Significance
The torsional stress to chromatin structure plays an important role in eukaryotic cell where DNA-binding proteins such as RNA polymerase and chromatin remodeling complexes actively apply torsional stress. Under torsional stress, chromatin keeps balance between the integrity of the eukaryotic genome for storage of the genetic information and the accessibility of the DNA for gene expression. Using all-atom molecular dynamics simulations, we found that positive torsion facilitated asymmetric unwrapping of the DNA without a large extension of the DNA, while negative torsion suppressed unwrapping. Our results suggest that the stability of individual nucleosomes in chromatin can be changed or tuned by torsional stress, thereby DNA accessibility of regulatory proteins can be changed to regulate gene expression.
Keywords: nucleosome, unwrapping, DNA, bendability, torsional stress
Abstract
Torsional stress has a significant impact on the structure and stability of the nucleosome. RNA polymerase imposes torsional stress on the DNA in chromatin and unwraps the DNA from the nucleosome to access the genetic information encoded in the DNA. To understand how the torsional stress affects the stability of the nucleosome, we examined the unwrapping of two half superhelical turns of nucleosomal DNA from either end of the DNA under torsional stress with all-atom molecular dynamics simulations. The free energies for unwrapping the DNA indicate that positive stress that overtwists DNA facilitates a large-scale asymmetric unwrapping of the DNA without a large extension of the DNA. During the unwrapping, one end of the DNA was dissociated from H3 and H2A-H2B, while the other end of the DNA stably remained wrapped. The detailed analysis indicates that this asymmetric dissociation is facilitated by the geometry and bendability of the DNA under positive stress. The geometry stabilized the interaction between the major groove of the twisted DNA and the H3 αN-helix, and the straightened DNA destabilized the interaction with H2A-H2B. Under negative stress, the DNA became more bendable and flexible, which facilitated the binding of the unwrapped DNA to the octamer in a stable state. Consequently, we conclude that the torsional stress has a significant impact on the affinity of the DNA and the octamer through the inherent nature of the DNA and can change the accessibility of regulatory proteins.
The genome DNA of eukaryotes is stored compactly in a nucleus and folded into a higher order structure called chromatin to protect the DNA from being damaged by radiation and so on. At the same time, decompaction of the chromatin structure is also required for the control of gene regulation in cellular processes for transcription, replication, repair, and recombination. Nucleosome is the fundamental structural unit of chromatin and is composed of histone proteins and DNA. Its crystal structure revealed that within the nucleosome, 146 or 147 base pairs of DNA wrap 1.75 times around an octameric histone core (1). Although the nucleosome is stable in physiological conditions, thermal fluctuations cause spontaneous partial unwrapping of nucleosomal DNA from the histone core. Experiments have found a dynamic motion between an open state with partially unwrapped DNA and a closed state with fully wrapped DNA (2, 3), which is often referred to as the breathing motion of the nucleosome. Förster resonance energy transfer (FRET) experiments (4–6) showed that in the absence of salt, the outer superhelical turn of nucleosomal DNA (hereinafter referred to as “the outer turn of DNA”) within the nucleosome repeatedly unwraps and rewraps.
In addition to the spontaneous unwrapping of the outer turn of DNA, some DNA-binding proteins, such as transcribing RNA polymerase (RNAP) (7, 8) and chromatin remodelers (9, 10), actively apply forces to unwrap or rewrap nucleosomal DNA. During transcription, RNAP moves along the DNA helix and generates positive stress on the DNA in front of the RNAP and negative stress behind it. Consequently, DNA downstream and upstream of the polymerization site are overwound (either overtwisted or positively supercoiled) and underwound (either undertwisted or negatively supercoiled), respectively (11). As nucleosome core particles prefer to bind negatively supercoiled DNA, overwound DNA ahead of the RNAP would destabilize the DNA–histone or histone–histone interactions in the nucleosome, and underwound DNA behind the RNAP would promote reassembly of nucleosomes (12, 13).
Single-molecule experiments have shown that torsional stress by mechanical force has a significant impact on the stability of the nucleosome (5, 14, 15). Another experiment by scanning force microscopy showed that overwound DNA led to larger nucleosome opening angles than relaxed or underwound DNA (16), suggesting that the overwound DNA facilitates the unwrapping of the outer turn of DNA.
However, the mechanism of unwrapping and rewrapping the outer turn of DNA is not well known at the atomic level. In order to understand how the torsional stress on the outer turn of DNA affects the stability of the nucleosome, we carried out all-atom molecular dynamics (MD) simulations of the nucleosome under torsional stress. Previously, we investigated unwrapping of two half superhelical turns of nucleosomal DNA from either end of the DNA (hereinafter referred to as “two outer half turns of DNA”) without torsional stress (17–20). The free energy under torsional stress in this article will give further insight into understanding the unwrapping of the outer turn of DNA.
Results
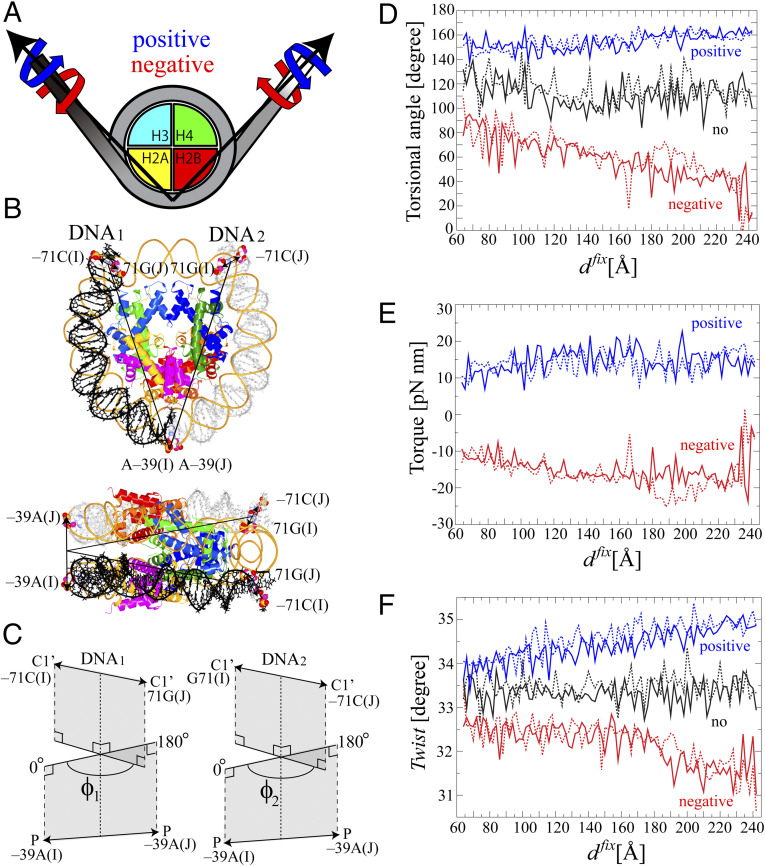
We applied positive or negative torsional stress at the third base pair from each end of the DNA to suppress base pair breakup at either end of the DNA as shown in Fig. 1 (see SI Appendix for the setup and the procedure of the simulations in SI Appendix, Fig. S1). Fig. 1A shows the schematic picture of the nucleosome with the direction of the positive and negative stress during the umbrella sampling simulations, while the atoms on which the stress was imposed are shown in the space-filled model (Fig. 1B). The definition of the torsion to describe the torsional stress is shown in Fig. 1C. As the unwrapping of the DNA proceeds, the torsion increased and decreased under positive and negative stress, respectively (Fig. 1D), according to the torsional stress imposed by Eq. 2. The magnitude of the applied torque was mostly within 10 to 20 pN nm (Fig. 1E), which is comparable with the Escherichia coli RNAP stall torque of ∼11 pN nm (8). DNA twist gradually increased and decreased upon addition of the torsional stress (Fig. 1F). The values of the twist, ∼33.0° ± 2.0°, were within the range where the elastic property of the DNA is maintained (21–23).
