Significance
Persisters represent a bacterial subpopulation that survive high antibiotic concentrations without being resistant. Their role in clinics in persistent infections and their molecular and functional landscape is not fully established. Staphylococcus aureus is a pathobiont that causes severe invasive infections often difficult to treat. Here, we assessed S. aureus recovered directly from persistent infections and show that host-mediated stress and antibiotic exposure promoted persister formation. Using a multiomics approach and enrichment of persisters, we were able to draw a molecular atlas of persisters, to correlate accumulation of insoluble proteins and ATP depletion with dormancy and persistence. Our results give insights into the molecular profile of bacterial persisters and provide a guide for therapy optimization for persistent S. aureus infections.
Keywords: Staphylococcus aureus, antibiotic persistence, persisters, antimicrobial therapy, persistent infection
Abstract
Staphylococcus aureus causes invasive infections and easily acquires antibiotic resistance. Even antibiotic-susceptible S. aureus can survive antibiotic therapy and persist, requiring prolonged treatment and surgical interventions. These so-called persisters display an arrested-growth phenotype, tolerate high antibiotic concentrations, and are associated with chronic and recurrent infections. To characterize these persisters, we assessed S. aureus recovered directly from a patient suffering from a persistent infection. We show that host-mediated stress, including acidic pH, abscess environment, and antibiotic exposure promoted persister formation in vitro and in vivo. Multiomics analysis identified molecular changes in S. aureus in response to acid stress leading to an overall virulent population. However, further analysis of a persister-enriched population revealed major molecular reprogramming in persisters, including down-regulation of virulence and cell division and up-regulation of ribosomal proteins, nucleotide-, and amino acid-metabolic pathways, suggesting their requirement to fuel and maintain the persister phenotype and highlighting that persisters are not completely metabolically inactive. Additionally, decreased aconitase activity and ATP levels and accumulation of insoluble proteins involved in transcription, translation, and energy production correlated with persistence in S. aureus, underpinning the molecular mechanisms that drive the persister phenotype. Upon regrowth, these persisters regained their virulence potential and metabolically active phenotype, including reduction of insoluble proteins, exhibiting a reversible state, crucial for recurrent infections. We further show that a targeted antipersister combination therapy using retinoid derivatives and antibiotics significantly reduced lag-phase heterogeneity and persisters in a murine infection model. Our results provide molecular insights into persisters and help explain why persistent S. aureus infections are so difficult to treat.
Staphylococcus aureus is a human pathobiont that intermittently or permanently colonizes ∼50% of the human population. Colonization increases the risk of staphylococcal infections (1), ranging from skin and soft-tissue infections to life-threatening infections, including bacteraemia and endocarditis (2). Chronic S. aureus infections require prolonged antibiotic treatment and extended hospitalization, thereby increasing healthcare-associated costs (3, 4).
Grown immediately after isolation from infection sites, S. aureus can display colony radius heterogeneity also known as small colony variants (SCVs) (5, 6). Most small colonies revert to a normal colony size when cultured under laboratory conditions with no apparent genetic determinant and are here referred to as nonstable small colonies (nsSCs) (7). Factors, such as acidic pH, antibiotic exposure, and an intracellular or abscess milieu, can exacerbate colony radius heterogeneity on agar plates (6–9).
We recently demonstrated that nsSCs result from a prolonged bacterial lag-phase and represent a subpopulation of persisters (6). Persisters are nongrowing bacteria that are able to survive high concentrations of antibiotics, to which they remain nonetheless fully susceptible (10–13). Besides spontaneous occurrence of a small fraction of persisters, many factors such as acidic pH, starvation, intracellular milieu, and drugs can trigger antibiotic persistence, e.g., by inducing tolerance by lag (10, 14–17). In patients, these nondividing bacteria may subsequently resume growth and cause recurrent infections (18). Antibiotic persistence can moreover facilitate the evolution of drug resistance, rendering elimination of persisters paramount (19–21).
Most research on antibiotic persistence has been carried out in vitro in nutrient-rich media, which poorly mimics the host environment. Moreover, domesticated laboratory strains rather than freshly isolated clinical strains (22) were used and only a few studies have investigated bacterial persistence in the clinical setting (20, 23, 24).
Given the increasing prevalence of antibiotic resistance and the high incidence of relapsing and persistent infections worldwide (10, 25), we investigated the molecular basis of phenotypic heterogeneity underlying antibiotic persistence and host–pathogen interactions using S. aureus patient-derived isolates. We used multiomics analyses and phenotypic profiling to generate an atlas of molecular changes in an acid-stress-exposed and in a persister-enriched S. aureus population. Additionally, we determined potential dormancy characteristics like adenosine triphosphate (ATP) levels, aconitase activity, and accumulation of insoluble proteins in a persister-enriched S. aureus population. To assess treatment optimization for persistent S. aureus infections, we tested the efficiency of retinoid derivatives in antibiotic combination therapy regimens.
Results
Clinical Case.
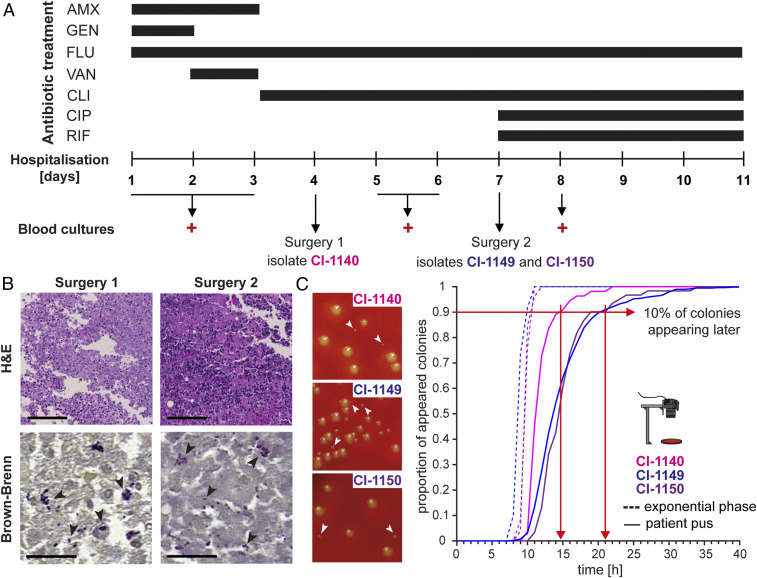
A 65-y-old previously healthy male presented to the emergency room with back pain. Clinical examination showed no sensorimotor deficits. Inflammatory markers were normal, and he was treated with analgesics and physiotherapy. However, the patient developed dysarthria and cranial computed tomography revealed a subacute cerebral ischemia in the right parietal lobe. Due to neurological deterioration and multiorgan failure, the patient was transferred to the intensive care unit. Vegetation on the mitral valve consistent with native valve endocarditis was detected. An empirical antimicrobial therapy with amoxicillin (AMX), flucloxacillin (FLU), and gentamicin (GEN) was initiated (Fig. 1A). Treatment was switched to FLU and vancomycin (VAN) and then narrowed to FLU after the isolate was confirmed as methicillin-sensitive S. aureus (MSSA). Blood tests showed elevated levels of C-reactive protein (CRP), white blood cells, and neutrophils (SI Appendix, Fig. S1A and Table S1). Radiological evaluation revealed an epidural and several prevertebral abscesses (SI Appendix, Fig. S1B). Blood cultures remained positive. Thus, clindamycin (CLI) was added to the antibiotic regimen and 4 and 7 days after therapy onset the large epidural abscess was evacuated (Fig. 1A). After the second surgery, the antibiotic regimen was escalated to include ciprofloxacin (CIP) and rifampicin (RIF) (Fig. 1A). Tissue histology showed immune cell infiltration (Fig. 1B, top panel) and confirmed the presence of gram-positive cocci (Fig. 1B, bottom panel). Despite extensive debridement and targeted antimicrobial treatment, the patient died 11 days after hospitalization (Fig. 1A).
Fig. 1.
Clinical course, treatment, and lag-time analysis of S. aureus derived directly from a difficult-to-treat infection. (A) Overview of the clinical course, antibiotic treatment regimen, and surgical interventions. Positive blood cultures and isolated S. aureus strains are indicated. (B) Histological staining of patient tissue samples. Hematoxylin and eosin (H&E) staining (top panel) shows immune cell infiltration; Brown-Brenn staining (bottom panel) visualizes bacteria. (Scale bar for H&E, 100 µm; for Brown-Brenn, 20 µm.) (C) Representative images of colony radius heterogeneity of bacteria plated immediately after surgery (left panel). Proportion of appeared colonies over time, when plating bacteria directly from patient pus (continuous line) and upon recultivation in nutrient-rich medium to exponential phase (segmented line) (right panel). Red arrows indicate time point at which 90% of the colonies appeared.
