Significance
Meiotic recombination is regulated by both genetic and epigenetic factors such as DNA methylation. In maize, we found that the mop1 mutation removes CHH (where H = A, T, or C) methylation that is immediately adjacent to sites of frequent recombination in both chromosomal arms and pericentromeric regions. We further found that the mop1 mutation increased meiotic recombination frequencies in chromosomal arms but decreased them in pericentromeric regions. Our data demonstrate that although CHH methylation is present at a much lower level than CG and CHG methylation, it has a substantial effect on recombination frequencies, suggesting an important role for RNA-directed DNA methylation in meiotic recombination in maize.
Keywords: meiotic recombination, DNA methylation, mop1, lbl1
Abstract
Meiotic recombination is a fundamental process that generates genetic diversity and ensures the accurate segregation of homologous chromosomes. While a great deal is known about genetic factors that regulate recombination, relatively little is known about epigenetic factors, such as DNA methylation. In maize, we examined the effects on meiotic recombination of a mutation in a component of the RNA-directed DNA methylation pathway, Mop1 (Mediator of paramutation1), as well as a mutation in a component of the trans-acting small interference RNA biogenesis pathway, Lbl1 (Leafbladeless1). MOP1 is of particular interest with respect to recombination because it is responsible for methylation of transposable elements that are immediately adjacent to transcriptionally active genes. In the mop1 mutant, we found that meiotic recombination is uniformly decreased in pericentromeric regions but is generally increased in gene rich chromosomal arms. This observation was further confirmed by cytogenetic analysis showing that although overall crossover numbers are unchanged, they occur more frequently in chromosomal arms in mop1 mutants. Using whole genome bisulfite sequencing, our data show that crossover redistribution is driven by loss of CHH (where H = A, T, or C) methylation within regions near genes. In contrast to what we observed in mop1 mutants, no significant changes were observed in the frequency of meiotic recombination in lbl1 mutants. Our data demonstrate that CHH methylation has a significant impact on the overall recombination landscape in maize despite its low frequency relative to CG and CHG methylation.
Meiosis is a specialized type of cell division during which a single round of DNA replication is coupled to two rounds of chromosome segregation (1, 2). During meiotic prophase I, a critical step is homologous recombination, which promotes homologous pairing and generates crossovers (COs) (3, 4). Meiotic recombination is initiated by the formation of DNA double-strand breaks (DSBs) caused by the topoisomerase-like proteins, SPO11-1 and SPO11-2, as well as several accessory proteins (5, 6). DSBs are then processed into single-strand DNA tails that are associated with RAD51 and DMC1 recombinase proteins, which mediate interhomolog strand invasion (5–8). As a consequence of homologous recombination, COs and noncrossovers (NCOs) are formed (9, 10). In most organisms examined, only a small fraction of DSBs result in COs. For example, ∼300 to 500 DSBs are estimated to form in maize, which are only resolved as ∼20 COs per meiosis (11, 12). Mechanisms that determine whether DSBs result in COs or NCOs are poorly understood. At least three such mechanisms exist: CO assurance, CO interference, and CO homeostasis. CO assurance ensures formation of at least one obligatory CO per homologous chromosome pair, which is required for chromosome orientation and segregation (13). CO interference impacts CO distribution by preventing multiple CO formation in close proximity (14). Class I COs are interference sensitive, and their formation depends on ZMM proteins (Zip1, Zip2, Zip3, Zip4, Msh4, Msh5, Spo16, and Mer3) (15). In contrast, class II COs, which require the proteins Mus81 and Mms4 in Saccharomyces cerevisiae and Arabidopsis, are interference insensitive and result in randomly distributed COs (16–18). CO homeostasis maintains stable CO numbers even if the variation in the number of DSBs is substantial (19). Although CO homeostasis is robust in yeast and mice (19, 20), its effects in maize appear to be limited (12).
COs are not uniformly distributed along chromosomes. Rather, they are enriched in hotspots, which are controlled by both genetic and epigenetic regulation. In contrast to a number of well-conserved genes required for recombination, epigenetic factors that influence recombination are both elusive and diverse among species. In budding yeast, CO hotspots have no sequence preference but are enriched in nucleosome-depleted regions within promoters near H3K4me3 marks (21, 22). In mammals, COs occur more frequently in degenerate DNA sequence motifs that are targeted by a rapidly evolving zinc-finger protein PRDM9 (23). Mammalian COs are also enriched with H3K4me3 marks, but located farther away from promoter regions than those in yeast (24, 25). In plants, CO hotspots are observed at gene transcription start sites (TSSs) or transcription termination sites (TTSs) where they are enriched with H2A.Z nucleosomes and have DNA hypomethylation and decreased nucleosome occupancy (26–28). In plants with large genomes most COs are located in distal chromosomal regions. Conversely, COs are suppressed in heterochromatic centromere and pericentromeric regions (22, 24, 29, 30).
Heterochromatin, often composed of repeats or transposable elements (TEs), is associated with certain histone modifications and DNA methylation. In plant genomes, DNA methylation occurs in CG, CHG, and CHH (where H = A, T, or C) sequence contexts (31). In Arabidopsis, CG methylation is maintained through DNA replication by METHYLTRANSFERASE1 (MET1) and chromatin-remodeling proteins such as DECREASED DNA METHYLATION1 (DDM1) (31). Non-CG methylation is maintained by CHROMOMETHYLASE1 (CMT1), CMT2, and CMT3 in conjunction with the histone methyltransferase KRYPTONITE (31, 32). In addition, methylation at CHH sites and some CG and CHG sites can be de novo targeted by the RNA-directed DNA methylation (RdDM) pathway, which involves generation of small interfering RNAs (siRNAs) and induction of DNA methylation (33, 34). In Arabidopsis, loss of methylation in ddm1 and met1 mutants results in increased recombination in euchromatic regions of the genome, but causes no change or even a counterintuitive decrease in the number of COs in heterochromatic regions (35–38). Mutations in the homologs of MET1 and DDM1 are lethal in maize (39), suggesting the influence of DNA methylation may be more significant in species with much larger genomes. Because of this, information regarding how DNA methylation changes in any sequence context impact recombination is scarce in maize. The maize genome is 15 times larger than that of Arabidopsis, and most of it (>85%) is composed of densely methylated TEs (40). Thus, maize represents a suitable model for elucidating how a large genome maintains its genome integrity and at the same time, regulates meiotic recombination distribution through DNA methylation.
