Significance
Using in vitro and in vivo approaches, this report demonstrated the increased METTL3 expression and m6A hypermethylation in response to disturbed flow and oscillatory shear stress. METTL3 hypermethylates m6A sites at various downstream targets, up-regulates NLRP1 and down-regulates KLF4, and elicits atherogenic responses. Collectively, METTL3 and m6A hypermethylation epigenetically regulates the atherogenic gene expression and phenotypes in the initiation of atherosclerosis. Our data may enlighten the field of RNA epigenetics in the pathogenic mechanisms of heart diseases.
Keywords: N6-methyladeosine RNA methylation, METTL3, shear stress, atherosclerosis, oscillatory flow
Abstract
Atherosclerosis is characterized by the plaque formation that restricts intraarterial blood flow. The disturbed blood flow with the associated oscillatory stress (OS) at the arterial curvatures and branch points can trigger endothelial activation and is one of the risk factors of atherosclerosis. Many studies reported the mechanotransduction related to OS and atherogenesis; however, the transcriptional and posttranscriptional regulatory mechanisms of atherosclerosis remain unclear. Herein, we investigated the role of N6-methyladenosine (m6A) RNA methylation in mechanotransduction in endothelial cells (ECs) because of its important role in epitranscriptome regulation. We have identified m6A methyltransferase METTL3 as a responsive hub to hemodynamic forces and atherogenic stimuli in ECs. OS led to an up-regulation of METTL3 expression, accompanied by m6A RNA hypermethylation, increased NF-κB p65 Ser536 phosphorylation, and enhanced monocyte adhesion. Knockdown of METTL3 abrogated this OS-induced m6A RNA hypermethylation and other manifestations, while METTL3 overexpression led to changes resembling the OS effects. RNA-sequencing and m6A-enhanced cross-linking and immunoprecipitation (eCLIP) experiments revealed NLRP1 and KLF4 as two hemodynamics-related downstream targets of METTL3-mediated hypermethylation. The METTL3-mediated RNA hypermethylation up-regulated NLRP1 transcript and down-regulated KLF4 transcript through YTHDF1 and YTHDF2 m6A reader proteins, respectively. In the in vivo atherosclerosis model, partial ligation of the carotid artery led to plaque formation and up-regulation of METTL3 and NLRP1, with down-regulation of KLF4; knockdown of METTL3 via repetitive shRNA administration prevented the atherogenic process, NLRP3 up-regulation, and KLF4 down-regulation. Collectively, we have demonstrated that METTL3 serves a central role in the atherogenesis induced by OS and disturbed blood flow.
Vascular endothelial cells lining the inner surface of blood vessels are exposed to different types of shear flow, including pulsatile and oscillatory flows. These patterns of blood flow and the associated shear stresses induce mechanotransduction in the endothelial cells (ECs) to modulate vascular functions and homeostasis (1). Pulsatile shear stress (PS) occurs at straight vessels and maintains vascular homeostasis (2). In contrast, the oscillatory shear stress (OS) at vessel branch points and curvatures contributes to inflammatory and atherogenic responses (2). These patterns of shear stress acting on ECs have been shown to be key determinants for the balance between resistance and susceptibility to cardiovascular diseases (3). Several lines of evidence have demonstrated that OS can sensitize the endothelium and promote atherogenesis. Endothelial transcription factors, coactivators, and repressors are all responsive to shear stresses to modulate the atherogenic processes (4). Epigenetic modifications are sensitive to flow patterns to regulate endothelial gene expression via epigenomic DNA methylation, posttranscriptional histone modifications, and other mechanisms involving noncoding RNAs (4, 5). However, the precise mechanisms of epigenetic modifications in shear stress-induced EC gene expression patterns and phenotypes remain unclear.
There is increasing evidence that posttranscriptional epitranscriptomic RNA modifications play important roles in modulating cellular functions, including stem cell differentiation (6), tumorigenesis (7), and stress-related responses (8). N6-methyladenosine (m6A) RNA methylation, the most abundant modification of mammalian messenger RNA (mRNA), is a reversible chemical modification regulated by the coordinated actions of “writer” methyltransferases and “eraser” demethylases (9). The “reader” proteins then recognize the m6A marks on mRNA to modulate RNA decay, splicing, translation, and localization. METTL3 is a key member of m6A “writer” methyltransferase complexes that have been reported to regulate various processes such as endothelial-to-hematopoietic transition (EHT) (10), T cell homeostasis (11), and stress response in the brain (12). METTL3-dependent m6A methylation in the heart has been shown to play a crucial role in cardiomyocyte hypertrophy and other heart diseases (13). However, the epitranscriptomic role of METTL3-dependent m6A modification in the shear stress modulation of EC functions under normal and disease conditions (e.g., atherosclerosis) remains to be determined.
In the present study, we analyzed the m6A RNA methylation status in experimental atherogenic models in which ECs were either exposed to OS condition in a shear flow device or treated with the proatherogenic cytokine tumor necrosis factor-alpha (TNF-α). The effects on epitranscriptome m6A writer were compared between normal and atherogenic conditions, including determinations in different aortic regions exposed to PS and OS, and in normal and atherosclerotic aortas. Using the next generation sequencing (NGS) and m6A-enhanced cross-linking and immunoprecipitation (eCLIP) technologies, the downstream molecules hypermethylated by the major m6A writer were determined. The changes in m6A modifications, m6A writer, and downstream targets were further confirmed in the mouse atherosclerosis model induced by partial ligation of carotid artery. Our findings reveal a mechanism involving m6A modifications and aberrant expression of epitranscriptome m6A writer that may contribute to the atherogenic gene expression and phenotypes in the initiation of atherosclerosis.
Results
