Significance
Antibodies are crucial for the immune response against bacteria. To drive bacterial killing, antibodies should bind to the bacterial cell and induce the complement reaction. This requires target-bound IgGs to form hexameric IgG platforms that are kept together by noncovalent Fc-Fc interactions. Interestingly, pathogenic bacteria produce IgG-binding molecules that bind specifically to the Fc region needed for hexamerization. Here we demonstrate that staphylococcal protein A (SpA) from Staphylococcus aureus specifically blocks formation of IgG hexamers and downstream activation of complement. Furthermore, we show that IgG3 antibodies (which are not recognized by SpA) have superior capacity to activate complement and induce killing of S. aureus by human phagocytes. These insights provide a crucial rationale for optimizing antibody therapies against S. aureus.
Keywords: antibodies, complement, IgG hexamerization, staphylococcal protein A, Staphylococcus aureus
Abstract
Immunoglobulin (Ig) G molecules are essential players in the human immune response against bacterial infections. An important effector of IgG-dependent immunity is the induction of complement activation, a reaction that triggers a variety of responses that help kill bacteria. Antibody-dependent complement activation is promoted by the organization of target-bound IgGs into hexamers that are held together via noncovalent Fc-Fc interactions. Here we show that staphylococcal protein A (SpA), an important virulence factor and vaccine candidate of Staphylococcus aureus, effectively blocks IgG hexamerization and subsequent complement activation. Using native mass spectrometry and high-speed atomic force microscopy, we demonstrate that SpA blocks IgG hexamerization through competitive binding to the Fc-Fc interaction interface on IgG monomers. In concordance, we show that SpA interferes with the formation of (IgG)6:C1q complexes and prevents downstream complement activation on the surface of S. aureus. Finally, we demonstrate that IgG3 antibodies against S. aureus can potently induce complement activation and opsonophagocytic killing even in the presence of SpA. Together, our findings identify SpA as an immune evasion protein that specifically blocks IgG hexamerization.
Antibodies play a key role in the human immune response against bacterial infections. While antibodies can bind and neutralize bacterial virulence factors, they can also signal to components of the innate immune system and induce bacterial killing. To do so, antibodies bind bacterial cells via their variable (Fab) region and subsequently trigger Fc-mediated effector functions (1). The complement system, a large network of plasma proteins, forms an important effector of antibody-dependent immune protection against invading bacteria. An activated complement cascade results in efficient decoration of bacteria with C3-derived molecules that are essential to trigger highly effective phagocytic uptake via complement receptors on phagocytes. Furthermore, complement generates chemoattractants and induces direct killing of gram-negative bacteria. Because effective complement activation is an important effector mechanism of therapeutic antibodies in cancer (2), the ability of complement to kill bacteria could also be exploited for antibacterial therapies against (antibiotic-resistant) pathogens (3–5).
The antibody-driven, “classical” complement pathway is initiated when circulating C1 complexes are recruited to antibody-labeled target surfaces (6). The most abundant antibody isotype in serum is immunoglobulin (Ig) G, which is subdivided into subclasses IgG1, IgG2, IgG3, and IgG4 in order of decreasing abundance. IgG antibodies can bind surface antigens via their Fab regions and subsequently recruit C1 via their Fc region (SI Appendix, Fig. S1A). The C1 complex consists of three large units: C1q, C1r, and C1s. C1q comprises the antibody recognition unit of the C1 complex and is composed of six globular heads connected by collagen-like stalks. On binding of C1q, its associated proteases C1r and C1s are activated to cleave other complement proteins that together form enzymes on the surface that catalyze the covalent deposition of C3b molecules onto the bacterial surface (SI Appendix, Fig. S1A). C3b molecules are recognized by complement receptors on phagocytes (neutrophils, macrophages), which engulf and digest bacteria intracellularly. The deposition of C3b also results in amplification of the complement cascade and activation of downstream complement effector functions.
In recent years, it has become clear that efficient binding of C1 to target-bound IgG molecules requires IgGs to form ordered hexameric ring structures (7, 8). Cryo-electron tomography and atomic force microscopy studies revealed that the six globular heads of C1q can simultaneously bind to each of the six IgG molecules that form a hexameric binding platform (7) (SI Appendix, Fig. S1B). The formation of these hexamers is induced on antibody binding to surface-bound antigens and driven by noncovalent interactions between the Fc regions of neighboring IgG molecules (9) (Fig. 1A).
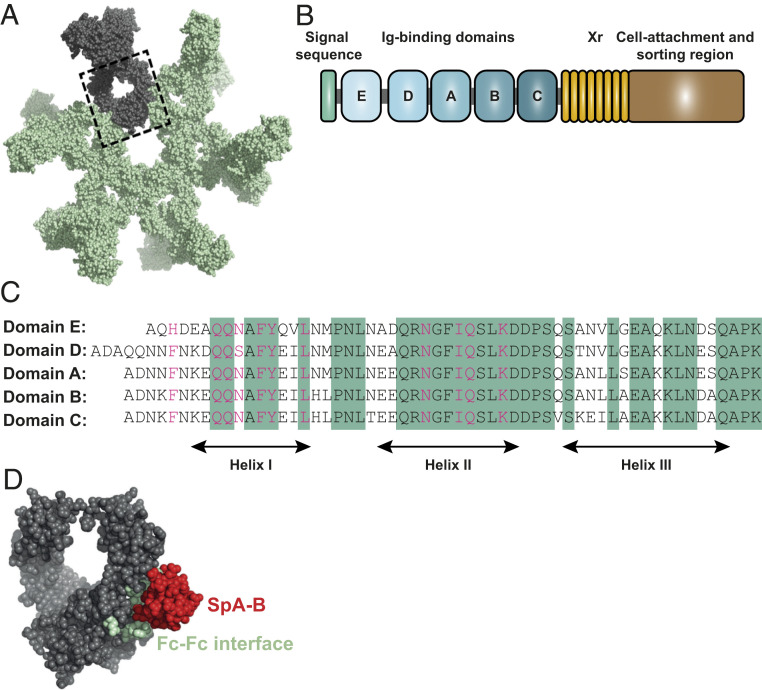
Fig. 1.
The Ig-binding domains of SpA bind to residues of the IgG-Fc region that are involved in IgG hexamerization. (A) IgG hexamer crystal packing of IgG1-b12 (Protein Data Bank [PDB] ID code 1HZH). A single IgG is depicted in gray, and the IgG-Fc domain is enclosed in the dashed box. (B) Schematic representation of SpA organization. SpA consists of a signal sequence, five Ig-binding domains (E, D, A, B, and C), an Xr region (octapeptide repeats variable in number), and a cell wall attachment and sorting region that includes a constant Xc region, the LPETG motif, a hydrophobic anchor, and positively charged residues. (C) Sequence alignment of the five highly homologous Ig-binding domains of SpA. The amino acid residues conserved in all five domains are highlighted in green. The residues involved in the interaction with the Fc region of IgG are shown in pink. (D) Space-filling presentation depicting the Fc domain of IgG1-b12 and its interaction with SpA-B (PDB ID code 1FC2; the complementary Fc docking domain for SpA-B is hidden). The residues involved in Fc-Fc interactions required to form the IgG hexameric ring are depicted in green, and the crystal structure of SpA-B is shown in red.
Interestingly, some bacteria produce IgG-binding molecules that recognize the Fc domain of IgGs (10). The best known of these is staphylococcal protein A (SpA), a 42-kDa protein that has a high affinity for the Fc region of IgG and thus is commonly used as a tool in affinity chromatography to purify monoclonal antibodies. SpA is produced by Staphylococcus aureus, an important human pathogen that is the main cause of serious hospital-acquired infections, such as bacteremia, sepsis, and endocarditis (11). Due to the dramatic increase in antibiotic resistance and the lack of proper vaccines, physicians are frequently left with no useful or suboptimal alternatives when treating these infections.
SpA is considered an important virulence factor (12, 13) and vaccine candidate (14, 15). The protein is abundantly present on the bacterial cell wall (16–18) but is also released in the extracellular environment (19, 20). SpA is composed of a signal sequence, five sequential Ig-binding domains (denoted E, D, A, B, and C), an Xr region and a cell wall attachment and sorting region (21) (Fig. 1B). Each of the five repeating Ig-binding domains adopts a three-helical structure that can bind to the Fc region of IgG via helices I and II (22) and the Fab region of the VH3 type family of antibodies via helices II and III (23–25). The binding of SpA to Ig-Fc regions is considered to protect S. aureus from phagocytic killing (14) while cross-linking of Ig-Fab regions triggers the proliferation and apoptotic collapse of B cells (26). The five Ig-binding domains are highly homologous, sharing 74 to 91% of their amino acid sequence relative to the A domain (27) (Fig. 1C). It has been demonstrated that the binding interface of the B domain of SpA (SpA-B) and IgG1-Fc involves 11 amino acid residues of SpA-B and 9 residues of IgG1-Fc (22). Interestingly, SpA binds to all IgG subclasses except IgG3, due to a substitution in one of the nine Fc-contact residues in IgG3 (His435 in IgG1 becomes Arg435 in IgG3) (28). The residue Arg435 has been suggested to cause steric hindrance to SpA when binding to the IgG3-Fc (29). The reported crystal structure of SpA-B and its IgG-Fc interaction site are depicted in Fig. 1D. Of note, SpA binds to the same interface where the IgG Fc-Fc interactions take place to form the hexameric IgG platform required for complement activation.
