Significance
We discovered that the Drosophila transcription factor, Cabut, an ortholog of human KLF-10 and KLF-11, is capable of trans-activating a large set of cell cycle genes that are also E2F1 targets. Thereby, Cabut can drive ectopic cell cycles in many Drosophila cell types. That Cabut has E2F-like activity is especially interesting when one considers that the E2Fs, which are the best characterized transcriptional regulators of the eukaryotic cell cycle, are not actually essential for cell cycle gene transcription or cell cycle progression. Hence, it is clear that other transcription factors must also regulate the transcription of the 300 to 400 genes necessary for DNA replication and mitosis. Cabut appears to be one these missing transcription factors that can complement E2F.
Keywords: Cabut (Cbt), cell cycle exit, E2F1, intestinal stem cell (ISC), proliferation
Abstract
Using a gain-of-function screen in Drosophila, we identified the Krüppel-like factor Cabut (Cbt) as a positive regulator of cell cycle gene expression and cell proliferation. Enforced cbt expression is sufficient to induce an extra cell division in the differentiating fly wing or eye, and also promotes intestinal stem cell divisions in the adult gut. Although inappropriate cell proliferation also results from forced expression of the E2f1 transcription factor or its target, Cyclin E, Cbt does not increase E2F1 or Cyclin E activity. Instead, Cbt regulates a large set of E2F1 target genes independently of E2F1, and our data suggest that Cbt acts via distinct binding sites in target gene promoters. Although Cbt was not required for cell proliferation during wing or eye development, Cbt is required for normal intestinal stem cell divisions in the midgut, which expresses E2F1 at relatively low levels. The E2F1-like functions of Cbt identify a distinct mechanism for cell cycle regulation that may be important in certain normal cell cycles, or in cells that cycle inappropriately, such as cancer cells.
Accurate control of cell proliferation is important for the generation of properly sized and organized tissues and organs during development, and to maintain homeostasis during tissue maintenance in adults. Exit from the cell cycle is regulated such that cells typically exit into G1 upon terminal differentiation. Loss of this coordination or reversal of cell cycle exit can result in tissue dysplasia, developmental defects, and diseases such as cancer. Work in many systems investigating the role of negative regulators of G1 during cell cycle exit has demonstrated that members of the retinoblastoma (Rb) family of tumor suppressors and cyclin-dependent kinase inhibitors (CKIs) are required for cell cycle exit in some cell types, but are not required in others (1–4). Research focused on positive regulators of the cell cycle and has shown that the inhibition of the E2F1/dimerization partners (DP) transcription factors and G1 cyclin/cyclin-dependent kinase (Cdk) activities are key events in proper cell cycle exit (5–12). Recent studies also highlight the role of chromatin modification in establishing the postmitotic state during cell differentiation (13–15). Given that Rb and CKI family genes are not uniformly required for cell cycle exit, some of the mechanisms by which E2F1/DP and cyclin/Cdk activities are controlled upon exit remain unknown.
Most cell types in higher animals normally exit the cell cycle in G1, implicating the inhibition of S phase entry as the key regulated step. In eukaryotic cells, G1 to S phase progression is promoted by activating E2F transcription factors and their DPs. E2F/DP complexes control the expression of genes required for cell cycle progression, such as those involved in DNA replication and mitosis (16–20). The specific E2F subunit incorporated into a complex determines its function as either a transcriptional activator or constitutive repressor. Activating E2F complexes can also be converted to repressors when bound by Rb proteins (21, 22), and this association is lost upon the phosphorylation of the Rb proteins by either Cyclin D (CycD)/Cdk4 or Cyclin E (CycE)/Cdk2 (23, 24). CKIs also participate in this regulation. CycE activity is restrained by members the Cip/Kip family of CKIs (p21, p27, and p57), while CycD activity is restrained by Ink-type CKIs (p15, p16, p18, and p19) (4, 12, 25, 26). Nevertheless, as CycE is a canonical transcriptional target of the activating E2Fs (E2F1, -2, -3), the E2F–CycE–RB interaction can generate a positive feedback loop for G1/S progression (27, 28), and this loop must be broken to allow exit into G1 phase.
Drosophila has been a key model organism for studying the cell cycle, in part because the fly genome contains fewer copies of cell cycle regulatory genes than mammalian genomes. Drosophila have one activating E2F (E2f1), one repressive E2F (E2f2), and a single DP gene (Dp), while humans have eight known E2Fs and three DP genes (8, 29–32). Furthermore, Drosophila have only two Rb genes (Rbf, Rbf2) (21), a single Cyclin D gene (CycD) (33), a single Cyclin E gene (CycE) (34), a single Cip/Kip-type CKI, Dacapo (dap) (30, 35, 36), and no Ink-type CKI. Investigations of cell cycle exit in Drosophila have been facilitated by the remarkably synchronous timing of cell cycle exit in both the pupal eye and wing blade. In both organs, cells complete all divisions by 24 h after puparium formation (APF, at 25 °C) and nearly 100% of the cells exit the cell cycle with a G1 DNA content (6, 37–39).
Genetic studies in Drosophila demonstrate that the negative regulators Rbf, Rbf2, E2F2, and Dap are only partially responsible for promoting cell cycle exit, and play a more significant role in cell cycle exit in the developing eye than in the developing wing (5, 10, 20, 40, 41). We previously investigated the ability of positive cell cycle regulators to bypass cell cycle exit and identified a double assurance mechanism that limits continued cycling in the fly eye and wing. This work demonstrated that overexpression of either E2F1 or CycE caused a single extra cell cycle, and that their activities are independently constrained for proper exit, in part by Rbf- and Dap-independent mechanisms (5). Follow-up studies described the relationship between E2F1 and CycE after cell cycle exit (20), and the dynamic changes in chromatin structure that occur during differentiation (13), but precisely how the cell cycle control apparatus is constrained to ensure exit into G1 is still poorly understood in this and other developmental contexts.
To further understand the events regulating cell cycle exit, we took a gain-of-function approach to identify genes that were capable of delaying cell cycle exit when overexpressed, potentially by increasing the activity of E2F1 and CycE. Using this approach, we identified the Sp/Krüppel-like factor (KLF) family transcription factor Cabut (cbt), also known as dTIEG (42), as a potent cell cycle activator capable of delaying cell cycle exit when overexpressed. As detailed below, Cbt causes ectopic proliferation not by activating E2F1 or CycE, but instead by virtue of having its own E2F1-like transcriptional activity.
Results
PCNA-miniwhite+: A Reporter to Detect Ectopic E2F1 Activity as Eye Color.
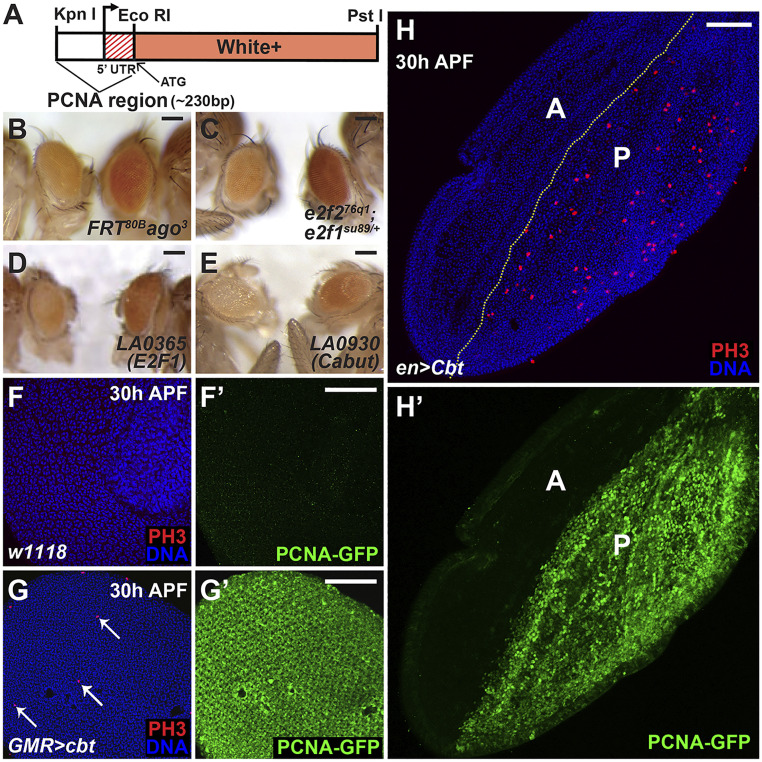
To identify new regulators of cell cycle exit, we designed a screen that detects ectopic E2F1 activity and cellular proliferation in the eye after normal cell cycle exit. To do this we generated the PCNA-miniwhite+ reporter gene, which fuses an E2F1-responsive fragment from the proliferating cell nuclear antigen (PCNA) promoter (43) to the miniwhite+ protein coding sequence (Fig. 1A). The pheno-critical period for white gene activity is 24 to 48 h APF (44), a time when all cells in the pupal eye have normally exited the cell cycle and transcription from the PCNA promoter and most other E2F1 target genes decreases (5). w− flies carrying the pBac[eCFP PCNA-miniwhite+] reporter gene normally had pale yellow eye color, likely due to the perdurance of White protein produced prior to cell cycle exit. We expected that when the silencing of E2F1 activity was abrogated or delayed, the eye would maintain PCNA promoter activity after 24 h APF, resulting in sufficient expression of white during the pheno-critical time to generate orange or red eye color. We tested this by crossing PCNA-miniwhite+ into several mutant backgrounds. Cells mutant for the ubiquitin-ligase archipelago (ago) maintain CycE activity, and consequently delay cell cycle exit (35) and the silencing of E2F1 transcriptional activity in the eye. Accordingly, ago mutant cells generated clonally in w− PCNA-miniwhite+ eyes displayed red eye color (Fig. 1B). We also tested whether maintaining E2F1 activity by decreasing repressive E2F function could generate red eyes in PCNA-miniwhite+ flies. Thus, a single copy of the e2f1su89 allele, which is unable to bind Rbf (40), in an E2f2−/− background also resulted in red eye color due to increased PCNA-miniwhite+ expression (Fig. 1C). We surmise that the PCNA-miniwhite+ gene can function as a reporter of ectopic E2F1 or CycE activity and cell proliferation.
Fig. 1.
A screen to identify regulators of the cell cycle identifies Cabut. (A) Schematic of the PCNA-miniwhite+ reporter gene. (B–E) Drosophila carrying the PCNA-miniwhite+ reporter and other transgenes or mutations, with age- and sex-matched controls on the left. (B) FRT82B ago3 mutant clones generated by ey-FLP have red eye pigmentation with PCNA-miniwhite+. (C) e2f276q1; e2f1su89/+ mutant eyes are red with PCNA-miniwhite+. (D and E) LA enhancer-promoter lines predicted to express either E2f1 (LA0365) (D) or cabut (LA0930) (E) crossed to GMR-Gal4(ey-CFP); PCNA-miniwhite+ (Right) have increased red eye pigmentation. (F) Thirty hours APF, control eyes did not undergo ectopic mitoses (anti-PH3 immunostaining, red) and showed no PCNA-GFP expression (F′). (G and H) Thirty hours APF, eyes and wings with cbt expression by GMR-Gal4 and en-Gal4, respectively, demonstrated ectopic mitoses (red, G and H) and PCNA-GFP expression (green, G′ and H′). Arrows in G indicated the mitotic (PH3+) cells. “A” and “P” shown in (H and H′) marked the anterior and posterior compartments of the pupal wing. Nuclei were stained with Hoechst (blue) in F, G, and H. (Scale bars, 50 μm.)
Identification of Genes Causing Ectopic Proliferation.
To identify genes involved in regulating cell cycle exit, we combined PCNA-miniwhite+ with GMR-Gal4 marked by ey-CFP and screened the University of California, Los Angeles collection of unidirectional UAS element insertion lines (LA lines, from John Merriam, University of California, Los Angeles, CA). We screened 400 nonredundant lines, and obtained 15 positive hits with increased eye pigmentation (SI Appendix, Table S1). Included in these hits were lines predicted to overexpress E2f1 (Fig. 1D) and CycE, demonstrating the ability of the approach to detect ectopic proliferation. Activation of LA0930 by GMR-Gal4 also caused red eye color when compared to age- and sex-matched sibling controls (Fig. 1E). The nearest downstream annotated gene to the mapped LA0930 insertion site encodes the transcription factor Cbt, suggesting that cbt overexpression was responsible for activation of the PCNA promoter. Prior studies of Cbt had identified it as a member of the SP/KLF family of transcription factors. Cbt has been proposed to be involved in Decapentaplegic (Dpp) signaling during embryonic dorsal closure and in the larval wing disk (45–48), to associate with the Yorkie (Yki) transcription factor (49), and to coordinate energy metabolism during sugar sensing (50).
We confirmed that overexpressed cbt activated the PCNA promoter by analyzing the effect of a UAS-cbt transgene on another E2F1-responsive reporter, PCNA-GFP (43), which contains the same PCNA promoter fragment as PCNA-miniwhite+. Cbt expression controlled by GMR-Gal4 resulted in ectopic PCNA-GFP expression at 30 h APF in the pupal eye (Fig. 1G′). Cbt expression in the posterior wing compartment, driven by engrailed (en)-Gal4 also induced strong ectopic PCNA-GFP expression (Fig. 1H′), demonstrating that Cbt activates the PCNA promoter in multiple tissues.
Cbt Causes an Ectopic Cell Cycle.
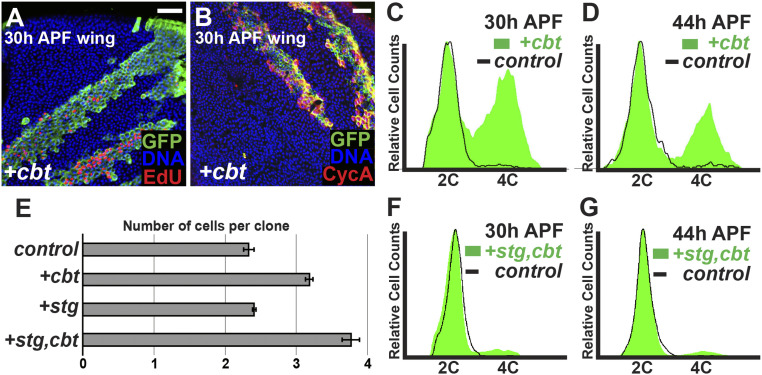
Immunostaining for phospho-histone H3 (PH3), a marker of mitosis, showed that ectopic Cbt not only promoted PCNA expression, but also triggered ectopic cell divisions in both the wing and eye at 30 h APF (Fig. 1 G and H). To determine whether these ectopic mitoses represented complete extra cycles, we generated GFP-labeled clones using the hs-flp;tub > CD2 > Gal4;tub-Gal80ts method, which allows the conditional expression of UAS-linked transgenes (51). Clones expressing UAS-cbt were generated 48 to 72 h after embryo deposition (AED), in animals raised at 18 °C to prevent Gal4-mediated expression. These animals were shifted to 29 °C at 0 h APF, thereby inactivating Gal80ts and activating Gal4 and UAS-cbt expression. Cbt expression in clones caused ectopic mitoses, ectopic DNA replication visualized by EdU labeling (Fig. 2A and SI Appendix, Fig. S1 A and A′) and ectopic S and G2 phases, as visualized by anti-Cyclin A (CycA) immunostaining (Fig. 2B and SI Appendix, Fig. S1 B and B′) at time points after normal cell cycle exit. The presence of cells in different cell cycle phases in cbt-expressing clones indicated an induction of ectopic cell proliferation.
Fig. 2.
Cbt causes an ectopic cell cycle in pupal wings. (A and B) Thirty hours APF, wings with GFP+-marked clones (green) expressing cbt by hs-flp; tub > CD2 > Gal4;tub-Gal80ts. Cbt induced ectopic DNA replication by EdU incorporation (red) and ectopic S and G2 phases by anti-CycA staining (red). Nuclei were stained with Hoechst (blue). (Scale bars, 20 μm.) (C, D, F, and G) FACS of ap-Gal4;tub-Gal80ts wings with GFP labeled cbt expression (C and D) or cbt+stg coexpression (F and G) at 30 h APF (C and F) and 44 h APF (D and G). (E) Number of cells per clone for a given genotype, with clonal induction at 0 h APF.
To further assess cell cycle phasing in cbt overexpressing cells, we used FACS to measure DNA content. In this case, we used apterous (ap)-Gal4 tub-Gal80ts to induce UAS-GFP and UAS-cbt in the dorsal wing blade at 0 h APF, while the ventral wing blade served as a GFP− wild-type control. At 30 h APF, a significant proportion of GFP+ cbt-expressing cells were present in S and G2 phases, while essentially all wild-type non-GFP–expressing cells had exited the cell cycle into G1 phase (Fig. 2C). We also analyzed pupal eyes and wings expressing cbt at 44 h APF. We did not see ectopic mitoses or DNA replication at this timepoint, indicating that cell cycle exit had occurred prior. However, FACS analysis at 44 h APF showed that many cbt expressing cells still had a G2 DNA content (Fig. 2D). Given that no DNA replication was seen in cbt-expressing cells after 36 h APF, our results indicate that Cbt induced a single ectopic S phase in a large proportion of cells, followed by an ectopic M phase in a smaller proportion.
Another possible explanation for the phenotype observed upon cbt overexpression is a developmental delay, such that cells divide the appropriate number of times but do so more slowly than in the wild-type, consequently cycling longer into the pupal stage. To address this possibility, we used clonal labeling to count the number of cell divisions in pupal wings (5). Using the hs-flp;tub > CD2 > Gal4;tub-Gal80ts system, labeled clones were generated at 0 h APF and wings were dissected and fixed between 40 and 44 h APF. More than 100 clones per genotype were scored. GFP+ control clones contained on average 2.33 ± 0.07 cells per clone, illustrating the single cell division that occurs between 0 and 24 h APF. Clones expressing cbt contained an average of 3.17 ± 0.05 cells per clone (Fig. 2E), consistent with our conclusion that overexpressed Cbt induced a single ectopic cell cycle in ∼50% of cells.
Coexpression of String with Cbt Bypasses Exit into G2.
A significant proportion of cbt-expressing cells exited the cell cycle with a G2 DNA content. One potential explanation for this is that Cbt activity cannot prevent the down-regulation of one of more factors involved in M phase progression, which occurs at 24 h APF in cells exiting the cell cycle normally. We addressed this possibility by coexpressing cbt with the M phase activator String (Stg), a Cdc25-type phosphatase that activates Cdk1 and triggers mitosis in most Drosophila cells (52–54). In 30-h APF wings, coexpression of cbt with stg resulted in a majority of cells with a G1 DNA content (Fig. 2F). Wings coexpressing cbt and stg at 44 h APF also contained fewer G2 cells than wings expressing cbt alone (Fig. 2 D and G). These results suggest that the addition of stg to cbt overexpression allows a greater number of cells to complete an entire ectopic cell cycle prior to exiting the cell cycle. To confirm that ectopic cell divisions still occurred during cbt+stg coexpression, we generated GFP-labeled clones at 0 h APF and scored clone size at 40 to 44 h APF in pupal wings. While clones expressing cbt alone contained an average of 3.17 ± 0.05 cells per clone, clones coexpressing cbt and stg contained and average of 3.77 ± 0.12 (Fig. 2E). This suggests that although cbt is able to induce ectopic proliferation, it does so without inducing sufficient amounts of Stg activity to drive all cells through an extra cycle.
Cbt Promotes G1/S Progression in Proliferating Cells.
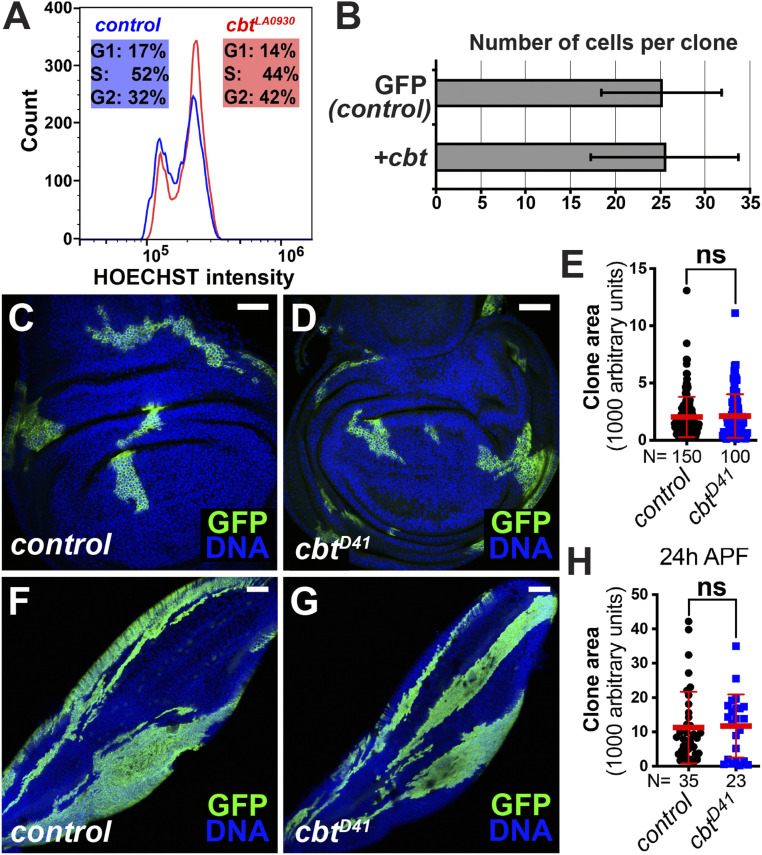
To better understand the effects of Cbt as a cell cycle regulator, we analyzed the effect of cbt overexpression in proliferating cells. GFP-labeled cbt-expressing clones were induced using the hs-flp;act > CD2 > Gal4 system at 48 h AED, and clones were analyzed 72 h later in the third-larval instar wing discs, when most cells still proliferate. FACS analysis showed that cbt expressing cells were slightly more likely to be in G2 phase than control cells, further indicating a role for Cbt in G1/S progression (Fig. 3A). When we analyzed these clones to determine if cbt expression affected the number of cell divisions, we found that there was no significant effect on the number of cells per clone (Fig. 3B). These data indicate that cbt overexpression promotes G1/S in proliferating wing cells without affecting overall cell doubling times. This is consistent with the effects of other positive G1/S regulators in the wing, like Cyclin E, which can accelerate G1/S progression without affecting the overall number of cell cycles (55).
Fig. 3.
Cbt promotes G1/S progression in proliferating cells. (A and B) GFP+-labeled clones (green) expressing cbt were induced at 48 h AED by hs-flp;act > CD2 > Gal4 and analyzed at the third-larval instar. (A) Cells expressing cbt (cbtLA0930) were shifted from G1 and S phase to G2, relative to GFP− control cells. Cell counts per experiment were ∼10,000. Samples were run on a Cytoflex flow cytometer. (B) cbt expression (UAS-cbt) did not cause an increase number of cells per clone compared to wild-type controls. (C–H) cbtD41/D41 mutant clones were generated by MARCM system at 48 h AED and analyzed in the third-instar wing discs (C–E) or 24-h APF wings (F–H). Representative images showed the clone areas of wild-type control (C and F) and cbtD41/D41 mutant (D and G). Clones were labeled by GFP. Nuclei were stained with DAPI (blue). (E and H) Quantification of MARCM clone areas (mean ± SD, t test, nonsignificant [ns] P > 0.05). Each dot represents one sample. (Scale bars, 40 μm in C and D; 80 μm in F and G.)
Cbt Is Dispensable for Wing Cell Proliferation.
Next, we asked whether the loss of cbt function caused a proliferative defect in cycling cells. To address this question, we required a null cbt mutant. However, all of the available deletions that eliminate cbt also disrupt an adjacent gene, Mediator complex subunit 15 (MED15) (SI Appendix, Fig. S2A) (45). Given this, we generated two new cbt mutants, cbtD41 and cbtD148, by P-element excision (SI Appendix, Fig. S2A). Both alleles are small deletions within cbt that do not impinge on MED15. Using complementation tests, we determined that cbtD41 is a null cbt allele while cbtD148 is not (SI Appendix, Fig. S2B). Lethal phase tests showed that most homozygous cbtD41/D41 mutants died during the second-larval instar, and that ∼5% survived past the third larval instar but died in the pupal stage (SI Appendix, Fig. S2C). Furthermore, by applying the Fly-FUCCI system (56), we found that the homozygous third-instar cbtD41/D41 larvae had small wing imaginal discs with disrupted cell cycle progression (SI Appendix, Fig. S2D), suggesting that Cbt is essential for normal tissue growth. To more accurately test whether Cbt is required for cell cycle progression in the wing, we performed mosaic analysis with a repressible cell marker (MARCM) clonal assays using the cbtD41/D41 allele. Cell clones homozygous for cbtD41 did not show growth defects in either third instar larval wing discs (Fig. 3 C–E) or pupal wings (Fig. 3 F–H). This indicates that, in wing development, other cell cycle regulators (e.g., E2F1) may compensate for Cbt’s function in G1/S progression.
Cbt Regulates Intestinal Stem Cell Proliferation.
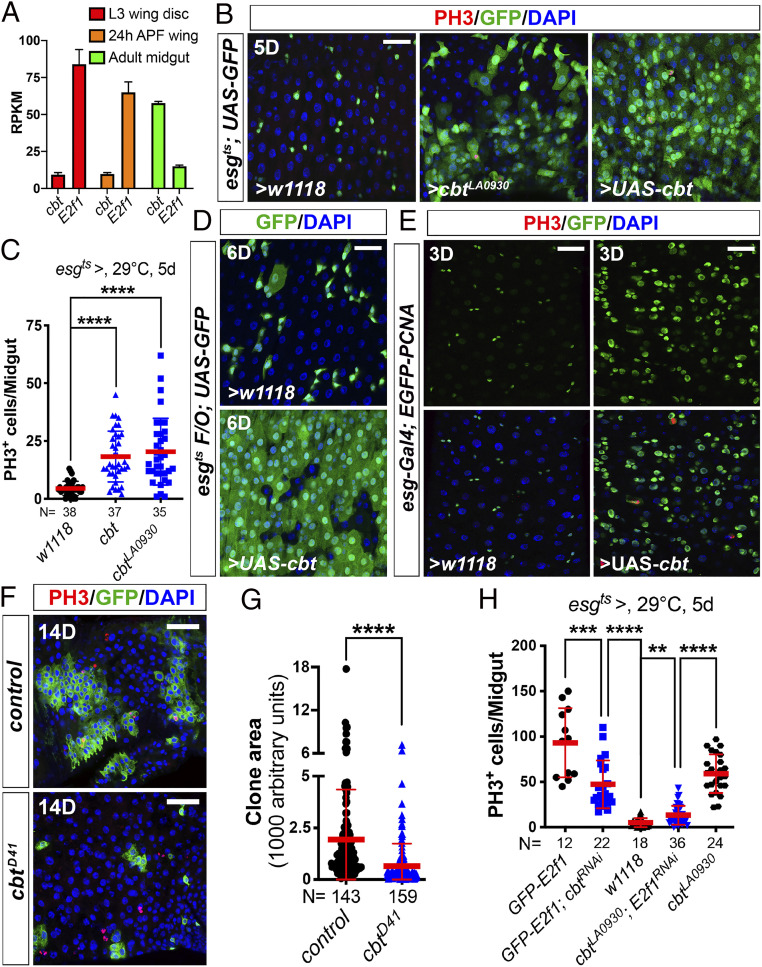
Although our tests showed that Cbt is not required for cell cycle regulation in the developing wing, we suspected that its cell cycle regulatory functions could be important in other tissues. For example, Cbt could be less relevant for cell cycle progression in cell types that have high levels of E2F1, and more important in tissues that have relatively less E2F1. To test this idea, we compared the relative mRNA expression levels of E2f1 and cbt between the wing and the adult midgut (intestine), a tissue that supports high levels of proliferation during epithelial regeneration. Using RNA-sequencing (RNA-seq) datasets from our laboratory and others (13), we found that cbt mRNA expression was much higher than E2f1 in the adult midgut, whereas the reverse was true in larval and pupal wings (Fig. 4A). These expression patterns suggested that Cbt could be important for cell proliferation in the midgut.
Fig. 4.
Cbt compensates E2F1 for intestinal stem cell proliferation. (A and B) The expression levels of cbt and E2f1 were checked using available RNA-seq datasets. The third-instar wing disk and pupal wing RNA-seq data (A) were published in Ma et al. (13). The midgut RNA-seq data (A) was generated in the B.A.E. laboratory. (B) Overexpression of cbt in progenitor cells was driven by esgts. Two- to 3-d-old adult females were shifted from 18 °C to 29 °C for 5 d before dissection. Midguts were stained with anti-GFP and anti-PH3 antibodies and DAPI. ISC mitoses were quantified by PH3+ cells. (Scale bar, 30 μm.) Quantification data shown in C represents the mean ± SD (t test, ****P < 0.0001). Each dot represents one sample. (D) UAS-cbt was overexpressed by the esgts F/O system. Esgts F/O > w1118 was used as control. Flies were raised at 18 °C and then shifted to 29 °C for 6 d before dissection. Midguts were stained with anti-GFP antibody and DAPI. (Scale bar, 30 μm.) (E) Overexpression of cbt in progenitor cells driven by esg-Gal4. Flies were raised at 29 °C. 3-d-old adult females were dissected and stained with anti-GFP and anti-PH3 antibodies and DAPI. These pictures were taken from the R5 region of the midguts with the same laser settings on the confocal microscope. (Scale bars, 30 μm.) (F) cbtD41/D41 mutant clones were generated using the MARCM system. Representative images showed the clone areas of wild-type control (F, Upper) and cbtD41/D41 mutant (Lower) clones 14 d after clone induction. MARCM clones were labeled by GFP. Mitotic cells were stained with anti-PH3 antibody. Nuclei were stained with DAPI (blue). (Scale bars, 40 μm.) (G) Quantification of MARCM clone areas: control (n = 143 clones) versus cbtD41/D41 mutant (n = 159 clones) (mean ± SD, t test, ****P < 0.0001). (H) Different genetic manipulations in progenitor cells driven by esgts. Two- to 3-d-old adult females were shifted from 18 °C to 29 °C for 5 d before dissection. Midguts were stained with anti-GFP and anti-PH3 antibodies and DAPI. ISC mitoses were quantified by PH3+ cells. Quantification data represent the mean ± SD (t test, **P < 0.01, ***P < 0.001, ****P < 0.0001). Each dot represents one sample.
Hence, we assessed cbt function in the adult midgut. Consistent with what we observed in the wing and eye, overexpression of cbt in midgut progenitor cells (intestinal stem cells [ISCs] and enteroblasts) using the esg-Gal4 tub-Gal80ts UAS-GFP (esgts) driver significantly increased ISC mitoses (Fig. 4 B and C). Using the esgtsF/O system (esg-Gal4 tubGal80ts UAS-GFP act > CD2 > Gal4 UAS-flp), which labels ISCs and all their clonal progeny after a temperature shift, we tested if cbt overexpression could influence gut epithelial turnover. Normally, the midgut epithelium renews in ∼12 d at 29 °C in females (57). However, in the cbt overexpression condition, virtually complete gut renewal was achieved by 6 d (Fig. 4D), indicative of accelerated stem cell divisions.
Next, to assess whether ectopic Cbt-driven ISC proliferation is related to the regulation of cell cycle genes, we examined the expression of PCNA using an EGFP-tagged PCNA transgenic line (58). We found that overexpression of cbt in ISCs significantly increased the levels of EGFP-PCNA (Fig. 4E), suggesting that Cbt’s ISC phenotypes are, at least in part, a result of the up-regulation of the cell cycle genes like PCNA. Furthermore, MARCM clonal analysis showed that cbtD41/D41 mutant clones grew significantly less than wild-type controls (Fig. 4 F and G). Thus, in contrast to the situation in the wing and eye, cbt is both limiting and required for normal ISC proliferation. To address the relationship of cbt to E2f1 in the midgut, we performed genetic epistasis experiments using RNAi. We found that depletion of E2f1 significantly repressed ISC mitoses driven by overexpressed cbt (Fig. 4H) and, conversely, that depletion of cbt also repressed E2F1-induced ISC proliferation (Fig. 4H). However, these repressive actions were not complete, suggesting a partial overlap of Cbt and E2F1 function.
Cbt and E2F1 Regulate a Shared Set of Cell Cycle Target Genes.
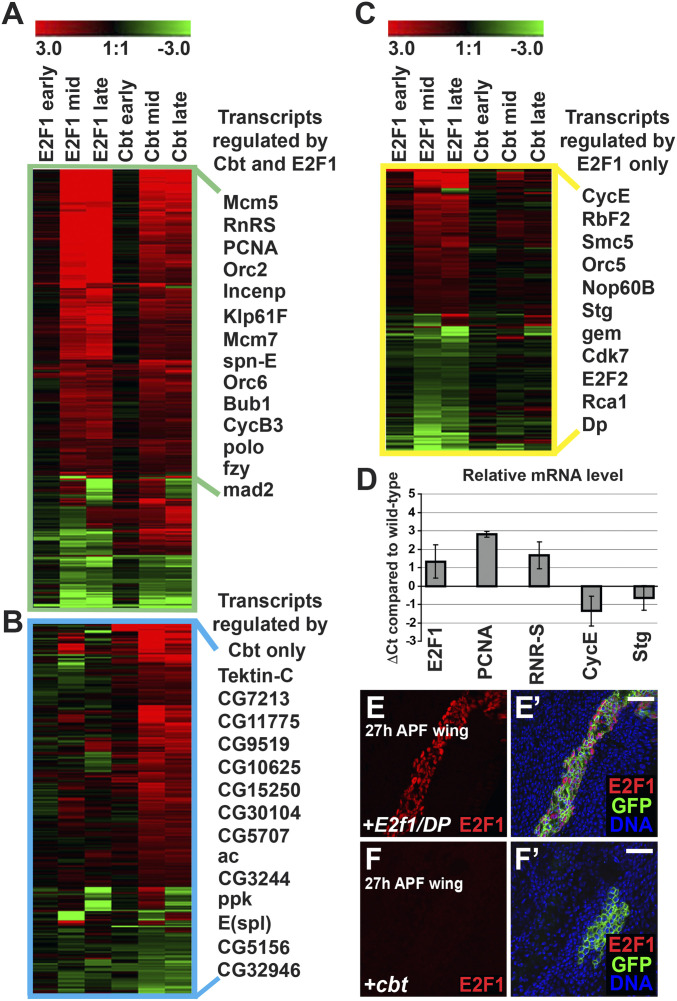
To gain insight into how cbt regulates G1/S progression, we used oligonucleotide expression arrays to determine the how overexpressed cbt alters gene expression in developing wings. After 24 h of cbt overexpression, RNA samples were taken from proliferating wing cells (third-larval instar, L3), at the time of normal cell cycle exit (24 h APF), and after cell cycling had normally ceased (36 h APF). We identified 598 transcripts (SI Appendix, Table S2) affected by cbt expression with a fold-change of 1.3 or greater (log2 ± 0.4 compared to controls, P < 0.05) compared to similarly staged control wings at both the 24-h and 36-h APF time points. We further compared the effects of cbt overexpression to the effects of E2f1/DP overexpression at 24 h and 36 h APF (20). Overall, we observed a remarkable similarity between the transcript changes in cbt- and E2f1/DP-expressing wings. We took a conservative approach in our comparison by requiring that transcripts be affected by both Cbt and E2F1 with a fold-change of 1.3 of greater (log2 ± 0.4 compared to controls, P < 0.05), at both 24 h and 36 h APF, to describe them as coregulated. Using this metric we identified 334 transcripts that were coregulated by both Cbt and E2F1. Many of these have previously been identified as E2F1 targets (16–19) (Fig. 5A). Comparison between Cbt- and E2F1-affected transcripts at 24 h APF gave a correlation coefficient of 0.43. There was a correlation coefficient of 0.57 between E2F1- and Cbt-affected transcripts at 36 h APF. Furthermore, comparison of transcripts affected at 24 h APF to those affected at 36 h APF gave a correlation coefficient of 0.39 (P < 1e−6), indicative of similar regulatory actions of Cbt and E2F1. Analysis of enriched gene ontology (GO) terms among these transcripts yielded the terms “cell cycle” (P < 5.28e−19), “chromosome segregation” (P < 6.48e−11), and “cell division” (P < 2.13e−10), further emphasizing the ability of Cbt to regulate the transcription of genes involved in cell cycle progression.
Fig. 5.
Cbt and E2F1 regulate a shared set of cell cycle genes. (A–C) Microarray analysis of gene expression in cbt or E2f1/DP expressing wings by ap-Gal4;tub-Gal80ts compared to controls (y, w, hs-flp). Heatmaps show transcript changes (color range indicates the log2 ratio of expression compares to controls). Transcripts were hierarchically clustered using Genesis software and with representative transcripts to the right. (A) Heatmap of the 334 transcripts significantly regulated by both E2F1/DP and Cbt. All transcripts with a fold-change of 1.3 or more (log2 ± 0.4, P < 0.05) at both 24 h (mid) and 36 h (late) APF time points are shown. (B) Heatmap of the 279 transcripts regulated by Cbt with a fold-change of 1.3 or more (log2 ± 0.4, P < 0.05) at both time points but not by E2F1/DP. (C) Heatmap of 200 of the >2,000 transcripts regulated by E2F1/DP with a fold-change of 1.3 or more (log2 ± 0.4) but not by Cbt at both time points. (D) RT-qPCR quantification of gene expression in wings expressing cbt compared to controls. Cbt was induced at 0 h APF by ap-Gal4;tub-Gal80ts and levels of transcript expression at 30 h APF was quantified by the ΔΔCt method (97). Wild-type expression at 30 h APF is equivalent to 0Ct while comparative changes in transcript level in cbt expressing wings was measured by ΔCt, equivalent to the log2 of the difference in transcript level. ΔCt values represent the average of three biological replicates and error bars demonstrate the range. (E–F′) GFP marked clones generated by hs-flp;tub > Gal4;tub-Gal80ts expressing E2f1/DP (E and E′) or cbt (F and F′) analyzed in 27-h APF wings. Overexpression of E2f1/DP strongly increased E2F1 protein detected by immunofluorescence (red), whereas overexpression of cbt did not. Nuclei were counter stained with Hoechst 33258 (blue). (Scale bars, 20 μm.)
Cbt also regulated 279 transcripts that were not significantly regulated by E2F1 at both 24- and 36-h APF time points (Fig. 5B). These transcripts were not significantly enriched for any GO term and many of these genes are uncharacterized. This indicates that, although cbt can play a significant role in cell cycle progression, it likely has other functions unrelated to the cell cycle, as previously proposed (50, 59–62).
It should be noted that although ectopic cbt induced a significant number of genes known to be E2F1 targets, several well-validated E2F1 targets were not activated (Fig. 5C). Included in this group were cycE and stg, the rate-limiting factors for S and M phase progression in the wing, respectively (55, 63). This is consistent with the results we obtained by cbt and stg coexpression (Fig. 2 E–G). To confirm that these transcripts were not affected by cbt overexpression at cell cycle exit, we did reverse-transcription quantitative PCR (RT-qPCR) on mRNA from wings expressing cbt under ap-Gal4;tub-Gal80ts control at 30 h APF (Fig. 5D). In agreement with the microarray data, RT-qPCR showed that Cbt did not increase cycE or stg transcript levels, but that pcna and rnr-S levels were significantly increased. These results demonstrate a role for Cbt in regulating the expression of genes involved in cell cycle progression, but not in controlling the G1/S and G2/M rate-limiting factors, cycE and stg.
Cbt Acts Independently of E2F1.
We next assessed the relationship of Cbt and E2F1 in cell cycle progression. Although RT-qPCR data suggested that overexpressed cbt might promote E2f1 expression (Fig. 5D), assays using an enhancer trap allele of E2f1 (E2f1rM729) that is commonly used as a reporter for transcription of the E2f1 locus (64, 65) did not confirm this (SI Appendix, Fig. S3 A and A′). To analyze the effect of cbt on E2F1 protein levels, GFP-labeled overexpression clones were generated and analyzed at 27 h APF using immunofluorescence with an anti-E2F1 antibody. In both wings and eyes, clones overexpressing E2f1 had very high E2F1 protein levels (Fig. 5 E and E′ and SI Appendix, Fig. S3 B and B′), but cbt overexpression did not increase the expression E2F1 protein (Fig. 5 F and F′ and SI Appendix, Fig. S3 C and C′). To further test whether E2F1 levels were altered in the loss-of-function clones of cbt, we performed MARCM clonal assays using the cbtD41/D41 allele. Cell clones homozygous for cbtD41 did not show any alteration of E2F1 protein levels in third-instar wing or eye discs (SI Appendix, Fig. S3 D–E′′). These observations indicate that ectopic cbt promotes cell cycle progression without increasing E2f1 mRNA or protein expression.
Next, to further explore the relationship between cbt and E2f1 target genes, we carried out de novo motif discovery on our microarray data using Multiple Em for Motif Elicitation (MEME, PMID: 19458158) (66). This work identified a putative regulatory motif, GCAGYKGCAGCG (SI Appendix, Fig. S4A), that is overrepresented in the promoters of many Cbt-regulated genes (E-value: 1.6e-014), as well as in the set of Cbt and E2F1 coregulated genes (E-value: 6.1e-004). To evaluate whether this motif might be a Cbt-binding site, we used Cbt chromatin immunoprecipitation-sequencing (ChIP-seq) data from fly embryos from the modENCODE project (67), and carried out similar de novo motif discovery analysis of all Cbt binding regions. This identified a motif very similar to the one given by our own microarray data (SI Appendix, Fig. S4A). This further suggests that our analysis has identified true Cbt targets, and that GCAGYKGCAGCG is likely to be a Cbt binding site. As expected, this motif is present in the PCNA promoter (SI Appendix, Fig. S4B), consistent with the interpretation that pcna is a direct binding target of Cbt. In addition, to assess whether Cbt binding was observed in the promoters of the cell cycle genes that were activated by cbt overexpression, we examined the overlap of Cbt ChIP peaks published by Ruiz‐Romero et al. (49) and the Cbt target genes from our microarray data (SI Appendix, Table S2). Both of these datasets were generated from wing discs. Among the 554 genes (598 transcripts) that were transcriptionally regulated by Cbt, 86 (including PCNA) had Cbt binding sites (SI Appendix, Table S3). These 86 overlapping genes define likely direct transcriptional targets of Cbt. GO analysis showed that most of these genes are cell cycle genes (SI Appendix, Table S4). These results further emphasize the ability of Cbt to regulate the transcription of genes involved in cell cycle progression.
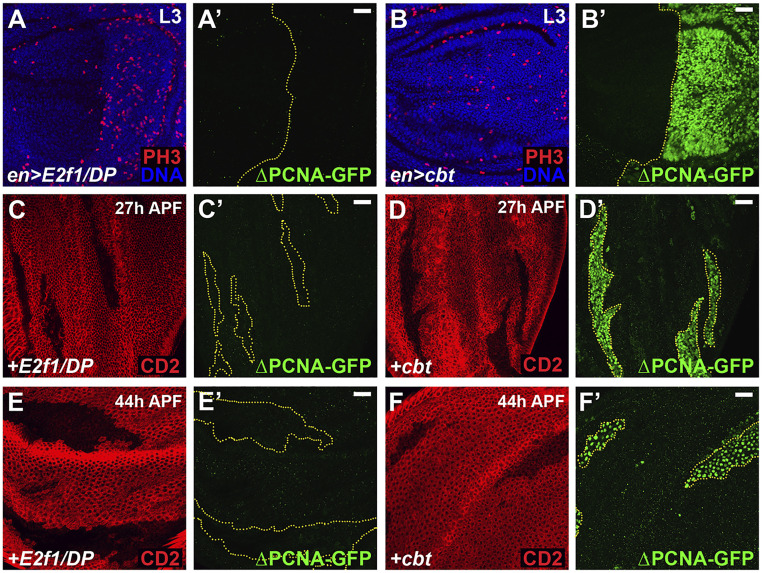
To better understand how cbt functions related to E2f1, we investigated control of the PCNA-GFP reporter gene, which is often used to assay E2F1 activity (43). The two E2F1 binding sites in this reporter are not completely necessary for pcna transcription, as GFP is still weakly transcribed in a version of the reporter mutated in these two E2F1 binding sites (ΔPCNA-GFP) (43). However, these E2F1 binding sites are required for high-level transcription in response to E2F1 in normal cells. Interestingly, ectopic expression of cbt in third-instar wing or eye discs activated ΔPCNA-GFP expression (Fig. 6 B and B′ and SI Appendix, Fig. S5 B and B′), whereas ectopic E2f1/DP did not (Fig. 6 A and A′ and SI Appendix, Fig. S5 A and A′). We also generated cbt or E2f1/DP overexpressing cell clones and analyzed these for ΔPCNA-GFP induction after cell cycle exit at 27 h and 44 h APF. In these cases as well, overexpressed E2f1/DP did not induce ΔPCNA-GFP (Fig. 6 C, C′, E, and E′ and SI Appendix, Fig. S5 C and C′), whereas overexpressed cbt did (Fig. 6 D, D′, F, and F′ and SI Appendix, Fig. S5 D and D′). Furthermore, we found that overexpression of dTIEG, the shorter isoform of Cbt that lacks 81 amino acids at the N terminus (SI Appendix, Figs. S2A and S6A), was unable to activate ΔPCNA-GFP in eye discs (SI Appendix, Fig. S6 B–C′). dTIEG could, however, activate the wild-type PCNA-GFP reporter (SI Appendix, Fig. S6 D–E′′). These results indicate that Cbt can activate the expression of some E2F1 target genes (e.g., pcna) independently of E2F1 binding sites, and that this requires specific sequences in the cbt N terminus.
Fig. 6.
Cbt activates the PCNA-GFP reporter independent of the E2F1 binding sites. (A and B) en-Gal4 was used to express either E2f1/DP (A) or cbt (B) in the proliferating cells of third-instar wing discs. Mitoses were detected by anti-PH3 antibody staining (A and B) and expression of a PCNA-GFP reporter mutated in the E2F1 binding sites (ΔPCNA-GFP) was analyzed (A′ and B′). Nuclei were stained with Hoechst (blue). (C–F) Clones marked by the absence of CD2 (red) expressing either E2f1/DP (C and E) or cbt (D and F) were induced at 48 to 72 h AED by the hs-flp;act > CD2 > Gal4 and examined at the given times for ΔPCNA-GFP expression. E2f1/DP expression did not activate the ΔPCNA-GFP reporter at any time point (A, C, and E). Cbt strongly activated the ΔPCNA-GFP reporter at all time points (B, D, and F) (Scale bars, 20 μm.)
Cbt Drives Ectopic Proliferation Independently of Dp.
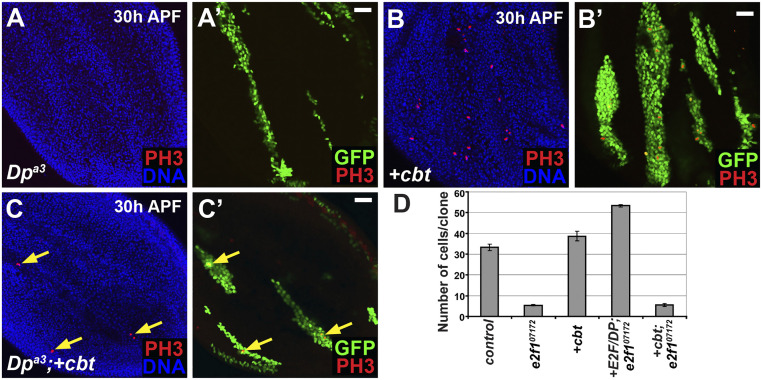
Finally, we investigated whether Cbt requires E2F1 activity at all to delay cell cycle exit. In Drosophila, loss of Dp is believed to remove all E2F transcriptional activity, both activating and repressive, yet cells mutant for Dp cycle and exit the cell cycle relatively normally (20, 41). We used the MARCM system to generate cbt overexpressing cells that were also homozygous null for Dp, and analyzed these for ectopic mitoses. Clones were induced between 48 and 72 h AED and analyzed in pupal wings at 30 h APF. Ectopic mitoses were evident (Fig. 7 A–C). The ability of Cbt to drive ectopic proliferation in Dp mutant cells, which should lack both E2F activating and repressive activities, suggests that the Cbt-induced cell cycling does not require E2F1 activity. We also tested whether ectopic cbt could rescue the effects of loss of E2f1 in proliferating wing cells. We used the MARCM system to generate GFP-labeled E2f1 mutant clones that overexpressed cbt in imaginal wing discs at 72 h AED, and scored the number of cells per clone 48 h later. In this case, cbt overexpression did not rescue the cell cycle arrest that occurs in E2f17172 mutant cells, whereas the overexpression of E2f1/DP complexes in these cells did rescue them (Fig. 7D). Cell cycle arrest in E2f1 mutant cells is believed to result from an increase in E2F2- and Rbf-mediated transcriptional repression (32), which is evidently dominant to overexpressed cbt.
Fig. 7.
Cbt does not require E2F1 activity to drive proliferation but cannot rescue the loss of e2f1 (A–C) GFP+-labeled clones generated by the MARCM system at 48 to 72 h AED and analyzed at 30 h APF in pupal wings for ectopic mitoses by anti-PH3 staining (red). Nuclei were stained with Hoechst (blue). (A) Clones mutant for Dpa3−/− did not undergo ectopic mitoses. (B) Ectopic mitoses were present in clones expressing cbt. (C) Clones mutant for Dpa3−/− and expressing cbt also demonstrated ectopic mitoses (arrows). (D) Using the MARCM system GFP-labeled clones were induced at 72 h AED and number of cells per clone in wing discs was quantified 48 h later. There was no significant difference between e2f17172 clones and e2f17172 clones expressing cbtLA0930. (Scale bars, 20 μm.)
Discussion
For many years, it has been known that Drosophila cells lacking all E2F1 activating and repressive activity (Dp mutant or e2f1/e2f2 double-mutant cells) can express cell cycle genes sufficiently to support extensive cell proliferation (32, 68, 69). Thus, despite the fact the E2Fs are by far the best-characterized and most-specific cell cycle regulatory transcription factors, there must be other transcription factors that can fill E2F1’s role. In Drosophila, DREF has been proposed to be one such factor (70), and the DREAM complex also plays an important role, albeit in association with E2F/Dp and RB family factors (71). In this study we present the SP/KLF-like factor Cbt as a cell cycle regulatory transcription factor that has E2F1-like, but E2F1-independent, activity. Based on multiple lines of evidence, we propose that Cbt activity directs the expression of cell cycle genes independently of both E2F1 activity and the E2F1 binding sites in the promoters of its targets. The ability of Cbt to activate its targets independently of E2F1 binding sites appears to lie in sequences in its N terminus, which are present only in the longer of the two Cbt protein isoforms expressed in Drosophila (SI Appendix, Figs. S2A and S6A). Interestingly, it is these N-terminal sequences that make Cbt more orthologous to human KLF10 and KLF11 than to other KLF genes (SI Appendix, Fig. S6A). Our findings suggest a model in which Cbt provides cells with an additional pathway by which they can control the transcription of cell cycle genes. By cross-comparing published Cbt ChIP-seq data (49) and E2F1 ChIP-chip data (72), we found a significant overlap (P < 10−27, hypergeometric test) that 117 genes are mutual targets of both Cbt and E2F1 (SI Appendix, Table S5). GO analysis suggested that these 117 genes mediate a broad range of functions, including DNA replication, cell cycle, and multiple metabolic processes (SI Appendix, Table S6), further suggesting that Cbt is a critical cell cycle regulator that can complement E2F1 in many cellular contexts.
In the developing wing and eye the potent activity of E2F1 in proliferating cells appears to mask the ability of Cbt to regulate its cell cycle targets, but when E2F1 transcriptional activating activity is diminished, as occurs at cell cycle exit in the wing and eye discs, or at steady-state in the adult midgut, ectopic Cbt activity is capable of driving inappropriate cell cycles. Interestingly however, Cbt was not capable of promoting the expression of either CycE or Stg in the wing, the two factors previously shown to be proximal rate-limiting regulators of G1/S and G2/M progression in the developing fly wing and eye. Because of this, we propose that, in the pupal wing and eye, cbt overexpression may induce ectopic proliferation by providing higher than normal levels of a set of downstream genes that are required for DNA replication and mitosis (e.g., Mcm5, -7, Orc2, -6, CycB3, fzy) and that this increase in substrate availability permits residual CycE/Cdk2 and Stg/Cyclin/Cdk1 activity to drive additional S and M phases.
Beyond E2F1, another regulatory pathway that plays a fundamental role for cell cycle progression is the Hippo pathway. Yki, the downstream transcriptional activator of the Hippo pathway, controls multiple cell cycle regulators’ expression (e.g., E2F1 and CycE) (73, 74). A previous study (49) proposed that Cbt associates with Yki to promote cell proliferation. Ruiz-Romero et al. (49) showed that Diap1-GFP, a Yki activity reporter, is decreased in cbt mutant (dTIEGS14/S14) clones, suggesting that cbt is required for Yki activation. However, this conclusion is confounded by the fact that the dTIEGS14 allele used in that paper is a large deletion that removes the DNA encoding both cbt and MED15. We reexamined the regulation of Yki activity by Cbt using our cbtD41 allele, which doesn’t impact MED15. In cbtD41/D41 mutant clones, the expression of the Yki target, Diap1, was normal (SI Appendix, Fig. S7 A and A′). However, knockdown of MED15 did significantly suppress the expression of Diap1 (SI Appendix, Fig. S7 B and B′), indicating that the decrease in Diap1-GFP in dTIEGS14 mutant cells was due to the loss of MED15 rather than cbt. Hence, we conclude that Cbt regulates cell proliferation independently of the Hippo pathway.
In mammals, the cbt homologs KLF10 and KLF11 are rapidly expressed following induction of TGF-β signaling, and interact with Smad family members to function as effectors of TGF-β signaling (42, 75–82). In this study, we identified a putative regulatory motif of GCAGYKGCAGCG that resembles a Mad-like motif (SI Appendix, Fig. S4C), consistent with the established relationship in both Drosophila and mammals between Cbt and the TGF-β signaling pathway (42, 45, 46, 83). Further work exploring the relationships between Cbt, E2F1, and the TGF-β signaling pathway may help elucidate an unsuspected relationship between this key developmental signaling pathway and cell cycle control.
The ability of Cbt to induce ectopic cell proliferation in the wing, eye, and gut suggests that it could have oncogenic function. Indeed, ectopic activity of several members of the SP/KLF family has been associated with a variety of cancerous phenotypes (84–88). Although the most immediate mammalian homologs of cbt, KLF10 and KLF11 (members of the TIEG family), are known primarily as cell cycle repressors (42), KLF10 overexpression has nevertheless also been detected in several cancer cell lines, including renal clear cell carcinoma and glioblastoma (89). In these cancers, KLF10 activity is thought to increase TGF-β1 signaling and thereby to promote adhesion, migration, and regeneration in epithelial cell types (89, 90). This is consistent with data showing that overexpression of KLF10 in certain cell types has effects resembling those caused by activation of TGF-β signaling, and suggests a positive feedback loop between KLF10 and TGF-β signaling (78, 79, 82, 91). Furthermore, when we refer to patient data from The Cancer Genome Atlas, we notice that among 220 colorectal adenocarcinoma patients with mutations and copy number variations, 9.5% of them have KLF10 gene amplification or KLF10 ectopic overexpression. This is in line with our oncogenic phenotype of Cbt in fly gut. Similarly, high KLF11 expression has been associated with gastric and breast cancers (92, 93). How these connections might play out in terms of cell cycle control is an interesting topic for future studies.
Materials and Methods
Fly Strains.
Please see SI Appendix, Supplementary Materials and Methods for details on fly strains.
Cbt Mutagenesis.
A P-element insertion, P[GawB]cbt[NP5201] (Kyoto # 104895), resides in the first intron of the cbt gene. After the mobilization of P[GawB]cbt[NP5201] driven by Δ2-3 transposase, the cbt deletions were screened using the following primers: Fwd: TGGTTGCTCCACTGCCGATGACG; Rev: CCACTCATCAAGCAAAAAACATTCC.
Two mutants, cbtD41 and cbtD148, were obtained from screening. Genomic PCR and subsequent sequencing showed that cbtD41 had a 900-bp deletion, which impaired the coding region of all three cbt transcript isoforms (RA, RB, and RC). cbtD148 had a 1,018-bp deletion, which deleted 59 bp of the first exon of cbt-RA but didn’t alter the exons of cbt-RB or cbt-RC.
Generation of PCNA-miniwhite+ and GMR-Gal4(ey-CFP).
The PCNA promoter was amplified from genomic DNA using the following primer pair: Fwd: CCCAAGCTTTCCAAACCAGTTGGCAGGCCGC and Rev: CATGAATTCTGTGTTTTATTATTTAAATACTGATGACG. The amplified PCR product was digested using HindIII and EcoRI and cloned into pBlueScript II HindIII/EcoRI sites. The resulting vector is pBS-PCNA. The miniwhite+ gene was amplified from pUAST using the following primer pair: Fwd: CCGGAATTCATGGGCCAAGAGGATCAGGAG and Rev: AACTGCAGCCGAATTAATTCTAGTTCCAG. The amplified PCR product was digested using EcoRI and PstI and cloned into pBlueScript II EcoRI/PstI sites. The resulting vector is pBS-miniwhite+. An EcoRI/PstI fragment from pBS-miniwhite+ containing the miniwhite+ coding sequence was cloned into EcoRI/PstI sites of pBS-PCNA. The resulting vector is pBS-PCNA-miniwhite+. SmaI/XhoI fragment from pBS-PCNA-miniwhite+ containing PCNA-miniwhite+ was cloned into the piggyBac vector pBSII-ITR1.1k-ECFP to generate pBac.ECFP. PCNA-miniwhite+.PiggyBac transformants were generated by coinjecting pBac.ECFP.PCNA-miniwhite+ with pBSII-Act5c-orf (piggyBac transposase). To generate GMR-Gal4(ey-CFP), an XhoI/HindIII (blunted) fragment from pGMR containing GMR, hsp70Bb and hsp70Ab was cloned into the XhoI/StuI sites of pBac-ECFP, the resulting vector is pBac-GMR-ECFP. A BglII/BamHI fragment from pGMR-Gal4 containing the Gal4 coding sequence was cloned into the BglII site of pBac-GMR-ECFP, between hsp70Ab and hsp70Ab sequences, the resulting vector is pBac-GMR-Gal4-ECFP.
Immunostaining.
Please see SI Appendix, Supplementary Materials and Methods for details on immunostaining.
Clonal Analysis.
Overexpression clones we generated by either the heat-shock hs-flp;act > CD2 > Gal4 method or the hs-flp;tub > CD2 > Gal4 method with tub-Gal80ts (51) inactivated at 29 °C. Hours APF represent equivalent time at 25 °C and adjusted appropriately as described in Buttitta et al. (5). Larvae were heat-shocked for 10 min at 37 °C between 48 and 72 h AED. Loss-of-function clones (or appropriate controls) were either generated by mitotic recombination (94) or by MARCM (95). Larvae were heat-shocked for 45 min-1 h between 48 and 72 h AED, staged at white prepupae (if necessary), and dissected at the designated time point. For the MARCM clone assay performed in the midgut, 2- to 3-d-old female flies were heat-shocked for 20 min at 34 °C and dissected at the designated time point.
Clone Cell Counts.
Please see SI Appendix, Supplementary Materials and Methods for details on cell counts.
Flow Cytometry.
Dissociation of cells from staged and dissected larvae and pupae was performed as described in refs. 5, 63, and 96. Cell counts per experiment were ∼20,000 except in tissues 44 h APF, which had cell counts ∼10,000. Each experiment was performed at least three times and representative examples are shown. Experiments were performed on a Vantage2 cell sorter and analyzed using CellQuest (Becton-Dickinson).
Microarrays.
Please see SI Appendix, Supplementary Materials and Methods for details on microarrays.
RT-qPCR.
Please see SI Appendix, Supplementary Materials and Methods for details on RT-qPCR.
Statistical Analysis.
Statistical analyses were performed using the GraphPad Prism 8. Statistical significance (P values) of experiments were calculated by unpaired two-tailed Student’s t test. Statistical significance was denoted as follows: nonsignificant (ns) P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Supplementary Material
Acknowledgments
We thank Harene Venghatakrishnan for assistance with the experiments reported in SI Appendix, Fig. S6 B–E′′; J. Merriam, N. Paricio, R. Duronio, S. Di Talia, X. Bi, and I. Rodriguez for flies and/or antibodies; the Fred Hutchinson Cancer Research Center Imaging, Array, and Flow Cytometry Facilities for help with data acquisition and array hybridizations; to J. Davison and M. Morgan for help with statistical analysis; and the Van Gilst Laboratory for use of their equipment. This work was supported by the Huntsman Cancer Foundation and the Center for Genomic Medicine/Utah Genome Project at the University of Utah, and National Institutes of Health Grants R01 GM070887, R01 GM124434, and P30 CA042014 (to B.A.E.). A.J.K. was supported by Developmental Biology Training Grant T32 HDO7183. L.A.B. was supported by Leukemia & Lymphoma Society Special Fellowship (LLS#3370-09) and NIH K99 GM086517.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015675118/-/DCSupplemental.
Data Availability.
All study data are included in the article and SI Appendix.
References
- 1.Vidal A., Koff A., Cell-cycle inhibitors: Three families united by a common cause. Gene 247, 1–15 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Foijer F., Wolthuis R. M., Doodeman V., Medema R. H., te Riele H., Mitogen requirement for cell cycle progression in the absence of pocket protein activity. Cancer Cell 8, 455–466 (2005). [DOI] [PubMed] [Google Scholar]
- 3.Du W., Dyson N., The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. EMBO J. 18, 916–925 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pennycook B. R., Barr A. R., Restriction point regulation at the crossroads between quiescence and cell proliferation. FEBS Lett., 10.1002/1873-3468.13867 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Buttitta L. A., Katzaroff A. J., Perez C. L., de la Cruz A., Edgar B. A., A double-assurance mechanism controls cell cycle exit upon terminal differentiation in Drosophila. Dev. Cell 12, 631–643 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Buttitta L. A., Edgar B. A., Mechanisms controlling cell cycle exit upon terminal differentiation. Curr. Opin. Cell Biol. 19, 697–704 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller J. P., Yeh N., Vidal A., Koff A., Interweaving the cell cycle machinery with cell differentiation. Cell Cycle 6, 2932–2938 (2007). [DOI] [PubMed] [Google Scholar]
- 8.van den Heuvel S., Dyson N. J., Conserved functions of the pRB and E2F families. Nat. Rev. Mol. Cell Biol. 9, 713–724 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Onoyama I., Nakayama K. I., Fbxw7 in cell cycle exit and stem cell maintenance: Insight from gene-targeted mice. Cell Cycle 7, 3307–3313 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Firth L. C., Baker N. E., Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev. Cell 8, 541–551 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Sun D., Buttitta L., States of G0 and the proliferation-quiescence decision in cells, tissues and during development. Int. J. Dev. Biol. 61, 357–366 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Grant G. D., Cook J. G., The temporal regulation of S phase proteins during G1. Adv. Exp. Med. Biol. 1042, 335–369 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma Y., McKay D. J., Buttitta L., Changes in chromatin accessibility ensure robust cell cycle exit in terminally differentiated cells. PLoS Biol. 17, e3000378 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung T. H., Rando T. A., Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 14, 329–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swygert S. G., et al. , Condensin-dependent chromatin compaction represses transcription globally during quiescence. Mol. Cell 73, 533–546.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimova D. K., Stevaux O., Frolov M. V., Dyson N. J., Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17, 2308–2320 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevaux O., Dyson N. J., A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14, 684–691 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Ishida S., et al. , Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21, 4684–4699 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polager S., Kalma Y., Berkovich E., Ginsberg D., E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21, 437–446 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Buttitta L. A., Katzaroff A. J., Edgar B. A., A robust cell cycle control mechanism limits E2F-induced proliferation of terminally differentiated cells in vivo. J. Cell Biol. 189, 981–996 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dyson N., The regulation of E2F by pRB-family proteins. Genes Dev. 12, 2245–2262 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Dyson N. J., RB1: A prototype tumor suppressor and an enigma. Genes Dev. 30, 1492–1502 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du W., Pogoriler J., Retinoblastoma family genes. Oncogene 25, 5190–5200 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narasimha A. M., et al. , Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. eLife 3, e02872 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherr C. J., Roberts J. M., CDK inhibitors: Positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Otto T., Sicinski P., Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer 17, 93–115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimova D. K., Dyson N. J., The E2F transcriptional network: Old acquaintances with new faces. Oncogene 24, 2810–2826 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Du W., Vidal M., Xie J. E., Dyson N., RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10, 1206–1218 (1996). [DOI] [PubMed] [Google Scholar]
- 29.Sawado T., et al. , dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 251, 409–415 (1998). [DOI] [PubMed] [Google Scholar]
- 30.de Nooij J. C., Letendre M. A., Hariharan I. K., A cyclin-dependent kinase inhibitor, Dacapo, is necessary for timely exit from the cell cycle during Drosophila embryogenesis. Cell 87, 1237–1247 (1996). [DOI] [PubMed] [Google Scholar]
- 31.Duronio R. J., O’Farrell P. H., Developmental control of the G1 to S transition in Drosophila: Cyclin E is a limiting downstream target of E2F. Genes Dev. 9, 1456–1468 (1995). [DOI] [PubMed] [Google Scholar]
- 32.Frolov M. V., et al. , Functional antagonism between E2F family members. Genes Dev. 15, 2146–2160 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Finley R. L. Jr, Thomas B. J., Zipursky S. L., Brent R., Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc. Natl. Acad. Sci. U.S.A. 93, 3011–3015 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoblich J. A., et al. , Cyclin E controls S phase progression and its down-regulation during Drosophila embryogenesis is required for the arrest of cell proliferation. Cell 77, 107–120 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Moberg K. H., Bell D. W., Wahrer D. C., Haber D. A., Hariharan I. K., Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413, 311–316 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Lane M. E., et al. , Dacapo, a cyclin-dependent kinase inhibitor, stops cell proliferation during Drosophila development. Cell 87, 1225–1235 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Schubiger M., Palka J., Changing spatial patterns of DNA replication in the developing wing of Drosophila. Dev. Biol. 123, 145–153 (1987). [DOI] [PubMed] [Google Scholar]
- 38.Milán M., Campuzano S., García-Bellido A., Cell cycling and patterned cell proliferation in the Drosophila wing during metamorphosis. Proc. Natl. Acad. Sci. U.S.A. 93, 11687–11692 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolff T., Ready D., “Pattern formation in the Drosophila retina” in The Development of Drosophila Melanogaster, Bate M., Arias A., Eds. (Cold Spring Harbor Press, Cold Spring Harbor, NY, 1993), pp. 1277–1326. [Google Scholar]
- 40.Weng L., Zhu C., Xu J., Du W., Critical role of active repression by E2F and Rb proteins in endoreplication during Drosophila development. EMBO J. 22, 3865–3875 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frolov M. V., Moon N. S., Dyson N. J., dDP is needed for normal cell proliferation. Mol. Cell. Biol. 25, 3027–3039 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spittau B., Krieglstein K., Klf10 and Klf11 as mediators of TGF-beta superfamily signaling. Cell Tissue Res. 347, 65–72 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Thacker S. A., Bonnette P. C., Duronio R. J., The contribution of E2F-regulated transcription to Drosophila PCNA gene function. Curr. Biol. 13, 53–58 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Steller H., Pirrotta V., Expression of the Drosophila white gene under the control of the hsp70 heat shock promoter. EMBO J. 4, 3765–3772 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez I., Drosophila TIEG is a modulator of different signalling pathways involved in wing patterning and cell proliferation. PLoS One 6, e18418 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muñoz-Descalzo S., Terol J., