Significance
Historically, it has been understood that for gene expression in eukaryotes, each messenger RNA encodes a single protein. With the recent development of technologies to sequence full-length transcripts en masse, we have discovered hundreds of examples in two species of green algae where two, three, or more proteins are translated from a single transcript. These “polycistronic” transcripts are found in diverse species throughout the green algal lineage, which highlights their biological importance. We have leveraged these findings to coexpress pairs of genes on polycistronic transcripts in vitro, which should facilitate efforts to engineer algae for research and industrial applications.
Keywords: transcriptome, bicistronic, dicistronic, leaky ribosome scanning, uORFs
Abstract
Polycistronic gene expression, common in prokaryotes, was thought to be extremely rare in eukaryotes. The development of long-read sequencing of full-length transcript isomers (Iso-Seq) has facilitated a reexamination of that dogma. Using Iso-Seq, we discovered hundreds of examples of polycistronic expression of nuclear genes in two divergent species of green algae: Chlamydomonas reinhardtii and Chromochloris zofingiensis. Here, we employ a range of independent approaches to validate that multiple proteins are translated from a common transcript for hundreds of loci. A chromatin immunoprecipitation analysis using trimethylation of lysine 4 on histone H3 marks confirmed that transcription begins exclusively at the upstream gene. Quantification of polyadenylated [poly(A)] tails and poly(A) signal sequences confirmed that transcription ends exclusively after the downstream gene. Coexpression analysis found nearly perfect correlation for open reading frames (ORFs) within polycistronic loci, consistent with expression in a shared transcript. For many polycistronic loci, terminal peptides from both ORFs were identified from proteomics datasets, consistent with independent translation. Synthetic polycistronic gene pairs were transcribed and translated in vitro to recapitulate the production of two distinct proteins from a common transcript. The relative abundance of these two proteins can be modified by altering the Kozak-like sequence of the upstream gene. Replacement of the ORFs with selectable markers or reporters allows production of such heterologous proteins, speaking to utility in synthetic biology approaches. Conservation of a significant number of polycistronic gene pairs between C. reinhardtii, C. zofingiensis, and five other species suggests that this mechanism may be evolutionarily ancient and biologically important in the green algal lineage.
The term “polycistronic” describes the situation in which two (bicistronic/dicistronic), three (tricistronic), or more separate proteins are encoded on a single molecule of messenger RNA (mRNA). In prokaryotes, polycistronic expression is common. Prokaryotic genes, usually with a shared function or pathway, are clustered into operons that are cotranscribed to generate polycistronic mRNAs. Similarly, mitochondria and plastids, which evolved from prokaryotes, express many of their genes on polycistronic transcripts. Many viral genomes express multiple genes on a common transcript in order to maximize the coding potential of their extremely compact genomes. In contrast, the paradigm for protein expression from nuclear genes in eukaryotes has been that genes are expressed monocistronically; that is, each transcript carries a single protein-coding open reading frame (ORF).
Over the last few decades, a growing number of exceptions to the paradigm of monocistronic gene expression in eukaryotes have been identified, which largely fall into two categories. The first type, described in kinetoplastids and nematodes, features operons of several genes that are transcribed polycistronically before undergoing cleavage and 5′ trans splicing of a “spliced leader” sequence to produce monocistronic mature mRNAs (1). The second type of polycistronic expression includes cases in which the mature mRNA is polycistronic (most often bicistronic), examples of which include the SNURF-SNRPN locus in humans and the tomPRO1 locus in tomato (2, 3). In Drosophila, the discovery of a dicistronic heat shock protein locus in 1988 paved the way for discovery of many more polycistronic loci: at least 168 as of 2015 (4, 5). In this work, we focus on the latter category of polycistronic nuclear gene expression.
Recently, the discovery of polycistronic loci has accelerated with the availability of new methodologies for transcript sequencing and annotation that rely on long reads of whole-transcript isomers on the PacBio and Oxford Nanopore platforms (hence, Iso-Seq). For instance, transcriptome sequencing in the fungus, Plicaturopsis crispa, revealed 314 loci where two or more ORFs were coexpressed on polycistronic transcripts (6). Similarly, a recent study in the tree cotton plant, Gossypium arboreum, used Iso-Seq for structural annotation and found 1,115 loci that exhibited evidence of polycistronic expression (7). In both studies, polycistronic expression at these loci was not exclusive (i.e., the genes that were observed on polycistronic transcripts were also identified on monocistronic transcripts). Neither study provided evidence for production of distinct polypeptides from the corresponding ORFs.
Within the Archaeplastida, the green algae (Chlorophyta) diverged from the lineage of land plants (Streptophyta) more than a billion years ago. Green algal species, such as Chlamydomonas reinhardtii, have been especially valuable reference organisms for understanding chloroplast biology (8). Recently, we developed Chromochloris zofingiensis as another reference organism for dissecting central carbon metabolism, nutrient physiology, and signaling (9, 10). Although the evolutionary lineages that include C. zofingiensis and C. reinhardtii likely diverged in the Precambrian (i.e., >541 My ago) (11), both species share a number of characteristics that make them valuable for discovery research. While both species have high-quality, chromosome-scale genome assemblies (9, 12), functional and systems biology studies are hindered by incomplete or inaccurate structural gene annotations. In an effort to improve these, we sought to describe the transcriptomes of both species with Iso-Seq. This analysis revealed pervasive polycistronic transcripts in both species. We observed 173 exclusively polycistronic loci in C. zofingiensis and 87 in C. reinhardtii. Many more loci were incompletely polycistronic (i.e., both monocistronic and polycistronic transcripts were evident). Many of the polycistronic loci are evolutionarily conserved between C. reinhardtii and C. zofingiensis and in other chlorophytes. In this work, we employ a variety of complementary in vivo and in vitro approaches to validate that hundreds of proteins in these two chlorophyte species are translated from polycistronic transcripts.
Results
Identification of Polycistronic Expression in Two Diverged Green Algal Species.
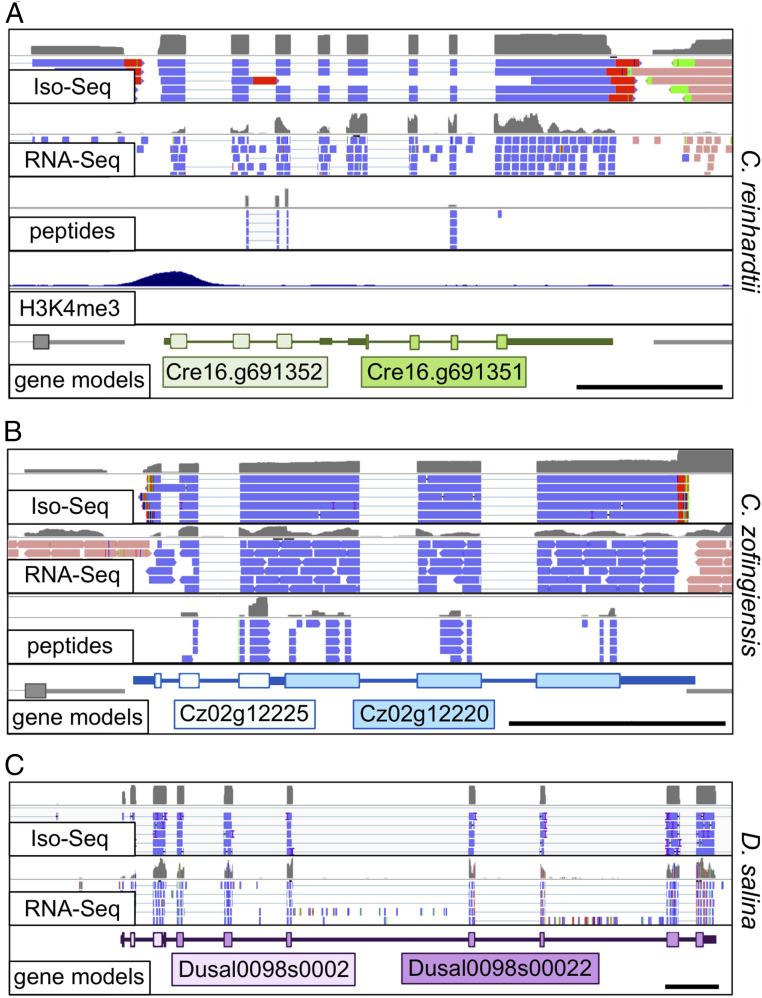
Analysis of long-read, single-molecule sequences of mRNA from C. reinhardtii and C. zofingiensis revealed hundreds of loci in which Iso-Seq reads overlapped with two or more ORFs. After extensive manual curation, the list was pared to 87 loci in C. reinhardtii and 173 loci in C. zofingiensis, in which two or more genes (up to six) were consistently and exclusively found to be associated with a single transcript (Dataset S1). Since the data on hand do not cover all genes in the present structural annotations (e.g., C. zofingiensis coverage is ∼74%) and only loci that were supported by multiple Iso-Seq reads were considered, the numbers of polycistronic loci described here represent a minimum estimate. Browser views of example bicistronic gene pairs in C. reinhardtii and C. zofingiensis are presented in Fig. 1 A and B, respectively. In addition to these loci, we noted many other loci in which one or more ORFs were alternatively polycistronic and monocistronic. For C. reinhardtii, we identified as many loci that were fully polycistronic as were partially so. In this study, we focus on the exclusively polycistronic transcripts in the two divergent species to assess whether they represent genuine polycistronic genes, as opposed to artifacts of the Iso-Seq methodology. Several criteria establish the authenticity of the polycistronic mRNAs.