Here we investigated the impact of IgG-Fc binding properties of SpA on the assembly of IgG molecules into hexamers both in solution and on antigenic surfaces. We show that SpA blocks the formation of the hexameric C1q binding platform and as a result inhibits IgG-dependent complement activation and opsonophagocytic killing (OPK) of S. aureus. Our data provide an important contribution to the understanding of molecular mechanisms of complement evasion, which is crucial for the intelligent design of new therapeutic strategies to tackle infectious diseases.
Results
SpA Binds to the Fc Region of IgG1, IgG2, and IgG4 with 1:1 Stoichiometry.
To investigate whether the interaction between SpA and IgG-Fc could affect IgG-dependent complement activation, we first examined the IgG-binding properties of SpA. Most S. aureus strains express SpA with five highly homologous Ig-binding domains (A to E; Fig. 1C) that can bind to the Fc region of IgG. However, the single B-domain of SpA has been extensively used for structural and biochemical studies (22, 30), and thus here we tested both a soluble SpA construct containing all five domains (SpA) and a SpA construct consisting solely of the B-domain (SpA-B).
To exclude potential interactions between the SpA proteins and IgG Fab domains, we used human monoclonal IgGs that do not bind SpA via the Fab region. This was verified by comparing the binding of these antibodies to beads coated with different forms of SpA-B (SI Appendix, Fig. S2A). While SpA-B domain wild type (SpA-B) or SpA-B domain with abolished Fab-binding but intact Fc-binding properties (SpA-BAA; D36A and D37A mutations) bound to IgG1 antibodies, SpA-B domain with abolished Fc-binding but intact Fab-binding properties (SpA-BKK; Q9K and Q10K mutations) did not interact with IgG1 (SI Appendix, Fig. S2A and Table S1). As anticipated, none of the SpA-B forms bound to IgG3 antibodies (SI Appendix, Fig. S2A). As a reference binding experiment for all antibodies, including IgG3, we used protein G beads (protein G binds to all IgG subclasses, including IgG3) and also measured binding of a VH3 family antibody (anti-Hla IgG1) as a control for Fab binding to SpA-BKK. Of note, we further confirmed by native mass spectrometry (native MS) that the KK mutations of SpA-BKK effectively impaired binding to Fc domains (SI Appendix, Fig. S2 B and C). Native MS analyzes masses of intact protein complexes in a native state, allowing noncovalent interactions to remain intact (31, 32).
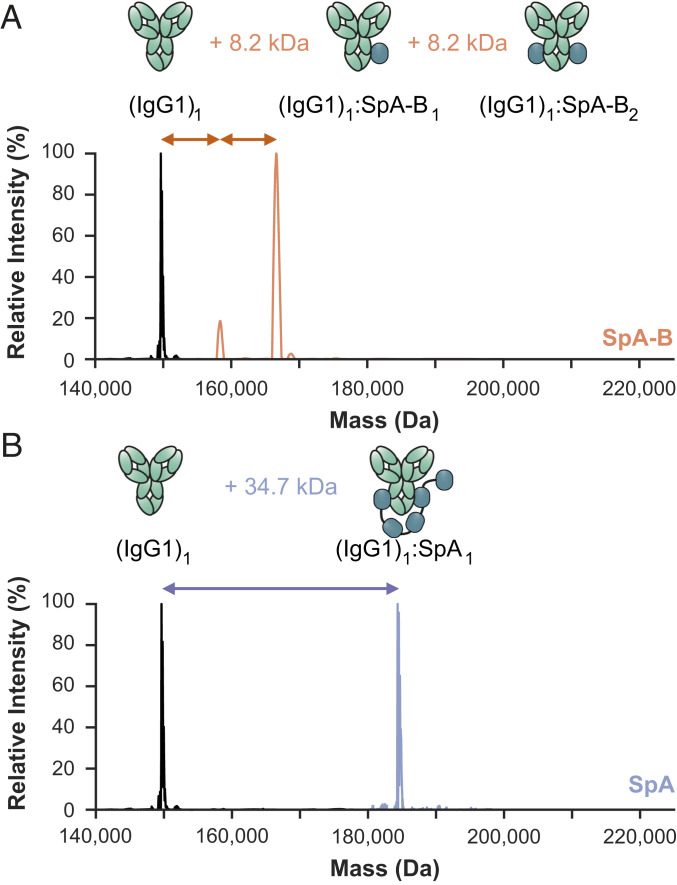
We used enzyme-linked immunosorbent assay and native MS to study the interactions between the SpA constructs and human IgGs. In line with previous studies, our results indicate that both SpA constructs bound strongly to the Fc region of all human IgG subclasses, except IgG3 (Fig. 2 and SI Appendix, Fig. S3 and Table S2). Interestingly, the binding stoichiometry differed between SpA and SpA-B. As expected with the presence of two identical binding sites on the IgG-Fc, single-domain SpA-B could—and indeed did—bind to IgG molecules with a stoichiometry of 2:1 (Fig. 2A). SpA, however, although containing five IgG-binding domains, was found to bind principally to a single IgG molecule with a stoichiometry of 1:1 (Fig. 2B). This suggests that full-length SpA may bind both IgG-Fc binding sites simultaneously.
Fig. 2.
Single-domain SpA-B binds IgG1 with 2:1 stoichiometry, whereas five-domain SpA binds IgG1 with 1:1 stoichiometry. (A and B) Deconvoluted native mass spectra show that the mass of IgG1 (black) is shifted when incubated with SpA-B (orange) (A) or SpA (blue) (B). This shift corresponds to binding of one or two copies of SpA-B to an IgG1 molecule and of only one SpA molecule to a single IgG1 molecule. The cartoon showing (IgG1)1:SpA1 binding is speculative, as it is still unknown which SpA domain interacts with which binding site on the IgG molecules.
SpA Binding Prevents IgG Molecules from Oligomerizing.
Next, we evaluated whether SpA binding could reduce IgG hexamerization in solution. Although the formation of IgG oligomers is a surface phenomenon that requires IgG binding to antigenic surfaces, this process can be mimicked in solution by introducing three mutations in the Fc region of IgG that enhance Fc-Fc interactions (7, 32). Combined mutation of residues E345R (Glu345→Arg), E430G (Glu430→Gly), and S440Y (Ser440→Tyr) results in IgG-RGY, which readily forms hexamers in solution in a dynamic equilibrium (7, 32). Here we used native MS to investigate how SpA affects the monomer-hexamer equilibrium of IgG-RGY. As previously reported (7, 32), native mass spectra of IgG1-RGY showed the presence of monomeric [denoted (IgG1)1] and hexameric [(IgG1)6] species, with intermediate states observed at lower abundance (Fig. 3A and SI Appendix, Table S3). For IgG subclasses that bind SpA, the relative abundance of IgG oligomers was drastically reduced on incubation with either SpA-B or SpA (Fig. 3 A and B and SI Appendix, Table S3). Under these conditions, we observed strong binding of SpA-B/SpA to IgG monomers but not to the hexameric IgG species (SI Appendix, Table S3).
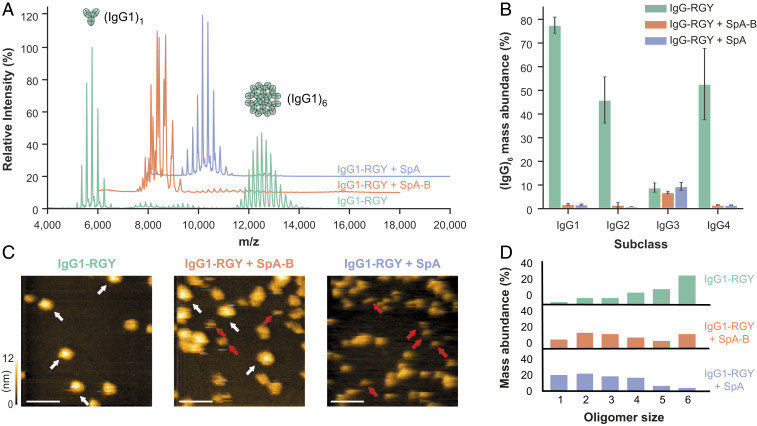
Fig. 3.