Atherogenic Oscillatory Shear Flow Increases METTL3-Dependent m6A Methylation.
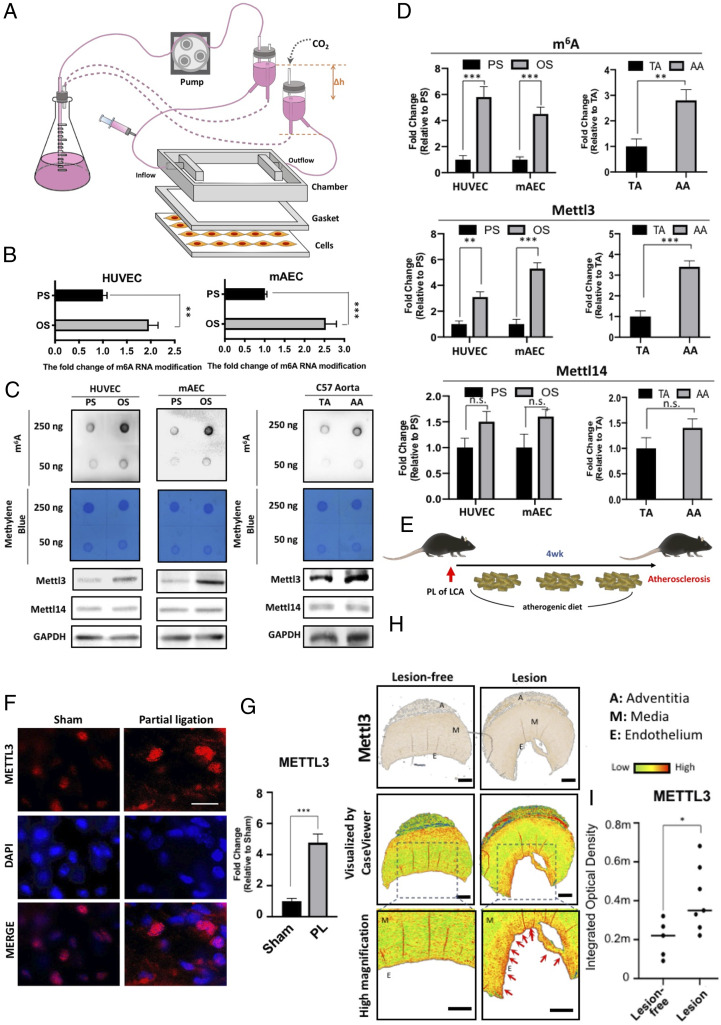
To investigate the role of m6A modification in EC response to different patterns of shear stress, human umbilical vein endothelial cells (HUVECs) and mouse aortic endothelial cells (MAECs) were exposed to pulsatile shear flow (PS) at a shear stress of 12 ± 4 dyn/cm2 or oscillatory shear flow (OS) at a shear stress of 0.5 ± 4 dyn/cm2 in a parallel-plate flow chamber for 48 h (Fig. 1A). MAECs (catalog no. T4592) was purchased from Applied Biological Materials Inc., and HUVECs (catalog no. C-12206) were obtained from Promocell. m6A modification of mRNA quantified by liquid chromatography-tandem mass spectrometry (LCMS) was found to be 2–2.5 times more abundant in both HUVECs and HAECs subjected to OS, in comparison to PS (Fig. 1B). Furthermore, the m6A level in both cell lines increased markedly under OS as compared to PS, when measured by dot blot (Fig. 1C; specificity validation: SI Appendix, Fig. S1B). We made in vivo examination of ECs in the straight part of the thoracic aorta (TA), which is exposed to PS flow, and the inner and outer curvatures of the aortic arch (AA), which is exposed to OS (SI Appendix, Fig. S1A). Indeed, the content of m6A was higher in AA than the straight TA (Fig. 1C). Since m6A marks are imposed by the methyltransferase complex, we measured the levels of its two major components, METTL3 and METTL14. The protein levels of METTL3, the major catalytic subunit of the methyltransferase responsible for m6A modification, were up-regulated in both HUVEC and MAEC in response to OS as compared to PS (Fig. 1C and D). The METTL3 increased at both mRNA and protein levels in a shear pattern and time-dependent manners over 48 h (SI Appendix, Fig. S1 C and D). In contrast, the protein level of METTL14, another methyltransferase subunit, remained unchanged (Fig. 1C). Consistent with these results, METTL3, but not METTL14, was up-regulated in AA as compared to TA (Fig. 1C and D). Furthermore, en face immunofluorescence staining shows that, in control untreated C57BL/6 mice, the expression levels of METTL3 was higher in the endothelium of the inner AA than that of the outer AA or TA (SI Appendix, Fig. S1D). Immunohistochemistry also revealed the higher METTL3 expression in the endothelium of the inner AA than that of the outer AA (SI Appendix, Fig. S1 C and D). Moreover, we experimentally created the atheroprone OS condition in the left carotid artery (LCA) by partial vessel ligation in high-cholesterol atherogenic diet-fed ApoE−/− mice, with the sham-operated LCA in another intact mice as a sham control (Fig. 1E). En face immunostaining of METTL3 revealed its marked up-regulation in the partially ligated LCA as compared to the control sham-operated LCA (Fig. 1 F and G). Finally, we performed immunofluorescent staining of METTL3 in the human paraffin-embedded tissue array histological samples of lesion-free aorta and aorta affected by atherosclerosis. METTL3 signal level was significantly higher in the aorta affected by atherosclerosis, as compared to the lesion-free aorta (Fig. 1 H and I; specificity validation of METTL3: SI Appendix, Fig. S1H). To summarize, we have demonstrated in both in vitro and in vivo systems that atherogenic OS was associated with increases in m6A RNA methylation and the level of METTL3, the catalytic subunit of the methyltransferase responsible for imposition of m6A marks.
Fig. 1.
OS induces METTL3-dependent m6A methylation. (A) Scheme of the parallel-plate flow chamber for generating OS and PS at shear stresses of 0.5 ± 4 dyn/cm2 and 12 ± 4 dyn/cm2, respectively. (B) LCMS analysis of the content of m6A-modified mRNA in MAECs and HUVECs under PS or OS. Mean relative values are shown with SD error bars, **P < 0.05, ***P < 0.005. (C) Dot blot analysis of the contents of m6A-modified mRNA (Upper) and Western blot (Lower) of METTL3 and METTL14 protein expression in HUVECs and MAECs exposed to PS and OS, and in TA and AA from WT C57BL/6J mice. (D) Quantification of dot blot and Western blot. The mean values relative to PS or OS are shown with SD error bars, ***P < 0.005; NS, not significant. (E) Scheme for the induction of atherosclerosis in high-cholesterol atherogenic diet-fed ApoE−/− mice with partial ligation of left coronary artery. (F) En face immunofluorescence staining of METTL3 in the endothelial layer of PL LCA and sham control LCA. (G) Quantification showing mean fluorescence intensity values relative to sham control LCA with SD error bars, ***P < 0.005. (H, Top) Immunohistochemistry staining of the human paraffin-embedded tissue array of lesion-free aorta and aorta affected by atherosclerosis. (Middle) CaseViewer software was used to visualize METTL3-positive staining in these immunohistochemistry samples. The images were further zoomed in to show the endothelium with higher magnification. (I) The quantification showing METTL3 expression in the paraffin-embedded cross-sections of the human artery walls of the indicated samples. Quantification of METTL3 signal expressed as median and quartile integrated optical density (IOD).
METTL3 Mediates Proinflammatory Effects in Endothelial Cells in Response to Atherogenic Flow.
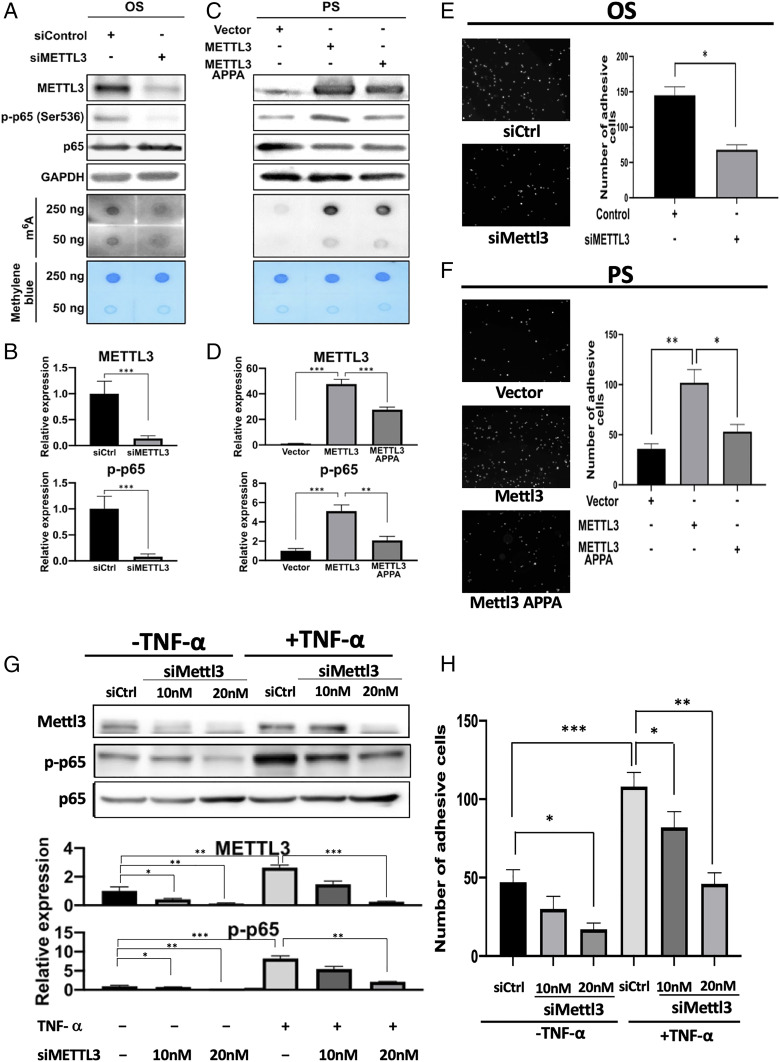
Atherosclerosis is a chronic inflammatory disease characterized by increased secretion of inflammatory cytokines and activation of NF-κB signaling, which has the phosphorylation of RelA/p65 member of NF-κB family as a key event. To investigate the effects of METTL3 on proatherogenic inflammatory processes, HUVECs with METTL3 knockdown by small interfering RNA (siRNA) was subjected to OS, and the m6A RNA methylation and RelA/p65 Ser536 phosphorylation (p-p65) were examined by dot blot and Western blot, respectively. METTL3 knockdown resulted in decreases in m6A RNA levels and RelA/p65 phosphorylation (Fig. 2 A and B). In a reverse experiment, METTL3 and its catalytic mutant form, METTL3 APPA (SI Appendix, Fig. S2A), were overexpressed in HUVECs exposed to nonatherogenic PS. The overexpression of METTL3 resulted in increases in m6A RNA levels and RelA/p65 phosphorylation (Fig. 2 C and D). The overexpression of METTL3 APPA resulted in significantly less efficient phosphorylation of RelA/p65 (Fig. 2 C and D).
Fig. 2.