Fig. 1.
The system of the nucleosome for simulations under torsional stress. We used a crystal structure of a nucleosome with α-satellite palindromic 147 bp DNA (Protein Data Bank code: 1kx5) (46). The histone tails were truncated to facilitate the unwrapping of the DNA. Consequently, residues 46 to 132 in H3, 25 to the C terminal in H4, 16 to 114 in H2A, and 32 to 121 in H2B were used. (A) Schematic representation of the system of the nucleosome for torsional stress. The axes of the torsional stress are shown as arrows. The torques are applied to the ends of the DNA anticlockwise and clockwise along the axes under positive (blue) and negative (red) stress, respectively. (B) The initial structure of the nucleosome is shown in tube model with the view from top and side. The histone proteins, H3, H4, H2A, and H2B, are shown in blue, green, yellow, and red, respectively. DNA1 and DNA2 are shown in black and gray, respectively. The base pairs on which the torsional restraints were imposed, −71 C:G 71 in DNA1 or 2 and −39 A in DNA1 or 2 located opposite the dyad, are shown in the space-filled model. (C) The definition of the torsional angles of ϕ1 and ϕ2. The torsional angle of ϕ1 is calculated using the positions of two pairs of atoms. The first pair is the two C1’ atoms of −71 C in chain I and 71 G in chain J, and the other pair is the two phosphorus atoms of −39 A in chain I and chain J located opposite the dyad. The axis of torsion was defined as the line passing through the two midpoints between the two C1’ atoms and between the two phosphorus atoms. Then, the torsional angle of ϕ1 is defined as the angle between a plane which includes the two C1’ atoms and the axis and another plane which includes the two phosphorus atoms and the axis. The torsional angle of ϕ2 is defined in a similar way to the torsional angle of ϕ1. (D) Torsional angle, (E) torque, and (F) Twist in DNA1 and DNA2 are plotted against dfix during the last 50 ns of the umbrella sampling simulations. These parameters show the structural response of each of 90 replicas to the torsional restraint. Twist of the base pair step parameter was analyzed for the third to 35th base pairs from the end of the DNA1 or 2. The effect of the torsional stress on the DNA was incorporated as follows in a similar way to the procedure used by refs. 21, 22: for the i-th window. Both and were set at 0° and 180° for the generation of positive and negative stress, respectively. Torque was calculated by . Details are given in Materials and Methods and SI Appendix, Supplementary Methods.
Free Energy for Unwrapping the DNA.
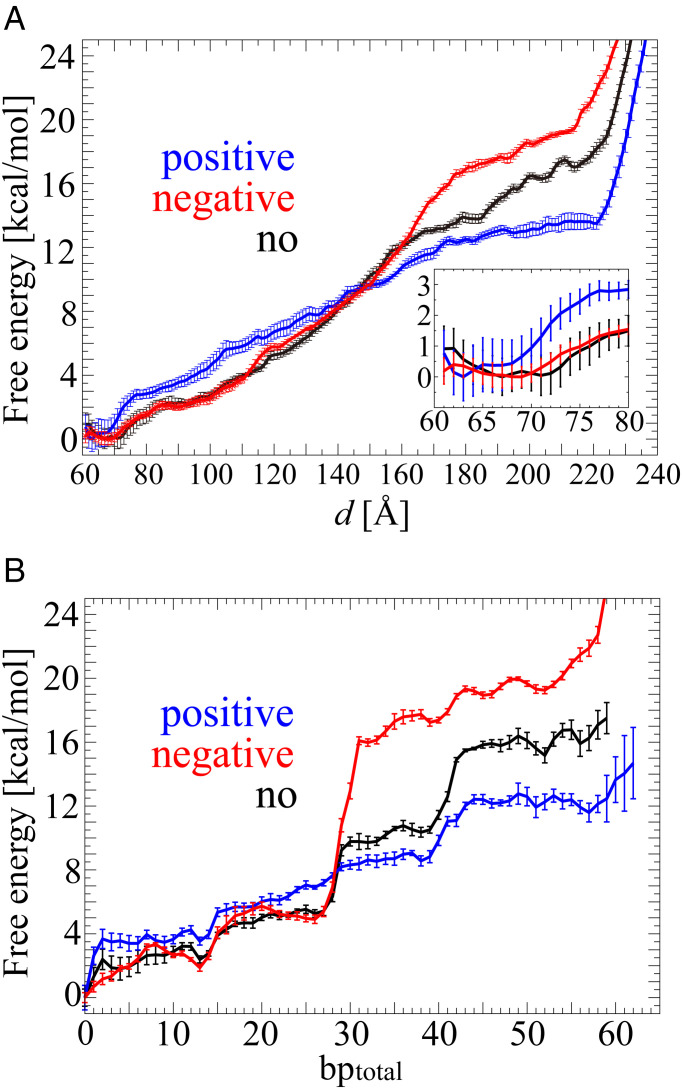
We calculated the free energy for unwrapping the DNA against the DNA end-to-end distance d using the weighted histogram analysis method (WHAM) algorithm (24, 25) (Fig. 2A). The free energies had a global minimum free energy of 0.0 kcal/mol when d = 63, 69, and 67 Å under positive, negative, and no stress, respectively. Up to d = 145 Å, the free energy under positive stress was larger than those under negative and no stress, indicating more free energy is required to extend the distance between two DNA ends under positive stress. In particular, the differences in the free energy up to that distance among the three torsional conditions come from the sharper rise of the free energy from d = ∼63 to 76 Å under positive stress compared with those from d = ∼70 to 85 Å under negative and no stress, where the DNA started to dissociate from the H3 αN-helix. This indicates that the interaction between the DNA and the H3 αN-helix was stronger under positive stress. A relatively moderate increase in the free energy followed the sharp rise. The gradient of the free energy under positive stress was nearly constant up to d = 170 Å, while those under negative and no stress gradually increased along d. Eventually, free energies became larger than that for positive stress at d = ∼150 Å. When two outer half turns of DNA were fully unwrapped, the free energies were 13.5 kcal/mol at d = 220 Å, 19.5 kcal/mol at d = 214 Å, and 17.0 kcal/mol at d = 214 Å under positive, negative, and no stress, respectively. After those distances, the free energies rapidly increased because the DNA started to be stretched.
Fig. 2.
Free energies for unwrapping the DNA against d (A) and bptotal (B) under positive (blue), negative (red), and no (black) torsional stress. The averages of the SEMs in the free energy along d (within the range of sufficient sampling data) were 0.41, 0.24, and 0.31 kcal/mol under positive, negative, and no stress, respectively. Inset: enlargement of the free energy from d = 60 to 80 Å. The averages of the SEMs in the free energy along bptotal were 0.45, 0.30, and 0.38 under positive, negative, and no stress, respectively. The SEMs are shown in error bars along the free energy.