Growth-Resumption Delays Reflect a Nondividing Subpopulation.
We received the pus samples immediately after evacuation of the abscesses on days 4 and 7. These samples were directly processed and plated onto agar plates. Automated agar-plate imaging was used to analyze colony growth dynamics and to determine bacterial lag times (6, 26). We observed a broad heterogeneity in colony radii at 24 h for bacteria isolated during both first (clinical isolate [CI]-1140) and second surgery (CI-1149 and CI-1150) (Fig. 1C, left panel). Additionally, we found a substantial shift toward prolonged lag times in bacteria cultured directly from the patient’s pus (10% of the colonies appearing after 14 h for CI-1140 and 21 h for CI-1149 and CI-1150) compared with the same bacteria grown exponentially in nutrient-rich medium (100% of the colonies were already present after 10 h) (Fig. 1C, right panel and SI Appendix, Fig. S1C), highlighting that the environment within the host including antibiotic treatment affects bacterial heterogeneity and can lead to prolonged bacterial lag times, indicating a substantial persister subpopulation.
Genomic Characterization of S. aureus Clinical Isolates.
Multilocus sequence type analysis identified the three isolates as sequence type 8 (ST8). To investigate the possibility of in host evolution over the short infection period, we performed whole genome sequencing of all clinical isolates (CI-1140, CI-1149, and CI-1150) (27). Variant calling and accessory gene content analysis indicated that the three clinical isolates were genetically identical and that the detected lag phenotypes were triggered by the in host environment and were not based on genetic differences (SI Appendix, Fig. S2A). Thus, we used isolate CI-1149 for further experiments. The S. aureus ST8 lineage is globally disseminated and includes the hypervirulent USA300 methicillin-resistant clone. Phylogenetic comparison of the clinical isolate CI-1149 with the laboratory strains Cowan I, USA300_JE2, and USA300_11358 and the reference strain USA300_FPR3757 (28) confirmed closer relatedness of CI-1149 to the USA300 clone compared with Cowan I. However, over 1,000 single nucleotide polymorphisms (SNPs) were found to distinguish CI-1149 from USA300_JE2 (SI Appendix, Fig. S2A), and comparative genomics revealed the absence of characteristic USA300 elements in CI-1149—the staphylococcal cassette chromosome mec (SCCmec), the arginine catabolic mobile element (ACME), and the Panton-Valentine leucocidin (PVL) genes lukF and lukS (SI Appendix, Fig. S2 B and C). This PVL-negative profile is typical for ST8 strains circulating in Europe (29).
To explore the ability of drug-sensitive strains to survive antibiotic challenge as an underlying cause of relapsing infections in general, we included three additional MSSA isolates derived from other difficult-to-treat S. aureus infections with ongoing bacteraemia (SI Appendix, Table S1). These additional MSSA isolates were of different genetic backgrounds (ST398: CI-2015 and CI-2857 and ST45: CI-2134) (SI Appendix, Table S2). While virulence profiling of the clinical isolate CI-1149 showed high similarity to the USA300 strains (SI Appendix, Fig. S2C), the other clinical isolates (CI-2015, CI-2857, and CI-2134) were lacking lukE/D as well as lukS/F-PV and serine protease-like proteins (splA/B/C/D/E/F).
Phenotypic Plasticity in Response to Stress Contributes to Antibiotic Persistence.
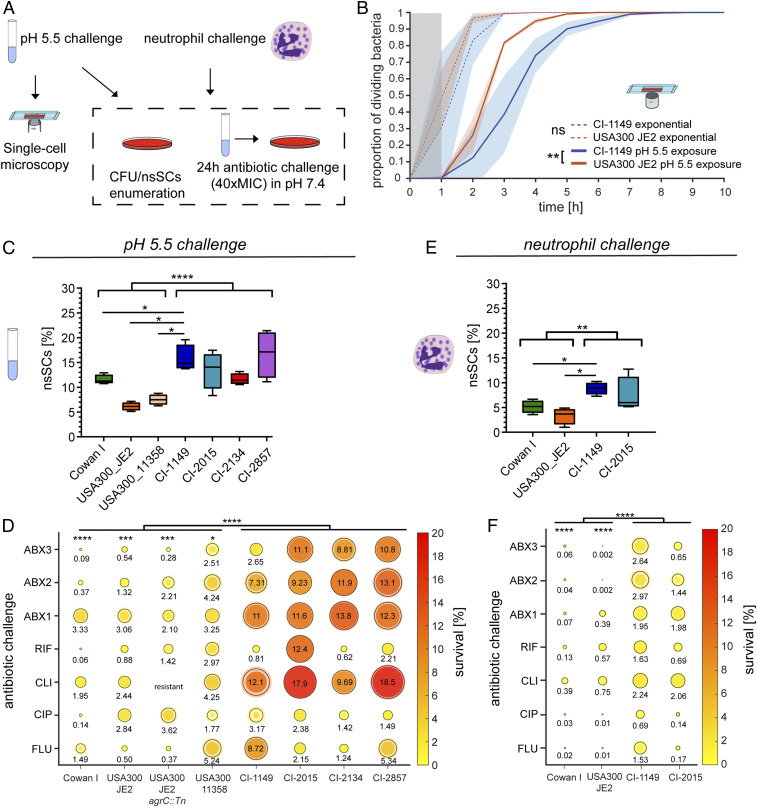
We mimicked abscess conditions by exposing bacteria to acidic pH or neutrophils and identified a subpopulation of dormant cells that showed an increased lag time, giving rise to nsSCs on agar plates (Fig. 2A). To verify that colony appearance time reflected the single bacterial cells’ lag time, we monitored time to first cell division after change to favorable growth conditions using time-lapse microscopy. The single bacterial cells’ lag time was assessed for both acidic pH-exposed and exponential growth-phase bacteria. No significant differences in time to first cell division in nutrient-rich medium were found between CI-1149 and the ST8 strain USA300_JE2 in exponential growth phase. In contrast, upon acidic pH exposure, CI-1149 showed an increased lag time of up to 7 h, compared with 5 h for USA300_JE2 (Fig. 2B). Congruently, CI-1149 was associated with higher proportions of nsSCs (two- to threefold) than the laboratory strains USA300_JE2, USA300_11358, and Cowan I after exposure to acidic pH or neutrophils. Overall, strains isolated from persistent infections (CI-1149, CI-2015, CI-2134, and CI-2857) showed higher levels of nsSCs (two- to threefold) than the reference strains, reflecting a higher proportion of dormant bacterial cells (Fig. 2 C and E).
Fig. 2.
Acidic pH and neutrophil exposure increase bacterial lag time and result in antibiotic persistence. (A) Scheme of experimental procedure to determine CFUs, nsSCs, and bacterial single-cell lag times. (B) Single-cell microscopy of bacteria growing in exponential phase in nutrient-rich medium as well as after 3 d of pH 5.5 exposure. N = 3 biological replicates. Shaded areas depict the SD. USA300_JE2 WT exponential n = 660 bacterial cells, USA300_JE2 pH 5.5 n = 662, CI-1149 exponential n = 500, and CI-1149 pH 5.5 n = 335; gray zone marks the period at the beginning of the experiment, where cell divisions could occur, but would not be observed. Statistical significance was determined at 80% growing bacteria. (C) Proportion of nsSCs in clinical S. aureus strains and laboratory strains after 3 d of pH 5.5 exposure. N = 4 biological replicates. (D) Antibiotic challenge survival assay of the isolate CI-1149, laboratory strains and other clinical isolates. Following 3 d of pH 5.5 exposure, bacterial survival in FLU, CLI, CIP, RIF, or the combinations FLU+CLI (ABX1), FLU+CLI+CIP (ABX2), and FLU+CLI+CIP+RIF (ABX3) at 40× MIC in pH 7.4 was analyzed after 24 h and calculated relative to the initial inoculum. Shaded halos show standard error of mean (SEM) of four biological replicates. (E) Proportion of nsSCs in clinical S. aureus strains and laboratory strains after 5 h exposure to human neutrophils. N = 4 biological replicates for bacteria and N = 3 for healthy donors. (F) Antibiotic challenge survival assay of the clinical isolates CI-1149 and CI-2015, and laboratory strains. Bacterial survival after 24 h in FLU, CLI, CIP, RIF, or combinations (ABX1 to 3) at 40× MIC in pH 7.4 after 5 h of exposure to neutrophils was analyzed and calculated relative to the initial inoculum. Shaded circles show SEM of four biological replicates for bacteria and N = 3 for healthy donors. Asterisks above bars indicate significant difference to CI-1149. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
To determine whether high proportions of late-growing bacteria correlated with increased antibiotic persistence, we challenged acidic pH preexposed bacteria with either single or antibiotic combinations—ABX1 (FLU+CLI), ABX2 (FLU+CLI+CIP), or ABX3 (FLU+CLI+CIP+RIF)—mimicking the treatment the patient received. Isolate CI-1149 showed increased antibiotic survival compared with the laboratory strains (Fig. 2D). Only the RIF-treated CI-1149 sample exhibited no increased antibiotic persistence compared with the laboratory strains, without developing RIF resistance. We tested whether the other clinical isolates derived from difficult-to-treat infections displayed a similar phenotype as CI-1149. Indeed, we found an elevated ability of the additional clinical isolates to survive antibiotic challenges after acidic pH exposure similar to CI-1149 (Fig. 2D), with CI-2015 displaying even higher persistence levels to CLI and RIF than CI-1149. To evaluate the potential effect of the accessory gene regulator (Agr) system on antibiotic persistence, we tested the USA300_JE2 agrC mutant (Fig. 2D). We did not see an increase in survival in the agrC mutant compared with its wild type, consistent with the results observed for the partially functional Agr system in Cowan I.