METTL3 mediates proinflammatory effects in ECs. (A and B) Western blot (A) and its quantification expression (B) of METTL3, p65 and p-p65 (Upper), with dot blot analysis of the contents of m6A-modified RNA (Lower in A) in HUVECs treated with OS and METTL3 knockdown. (C and D) Western blot (C) and its quantification of expressions (D) of METTL3, p65 and p-p65 (Upper), dot blot analysis of the content of m6A-modified RNA (Lower in C) in HUVECs treated with PS and overexpression of METTL3 or its catalytic mutant METTL3 APPA. Monocyte adhesion assay: HUVECs treated with OS and METTL3 knockdown (E), and with PS and overexpression of METTL3 or its catalytic mutant METTL3 APPA (F). Quantification (Upper) and the representative fluorescent images of monocyte adhesion of five images are shown. SD: error bars, *P < 0.05, **P < 0.005. (G) Western blot (Upper) and quantification (Lower) of expressions of METTL3, p65, and p-p65 in HUVECs with METTL3 knockdown by siMETTL3 at 5 μg (#1) or 10 μg (#2) in the absence or presence of TNF-α stimulation. (Lower) Quantification of the blots with indicated treatment. (H) Monocyte adhesion assay performed on HUVECs subjected to METTL3 knockdown by siMETTL3 at 5 μg (#1) or 10 μg (#2) with or without TNF-α stimulation. Data shown as mean number of adherent monocytes from six images, SD error bars, *P < 0.05.
Adhesion of monocytes to ECs is a key pathological event in atherogenesis. Therefore, we examined the adhesion of fluorescently labeled monocytes to HUVECs under OS with knockdown of METTL3 and under PS with overexpression of METTL3 or METTL3 APPA. The knockdown of METTL3 in HUVECs exposed to OS markedly reduced the adhesion of monocytes (Fig. 2E). The overexpression of METTL3 in HUVECs subjected to PS resulted in significantly increased adhesion of monocytes, while overexpression of catalytically inactive METTL3 APPA failed to elicit the same effect (Fig. 2F). TNF-α is an inflammatory cytokine and an upstream regulator of the NF-κB pathway that is believed to play a crucial role in the development of atherosclerosis lesion. LCMS analysis showed increased m6A modification of mRNA upon TNF-α stimulation in both HUVECs and HAECs (SI Appendix, Fig. S2 B and C). We next investigated the role of METTL3 in TNF-α–mediated proatherogenic inflammatory responses in ECs. The treatment of HUVECs with TNF-α resulted in activation of the NF-κB pathway as indicated by the increased level of p-p65 (Fig. 2G). Knockdown of METTL3 significantly attenuated the TNF-α–mediated phosphorylation of p65 (Fig. 2G). While TNF-α caused increased adhesion of monocytes, METTL3 knockdown significantly reduced monocyte adhesion in both TNF-α–treated and TNF-α–free groups of HUVECs (Fig. 2H and SI Appendix, Fig. S2D). These results indicate that METTL3 is a mediator of the proatherogenic inflammatory effects in ECs subjected to disturbed flow (SI Appendix, Fig. S2E).
METTL3 Hypermethylated the m6A Sites of Downstream mRNA Targets Involved in Proatherogenic Process.
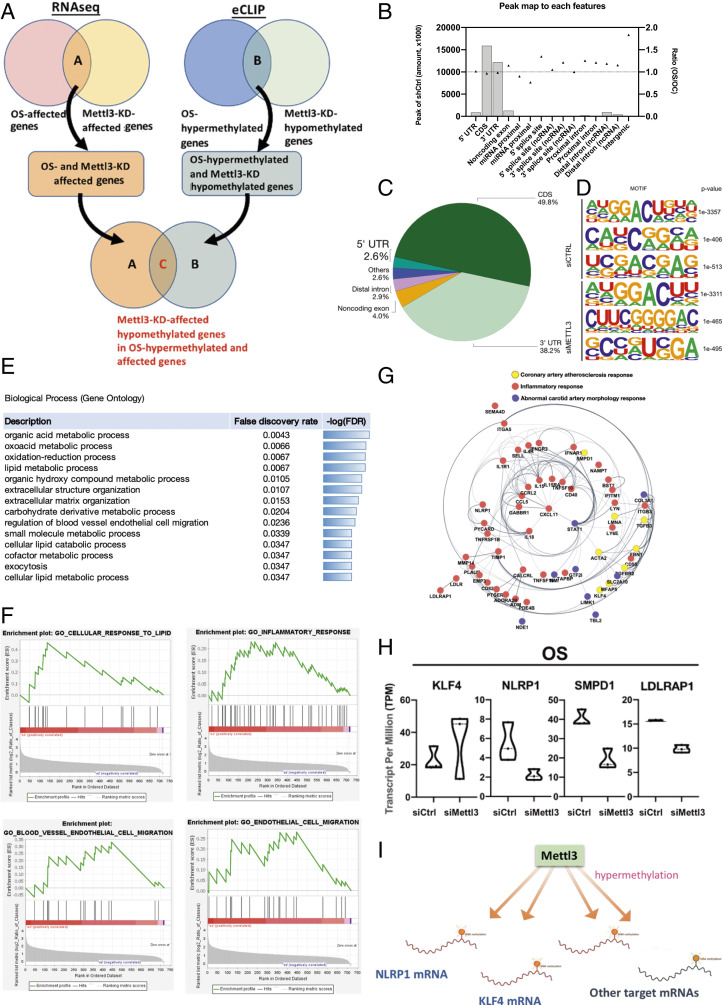
Since METTL3 mediates the proinflammatory effects in ECs subjected to atherogenic flow, we worked on identifying the targets of METTL3-mediated RNA methylation. First, we performed RNA-sequencing (RNA-seq) and m6A-enhanced cross-linking and immunoprecipitation (eCLIP) analysis (14) in HUVECs exposed to OS with or without knockdown of METTL3, in comparison to PS. The RNA-seq results revealed that the expressions of a total of 278 genes were affected by METTL3 knockdown under OS. eCLIP results revealed a total of 441 genes containing m6A sites that were hypomethylated by METTL3 knockdown under OS (Fig. 3A). We overlaid the RNA-seq data of METTL3 knockdown-affected genes with the eCLIP data to show the METTL3 knockdown-induced hypomethylated m6A sites. Overall, our data showed that a total of 47 genes were identified as OS-hypermethylated and OS-affected genes that can be affected and hypomethylated by METTL3 knockdown. (Fig. 3A).
Fig. 3.
Identification of downstream m6A methylation sites of METTL3 in OS-treated HUVECs using NGS-based approaches and eCLIP analysis. (A) Venn diagrams showing the corresponding gene numbers of genes affected by OS and METTL3-knockdown (Upper Left) and the gene numbers of OS-hypermethylated genes and METTL3-knockdown hypomethylated genes (Lower). The overlap between OS-affected genes that were also affected by Mettl3-knockdown (label A) and OS-hypermethylated genes that were hypomethylated by Mettl3-knockdown (label B) represents the total m6A sites that were OS-affected and OS-hypomethylated by METTL3 knockdown (label C). (B) Bar charts showing the peaks that map to each feature and the ratio of shMettl3-treated HUVECs over shCtrl-treated HUVECs exposed to OS. (C) The pie chart depicts the relative frequency of peaks that map to each feature type, with a peak log2-fold enrichment ≥3 and P value ≤0.001. (D) HOMER identifies enriched motifs in CLIP-seq peaks. (E) The GO term enrichment analysis shows the total m6A sites affected and hypomethylated by METTL3 knockdown. Gene ratio is the number of genes annotated to the GO term in the input list of genes. Bg ratio is the number of genes annotated to that GO term in the entire background set. (F) GSEA of the total m6A sites that were affected and hypomethylated by METTL3 knockdown. (G) Identification of the network genes that are involved in the pathways related to total m6A sites that were affected and hypomethylated by METTL3 knockdown in OS-treated HUVECs. (H) RNA-seq profiles showing the transcript expressions of KLF4, NLRP1, SMPD1, and LDLRAP1. (I) Scheme illustrates that Mettl3 hypermethylates downstream mRNAs including KLF4, NLRP1, and other mRNAs in OS-exposed HUVECs.