To interpret the unwrapping process from a structural viewpoint, we changed the perspective of the free energy as a function of the total number of unwrapped base pairs (hereafter referred to as bptotal) using SI Appendix, Eqs. S7 and S8 (Fig. 2B). The nucleosome under positive, negative, and no stress had a global free energy minimum of 0.0 kcal/mol at bptotal = 0. In all cases, the free energies for positive, negative, and no stress reached 12.0, 19.3, and 15.2 kcal/mol at bptotal = ∼52, respectively, when the unwrapping of two outer half turns of DNA was completed. During the unwrapping, distinctive increases in the free energy were observed at bptotal = 0, ∼13, 26, 39, and 52 under all the three cases. A remarkably sharp increase in free energy was observed at bptotal = 26 under negative and no stress but not under positive stress.
Large-Scale Asymmetric Unwrapping of the DNA under Positive Stress.
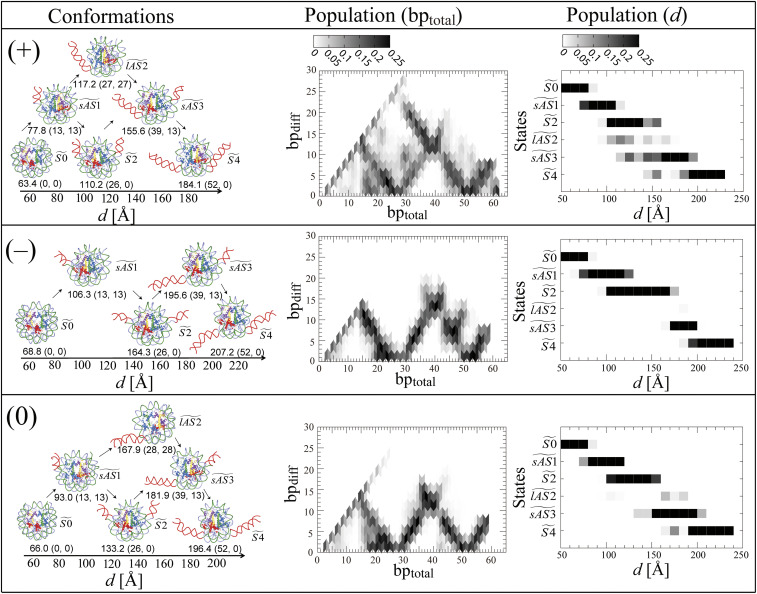
Next, we examined how the DNA unwrapping process advanced. To address it, we counted the difference in the number of unwrapped DNA base pairs, bpdiff, which is defined as |bp1 − bp2|, where bp1 and bp2 denote the number of unwrapped base pairs from DNA1 and DNA2, respectively (hereafter bp is used for the number of unwrapped base pairs from one end if it is not necessary to distinguish between the two ends of the DNA) (Fig. 3).
Fig. 3.
Unwrapping paths under positive (+), negative stress (−), and no stress (0). The left-hand column shows representative conformations observed with d and (bptotal, bpdiff) in parentheses. bptotal is the sum of bp1 and bp2, where bp1 and bp2 are numbers of unwrapped base pairs at the ends of DNA1 and DNA2, respectively. bpdiff is the difference between bp1 and bp2, defined as | bp1 – bp2 |. The unwrapped DNA is shown in red. The orientation of each conformation is different from the others for the sake of visual clarity. The arrows indicate possible small- and large-scale asymmetric unwrapping of the DNA from states to (see also Fig. 4A). The middle column shows the average population of conformations as a function of bptotal and bpdiff. The average was calculated according to SI Appendix, Eq. S8. The population is normalized to the total population of bpdiff with the same bptotal. The values of bpdiff more than 0.25 are set at 0.25. The right-hand column shows the average populations of six conformational states, , , , , and , against d. The average was calculated according to SI Appendix Eq. S8 at the interval of 10 Å with d for each state and the remaining state. The population for each state is normalized to the total population of these states with the same d. The values of any population with more than 0.25 are set at 0.25.
We observed that there are small-scale and large-scale asymmetric paths (where one end is unwrapped a lot but the other end is not so much) for the unwrapping of the DNA in the figure. The branches of the transitional paths were observed at bptotal = ∼13, 26, 39, and 52, corresponding to bptotal where the free energy increases distinctively in Fig. 2B.
To understand the feature of these conformations at the branches of the transitional paths, six conformational states of , , , , , and were defined according to bptotal and bpdiff as shown in Fig. 4A. The representative conformations, corresponding to these states under positive (+), negative (−), and no stress (0), are shown in the left-hand column of Fig. 3, respectively. contains conformations around (bptotal, bpdiff) = (0, 0), the initial state where the DNA is tightly wrapped (symmetric 0); is around (bptotal, bpdiff) = (∼13, ∼13), a state where one end of the DNA dissociates from H3 (small-scale asymmetric 1); is around (bptotal, bpdiff) = (∼26, ∼0), a state where both ends of DNA1 or 2 disconnect from H3 (symmetric 2); is around (bptotal, bpdiff) = (∼26, ∼26), a state where one end of the DNA dissociates from H3 and H2A-H2B, but the other end of the DNA remains unwrapped (large-scale asymmetric 2); is around (bptotal, bpdiff) = (∼39, ∼13), a state where one end of the DNA dissociates from H3 and H2A-H2B, and the other end of the DNA dissociates from H3 (small-scale asymmetric 3); and is around (bptotal, bpdiff) = (∼52, ∼0), the final state of the unwrapping of two outer half turns of DNA where both ends of the DNA dissociate from H3 and H2A-H2B (symmetric 4). can be reached from by unwrapping the other end of the DNA from H3 or from by the continuous unwrapping of one end of the DNA from H2A-H2B.
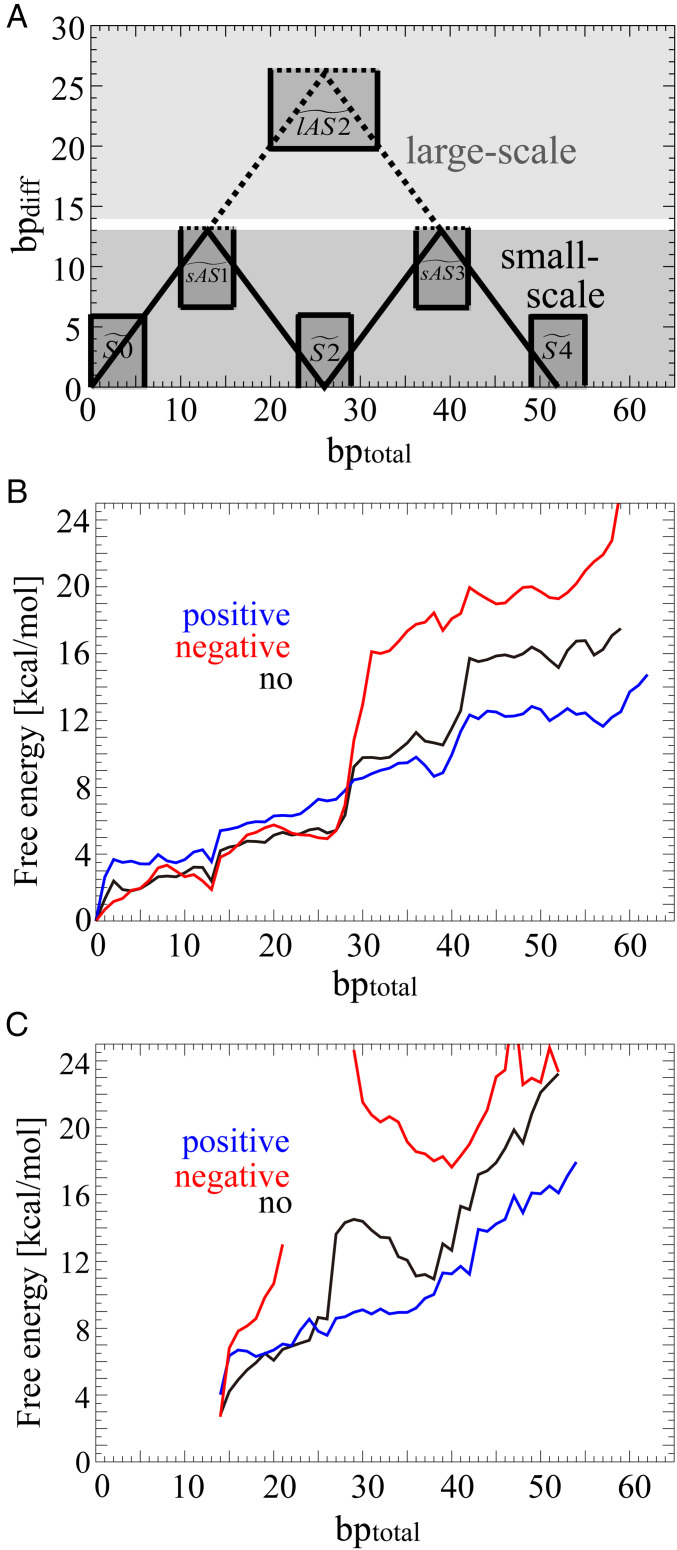
Fig. 4.