Fig. 1.
Browser view of polycistronic loci in three algal species. Presented here is a display of sequencing data from single-molecule, long-read sequencing of mRNA (Iso-Seq), short-read sequencing of mRNA (RNA-Seq), mass spectrometry analysis of the proteome (peptides), and ChIP-Seq analysis with an H3K4me3 pull down (H3K4me3). Data from each of these analyses were aligned to the appropriate genome assembly for three distantly related algal species: (A) C. reinhardtii, (B) C. zofingiensis, and (C) D. salina. For Iso-Seq, RNA-Seq, and peptides, the strand is indicated by color: plus strand is light blue, and minus strand is pink. Mismatches relative to the genome assembly, including poly(A) tails, are color coded: A = red, C = orange, G = blue, and T = green. Total coverage for each track is shown above reads in gray. For gene models, a thick line indicates the ORF, an intermediate line indicates UTRs, and a thin line indicates introns. (Scale bars: 1 kb.)
Polycistronic Genes Are Smaller and More Closely Spaced than Are Monocistronic Genes.
First, we compared the properties of the candidate polycistronic loci relative to monocistronic ones. In C. reinhardtii, both the upstream ORFs (median = 600 nucleotides [nt]) and the downstream ORFs (median = 852 nt) are significantly smaller than the ORFs of the monocistronic genes (median = 1,509 nt) (SI Appendix, Fig. S1A). Similar results were observed in C. zofingiensis for upstream ORFs (median = 444 nt), downstream ORFs (median = 876 nt), and monocistronic ORFs (median = 1,245 nt) (SI Appendix, Fig. S1C). However, the combined protein-coding capacity of the polycistronic transcripts (calculated by summing the lengths of all ORFs encoded by a polycistronic transcript) was comparable with that of the monocistronic ORFs in C. reinhardtii (median = 1,539 nt) and larger than the monocistronic ORFs in C. zofingiensis (median = 1,527 nt). Next, we quantified the inter-ORF distance for colinear genes (defined here as genes on the same strand of the same chromosome with ≤20,000-nt separation between ORFs) and plotted the distribution of these for monocistronic and polycistronic gene pairs (SI Appendix, Fig. S1 B and D). Polycistronic gene pairs were dramatically closer to each other in both C. reinhardtii (median = 260 nt) and C. zofingiensis (median = 212 nt) as compared with other colinear gene pairs (median = 3,410 and 3,264 nt for C. reinhardtii and C. zofingiensis, respectively). While polycistronic ORFs were more closely spaced than are other colinear genes, uORFs (small noncoding ORFs found in the 5′ untranslated regions [UTRs] of genes) were found to be even closer on average to their corresponding primary ORFs in C. reinhardtii (median = 139 nt) and in C. zofingiensis (median = 151 nt) (SI Appendix, Fig. S1 B and D).
Stop Codon Usage and Reading Frame Are Consistent with Separate ORFs.
ORFs are delineated by start and stop codons. We considered the possibility that multiple ORFs within a transcript might actually encode a single protein by means of stop codon read through. The stop codon of the upstream gene is one factor that separates the upstream ORF from the downstream one. Therefore, we examined the proportions of ochre, amber, and opal stop codons for polycistronic upstream and downstream genes and compared these with the proportions for monocistronic genes (SI Appendix, Fig. S2 A and C). Opal stop codons were employed in nearly half of all ORFs, with only minor differences between the polycistronic and monocistronic genes, and the proportion of each type of stop codon used in the upstream polycistronic genes is not significantly different from that used in other genes in either species. Another factor relevant for read through is a shared reading frame. Two ORFs are considered to be in frame if the inter-ORF sequence between them is evenly divisible by three. When we assessed the relative reading frames of the upstream ORF compared with the downstream one, we found that ∼1/3 of the ORFs were in frame for both algae (SI Appendix, Fig. S2 B and D), which would be expected by chance. Taken together, these patterns argue against the stop codon read-through hypothesis.
Genes in Polycistronic Loci Are Highly Coexpressed, with a Shared Promoter and Polyadenylated [Poly(A)] Tail.
Genuinely polycistronic mRNAs should result from a single promoter upstream of the most 5′ located ORFs, whereas artifactual polycistronic transcripts (resulting from errors in reverse transcription and library preparation) would result from independent promoters for each gene. We used three criteria to support the former situation.
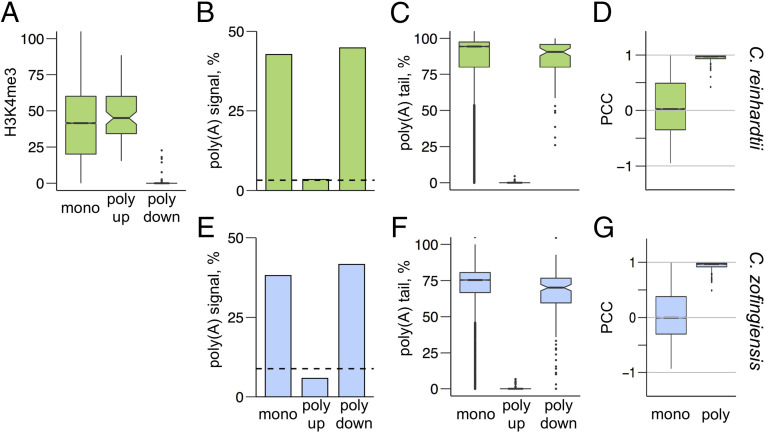
First, we sought to map promoter regions of candidate polycistronic transcripts using chromatin immunoprecipitation and sequencing (ChIP-Seq) to detect trimethylation of lysine 4 on histone H3 (H3K4me3), which is a highly stable epigenetic marker for transcription start sites in C. reinhardtii (13). The coverage of immunoprecipitated sequencing reads was compared with the coverage of input sequencing reads and used to calculate a score of H3K4me3 enrichment for each nucleotide in the genome. This is plotted in Fig. 1A as the “H3K4me3” track. The mean H3K4me3 enrichment score was calculated for the first 500 nt of each gene, and the distribution of these scores was plotted as a box plot for monocistronic, polycistronic upstream, and polycistronic downstream genes (Fig. 2A). The mean score for polycistronic upstream genes (47.7) was not significantly different from that for monocistronic genes (40.1). In contrast, polycistronic downstream genes had a dramatically lower mean score of 1.0. This evidence is consistent with transcription initiation occurring exclusively at the start of the upstream gene for these 87 loci.
Fig. 2.
Evidence of polycistronic expression. (A) ChIP-Seq was performed on C. reinhardtii DNA with an antibody to H3K4me3 to identify transcription start sites. A score of H3K4me3 marks relative to input was calculated for each nucleotide in the genome. The mean score for the 500 nt at the 5′ end of each gene model was calculated, and the distribution of these scores is plotted as a box plot for all monocistronic (“mono,” n = 17,594), polycistronic upstream (“poly up,” n = 87), and polycistronic downstream (“poly down,” n = 87) genes. (B) The presence of a UGUAA polyadenylylation signal sequence within the final 100 nt of each computationally annotated gene model was determined for C. reinhardtii for monocistronic (n = 17,594), polycistronic upstream (n = 87), and polycistronic downstream (n = 87) genes. The expected frequency of that sequence within a random 100-nt sequence with the same GC content is plotted as a dashed line. (C) Poly(A) tails were identified by the presence of eight or more sequential A’s in the Iso-Seq reads. The coverage of poly(A)-containing reads was compared with the total coverage of Iso-Seq reads within the 3′-terminal 1,000 nt of each gene model. The distribution of this poly(A)-containing coverage for genes with ≥10 Iso-Seq reads is plotted in box plots for monocistronic (n = 11,658), polycistronic upstream (n = 79), and polycistronic downstream (n = 83) genes for C. reinhardtii. (D) Colinear gene pairs (adjacent genes on the same strand of the same chromosome with ≤20,000 nt between ORFs) were identified, and a Pearson's correlation coefficient (PCC) was calculated for each gene pair across a range of RNA-Seq samples. The distributions of PCC values for C. reinhardtii for monocistronic (n = 10,884) and polycistronic (“poly,” n = 84) gene pairs are plotted as a box plot. (E) An analysis of poly(A) signal sequences was performed on C. zofingiensis for monocistronic (n = 13,585), polycistronic upstream (n = 173), and polycistronic downstream (n = 173) genes as in B. (F) An analysis of poly(A) tailing was performed on C. zofingiensis for monocistronic (n = 11,476), polycistronic upstream (n = 142), and polycistronic downstream (n = 150) genes as in C. (G) An analysis of coexpression was performed on C. zofingiensis for monocistronic (n = 12,284) and polycistronic (n = 215) gene pairs as in D. For box plots, whiskers indicate 1.5 times the interquartile range, and notches indicate the confidence interval of the median. Outliers are plotted as individual points.
Second, we surveyed the occurrence of poly(A) tails and polyadenylation signal sequences associated with each transcript. If a pair of colinear genes is exclusively expressed as a polycistronic transcript, it would be expected that the downstream but not the upstream gene would have a poly(A) tail. By this logic, upstream genes in polycistronic gene pairs would be expected to have fewer polyadenylation signal sequences than the corresponding downstream genes. The most frequently used signal for C. reinhardtii is “UGUAA” (14). To determine if the same polyadenylation signal sequence was used by C. zofingiensis, we quantified all 5-mers within the 3′ termini of C. zofingiensis transcripts. The same sequence, UGUAA, was observed at more than double the frequency of any other 5-mer. All genes were scored for the presence of UGUAA within the final 100 nt of the annotated transcript sequence (i.e., from gene models that had been computationally generated to produce classical monocistronic transcripts for each ORF). This generous range was used because the Iso-Seq data indicate substantial alternative poly(A) tailing of transcripts, and we wanted to capture putative polyadenylation signal sequences upstream of the annotated 3′ ends of the transcripts. The fraction of genes with a polyadenylation signal was sorted into polycistronic upstream, polycistronic downstream, and monocistronic (i.e., the remaining) genes (Fig. 2 B and E). Given the relative guanine-cytosine (GC) content of the two species, we calculated the expected frequency of a UGUAA 5-mer to occur by chance in a sequence of 100 nt, which is presented in the figure as a dashed line. The actual frequency of polyadenylation signals in polycistronic downstream genes is nearly identical to the frequency in monocistronic genes. In contrast, the frequency of polyadenylation signals in polycistronic upstream genes was dramatically lower and less than or equal to the frequency expected by random chance.
Next, we used Iso-Seq data to assess transcript polyadenylation. The 100 nt immediately upstream of a stretch of eight or more A’s were computationally isolated from the untrimmed Iso-Seq reads, mapped to the genome, and quantified relative to the total number of Iso-Seq reads that mapped to the same loci (Fig. 2 C and F).
Consistent with the idea that the 87 loci in C. reinhardtii and the 173 loci in C. zofingiensis are expressed as polycistronic transcripts with a single 3′ poly(A) tail, we observed almost no poly(A)-adjacent reads mapping to the 3′ ends of the upstream genes (0.1%). In contrast, we observed comparable numbers of poly(A)-adjacent reads in polycistronic downstream genes (85.6%) as was observed for monocistronic genes (86.8%).
Third, we estimated the abundance of transcripts for each gene individually (i.e., regardless of polycistronic or monocistronic expression) from RNA-Seq datasets. For a true polycistronic mRNA, we expect nearly identical abundance estimates for upstream and downstream genes. To test this, we calculated Pearson Correlation Coefficient (PCC) values to compare the similarity in transcript abundance estimates for polycistronic gene pairs across a wide range of conditions. For comparison, we also calculated PCC values for all colinear gene pairs (Fig. 2 D and G). The median PCC value for the polycistronic gene pairs was 0.97 for both C. reinhardtii and C. zofingiensis (i.e., nearly perfect correlation). PCC values for the other colinear gene pairs were widely distributed between +1 and –1, with a median value of 0.02 for C. reinhardtii and 0.003 for C. zofingiensis.
From these results (a single promoter, a single poly[A] tail, and equal abundance of transcripts for each ORF), we conclude that the 87 transcripts in C. reinhardtii and the 173 transcripts in C. zofingiensis are authentic and exclusively polycistronic.
Identification of Peptides from Upstream and Downstream Genes Validates That both ORFs Are Translated.
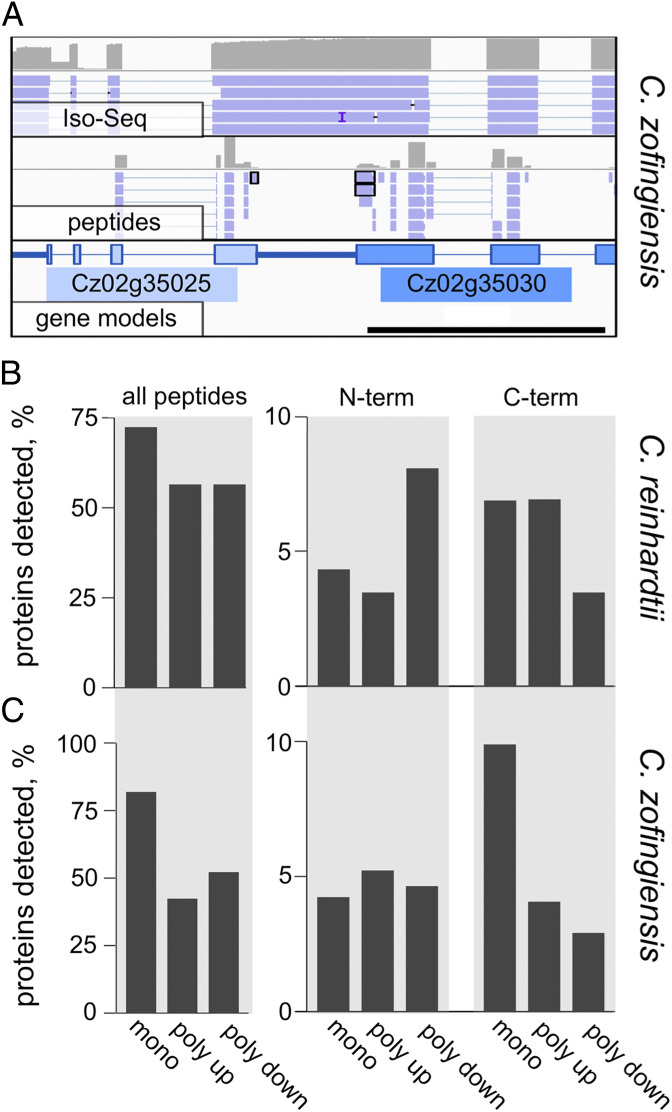
Having demonstrated that the upstream and downstream polycistronic genes are cotranscribed to produce a common mRNA, we asked whether both ORFs are translated. We queried pools of proteomics data for both C. reinhardtii and C. zofingiensis to identify peptides corresponding to proteins derived from any of the ORFs within polycistronic mRNAs (Dataset S2). The proteomic libraries used for this study were prepared from trypsin-digested total protein extracts. In addition to identifying internal peptides, we also identified N-terminal peptides (those with an N-terminal Met that is not immediately downstream of an Lys or Arg codon in the predicted ORF) or C-terminal peptides (those with a C-terminal residue that is adjacent to a stop codon in the predicted ORF). An example of a polycistronic locus from C. zofingiensis in which multiple distinct peptides were found from both the upstream ORFs and downstream ORFs is presented in Fig. 3A. Not only do the peptides validate that both ORFs are translated in vivo, they also confirm that the two proteins are translated separately, as demonstrated by the C-terminal peptide for the upstream ORF and N-terminal peptide for the downstream ORF.
Fig. 3.
Proteomic analysis validates expression of polycistronic ORFs. Peptides from the proteomes of C. reinhardtii and C. zofingiensis were identified by mass spectrometry of trypsin-digested cell lysates. (A) In order to visualize the peptides identified by this method, the sequences of identified peptides were “reverse translated” into nucleotide sequences in silico and mapped to the appropriate genome. A polycistronic gene pair from C. zofingiensis is presented with peptide and Iso-Seq data plotted against the gene models as in Fig. 1. A C-terminal peptide from the upstream gene and two N-terminal peptides from the downstream gene are highlighted. (B) The percentages of C. reinhardtii genes whose gene product was detected by at least one unambiguously assigned peptide for monocistronic (mono, n = 17,594), polycistronic upstream (poly up, n = 87), and polycistronic downstream (poly down, n = 87) genes are presented in Left under “all peptides.” The subsets of gene products that were detected by an N-terminal or C-terminal peptide are presented under columns labeled “N-term” (Center) and “C-term” (Right), respectively. (C) The percentages of detected proteins from C. zofingiensis are plotted exactly as described for B for monocistronic (n = 13,585), polycistronic up (n = 173), and polycistronic down (n = 173) genes.