Based upon the eCLIP data, the largest percentage (49.8%) of identified m6A sites were located within the coding sequences (CDS), 2.9% covered distal intron, 4% covered noncoding exon, 38.2% were located in the 3′ UTRs, and about 3% in the 5′ UTRs (Fig. 3A). The methylated adenosines were predominantly located within a number of distinct sequence motives (Fig. 3B). The locations of methylated adenosines were not modified by METTL3 knockdown (Fig. 3 C and D and SI Appendix, Fig. S3A). Gene Ontology (GO) analysis revealed that the 278 genes were affected by METTL3 knockdown under OS were the major elements responsible for genes important for the regulation of EC migration, cellular lipid catabolic process, and extracellular matrix organization and structure organization (Fig. 3E). Using gene set enrichment analysis (GSEA), we observed that the genes were affected by METTL3 knockdown under OS are primarily the genes that are up-regulated in cellular response to lipids, inflammatory responses, blood vessel endothelial cell migration, and EC migration (Fig. 3F and SI Appendix, Fig. S3B).
We further elucidated the key factors that contribute to posttranslational modification to gain additional insights into the functional consequences of differential expression patterns of downstream proteins of METTL3 in ECs under OS. Using gene network analysis with the IPA software package to construct network modules from the 47 genes containing m6A sites that were hypomethylated by METTL3 knockdown under OS, we found that KLF4 and NLRP1 are potentially the key factors regulating the pathways related to METTL3-mediated hypermethylation and OS-induced atherogenic events. (Fig. 3G). Among these 47 genes (SI Appendix, Table S1), the top 4 most significantly affected by METTL3 knockdown are KLF4, NLRP1, SMPD1, and LDLRAP1 (Fig. 3H). Compared with PS-treated cells, the eCLIP abundance of NLRP1 and KLF4 were enriched in OS-treated cells with siCtril, and were diminished by METTL3 knockdown (SI Appendix, Fig. S3C). Collectively, METTL3 hypermethylates several target mRNAs under OS and, consequently, up-regulates NLRP1 and down-regulates KLF4 (Fig. 3I).
Identification of m6A Reader Proteins that Mediate the Effect of METTL3-Dependent Methylation of NLRP1 and KLF4 mRNAs.
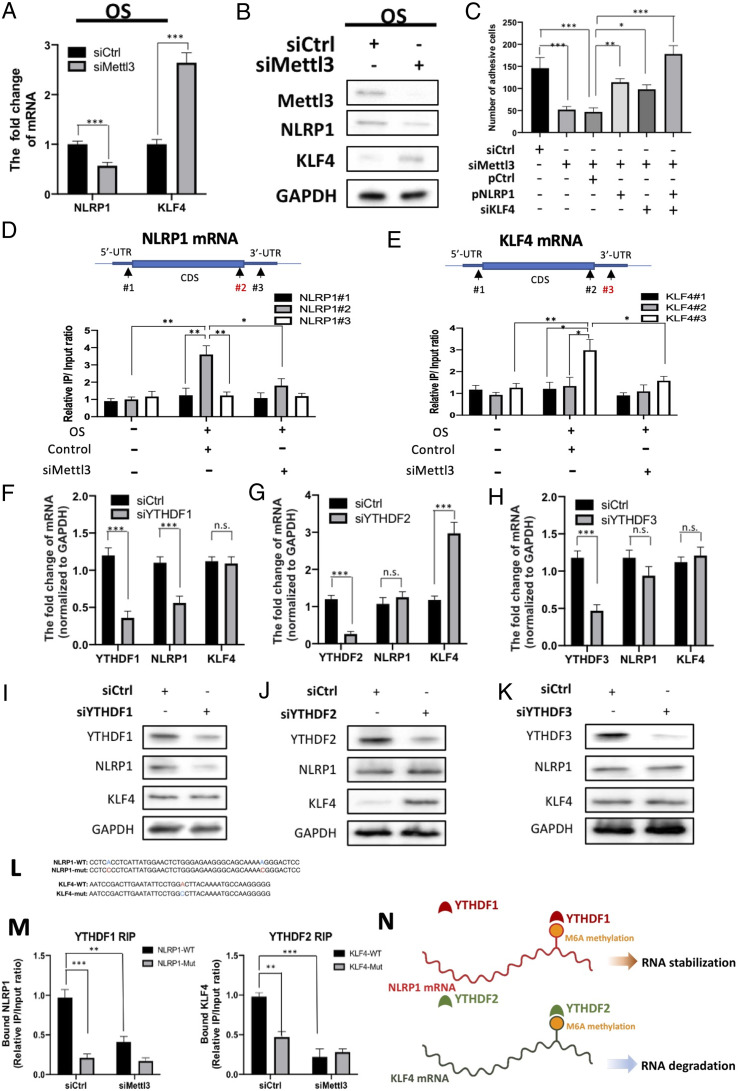
We next validated the expressions of NLRP1 and KLF4 by qRT-PCR and Western blot. Similar to the findings of RNA-seq, the knockdown of METTL3 under OS resulted in down-regulation of NLRP1 and up-regulation of KLF4 at mRNA (Fig. 4A) and protein (Fig. 4B) levels, indicating that METTL3 may regulate either the stability or degradation of these mRNAs. While monocyte adhesion was reduced by METTL3 knockdown, it was increased by the concomitant overexpression of NLRP1 and/or knockdown of KLF4, thus emphasizing the crucial role of these two factors in regulating monocyte adhesion (Fig. 4C). To elucidate the location of m6A methylation sites on the mRNAs that were hypermethylated by OS, we performed RNA immunoprecipitation (RNA-IP) to immunoprecipitate the hypermethylated RNA fragments and amplified these crucial regions with the indicated primers. For NLRP1 mRNA, OS induced a substantial hypermethylation on the CDS near the stpp codon, but not on the 5′ UTR or 3′ UTR. Knockdown of METTL3 abolished the hypermethylation on the CDS (Fig. 4D). For KLF4 mRNA, OS led to hypermethylation on the 3′ UTR that can be erased by METTL3 knockdown (Fig. 4E). These data indicate that METTL3 predominantly hypermethylated m6A sites at the CDS regions near the 3′ UTR in response to disturbed flow.
Fig. 4.
Identification of m6A reader proteins that recognize METTL3-dependent methylation on NLRP1 and KLF4 mRNAs. (A) qRT-PCR analysis showing NLRP1 and KLF4 mRNA expressions in OS-treated HUVECs with or without Mettl3 knockdown. siCtrl, scrambled siRNA control. Mean values relative to siCtrl are shown with SD error bars, n = 3, ***P < 0.001. (B) Western blot showing NLRP1 and KLF4 proteins in OS-treated HUVECs with indicated treatment. (C) Monocyte adhesion assay performed on OS-treated HUVECs with or without Mettl3 knockdown, and with or without the concomitant NLRP1 overexpression (pNLRP1) and/or KLF4 knockdown (siKLF4). Data shown as means with SD error bars, n = 6, *P < 0.05, **P < 0.005, ***P < 0.001. (D and E) RNA-IP analysis showing the location of given m6A methylation sites in NLRP1 (D) or KLF4 mRNA (E) OS-treated HUVECs with indicated treatment. (F–H) qRT-PCR analysis showing the expression of YTHDF family members, NLRP1, and KLF4 mRNA in OS-treated HUVECs upon knockdown of YTHDF1 (F), YTHDF2 (G), and YTHDF3 (H). Data expressed as mean fold changes relative to control with SD error bars, n = 3, ***P < 0.001, n.s., not significant. (I–K) Western blot of the expression of YTHDF family members, NLRP1, and KLF4 proteins upon the knockdown of YTHDF1 (I), YTHDF2 (J), or YTHDF3 (K) in OS-treated HUVECs. (L) Sequences of NLRP1 and KLF4 RNA fragments with either WT or mutated (mut) m6A sites. (M) RNA-IP assay showing the binding affinity between METTL3-hypermethylated target mRNAs and YTHDF family members with or without METTL3 knockdown. Synthesized RNA oligos of NLRP1 and KLF4 with either WT or mutated (Mut) m6A sites were introduced by electroporation, and the binding affinity between NLRP1 (Left) and YTHDF1 (Right), and between KLF4 and YTHDF2, were assessed. Data expressed as mean m6A enrichment relative to WT with SD error bars, n = 3, ***P < 0.001 (Student’s t test). (N) Scheme showing that YTHDF family members recognize METTL3-mediated m6A methylation on NLRP1 and KLF4 mRNAs under OS.