Free energies for the small- and large-scale asymmetric unwrapping of the DNA against bptotal. According to the classification of the probability distribution (A), the free energies of the small- (B) and large-scale (C) unwrapping are shown in blue, red, and black under positive, negative, and no stress, respectively. The probability distribution calculated according to SI Appendix, Eq. S2 was classified as that in the small-scale asymmetric unwrapping of the DNA when bpdiff ≤ 13 (dark gray area) and as that in the large-scale asymmetric unwrapping of the DNA when bpdiff ≥ 14 (thin gray area). Definition of the six states of , , , , , and are also shown to be (0 ≤ bptotal ≤ 6, 0 ≤ bpdiff ≤ 6), (0 ≤ bptotal ≤ 16, 7 ≤ bpdiff), (23 ≤ bptotal ≤ 29, 0 ≤ bpdiff ≤ 6), (20 ≤ bptotal ≤ 32, 20 ≤ bpdiff), (36 ≤ bptotal ≤ 42, 7 ≤ bpdiff), and (49 ≤ bptotal ≤ 55, 0 ≤ bpdiff ≤ 6), respectively. The solid lines connecting , , , and and the dotted lines connecting , , and denote representative transitional paths for small- and large-scale asymmetric unwrapping, respectively. Free energies of the small- and large-scale unwrapping were calculated according to SI Appendix, Eq. S7. The ranges of bptotal for the free energies of the large-scale unwrapping were considered to be 14 to 54 under positive, 14 to 21 and 29 to 52 under negative, and 14 to 52 under no stress.
The middle and right-hand columns of Fig. 3 show the populations of these conformational states against bp and d, respectively. The right-hand column of Fig. 3 shows that the populations of , , and were remarkably different in the range of d = 100 to 180 Å among the three stress conditions. Under negative stress, the state from d = 120 to 170 Å is mainly located at , where (bptotal, bpdiff) is (∼26, ∼0). SI Appendix, Fig. S2 shows representative conformations in the range of d from 130 to 170 Å (free energy from 6.9 to 15.0 kcal/mol), where is exclusively dominant. The exclusive existence of in the wide ranges of d and the free energy is the cause of the remarkably sharp rise of the free energy with regard to bptotal under negative stress. In contrast to negative stress, the range of the exclusive existence of was narrower from d = 120 to 130 Å under no stress, and there was no exclusive existence of under positive stress.
Comparing the populations of the states among the three stress conditions, the large-scale asymmetric unwrapping of the DNA clearly appeared only under positive stress. Furthermore, compared with no stress, the populations against d have shifted to the left under positive stress, while they have shifted to the right under negative stress. These results indicate that the process of unwrapping the DNA proceeded faster and slower against d under positive and negative stress, respectively. Under positive stress, it is possible to reach conformation even at d = ∼150 Å. Moreover, the large-scale asymmetric unwrapping at was observed in the wider region, d = ∼100 to 170 Å, under positive stress, while was observed in two places, d = ∼160 to 190 Å, under no stress. Consequently, positive stress facilitated the unwrapping of the DNA, while negative stress suppressed it.
The Free Energies for the Small- and Large-Scale Asymmetric Unwrapping of the DNA.
We calculated free energies for the small- and large-scale asymmetric unwrapping of the DNA using SI Appendix, Eq. S8 (Fig. 4 B and C). The probability distribution for the small- and large-scale asymmetric unwrapping of the DNA were classified with bpdiff ≤ 13 and bpdiff ≥ 14 (Fig. 4A), respectively. ΔGlAS2/S2, the difference between GlAS2 (the free energy in ) and GS2 (the free energy in ), decides the transitional paths for the large- and small-scale asymmetric unwrapping. ΔGlAS2/S2 (= GlAS2 − GS2) were estimated to be about 2.0, no data, and 9.2 kcal/mol under positive, negative, and no stress, respectively (SI Appendix, Fig. S3), indicating that the large-scale asymmetric unwrapping of the DNA is facilitated under positive stress. It should be noted that positive stress makes ΔGlAS2/S2 smaller by increasing GS2 (, a higher value of 7.2 kcal/mol than that of 5.3 kcal/mol for no stress) and decreasing GlAS2 (, a lower value of 9.2 kcal/mol than that of 14.5 kcal/mol for no stress), which both facilitate the large-scale asymmetric unwrapping of the DNA.
Positive Stress Utilizes the Geometry of the DNA (Minor Groove) to Stabilize the DNA–H3 Interaction.
The crystal structure of the nucleosome (1) used as the initial atomic model shows the minor grooves of the DNA at superhelical location (SHL) = ±6.5 partially interact with the H3 αN-helix. Under positive stress, both of the minor grooves of the twisted DNA rotated and further strengthened the interaction with the H3 αN-helix from d = 63 to ∼76 Å (SI Appendix, Fig. S4 A–D) on both sides of the DNA. A sharp increase in the free energy of 3.6 kcal/mol from d = 63 to ∼76 Å (Fig. 2 A, Inset) would be required to break the stable interaction between the minor groove of the DNA and the H3 αN-helix. In contrast to positive stress, the frequency of simultaneous contact on both sides of the DNA was much smaller under negative stress (SI Appendix, Fig. S4 A and B).
Next, we measured how long contact between the DNA and the H3 αN-helix was maintained in the small-scale asymmetric unwrapping of the DNA from to through . From to , contact was retained for the longest under positive stress (SI Appendix, Fig. S4E). This indicates that contact between the twisted minor groove of the DNA and the H3 αN-helix was stronger, and another high energy of 3.6 kcal/mol (Fig. 4B) (see also in SI Appendix, Fig. S3) would be used for breaking the remaining interaction with H3 αN-helix when the small-scale asymmetric unwrapping advances.
Positive Stress Utilizes the Inherent Dynamic Nature of the DNA (Bendability and Flexibility) to Destabilize the DNA–H2A-H2B Interaction.