Binding of SpA-B or SpA blocks IgG-RGY monomers from assembling into higher-order oligomers. (A) Native mass spectra of IgG1-RGY in the absence (green) and presence of SpA-B (orange) or SpA (blue). (B) Hexamer relative mass abundance of IgG-RGY subclasses in the absence and presence of SpA-B or SpA, assessed by native MS. Error bars indicate the SD over three replicate samples. Although RGY mutations effectively induce hexamer formation of IgG1, IgG2, and IgG4, IgG3-RGY had a lower propensity to form hexamers in solution. (C) HS-AFM images of IgG1-RGY on DNP-SLBs in the absence and presence of SpA-B or SpA, after preincubation in solution. White arrows indicate (IgG1)6; red arrows, (IgG1)1. (Scale bars: 100 nm.) (D) IgG oligomer distribution of IgG1-RGY alone (n = 372) and in the presence of SpA-B (n = 697) or SpA (n = 386) on DNP-SLBs after preincubation in solution. The histogram displays the fraction of IgGs constituting the respective oligomer species. Oligomer distribution was quantified by force-induced dissociation. n refers to the number of individually characterized IgGs. Statistical significance between the three experiments was evaluated with the two-tailed Mann–Whitney U test. Control vs. SpA-B, P < 0.00001; control vs. SpA, P < 0.00001; SpA-B vs. SpA, P < 0.001.
Native MS measurements were corroborated by high-speed atomic force microscopy (HS-AFM) experiments on 2,4-dinitrophenol (DNP)-labeled lipid-containing supported lipid bilayers (DNP-SLBs). Preincubation of the DNP-SLBs with anti-DNP antibody IgG1-RGY alone or in combination with SpA-B or SpA resulted in distinctive distributions of different-sized IgG1-RGY oligomers (Fig. 3C). Examination of oligomer size and overall IgG1 surface density by force-induced oligomer dissociation experiments (8) allowed us to compile quantitative oligomer distributions (Fig. 3D). Preincubation of anti-DNP IgG1-RGY with multidomain SpA led to a drastic reduction in higher-order IgG1-RGY oligomers on the DNP-SLB surface (Fig. 3D), in agreement with our finding in the native MS experiments (Fig. 3B). However, while the effect of both SpA constructs on the IgG1-RGY hexamer population in solution was comparable (Fig. 3B), we observed only a ∼50% reduction of IgG1-RGY hexamers when IgG1-RGY was preincubated with the SpA-B domain on DNP-SLBs (Fig. 3D). Altogether, these data suggest that SpA binds competitively to the Fc-Fc interaction site on IgG monomers, which effectively prevents IgGs from forming higher-order IgG oligomers.
SpA Prevents the Formation of (IgG)6:C1q Complexes in Solution.
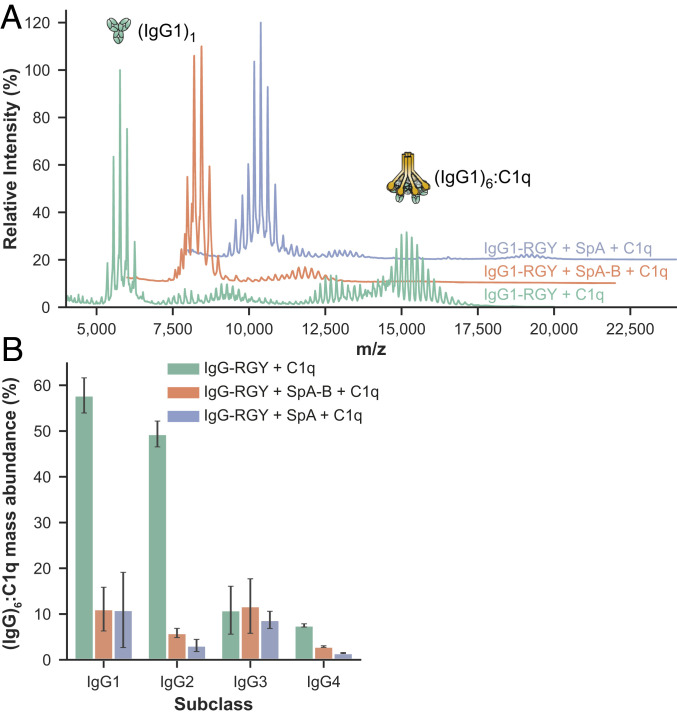
Having demonstrated that SpA blocks IgG hexamerization in solution, we next explored whether SpA affects the formation of (IgG)6:C1q complexes. We used native MS to study the formation and behavior of these complexes in the presence and absence of SpA. In agreement with earlier data (32), native mass spectra of IgG1-RGY incubated with C1q showed clearly distinguishable species with masses corresponding to (IgG1)1, (IgG1)6, and (IgG1)6:C1q (Fig. 4A and SI Appendix, Table S4). The experiments to study the effect of SpA were performed in two ways. When SpA or SpA-B was added to IgG1-RGY preincubated with C1q, the abundance of (IgG1)6:C1q complexes was strongly reduced (Fig. 4A and SI Appendix, Table S4). Likewise, after preincubation of IgG1-RGY with the SpA constructs, the addition of C1q did not lead to detectible levels of (IgG1)6:C1q complexes. Thus, regardless of the order of mixing of IgG1-RGY, C1q, and SpA, we principally detected complexes of monomeric IgG bound to SpA constructs and free unbound C1q. Similar to IgG1-RGY, both SpA constructs also prevented the assembly of (IgG)6:C1q complexes for IgG2-RGY and IgG4-RGY (Fig. 4B). Overall, these results suggest that SpA prevents the binding of C1q to IgG by blocking formation of the hexameric IgG platform in solution.
Fig. 4.
SpA-B and SpA prevent the assembly (IgG)6:C1q complexes in solution. (A) Native mass spectra of IgG1-RGY:C1q in the absence (green) and the presence of SpA-B (orange) or SpA (blue). (B) The relative mass abundance of (IgG)6:C1q complexes in the absence and presence SpA-B or SpA, assessed by native MS. Error bars indicate the SD over three replicate samples. We observed lower amounts of (IgG)6:C1q complexes using RGY mutants of IgG3 and IgG4. This is because, compared with the other subclasses, IgG3 mutants have a lower hexamerization propensity, and IgG4 has lower affinity for C1q.
SpA Inhibits Binding of C1q and C1 to Antigen-Bound IgGs on Target Surfaces.
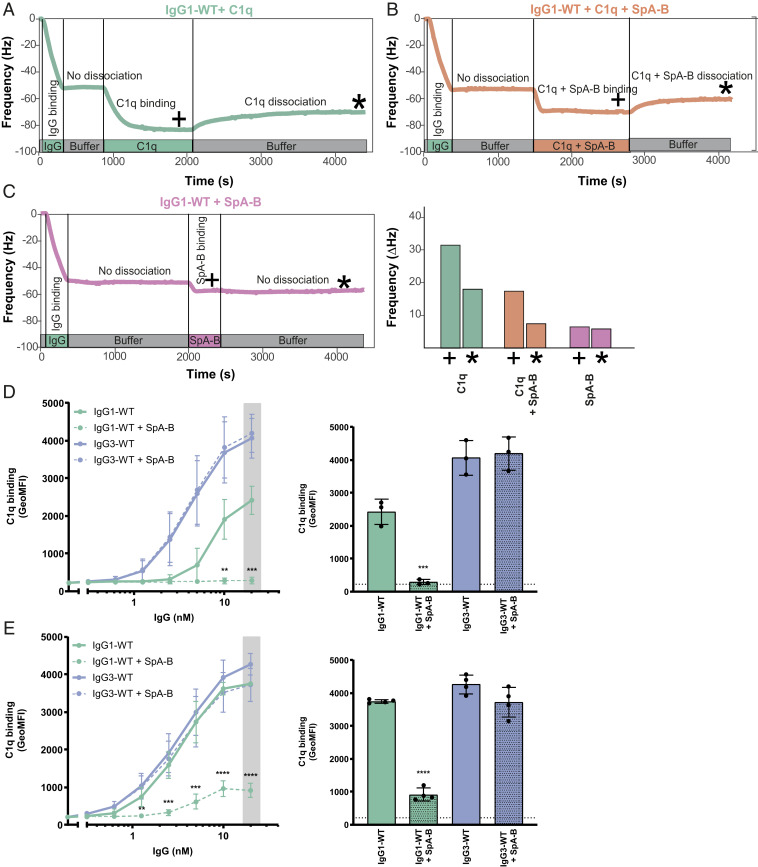
Next, we assessed how these observations for stabilized hexamers of mutant IgG-RGY in solution compare to the behavior of wild-type IgGs bound to antigenic surfaces. Since SpA and SpA-B block the formation of IgG hexamers through the same mechanism, we focused the following experiments on SpA-B. Using a quartz crystal microbalance (QCM) that we have previously used to quantitatively study binding and oligomerization of IgG1 to antigenic SLBs (9), we assessed the association of wild-type IgG1 to similar DNP-SLBs as used for HS-AFM experiments and followed the subsequent binding of C1q in the presence or absence of SpA-B (Fig. 5 A and B). Monitoring the resonance frequency change of the DNP-SLB–covered SiO2-coated quartz crystal, which is proportional to the change in bound mass, yields characteristic binding curves of anti-DNP IgG1-WT interacting with DNP-SLBs (Fig. 5 A–C, green time interval). As is evident from the constancy of the sensorgrams in the subsequent time interval (Fig. 5 A–C; gray time interval), removal of IgG1-WT from the running buffer did not induce the dissociation of considerable amounts of IgG1-WT, reflecting stable binding. After having established equal amounts of surface-bound IgGs in all three experiments, we added C1q in the absence (Fig. 5A, green time interval) and the presence of SpA-B (Fig. 5B, orange time interval), which resulted in distinct binding curves. While C1q robustly associated to IgG1-WT on our antigenic membranes (Fig. 5A), simultaneous incubation of C1q with SpA-B strongly reduced the binding signal (Fig. 5B). Addition of SpA-B alone (without C1q; Fig. 5C) resulted in a strong association of SpA-B to IgG1-WT with no detectable dissociation when SpA-B was removed from the running buffer, indicative of a high-affinity interaction. Notably, the remaining frequency shift (and thus the associated mass) at the end of the dissociation phase equaled the corresponding frequency shift for the C1q + SpA-B mixture, suggesting that the presence of SpA-B effectively precludes stable C1q binding (Fig. 5B).
Fig. 5.
SpA-B inhibits binding of C1q to antigen-bound IgGs on target surfaces. (A–C) QCM sensorgrams of C1q alone (A), C1q and SpA-B (B), or SpA-B alone (C) binding to anti-DNP wild-type IgG1 antibodies (IgG1-WT) bound to DNP-SLBs. Binding of C1q (14 nM) was followed in real time in the absence or presence of SpA-B. Binding of SpA-B alone was monitored in a similar experiment without C1q. Bars represent the respective equilibrium level (+) and the level at the end of the dissociation phase (*). (D and E) C1q binding on IgG1-WT and IgG3-WT bound to DNP-coated beads after incubation of C1q (D) or C1 complex (E) in the absence (solid lines) or presence (dotted lines) of SpA-B, detected with FITC-conjugated rabbit F(ab’)2 anti-human C1q by flow cytometry. Bars represent the same data for the 20 nM IgG concentration only, and the black dotted line shows the background fluorescence from beads not incubated with IgG. Data are presented as geometric mean fluorescence intensity (GeoMFI) ± SD of three or four independent experiments. Statistical analysis was performed using an unpaired two-tailed t test to compare buffer and SpA-B conditions. **P < 0.01; ***P < 0.001; ****P < 0.0001.
QCM observations were corroborated by flow cytometry analyses, in which DNP-coated beads (33) were first labeled with anti-DNP IgG1-WT or IgG3-WT as a control. As expected, the presence of SpA-B inhibited C1q binding to IgG1-WT–labeled beads, but binding to IgG3-WT was not altered (Fig. 5D). As a control, we confirm that binding of IgG1-WT and IgG3-WT to DNP-coated beads was similar (SI Appendix, Fig. S4A), while SpA-B could bind only to IgG1-WT–loaded target antigen (SI Appendix, Fig. S4B). Interestingly, SpA-B also blocked C1q binding when incubated after IgG1-WT:C1q complexes were formed (SI Appendix, Fig. S4C).
Because the classical complement pathway is initiated by C1q that is in complex with C1r and C1s proteases, we repeated the same experiments using C1qr2s2 complexes instead of C1q alone. Also, for this fully assembled C1 complex, we observed that SpA-B strongly reduced C1q binding to DNP-bound IgG1-WT, and that its presence did not affect C1q binding to IgG3-WT (Fig. 5E and SI Appendix, Fig. S4D). Finally, we confirmed that similar to SpA-B, SpA also largely reduced C1q binding to surface-bound IgGs on both bead surfaces (SI Appendix, Fig. S5A) and lipid membranes (SI Appendix, Fig. S5B). Altogether, these data show that both SpA constructs efficiently block C1q binding to surface-bound antibodies of the IgG1 subclass.
SpA Inhibits C1q Binding and Complement Activation on S. aureus.
We next determined whether SpA could affect antibody-dependent complement activation on the surface of S. aureus. To prevent cell surface SpA (and Sbi, another Ig-binding protein of S. aureus) (34) from binding the antibody’s Fc region, we here used a strain devoid of SpA and Sbi (NewmanΔspa/sbi). NewmanΔspa/sbi was labeled with a human monoclonal antibody directed against wall teichoic acid (WTA) (35), a highly abundant anionic glycopolymer that is covalently anchored to the peptidoglycan layer (36). Because anti-WTA antibody (clone 4497) belongs to VH3-type family (37), it is expected to bind to SpA-B or SpA via Fab region. Nevertheless, we showed that anti-WTA IgG1 antibodies do not bind to beads coated with SpA-BKK (an SpA-B mutant that binds only to the IgG-Fab region) (SI Appendix, Fig. S6A) and further confirmed by native MS that SpA-B and SpA bind only to the Fc region of this antibody (SI Appendix, Fig. S6 B–E).
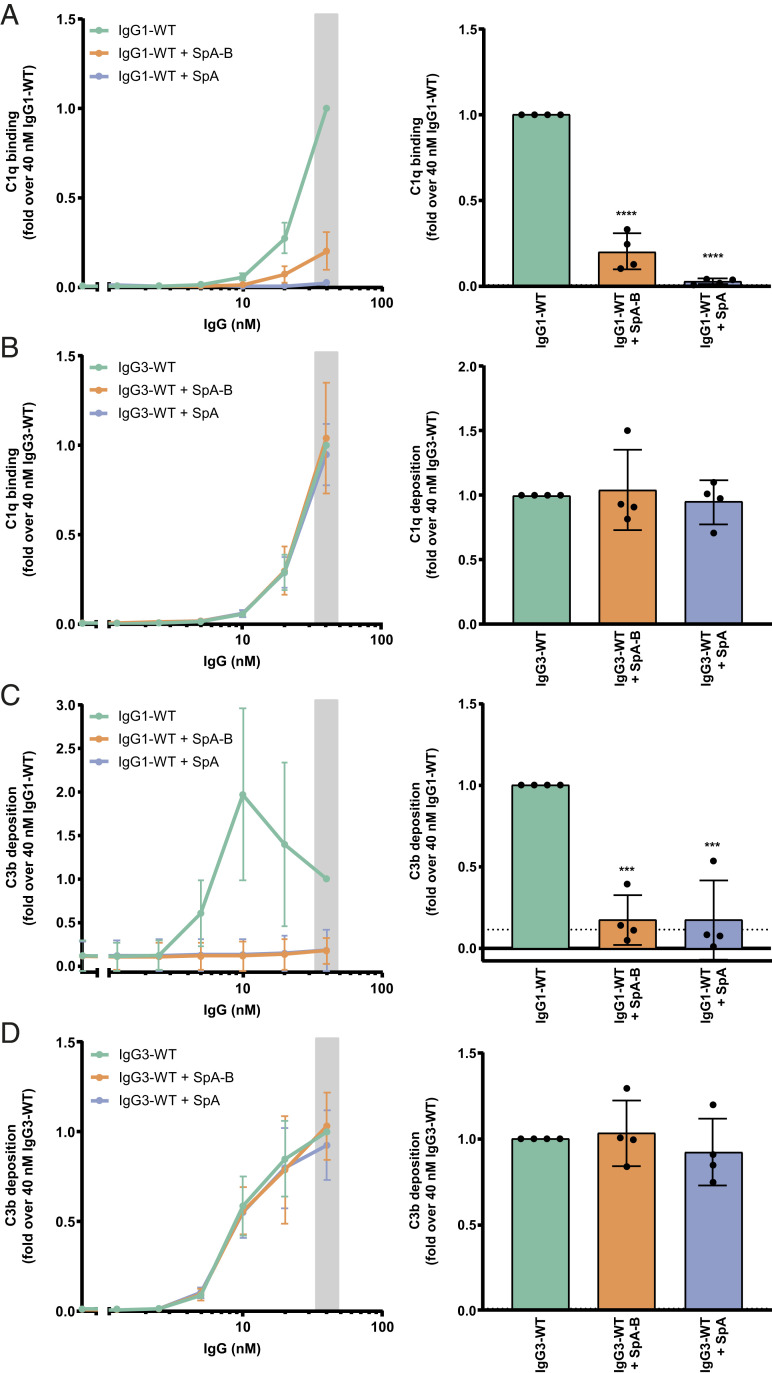
To measure C1q binding and downstream complement activation on the bacterial surface, we incubated IgG-labeled bacteria with human serum in combination with buffer, SpA-B, or SpA. Because serum contains not only complement proteins, but also many different antibodies, including IgGs that do not bind SpA (IgG3) and antibodies that bind SpA via their Fab region (VH3-type family Igs), we used serum depleted of naturally occurring antibodies (ΔIgG/IgM serum). In this way, we could exclusively determine the effect of SpA on complement activation by monoclonal anti-WTA antibodies. In agreement with the results from experiments using DNP-beads, we observed that both SpA constructs strongly reduced C1q binding on S. aureus labeled with anti-WTA IgG1 antibodies (Fig. 6A). When the bacteria were labeled with anti-WTA IgG3, C1q binding was not inhibited by the presence of SpA-B or SpA (Fig. 6B).