Considering all polycistronic loci, we detected at least one unambiguously assigned peptide from 56% of the upstream ORFs and 56% of the downstream ORFs for C. reinhardtii (Fig. 3B). For C. zofingiensis, we detected peptides from 42% of the upstream ORFs and 52% of the downstream ORFs (Fig. 3C). This is less than the percentage of monocistronically expressed proteins that were detected: 72 and 82% for C. reinhardtii and C. zofingiensis, respectively. However, the polycistronically expressed proteins are significantly smaller than monocistronic proteins (SI Appendix, Fig. S1 A and C), and smaller proteins are detected at a lower frequency than are larger proteins (SI Appendix, Fig. S3). For example, in C. zofingiensis, 64% of ORFs had a detectable gene product in the proteomics data at the median size for polycistronic upstream ORFs (444 nt) vs. 86% at the median size for all ORFs (1,245 nt).
The percentage of polycistronic proteins that could be identified by an N-terminal or C-terminal peptide was also examined. In C. reinhardtii, an N-terminal peptide was detected for 8% of the polycistronic downstream ORFs (compared with 4% of monocistronic ORFs), and a C-terminal peptide was detected for 7% of the polycistronic upstream ORFs (compared with 7% for monocistronic ORFs) (Fig. 3B). In C. zofingiensis, 5% of polycistronic downstream ORFs were identified by an N-terminal peptide, and 4% of polycistronic upstream ORFs were identified by a C-terminal peptide (Fig. 3C). These results are consistent with independent translation of two separate ORFs, as opposed to posttranslational cleavage of a single polypeptide.
Polycistronic Translation Is Recapitulated In Vitro and Can Be Used for Synthesis of Reporters and Selectable Markers.
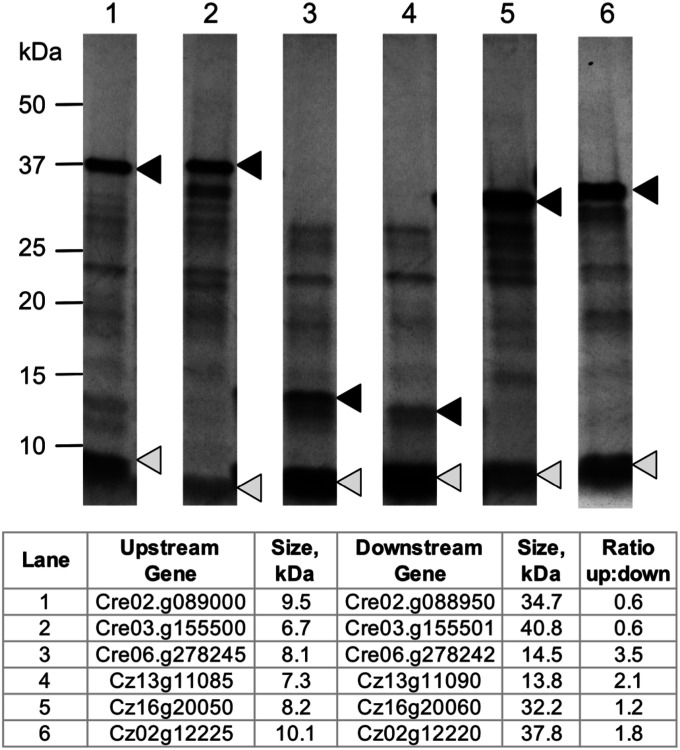
The proteomic data above validate the polycistronic functionality of the mRNAs in vivo for at least two species of green algae. To assess whether polycistronic mRNAs can be translated in classic in vitro systems, we generated constructs encoding several polycistronic gene pairs and offered them as templates for coupled in vitro transcription and translation reactions (Dataset S3). The radiolabeled translation products from wheat germ extract are presented in Fig. 4. We identified pairs of translation products at or near the predicted sizes (<20% variation from predicted size) corresponding to the ORFs for six constructs: three from C. reinhardtii and three from C. zofingiensis. The ratio of the abundance of the upstream gene product relative to the downstream one was calculated for each pair and averaged 1.6:1 for these six loci (maximum = 3.5:1; minimum = 0.6:1). As a point of comparison, the ratio of gene products expressed from an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES)-containing bicistronic vector is comparable at 0.7:1 (SI Appendix, Fig. S4).
Fig. 4.
In vitro transcription and translation of polycistronic loci. RNAs corresponding to polycistronic transcripts were synthesized from corresponding DNA templates (Methods) and translated in vitro in wheat germ extracts containing radiolabeled methionine (Met). The products were separated by denaturing polyacrylamide gel electrophoresis and visualized by fluorography. Upstream gene products are indicated by white triangles, and downstream gene products are indicated by black triangles. The polycistronic gene pairs and their expected sizes are presented as a table. Gene identifications from C. reinhardtii begin with “Cre,” and gene identifications from C. zofingiensis begin with “Cz.” The intensities of each band were normalized relative to the number of Met, and the ratios of the upstream to the downstream gene product for each pair are presented in the accompanying table.
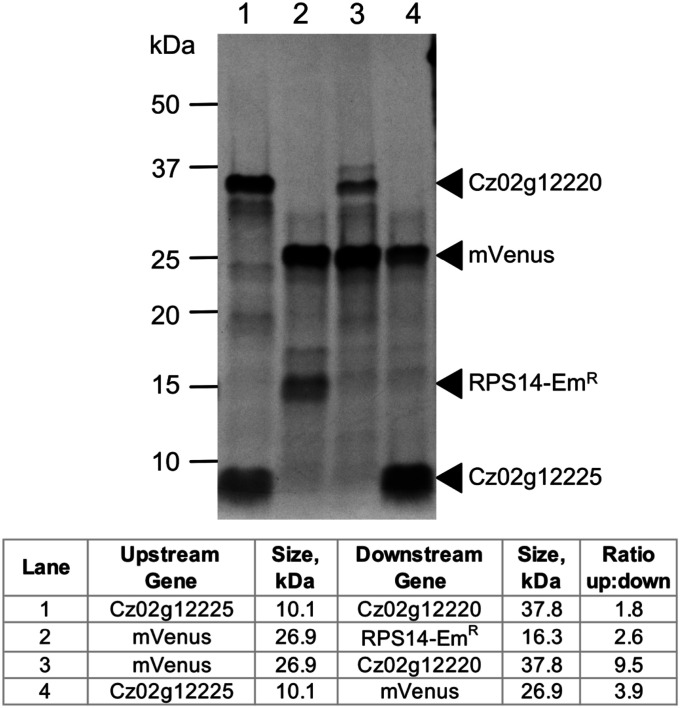
To distinguish whether foreign sequences could be translated from these mRNAs, we replaced the upstream ORFs and/or downstream ORFs with a gene encoding a reporter protein (mVenus, derived from yellow fluorescent protein) or a drug-selectable marker protein (ribosomal protein RPS14-EmR, which confers resistance to the drug emetine). Again, we noted correct synthesis of mVenus from either the upstream or downstream position in the polycistronic mRNA from C. zofingiensis Cz02g12225/Cz02g12220 (Fig. 5, lanes 3 and 4). Similarly, the inter-ORF region from a bicistronic gene pair in C. reinhardtii (Cre10.g466000/Cre10.g465950) was sufficient to coexpress both mVenus and RPS14-EmR (Fig. 5, lane 2).
Fig. 5.
Polycistronic expression of exogenous reporter and drug-selectable proteins. Proteins were in vitro translated from polycistronic transcripts exactly as in Fig. 4. Proteins in lane 1 were translated from the endogenous sequence of a bicistronic locus in C. zofingiensis as a control (same as Fig. 4, lane 6). In lanes 3 and 4, either the upstream ORF or the downstream ORF of that locus was replaced with a yellow fluorescent protein derivative, mVenus. In lane 2, both the upstream ORF and the downstream ORF of a bicistronic locus from C. reinhardtii (Cre10.g466000/Cre10.g465950) were replaced with mVenus and RPS14-EmR, which confers resistance to the drug emetine. The intensities of each band were normalized relative to the number of methionine, and the ratios of the upstream to the downstream gene product for each pair are presented in the accompanying table.
Role of Kozak-Like Sequence.