Not only the DNA–H3 interaction but also the DNA–H2A-H2B interaction should affect the free energies at the states of , , and . Here, we analyzed two physical quantities of the DNA as the enthalpic contribution: 1) the number of contacts between the DNA and the octamer and 2) the local bending angles for base pairs of DNA1 or 2. One may expect that positive and negative stress change the pattern of the contact between the minor groove of the DNA at SHL = ± 5.5 and the H2A-H2B L1-L2 loops as was found in contact between the minor groove of the DNA at SHL ± 6.5 and the H3 αN-helix. However, the number of contacts did not show any clear difference (SI Appendix, Fig. S5). This is thought to be because the minor groove of the DNA at SHL = ± 5.5 has already been facing the H2A-H2B L1-L2 loops under no stress. This implies that the strength of the interfacial interaction between the DNA and the H2A-H2B L1-L2 loops did not change under stress and did not contribute to the difference in their free energies.
SI Appendix, Fig. S6 shows that the local bending angles decreased as the unwrapping of the DNA proceeded under positive stress, while they increased under negative stress. Compared with the bend angles under no stress, the bends of the base pair steps of −71 C:G 71/−70 A:T 70, −68 T:A 68/−67 A:T 67, −60 T:A 60/−59 G:C 59, −58 C:G 58/−57 A:T 57, and −51 T:A 51/−50 A:T 50 increased by ∼2.0° under negative stress and decreased by ∼1.0° under positive stress. The pyrimidine–purine steps such as TA and CA dinucleotide steps have been generally found to be more flexible and more easily bent than other steps (26, 27). Consequently, stress induces conformational changes at these dinucleotide steps; positive stress makes unwrapped DNA straighter, and negative stress makes it more bent.
As for the entropic contribution, we examined the conformational entropies of DNA against bp according to SI Appendix, Eq. S13 as shown in SI Appendix, Fig. S7. The entropy under positive stress was lower than under no stress, probably because the unwrapped DNA under positive stress is straightened, and the DNA became less flexible. In contrast, the entropy of the DNA under negative stress was larger than that under no stress. This indicates that the DNA became more flexible, probably due to the partial deformation of the DNA. These results indicate that the gain in the conformational entropy, which decreased the free energy for unwrapping the DNA, increased in order under positive, no, and negative stress, while the internal energy required to deform the DNA for unwrapping, which increased the free energy, increased in order under positive, no, and negative stress. For example, the DNA of the representative conformation at d = ∼164 Å under negative stress was sharply bent and partially underwound (the left-hand column of Fig. 3 for negative stress or SI Appendix, Fig. S2C). It should be noted that the conformation was located at a steep gradient of the free energy at d = ∼150 to 180 Å (or at the end of the state of ), where the force to unwrap was estimated to be ∼17 pN (Fig. 2A). The maximum force was estimated to be 27 pN between d = 163 and 165 Å. This value corresponds to the force of ∼30 pN when a single DNA starts to unwind (28). The deformation at d = ∼165 Å under negative stress is thought to be because the increased flexibility of the DNA enabled the DNA to deform against the extension of d. In contrast to negative stress, maximum forces of 14 and 24 pN were also observed in the end of at d = 155 and 159 Å under positive and no stress, respectively, where any DNA melting as was observed under negative stress was not observed.
Therefore, the internal energy of the DNA for unwrapping is mainly thought to cause the difference in the free energies of the states of , , and .
Discussion
Comparison with the Experimental and Computational Data.
The free energy had a local minimum at bptotal = 13, which can explain why the partially unwrapped nucleosome structures have been observed by cryogenic-electron microscope (cryo-EM) (29). The free energy for the entire unwrapping of two outer half turns of DNA was estimated to be ∼15 kcal/mol without torsional stress. This value is higher than the experimental values which range ∼5 to 12 kcal/mol; however, these values are likely to depend on how the experimental data were handled (4, 5, 30, 31).
The free energy in total indicates that positive torsion facilitated unwrapping the DNA, while negative torsion suppressed unwrapping (or facilitated rewrapping) the DNA. This is consistent with the experimental data which showed strong destabilization of the nucleosome under a positive torque (12, 13). This also supports the “twin-helix” domain model (11) that overtwisted DNA ahead of the RNAP disassembles the nucleosome, and undertwisted DNA behind the RNAP reassembles the nucleosome (12, 13).
Large-scale asymmetric unwrapping of nucleosomal DNA has been observed by experiments of time-resolved small-angle X-ray scattering (32, 33), FRET (34), high-speed atomic force microscopy (35), and cryo-EM (29). Large-scale asymmetric unwrapping was also observed in transcription (36) and remodeling (10) processes. Our previous all-atom MD simulation of the nucleosome without torsional stress (20) has observed small-scale asymmetric unwrapping of the DNA in and . In addition to and , MD simulations of the nucleosome using coarse-grained models have observed large-scale unwrapping in (37, 38).
The increased bendability of the DNA under negative stress is likely to be a cause for the stabilization of the nucleosome. In general, the high-affinity sequences such as AT-rich sequences bend toward the minor groove and have higher bendability (34, 39, 40). Decrease and increase in the bending angle of a single DNA under positive and negative torsional stress, respectively, have been observed by all-atom MD simulations (21–23). These previous reports are consistent with our present results.
Interpretation of Experimental Data by Single-Molecule Experiments.
The stabilization of the nucleosome under positive and negative stress was experimentally studied (14, 41, 42). Sheinin et al. used an angular optical trapping method to measure the extension of the DNA with the mononucleosome under extension force and torque (14). The occurrence of the release of the outer turn of DNA increased under positive and negative stress and was interpreted as the interaction between the outer turn of DNA and H2A-H2B being stabilized under positive and negative stress (14). Bancaud et al. (41) applied the same technique to single nucleosome arrays to measure the extension of the arrays under extension force and rotation. The rotation-extension curve under positive and negative stress was interpreted as being caused by the positively and negatively crossed linker DNAs at the entry/exit of the nucleosomes in a tightly wrapped state (42). The studies by Sheinin et al. and Bancaud et al. are consistent with each other in terms of the stability of the nucleosome under stress. However, the cause of the stability was considered differently. Sheinin et al. (14) considered the stabilization of the DNA–H2A-H2B interaction along the reaction coordinate of the unwrapped base pairs of the DNA while Bancaud et al. (41) considered the stabilization of the crossed linker DNA along the reaction coordinate of the supercoil density.
Sheinin et al. (14) analyzed the number of released base pairs of the DNA (including a high-affinity 601-positioning element) on both ends from the force-extension curve by implicitly assuming that the nucleosome unwraps symmetrically. In a FRET experiment, however, Ngo et al. (34) showed asymmetric unwrapping of the nucleosome with a 601-positioning sequence (under no stress) and suggested that it was due to the difference in rigidity between the first and second halves of the DNA sequence. Asymmetric unwrapping of the nucleosome with the sequence of Windom 601L has also been observed (under no stress) at a high ionic concentration of more than 1 M MgCl2 by all-atom MD simulations (43). Asymmetric unwrapping of DNA with an asymmetric sequence would be more likely to occur than for DNA with a symmetric sequence as was used in our study.
Consequently, the force-extension curves might need to be reinterpreted by taking into consideration other factors such as linker DNA crossing, the DNA–H3 αN-helix interaction, and asymmetric unwrapping of the nucleosome.
Proposed Model for Asymmetric Unwrapping of Nucleosomal DNA.
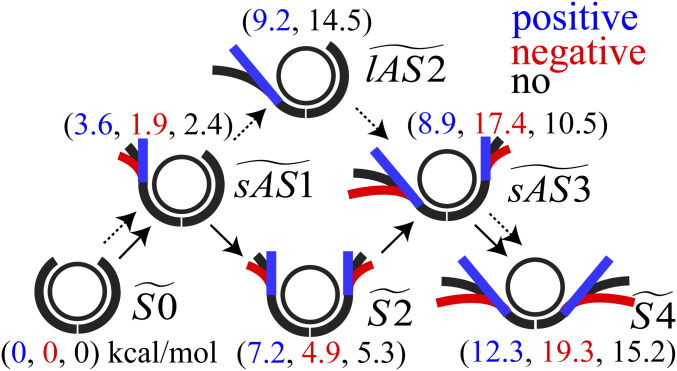
We propose a model for the small- and large-scale asymmetric unwrapping of nucleosomal DNA as follows (Fig. 5).