m6A methylation can have different functional consequences on mRNAs that are mediated by the binding of different YTH family m6A reader proteins. To identify which YTH family m6A reader proteins were responsible for recognizing the METTL3-mediated hypermethylation on NLRP1 and KLF4 mRNAs, we knocked down various members of the YTH family in OS-treated HUVECs. The knockdown of YTHDF1, but not YTHDF2 or YTHDF3, resulted in significant down-regulation of NLRP1 mRNA (Fig. 4 F–H). However, KLF4 mRNA was significantly up-regulated by the knockdown of YTHDF2 (Fig. 4G), but not other YTHDF family members (Fig. 4 F and H). Consistent with these findings, the knockdown of YTHDF1 resulted in down-regulation of NLRP1, and the knockdown of YTHDF2 resulted in up-regulation of KLF4 at the protein levels in OS-treated HUVECs (Fig. 4 I–K). To validate the binding of YTHDF proteins to NLRP1 and KLF4 mRNAs, we designed RNA fragments of NLRP1 and KLF4 with either WT or mutated m6A sites based upon our eCLIP results, with or without METTL3 knockdown (Fig. 4 L and M and SI Appendix, Fig. S4). Subsequently, these RNA fragments with distinct m6A sites were exogenously introduced into HUVECs with or without METTL3 knockdown, and the OS-treated HUVECs were then subjected to RNA-IP analysis. METTL3 knockdown markedly diminished the interaction between YTHDF1 and NLRP1 mRNA and that between YTHDF2 and KLF4 mRNA. Compared to WT m6A sites, the mutated m6A sites also abrogated the interactions between YTHDF1 and NLRP1 mRNA and between YTHDF2 and KLF4 mRNA (Fig. 4M). These results suggest that METTL3-mediated hypermethylation contributes to the stabilization of NLRP1 mRNA and the degradation of KLF4 mRNA under OS, through the recognition of m6A hypermethylation by YTHDF1 and YTHDF2 reader proteins, respectively (Fig. 4N).
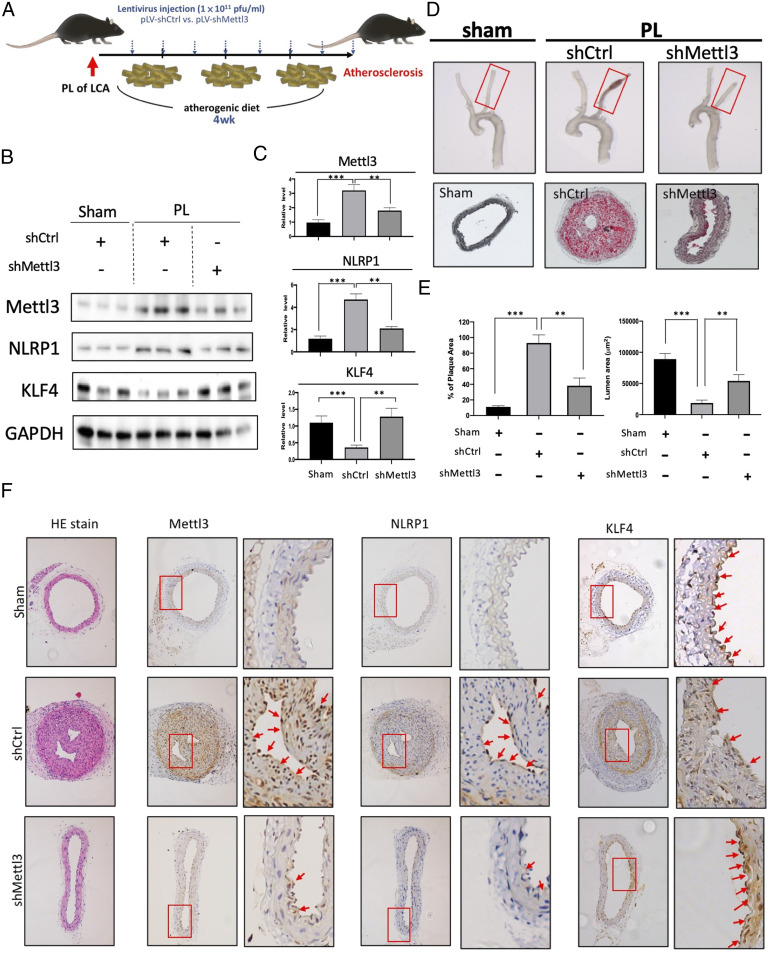
Knockdown of the Mettl3 Gene Inhibits Atherogenesis in a Mouse Model.
To demonstrate the in vivo relevance of the METTL3-mediated NLRP1 and KLF4 m6A hypermethylation and its functional consequences found in vitro, we investigated the role of METTL3 in OS-induced responses in vascular ECs in vivo. We used an atherogenic partial arterial ligation (PL) in high-cholesterol atherogenic diet-fed ApoE-deficient (ApoE−/−) mouse model, in which the LCA was partially ligated to create flow disturbance and enhanced-lesion formation (15). Lentiviral vector (pLV) encoding Mettl3-targeting shRNA (shMettl3) or scrambled control shRNA were administrated twice per week over the 4-wk experimental course (Fig. 5A). The LCAs of the sham-operated mice were used in parallel as the sham control. Consistent with the findings in cultured HUVECs and MAECs that OS increased m6A modifications (Fig. 1A), METTL3 and NLRP1 were up-regulated and KLF4 was down-regulated in the LCAs with PL, when compared with the sham-operated control LCA (Sham LCA) (Fig. 5 B and C). Importantly, multiple administrations of shMettl3 significantly reversed the effects of PL (Fig. 5 B and C). In gross necropsy, compensatory arterial expansion was observed at the sites of lesions in the PL compared with sham-operated control LCA (Fig. 5D). shMettl3 effectively prevented the arterial expansion in the PL (Fig. 5D). We further examined the plaque formation using Oil Red O staining and lumen size measurements under the indicated experimental conditions (Fig. 5 D and E). PL led to severe atherosclerotic lesions with neural fat-containing plaque formation and decreased lumen size (Fig. 5 D and E). Importantly, Mettl3 knockdown effectively abrogated the plaque formation and prevented the lumen stenosis (Fig. 5C). We further performed immunohistochemistry staining to examine the expression of METTL3 and its targeted genes in the same model of atherogenesis with or without shMettl3 administration. As shown in Fig. 5D, PL induced plaque formation and decreased arterial lumen size, with the concurrent up-regulations of METTL3 and NLRP1 (Fig. 5F, shCtl). ShMettl3 abolished the PL-induced plaque formation and decreased METTL3 and NLRP1 expressions (Fig. 5D, shMettl3). Taken together, these results demonstrated that the knockdown of Mettl3 inhibited atherosclerotic lesion formation in the mouse model.
Fig. 5.
Mettl3 knockdown inhibits atherogenesis in vivo. (A) The experimental design for Mettl3 gene knockdown expression in the partial ligation (PL)-induced high-cholesterol atherogenic diet-fed ApoE−/− mouse model of atherosclerosis. Atherosclerotic lesions were induced by PL over 4 wk. Lentivirus encoding control shRNA (pLV-shCtrl) or shRNA targeting Mettl3 (pLV-shMettl3) was repetitively administered twice per week. (B) Western blot and its quantification (C) showing the expression of METTL3, NLRP1, and KLF4 proteins in the arterial endothelium from mice with indicated treatments. (D) Gross necropsy (Upper) of LCAs from ApoE−/− mice with indicated treatments. Plaque formation and the changes in lumen size (Center) were evaluated using Oil Red O staining. (E) The formation of Oil Red O-stained plaques and quantitation of lumen area. (F) Hematoxylin and eosin staining and immunohistochemistry analysis of METTL3 and NLRP1 proteins in the cross-sections from sham-operated LCA and PL LCA with indicated treatments. Boxes highlighted the expression patterns of METTL3, NLRP1, and KLF4 in the endothelium from LCA.