In maize, TEs typically have high levels of CG and CHG methylation, with a low level of CHH methylation that peaks at the edges of TEs (41, 42). It has been shown that Mop1 (Mediator of paramutation1), a component of the RdDM pathway, is required to maintain MuDR transposon methylation and silencing (43). Mop1 is a sequence ortholog of RNA dependent RNA polymerase 2 (RDR2) in Arabidopsis, which is responsible for maintaining transcriptional gene silencing (TGS) (44). MOP1 converts single-strand transcripts into double-strand RNAs that are then cleaved and processed into an abundant class of 24-nucleotide (nt) small RNAs (42, 45). These 24-nt siRNAs can recruit histone modifiers and DNA methyltransferases back to the original DNA sequences to maintain methylation of TEs (33, 34, 45). This pathway is thought to reinforce methylation at regions immediately adjacent to genic regions in high TE content genomes such as that of maize (41, 42, 46). These regions, referred to as “CHH islands,” are thought to serve as boundaries between deeply heterochromatic intergenic regions and the open chromatin associated with expressed genes (42). Genomewide, the mop1 mutation results in a loss of methylation at silenced TEs in regions of the genome that are immediately adjacent to transcriptionally active genes in the maize genome (42), making this mutant an ideal model for exploring the relationship between DNA methylation and recombination. Although the level of CHH methylation is globally low, recent research in Arabidopsis demonstrates that redistribution of COs toward subtelomeric chromosomal ends was observed in the neddylation/rubylation pathway mutant axr1. This redistribution is correlated with DNA hypermethylation of TEs in the CHH context in the mutant (47), suggesting a connection between CHH methylation and meiotic recombination. In Arabidopsis, in addition to the RdDM pathway, a second, related small RNA silencing pathway involves SUPPRESSOR OF GENE SILENCING3 (SGS3), which works together with RNA-dependent RNA polymerase 6 (RDR6) to generate 21- to 22-nt trans-acting small interference RNAs (tasiRNAs) (48–50). The major role of the 21- to 22-nt tasiRNAs generated by SGS3/RDR6 functions in posttranscriptional gene silencing (PTGS), but recent studies have revealed that some of these 21- to 22-nt tasiRNAs can also participate in TGS through a noncanonical RDR6-dependent RdDM pathway (49, 51–53).
To understand the role that the canonical RdDM and noncanonical RDR6-dependent RdDM pathways may play in meiotic recombination, we investigated the recombination frequencies in maize mutants of Mop1 and Lbl1 (Leafbladeless1), an ortholog of Arabidopsis SGS3, compared to their wild-type or heterozygous siblings. Our results show that the mop1 mutation does not significantly alter overall meiotic recombination frequencies, but, rather, leads to a redistribution of recombination events along the chromosomes. In contrast, no significant differences were observed in the frequency of meiotic recombination in either the chromosomal arms or the pericentromeric regions between lbl1 mutants and wild-type plants. Together, our data show that removal of DNA methylation in regions near genes can have global effects on the frequency of recombination. Our data also suggest that different epigenetic pathways may have distinct roles in the process of meiotic recombination.
Results
The mop1 Mutation Increased Meiotic Recombination Frequencies in Chromosomal Arms but Decreased Them in Pericentromeric Regions.
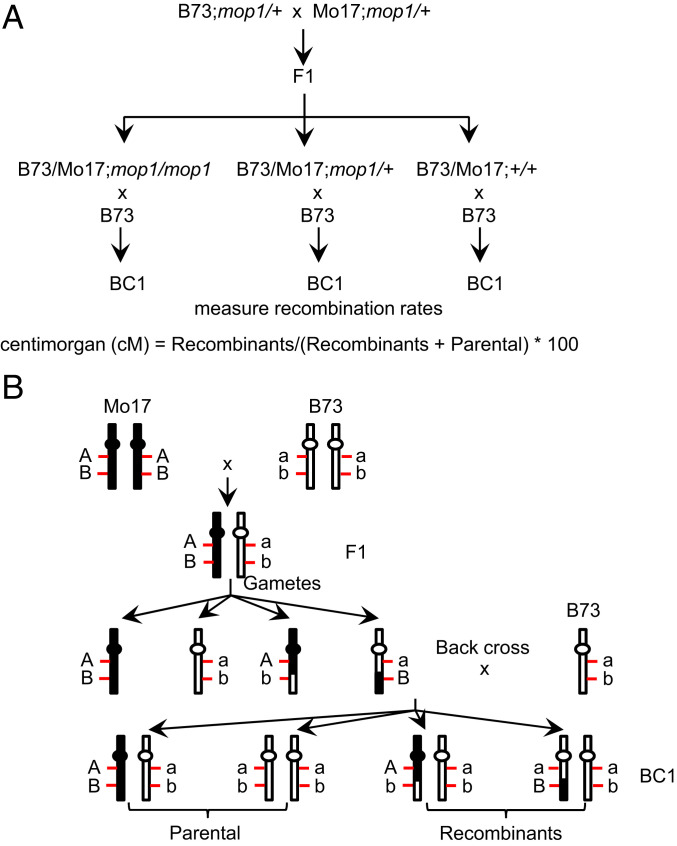
To understand the role of MOP1 in meiotic recombination, we crossed mop1 heterozygous plants in the B73 and Mo17 inbred backgrounds to each other (B73;mop1/+ × Mo17;mop1/+). We then backcrossed the F1 mop1 homozygous, heterozygous, and wild-type plants as females to B73 as male. This resulted in three backcross (BC1) populations derived from each F1 genotype (Fig. 1A). We used previously identified InDel (insertion and deletion) polymorphisms between these two maize backgrounds (54) to detect the recombinants in the BC1 populations to compare the meiotic recombination frequency in the F1 individuals that are B73/Mo17;mop1/mop1, B73/Mo17;mop1/+ or B73/Mo17;+/+ (Fig. 1B).
Fig. 1.
Strategy to construct the backcross (BC1) populations and measure recombination frequency. (A) Genetic pipeline to construct backcross populations from different F1 genotypes. B73 and Mo17 are two maize inbred lines. (B) Schematic diagram to measure recombination frequency. A (a) and B (b) represent polymorphic markers between the B73 and Mo17 genomes.
In many organisms, meiotic recombination is unevenly distributed along chromosomes, and recombination in most pericentromeric regions is suppressed (22, 24, 29, 30). Thus, we separated the 10 maize chromosomes into representative chromosomal arms and pericentromeric regions based on the gene and transposable element density as well as recombination frequency per megabase (Mb) of DNA as determined by 6,257 genetic markers in the previously described integrated map (SI Appendix, Fig. S1 and Table S1) (55). The maize genome (version 4) harbors 39,005 protein encoding genes on the 10 chromosomes, and of these genes, 27,044 (69.3%) are located in the chromosomal arms. The gene densities and gene numbers are both significantly higher in the chromosomal arms than they are in the pericentromeric regions (SI Appendix, Table S1). In addition, the recombination frequency (cM/Mb, centimorgan per megabase) is 10 times higher in the chromosomal arms than that in the pericentromeric regions in maize, indicating that meiotic recombination varies dramatically in different genomic regions. This is in part due to the fact that the vast majority of recombination in maize occurs in and around genes, which are less dense in the pericentromeric regions (11.29 genes/Mb in the pericentromeric regions versus 26.54 genes/Mb in the chromosomal arms, a ratio of 0.43). However, we also found that recombination rates per gene are significantly reduced in the pericentromeric regions (0.02 cM/gene in the pericentromeric regions versus 0.08 cM/gene in the chromosomal arms, a ratio of 0.21, P < 0.0001). We selected a set of 20 representative marker pairs in both the chromosomal arms and the pericentromeric regions from each of the 10 maize chromosomes to compare the recombination frequency between mop1 and wild-type plants (SI Appendix, Fig. S2 and Dataset S1) (54).
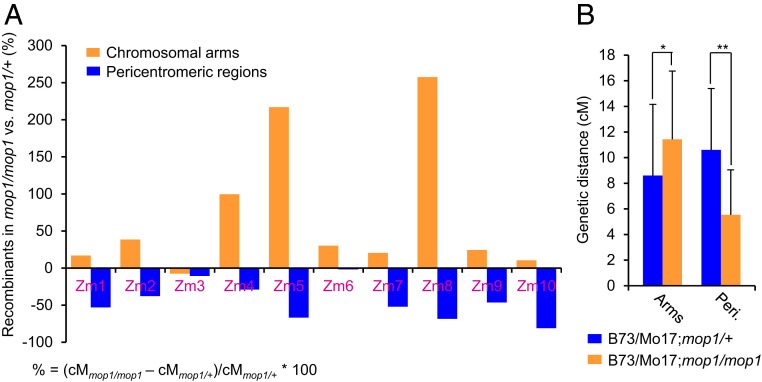
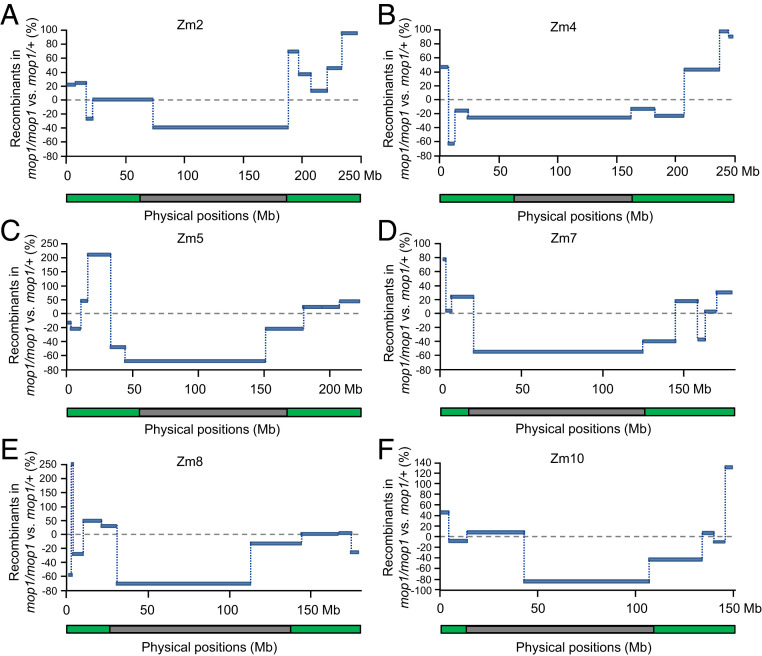
We first looked at the 142 and 126 BC1 progenies derived from the B73/Mo17;mop1/mop1 and B73/Mo17;mop1/+ F1 sibling individuals. For 9 of the 10 marker pairs located in the chromosomal arms that we examined, recombination was increased in the mop1 mutant (Fig. 2A and Dataset S2). The increase was largest on chromosomes 8, 5, and 4, with percentage increases of 257.4%, 216.9%, and 99.5%. In contrast, recombination frequencies in the pericentromeric regions of all of the 10 maize chromosomes were uniformly decreased in the mop1 mutant, with the larger effects on chromosomes 10, 8, and 5, with percentage decreases of 81.0%, 68.5%, and 66.8% (Fig. 2A and Dataset S2). The overall average genetic distances (cM) were significantly increased in the chromosomal arms (P < 0.05) but decreased in the pericentromeric regions (Fig. 2B, P < 0.01, Student’s paired t test), suggesting that mop1 influences recombination differently in these two distinct genomic regions. To gain additional resolution, we designed an additional 42 markers that made it possible to survey 6 of 10 entire maize chromosomes (Dataset S1). As shown in Fig. 3 and Dataset S3, with few exceptions, recombination frequencies were increased in the mop1 mutant in the chromosomal arms and were uniformly decreased in the pericentromeric regions. To confirm our results, we investigated a second pair of BC1 families, which included 133 and 155 progenies derived from the B73/Mo17;mop1/mop1 and B73/Mo17;+/+ F1 sibling individuals crossed as females (SI Appendix, Table S2). The recombination frequencies in the pericentromeric regions of the four chromosomes were consistently decreased in the mop1 mutant of all of the examined regions. For the eight regions that are located in the chromosomal arms, recombination frequencies were increased in four regions, but decreased in the other four regions in the mop1 mutant (SI Appendix, Table S2). Overall, the data suggest that the mop1 mutation increases recombination frequencies in chromosomal arms and decreases them in pericentromeric regions (SI Appendix, Table S2).
Fig. 2.
Meiotic recombination rates were increased in chromosomal arms but deceased in pericentromeric regions in mop1. (A) The changes of recombination rates in different genomic regions when comparing B73/Mo17;mop1/mop1 and B73/Mo17;mop1/+ in the 10 maize chromosomes. The y axis represents the percent change in recombination in B73/Mo17;mop1/mop1 compared with B73/Mo17;mop1/+. % = (cMmop1/mop1 – cMmop1/+)/cMmop1/+ × 100. The x axis represents the 10 maize chromosomes. (B) Comparison of the average genetic distance (cM) in B73/Mo17;mop1/mop1 and B73/Mo17;mop1/+ intervals. *P < 0.05; **P < 0.01. Student’s paired t test.
Fig. 3.
Meiotic recombination rates along six entire maize chromosomes. (A) Chromosome 2. (B) Chromosome 4. (C) Chromosome 5. (D) Chromosome 7. (E) Chromosome 8. (F) Chromosome 10. InDel markers were used to genotype the backcross populations derived from B73/Mo17;mop1/mop1 and B73/Mo17;mop1/+ F1 plants. The y axis represents the percentages of changes of B73/Mo17;mop1/mop1 compared with B73/Mo17;mop1/+. % = (cMmop1/mop1 – cMmop1/+)/cMmop1/+ × 100). The x axis represents the physical distances along each chromosome. Green and gray bars indicate the chromosomal arms and pericentromeric regions of each chromosome.
Overall Class I CO Precursors Are Unchanged but Occur More Frequently in Chromosomal Arms in the mop1 Mutant.
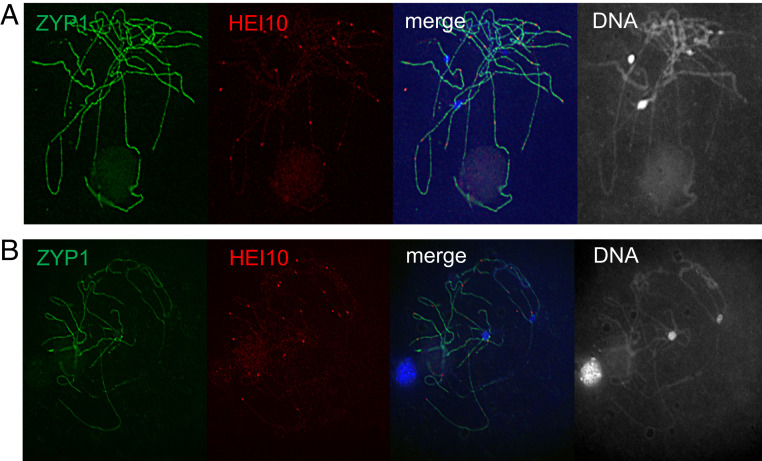
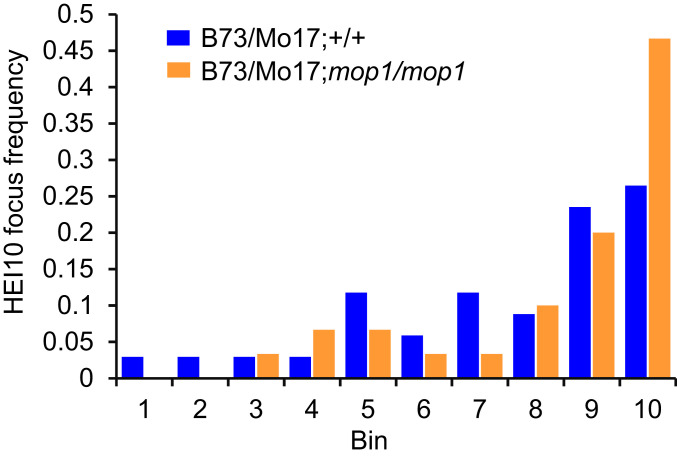
To further investigate changes in recombination distribution observed in mop1 mutants, we analyzed COs cytologically through immunolocalization using an antibody against maize HEI10. In Arabidopsis, HEI10 is a ZMM protein that is functionally related to yeast Zip3 ZMM, which generates continuity between early recombination intermediates and class I COs. HEI10 specifically promotes class I CO formation and marks 85 to 90% of total COs (56–60). Out of a total of 24 wild-type and 21 mutant meiocytes, we observed an average of 20.5 and 19.2 HEI10 foci (SI Appendix, Table S3), an example of which is shown in Fig. 4, suggesting that mop1 does not dramatically alter the overall number of class I CO precursors. To elucidate CO distribution along chromosomes, we traced chromosome 6, which can be recognized easily in our chromosome spreads because the short arm of chromosome 6 contains the ribosomal DNA (rDNA) array that is associated with the nucleolus (Fig. 4). We analyzed 34 and 30 HEI10 signals on chromosome 6 from 21 wild-type and 24 mop1 meiocytes at the pachytene stage, respectively. Signals representing HEI10 foci were assigned to 10 bins along chromosome 6 from the short to the long arms. Interestingly, we found that distribution of HEI10 signals is shifted toward the end of chromosome 6 in mop1 meiocytes as 46.7% (14/30) of HEI10 detected in the last bin, representing the most distal region of the long arm of this chromosome (Fig. 5). In contrast, only 26.5% (9/34) HEI10 foci in wild-type meiocytes were assigned to this bin. The absence of an increase of recombination within the short arm (bin 1) of chromosome 6 is likely due to the very small quantity of euchromatin on this arm (Fig. 5 and SI Appendix, Fig. S2). These observations are consistent with our genetic recombination data and suggest that rather than altering the overall recombination frequency, the mop1 mutation results in a redistribution of recombination toward the chromosomal distal regions of the chromosome and away from pericentromeric regions. Based on these observations, we hypothesize that an increase in recombination in the euchromatic distal portion of chromosomes results in CO suppression in the heterochromatic proximal portion of the chromosomes.
Fig. 4.
The numbers of class I crossover precursors are unchanged in mop1. Immunolocalizations were performed using ZYP1 and HEI10 antibodies, which mark chromosomes and class I crossover precursors, respectively. (A) Wild type, B73/Mo17;+/+. (B) mop1 mutant, B73/Mo17;mop1/mop1.
Fig. 5.
HEI10 foci occur more frequently in chromosomal distal ends in mop1. Chromosome 6 was divided into 10 equally sized bins from the short to the long arm. The HEI10 focus frequency was calculated by the number of HEI10 foci in each bin out of the total HEI10 foci in all the 10 bins.
The mop1 Mutation Dramatically Reduces CHH Methylation Near Genes.
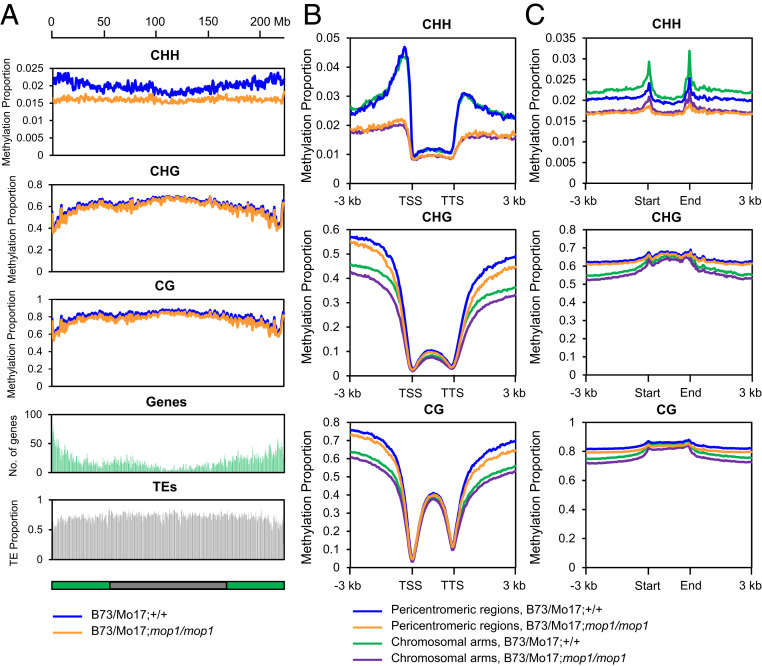
To address the causes of the recombination changes we observed in mop1 mutants, we collected 1.5- to 2.0-mm-long immature anthers, which contain meiocytes undergoing meiosis I (61) from F1 plants and performed high-throughput whole genome bisulfite sequencing (WGBS) to determine the level of DNA methylation of B73/Mo17;mop1/mop1 mutant and wild-type plants. We obtained a total of 301 million paired end reads from four samples representing two biological replicates (SI Appendix, Table S4). We mapped the WGBS reads to the B73 reference genome and observed that CG and CHG methylation are higher in the pericentromeric regions than those in the chromosomal arms. In contrast, CHH methylation is lower in the pericentromeric regions than that in the chromosomal arms in wild-type anthers. In addition, we observed slight decreases of CG and CHG methylation but a dramatic decrease of CHH methylation in the mop1 mutant of all 10 chromosomes (Fig. 6A and SI Appendix, Fig. S3).
Fig. 6.
CHH (H = A, T, or C) methylation is largely reduced in mop1 in immature anthers. (A) Chromosome distribution of DNA methylation, genes, and TEs, measured in 1-Mb windows with 500-kb shifts. (B) Patterns of methylation in and flanking genes. (C) Patterns of methylation in and flanking TEs. DNA methylation levels were calculated in 50-bp windows in the 3-kb upstream and downstream regions of the genes/TEs located in pericentromeric regions and chromosomal arms. Each gene/TE sequence was divided into 40 equally sized bins to measure the gene/TE body methylation. Bin sizes differ from gene/TE to gene/TE because of the different lengths of genes/TEs. Methylation for each sample was calculated as the proportion of methylated C over total C in each sequence context (CG, CHG, and CHH, where H = A, T, or C) averaged for each window. The average methylation levels were determined by combining two biological replicates for each genotype.
Previous research had shown that MOP1 is one of the major components in RdDM, and that the mop1 mutant primarily affects CHH islands near genes (39, 42, 46). Therefore, we plotted the DNA methylation level of CG, CHG, and CHH within gene bodies and 3 kb upstream of TSSs and 3 kb downstream of TTSs. We then separated the genes into pericentromeric regions and chromosomal arms and compared the DNA methylation levels between mop1 mutants and wild-type siblings. In wild-type anthers, both CG and CHG methylation are higher in the pericentromeric regions than in the chromosomal arms in the 3-kb up- and downstream regions around genes, and are slightly reduced in mop1 mutants in these regions. Consistent with previous results, CHH methylation was substantially and equally reduced in both of these genomic regions in the mutant (Fig. 6B). The peaks in the upstream and downstream regions, previously referred to as CHH islands (41, 42), are largely removed in the mutant, confirming a significant role for MOP1 in de novo CHH methylation. Previous research has shown that pericentromeric regions are enriched in H3K9me2 and H3K27me2 and CG and CHG methylation but are depleted of both CHH methylation and small RNAs (46). Our data show that CHH islands close to genes in both the pericentromeric regions and chromosomal arms are largely removed in the mop1 mutant (Fig. 6B). This suggests that although pericentromeric regions are less open and have limited accessibility, there are still many active genes in these regions that have the same characteristics as genes in chromosomal arms with respect to these CHH islands. Additionally, it should be noted that even in chromosomal arms as we have defined them, gene density is low relative to other grasses such as rice and sorghum (62, 63), and there are often long stretches of heterochromatic nested TEs between genes in maize.
We next investigated the DNA methylation level of CG, CHG, and CHH within TE bodies and their flanking regions. No obvious difference of CG and CHG methylation was observed between DNA from wild-type and mop1 anthers, but CHH methylation was notably decreased in mop1 in TE bodies and in the 3-kb upstream and downstream regions. Interestingly, there are two small peaks of CHH methylation at the edges of TEs, which are reduced in mop1 (Fig. 6C). Together these data suggest that the redistribution of COs observed in mop1 may result from the hypomethylation of TEs, particularly CHH hypomethylation in TEs that are adjacent to genes.
The lbl1 Mutation Has No Significant Effects on Meiotic Recombination.
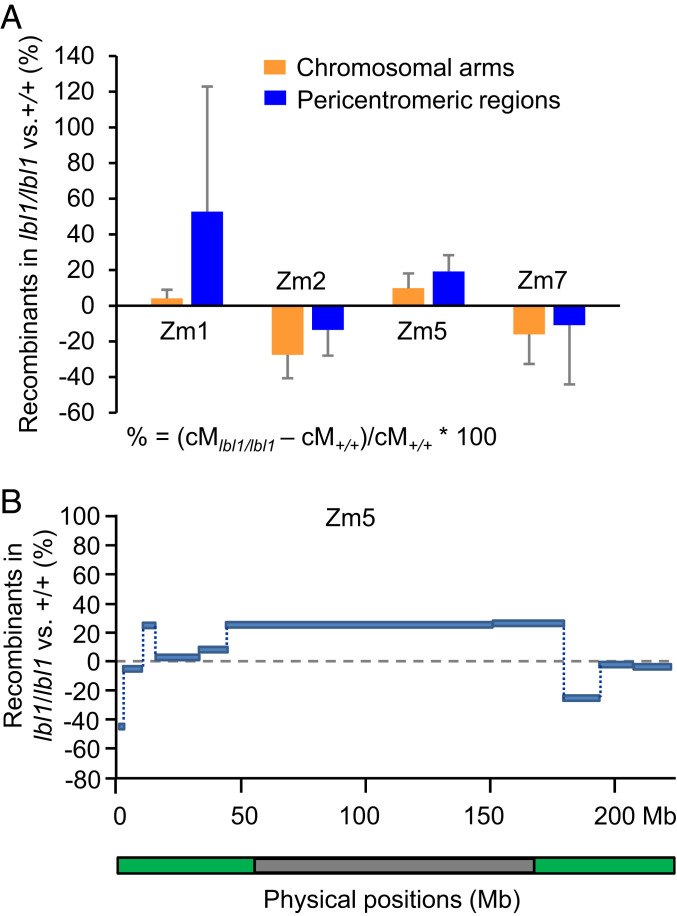
In addition to RdDM, also known as TGS, another important gene silencing pathway is PTGS, which generally involves 21- to 22-nt siRNAs (64). Recent research has revealed that 21- to 22-nt siRNAs associated with PTGS can also participate in RdDM via what has been referred to as the RDR6 RdDM pathway (51, 52). In maize, a key component of this pathway is Lbl1, a homolog of SGS3 in Arabidopsis (48–50). To determine whether the RDR6-dependent RdDM pathway affects meiotic recombination in maize, we compared the recombination frequency between lbl1 mutants and the wild-type siblings. We followed the same strategy that we used for mop1 mutants. We crossed lbl1 heterozygous plants in the B73 and Mo17 inbred backgrounds to each other (B73;lbl1/+ × Mo17;lbl1/+). We then backcrossed the F1 lbl1 homozygous, heterozygous and wild-type plants to B73 to produce BC1 populations in order to measure the recombination frequency for each F1 genotype using the same InDel markers that we used for mop1. Given that homozygous lbl1 is male sterile (48), all the F1 genotypes used here to produce BC1 populations were crossed as females. We measured the recombination frequencies of the chromosomal arms and pericentromeric regions of chromosomes 1, 2, 5, and 7 for two biological replicates (Dataset S4). As shown in Fig. 7A and Dataset S4, no consistent differences were observed between lbl1 mutants and wild-type siblings for either of the two replicates. To confirm our results, we also examined recombination along the entire length of chromosome 5. No obvious changes of recombination frequencies were observed between lbl1 mutants and wild types (Fig. 7B and SI Appendix, Table S5). Together, these data suggest that the lbl1 mutation does not have global effects on meiotic recombination. However, it remains a possibility that this mutation may affect recombination at specific sites at which DNA methylation has been largely reduced in the mutants.
Fig. 7.
Meiotic recombination rates are not significantly changed in lbl1. (A) The changes of recombination rates in different genomic regions between B73/Mo17;lbl1/lbl1 and B73/Mo17;+/+ of four maize chromosomes. (B) Changes in meiotic recombination along chromosome 5 in lbl1 mutants compared to the wild-type control. The y axis represents the percentages of recombination changes of B73/Mo17;lbl1/lbl1 compared with B73/Mo17;+/+. % = (cMlbl1/lbl1 – cM+/+)/cM+/+ × 100. Green and gray bars indicate the chromosomal arms and the pericentromeric regions of the chromosome.
Discussion
In this study, we investigated the recombination frequencies in two representative mutants that affect small RNA pathways in maize, mop1 and lbl1. We demonstrate that removal of CHH methylation in mop1 results in a redistribution of recombination toward the distal regions of the chromosomes and away from pericentromeric regions, a similar phenomenon as was observed in met1 mutants in Arabidopsis (36–38). In that species, MET1 is responsible for CG methylation throughout the genome and constitutes 30.5% of the DNA methylation in that species (65). In contrast, in maize, MOP1 primarily functions in CHH methylation at RdDM loci in maize, which constitutes only ∼2% of the methylation in the maize genome (46). In the pericentromeric regions of the maize genome, there is very high DNA methylation at CG and CHG (∼90% CG methylation and ∼75% CHG methylation), and very little CHH methylation (∼2%) (SI Appendix, Fig. S3) (46). In the chromosomal arm regions near genes, CHH methylation can be as high as 5%, but even in these regions, it is still not comparable with CG (64%) and CHG methylation (46%) (Fig. 6). Despite the relatively subtle effects that the mop1 mutant has on global levels of DNA methylation, we observed a substantial effect on recombination frequencies, suggesting an important role for RdDM in meiotic recombination in maize. Notably, the loss of CHH islands may trigger additional loss of CHG methylation in the up- and downstream regions near genes (Fig. 6), which may also contribute to the changes of recombination frequency in mop1 mutants. We find that 11,961 (30.7%) genes are located in centromeric and pericentromeric heterochromatin as defined here (SI Appendix, Fig. S1 and Table S1). Many of these genes harbor CHH islands in the flanking regions that are dependent on Mop1. This suggests that genes in these regions do not differ fundamentally with respect to the role that RdDM plays in providing a boundary between genes and adjacent TEs. This also suggests that although recombination is generally suppressed in heterochromatin (SI Appendix, Fig. S2), there may be some local recombination hotspots that mop1 has effects on.
The maize genome is relatively large (∼2.3 Gb) and has a large number of TEs in both pericentromeric regions and chromosomal arms (SI Appendix, Table S1) (66). In both regions, the majority (85.5%) of genes are located within 1 kb of transposons (42). The majority of these transposons are class II DNA TEs, and CHH islands tend to form at the edges of these elements (42). Given that these CHH islands are enriched near transcriptionally active genes (42), they tend to be adjacent to recombination hotspots, which are also enriched at genes (55). Because DNA methylation locally inhibits CO hotspots (67), we hypothesized that removal of CHH methylation in mop1 would increase recombination frequency. This appears to be true in chromosomal arms, but the opposite appears to be the case in pericentromeric regions, which have depressed recombination in mop1 mutants. We hypothesize that the redistribution of COs observed in mop1 is caused by CO interference and constant CO numbers. Removal of DNA methylation could make the DNA more accessible to homologous recombination enzymes in chromosomal arms, fostering DSBs, homology searches, strand invasion, and exchange. In contrast, although methylation has also been removed from genes located in pericentromeric regions, it is possible that additional chromatin modifications in these regions may block or delay either the initial DSBs or subsequent recombination. Thus, hypomethylation may be sufficient to trigger the changes in chromosomal arms that allow enhanced meiotic recombination but not sufficient to trigger these changes in heterochromatin. In addition, early CO in chromosomal arms may prevent CO formation in pericentromeric regions due to CO interference. It is known that replication occurs later in pericentromeric regions in Arabidopsis (68). Thus, it is possible that a loss of DNA methylation earlier in chromosomal arms results in an increase in the recombination frequency early (38). Given that COs interfere with each other to maintain stable CO numbers, this may result in reduced COs in pericentromeric regions.
The overall effect of mop1 on recombination frequency was not dramatic, but it was consistent among different chromosomes, and was similar in scale to what has been observed in met1 mutants in Arabidopsis (36, 38) (SI Appendix, Figs. S2 and S3). Global reduction in DNA methylation levels are likely to influence recombination patterns via changes in chromatin structure (28, 38), and thus, the relationship between changes in DNA methylation and changes in recombination may be only indirect (28). Although possible, we suggest this is less likely in the case of the mop1 mutant because of the relatively subtle effects that this mutant has. In contrast to met1 and ddm1 mutants, which have dramatic effects on both methylation and patterns of histone modification throughout plant genomes, the effects of mop1 are relatively discrete and precisely targeted. Thus, the proximate effects of this mutant are likely to be local rather than global, although the overall effects can clearly be global. We also observed a few exceptions in the chromosomal arms showing decreased recombination frequencies in mop1 mutants. Given that the maize genome harbors a large number of TEs even in the chromosomal arms, we suspect those regions may behave more like pericentromeric regions than chromosomal arms.
Our analysis of recombination is informative, but due to the large regions and limited number of progenies assayed, the resolution of our data is too low to determine the specific genomic and epigenomic features influencing meiotic COs in mop1 mutants. Higher resolution recombination data will allow us to observe changes in or near individual genes, which is where most recombination occurs in maize (42). We hypothesize that for recombination to be altered by the mop1 mutant at a given gene or run of genes, that gene or genes should have CHH islands that are eliminated in this mutant.
Materials and Methods
Plant Materials and Growing Conditions.