Since all the clinical isolates were obtained from abscesses, which consist mainly of neutrophils, we tested in a second step the clinical strains’ ability to withstand neutrophils. CI-1149 and USA300_JE2 were more cytotoxic to human neutrophils than Cowan I, and CI-1149 survived neutrophil killing better than Cowan I (SI Appendix, Fig. S3 A and B). Despite its cytotoxicity, CI-1149 showed better intracellular survival than USA300_JE2 and Cowan I (SI Appendix, Fig. S3C). To determine whether exposure to neutrophils leads to an increased lag-time heterogeneity and antibiotic persistence, we measured colony radius heterogeneity in CI-1149 and CI-2015, which showed the highest levels of acidic pH-triggered antibiotic persistence (Fig. 2D) and two laboratory strains. Again, the clinical isolates showed a higher colony radius heterogeneity compared with the laboratory strains (Fig. 2E). Challenging bacteria preexposed to neutrophils with antibiotics (40× minimum inhibitory concentration [MIC]) resulted in elevated global antibiotic survival levels (1 to 3 logs higher) in CI-1149 and CI-2015 compared with USA300_JE2 and Cowan I (Fig. 2F). These results highlight the plasticity of clinical isolates derived directly from difficult-to-treat infections to respond to stressors, not limited to abscesses, leading to increased phenotypic heterogeneity and antibiotic survival.
CI-1149 Shows Increased Lag Time and Antibiotic Persistence in Mice.
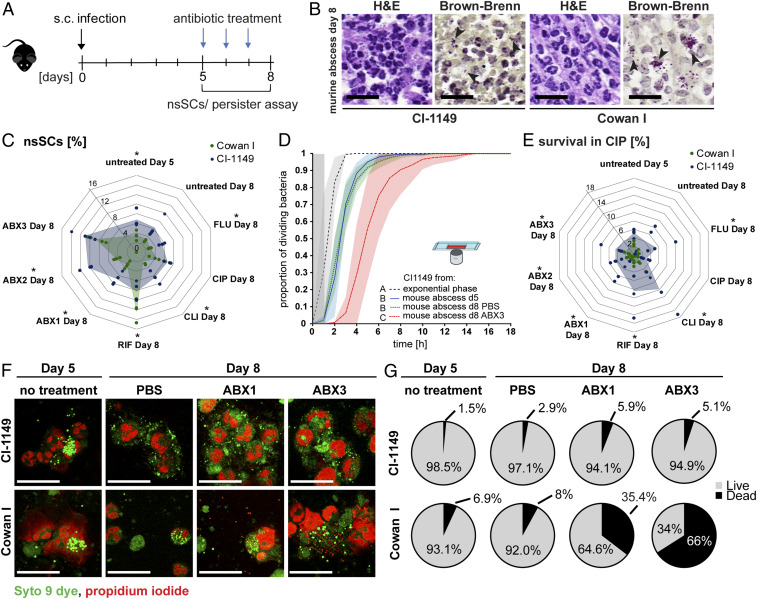
To test whether a subpopulation of persisters arises in vivo, we infected mice with either the clinical isolate CI-1149 or Cowan I mimicking the patient’s infection condition in a murine abscess model (6, 30). Five days postinfection (pi), mice were treated with antibiotics over 3 d (Fig. 3A). Histology of the murine abscesses showed neutrophil infiltration as well as bacterial aggregates for both strains, mirroring the findings observed in the patient (Figs. 1B and 3B).
Fig. 3.
Increased lag time and formation of persisters in mice. (A) Scheme of the murine infection model. (B) Representative images of H&E and Brown-Brenn stained murine abscess tissue biopsies. (Scale bars, 20 µm.) Arrows indicate bacterial aggregates. (C) Proportion of nsSCs of CI-1149 and Cowan I after sampling from murine abscesses and different treatments. Net lines show percentages of nsSCs (as indicated). Shaded area indicates mean survival of four biological replicates. (D) Single-cell microscopy of CI-1149 during exponential growth and after harvesting from murine abscesses. N = 3 to 8 mice per condition; shaded areas depict the SD; exponential phase n = 500 bacterial cells, day 5 abscess n = 2,466, day 8 abscess PBS n = 1,052, and day 8 abscess ABX3 n = 321; gray zone marks the period at the beginning of the experiment where cell divisions could occur, but would not be observed. (E) Antibiotic challenge survival assay of bacteria harvested from murine abscesses and exposed to CIP (40× MIC) for 24 h. Survival was calculated relative to the initial inoculum. Shaded area shows the mean of four biological replicates. (F) Representative images of live (green)- dead (red)-stained murine abscess pus. (Scale bars, 25 µm.) (G) Quantification of live/dead staining. Eight to 12 images per condition (in total 187 to 1,046 bacteria) were analyzed. N = 4 mice per condition. *P < 0.05.
CI-1149 isolated from murine abscesses showed a higher proportion of nsSCs than Cowan I in most of the tested conditions (Fig. 3C). Additionally, using single-cell microscopy, we confirmed that the observed lag time on agar plates was also reflected by single bacterial cell lag times (Fig. 3D and SI Appendix, Fig. S3E). We demonstrated that increased lag time correlated with antibiotic persistence in a bacterial survival assay (Fig. 3E and SI Appendix, Fig. S3F) against CIP and FLU, where the patient received FLU over the entire treatment period and CIP for the last five days. CI-1149 survived the 24 h antibiotic challenge significantly better than Cowan I, with 4 to 18% of survivors, depending on the antibiotic used to treat the mice (Fig. 3E). Confocal microscopy revealed a higher proportion of live CI-1149 (94.9%) in murine pus compared with Cowan I (34%) after antibiotic treatment (ABX3) in mice (Fig. 3 F and G), further demonstrating elevated antibiotic persistence of this clinical isolate in vivo.
Acid Stress Induces Metabolic and Virulence Changes in CI-1149.
The pH in an abscess typically ranges from 5.5 to 7.5 (31, 32). To investigate the effect of acid stress on bacteria, we analyzed proteomic and transcriptomic changes in CI-1149 grown in pH 5.5 versus pH 7.4 using mass spectrometry (MS) (33) and RNA sequencing (RNA-seq) (34). MS analysis revealed increased expression of citrate cycle (TCA) proteins, metabolic proteins, and virulence factors after acid stress. Decreased expression was observed for ABC transporters, adhesion proteins, and among others, the two-component systems SaeS/R, and the amino acid biosynthesis protein ThrC (SI Appendix, Fig. S4 A and B). RNA-seq showed that transcripts involved in translation were generally up-regulated and those involved in carbohydrate and amino acid metabolism were mostly down-regulated in CI-1149 after pH 5.5 stress (SI Appendix, Fig. S4 C–E).
Comparison with USA300_JE2 revealed a significant down-regulation of quorum-sensing pathways and an up-regulation of metabolic enzymes in CI-1149 after pH 5.5 stress (SI Appendix, Fig. S5 A, C, and E). Comparison with Cowan I revealed differences in virulence, translation, ribosomal proteins, DNA replication, and repair proteins (SI Appendix, Fig. S5 B, D, and F). Generally, the MS analysis showed that CI-1149 up-regulated its energy metabolism to a higher extent after acidic pH stress than the laboratory strains.