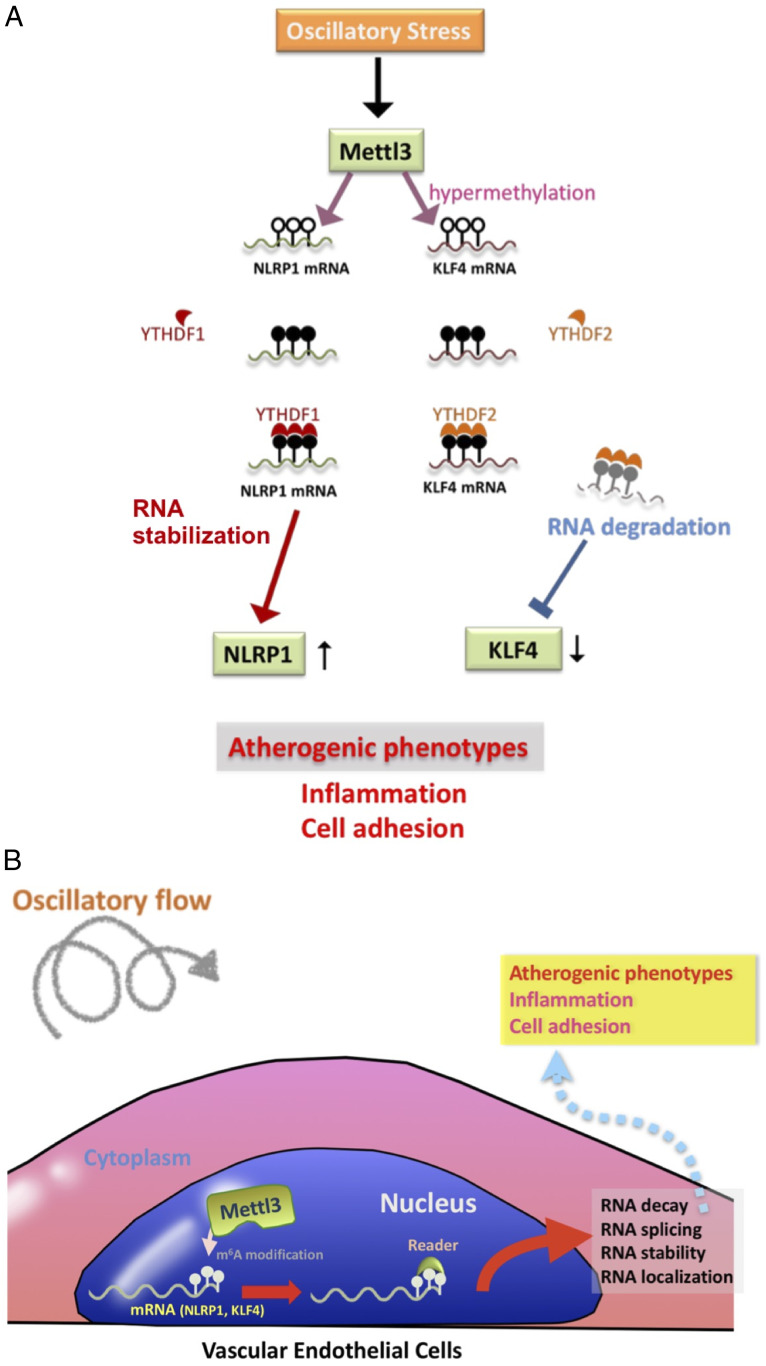
Collectively, our findings have demonstrated an OS-elicited atherogenic mechanism involving METTL3 up-regulation and m6A RNA hypermethylation in vascular ECs. Using bioinformatic approach and m6A eCLIP assay, we identified NLRP1 and mRNAs as the targets of METTL3-mediated hypermethylation. The m6A-hypermethylated NLRP1 mRNA is specifically recognized by the reader YTHDF1 to enhance the translation of NLRP1 protein. In contrast, the m6A-hypermethylated KLF4 mRNA is recognized by YTHDF2 to promote RNA degradation (Fig. 6A). Overall, our results demonstrated that the OS-induced METTL3 leads to EC atherogenic responses by increasing NLRP1 and decreasing KLF4 through m6A modifications (Fig. 6B).
Fig. 6.
Scheme illustrating the role of METTL3-dependent m6A RNA modifications in OS-induced proatherogenic events. The working models of METTL3-dependent m6A RNA modifications in OS-induced proatherogenic events are shown. (A) OS up-regulates METTL3 and induces a METTL3-dependent m6A RNA hypermethylation in downstream mRNA targets, e.g., NLRP1 and KLF4. For NLRP1 mRNA, the METTL3-mediated hypermethylation is recognized by YTHDF1, leading to enhanced translation of NLRP1. For KLF4, YTHDF2 recognizes the methylation sites and further lead to the degradation of KLF4 mRNA. (B) Overall, m6A RNA modifications may induce RNA decay, splicing, stability, and their localization; further modulate downstream protein expression; and eventually contribute to proatherogenic events.
Discussion
The distribution of atherosclerotic lesions in the human vascular system is highly focal. It is widely accepted that the primary stimulus for atherosclerotic lesion formation is the local activation of ECs and an increase of permeability, which may be triggered by the hemodynamic forces at sites of blood vessel bifurcations and bends (16). Previous studies have demonstrated the involvements of various microRNAs (miRNAs), such as miR-92a (17, 18), miR-10a (19), and miR-146a (20) in coordinating EC responses of various atherogenic stimuli. We have also shown that, through modulating the maturation of miR-93 and miR-484 that target KLF2 and eNOS mRNAs, the RNA binding protein nucleolin regulates EC health and the progression of coronary artery diseases, implicating the differential regulations of miRNA biogenesis in ECs under the atheroprone oscillatory shear vs. atheroprotective pulsatile shear (21). Epigenetic regulations, including DNA methylation (22) and histone modification (16), have also been implicated in the responses of ECs to flow conditions. Recently, m6A methylation of RNA has come into prominence as another important epigenetic mechanism controlling various biological processes. However, the involvement of m6A RNA epigenetics in atherosclerosis has not been systematically investigated.
In this study, we demonstrated increases of m6A content and METTL3 expression in atheroprone conditions (Fig. 1). Importantly, this phenomenon was demonstrated in different models of atherogenesis, both in vitro and in vivo in human and mouse systems. The parallel plate flow system is a widely used method for the investigation of fundamental mechanisms of atherogenesis, such as flow-dependent cytoskeleton remodeling (23, 24). It allows the application of physiological PS and pathological OS on cultured ECs in a controlled manner. By using this system, we showed that OS significantly elevated the levels of hypermethylated m6A mRNA and METTL3, the catalytic subunit of m6A methyltransferase (Fig. 1A). This observation was further corroborated by the findings of increased expression of METTL3 in human and mouse artery regions exposed to disturbed flow, as well as human atherosclerotic vessels (Fig. 1 B and C). Furthermore, our study on carotid ligation mouse model that mimics atheroprone conditions in vivo (15) also indicated the involvement of m6A RNA modification and METTL3 in atherogenesis.
In the m6A writer complex, METTL3 is the subunit with enzymatic activity to catalyze m6A methyltransferase. With the help from METTL14, which binds the target RNAs, it acts as a global regulator of RNA methylation. The specificity of its action depends on the motives in different regions of mRNAs. Its action is associated with cell state transitions. When a cell loses its homeostasis, METTL3 triggers global mRNA methylation in the cell to determine its functions and fate by modulating diverse subsets of mRNAs to different fates. Activation of inflammatory response is a hallmark of atherosclerosis (25). We found that under OS conditions, a proinflammatory response, including monocyte recruitment and NLRP1 elevation, was triggered in ECs. Previous studies showed that disturbed flow activates several pathways that lead to transcriptional activation of NF-κB, resulting in enhanced inflammatory response (26, 27). Notably, we observed that the knockdown of METTL3 expression in ECs under OS condition caused suppression of RelA/p65 phosphorylation, which is required for NF-κB activity (Fig. 2B); in contrast, overexpression of METTL3 in ECs under PS condition resulted in an increase of RelA/p65 phosphorylation (Fig. 2A). We showed that the elevated METTL3 under OS induced proinflammatory mechanisms and that suppressing METTL3 diminished the OS-induced inflammation response of ECs. Further development of METTL3 targeting strategy, such as using AAV delivery vehicles, has the potential of leading to a novel therapeutic method for atherosclerosis patients.
m6A marks are imposed and read on a global scale to regulate transitions of cellular states. We applied eCLIP to delineate the subset of mRNAs with elevated m6A modification levels under OS condition. Among the targets of m6A methylation under OS, we identified KLF4 and NLRP1 as downstream effectors regulating opposing cell functions under OS condition. NLRP1 is the first protein shown to form inflammasomes, and it has been suggested to have a key role in shifting the ECs to the proinflammatory state (28). KLF4 is a transcription factor regulating endothelial function and promoting vascular homeostasis (29). Unlike NLRP1, which induces inflammation response, KLF4 inhibits inflammation in endothelial cells such as HUVEC and those in human coronary artery (30, 31). METTL3-mediated m6A modifications on NLPR1 and KLF4 mRNA result in an increase of NLRP1 and a decrease of KLF4 expression level. The diverse m6A-mediated mRNA regulations may derive from the involvements of different m6A reader proteins binding to the two transcripts. YTHDF1 binds to the m6A-modified NLRP1 and stabilizes its mRNA stability, whereas YTHDF2 binds to the m6A-modified KLF4 and induces the degradation of KLF4 mRNAs. In this model, METTL3 triggers RNA modification on its downstream mRNA transcripts and orchestrates different m6A readers to determine the fates of these transcripts, thus promoting monocyte adhesion and enhancing proinflammatory pathways. The details of the RNA epigenetic regulation of NLRP1 and KLF4, as well as the downstream molecular mechanisms in regulating endothelial inflammation response, still need further exploration.