Fig. 6.
SpA decreases IgG-mediated C1q binding and downstream complement on S. aureus. (A and B) C1q binding on anti-WTA wild-type IgG1 (IgG1-WT) (A) or IgG3 antibodies (IgG3-WT) (B) bound to the NewmanΔspa/sbi surface after incubation of bacteria with 1% ΔIgG/IgM human serum in the absence (green) or presence of SpA-B (orange) or SpA (blue), detected with a chicken anti-human C1qA antibody by flow cytometry. (C and D) C3b deposition on the NewmanΔspa/sbi surface after incubation of bacteria with IgG1-WT (C) or IgG3-WT (D), 1% ΔIgG/IgM human serum and buffer (green), and SpA-B (orange) or SpA (blue), detected with a monoclonal murine anti-human C3d antibody by flow cytometry. Data are presented as mean ± SD fold change over 40 nM concentration of IgG control of at least three independent experiments. Bars represent the same data for the 40 nM IgG concentration only, and the black dotted line shows the background fluorescence from bacteria not incubated with IgG. Statistical analysis was performed using one-way ANOVA to compare buffer condition with SpA-B and SpA conditions. ***P < 0.001; ****P < 0.0001.
To determine whether the inhibition of C1q binding leads to downstream inhibition of the complement cascade, we also measured the deposition of C4b and C3b. The binding of C1q to target-bound antibodies activates its attached C1r/C1s proteases that cleave C4 and C2 and generate the C3 convertase (C4b2b) (38). Subsequently, the covalently attached C3 convertase catalyzes the deposition of C3b on the target surface (38). On incubation of IgG1-labeled S. aureus with human (Ig-depleted) serum and SpA-B or SpA, surface deposition of C4b and C3b was completely abolished (SI Appendix, Fig. S7A and Fig. 6C). As anticipated, C4b and C3b deposition on NewmanΔspa/sbi labeled with anti-WTA IgG3 antibodies remained unchanged in the presence of SpA-B or SpA (SI Appendix, Fig. S7B and Fig. 6D). Of note, when the C3b deposition assay was repeated to include antibodies recognizing the hapten DNP as an isotype control, we did not detect C3 products on the surface of S. aureus. Only incubation with S. aureus–recognizing anti-WTA antibodies led to detectable C3b levels, confirming that this signal reflects the presence of covalently bound C3 products that were deposited on complement activation (SI Appendix, Fig. S8). Altogether, these findings demonstrate that SpA effectively prevents C1q recruitment and downstream activation of the complement cascade on S. aureus.
SpA Decreases OPK of S. aureus by IgG1 but Not IgG3.
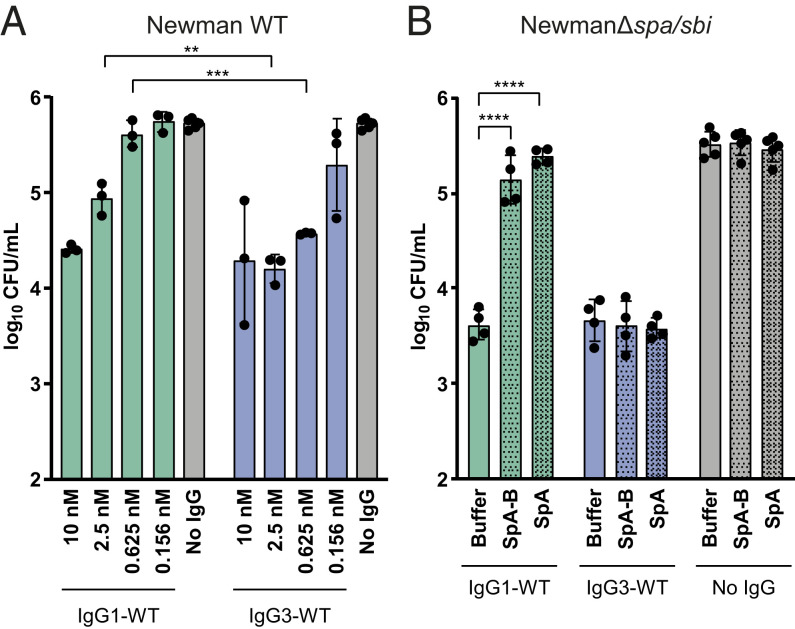
Finally, we evaluated whether the inhibitory effect of SpA on complement activation could result in less killing of S. aureus by neutrophils. Neutrophils are the first cells to be recruited from the blood to the site of infection, where they engulf and internalize bacteria via phagocytosis and subsequently kill them by exposure to antimicrobial agents such as antimicrobial peptides, reactive oxygen species and enzymes (39). Previous work from our group demonstrated that the deposition of C3-derived opsonins greatly enhances the phagocytic uptake of S. aureus (40), because C3b molecules can be recognized by complement receptors on phagocytes. To compare the OPK activity of anti-WTA IgG1 and IgG3 antibodies, we used a wild-type S. aureus strain (Newman WT) that expresses both cell-wall anchored SpA and cell-associated Sbi. The Newman WT were incubated with IgG1 or IgG3 and 1% ΔIgG/IgM serum as a complement source and then mixed with freshly isolated human neutrophils to enable phagocytosis and killing. We found that IgG3 antibodies more potently induced Newman WT killing than IgG1 antibodies (Fig. 7A).
Fig. 7.
SpA decreases OPK killing of S. aureus by IgG1 but not IgG3. (A) CFU enumeration of Newman WT after incubation with IgG1-WT (green), IgG3-WT (blue), or no IgG (gray) in presence of 1% ΔIgG/IgM human serum, followed by incubation with human neutrophils. (B) CFU enumeration of NewmanΔspa/sbi after incubation with 2.5 nM anti-WTA IgG1-WT (green), IgG3-WT (blue), or no IgG (gray) in the presence of 1% ΔIgG/IgM human serum and buffer, SpA-B, or SpA, followed by incubation with human neutrophils. Data are presented as log10 CFU/mL ± SD of at least three independent experiments. Statistical analysis was performed using an unpaired two-tailed t test to compare IgG1-WT and IgG3-WT conditions (A) or one-way ANOVA to compare buffer condition with SpA-B and SpA conditions (B). **P < 0.01; ***P < 0.001; ****P < 0.0001.
To determine whether this is indeed caused by the interaction of SpA with IgG1 antibodies, we also performed killing of NewmanΔspa/sbi in the absence or presence of exogenous SpA-B or SpA. While anti-WTA IgG1 antibodies induced OPK of S. aureus (Fig. 7B), we observed that the addition of soluble SpA-B or SpA blocked OPK (Fig. 7B). As expected, killing of S. aureus in the presence of IgG3 remained unaffected in the presence of SpA (Fig. 7B). Overall, these data suggest that both soluble and cell-exposed SpA reduce the OPK of S. aureus via IgG1 but not via IgG3.
Discussion
Antibody-dependent complement activation is an important immunologic mechanism to accelerate bacterial killing (1). To effectively trigger complement, antibodies should bind bacterial cells and subsequently form oligomeric IgG clusters that recruit C1 (7, 8). In this paper, we identify SpA from S. aureus as an example of a bacterial immune evasion molecule that specifically blocks antibody clustering by inhibiting IgG Fc-Fc contacts. These findings are of relevance to the basic pathophysiology of staphylococcal infections, and also to the development of immune therapies against S. aureus. Furthermore, SpA could be used as a tool to better understand the role of IgG clustering in various disease processes in which antibodies and complement are involved.
Our study demonstrates that by binding to the Fc region of IgG, soluble SpA blocks IgG hexamerization, resulting in inhibition of C1q recruitment, downstream complement activation on the S. aureus surface, and bacterial killing by human phagocytes. Although the interference of SpA with the classical complement pathway has been noted previously, the exact molecular mechanism has remained elusive and there has been debate about its role as an activator or inhibitor (41–45) of the complement system. Thanks to recent insights into IgG oligomerization (7) and more advanced methods for directly visualizing this process, we have been able to unravel the mechanism of SpA-dependent complement inhibition in highly purified conditions. The binding of SpA to the Fc region of monomeric, but not hexameric, IgG-RGY species suggests that SpA binds exclusively to free, unoccupied Fc regions. Thus, by binding to monomeric IgGs, SpA likely prevents the transition to the hexameric state.
Since S. aureus strains produce SpA with four or five Ig-binding domains (46), we compared the complement inhibitory activity of SpA with five Ig-binding domains versus a single SpA-B domain. AFM experiments suggest that SpA more effectively blocks hexamerization of IgG molecules on antigenic membranes than SpA-B. Because multidomain SpA is likely to bind IgG bivalently, it dissociates at a lower rate than monovalently bound SpA-B. The resulting binding affinity advantage of SpA over SpA-B allows it to compete more effectively with the IgG Fc-Fc interactions. Indeed, multidomain SpA was more effective than SpA-B in inhibiting C1q binding to target-bound IgGs, which is consistent with the notion that multivalent SpA molecules have a complement inhibitory advantage over a single SpA domain. However, on measuring downstream complement activation, the inhibitory advantage of SpA over a single SpA domain was less evident. We speculate that the requirement for multiple domains may be even more important for complement inhibition by cell-anchored SpA. Since the majority of SpA produced by S. aureus is anchored to the bacterial cell wall, multiple SpA domains may be needed to provide the molecule with sufficient length and flexibility to bind Fc domains of bacterium-bound IgG. Although the majority of SpA is cell-anchored, 6.5 to 15% is secreted before sorting (20) or released from the cell wall after enzymatic cleavage by LytM (19). We observed that the concentration of recombinant multidomain SpA necessary to decrease C1q deposition on an antigenic surface is lower than the amount reportedly secreted by Newman or USA300 strains in vitro (47).
Besides SpA, there are other bacterial proteins that bind to Ig (10). The best known are protein G (from group C and G streptococci), M and M-like proteins (group A streptococci), protein L (Peptostreptococcus magnus), Sbi, and SSL10 (S. aureus). Protein G, protein M, M-like proteins, and Sbi all bind the Fc domain of IgGs in the vicinity of the SpA-binding site (48–50), suggesting that these IgG-Fc–binding proteins may be able to block IgG hexamerization as well.
Overall, our data provide insights crucial for the development of effective immune therapies against S. aureus. It is well recognized that both neutrophils and complement play essential roles in the killing of S. aureus. While neutrophils can engulf S. aureus directly, previous work from our group demonstrated that the decoration of bacteria with C3-derived opsonins strongly enhances effective phagocytosis (40). Therefore, the identification of antibacterial antibodies with strong complement-activating potential provides an interesting approach to boost the host immune system and prevent or treat these infections. From this study, it is now clear that such approaches can be more successful if we take the SpA-dependent antibody modulation into account. Although most active and passive immunization approaches to develop or induce antibodies targeting S. aureus surface components (e.g., capsule polysaccharide, lipoteichoic acid, various surface adhesins) have failed at the clinical trial stage (5), we propose that such strategies could be more effective when also blocking the effect of SpA. In this context, although our comparison of IgG1’s and IgG3’s potency in enabling OPK of S. aureus does not allow discrimination of the inhibitory effect of SpA on FcγR-mediated from complement-mediated OPK, it suggests that monoclonal antibodies targeting surface components of S. aureus should be developed as IgG3 (or variants thereof) that are not targeted by SpA. Alternatively, any amino acid modification in IgG1 or IgG2 subclasses that prevents SpA binding to IgG, but not IgG hexamerization, would be of value. We also propose that the use of SpA as a vaccine antigen or of monoclonal antibodies directed against Fc-binding domains of SpA might be needed to prevent the anticomplement effect of SpA and thereby increase the chance of bacterial clearance.
Furthermore, SpA could also be used as a research tool to specifically examine the role of IgG hexamerization in various disease processes. For instance, SpA or a single domain of SpA (mutated to bind only the IgG-Fc region) could be used to study whether antibodies induced during an infection require Fc-Fc contacts to induce complement activation on the invading pathogen. In fact, our data with IgG1 antibodies against WTA suggest that naturally induced antibodies against S. aureus indeed require Fc-Fc contacts to induce complement activation on the surface of bacteria. Although we used monoclonal antibodies, it is well recognized that IgG1 antibodies against WTA are produced during an S. aureus infection in vivo (51–54). Also, given that excessive activation of complement has been associated with clinical manifestation of several autoimmune diseases, SpA could be used to study whether autoreactive antibodies induce IgG clustering on altered host cells. Finally, SpA, or molecules targeting the SpA-binding site in IgG, can potentially block unwanted antibody responses. Indeed, the therapeutic potential of SpA has been tested for treatment of autoimmune disorders (55–57), although the rationale for using SpA was based on its effectiveness as a B cell superantigen.
In conclusion, the identification of SpA as a biological inhibitor of IgG hexamerization will increase our understanding of antibody-dependent immunologic mechanisms and may help accelerate the development of immune interventions in infection and inflammation.
Materials and Methods
More details on the materials and methods used in this study are provided in SI Appendix, Materials and Methods.
Native MS.
Native MS analyses were performed on a modified LCT time-of-flight instrument (Waters) or a standard Exactive Plus EMR Orbitrap instrument (Thermo Fisher Scientific). Before analysis, buffers were exchanged to 150 mM ammonium acetate (pH 7.5) through six consecutive dilution and concentration steps at 4 °C using Amicon Ultra centrifugal filters with a 3-kDa or 10-kDa molecular weight cutoff (Merck). Protein complexes were assembled by mixing the subcomponents at the desired molar ratios, followed by incubation at room temperature (RT) for at least 30 min. For experiments studying the effect of SpA constructs, the incubation step with SpA proceeded for at least 3 h at 37 °C due to the relatively slow disassembly rate of the IgG-RGY hexamers. IgG-RGY hexamers were measured at a total IgG concentration of 2 µM in the presence or absence of 10 µM SpA-B or SpA (ProSpec). For measurements of (IgG)6:C1q complexes, 0.5 µM C1q (Complement Technology) was used. Samples were loaded into gold-coated borosilicate capillaries (prepared in house) for direct infusion from a static nano-electrospray ionization source. Relative abundances of protein complexes were determined using an in-house script that sums and compares ion intensities of the different species, similar to a previously described method (58). Deconvoluted mass spectra were generated by Bayesian deconvolution using UniDec (59).
HS-AFM.
HS-AFM (Research Institute of Biomolecule Metrology) was conducted in tapping mode at RT in buffer, with free amplitudes of 1.5 to 2.5 nm and amplitude set points >90%. Silicon nitride cantilevers with electron beam-deposited tips (USC-F1.2-k0.15; Nanoworld AG), nominal spring constants of 0.15 N m−1, resonance frequencies around 500 kHz, and a quality factor of ∼2 in liquids were used. Imaging was performed in buffer 1 (10 mM Hepes, 150 mM NaCl, and 2 mM CaCl2, pH 7.4). All IgGs were diluted and incubated in the same buffer.
DNP-labeled SLBs for HS-AFM were prepared on muscovite mica. The liposomes were incubated on the freshly cleaved surface (500 µg mL−1 in buffer 1), placed in a humidity chamber to prevent evaporation, and heated to 60 °C for 30 min. Then the temperature was gradually cooled to RT within 30 min, followed by exchanging the solution with buffer 1. After 10 min of equilibration at RT and 15 more buffer exchanges, the SLB was ready for imaging. In order to passivate any exposed mica, SLBs were incubated with 333 nM IgG1-b12 (irrelevant human IgG1 control antibody against HIV-1 gp120) (60) for 10 min before the molecules of interest were added.
QCM.
QCM experiments were done using a two-channel QCM-I system (MicroVacuum). AT cut SiO2-coated quartz crystals with a diameter of 14.0 mm and a resonance frequency of 5 MHz were used (Quartz Pro AB). All sensorgrams were recorded on the first, third, and fifth harmonic frequencies. The data shown are related to the third harmonic. Before each set of experiments, the SiO2-coated crystals were cleaned by immersion in 2% sodium dodecyl sulfate (SDS) for 30 min, followed by thorough rinsing with Milli-Q H2O. The chips were dried in a gentle stream of N2 and oxidized using air plasma (4 min at 80 W), then mounted in the measurement chamber. The sensor surface was further cleaned by a flow of 2% SDS at 250 µL min−1 for 5 min, followed by Milli-Q H2O at 250 µL min−1 for 5 min directly before the measurements. Finally, IgG-free buffer was injected for equilibration at 50 µL min−1. All subsequent injections were performed at 50 µL min−1. To generate DNP-SLBs on QCM chips, the DPPC:DPPE:DNP-cap-DPPE liposome stock solution was heated to 60 °C for 30 min and then slowly cooled to RT within 30 min. The solution was ready for injection after dilution to 200 µg/mL with buffer 1. DNP-SLB formation was typically complete after 30 min at 50 µL min−1, after which the flow medium was changed to buffer 1 for equilibration.
DNP-Coated Beads Assays.
