thread spray method, and vice versa (Figure S8). Result of these experiments showed that the presence of blood can be detected on a single fiber using thread spray irrespective of whether Bluestar® analysis was performed first (Table S2). Perhaps a more challenging scenario would be the case where an assailant(s) had washed textiles/fabrics prior to forensic analysis. To investigate the viability of thread spray MS as a suitable sensitive forensic method, we simulated this scenario: a piece of fabric containing whole blood was machine washed, after which a single thread was pulled from the cleaned fabric. Interestingly, thread spray MS analysis of the washed cloth showed traces of heme on the evidentiary garment containing both human and canine blood (Figure S9, Table S3–S4). Similarly, the Bluestar® presumptive test was performed to detect the presence Fe from both human and canine bloods in the washed fabric. The chemiluminescence signal was found diffused throughout the fabric and not localized in the region of the bloodstain (Figure S9). Such a result leads to ambiguity in interpretation since the observed signal can originate from traces of bleach from the soap used for washing or a diffused Fe concentration. Clearly, the molecular information derived from the thread spray experiment is more advantageous in reducing false positive result. In the case of no visual bloodstain evidence, the thread spray method can be combined with other forensic data to reconstruct potential blood spatter scenes. Since threads can be pulled from evidentiary garments, the thread spray MS approach will allow investigators to get a comprehensive view of the surface with ease without analyzing the entire garment, which could be laborious, destructive, and time consuming. The ability to perform this experiment without sample preparation and within a short amount of time (<1 min) add extra flexibility to the overall forensic detection procedure. The observed selectivity is also expected to reduce false negatives from cleaning solutions, cosmetics, and certain vegetables.
CONCLUSION
Thread spray ionization can play a pivotal role in the field of forensic science through the sensitive detection and identification of blood. We have demonstrated thread spray to be an effective method for sampling large proteins and smaller organic compounds (e.g., lipids and heme) through substrate-based ambient ionization when using denaturing spray solvents. The simple sample preparation workflow, high selectivity and sensitivity, and mass accuracy give it an advantage over other presumptive tests that are based solely on the detection of a luminescent signal. Moreover, through its ability to provide information on smaller endogenous compounds like heme and lipids, we envision that this platform can be coupled to portable MS to allow on-site analysis of blood samples, thus eliminating the need for storage and sample contamination during transportation. With bloodstain absorbed into the fibrous material, the use of other (traditional) ambient ionization methods would require sample collection/processing before analysis. Thread spray analysis requires no further sample processing thus evidentiary fibers can be analyzed directly by mass spectrometry. The ease with which proteins can be detected from the thread spray method is unique among other ambient ionization methods such as direct analysis in real time and desorption electrospray ionization. One possible disadvantage of thread spray is that optimization of spray solvent composition is needed to overcome analyte-fiber interactions to enable subsequent ionization of the desorbed analyte. However, we believe other ambient ionization methods may suffer similar challenges in analyte desorption. Combining thread spray with other commercially available forensic presumptive tests reduces the possibility of false positives and adds a layer of molecular specificity. This study sheds new insights on forensic serology and demonstrates that thread spray has high potential for rapid forensic analysis of untreated whole blood samples in various absorbent materials.
Supplementary Material
Acknowledgements
This research was supported by the National Institute of Allergy and Infectious Diseases (Award Number R01-AI-143809). The authors would like to acknowledge the Center of Electron Microscopy and Analysis (CEMAS) at The Ohio State University for their help in the utilization of the electron microscope.
Footnotes
Conflicts of interest
There are no conflicts to declare.