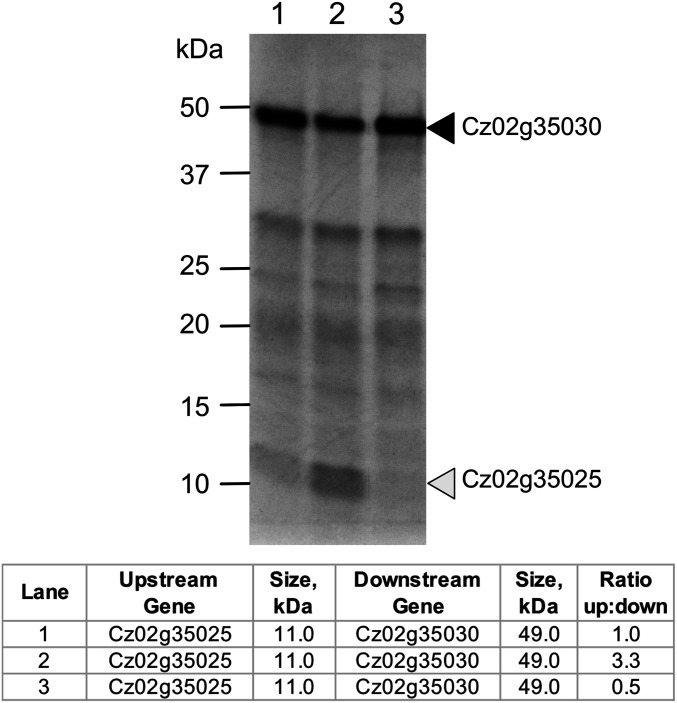
We used the in vitro translation system to test whether the synthesis of the downstream ORF depends on the synthesis of the upstream one. One mechanism for assessing this is to modify the Kozak-like sequence of the upstream ORF. We modified the endogenous sequence (ACA CCT GTC ATG CTG) associated with ORF Cz02g35025 to be stronger (ACA GCC ACC ATG CTG) or weaker (AGG GAG TTT ATG TTG) Kozak-like sequences (based on computational analysis of all Kozak-like sequences in C. zofingiensis). The endogenous sequence produced a 1:1 ratio of upstream and downstream products (Fig. 6 and SI Appendix, Fig. S5). Strengthening the Kozak sequence changed the ratio to 3.3:1, and weakening it changed the ratio to 0.5:1.
Fig. 6.
Manipulating the upstream Kozak-like sequence alters expression. Three different versions of a polycistronic locus from C. zofingiensis were synthesized and subjected to in vitro coupled transcription and translation as in Fig. 4. Each construct contained the same ORFs and inter-ORF sequence for gene 1 (Cz02g35025, 11.0 kDa) and gene 2 (Cz02g35030, 49.0 kDa). Only the nucleotides proximal to the first start codon were altered between the constructs. The construct in lane 1 contained the endogenous Kozak-like sequence, while the constructs in lanes 2 and 3 contained a stronger or weaker Kozak-like sequence, respectively. The intensities of each band were normalized relative to the number of methionine, and the ratios of the upstream to the downstream gene product for each reaction are presented in the accompanying table. Different exposures of this gel are presented in SI Appendix, Fig. S5.
Polycistronic Loci Are Conserved in the Green Algal Lineage.
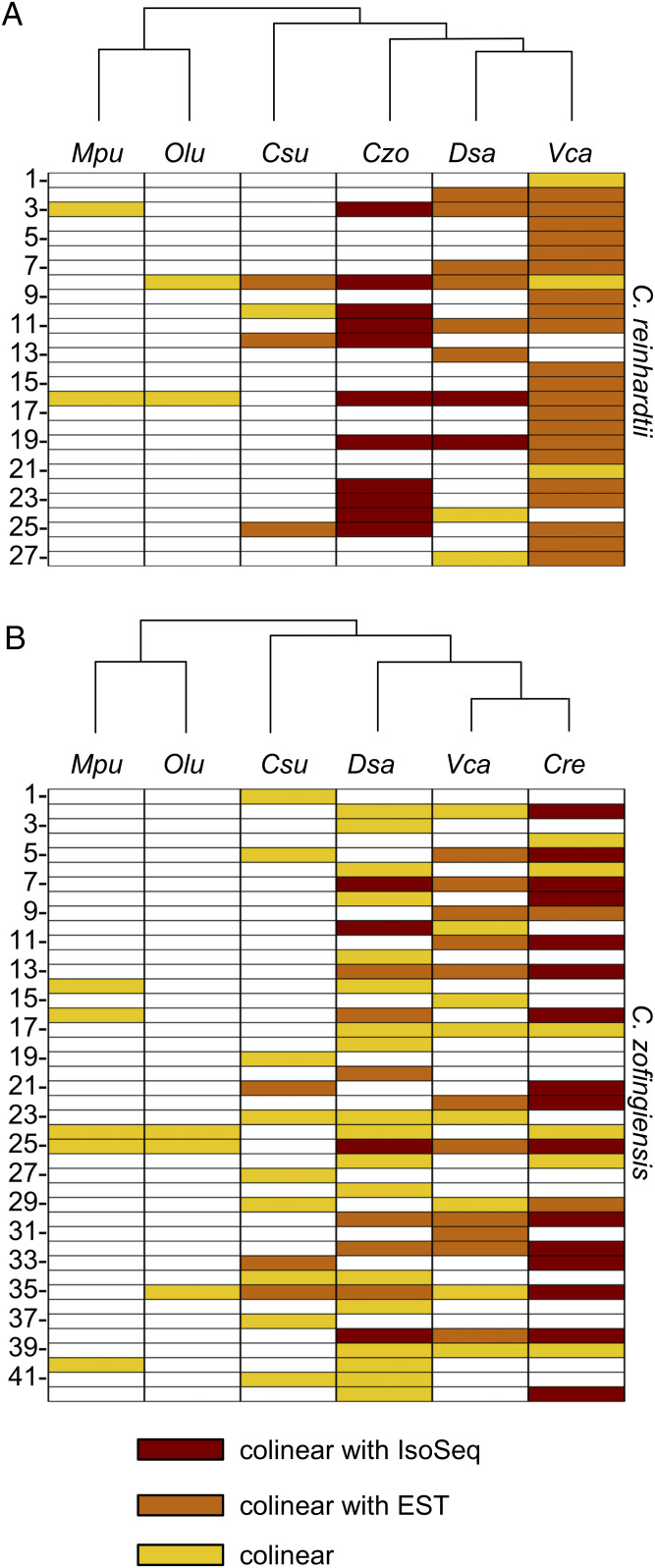
When genetic features are conserved between species that diverged hundreds of millions of years ago, those features are likely to play an important role in the physiology of those species. Given that polycistronic expression is pervasive in two chlorophytes, we wished to determine if the phenomenon extends beyond those species. The protein sequences encoded by polycistronic loci in C. reinhardtii and C. zofingiensis were used as queries in a search for candidate polycistronic loci in five other chlorophyte species: Coccomyxa subellipsoidea, Dunaliella salina, Ostreococcus lucimarinus, Micromonas pusilla, and Volvox carteri (Dataset S4). A phylogenetic tree demonstrating the evolutionary distance between these species is presented in SI Appendix, Fig. S6. For 27 of 87 polycistronic loci in C. reinhardtii, at least one other chlorophyte species had a pair of colinear ORFs (neighboring ORFs on the same strand) with significant sequence similarity (bit score ≥30) to the corresponding pair in C. reinhardtii (Fig. 7A). Seven loci from C. reinhardtii had matches in three or more species, with as many as 24 matches to V. carteri, the most closely related species in the study set. When sequences from C. zofingiensis were used as the query, 42 of 173 polycistronic loci had pairs of colinear hits in at least one other species (Fig. 7B), with the most hits, 28, shared with D. salina.
Fig. 7.
Conservation of polycistronic loci in other chlorophytes. Pairs of protein sequences encoded on polycistronic transcripts in C. reinhardtii and C. zofingiensis were used as query sequences to search for potential conserved polycistronic loci in other chlorophyte species. Pairs of proteins from 27 of 87 (31%) polycistronic loci in C. reinhardtii had significant sequence similarity (bit score ≥30) to pairs of proteins encoded by colinear ORFs (i.e., adjacent ORFs on the same reading strand) in at least one other chlorophyte species. For C. zofingiensis, this was true for 42 of 173 (24%) polycistronic loci (Dataset S4 has details). These results are summarized for (A) C. reinhardtii and (B) C. zofingiensis, where each row represents a polycistronic locus and each column represents a different chlorophyte species. Yellow bars denote that a colinear pair of ORFs was found in that species with significant similarity to a pair of polycistronic ORFs from C. reinhardtii or C. zofingiensis. For some colinear pairs of ORFs, there was additional Iso-Seq or EST data showing polycistronic transcription of the two ORFs. These are indicated in red and orange, respectively. Columns are ordered by the phylogenetic tree above each panel (SI Appendix, Fig. S6 has details). Species are labeled according to the following code: Cre, C. reinhardtii; Csu, C. subellipsoidea; Czo, C. zofingiensis; Dsa, D. salina; Mpu, M. pusilla; Olu, O. lucimarinus; Vca, V. carteri.