The mop1 and lbl1 mutants were introgressed into the B73 and Mo17 backgrounds for at least seven generations. The plant materials were grown in greenhouse condition (82 °F/72 °F day/night, 16 h light, 8 h dark) in winters and on the agricultural research farms during summers at Purdue University and Miami University. In each experiment, mutant plants were planted and grown together with their wild-type control plants to avoid variability in environmental conditions and crossed as females to B73. The crosses are illustrated in Fig. 1A.
Detection of Recombination Frequency.
All the polymorphic markers used in this research are previously identified InDel markers (54). PCR primers used to detect these markers were designed using Primer3Plus: http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi. The markers were confirmed by PCR amplification using B73, Mo17, and B73/Mo17 F1 DNA. We selected the InDel markers to represent the chromosomal arms and pericentromeric regions of each chromosome, which were defined based on the gene and TE densities, as well as recombination frequency per physical distance as determined by the integrated genetic map of the reference genome (55), as shown in SI Appendix, Fig. S1. BC1 populations were planted in the greenhouse, and leaf 2 of each BC1 individual was collected for DNA extraction using a modified cetyl trimethylammonium bromide (CTAB) method. Recombination frequency was determined by looking at BC1 progeny derived from at least two F1 mutant and two F1 wild-type or heterozygous individuals using the InDel markers described above. Genetic distance (cM) was measured in the BC1 populations to compare the meiotic recombination frequency in the F1 mutant and heterozygous and homozygous wild-type siblings (Fig. 1B and Dataset S1).
Immunostaining and CO Analyses.
Immature tassels of B73/Mo17;mop1/mop1 and B73/Mo17;+/+ were carefully removed from 4- to 6-wk-old plants and fixed in Farmer’s fixative. Meiotic anthers (1.5 to 2.0 mm), in which meiocytes are undergoing meiosis I, were dissected and meiocytes were squashed on poly-l-lysine-coated slides. Immunostaining was performed as previously described (69). Anti-ZYP1 and anti-HEI10 antibodies were used at 1:200 dilution (70). Slides were imaged using a Deltavision core microscope (GE Healthcare) and image analyses was performed by ImageJ.
DNA Methylation.
The immature anthers (1.5 to 2.0 mm) of B73/Mo17;mop1/mop1 and B73/Mo17;+/+ F1 plants were collected on the agricultural research farm at Miami University, flash frozen in liquid nitrogen, and preserved at −80 °C. DNA was extracted from frozen tissue using the modified CTAB method. Due to the low amount of DNA, high-throughput low input WGBS was performed at Novogene and 150-bp paired end reads were generated (SI Appendix, Table S4). Read quality was assessed by FASTQC. Low quality and adapter sequences were filtered and trimmed. The remaining paired end reads were further trimmed with 10 bp of the end of read 1, and 10 bp of the beginning of read 2. The clean reads were then mapped to the B73 reference genome v4 (66) using Bismark (71) allowing one mismatch in the seed sequence (-N 1 -I 50). PCR duplicates were removed using Picardtools. Only uniquely mapped and properly paired reads were retained for methylation calls. Alignments were performed to identify the methylated cytosines using the Bismark methylation extractor with default parameters. The numbers of methylated and unmethylated reads were counted using bismark2bedGraph and coverage2cytosine in Bismark. The proportion of each type of methylation (CG, CHG, and CHH) was determined as absolute methylation levels in each 50-bp window of the 3-kb upstream and downstream regions of the protein encoding genes and TEs (72). For gene and TE body methylation, each gene and TE sequences were divided into 40 equally sized bins, and the absolute methylation levels were calculated in each bin (73).
Supplementary Material
Acknowledgments
We thank all current and past undergraduate students working in the M.Z. laboratory for generating the recombination frequency data. We appreciate the helpful comments and suggestions by the two anonymous reviewers. This work was supported by the National Institute of General Medical Sciences (NIGMS) of the NIH under Award R15GM135874, Miami University startup funds to M.Z., Institutional Development Award from the NIGMS under Grant P20GM103476 to D.W., and NSF Grant DBI-1237931 to D.L.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009475118/-/DCSupplemental.
Data Availability.
The whole genome bisulfite sequencing data of 1.5- to 2.0-mm F1 anthers of mop1 mutants and the wild-type siblings have been deposited in the National Center for Biotechnology Information Sequence Read Archive under associate code PRJNA681556 and accession nos. SRR13167029–SRR13167032.
References
- 1.Villeneuve A. M., Hillers K. J., Whence meiosis? Cell 106, 647–650 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Hunter N., Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lambing C., Franklin F. C., Wang C. R., Understanding and manipulating meiotic recombination in plants. Plant Physiol. 173, 1530–1542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang S., Kleckner N., Zhang L., Crossover maturation inefficiency and aneuploidy in human female meiosis. Cell Cycle 16, 1017–1019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keeney S., Neale M. J., Initiation of meiotic recombination by formation of DNA double-strand breaks: Mechanism and regulation. Biochem. Soc. Trans. 34, 523–525 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Vrielynck N., et al. , A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351, 939–943 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Kurzbauer M. T., Uanschou C., Chen D., Schlögelhofer P., The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell 24, 2058–2070 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M. S., Grubb J., Zhang A., Rust M. J., Bishop D. K., Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLoS Genet. 11, e1005653 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingsworth N. M., Brill S. J., The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev. 18, 117–125 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockmill B., et al. , High throughput sequencing reveals alterations in the recombination signatures with diminishing Spo11 activity. PLoS Genet. 9, e1003932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franklin A. E., et al. , Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11, 809–824 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidhu G. K., et al. , Recombination patterns in maize reveal limits to crossover homeostasis. Proc. Natl. Acad. Sci. U.S.A. 112, 15982–15987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones G. H., Franklin F. C., Meiotic crossing-over: Obligation and interference. Cell 126, 246–248 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Zhang L., Liang Z., Hutchinson J., Kleckner N., Crossover patterning by the beam-film model: Analysis and implications. PLoS Genet. 10, e1004042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitby M. C., Making crossovers during meiosis. Biochem. Soc. Trans. 33, 1451–1455 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Berchowitz L. E., Francis K. E., Bey A. L., Copenhaver G. P., The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 3, e132 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de los Santos T., et al. , The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164, 81–94 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins J. D., Buckling E. F., Franklin F. C., Jones G. H., Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Martini E., Diaz R. L., Hunter N., Keeney S., Crossover homeostasis in yeast meiosis. Cell 126, 285–295 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cole F., et al. , Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerton J. L., et al. , Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 97, 11383–11390 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan J., et al. , A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 144, 719–731 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grey C., Baudat F., de Massy B., PRDM9, a driver of the genetic map. PLoS Genet. 14, e1007479 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myers S., Bottolo L., Freeman C., McVean G., Donnelly P., A fine-scale map of recombination rates and hotspots across the human genome. Science 310, 321–324 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Brick K., Smagulova F., Khil P., Camerini-Otero R. D., Petukhova G. V., Genetic recombination is directed away from functional genomic elements in mice. Nature 485, 642–645 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi K., et al. , Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat. Genet. 45, 1327–1336 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Li L., Yan J., Dissecting meiotic recombination based on tetrad analysis by single-microspore sequencing in maize. Nat. Commun. 6, 6648 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kianian P. M. A., et al. , High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9, 2370 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraut L., et al. , Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 7, e1002354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mézard C., Jahns M. T., Grelon M., Where to cross? New insights into the location of meiotic crossovers. Trends Genet. 31, 393–401 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Law J. A., Jacobsen S. E., Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jackson J. P., Lindroth A. M., Cao X., Jacobsen S. E., Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560 (2002). [DOI] [PubMed] [Google Scholar]