RNA-seq identified many more differentially expressed pathways than the MS analysis. Down-regulation of quorum-sensing pathways was confirmed; additionally, transporters, and signaling and cellular process pathways were also down-regulated at the transcript level in CI-1149 compared with the laboratory strains after acid stress, whereas transcripts for translation, ribosomes, chaperones, and DNA repair were found to be up-regulated (SI Appendix, Fig. S6).
Persister Enrichment Reveals Protein Networks Involved in Antibiotic Persistence.
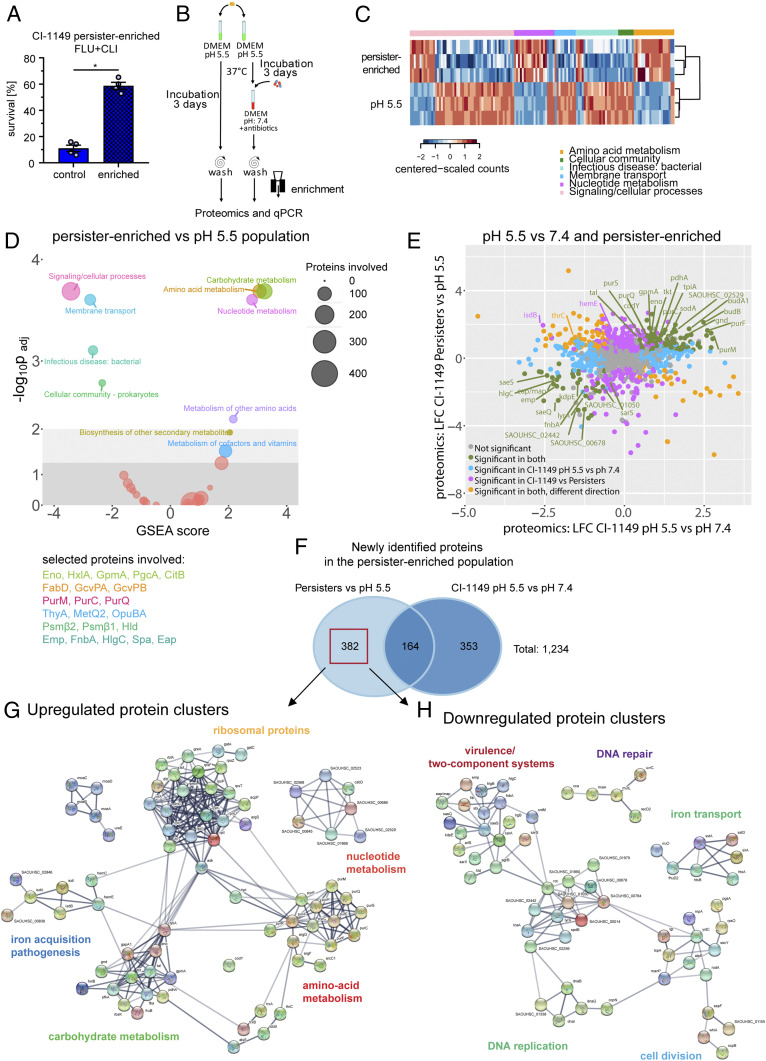
The population of acidic pH-exposed CI-1149 was enriched for persisters by selection of live bacteria after antibiotic exposure to avoid confounding signals from dead or dying bacteria. Subsequently, to test for the increased persister levels, we challenged the CI-1149 persister-enriched population with FLU+CLI and found the persister proportion to be enriched sixfold (Fig. 4A).
Fig. 4.
Molecular signature of a S. aureus persister-enriched subpopulation. (A) Antibiotic survival assay, involving 24 h exposure to FLU+CLI (40× MIC) for a persister-enriched population and pH 5.5-stressed bacteria. Survival was calculated relative to the initial inoculum. N = 4 biological replicates. Error bars depict SEM. (B) Scheme for MS analysis of persister-enriched S. aureus. (C) Heatmap showing three biological replicates for selected significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways from D. (D) GSEA on KEGG pathways from persister-enriched CI-1149 vs. pH 5.5-stressed bacteria. Circles’ diameter indicates number of involved proteins. Selected proteins are displayed in the corresponding pathway color. (E) Dot plot comparing proteomic changes in persister-enriched and pH 5.5-stressed CI-1149 vs. pH 5.5 stress compared with pH 7.4 growth. LFC > 1 (log2 fold change; fold change >2), P < 0.05. (F) Venn diagram showing overlaps and uniquely identified proteomic changes in persister-enriched CI-1149 vs. pH 5.5-stressed bacteria compared with pH 5.5 vs. pH 7.4 growing bacteria. Red box indicates uniquely differentially expressed proteins in the persister-enriched population that were further analyzed via the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING). (G) STRING analysis on up-regulated proteins found in the persister-enriched population identifies up-regulated protein–protein interaction networks. (H) STRING analysis on down-regulated proteins found in the persister-enriched population identifies down-regulated protein–protein interaction networks.
Following enrichment, we conducted a proteomic analysis and compared the proteome profile of the persister-enriched population with that of the pH-stressed population (Fig. 4B) (33). Gene set enrichment analysis (GSEA) carried out to check the broader functions of the differentially expressed proteins, revealed that the persister-enriched population showed decreased expression levels of proteins involved in quorum sensing, membrane transport, virulence factors, and signaling and cellular processes. On the contrary, we found an up-regulation of amino acid metabolic proteins and nucleotide metabolic pathways in the persister-enriched population (Fig. 4 C and D), suggesting that persisters are not completely metabolically inactive.
We identified 289 differentially expressed proteins in the persister-enriched population that were not detected by MS in the entire pH 5.5-stressed population (Fig. 4E). These included down-regulated proteins involved in virulence and quorum sensing, DNA replication and recombination, and cell wall synthesis. Among the small number of up-regulated proteins were HemE, PurA, and PknB. We identified an additional 93 proteins in the persister-enriched population that were regulated in a different direction in the entire pH 5.5-exposed population, giving an overall total of 382 differentially expressed proteins in the persister-enriched population compared with the entire pH 5.5-stressed population (Fig. 4 E and F). Another 164 proteins were found to be regulated in the same direction after pH 5.5 stress compared with pH 7.4 growth (Fig. 4E). Notably, the pur-operon, the stringent response protein CodY, the adhesion protein FnbA, and the histidine kinase SaeS were identified in both persister-enriched and the pH 5.5-stressed populations (SI Appendix, Fig. S7 A and B), suggesting a general role in the response to acidic pH stress.
In silico interaction analysis was used to dissect whether the protein interactions of differentially expressed proteins in the persister population contributed to specific adaptive changes (Venn diagram showing 382 proteins in Fig. 4F). Among the up-regulated proteins in the persister-enriched population we identified networks of ribosomal proteins, proteins involved in nucleotide metabolism, amino acid metabolism, protein folding, iron acquisition/pathogenesis, and carbohydrate metabolism (Fig. 4G), suggesting that these networks are potentially involved in persister formation and maintenance. Among down-regulated proteins in the persister-enriched population, networks included DNA repair and replication, cell division, virulence, and two-component systems (Fig. 4H), highlighting that target inactivation/down-regulation favors the inherent antibiotic tolerance of persisters. Similar molecular signatures were identified in persister-enriched populations of other clinical isolates from persistent infections (CI-2015 and CI-2134) (SI Appendix, Figs. S7 F, G, and H and S8 A and B), but not of laboratory strains (Cowan I and USA300_JE2) (SI Appendix, Fig. S8 C, D, and G), suggesting that these changes may be characteristics of in-host adapted S. aureus strains (35). Together, our findings emphasize the relevance of performing experiments with recently isolated strains rather than only with highly passaged laboratory strains.
The Reversible Physiological State of Persisters.