Streptavidin beads (Dynabeads M-270; Invitrogen) were washed in PBS-TH (phosphate-buffered saline [PBS], 0.05% [vol/vol] Tween-20, and 0.5% human serum albumin [HSA]) and incubated (diluted 100×) with 1 µg/mL biotinylated DNP (DNP-PEG2-GSGSGSGK(Biotin)-NH2; Pepscan Therapeutics) in PBS-TH for 30 min at 4 °C with shaking. For each condition, 0.5 µL of beads was used (∼3 × 105 beads/condition). After two washes with PBS-TH, DNP-coated beads were incubated with 20 nM anti-DNP IgG or twofold serial dilutions of anti-DNP IgG (starting from 20 nM IgG) for 30 min at 4 °C with shaking (±700 rpm). The following incubation steps were performed under shaking conditions (±700 rpm) for 30 min at 4 °C unless stated otherwise. IgG-bound DNP-coated beads were washed twice with VBS-TH (Veronal buffered saline [pH 7.4], 0.5 mM CaCl2, 0.25 mM MgCl2, 0.05% [vol/vol] Tween-20, and 0.5% HSA) and incubated with 1.3 nM of purified C1q or C1 (Complement Technology) alone, in combination with, or followed by incubation of 200 nM or fourfold dilutions (starting from 1 µM) of recombinant SpA-B or SpA (ProSpec) in VBS-TH at 37 °C. Finally, the beads were washed twice with PBS-TH and incubated with 4 µg/mL fluorescein isothiocyanate (FITC)-conjugated rabbit F(ab′)2 anti-human C1q. C1q binding to the beads was detected using flow cytometry (BD FACSVerse), and data were analyzed based on a single bead population using FlowJo software.
Complement Deposition Assays on S. aureus Surface.
S. aureus NewmanΔspa/sbi strain was fluorescently labeled by transformation with the pCM29 plasmid, constitutively expressing mAmetrine under regulation of the sarA promotor, as described previously (61, 62). Bacteria were grown overnight in Todd Hewitt broth (THB) plus 10 µg/mL chloramphenicol, diluted to an OD600 of 0.05 in fresh THB plus chloramphenicol, and cultured until midlog phase (OD600 = 0.5). Cells were collected, washed, resuspended in RPMI-H medium (RPMI + 0.05% HSA), and aliquoted at −20 °C.
Similar to the Dynabeads assays, all incubation steps were performed under shaking conditions (±700 rpm) for 30 min at 4 °C (unless stated otherwise), followed by a single wash with RPMI-H by centrifugation. Bacteria (7.5 × 105 CFU) were incubated with twofold titration (starting from 40 nM) IgG in RPMI-H, followed by incubation with 1% ΔIgG/IgM serum in combination with buffer and 200 nM of SpA-B or SpA in RPMI-H for 30 min at 37 °C. For C1q detection, bacteria were incubated with 0.5 µg/mL chicken anti-human C1qA (Sigma-Aldrich), followed by incubation with phycoerythrin-conjugated donkey F(ab′)2 anti-chicken (Jackson ImmunoResearch), diluted 1:500 in RPMI-H. For C4b and C3b detection, 1 µg/mL murine anti-C4d (Quidel) or anti-C3d (Quidel) antibody was incubated with bacteria, respectively. Subsequently, bacteria were incubated with FITC-conjugated goat F(ab′)2 anti-mouse (Dako), diluted 1:100 in RPMI-H. After labeling, samples were fixed with 1% paraformaldehyde in RPMI-H, and the binding of C1q, C4b, and C3b to bacteria was detected using flow cytometry (BD FACSVerse). Data were analyzed using FlowJo software.
OPK of S. aureus by Neutrophils.
Human neutrophils were purified from blood of healthy donors by the Ficoll/Histopaque density gradient method (63). S. aureus Newman WT and NewmanΔspa/sbi constitutively expressing mAmetrine were freshly grown to midlog phase, washed, and opsonized as follows. Newman WT were incubated with fourfold titration (starting from 10 nM) of anti-WTA IgG1 or IgG3 and 1% ΔIgG/IgM serum in Hank’s balanced salt solution (HBSS) + 0.1% HSA (HBSS-H). NewmanΔspa/sbi were incubated with 2.5 nM anti-WTA IgG1 or IgG3 and 1% ΔIgG/IgM serum in the absence or presence of exogenous SpA-B or SpA (200 nM) in HBSS-H. After 30 min at 37 °C, the bacteria (8.5 × 105 CFU) were incubated with freshly isolated neutrophils for 90 min under 5% CO2 at 37 °C, at a 1:1 bacteria:cell ratio. Subsequently, neutrophils were lysed with cold 0.3% (wt/vol) saponin in water for up to 15 min on ice, and samples were serially diluted in PBS and plated in duplicate onto TSA plates. The plates were incubated overnight at 37 °C, and viable bacteria were quantified by CFU enumeration.
Ethical Statement.
Human serum and blood were obtained from healthy donors after informed consent was obtained from all subjects, in accordance with the Declaration of Helsinki. Approval from the Medical Ethics Committee of the University Medical Center Utrecht was obtained (METC protocol 07-125/C, approved March 1, 2010).
Supplementary Material
Acknowledgments
We thank Annette Stemerding for fruitful discussions. This research is supported by the Dutch Technology Foundation STW, which is part of the Netherlands Organisation for Scientific Research (NWO), and which is partly funded by Ministry of Economic Affairs (TTW-NACTAR Grant #16442 [to AJRH and SHMR]). This work was supported by the European Union’s Horizon 2020 research program H2020-MSCA-ITN (675106, to J.A.G.v.S. and F.B.), and a European Research Council (ERC) Starting Grant (639209, to S.H.M.R.). M.A.d.B. and A.J.R.H. further acknowledge funding for the large-scale proteomics facility, the Netherlands Proteomics Center, through the X-omics Road Map program (Project 184.034.019) and the EU Horizon 2020 program Epic-XS (Project 823839). A.J.R.H. and S.H.M.R. acknowledge the Utrecht University Molecular Immunology Hub. J.P. receives support from the European Fund for Regional Development (EFRE, IWB2020), the Federal State of Upper Austria, and the Austrian Science Fund (FWF, P33958 and P34164).
Footnotes
Competing interest statement: A.R.C. participated in a postgraduate studentship program at GlaxoSmithKline (GSK). F.J.B., J.A.G.v.S., K.P.M.v.K., J. Schuurman, and S.H.M.R. are listed as coinventors on a patent describing antibody therapies against Staphylococcus aureus. F.J.B., R.N.d.J., and J. Schuurman are Genmab employees. F.B. is an employee of the GSK group of companies and is a coinventor on patents for S. aureus vaccine candidates.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2016772118/-/DCSupplemental.
Data Availability.
All study data are included in the main text and SI Appendix.