Fig. 5.
Schematic representation of the model of the transitional paths for the small- and large-scale asymmetric unwrapping of two outer half turns of DNA under torsional stress. The unwrapping of two outer half turns of DNA under positive, negative, and no stress is schematically shown in blue, red, and black, respectively. The transitional states are labeled by , , , , , and . The likelihood of the transition from wrapped to unwrapped DNA from to is schematically shown by the solid and dotted arrows for the small- and large-scale asymmetric unwrapping of the DNA, respectively. The values in parentheses are values of the free energies in each state.
First, the positive torsional stress overtwists the DNA, while the negative torsional stress undertwists the DNA. Second, the minor groove of the DNA at SHL = ± 6.5 rotates and further strengthens the interaction with the H3 αN-helix under positive stress, which stabilizes the nucleosome and suppresses the transition from to . Third, the change in the bendability and flexibility of the DNA influences the free energy for the unwrapping of the DNA. Overtwisted DNA under positive stress is straightened. The straightened DNA would increase the bending energy if the DNA retained the bent form to maintain its contact with H2A-H2B. This high bending energy in the DNA would facilitate the unwrapping of the DNA to relax the bent form of the DNA from to . In contrast, the DNA under negative stress is more bendable and would not require as much internal bending energy to wrap the DNA as under positive stress. It is notable that our simulation indicates that the change in the affinity of the DNA and H2A-H2B under positive and negative stress is not because of the change in the interfacial direct interaction between the DNA and H2A-H2B but because of the change in the inherent dynamic nature of the DNA with regard to its bendability and flexibility.
Finally, ΔGlAS2/S2 decides the transitional path for the large- and small-scale asymmetric unwrapping. In terms of DNA–histone interactions, ΔGlAS2/S2 can be regarded as ΔG(H2A-H2B)/H3, the difference between GH2A-H2B (the free energy for unwrapping one end of the DNA from H2A-H2B) and GH3 (the free energy for unwrapping the other end of the DNA from H3). Under positive stress, ΔG(H2A-H2B)/H3 is relatively small, 2.0 kcal/mol. Thus, after one end of the DNA unwraps from H3, further unwrapping of the same end of the DNA from the H2A-H2B is likely to occur within the thermal fluctuation of kBT = ∼0.6 kcal/mol. In contrast to positive stress, ΔG(H2A-H2B)/H3 for negative and no stress are so large that the large-scale asymmetric unwrapping of the DNA from H2A-H2B would be strongly suppressed. In fact, when one end of the DNA unwraps from H3, the other end of the DNA is more likely to start unwrapping. Consequently, the large- and small-scale asymmetric unwrapping of the DNA is facilitated under positive and negative stress, respectively.
The idea that the magnitude of the difference between GH2A-H2B and GH3, ΔG(H2A-H2B)/H3, plays a role in the small- and large-scale asymmetric unwrapping would be applicable not only to the torsional stress but also other factors, such as the histone tails (19, 37, 44) (which we did not consider in this study), the DNA sequence (34), and histone modifications.
The Efficient Unwrapping of Nucleosomal DNA in Chromatin under Positive Stress.
The free-energy landscapes against d were similar under the three different stress conditions in the sense that the free energies gradually increased (Fig. 2). The populations of the transitional states from to , however, were clearly different among the conditions as shown in Fig. 3. Under positive stress, the nucleosome can reach even at d less than ∼100 Å. This indicates that the unwrapping of the DNA from H2A-H2B can occur without a large extension of the DNA under positive stress. This efficient unwrapping of the DNA would allow proteins to access the exposed DNA even when nucleosomes are in condensed chromatin.
Computational Limitation.
To check the reproducibility of our results, we compared the free-energy curve with a previously reported free energy (20). In the previous study, the MD simulation was carried out at a concentration of 120 mM NaCl in a simulation box of 150 Å × 150 Å × 150 Å to obtain the unwrapping free energy under no stress. The SD between these independent studies was calculated to be 0.40 kcal/mol (SI Appendix, Fig. S8). We also examined the deviations of the free energy in this study. SI Appendix, Fig. S9 shows the free-energy curves at intervals of 10 ns for the last 50 ns in the umbrella sampling simulation. The SDs for the five free-energy curves were calculated to be 0.91, 0.54, and 0.70 kcal/mol under positive, negative, and no stress, respectively. Consequently, the difference in the free energies in these studies under no stress, 0.40 kcal/mol, was within the SD of the sampled free-energy curves, 0.70 kcal/mol. These results suggest that the free energy in this study (at least under no stress) is reproducible within these error ranges.
As the DNA sequence used in our study was symmetric, the distribution of the unwrapped DNA should also be symmetric, which has been shown in the Monte Carlo simulation of the asymmetric unwrapping at the outer and inner turns of DNAs with a coarse-grained model (45). As shown in SI Appendix, Fig. S10, we measured (bp1 − bp2)/(bp1 + bp2) as an indicator of the extent of the asymmetry (0.0: completely symmetric and ±1.0: completely asymmetric). The figure shows that once the nucleosome unwrapping became asymmetric in one direction (e.g., bp1 >> bp2), it did not become asymmetric in the other direction (bp1 << bp2) within 100-ns simulations. This indicates that the 100-ns simulation per window may not be enough to sample asymmetric distribution in both directions. However, the overall distribution of conformations from all the replicas was fairly even on both ends, indicating that the conformation sampling along the reaction coordinate is sufficient at least to show the facilitating of the asymmetric unwrapping of the DNA under positive stress.
It should be noted that there is a difference between the systems studied in single-molecule experiments and our study. In our study, the linker DNA was not included; however, a long linker DNA was in principle included in the experiments. Simulation of the nucleosome with long linker DNA under torsional stress would enable direct comparison and give more detailed information about the unwrapping of the nucleosome.
Despite the limitation of not including linker DNA in the model to reduce the computational time, we believe the free-energy landscape of the unwrapping of two outer half turns of DNA in our study gives insight into the symmetric and asymmetric unwrapping of two outer half turns of DNA under torsional stress.
Conclusion
Our simulation showed that torsional stress can regulate the transitional paths of the small- and large-scale asymmetric unwrapping of two outer half turns of DNA through the geometry of the DNA (minor groove) and the inherent dynamic nature of the DNA (bendability and flexibility). We demonstrated that under positive stress, the strengthened interaction between the twisted minor groove of the DNA and the H3 αN-helix, and the weakened interaction between the straightened DNA and the H2A-H2B L1-L2 loops, both facilitated the large-scale asymmetric unwrapping of two outer half turns of DNA. In contrast, the undertwisted DNA generated by negative stress made the DNA bent and flexible, which suppressed unwrapping. Our data suggests that DNA accessibility in the chromatin can be regulated by enhancing or suppressing asymmetric unwrapping without a large extension of the DNA through torsional stress generated by proteins such as RNAP and chromatin remodeling complexes, which may explain why genes in condensed chromatin states can be expressed.
Materials and Methods
Atomic Model.