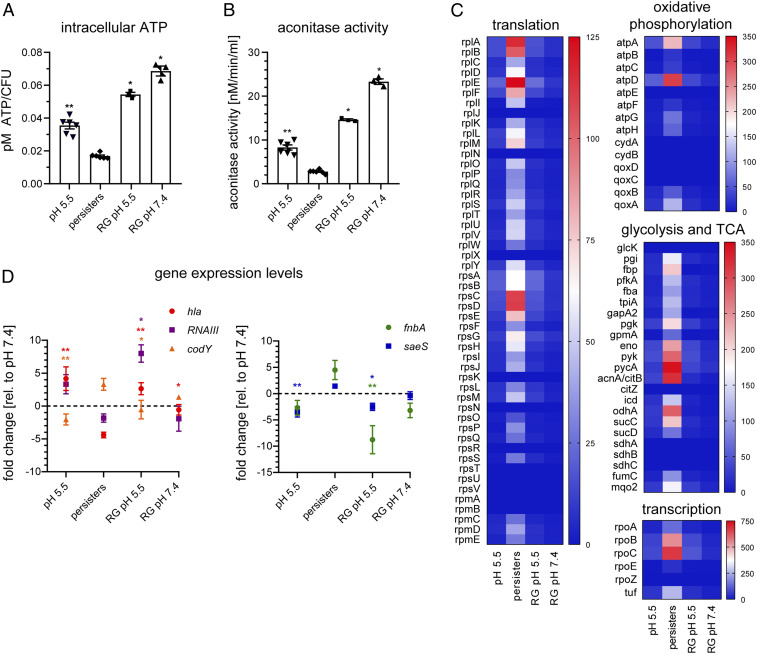
We investigated the physiological state of the persister-enriched population and determined whether this dormant subpopulation reverts to its virulent phenotype. We measured aconitase activity, ATP levels, and accumulation of insoluble proteins that might determine dormancy depth and antibiotic persistence. In addition, persisters were assessed for their ability to regrow and to regain a virulent phenotype, critical features in persistent infections. We found that the persister-enriched population showed significantly reduced ATP levels (∼50%) compared with the pH 5.5-stressed population (Fig. 5A and SI Appendix, Figs. S7 D and S8 F). Upon regrowth, persister cells reestablished normal intracellular ATP levels, indicating reversion to an actively growing phenotype after stress removal (Fig. 5A).
Fig. 5.
Reversible metabolic down-regulation, ATP depletion, and accumulation of insoluble proteins in persisters. (A–D) CI-1149 grown in either DMEM pH 5.5 for 3 d, persister-enriched or persisters regrown in pH-adjusted DMEM (RG pH 5.5 or 7.4) to exponential phase. (A) Intracellular ATP concentration was determined using a BacTiter-Glo cell viability assay. N = 3 to 6 biological replicates. (B) Aconitase activity was quantified using an aconitase assay kit. N = 3 to 6 biological replicates. (C) Heatmaps show insoluble proteins, identified by MS, of pH 5.5-stressed bacteria, enriched persisters and regrown persisters in DMEM pH 5.5 and pH 7.4, involved in several biological processes. Heatmap scale indicates protein abundance. N = 3. (D) Quantitative real-time PCR of transcripts of hla, RNAIII, codY, fnbA, and saeS. Data, expressed in fold changes vs. DMEM pH 7.4 grown bacteria, are means of three biological replicates. Statistical significance was determined by one-way ANOVA with Dunnett’s posttest. *P < 0.05, **P < 0.01. Error bars depict SEM.
To test whether the increased abundance of aconitase that we detected in the persister-enriched population corresponds to higher enzymatic activity and contributes to energy production, we measured aconitase activity in the different bacterial populations. In accordance with the decreased ATP levels, we observed significantly reduced aconitase activity (<30%) in the persister-enriched population compared with the pH 5.5-stressed population, which reverted to normal levels during regrowth (Fig. 5B).
Protein aggregation has recently been associated with dormancy and antibiotic tolerance in Escherichia coli (36), but has not been identified so far as a factor linked to antibiotic persistence in S. aureus. We found that persister-enriched S. aureus populations contained increased levels of insoluble cytosolic proteins, including those involved in transcription, translation, and energy production, hand in hand with decreased energy levels in the bacteria (37). Regrowth of the bacteria led to a reduction of these insoluble proteins (Fig. 5C and SI Appendix, Fig. S9). Regrowth in acidic pH medium not only led to elevated ATP levels, increased aconitase activity, and reduced levels of insoluble proteins, but also resulted in the up-regulation of the virulence genes RNAIII and hla and the down-regulation of the adhesin fnbA and the stringent response regulator codY (Fig. 5D).
Enhanced Antibiotic-Mediated Bacterial Killing with Retinoid Derivatives In Vitro.
The retinoid derivative CD1530 has been shown to have a synergistic effect with gentamicin on S. aureus clearance by integrating into the bacterial membrane, rendering it permeable (38). We tested two retinoid derivatives, adapalene and isotretinoin, for their ability to reduce S. aureus persisters. These drugs, both currently used for acne treatment (39, 40), had no antimicrobial activity per se (SI Appendix, Fig. S10 A and B). However, when combined with conventional antibiotics they improved clearance of the S. aureus strains CI-1149 and Cowan I (SI Appendix, Fig. S10 A and B). We also tested CD1530, which shows antimicrobial activity (Fig. 6A and SI Appendix, Fig. S10C), for its ability to kill CI-1149 and Cowan I persisters. When tested on pH 5.5 and pH 7.4 grown bacteria, CD1530 was more effective in reducing bacterial numbers than conventional antibiotics (Fig. 6A and SI Appendix, Fig. S10 C, G, and H). We observed an improved antimicrobial effect when CD1530 was used in combination with conventional antibiotics in vitro (SI Appendix, Fig. S11).
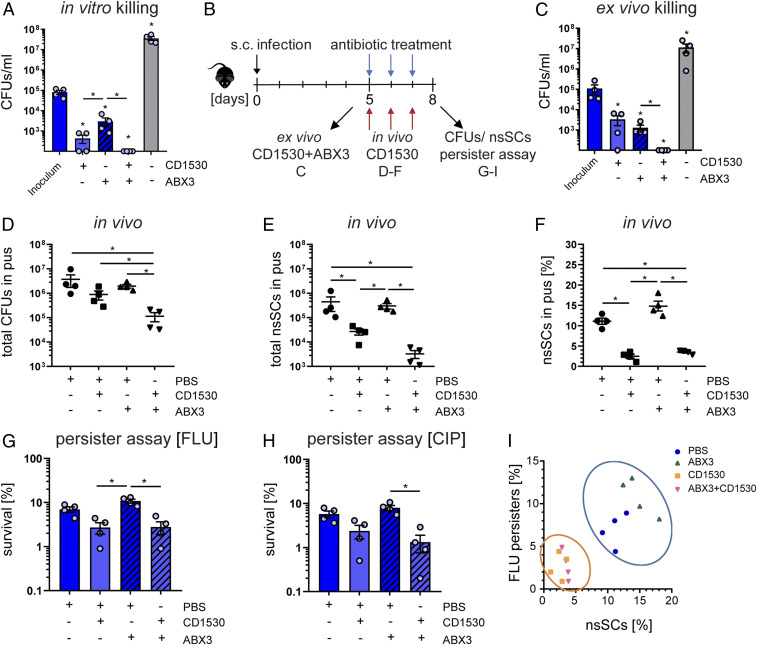
Fig. 6.
S. aureus persister levels are reduced in the presence of retinoid derivatives during antibiotic treatment. (A) In vitro killing of pH 5.5-stressed bacteria using the retinoid derivative CD1530 (20 µg/mL), ABX3 (40× MIC) alone, or CD1530 together with ABX3. Bacterial survival was assessed after 24 h. N = 4. (B) Overview of the murine abscess model. (C) Ex vivo killing assay of bacteria harvested from murine abscesses using the retinoid derivative CD1530 (20 µg/mL) alone, ABX3 (40× MIC), or CD1530 together with ABX3. Bacterial survival was assessed after 24 h. Bacteria from four independent mice were used. (D) Total CFU counts in mouse abscess pus at day 8. N = 4. (E) Total nsSCs counts in mouse abscess pus at day 8. (F) Percentage of nsSCs in mouse abscess pus at day 8. N = 4. (G) Ex vivo antibiotic challenge with FLU (40× MIC) for 24 h. Bacterial survival was determined relative to the inoculum. Bacteria from four independent mice were used. (H) Ex vivo antibiotic challenge with CIP (40× MIC) for 24 h. Bacterial survival was determined relative to the inoculum. (I) Correlation between the proportion of nsSCs and the proportion of persisters (FLU). CD1530 exposure reduced both nsSCs and persisters (FLU challenge). Each symbol represents one biological replicate. *P < 0.05; Error bars depict SEM.
A CD1530/Antibiotic Combination Reduces Bacterial Load and Persisters in Mice.
We further tested the efficacy of the CD1530/antibiotic combination in a murine infection model. Mice were infected subcutaneously and monitored for 5 d prior to antibiotic treatment in order to ensure abscess formation (Fig. 6B). First, abscesses were harvested, and isolated bacteria challenged with either CD1530, conventional antibiotics, or a combination of both for 24 h ex vivo, to test for bactericidal activity (Fig. 6C). Consistent with the in vitro results, we found a significant reduction of the bacterial load with CD1530 alone, but not to a greater extent than conventional antibiotic therapy. However, the bacterial load was reduced below our detection limit (of 100 colony-forming units [CFUs]/mL) using CD1530 in combination with conventional antibiotics (Fig. 6C).