References
- 1.Lu L. L., Suscovich T. J., Fortune S. M., Alter G., Beyond binding: Antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee C. H., et al. , IgG Fc domains that bind C1q but not effector Fcγ receptors delineate the importance of complement-mediated effector functions. Nat. Immunol. 18, 889–898 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laxminarayan R., et al. , Antibiotic resistance—the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Theuretzbacher U., Piddock L. J. V., Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe 26, 61–72 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Sause W. E., Buckley P. T., Strohl W. R., Lynch A. S., Torres V. J., Antibody-based biologics and their promise to combat Staphylococcus aureus infections. Trends Pharmacol. Sci. 37, 231–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaboriaud C., et al. , Structure and activation of the C1 complex of complement: Unraveling the puzzle. Trends Immunol. 25, 368–373 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Diebolder C. A., et al. , Complement is activated by IgG hexamers assembled at the cell surface. Science 343, 1260–1263 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strasser J., et al. , Unraveling the macromolecular pathways of IgG oligomerization and complement activation on antigenic surfaces. Nano Lett. 19, 4787–4796 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Strasser J., et al. , Weak fragment crystallizable (Fc) domain interactions drive the dynamic assembly of IgG oligomers upon antigen recognition. ACS Nano 14, 2739–2750 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Sidorin E. V., Solov’eva T. F., IgG-binding proteins of bacteria. Biochemistry (Mosc.) 76, 295–308 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Lowy F. D., Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Kim H. K., Kim H. Y., Schneewind O., Missiakas D., Identifying protective antigens of Staphylococcus aureus, a pathogen that suppresses host immune responses. FASEB J. 25, 3605–3612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmqvist N., Foster T., Tarkowski A., Josefsson E., Protein A is a virulence factor in Staphylococcus aureus arthritis and septic death. Microb. Pathog. 33, 239–249 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Falugi F., Kim H. K., Missiakas D. M., Schneewind O., Role of protein A in the evasion of host adaptive immune responses by Staphylococcus aureus. mBio 4, e00575-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H. K., Cheng A. G., Kim H. Y., Missiakas D. M., Schneewind O., Nontoxigenic protein A vaccine for methicillin-resistant Staphylococcus aureus infections in mice. J. Exp. Med. 207, 1863–1870 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatlin C. L., et al. , Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics 6, 1530–1549 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Ventura C. L., et al. , Identification of a novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PLoS One 5, e11634 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ythier M., et al. , Proteomic and transcriptomic profiling of Staphylococcus aureus surface LPXTG-proteins: Correlation with agr genotypes and adherence phenotypes. Mol. Cell. Proteomics 11, 1123–1139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Becker S., Frankel M. B., Schneewind O., Missiakas D., Release of protein A from the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111, 1574–1579 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O’Halloran D. P., Wynne K., Geoghegan J. A., Protein A is released into the Staphylococcus aureus culture supernatant with an unprocessed sorting signal. Infect. Immun. 83, 1598–1609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlén M., et al. , Complete sequence of the staphylococcal gene encoding protein A. A gene evolved through multiple duplications. J. Biol. Chem. 259, 1695–1702 (1984). [PubMed] [Google Scholar]
- 22.Deisenhofer J., Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry 20, 2361–2370 (1981). [PubMed] [Google Scholar]
- 23.Sasso E. H., Silverman G. J., Mannik M., Human IgM molecules that bind staphylococcal protein A contain VHIII H chains. J. Immunol. 142, 2778–2783 (1989). [PubMed] [Google Scholar]
- 24.Sasso E. H., Silverman G. J., Mannik M., Human IgA and IgG F(ab′)2 that bind to staphylococcal protein A belong to the VHIII subgroup. J. Immunol. 147, 1877–1883 (1991). [PubMed] [Google Scholar]
- 25.Graille M., et al. , Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: Structural basis for recognition of B-cell receptors and superantigen activity. Proc. Natl. Acad. Sci. U.S.A. 97, 5399–5404 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodyear C. S., Silverman G. J., Death by a B cell superantigen: In vivo VH-targeted apoptotic supraclonal B cell deletion by a Staphylococcal toxin. J. Exp. Med. 197, 1125–1139 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deis L. N., et al. , Multiscale conformational heterogeneity in staphylococcal protein a: Possible determinant of functional plasticity. Structure 22, 1467–1477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jendeberg L., et al. , Engineering of Fc(1) and Fc(3) from human immunoglobulin G to analyse subclass specificity for staphylococcal protein A. J. Immunol. Methods 201, 25–34 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Shah I. S., Lovell S., Mehzabeen N., Battaile K. P., Tolbert T. J., Structural characterization of the Man5 glycoform of human IgG3 Fc. Mol. Immunol. 92, 28–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gouda H., et al. , NMR study of the interaction between the B domain of staphylococcal protein A and the Fc portion of immunoglobulin G. Biochemistry 37, 129–136 (1998). [DOI] [PubMed] [Google Scholar]
- 31.Leney A. C., Heck A. J. R., Native mass spectrometry: What is in the name? J. Am. Soc. Mass Spectrom. 28, 5–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G., et al. , Molecular basis of assembly and activation of complement component C1 in complex with immunoglobulin G1 and antigen. Mol. Cell 63, 135–145 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Zwarthoff S. A., et al. , Functional characterization of alternative and classical pathway C3/C5 convertase activity and inhibition using purified models. Front. Immunol. 9, 1691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang L., Jacobsson K., Vasi J., Lindberg M., Frykberg L., A second IgG-binding protein in Staphylococcus aureus. Microbiology (Reading) 144, 985–991 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Lehar S. M., et al. , Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 527, 323–328 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Brown S., J. P. Santa Maria, Jr, Walker S., Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313–336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fong R., et al. , Structural investigation of human S. aureus-targeting antibodies that bind wall teichoic acid. MAbs 10, 979–991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law S. K. A., Dodds A. W., The internal thioester and the covalent binding properties of the complement proteins C3 and C4. Protein Sci. 6, 263–274 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nathan C., Neutrophils and immunity: Challenges and opportunities. Nat. Rev. Immunol. 6, 173–182 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Rooijakkers S. H. M., et al. , Immune evasion by a staphylococcal complement inhibitor that acts on C3 convertases. Nat. Immunol. 6, 920–927 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Sjöquist J., Stålenheim G., Protein A from Staphylococcus aureus. IX. Complement-fixing activity of protein A-IgG complexes. J. Immunol. 103, 467–473 (1969). [PubMed] [Google Scholar]
- 42.Kronvall G., Gewurz H., Activation and inhibition of IgG mediated complement fixation by staphylococcal protein A. Clin. Exp. Immunol. 7, 211–220 (1970). [PMC free article] [PubMed] [Google Scholar]
- 43.Stålenheim G., Castensson S., Protein A from Staphylococcus aureus conversion of complement factor C3 by aggregates between IgG and protein A. FEBS Lett. 14, 79–81 (1971). [DOI] [PubMed] [Google Scholar]
- 44.Stålenheim G., Protein A from Staphylococcus aureus XI. Fixation of human complement and of complement from guinea pig and rabbit. Acta Pathol. Microbiol. Scand. B Microbiol. Immunol. 79, 665–672 (1971). [PubMed] [Google Scholar]
- 45.Stålenheim G., Götze O., Cooper N. R., Sjöquist J., Müller-Eberhard H. J., Consumption of human complement components by complexes of IgG with protein A of Staphylococcus aureus. Immunochemistry 10, 501–507 (1973). [DOI] [PubMed] [Google Scholar]
- 46.Baum C., et al. , Non-spa-typeable clinical Staphylococcus aureus strains are naturally occurring protein A mutants. J. Clin. Microbiol. 47, 3624–3629 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoppenbrouwers T., et al. , Staphylococcal protein a is a key factor in neutrophil extracellular traps formation. Front. Immunol. 9, 165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atkins K. L., et al. , S. aureus IgG-binding proteins SpA and Sbi: Host specificity and mechanisms of immune complex formation. Mol. Immunol. 45, 1600–1611 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Sauer-Eriksson A. E., Kleywegt G. J., Uhlén M., Jones T. A., Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 3, 265–278 (1995). [DOI] [PubMed] [Google Scholar]
- 50.Frick I. M., et al. , Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc. Natl. Acad. Sci. U.S.A. 89, 8532–8536 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jung D.-J., et al. , Specific serum Ig recognizing staphylococcal wall teichoic acid induces complement-mediated opsonophagocytosis against Staphylococcus aureus. J. Immunol. 189, 4951–4959 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Lee J. H., et al. , Surface glycopolymers are crucial for in vitro anti-wall teichoic acid IgG-mediated complement activation and opsonophagocytosis of Staphylococcus aureus. Infect. Immun. 83, 4247–4255 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kurokawa K., Takahashi K., Lee B. L., The staphylococcal surface-glycopolymer wall teichoic acid (WTA) is crucial for complement activation and immunological defense against Staphylococcus aureus infection. Immunobiology 221, 1091–1101 (2016). [DOI] [PubMed] [Google Scholar]
- 54.van Dalen R., et al. , Do not discard Staphylococcus aureus WTA as a vaccine antigen. Nature 572, E1–E2 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Eftimiadi G., Vinai P., Eftimiadi C., Staphylococcal protein A as a pharmacological treatment for autoimmune disorders. J. Autoimmune Dis. 3, 40 (2017). [Google Scholar]
- 56.Bernton E., Gannon W., Kramer W., Kranz E., PRTX-100 and methotrexate in patients with active rheumatoid arthritis: A phase Ib randomized, double-blind, placebo-controlled, dose-escalation study. Clin. Pharmacol. Drug Dev. 3, 477–486 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Viau M., Zouali M., Effect of the B cell superantigen protein A from S. aureus on the early lupus disease of (NZBxNZW) F1 mice. Mol. Immunol. 42, 849–855 (2005). [DOI] [PubMed] [Google Scholar]
- 58.Wang G., Johnson A. J., Kaltashov I. A., Evaluation of electrospray ionization mass spectrometry as a tool for characterization of small soluble protein aggregates. Anal. Chem. 84, 1718–1724 (2012). [DOI] [PubMed] [Google Scholar]
- 59.Marty M. T., et al. , Bayesian deconvolution of mass and ion mobility spectra: From binary interactions to polydisperse ensembles. Anal. Chem. 87, 4370–4376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burton D. R., et al. , Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Pang Y. Y., et al. , agr-Dependent interactions of Staphylococcus aureus USA300 with human polymorphonuclear neutrophils. J. Innate Immun. 2, 546–559 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schenk S., Laddaga R. A., Improved method for electroporation of Staphylococcus aureus. FEMS Microbiol. Lett. 73, 133–138 (1992). [DOI] [PubMed] [Google Scholar]
- 63.Surewaard B. G. J., van Strijp J. A. G., Nijland R., Studying interactions of Staphylococcus aureus with neutrophils by flow cytometry and time lapse microscopy. J. Vis. Exp., e50788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the main text and SI Appendix.