The observation in the other chlorophyte species of colinear ORFs with high sequence similarity to polycistronic ORFs in C. reinhardtii and C. zofingiensis is suggestive, but not dispositive, that these ORFs are expressed on polycistronic transcripts in those other species. However, Iso-Seq data from one of the other chlorophyte species, D. salina, validated that conserved, colinear ORFs were indeed expressed on polycistronic transcripts for four loci. One such bicistronic locus in D. salina is presented in Fig. 1C.
For 23 conserved loci, expressed sequence tag (EST) data from C. reinhardtii, V. carteri, D. salina, and C. subellipsoidea confirmed that the colinear ORFs are found on polycistronic transcripts. Thus, for a subset of loci it appears that polycistronic expression has been conserved for more than 700 My of evolution in the green algal lineage.
Functional Significance of Polycistronic Expression.
Given that polycistronic gene expression is conserved, what could be the biological significance of expressing two or more ORFs from a single transcript? Are polycistronic ORFs functionally coupled? A good fraction of C. reinhardtii and C. zofingiensis proteins is not associated with reliable functional annotations (15), and an even higher proportion of proteins encoded by polycistronic transcripts is of unknown function relative to the whole proteome. Only 58 polycistronic gene products (33%) from C. reinhardtii and 125 gene products (32%) from C. zofingiensis are functionally annotated with a prediction in the Phytozome database, and 77 polycistronic gene products (44%) from C. reinhardtii and 167 (43%) from C. zofingiensis have a recognizable domain in the National Center for Biotechnology Information (NCBI) conserved domain database (SI Appendix, Fig. S7). As a result, most polycistronic transcripts (83% in C. reinhardtii and 79% in C. zofingiensis) encode one or more proteins of unknown function. Many of these “pioneer” proteins (i.e., novel proteins whose function remains to be discovered) are conserved: ∼44% of the C. reinhardtii and 60% of the C. zofingiensis proteins of unknown function share sequence similarity with at least one protein from an organism other than C. zofingiensis or C. reinhardtii, respectively.
To determine whether there is an identifiable functional link between cotranscribed gene products, we manually curated the 52 polycistronic transcripts whose ORFs contain at least one conserved domain (Dataset S5). We identified several polycistronic transcripts that may be involved in nucleic acid transactions. Of these, REX1 (Cre16.g683483/Cre16.g6834950) has been experimentally analyzed in C. reinhardtii (16). At this locus, a single transcript encodes two proteins, REX1-S and REX1-B, both of which are involved in DNA repair. The smaller of the two ORFs (Cre16.g683483) encodes REX1-S, which has sequence homology to Saccharomyces cerevisiae TFB5, a core subunit of the DNA repair/basal transcription factor II human (TFIIH) complex (17). This ORF is not annotated in the current C. reinhardtii gene annotations, which rely heavily on prediction algorithms that prioritize the longest ORF in a transcript. The REX1 bicistronic locus from C. reinhardtii is conserved in C. zofingiensis, C. subellipsoidea, and V. carteri (Fig. 7 A, line 25 and B, line 33). Another example is the Cz09g28170/Cz09g28180 locus encoding two proteins putatively involved in RNA degradation: an RNA helicase and a ribonuclease. Conserved domains in the proteins encoded at the Cz04g11210/Cz04g11215 and Cre12.g513245/Cre12.g513254 loci suggest roles in cell division. Cz04g11215 has a conserved SMC domain, which suggests it could be involved in chromosome segregation, while Cz04g11210 has some similarity to the PopZ cell pole-organizing protein. Cre12.g513254 contains a conserved domain that suggests it could be a metallo-beta-lactamase protein involved in DNA repair, and Cre12.g513245 may be involved in mitosis as a putative anaphase-promoting complex subunit 15 protein. Cz19g07080/Cz19g07100 may be involved in recombination. Cz19g07080 contains the alpha-helical domain of the GINS complex protein, Psf3, which is a subunit of a complex involved in chromosome replication, while Cz19g07100 contains similarity to the C-terminal catalytic domain of Cre recombinase.
One pair of proteins is very broadly conserved in diverse algal species. Other than C. reinhardtii (Cre06.g278242/Cre06.g278345) and C. zofingiensis (Cz13g11085/Cz13g11090), it is found in four of five other chlorophyte species that were examined (Fig. 7 A, line 8 and B, line 35). This bicistronic locus encodes two proteins that are likely to function in the mitochondrion. The upstream gene product appears to be a TOM22-like protein, most similar to TOM9.2 of Arabidopsis thaliana, which assists in TOM complex assembly (18). The downstream gene product is similar to SDHAF3 of Homo sapiens, which facilitates the assembly of succinate dehydrogenase (19). The SDHAF3-like proteins in C. reinhardtii and C. zofingiensis include a leucine/tyrosine/arginine (LYR) motif, which is present in accessory or assembly factors in the mitochondrion (20). The LYR motif is common in polycistronically expressed proteins. In addition to the SDHAF3 ortholog that is coexpressed with the TOM22-like protein, we identified four other LYR-motif protein subfamilies that are encoded on bicistronic transcripts (SI Appendix, Fig. S8). Three of these subfamilies have polycistronically expressed orthologs in both C. reinhardtii and C. zofingiensis. LYR-motif proteins, which are assembly factors for various target complexes, have been proposed to be involved in tuning mitochondrial respiration to nutrient status (21). Conservation of LYR-motif proteins in bicistronic transcripts may point to a conserved functional link with respect to assembly of respiratory complexes in algae.
Discussion
Discovery and Validation of Polycistronic Transcripts.
In this work, we describe the unexpected occurrence of hundreds of polycistronic mRNAs in two diverged green algae based on the findings that 1) two or more ORFs are encoded on a single transcript and 2) two or more ORFs in a common transcript are independently translated into proteins. With regard to the first point, polycistronic genes were identified originally by the presence of long transcripts spanning two or more genes using Iso-Seq data and then substantiated by multiple independent lines of evidence (summarized in Table 1), including H3K4me3 marks that identify a single promoter in C. reinhardtii for these polycistronic gene pairs (Fig. 2A), poly(A) sequence and polyadenylation signals downstream of only the most 3′ ORF (Fig. 2 B, C, E, and F), and near-perfect correlation of transcript abundance for each ORF in the polycistronic mRNA under a wide variety of conditions (Fig. 2 D and G). The possibility that the putative polycistronic loci described in this work were instead misidentified selenoproteins was also considered and rejected (described in SI Appendix, Supplementary Text). Taken together, these data argue against artifactual fusion of transcripts from adjacent genes.
Table 1.
Summary of observations and methodologies
| Observation | Methodology |
| Transcripts spanning two or more ORFs | Iso-Seq |
| Transcription start site at upstream gene only | ChIP-Seq with H3K4me3 antibody |
| Poly(A) signal at downstream gene only | Quantification of poly(A) signal sequence in gene models |
| Poly(A) tails on downstream gene only | Modified Iso-Seq |
| Coexpression of polycistronic genes | RNA-Seq |
| In vivo translation of upstream ORFs and downstream ORFs | Proteomics |
| Translation of two ORFs from one mRNA in heterologous system | In vitro transcription/translation |
With regard to the second point, we surveyed whole-cell proteomes to document that two or more ORFs in a common transcript are each translated into proteins. The capture of the N- and C-terminal peptides from some ORFs argues against splicing of the inter-ORF sequence to generate a single polypeptide. We did not recover peptides from every ORF in the set of polycistronic RNAs, but this is not unexpected given the lower depth of coverage in typical proteomics datasets, which is in addition biased against small proteins typical of the ORFs in the polycistronic mRNAs (SI Appendix, Figs. S1 A and C and S3). When the polycistronic genes were used to prime in vitro coupled transcription and translation, two proteins were indeed synthesized for six different constructs derived from both C. reinhardtii and C. zofingiensis sequences (Fig. 4). The change in ratio of translation product resulting from manipulating the Kozak-like sequence on the most 5′ ORF (Fig. 6) not only validates their independent synthesis, but also opens the door to engineering options in synthetic biology applications.
Mechanism of Polycistronic Expression.