Second, we used the combination of CD1530 with conventional antibiotics (ABX3) to treat mice with preformed abscesses and observed a significant reduction (10-fold) in bacterial load (Fig. 6D). CD1530 treatment also led to a significant reduction (100-fold) of late-growing bacteria, indicating a decreased persister fraction (Fig. 6 E and F), whereas conventional antibiotic therapy increased the proportion of late-growing bacteria, although not to a statistically significant extent (P = 0.7429) (Fig. 6 E and F). To investigate the effect of CD1530 on persisters, we performed an antibiotic killing assay on bacteria harvested from in vivo treated murine abscesses, using FLU (Fig. 6G and SI Appendix, Fig. S10E) and CIP (Fig. 6H andSI Appendix, Fig. S10F). Addition of CD1530 to the conventional antibiotic therapy in mice resulted in a significant decrease (10-fold) in persister levels. In both antibiotic challenges, we saw a reduction of bacterial survival when mice were also treated with CD1530 (Fig. 6 G and H). We confirmed these findings with another abscess-forming strain, Cowan I, as well as on persister-enriched populations, further corroborating the beneficial effect of CD1530 (SI Appendix, Fig. S10 G–O). Integrative analysis of persister levels and the proportion of late-growing bacteria showed that CD1530 had a reducing effect on nsSCs levels as well as on bacterial survival. Thus, a combination of retinoid derivatives, such as CD1530, with conventional antibiotics may offer considerable beneficial effects for the clinical treatment of bacterial infections, especially if these involve the presence of bacterial persisters (Fig. 6I and SI Appendix, Fig. S10O).
Discussion
This study shows that S. aureus sampled directly from a difficult-to-treat infection displayed both significant cell-to-cell lag-time variation and substantially increased lag times, indicating the presence of persister cells. Infection modeling reproduced the increased lag times as well as elevated levels of antibiotic persistence in the clinical isolate. This was also true for other strains from different genetic backgrounds isolated from various persistent infections. Multiomics analysis revealed dynamic cellular changes in CI-1149 after exposure to acidic pH leading to an overall metabolically active, virulent phenotype, and a subpopulation of persisters that showed reduced virulence factor expression and down-regulation of proteins involved in DNA replication and repair, cell division, and membrane transport. Our data show that pH-triggered S. aureus persisters accumulate insoluble proteins and have lower ATP levels along with reduced aconitase activity, potentially driving them into dormancy. Regrowth of these persisters in physiological medium led to a reversion of their phenotype, resulting in a virulent and metabolically active population. We further demonstrated an optimized treatment for persistent S. aureus infections associated with antibiotic tolerance and showed the effectiveness of retinoids combined with antibiotics in reducing S. aureus persisters.
Poor antibiotic penetration into the site of infection and consequent bacterial survival could partially explain the difficulties encountered in reducing the bacterial burden in the patient. However, lack of antibiotic penetration does not fully explain treatment failure in ongoing infections (41) and evidence supports a high rate of antibiotic penetration into abscesses (42). We found that antibiotics did penetrate into the abscess: colony growth was partially inhibited in the undiluted pus sample (SI Appendix, Fig. S1D). In host, CI-1149 displayed phenotypic plasticity, including dormant cells, and showed a high level of antibiotic persistence. One factor influencing bacterial stress response reactions in the host is acidic pH. Exposure to acidic pH, together with intracellular localization within neutrophils and abscesses, significantly prolonged the lag time in clinical isolates and less in laboratory strains. These results show how heterogeneity in a bacterial population correlates with bacterial survival after exposure to stress conditions. We examined the proteome and transcriptome of acidic pH-exposed bacteria to evaluate molecular changes that might contribute to the enhanced ability of CI-1149 to persist against antibiotic challenges. Studying both levels of regulation is informative because of the poor correlation between mRNA and protein levels due to differences in half-life, posttranscriptional regulation (43), or regulation by small RNAs (44). The production of Agr-controlled virulence factors (e.g., PSMα and α-hemolysin) increased upon acidic pH stress, as reported previously (45). Our results are indicative of a virulent state of acidic pH-exposed CI-1149 that might help the bacteria resist host immune clearance and facilitate the establishment as well as maintenance of an infection (46), which, in the case of our patient, could not be controlled despite extensive antibiotic use and repeated surgeries.
Persister enrichment enabled us to identify molecular changes exclusively in persisters. Proteomic analysis of persister-enriched CI-1149, CI-2015, and CI-2134 revealed a completely new range of proteins that were differentially expressed exclusively in the persister-enriched population and were not detected in the pH 5.5-stressed population. Expression of virulence factors, quorum-sensing proteins, and membrane transport proteins were down-regulated in the persister-enriched population, whereas amino acid, carbohydrate, and nucleotide metabolic pathways were up-regulated. Nucleotide metabolism, specifically purine metabolism, has previously been linked to rifampicin tolerance in S. aureus (47). Transcription of carB, coding for the large subunit of carbamoyl phosphate synthetase, an enzyme involved in pyrimidine and arginine synthesis, was significantly up-regulated in the persister-enriched population (1.7-fold). Disruption of carB led to a 2,500-fold reduction of survival in Pseudomonas aeruginosa after antibiotic treatment (48). De novo pyrimidine production may also play a role in S. aureus antibiotic persistence. Additionally, we found significantly reduced levels of PSMα1/4 and PSMβ1/2 in the persister-enriched population, consistent with the observation that PSMs have been linked to decreased persister formation in S. aureus (49).
We revealed accumulation of insoluble proteins, accompanied by ATP depletion in S. aureus persisters, phenotypes that were reversible upon regrowth. It has been shown that nutrient-starved E. coli form aggresomes driven by ATP depletion that indicate both dormancy depth and antibiotic persistence (36). ATP plays a critical role in maintaining proteostasis (50) and it has been recently demonstrated that ATP supports protein solubility and prevents macromolecular aggregation by acting as a hydrotrope (51). In addition, ATP fuels the protein quality-control system such as chaperones, foldases, and proteinases. Interestingly, we found that several proteases and chaperones such as ClpB, ClpL, ClpX, and GroS that play an important role in the clearance of protein aggregates and the resuscitation of persisters were up-regulated in persisters (52). We speculate that these chaperones enable the resuscitation of the bacteria with increasing ATP levels by becoming active and clearing such protein aggregates. Corrupting ClpP by acyldepsipeptide (ADEP4) leads to the ClpX-independent and uncontrolled proteolysis and ultimately to the killing of persisters (53). Nevertheless, it remains to be elucidated whether the accumulation of insoluble proteins is an active process leading to the antibiotic persistence phenotype, or merely a consequence of the reduced ATP levels. Low ATP levels have been linked to increased antibiotic persistence in S. aureus (54) and reactive oxygen species produced by macrophages can inactivate TCA-cycle enzymes and lead to antibiotic persistence (55). Although we detected increased levels of aconitase in the persister-enriched population, the functional analysis showed that aconitase is mainly inactive and accumulates in the insoluble protein fraction together with ribosomal proteins, transcription factors, and other metabolic enzymes. Accumulation of insoluble proteins accompanied by ATP depletion might play a crucial role in S. aureus antibiotic persistence, but further experiments are needed to test for their exact role in S. aureus persistence.
To improve treatment of persistent S. aureus infections, we tested retinoid derivatives alone and in combination with conventional antibiotics for their ability to reduce persister levels. The FDA-approved drugs adapalene and isotretinoin as well as CD1530 enhanced killing of S. aureus in combination with conventional antibiotics. CD1530 has been reported to reduce bacterial load in infections with the S. aureus strain MRSA-MW2 in combination with gentamicin (38). Here, we took it one step further and demonstrated that CD1530 not only reduces overall CFUs but specifically targets persisters in different S. aureus strains as well. Combinations of antibiotics and CD1530 in mice improved antibiotic killing, reduced persister levels as confirmed in persister assays, and decreased the proportion of nsSCs. CD1530 has been shown to penetrate and permeabilize the bacterial membrane without leading to direct cell lysis (38, 56). The planar aryl moiety of CD1530 that rigidifies the carboxylic acid facilitates the penetration into the lipid bilayers potentially leading to an increased passive diffusion of antibiotics and other molecules through the cell membrane (38). Additionally, retinoids can enhance the expression of cathelicidin antimicrobial peptide in dermal fibroblasts undergoing adipogenesis in response to infections, inhibiting the growth of S. aureus (57). In vivo, retinoids might target bacteria in different ways simultaneously—directly by membrane permeabilization and indirectly by inducing antimicrobial peptide expression by host cells. Our results suggest that CD1530 not only kills growing but can also target growth-arrested bacteria, reduce lag-time heterogeneity and persisters. Retinoids combined with conventional antibiotics may provide effective treatment for difficult-to-treat S. aureus infections with a high probability of an extensive persister subpopulation.
Materials and Methods
See SI Appendix, Materials and Methods for detailed descriptions of experimental conditions and data processing. Strains and primers used in this study are listed in SI Appendix, Tables S2 and S4. Clinical data and MICs are shown in SI Appendix, Tables S1 and S3.
Ethical Requirement.
Humans.
Isolation of human neutrophils and use of patient derived material (Business Administration System for Ethics Committees [BASEC] No. 2019–01735) was done in accordance with the Helsinki Declaration and approved by the Cantonal Ethical Research Committee Zurich. Informed patient consent was obtained.
Mice.
The protocols (ZH251/14 and ZH050/18) were approved by the institutional animal care and use committee of the University of Zurich and all experiments were conducted in approval of the Cantonal Veterinary Office Zurich.
Bacterial Growth Conditions.
For exponential growth conditions, bacteria were grown in tryptic soy broth (TSB) overnight (16 h), diluted, and regrown in TSB for 2 h. For pH-stress assays, S. aureus was inoculated into defined pH media at a starting OD600 of 0.2 and incubated at 37 °C and 5% CO2 for 3 d. NsSCs and antibiotic persistence were determined as described previously (8).
Neutrophil Infection Assays.
Human neutrophils were infected with S. aureus at a multiplicity of infection (MOI) of one for bacterial survival assay. To quantify antibiotic persistence, S. aureus were exposed to neutrophils (MOI1) for 5 h prior to the persister assay. To analyze intracellular survival of the bacteria, neutrophils were infected with S. aureus (MOI10) and bacterial load was quantified for several time points by plating serial dilutions and CFU enumeration. Bacterial cytotoxicity was assessed by measuring lactate dehydrogenase (LDH) release.
Persister Assay.
Bacteria were inoculated in DMEM pH 7.4 supplemented with antibiotics (40× MIC) as indicated. After 24 h, bacterial survival was determined by CFU enumeration and calculated relative to the inoculum. Exponential phase bacteria were used to test for baseline survival levels (SI Appendix, Fig. S3D).
Automated Agar-Plate Imaging and Single-Cell Time-Lapse Microscopy.
To determine colony growth dynamics, bacteria were plated onto agar plates, incubated at 37 °C, and imaged every 10 min for 48 h. Analysis of colony growth kinetics was performed with our in-house software “ColTapp” (6, 58). Bacterial single-cell lag times were quantified by time-lapse microscopy.
Murine Infection Model.
C57BL/6 mice were injected into the flanks with S. aureus to induce abscess formation as previously described (30). Mice were killed either 5 or 8 d pi depending on the treatment regimen. To test the antipersister activity of CD1530 (Sigma), mice were treated intraperitoneally with ABX3 or phosphate-buffered saline (PBS, Sigma) alone starting at day 5 pi. Additionally, the mice received either CD1530 (40 µg) or PBS subcutaneously in close proximity to the abscess for 3 d daily. Persister levels were determined by performing a persister assay with FLU or CIP. Abscess pus was stained using the bacterial live dead staining kit (Thermo Fisher) to analyze bacterial viability.
Persister Isolation and Enrichment.
S. aureus persisters were isolated from a FLU+CLI (40× MIC)-treated population using the MACSflex MicroBead Kit (Miltenyi Biotec). As previously described for E. coli persisters (59), propidium iodide (PI)-functionalized MicroBeads were used to capture dead bacterial cells and to enrich for surviving persisters (Fig. 4B).
Whole Genome Sequencing.
The genomes of the isolates (USA300_JE2, USA300_11358, Cowan I, CI-1140, CI-1149, CI-1150, CI-2015, CI-2857, and CI-2134) were sequenced on a MiSeq sequencer (Illumina). A detailed description of the genomic data analysis is available in SI Appendix, Materials and Methods.
RNA-Seq and Analysis.
Total RNA was isolated using the RNeasy Mini kit (Qiagen) following the manufacturer’s protocol. Vertis Biotechnologie AG, Freising, Germany, carried out further steps: rRNA depletion, RNA fragmentation, strand-specific cDNA library preparation, and Illumina NextSeq500 single-read sequencing (∼10 million reads per sample). A detailed description of the RNA-seq data analysis is available in SI Appendix, Materials and Methods. Selected transcripts (SI Appendix, Table S4) from S. aureus grown in DMEM pH 7.4 or pH 5.5 were assessed by qRT-PCR to confirm RNA-seq results (SI Appendix, Fig. S7C).
Proteomics.
S. aureus peptide samples were analyzed on a Q Exactive HF mass spectrometer equipped with a nano-electrospray ion source (Thermo Fisher Scientific). The Progenesis QI software (v2.0, Nonlinear Dynamics Limited) was used to extract peptide precursor ion intensities across all samples and the MaxQuant software (version 1.0.13.13) was used for protein identification.
ATP and Aconitase Activity Measurements.
Bacterial ATP levels were measured using a BacTiter-Glo kit (Promega) according to the manufacturer’s instructions. Bacterial samples were assayed for aconitase activity using an aconitase assay kit (Abcam) as per the manufacturer’s instructions.
Insoluble Protein Isolation.
Insoluble protein isolation from bacterial cells was modified from Tomoyasu et al. (60) and adapted for S. aureus. Isolated proteins were analyzed by mass spectrometry.
Statistical Analysis.
All experiments were analyzed for statistical significance by Mann–Whitney test (two tailed), unless otherwise indicated: *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. A detailed description of the statistical analysis of the RNA-seq and proteomics data are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was funded by the Swiss National Science Foundation (SNSF) Project Grant 31003A_176252 (to A.S.Z.), the SNSF Biobanking Grant 31BK30_185401 (to A.S.Z.), the Clinical Research Priority Program of the University of Zurich “Precision Medicine for Bacterial Infections” (to A.S.Z. and B.H.), Uniscientia Foundation Grant (to A.S.Z.), Swedish Society for Medical Research Foundation Grant P17-0179 (to S.M.S.), Promedica Foundation 1449/M (to S.D.B.), Svenska Kulturfonden Grant 106745 (to S.S.), Federation of European Microbiological Societies Meeting Attendance Grant FEMS-GO-2019-419, and Hartmann Müller Foundation travel grant (both to M.H.). We thank S. Dettwiler and F. Prutek for their technical help in tissue histology, the members of the A.S.Z. laboratory for comments on the manuscript, K. S. Kamath (Macquarie University Australia) for technical advice with proteomics analysis, and L. Andereggen for his help with the radiological images. We acknowledge Life Science editors for proofreading the manuscript. Imaging was performed with equipment maintained by the Centre for Microscopy and Image Analysis, University of Zurich. Proteomic analysis was performed at the proteomic core facility at the University of Basel, and we thank T. Bock for excellent technical support. We also thank the Functional Genomics Centre Zurich, University of Zurich/ETH Zurich, Y. Joho-Auchli, for technical assistance.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2014920118/-/DCSupplemental.
Data Availability.
Raw data have been deposited in European Nucleotide Archive (ENA): PRJEB37630 (27); Gene Expression Omnibus (GEO): GSE148024 (34); and PRIDE: PXD018372 (33), PXD018332 (37), and PXD022858 (35) and are publicly available. All other study data are included in the article and supporting information.