In conclusion, our study demonstrates an important role of m6A RNA modification in the OS-induced proinflammatory responses of ECs that may lead to atherosclerosis. We identify the m6A methyltransferase METTL3 as a key regulator that mediates the OS-induced m6A modification on RNA transcripts to regulate atherosclerotic inflammation. Suppression of METTL3 is able to inhibit the OS-induced m6A RNA hypermethylation, inflammatory response, and monocyte adhesion and reverse the changes of NLRP1 and KLF4 expression levels in ECs under OS condition. Our study provides experimental evidence in support of the involvement of RNA epigenetics in the cellular response to shear stress and the pathogenesis mechanism of atherosclerosis. The identification of METTL3 as the key regulator of OS-induced proinflammatory response of ECs suggests a potential therapeutic approach for atherosclerotic diseases by targeting METTL3.
Materials and Methods
Cell Culture and Flow Experiments.
MAECs (catalog no. T4592) was purchased from Applied Biological Materials Inc., and HUVECs (catalog no. C-12206) were obtained from Promocell. HUVECs and MAECs were cultured in DMEM (Sigma-Aldrich) containing 10% (vol/vol) Clonetics endothelial growth medium (Lonza), 10% fetal bovine serum (Gibco), 1% l-glutamate, and 1% sodium pyruvate. A parallel-plate flow chamber was used to impose fluid shear stress conditions as previously described (32). The flow channel in the chamber was created by a silicon gasket with dimensions of 2.5 cm in width (w), 5.0 cm in length, and 0.025 cm in height (h). The perfusion loop system was connected to the chamber with the glass seeded with cells and maintained under standard culture conditions (37 °C, 95% humidity, 5% CO2). The fluid shear stress (τ) imposed on the cells was calculated as τ = 6 Qμ/wh2, where Q is the flow rate and μ is the dynamic viscosity of the perfusate. Pulsatile shear flow (PS) has a shear stress of 12 ± 4 dynes/cm2, and oscillatory shear flow is at 0.5 ± 4 dynes/cm2.
LCMS.
Liquid chromatography-tandem mass spectrometry (LCMS) was used to analyze the ratio of m6A/A. mRNA was purified with Dynabeads (Thermo Fisher Scientific) and digested by nuclease P1 (Sigma-Aldrich) and alkaline phosphatase (Sigma-Aldrich). The resultant nucleoside samples were injected into LC-MS/MS (Agilent, LC/TQ). The results were analyzed using Analyst 1.6.1 software (SCIEX).
Dot Blot Assay.
For dot blot analysis, mRNA was isolated from total RNA using the PolyATtract mRNA Isolation System (Promega) according to the manufacturer’s protocol. mRNA was denatured by preheating at 95 °C for 3 min, chilled on ice for 1 min, spotted on a Hybond-N+ membrane (Cytiva), and fixed by UV–cross-linking. The membrane was incubated with the m6A antibody (SySy) overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibody, and the signal was detected by the ECL system (Cytiva) to determine the levels of m6A.
Electroporation in HUVECs.
Briefly, 3 × 106 cells were harvested and resuspended in electroporation buffer (Lonza). Subsequently, 5 μg of plasmid or siRNA was added into cells and mixed well. pcDNA-flag-METTL3 was a gift from Alessandro Fatica (Addgene no. 160250), and pcDNA-flag-METTL3/APPA was generated by site-directed mutagenesis. A-034 program and electroporation device (Lonza, Amaxa 4D-Nucleofector) were used to transfection, and cells were seed on collagen I-coating slide or culture plate. All siRNAs were purchased from Sigma-Aldrich and list below. (Mettl3: SASI_Hs01_00044321, KLF4: SASI_Hs01_00123156, NLRP1: SASI_Hs01_00151122, YTHDF1: SASI_Hs01_00233694, YTHDF2: SASI_Hs01_00133217, and YTHDF3: SASI_Hs01_00202278).
Immunofluorescence and Immunohistochemistry.
All procedures were followed previously described (33). All samples were incubated antibodies overnight after fixing and permeating. The following primary antibodies were used: CD31 (abcam, ab9498), Mettl3 (ABclonal, A8370), NLRP1 (Novus Biologicals, NBP1-54899), KLF4 (Cell Signaling Technology, 4038P). After staining, imageJ or Caseviewer (3DHISTECH Ltd.; https://www.3dhistech.com/solution/caseviewer/) were utilized to image quantification.
m6A-IP qPCR.
For m6A-IP, we followed previously protocol (34). In brief, RNA samples were fragmented by fragmentation reagent (Invitrogen, AM8740) and incubated with protein G dynabeads-conjugated m6A antibody (SYSY, 202003) overnight at 4 °C. Methylated RNAs were collected by elution buffer (6 mM m6A sodium salt, 50 mM Tris⋅HCl, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 U/μL RNase inhibitor), then RNAs were isolated by RNeasy kit (Qiagen), following One-Step Real-Time RT-PCR kit (Invitrogen, 12574018) was manipulated by quantification of m6A-methylated RNA.
m6A-eCLIP.
All experimental procedures were followed as described previously (14). In brief, 50 μg of total RNAs were purified, fragmented, cross-linked with m6A antibody, immunoprecipitated, and reversely transcribed to establish the cDNA library with 3′ adaptors for sequencing. The cross-linking procedure generated a C-to-T transition at m6A antibody-binding sites, which allowed us to map the mutations to known genomic sequences to identify the m6A sites with precision. The eCLIP service including RNA adapter ligation, reverse transcription, DNA adapter ligation, and PCR amplification were assisted and performed by Eclipse Bioinnovations Inc (https://eclipsebio.com/).
RNA-IP qRT-PCR Assay.
For RNA immunoprecipitation, all procedures were performed as described preciously (35). Before cell harvesting, cells were transfected with RNA fragments (synthesized oligos), including NLRP1-WT, NLRP1-Mut, KLF4-WT, and KLF4-Mut plasmids, respectively, using electroporation. Following cells were lysed by lysis buffer (20 mM Tris⋅HCl, 100 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 U/μL protease inhibitor, and 1 U/μL RNase inhibitor) after being treated with oscillatory stress. YTHDF1 (Proteintech) or YTHDF2 antibody (Proteintech) was added to RNA–protein complex and incubated for 4 h at 4 °C. Complex was incubated and rotated with protein G dynabeads (Invitrogen) for additional 4 h at 4 °C, then eluted RNAs by elution buffer (20 mM Tris⋅HCl, 100 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 1 U/μL RNase inhibitor, 0.1% sodium dodecyl sulfate, and 30 μg of proteinase K) for 30 min at 50 °C. Bound RNAs were isolated and detected by RNeasy kit (Qiagen) and One-Step Real-Time RT-PCR kit (Invitrogen, 12574018).
Monocyte Adhesion Assay.
Human THP-1 monocytes were cultured in RPMI medium 1640 and labeled with CellTracker Green CMFDA Dye (Thermo Fisher Scientific) in serum-free medium at 37 °C for 30 min followed by 10 min at 4 °C. Endothelial cells were incubated with the labeled THP-1 cells at a concentration of 5 × 105 cells/mL for 30 min at 37 °C. Nonadherent THP-1 cells were washed away with RPMI 1640, and the adherent monocytes were fixed with 4% paraformaldehyde for 5 min and counted using a fluorescence microscope.
Western Blot.
Endothelial cells were lysed in the RIPA buffer (25 mM Hepes, pH 7.4, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 125 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride. Equal amounts of protein were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred onto nitrocellulose membranes, blocked in 5% skimmed milk tris-buffered saline with Tween 20, and incubated with the primary antibodies overnight at 4 °C, followed by incubation with HRP-conjugated secondary antibody and detection of signal using the ECL system (Cytiva). The following primary antibodies were used: METTL3 (ABclonal, A8370), METTL14 (Novus Biologicals, NBP1-81392), NLRP1 (Novus Biologicals, NBP1-54899), KLF4 (Cell Signaling Technology, 4038P), and GAPDH (Cell Signaling Technology, 5174S).