In eukaryotes and the viruses that infect them, several methods of polycistronic expression are known or hypothesized. One mechanism, used by viruses such as EMCV, is via an IRES. For a polycistronic mRNA with an IRES, the ribosome can assemble at either the 5′ cap for translation of the upstream gene or on an IRES in the inter-ORF region for translation of the downstream gene. We cannot rule out the possibility that there are IRESs in green algal mRNAs. However, an analysis of the inter-ORF regions of the polycistronic loci of C. reinhardtii and C. zofingiensis with an IRES prediction tool found that these sequences had significantly lower IRES prediction scores than did empirically determined IRES sequences and were no more likely to function as IRESs than random stretches of intergenic sequence (SI Appendix, Supplementary Text and Fig. S10). In further support of the idea that algal inter-ORF sequences do not function as IRESs, work by Onishi and Pringle (22) demonstrated that a short stretch (6 to 10 nt) of unremarkable sequence placed between two ORFs was dramatically more effective at conferring expression of two proteins than any of six different known IRESs in a bicistronic expression vector assayed in C. reinhardtii in vivo. In light of this work, we suggest that Onishi and Pringle (22) inadvertently recapitulated polycistronic mRNAs that are a feature of gene structure and expression in C. reinhardtii. Further, based on our work and their work, we suggest that the complex secondary structure of an IRES may not only be ineffective in C. reinhardtii but may also inhibit the endogenous mechanisms underlying polycistronic expression.
In picornaviruses, polycistronic expression is mediated by a “2A element” (23). These elements are commonly used in expression vectors to produce multiple proteins from a single transcript. In these vectors, the 2A element is cloned in frame between pairs of ORFs. As the ribosome translates through the resulting mRNA, it fails to form a peptide bond at the C-terminal end of the 2A element while continuing to translate the rest of the transcript. The effect is that two or more separate polypeptides are translated from a single mRNA molecule. In C. reinhardtii and C. zofingiensis, the polycistronic ORFs that we identified are typically separated by one or more stop codons, and further, in 2/3 of the cases, the ORFs are out of frame (SI Appendix, Fig. S2). In addition, we observed the predicted N- and C-terminal peptides for many of the polycistronically encoded proteins (Fig. 5) and that altering the Kozak-like sequence of the upstream ORF affects the ratio of the two gene products (V). These observations support the model in which there is independent translation of two or more separate ORFs from polycistronic transcripts. Therefore, we rule out 2A-like elements as the primary mechanism driving polycistronic gene expression in green algae.
Two other mechanisms have been described that can facilitate the expression of multiple gene products from a single transcript. In the first, referred to as “posttermination reinitiation,” the ribosome terminates translation of the upstream gene, but the 40S subunit, if not the whole ribosome, remains bound to the mRNA (24). The ribosome can then reinitiate translation of a second ORF if a suitable start codon is nearby. In the second mechanism, called “leaky ribosomal scanning,” a ribosome scanning for a start codon will bypass the start codon of the upstream ORF at some frequency and begin translation at the start codon of a downstream ORF instead (25). The choice to start translation at a suboptimal Kozak-like sequence may be modulated in part by the availability of eukaryotic translation initiation factor 1 (26). These two models, posttermination reinitiation and leaky ribosome scanning, are not necessarily mutually exclusive, and our work does not definitively answer the question of which of these is at play in the green algae. However, by altering the Kozak-like sequence of the upstream gene in a bicistronic construct, we were able to affect the ratio of the upstream to downstream gene product in vitro (Fig. 6), which suggests that the sequence context of the start codons is an important factor. If posttranslational reinitiation was the dominant mechanism, altering the Kozak-like sequence of the upstream ORF should affect the overall expression of both proteins but not their ratio. Hence, our observations are more consistent with the model of leaky ribosome scanning.
In previous work, we and others described the occurrence of uORFs (i.e., short ORFs located in the 5′ UTRs of genes) as regulatory features that can inhibit protein synthesis (27, 28). At the CTH1 locus in C. reinhardtii, the extended isoform of the transcript, which is a poor template for translation, has 10 uORFs, while the translation permissive isoform has none (27). Genes with uORFs are common in C. reinhardtii; nearly three-quarters of annotated genes have one or more uORFs (29). While uORFs may inhibit translation of their corresponding primary ORF, they clearly do not abolish translation, or else, 75% of genes would be functionally silent. This suggests that C. reinhardtii has the ability to translate ORFs that are downstream of the first start codon in its transcripts; a feature that may well be an intrinsic component of gene expression. If C. reinhardtii and likely, other chlorophytes have evolved to translate ORFs that are downstream of a uORF, it is easy to imagine that this ability allows for the expression of two or more ORFs from the same transcript. It may be that polycistronic expression in the green algal lineage exists on a continuum with the phenomenon of uORFs.
Applications of Polycistronic Expression.
Green algae have been promoted as vehicles for the production of biofuels, pharmaceuticals, vaccines, and for toxic substance remediation (30–33). Many of these engineering efforts rely on expression of multiple transgenes (e.g., in a multistep metabolic pathway to avoid accumulation of a toxic intermediate) (34, 35). It can also be useful to produce two or more proteins in a particular stoichiometry (36), as in a heterodimer that requires equimolar production of two polypeptides (37). The focus in many algal systems has been on coexpression of a gene of interest with a selectable marker, which can be depressingly low (22). Polycistronic gene expression may be a valuable tool to achieve objectives for transgene expression. We documented the applicability of this tool in an in vitro translation system (Fig. 5). Specifically, we could synthesize a drug-selectable protein with a reporter protein together in a polycistronic construct.
In other references systems, such as mouse, the need for coexpression of two transgenes can be achieved by the use of an IRES in the expression vector (38). Unfortunately, many IRESs that function in other organisms do not appear to confer bicistronic expression in C. reinhardtii in vivo (22). Thus, the discovery that polycistronic expression is common in species such as C. reinhardtii and C. zofingiensis should be a great boon to those attempting to engineer expression of transgenes in green algae. The observation that replacing either the upstream or downstream ORF with an exogenous transgene does not abolish polycistronic expression suggests that there is nothing inherently special about the ORFs themselves for polycistronic expression. Lastly, by altering the Kozak-like sequence of the upstream gene, it is possible to affect the ratio of the two translation products. These findings should help accelerate progress in engineering algae.
Comparison with Polycistronic Expression in Other Species.
Polycistronic expression in trypanosomes and nematodes requires the trans-splicing of a spliced leader sequence upstream of each ORF. We observed no evidence of trans-splicing in the Iso-Seq data for either C. reinhardtii or C. zofingiensis. The Iso-Seq protocol was performed using poly(A)-selected mRNA and thus, represents a snapshot of all mature, polyadenylated mRNA that was present in the cell when the RNA was collected; trans-splicing, if it had been present, should have been readily observable as mismatched bases at the 5′ ends of transcripts in the Iso-Seq data when they were aligned to the genome assembly. Thus, the phenomenon described in this work appears to be wholly different from the polycistronic expression described in nematodes and trypanosomes.
Recently, polycistronic expression was observed in several species of fungi and in tree cotton (6, 7). In both studies, polycistronic expression was “incomplete.” Specifically, polycistronic loci were also expressed monocistronically. For the purpose of this work, we chose to focus on the 87 loci in C. reinhardtii and the 173 loci in C. zofingiensis for which the observed expression was exclusively polycistronic. However, it is worth noting that we identified at least 87 additional loci in C. reinhardtii in which both monocistronic and polycistronic expressions were observable. At these loci, some fraction of the Iso-Seq reads included two or more ORFs, but some additional fraction of Iso-Seq reads was smaller and included only the upstream or downstream ORF. The presence of both partially and completely polycistronic loci in the two chlorophyte species distinguishes this work from the prior studies in tree cotton and fungi (6, 7).
With the discovery of pervasive polycistronic expression in the chlorophytes of the Archaeplastida described here, the case can be made that polycistronic expression is important in plants (G. aboreum and several species of green algae), animals (Drosophila melanogaster), and fungi (P. crispa). Increasingly, it is becoming apparent that polycistronic expression plays an important role in eukaryotic, as well as prokaryotic, gene expression.
Methods
Identification and Characterization of Polycistronic Loci.
The transcriptomes of C. reinhardtii and C. zofingiensis were modified from previously available versions (v5.6 for C. reinhardtii and v5.2.3.2 for C. zofingiensis from https://phytozome.jgi.doe.gov/pz/portal.html) with input from RNA-Seq and Iso-Seq data as detailed in SI Appendix, Supplementary Methods. Candidate polycistronic loci were flagged if they met either of the following criteria: 1) significant overlap of greater than or equal to two genes on the same strand with Iso-Seq circular consensus sequence reads or 2) genes with greater than or equal to three introns in either the 5′ and 3′ UTR. Candidate loci were then manually examined on the Integrative Genomics Viewer (39) with Iso-Seq, RNA-Seq, H3K4me3 ChIP-Seq (for C. reinhardtii only), and proteomics data. Candidate polycistronic loci were then rejected or identified as fully polycistronic or partially polycistronic based on a thorough examination of this data. Polycistronic loci were characterized in terms of ORF size, ORF spacing, stop codon usage, reading frame relative to colinear genes, H3K4me3 marks, poly(A) signal sequences, poly(A) tailing, coexpression relative to colinear genes, detection of gene products by mass spectrometry analysis, synthesis of proteins by in vitro transcription/translation, and conservation in other Chlorophyte species. A detailed description of these analyses is provided in SI Appendix, Supplementary Methods.