References:
- 1.Virkler K and Lednev IK, Forensic Science International, 2009, 188, 1–17. [DOI] [PubMed] [Google Scholar]
- 2.Harbison S and Fleming R, RRFMS, 2016, 11. [Google Scholar]
- 3.Blum LJ, EsperanÇA P and Rocquefelte S, Canadian Society of Forensic Science Journal, 2006, 39, 81–99. [Google Scholar]
- 4.Vineyard AR, Hazelrigg EJ, Ehrhardt CJ and Connon CC, Journal of Forensic Sciences, 2019, 64, 878–887. [DOI] [PubMed] [Google Scholar]
- 5.Castelló A, Alvarez M and Verdú F, Canadian Society of Forensic Science Journal, 2002, 35, 113–121. [Google Scholar]
- 6.Morato NM, Pirro V, Fedick PW and Cooks RG, Anal. Chem, 2019, 91, 7450–7457. [DOI] [PubMed] [Google Scholar]
- 7.Durose M, 2014, 12. [PubMed]
- 8.Rentsch KM, TrAC Trends in Analytical Chemistry, 2016, 84, 88–93. [Google Scholar]
- 9.Frey BS, Damon DE and Badu‐Tawiah AK, Mass Spec Rev, 2019, mas.21601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swiner DJ, Jackson S, Burris BJ and Badu-Tawiah AK, Anal. Chem, 2020, 92, 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takats Z, Science, 2004, 306, 471–473. [DOI] [PubMed] [Google Scholar]
- 12.Cody RB, Laramée JA and Durst HD, Anal. Chem, 2005, 77, 2297–2302. [DOI] [PubMed] [Google Scholar]
- 13.Nemes P and Vertes A, Anal. Chem, 2007, 79, 8098–8106. [DOI] [PubMed] [Google Scholar]
- 14.Cotte-Rodríguez I, Takáts Z, Talaty N, Chen H and Cooks RG, Anal. Chem, 2005, 77, 6755–6764. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Ma X, Zhang S, Yang C, Ouyang Z and Zhang X, Analyst, 2009, 134, 176–181. [DOI] [PubMed] [Google Scholar]
- 16.Swiner DJ, Jackson S, Durisek GR, Walsh BK, Kouatli Y and Badu-Tawiah AK, Analytica Chimica Acta, 2019, 1082, 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberlin LS, Ferreira CR, Dill AL, Ifa DR and Cooks RG, Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids, 2011, 1811, 946–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saha S, Mandal MK and Hiraoka K, Anal. Methods, 2013, 5, 4731. [Google Scholar]
- 19.Ifa DR, Manicke NE, Dill AL and Cooks RG, Science, 2008, 321, 805–805. [DOI] [PubMed] [Google Scholar]
- 20.Ifa DR, Gumaelius LM, Eberlin LS, Manicke NE and Cooks RG, Analyst, 2007, 132, 461. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt EM, Franco MF, Regino KG, Lehmann EL, Arruda MAZ, de Carvalho Rocha WF, Borges R, de Souza W, Eberlin MN and Correa DN, Science & Justice, 2014, 54, 459–464. [DOI] [PubMed] [Google Scholar]
- 22.Eberlin LS, Haddad R, Sarabia Neto RC, Cosso RG, Maia DRJ, Maldaner AO, Zacca JJ, Sanvido GB, Romão W, Vaz BG, Ifa DR, Dill A, Cooks RG and Eberlin MN, Analyst, 2010, 135, 2533. [DOI] [PubMed] [Google Scholar]
- 23.Coon AM, Beyramysoltan S and Musah RA, Talanta, 2019, 194, 563–575. [DOI] [PubMed] [Google Scholar]
- 24.Musah RA, Cody RB, Dane AJ, Vuong AL and Shepard JRE, Rapid Communications in Mass Spectrometry, 2012, 26, 1039–1046. [DOI] [PubMed] [Google Scholar]
- 25.Lawton ZE, Traub A, Fatigante WL, Mancias J, O’Leary AE, Hall SE, Wieland JR, Oberacher H, Gizzi MC and Mulligan CC, J. Am. Soc. Mass Spectrom, 2017, 28, 1048–1059. [DOI] [PubMed] [Google Scholar]