- 33.Matzke M. A., Mosher R. A., RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Matzke M. A., Kanno T., Matzke A. J., RNA-directed DNA methylation: The evolution of a complex epigenetic pathway in flowering plants. Annu. Rev. Plant Biol. 66, 243–267 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Melamed-Bessudo C., Levy A. A., Deficiency in DNA methylation increases meiotic crossover rates in euchromatic but not in heterochromatic regions in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, E981–E988 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirouze M., et al. , Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 5880–5885 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yelina N. E., et al. , Epigenetic remodeling of meiotic crossover frequency in Arabidopsis thaliana DNA methyltransferase mutants. PLoS Genet. 8, e1002844 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yelina N. E., et al. , DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 29, 2183–2202 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu F. F., Dawe R. K., Gent J. I., Loss of RNA-directed DNA methylation in maize chromomethylase and DDM1-type nucleosome remodeler mutants. Plant Cell 30, 1617–1627 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnable P. S., et al. , The B73 maize genome: Complexity, diversity, and dynamics. Science 326, 1112–1115 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Gent J. I., et al. , CHH islands: De novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 23, 628–637 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li Q., et al. , RNA-directed DNA methylation enforces boundaries between heterochromatin and euchromatin in the maize genome. Proc. Natl. Acad. Sci. U.S.A. 112, 14728–14733 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodhouse M. R., Freeling M., Lisch D., Initiation, establishment, and maintenance of heritable MuDR transposon silencing in maize are mediated by distinct factors. PLoS Biol. 4, e339 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alleman M., et al. , An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 442, 295–298 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Nobuta K., et al. , Distinct size distribution of endogeneous siRNAs in maize: Evidence from deep sequencing in the mop1-1 mutant. Proc. Natl. Acad. Sci. U.S.A. 105, 14958–14963 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gent J. I., et al. , Accessible DNA and relative depletion of H3K9me2 at maize loci undergoing RNA-directed DNA methylation. Plant Cell 26, 4903–4917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Christophorou N., et al. , AXR1 affects DNA methylation independently of its role in regulating meiotic crossover localization. PLoS Genet. 16, e1008894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Timmermans M. C., Schultes N. P., Jankovsky J. P., Nelson T., Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125, 2813–2823 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Li H., Freeling M., Lisch D., Epigenetic reprogramming during vegetative phase change in maize. Proc. Natl. Acad. Sci. U.S.A. 107, 22184–22189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dotto M. C., et al. , Genome-wide analysis of leafbladeless1-regulated and phased small RNAs underscores the importance of the TAS3 ta-siRNA pathway to maize development. PLoS Genet. 10, e1004826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nuthikattu S., et al. , The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol. 162, 116–131 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuerda-Gil D., Slotkin R. K., Non-canonical RNA-directed DNA methylation. Nat. Plants 2, 16163 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Burgess D., Li H., Zhao M., Kim S. Y., Lisch D., Silencing of mutator elements in maize involves distinct populations of small RNAs and distinct patterns of DNA methylation. Genetics 215, 379–391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Settles A. M., et al. , Efficient molecular marker design using the MaizeGDB Mo17 SNPs and Indels track. G3 (Bethesda) 4, 1143–1145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S., et al. , Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5, e1000733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chelysheva L., et al. , The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 8, e1002799 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang K., et al. , The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 8, e1002809 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higgins J. D., Sanchez-Moran E., Armstrong S. J., Jones G. H., Franklin F. C., The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 19, 2488–2500 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Higgins J. D., et al. , Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell 24, 4096–4109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Falque M., Anderson L. K., Stack S. M., Gauthier F., Martin O. C., Two types of meiotic crossovers coexist in maize. Plant Cell 21, 3915–3925 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhai J., et al. , Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. U.S.A. 112, 3146–3151 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.International Rice Genome Sequencing Project , The map-based sequence of the rice genome. Nature 436, 793–800 (2005). [DOI] [PubMed] [Google Scholar]
- 63.Paterson A. H., et al. , The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556 (2009). [DOI] [PubMed] [Google Scholar]
- 64.Sijen T., et al. , Transcriptional and posttranscriptional gene silencing are mechanistically related. Curr. Biol. 11, 436–440 (2001). [DOI] [PubMed] [Google Scholar]
- 65.Niederhuth C. E., et al. , Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 17, 194 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiao Y., et al. , Improved maize reference genome with single-molecule technologies. Nature 546, 524–527 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zelkowski M., Olson M. A., Wang M., Pawlowski W., Diversity and determinants of meiotic recombination landscapes. Trends Genet. 35, 359–370 (2019). [DOI] [PubMed] [Google Scholar]
- 68.Lee T. J., et al. , Arabidopsis thaliana chromosome 4 replicates in two phases that correlate with chromatin state. PLoS Genet. 6, e1000982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chelysheva L. A., Grandont L., Grelon M., Immunolocalization of meiotic proteins in brassicaceae: Method 1. Methods Mol. Biol. 990, 93–101 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Ku J. C., et al. , Dynamic localization of SPO11-1 and conformational changes of meiotic axial elements during recombination initiation of maize meiosis. PLoS Genet. 16, e1007881 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krueger F., Andrews S. R., Bismark: A flexible aligner and methylation caller for bisulfite-seq applications. Bioinformatics 27, 1571–1572 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao M., Zhang B., Lisch D., Ma J., Patterns and consequences of subgenome differentiation provide insights into the nature of paleopolyploidy in plants. Plant Cell 29, 2974–2994 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schultz M. D., Schmitz R. J., Ecker J. R., ‘Leveling’ the playing field for analyses of single-base resolution DNA methylomes. Trends Genet. 28, 583–585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole genome bisulfite sequencing data of 1.5- to 2.0-mm F1 anthers of mop1 mutants and the wild-type siblings have been deposited in the National Center for Biotechnology Information Sequence Read Archive under associate code PRJNA681556 and accession nos. SRR13167029–SRR13167032.