References
- 1.Gorwitz R. J., Understanding the success of methicillin-resistant Staphylococcus aureus strains causing epidemic disease in the community. J. Infect. Dis. 197, 179–182 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Lowy F. D., Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Proctor R. A., Balwit J. M., Vesga O., Variant subpopulations of Staphylococcus aureus as cause of persistent and recurrent infections. Infect. Agents Dis. 3, 302–312 (1994). [PubMed] [Google Scholar]
- 4.Turner N. A., et al. , Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nat. Rev. Microbiol. 17, 203–218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Proctor R. A., et al. , Small colony variants: A pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat. Rev. Microbiol. 4, 295–305 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Vulin C., Leimer N., Huemer M., Ackermann M., Zinkernagel A. S., Prolonged bacterial lag time results in small colony variants that represent a sub-population of persisters. Nat. Commun. 9, 4074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuchscherr L., et al. , Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol. Med. 3, 129–141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leimer N., et al. , Nonstable Staphylococcus aureus small-colony variants are induced by low pH and sensitized to antimicrobial therapy by phagolysosomal alkalinization. J. Infect. Dis. 213, 305–313 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Onyango L. A., Hugh Dunstan R., Roberts T. K., Macdonald M. M., Gottfries J., Phenotypic variants of staphylococci and their underlying population distributions following exposure to stress. PLoS One 8, e77614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaban N. Q., et al. , Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigger J., Treatment of staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244, 497–500 (1944). [Google Scholar]
- 12.Lewis K., Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Lopatkin A. J., et al. , Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nat. Microbiol. 4, 2109–2117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dörr T., Vulić M., Lewis K., Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8, e1000317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gutierrez A., et al. , Understanding and sensitizing density-dependent persistence to quinolone antibiotics. Mol. Cell 68, 1147–1154.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Peyrusson F., et al. , Intracellular Staphylococcus aureus persisters upon antibiotic exposure. Nat. Commun. 11, 2200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levin B. R., et al. , A numbers game: Ribosome densities, bacterial growth, and antibiotic-mediated stasis and death. MBio 8, e02253-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libraty D. H., Patkar C., Torres B., Staphylococcus aureus reactivation osteomyelitis after 75 years. N. Engl. J. Med. 366, 481–482 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin-Reisman I., et al. , Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Liu J., Gefen O., Ronin I., Bar-Meir M., Balaban N. Q., Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science 367, 200–204 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Bakkeren E., et al. , Salmonella persisters promote the spread of antibiotic resistance plasmids in the gut. Nature 573, 276–280 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fux C. A., Shirtliff M., Stoodley P., Costerton J. W., Can laboratory reference strains mirror “real-world” pathogenesis? Trends Microbiol. 13, 58–63 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Dengler Haunreiter V., et al. , In-host evolution of Staphylococcus epidermidis in a pacemaker-associated endocarditis resulting in increased antibiotic tolerance. Nat. Commun. 10, 1149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barr D. A., et al. , Serial image analysis of Mycobacterium tuberculosis colony growth reveals a persistent subpopulation in sputum during treatment of pulmonary TB. Tuberculosis (Edinb.) 98, 110–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gollan B., Grabe G., Michaux C., Helaine S., Bacterial persisters and infection: Past, present, and progressing. Annu. Rev. Microbiol. 73, 359–385 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Levin-Reisman I., et al. , Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nat. Methods 7, 737–739 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Huemer M., Mairpady Shambat S.et al. , Stress response within the host affects antibiotic persistence and treatment outcome in S. aureus infections. European Nucleotide Archive (ENA). https://www.ebi.ac.uk/ena/browser/view/PRJEB37630. Deposited 7 April 2020.
- 28.Diep B. A., et al. , Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Strauß L., et al. , Origin, evolution, and global transmission of community-acquired Staphylococcus aureus ST8. Proc. Natl. Acad. Sci. U.S.A. 114, E10596–E10604 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ford C. W., Hamel J. C., Stapert D., Yancey R. J., Establishment of an experimental model of a Staphylococcus aureus abscess in mice by use of dextran and gelatin microcarriers. J. Med. Microbiol. 28, 259–266 (1989). [DOI] [PubMed] [Google Scholar]
- 31.Nekoofar M. H., et al. , pH of pus collected from periapical abscesses. Int. Endod. J. 42, 534–538 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Bessman A. N., Page J., Thomas L. J., In vivo pH of induced soft-tissue abscesses in diabetic and nondiabetic mice. Diabetes 38, 659–662 (1989). [DOI] [PubMed] [Google Scholar]
- 33.Huemer M., Mairpady Shambat S.et al. , Stress adaptation within the host affects antibiotic tolerance and treatment outcome in Staphylococcus aureus infections. Proteomics Identifications Database (PRIDE). https://www.ebi.ac.uk/pride/archive/projects/PXD018372. Deposited 4 April 2020.
- 34.Huemer M., Mairpady Shambat S.et al. , Stress adaptation within the host affects antibiotic persistence and treatment outcome in Staphylococcus aureus infections. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148024. Deposited 2 April 2020.
- 35.Huemer M., Mairpady Shambat S.et al. , Molecular Reprogramming and phenotype switching in Staphylococcus aureus lead to high antibiotic persistence and affect therapy success. Proteomics Identifications Database (PRIDE). https://www.ebi.ac.uk/pride/archive/projects/PXD022858. Deposited 2 December 2020. [DOI] [PMC free article] [PubMed]
- 36.Pu Y., et al. , ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 73, 143–156.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Huemer M., Mairpady Shambat S.et al. , Stress adaptation within the host effects antibiotic tolerance and treatment outcome in Staphylococcus aureus infections. Proteomics Identifications Database (PRIDE). https://www.ebi.ac.uk/pride/archive/projects/PXD018332. Deposited 2 April 2020.
- 38.Kim W., et al. , A new class of synthetic retinoid antibiotics effective against bacterial persisters. Nature 556, 103–107 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feneran A. N., Kaufman W. S., Dabade T. S., Feldman S. R., Retinoid plus antimicrobial combination treatments for acne. Clin. Cosmet. Investig. Dermatol. 4, 79–92 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leyden J., Stein-Gold L., Weiss J., Why topical retinoids are mainstay of therapy for acne. Dermatol. Ther. (Heidelb.) 7, 293–304 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conlon B. P., Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. BioEssays 36, 991–996 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Wagner C., Sauermann R., Joukhadar C., Principles of antibiotic penetration into abscess fluid. Pharmacology 78, 1–10 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Haider S., Pal R., Integrated analysis of transcriptomic and proteomic data. Curr. Genomics 14, 91–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waters L. S., Storz G., Regulatory RNAs in bacteria. Cell 136, 615–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weinrick B., et al. , Effect of mild acid on gene expression in Staphylococcus aureus. J. Bacteriol. 186, 8407–8423 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam K., Torres V. J., Staphylococcus aureus secreted toxins and extracellular enzymes. Microbiol. Spectr., 7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yee R., Cui P., Shi W., Feng J., Zhang Y., Genetic screen reveals the role of purine metabolism in Staphylococcus aureus persistence to rifampicin. Antibiotics (Basel) 4, 627–642 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cameron D. R., Shan Y., Zalis E. A., Isabella V., Lewis K., A genetic determinant of persister cell formation in bacterial pathogens. J. Bacteriol. 200, e00303-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu T., et al. , The Agr quorum sensing system represses persister formation through regulation of phenol soluble modulins in Staphylococcus aureus. Front. Microbiol. 8, 2189 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parry B. R., et al. , The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell 156, 183–194 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel A., et al. , ATP as a biological hydrotrope. Science 356, 753–756 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Rosenzweig R., Moradi S., Zarrine-Afsar A., Glover J. R., Kay L. E., Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339, 1080–1083 (2013). [DOI] [PubMed] [Google Scholar]
- 53.Conlon B. P., et al. , Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conlon B. P., et al. , Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1, 16051 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Rowe S. E., et al. , Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat. Microbiol. 5, 282–290 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fauvart M., Van den Bergh B., Michiels J., Stabbed while sleeping: Synthetic retinoid antibiotics kill bacterial persister cells. Mol. Cell 70, 763–764 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Liggins M. C., Li F., Zhang L. J., Dokoshi T., Gallo R. L., Retinoids enhance the expression of cathelicidin antimicrobial peptide during reactive dermal adipogenesis. J. Immunol. 203, 1589–1597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bär J., Boumasmoud M., Kouyos R. D., Zinkernagel A. S., Vulin C., Efficient microbial colony growth dynamics quantification with ColTapp, an automated image analysis application. Sci. Rep. 10, 16084 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sulaiman J. E., Hao C., Lam H., Specific enrichment and proteomics analysis of Escherichia coli persisters from rifampin pretreatment. J. Proteome Res. 17, 3984–3996 (2018). [DOI] [PubMed] [Google Scholar]
- 60.Tomoyasu T., Mogk A., Langen H., Goloubinoff P., Bukau B., Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40, 397–413 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Huemer M., Mairpady Shambat S.et al. , Stress adaptation within the host affects antibiotic persistence and treatment outcome in Staphylococcus aureus infections. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE148024. Deposited 2 April 2020.
Supplementary Materials
Data Availability Statement
Raw data have been deposited in European Nucleotide Archive (ENA): PRJEB37630 (27); Gene Expression Omnibus (GEO): GSE148024 (34); and PRIDE: PXD018372 (33), PXD018332 (37), and PXD022858 (35) and are publicly available. All other study data are included in the article and supporting information.