- 26.Fatigante WL, Mukta S, Lawton ZE, Bruno AM, Traub A, Gasa AJ, Stelmack AR, Wilson-Frank CR and Mulligan CC, J. Am. Soc. Mass Spectrom, 2020, 31, 336–346. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Liu J, Cooks RG and Ouyang Z, Angewandte Chemie International Edition, 2010, 49, 877–880. [DOI] [PubMed] [Google Scholar]
- 28.Jackson S, Swiner DJ, Capone PC and Badu-Tawiah AK, Analytica Chimica Acta, 2018, 1023, 81–88. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Lin F, Xu J and Xu W, Anal. Chem, 2015, 87, 3123–3128. [DOI] [PubMed] [Google Scholar]
- 30.Deng J, Yu T, Yao Y, Peng Q, Luo L, Chen B, Wang X, Yang Y and Luan T, Analytica Chimica Acta, 2017, 954, 52–59. [DOI] [PubMed] [Google Scholar]
- 31.Gómez-Ríos GA and Pawliszyn J, Angew. Chem. Int. Ed, 2014, 53, 14503–14507. [DOI] [PubMed] [Google Scholar]
- 32.Espy RD, Manicke NE, Ouyang Z and Cooks RG, Analyst, 2012, 137, 2344–2349. [DOI] [PubMed] [Google Scholar]
- 33.Ren Y, Wang H, Liu J, Zhang Z, McLuckey MN and Ouyang Z, Chromatographia, 2013, 76, 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Ju Y, Huang C and Wysocki VH, Anal. Chem, 2014, 86, 1342–1346. [DOI] [PubMed] [Google Scholar]
- 35.Simmons DA and Konermann L, Biochemistry, 2002, 41, 1906–1914. [DOI] [PubMed] [Google Scholar]
- 36.Wetter LR and Deutsch HF, J. Biol. Chem, 1951, 192, 237–242. [PubMed] [Google Scholar]
- 37.Malmgren L, Olsson Y, Olsson T and Kristensson K, Brain Research, 1978, 153, 477–493. [DOI] [PubMed] [Google Scholar]
- 38.Butler J, Chaseling J and Wright K, Journal of Forensic Sciences, 2019, 64, 1838–1843. [DOI] [PubMed] [Google Scholar]
- 39.Castelló A, Francés F and Verdú F, Talanta, 2009, 77, 1555–1557. [DOI] [PubMed] [Google Scholar]
- 40.Dai M and Huang G, Rapid Commun Mass Spectrom, , DOI: 10.1002/rcm.8759. [DOI] [PubMed] [Google Scholar]
- 41.Kaabia Z, Poirier J, Moughaizel M, Aguesse A, Billon-Crossouard S, Fall F, Durand M, Dagher E, Krempf M and Croyal M, Sci Rep, 2018, 8, 15893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doty KC and Lednev IK, Forensic Science International, 2018, 282, 204–210. [DOI] [PubMed] [Google Scholar]
- 43.McLaughlin G, Doty KC and Lednev IK, Forensic Science International, 2014, 238, 91–95. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Manicke NE, Yang Q, Zheng L, Shi R, Cooks RG and Ouyang Z, Anal. Chem, 2011, 83, 1197–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu H, Zhang H, Chingin K, Wei Y, Xu J, Ke M, Huang K, Feng S and Chen H, Anal. Chem, 2019, 91, 10532–10540. [DOI] [PubMed] [Google Scholar]
- 46.Sungur Ş and Gülmez F, Journal of Spectroscopy, 2015, 2015, 1–5. [Google Scholar]
- 47.Scimeca M, Bischetti S, Lamsira HK, Bonfiglio R and Bonanno E, Eur J Histochem, , DOI: 10.4081/ejh.2018.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellervik C and Vaught J, Clinical Chemistry, 2015, 61, 914–934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