Partial Carotid Ligation in High Cholesterol Diet-Fed ApoE−/− Mice.
Adult ApoE−/− mice were anesthetized with ketamine (100 mg/kg body weight)-xylazine (10 mg/kg body weight) mixture through intraperitoneal injection for the performance of partial carotid ligation, which is a model of acutely induced disturbed flow that leads to rapid endothelial dysfunction and atherosclerosis (36). The three branches in the LCA, including left external artery, left internal artery, and occipital artery (OA), were ligated by silk suture while leaving the superior thyroid artery intact. LCA from sham-operated mice was used the sham control. After operation, the ApoE−/− mice with either partial carotid ligation or sham operation were fed with a high-cholesterol atherogenic diet (up to 4 wk) to induce atherosclerosis in LCA. At various time points (1, 2, 3, and 4 wk), the mice were euthanized by CO2 inhalation. Perfusion of the arteries was performed to remove the residual blood, and the LCA samples were collected to conduct various experiments. For RNA collection and extraction, endothelial-enriched RNA was isolated by using insulin syringe to flush TRIzol reagent through the arterial lumen. For histological analysis, the arterial tissues were fixed with paraformaldehyde and cut longitudinally for en face staining.
Atherosclerotic lesions were visualized with en face Oil Red O staining. Briefly, mouse aorta was collected, fixed with formaldehyde, and further dissected to expose the lumen. Oil Red O was applied to stain the neutral lipids within the vessel lumen. Finally, the specimen was subjected to the microscopic examination under a dissection microscope to determine the plaque areas. ImageJ software was used for the analysis and quantification of the plaque area relative to the total vascular area.
Statistical Analyses.
All data were expressed as mean ± SD from at least three independent experiments. The differences in the variables between two groups were analyzed with an unpaired Student’s two-tailed t test. Differences among multiple groups were detected by one-way ANOVA, and when significance was observed, the criterion for significance was set as P < 0.05.
Supplementary Material
Acknowledgments
This study was funded by Ministry of Science and Technology Grants MOST 108-2320-B-010-019-MY3 and MOST 109-2327-B-010-007; University of California San Diego Grants HL108735, HL106579, and HL121365; Ministry of Health and Welfare Grants MOHW108-TDU-B-211-133001 and MOHW109-TDU-B-211-114001; Veterans General Hospital (VGH), National Taiwan Univesity Hospital (NTUH) Joint Research Program Grant VN109-16; VGH, Tri-Service General Hospital (TSGH), National Defense Medical Center (NDMC), Academia Sinica (AS) Joint Research Program Grants VTA107-V1-5-1, VTA108-V1-5-3, VTA109-V1-4-1; AS Clinical Research Center Grant IBMS-CRC109-P04; the “Cancer Progression Research Center, National Yang-Ming University” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan; and the Ministry of Education through the SPROUT Project–Center For Intelligent Drug Systems and Smart Bio-devices of National Chiao Tung University.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025070118/-/DCSupplemental.
Data Availability.
All study data are included in the article and/or supporting information.
References
- 1.Chiu J. J., Chien S., Effects of disturbed flow on vascular endothelium: Pathophysiological basis and clinical perspectives. Physiol. Rev. 91, 327–387 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajami N. E., et al. , Systems biology analysis of longitudinal functional response of endothelial cells to shear stress. Proc. Natl. Acad. Sci. U.S.A. 114, 10990–10995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neth P., Nazari-Jahantigh M., Schober A., Weber C., MicroRNAs in flow-dependent vascular remodelling. Cardiovasc. Res. 99, 294–303 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Geula S., et al. , Stem cells. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science 347, 1002–1006 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Dunn J., Simmons R., Thabet S., Jo H., The role of epigenetics in the endothelial cell shear stress response and atherosclerosis. Int. J. Biochem. Cell Biol. 67, 167–176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vu L. P., et al. , The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 23, 1369–1376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coots R. A., et al. , m(6)A facilitates eIF4F-independent mRNA translation. Mol. Cell 68, 504–514.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Hsu P. J., Chen Y. S., Yang Y. G., Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 28, 616–624 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C., et al. , m6A modulates haematopoietic stem and progenitor cell specification. Nature 549, 273–276 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Li H. B., et al. , m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 548, 338–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engel M., et al. , The role of m6A/m-RNA methylation in stress response regulation. Neuron 99, 389–403.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Al Rifai M., et al. , Factors of health in the protection against death and cardiovascular disease among adults with subclinical atherosclerosis. Am. Heart J. 198, 180–188 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorn L. E., et al. , The N6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation 139, 533–545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Nostrand E. L., et al. , Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu S., et al. , Vascular semaphorin 7A upregulation by disturbed flow promotes atherosclerosis through endothelial β1 integrin. Arterioscler. Thromb. Vasc. Biol. 38, 335–343 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He M., Martin M., Marin T., Chen Z., Gongol B., Endothelial mechanobiology. APL Bioeng. 4, 010904 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang F., et al. , MicroRNA-92a mediates endothelial dysfunction in CKD. J. Am. Soc. Nephrol. 28, 3251–3261 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang Y. J., et al. , Extracellular MicroRNA-92a mediates endothelial cell-macrophage communication. Arterioscler. Thromb. Vasc. Biol. 39, 2492–2504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee D. Y., et al. , MicroRNA-10a is crucial for endothelial response to different flow patterns via interaction of retinoid acid receptors and histone deacetylases. Proc. Natl. Acad. Sci. U.S.A. 114, 2072–2077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L. J., et al. , MicroRNA mediation of endothelial inflammatory response to smooth muscle cells and its inhibition by atheroprotective shear stress. Circ. Res. 116, 1157–1169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gongol B., et al. , Shear stress regulation of miR-93 and miR-484 maturation through nucleolin. Proc. Natl. Acad. Sci. U.S.A. 116, 12974–12979 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou J., Li Y. S., Wang K. C., Chien S., Epigenetic mechanism in regulation of endothelial function by disturbed flow: Induction of DNA hypermethylation by DNMT1. Cell. Mol. Bioeng. 7, 218–224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galbraith C. G., Skalak R., Chien S., Shear stress induces spatial reorganization of the endothelial cell cytoskeleton. Cell Motil. Cytoskeleton 40, 317–330 (1998). [DOI] [PubMed] [Google Scholar]
- 24.del Alamo J. C., Norwich G. N., Li Y. S., Lasheras J. C., Chien S., Anisotropic rheology and directional mechanotransduction in vascular endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 105, 15411–15416 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross R., Atherosclerosis–An inflammatory disease. N. Engl. J. Med. 340, 115–126 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Wang Y., et al. , Shear stress regulates the Flk-1/cbl/PI3K/NF-κB pathway via actin and tyrosine kinases. Cell. Mol. Bioeng. 2, 341–350 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y., et al. , Shear stress and VEGF activate IKK via the Flk-1/Cbl/Akt signaling pathway. Am. J. Physiol. Heart Circ. Physiol. 286, H685–H692 (2004). [DOI] [PubMed] [Google Scholar]
- 28.Bleda S., et al. , NLRP1 inflammasome, and not NLRP3, is the key in the shift to proinflammatory state on endothelial cells in peripheral arterial disease. Int. J. Cardiol. 172, e282–e284 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Bai L., Shyy J. Y. P., Shear stress regulation of endothelium: A double-edged sword. J. Transl. Int. Med. 6, 58–61 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Q., Meng Q., Qi M., Li F., Liu B., Shear-sensitive lncRNA AF131217.1 inhibits inflammation in HUVECs via regulation of KLF4. Hypertension 73, e25–e34 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Fan Y., et al. , Krüppel-like factors and vascular wall homeostasis. J. Mol. Cell Biol. 9, 352–363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J., et al. , BMP receptor-integrin interaction mediates responses of vascular endothelial Smad1/5 and proliferation to disturbed flow. J. Thromb. Haemost. 11, 741–755 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Chiou G. Y., et al. , Epigenetic regulation of the miR142-3p/interleukin-6 circuit in glioblastoma. Mol. Cell 52, 693–706 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Imam H., et al. , N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115, 8829–8834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peritz T., et al. , Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 1, 577–580 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Nam D., et al. , Partial carotid ligation is a model of acutely induced disturbed flow, leading to rapid endothelial dysfunction and atherosclerosis. Am. J. Physiol. Heart Circ. Physiol. 297, H1535–H1543 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.