Supplementary Material
Acknowledgments
This work was supported by US Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research Awards DE-SC0018301 for C. zofingiensis and DE-FC02-02ER63421 for C. reinhardtii. Proteomics analyses were performed under the Facilities Integrating Collaborations for User Science Program Proposals 49262, 49840, 49960, and 50797 and used resources at the US DOE Joint Genome Institute (JGI) and the Environmental Molecular Sciences Laboratory (EMSL; grid.436923.9), which are DOE Office of Science User Facilities. The work conducted by the DOE JGI is supported by Office of Science of the US DOE Contract DE-AC02-05CH11231. The work conducted by EMSL is supported by US DOE Office of Science Contract DE-AC05-76RL01830. In vitro translation analysis was supported in part by Office of Basic Energy Sciences of the US DOE Award DE-SC0017035. The ChIP-Seq analysis was supported by a European Molecular Biology Organization Fellowship ALTF 653-2013 (to D.S.). K.K.N. is an investigator at the Howard Hughes Medical Institute.
Footnotes
Competing interest statement: S.D.G. and S.S.M. have filed a disclosure entitled “Expressing Multiple Genes from a Single Transcript in Algae and Plants.”
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2017714118/-/DCSupplemental.
Data Availability.
Iso-Seq data are available from the US NCBI Short Read Archive (SRA; accession nos. PRJNA670202 for C. reinhardtii and PRJNA657905 for C. zofingiensis). RNA-Seq data are available from the NCBI Gene Expression Omnibus (accession nos. GSE112394 for C. reinhardtii and GSE92513 for C. zofingiensis). ChIP-Seq data from C. reinhardtii are available from NCBI SRA (accession no. PRJNA681680).
References
- 1.Lasda E. L., Blumenthal T., Trans-splicing. Wiley Interdiscip. Rev. RNA 2, 417–434 (2011). [DOI] [PubMed] [Google Scholar]
- 2.García-Ríos M., et al. , Cloning of a polycistronic cDNA from tomato encoding γ-glutamyl kinase and γ-glutamyl phosphate reductase. Proc. Natl. Acad. Sci. U.S.A. 94, 8249–8254 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray T. A., Saitoh S., Nicholls R. D., An imprinted, mammalian bicistronic transcript encodes two independent proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 5616–5621 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosby M. A.et al.; FlyBase Consortium , Gene model annotations for Drosophila melanogaster: The rule-benders. G3 (Bethesda) 5, 1737–1749 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pauli D., Tonka C. H., Ayme-Southgate A., An unusual split Drosophila heat shock gene expressed during embryogenesis, pupation and in testis. J. Mol. Biol. 200, 47–53 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Gordon S. P., et al. , Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS One 10, e0132628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang K., et al. , Multi-strategic RNA-seq analysis reveals a high-resolution transcriptional landscape in cotton. Nat. Commun. 10, 4714 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salomé P. A., Merchant S. S., A series of fortunate events: Introducing chlamydomonas as a reference organism. Plant Cell 31, 1682–1707 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth M. S., et al. , Chromosome-level genome assembly and transcriptome of the green alga Chromochloris zofingiensis illuminates astaxanthin production. Proc. Natl. Acad. Sci. U.S.A. 114, E4296–E4305 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roth M. S., et al. , Regulation of oxygenic photosynthesis during trophic transitions in the green alga Chromochloris zofingiensis. Plant Cell 31, 579–601 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Del Cortona A., et al. , Neoproterozoic origin and multiple transitions to macroscopic growth in green seaweeds. Proc. Natl. Acad. Sci. U.S.A. 117, 2551–2559 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merchant S. S., et al. , The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngan C. Y., et al. , Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat. Plants 1, 15107 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Shen Y., Liu Y., Liu L., Liang C., Li Q. Q., Unique features of nuclear mRNA poly(A) signals and alternative polyadenylation in Chlamydomonas reinhardtii. Genetics 179, 167–176 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blaby-Haas C. E., Merchant S. S., Comparative and functional algal genomics. Annu. Rev. Plant Biol. 70, 605–638 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Cenkci B., Petersen J. L., Small G. D., REX1, a novel gene required for DNA repair. J. Biol. Chem. 278, 22574–22577 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Vlček D., Sevcovicová A., Sviezená B., Gálová E., Miadoková E., Chlamydomonas reinhardtii: A convenient model system for the study of DNA repair in photoautotrophic eukaryotes. Curr. Genet. 53, 1–22 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Parvin N., et al. , TOM9.2 is a calmodulin-binding protein critical for TOM complex assembly but not for mitochondrial protein import in Arabidopsis thaliana. Mol. Plant 10, 575–589 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Dwight T., et al. , Analysis of SDHAF3 in familial and sporadic pheochromocytoma and paraganglioma. BMC Cancer 17, 497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angerer H., Eukaryotic LYR proteins interact with mitochondrial protein complexes. Biology (Basel) 4, 133–150 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mills C. A., Trub A. G., Hirschey M. D., Sensing mitochondrial acetyl-CoA to tune respiration. Trends Endocrinol. Metab. 30, 1–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onishi M., Pringle J. R., Robust transgene expression from bicistronic mRNA in the green alga Chlamydomonas reinhardtii. G3 (Bethesda) 6, 4115–4125 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luke G. A., et al. , Occurrence, function and evolutionary origins of ‘2A-like’ sequences in virus genomes. J. Gen. Virol. 89, 1036–1042 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson R. J., Hellen C. U. T., Pestova T. V., “Termination and post-termination events in eukaryotic translation” in Advances in Protein Chemistry and Structural Biology, Marintchev A., Ed. (Academic Press Inc., 2012), vol. 86, pp. 45–93. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M., Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fijalkowska D., et al. , eIF1 modulates the recognition of suboptimal translation initiation sites and steers gene expression via uORFs. Nucleic Acids Res. 45, 7997–8013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moseley J. L., et al. , Reciprocal expression of two candidate di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinnebusch A. G., Translational regulation of yeast GCN4. A window on factors that control initiator-trna binding to the ribosome. J. Biol. Chem. 272, 21661–21664 (1997). [DOI] [PubMed] [Google Scholar]
- 29.Cross F. R., Tying down loose ends in the Chlamydomonas genome: Functional significance of abundant upstream open reading frames. G3 (Bethesda) 6, 435–446 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demurtas O. C., et al. , A Chlamydomonas-derived Human Papillomavirus 16 E7 vaccine induces specific tumor protection. PLoS One 8, e61473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breuer G., Lamers P. P., Martens D. E., Draaisma R. B., Wijffels R. H., The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 124, 217–226 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Liu J., Mao X., Zhou W., Guarnieri M. T., Simultaneous production of triacylglycerol and high-value carotenoids by the astaxanthin-producing oleaginous green microalga Chlorella zofingiensis. Bioresour. Technol. 214, 319–327 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Ibuot A., Dean A. P., McIntosh O. A., Pittman J. K., Metal bioremediation by CrMTP4 over-expressing Chlamydomonas reinhardtii in comparison to natural wastewater-tolerant microalgae strains. Algal Res. 24, 89–96 (2017). [Google Scholar]
- 34.Schuetze T., Meyer V., Polycistronic gene expression in Aspergillus niger. Microb. Cell Fact. 16, 162 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGary K. L., Slot J. C., Rokas A., Physical linkage of metabolic genes in fungi is an adaptation against the accumulation of toxic intermediate compounds. Proc. Natl. Acad. Sci. U.S.A. 110, 11481–11486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang Y., et al. , Engineering and modulating functional cyanobacterial CO2-fixing organelles. Front. Plant Sci. 9, 739 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shimada H., Germana S., Hayashi H., Sachs D. H., LeGuern C., Expression of MHC class II DQ α/β heterodimers from recombinant polycistronic retroviral genomes. Surg. Today 33, 183–189 (2003). [DOI] [PubMed] [Google Scholar]
- 38.Bouabe H., Fässler R., Heesemann J., Improvement of reporter activity by IRES-mediated polycistronic reporter system. Nucleic Acids Res. 36, e28 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorvaldsdóttir H., Robinson J. T., Mesirov J. P., Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Iso-Seq data are available from the US NCBI Short Read Archive (SRA; accession nos. PRJNA670202 for C. reinhardtii and PRJNA657905 for C. zofingiensis). RNA-Seq data are available from the NCBI Gene Expression Omnibus (accession nos. GSE112394 for C. reinhardtii and GSE92513 for C. zofingiensis). ChIP-Seq data from C. reinhardtii are available from NCBI SRA (accession no. PRJNA681680).