We used a crystal structure of a nucleosome with α-satellite palindromic 147 bp DNA (Protein Data Bank code: 1kx5) (46). The histone tails were truncated to facilitate the unwrapping of the DNA. Consequently, residues 46 to 132 in H3, 25 to the C terminal in H4, 16 to 114 in H2A, and 32 to 121 in H2B were used. As the nucleosome core particle contains two copies of each octamer and the DNA has two ends, we distinguish them with suffixes “a” and “b” for the octamer and “1” and “2” for the ends of the DNA. In this study, nucleotides from −73 A to −39 A of chain I and nucleotides from 73 T to 39 T of chain J were assigned to DNA1, and nucleotides from 73 T to 39 T of chain I and nucleotides from −73 A to −39 A of chain J were assigned to DNA2 according to the nucleotide sequence in 1kx5 (Fig. 1B).
MD Simulations of the Systems in Water.
Simulations were carried out using an MD simulation program called SCUBA (20, 47–52) with the AMBER ff14SB (53), bsc1 (54), and ff99ions08 (55) force-fields for the octamer, DNAs, and ions, respectively. The structure of nucleosome was placed in a rectangular box of ∼125 Å × 245 Å × 155 Å (SI Appendix, Fig. S1A). ∼148,000 TIP3P water molecules (56) were added to surround the system. To neutralize the charges of the system, sodium ions were placed in the box, and then additional sodium and chloride ions were added at a concentration of 150 mM NaCl. In total, the system comprised ∼465,000 atoms. The system was equilibrated at a constant pressure of one bar and a temperature of 300 K for 10 ns. Details are given in SI Appendix, Supplementary Methods.
Adaptively Biased MD Simulation of the System.
To generate a series of configurations of the unwrapping of the nucleosome from tightly wrapped to unwrapped, the adaptively biased MD (ABMD) method (57) combined with the multiple-walker method (58) was carried out for ∼30 ns at a constant volume and a temperature of 300 K with six walkers (replicas) of the nucleosome, which were selected from the equilibration simulation. The reaction coordinate d was defined as the DNA near end-to-end distance, or the distance between the two centers of mass of the third base pair from each end of the DNA, −71 C:G 71 in DNA1 and 71 G:C −71 in DNA2. The value of the reaction coordinate in the initial structure was 70.7 Å. After the ABMD simulation was carried out, a series of 90 conformations at intervals of about 2.0 Å with d between 64 and 242 Å were selected from the conformational ensembles of the replicas. Details are given in SI Appendix, Supplementary Methods.
Umbrella Sampling Simulations under Torsional Stress and No Stress.
Using a series of 90 conformations in the ABMD simulation, umbrella sampling simulations were carried out for 100 ns to obtain the free energy for the unwrapping of nucleosomal DNA with torsional stress and no stress. The umbrella potential for the i-th window (i = 1, ..., Nwin = 90) used is
| [1] |
where is a fixed distance to maintain d within the range of 64 Å to 242 Å with intervals of 2 Å. is an arbitrary harmonic force constant, which was set at 0.2 kcal/mol/Å2. The trajectory for the last 50 ns was used for the analysis. The WHAM (24, 25) was used to refine the free-energy landscape. Details are given in SI Appendix, Supplementary Methods.
Torsional Stress in the Umbrella Sampling Simulations.
The effect of the torsional stress on the DNA for the i-th window was incorporated as follows in a similar way to the procedure used by refs. 21, 22:
| [2] |
where the torsional angles of ϕ1 and ϕ2 are defined using the C1’ atoms of the third base pairs from the ends of the DNA and the phosphorus atoms located opposite the dyad (Fig. 1 B and C). Both and were set at 0° and 180° for the generation of positive and negative stress, respectively. As appropriate values of the force constants k1i and k2i to produce torque within 10 to 20 pN nm were not known in advance, the values of k1i and k2i were adjusted for the first ∼25 ns. Since then, both of the force constants k1i and k2i were set at 2.0 × 10−2 × (i – 1) + 1.5 kcal/(mol rad2) for the positive stress and at 1.5 × 10−2 × (i – 1) + 0.50 kcal/(mol rad2) for the negative stress. Details are given in SI Appendix, Supplementary Methods.
The Number of Unwrapped Base Pairs.
Base pairs were defined as being unwrapped when the center of each base pair at the end of the DNA continuously deviated more than 4 Å from the center of the corresponding base pair in the reference structure, where DNA was fully wrapped. The energy-minimized structure of the nucleosome in water medium was used as the reference structure.
The Number of Contacts between the DNA and the Octamer.
Contacts were counted when any pair of atoms of the DNA and the octamer was located within a distance of 4 Å of each other.
Conformational Analysis of the DNA.
The conformational change of the DNA was measured using the base pair step parameters using program called 3DNA (59). Twist, Tilt, and Roll of the base pair step parameters were analyzed for the third to 35th base pairs from the end of the DNA1 or 2. The first and second base pairs at the end of DNA1 or 2 were not included in the analysis. The torsional stress would not influence these base pair steps because they behaved like a free end. The local bending angle was calculated as .
Conformational Entropy of the DNA.
The conformational entropies of the two ends of DNA1 or 2 were calculated using the quasiharmonic approximation (60). Details are given in SI Appendix, Supplementary Methods.
Computational Time.
All the simulations, requiring a total time of 27 μs, were mainly performed on the Cray XC40 supercomputer at Kyoto University and the SGI ICE X supercomputer at the Japan Atomic Energy Agency. The total computational time was more than 20 million central processing unit core hours.
Supplementary Material
Acknowledgments
We thank Yvonne Ishida for reading this manuscript carefully. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan as “Priority Issue on Post-K Computer” (Building Innovative Drug Discovery Infrastructure Through Functional Control of Biomolecular Systems) (Project ID: hp180191 and hp190171), “Program for Promoting Researches on the Supercomputer Fugaku” (Biomolecular dynamics in a living cell) (hp200135), the High Performance Computing Infrastructure system provided by Kyoto University (Project ID: hp180027, hp190007, and hp200029), and Japan Society for the Promotion of Science KAKENHI (18K06173 to H.I. and JP18H05534 to H.K.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020452118/-/DCSupplemental.
Data Availability.
Relevant conformations and trajectory files have been deposited in the Materials Cloud Archive 2020 (DOI:10.244335/materialscloud:r9-xt).
References
- 1.Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J., Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Gansen A., et al. , Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl. Acad. Sci. U.S.A. 106, 15308–15313 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G., Levitus M., Bustamante C., Widom J., Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Mihardja S., Spakowitz A. J., Zhang Y., Bustamante C., Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. U.S.A. 103, 15871–15876 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brower-Toland B. D., et al. , Mechanical disruption of individual nucleosomes reveals a reversible multistage release of DNA. Proc. Natl. Acad. Sci. U.S.A. 99, 1960–1965 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien F. T., van der Heijden T., Characterization of nucleosome unwrapping within chromatin fibers using magnetic tweezers. Biophys. J. 107, 373–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hodges C., Bintu L., Lubkowska L., Kashlev M., Bustamante C., Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science 325, 626–628 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J., Bai L., Wang M. D., Transcription under torsion. Science 340, 1580–1583 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Vugt J. J., et al. , Multiple aspects of ATP-dependent nucleosome translocation by RSC and Mi-2 are directed by the underlying DNA sequence. PLoS One 4, e6345 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tokuda J. M., et al. , The ATPase motor of the Chd1 chromatin remodeler stimulates DNA unwrapping from the nucleosome. Nucleic Acids Res. 46, 4978–4990 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L. F., Wang J. C., Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 84, 7024–7027 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teves S. S., Henikoff S., Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 21, 88–94 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark D. J., Felsenfeld G., A nucleosome core is transferred out of the path of a transcribing polymerase. Cell 71, 11–22 (1992). [DOI] [PubMed] [Google Scholar]
- 14.Sheinin M. Y., Li M., Soltani M., Luger K., Wang M. D., Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat. Commun. 4, 2579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaczmarczyk A., Meng H., Ordu O., Noort J. V., Dekker N. H., Chromatin fibers stabilize nucleosomes under torsional stress. Nat. Commun. 11, 126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elbel T., Langowski J., The effect of DNA supercoiling on nucleosome structure and stability. J. Phys. Condens. Matter 27, 064105 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Kono H., Shirayama K., Arimura Y., Tachiwana H., Kurumizaka H., Two arginine residues suppress the flexibility of nucleosomal DNA in the canonical nucleosome core. PLoS One 10, e0120635 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikebe J., Sakuraba S., Kono H., H3 histone tail conformation within the nucleosome and the impact of K14 acetylation studied using enhanced sampling simulation. PLoS Comput. Biol. 12, e1004788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Kono H., Distinct roles of histone H3 and H2A tails in nucleosome stability. Sci. Rep. 6, 31437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kono H., Sakuraba S., Ishida H., Free energy profiles for unwrapping the outer superhelical turn of nucleosomal DNA. PLoS Comput. Biol. 14, e1006024 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannan S., Kohlhoff K., Zacharias M., B-DNA under stress: Over- and untwisting of DNA during molecular dynamics simulations. Biophys. J. 91, 2956–2965 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liebl K., Zacharias M., Unwinding induced melting of double-stranded DNA studied by free energy simulations. J. Phys. Chem. B 121, 11019–11030 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Reymer A., Zakrzewska K., Lavery R., Sequence-dependent response of DNA to torsional stress: A potential biological regulation mechanism. Nucleic Acids Res. 46, 1684–1694 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar S., Bouzida D., Swendsen R. H., Kollman P. A., Rosenberg J. M., The weighted histogram analysis method for free energy calculations on biomolecules. I. The method. J. Comput. Chem. 13, 1011–1021 (1992). [Google Scholar]
- 25.Souaille M., Roux B., Extension to the weighted histogram analysis method: Combining umbrella sampling with free energy calculations. Comput. Phys. Commun. 135, 40–57 (2001). [Google Scholar]
- 26.Olson W. K., Gorin A. A., Lu X. J., Hock L. M., Zhurkin V. B., DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 95, 11163–11168 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujii S., Kono H., Takenaka S., Go N., Sarai A., Sequence-dependent DNA deformability studied using molecular dynamics simulations. Nucleic Acids Res. 35, 6063–6074 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gore J., et al. , DNA overwinds when stretched. Nature 442, 836–839 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Bilokapic S., Strauss M., Halic M., Histone octamer rearranges to adapt to DNA unwrapping. Nat. Struct. Mol. Biol. 25, 101–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson J. D., Widom J., Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 296, 979–987 (2000). [DOI] [PubMed] [Google Scholar]
- 31.Meng H., Andresen K., van Noort J., Quantitative analysis of single-molecule force spectroscopy on folded chromatin fibers. Nucleic Acids Res. 43, 3578–3590 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y., et al. , Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. Proc. Natl. Acad. Sci. U.S.A. 114, 334–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y., et al. , Revealing transient structures of nucleosomes as DNA unwinds. Nucleic Acids Res. 42, 8767–8776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngo T. T., Zhang Q., Zhou R., Yodh J. G., Ha T., Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell 160, 1135–1144 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyagi A., Ando T., Lyubchenko Y. L., Dynamics of nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry 50, 7901–7908 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Ramachandran S., Ahmad K., Henikoff S., Transcription and remodeling produce asymmetrically unwrapped nucleosomal intermediates. Mol. Cell 68, 1038–1053.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kenzaki H., Takada S., Partial unwrapping and histone tail dynamics in nucleosome revealed by coarse-grained molecular simulations. PLoS Comput. Biol. 11, e1004443 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., Zheng W., Papoian G. A., Wolynes P. G., Exploring the free energy landscape of nucleosomes. J. Am. Chem. Soc. 138, 8126–8133 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Ngo T. T., et al. , Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat. Commun. 7, 10813 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo D., et al. , MNase, as a probe to study the sequence-dependent site exposures in the +1 nucleosomes of yeast. Nucleic Acids Res. 46, 7124–7137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bancaud A., et al. , Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat. Struct. Mol. Biol. 13, 444–450 (2006). [DOI] [PubMed] [Google Scholar]
- 42.De Lucia F., Alilat M., Sivolob A., Prunell A., Nucleosome dynamics. III. Histone tail-dependent fluctuation of nucleosomes between open and closed DNA conformations. Implications for chromatin dynamics and the linking number paradox. A relaxation study of mononucleosomes on DNA minicircles. J. Mol. Biol. 285, 1101–1119 (1999). [DOI] [PubMed] [Google Scholar]
- 43.Winogradoff D., Aksimentiev A., Molecular mechanism of spontaneous nucleosome unraveling. J. Mol. Biol. 431, 323–335 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chakraborty K., Loverde S. M., Asymmetric breathing motions of nucleosomal DNA and the role of histone tails. J. Chem. Phys. 147, 065101 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Norouzi D., Zhurkin V. B., Dynamics of chromatin fibers: Comparison of Monte Carlo simulations with force spectroscopy. Biophys. J. 115, 1644–1655 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J., Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J. Mol. Biol. 319, 1097–1113 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Ishida H., Hayward S., Path of nascent polypeptide in exit tunnel revealed by molecular dynamics simulation of ribosome. Biophys. J. 95, 5962–5973 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishida H., Branch migration of Holliday junction in RuvA tetramer complex studied by umbrella sampling simulation using a path-search algorithm. J. Comput. Chem. 31, 2317–2329 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Ishida H., Matsumoto A., Free-energy landscape of reverse tRNA translocation through the ribosome analyzed by electron microscopy density maps and molecular dynamics simulations. PLoS One 9, e101951 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishida H., Essential function of the N-termini tails of the proteasome for the gating mechanism revealed by molecular dynamics simulations. Proteins 82, 1985–1999 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Ishida H., Matsumoto A., Mechanism for verification of mismatched and homoduplex DNAs by nucleotides-bound MutS analyzed by molecular dynamics simulations. Proteins 84, 1287–1303 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Ishida H., Kono H., H4 tails potentially produce the diversity in the orientation of two nucleosomes. Biophys. J. 113, 978–990 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maier J. A., et al. , ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivani I., et al. , Parmbsc1: A refined force field for DNA simulations. Nat. Methods 13, 55–58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joung I. S., Cheatham T. E. 3rd, Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B 112, 9020–9041 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 57.Babin V., Roland C., Sagui C., Adaptively biased molecular dynamics for free energy calculations. J. Chem. Phys. 128, 134101 (2008). [DOI] [PubMed] [Google Scholar]
- 58.Raiteri P., Laio A., Gervasio F. L., Micheletti C., Parrinello M., Efficient reconstruction of complex free energy landscapes by multiple walkers metadynamics. J. Phys. Chem. B 110, 3533–3539 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Lu X.-J., Olson W. K., 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 31, 5108–5121 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schlitter J., Estimation of absolute and relative entropies of macromolecules using the covariance matrix. Chem. Phys. Lett. 215, 617–621 (1993). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant conformations and trajectory files have been deposited in the Materials Cloud Archive 2020 (DOI:10.244335/materialscloud:r9-xt).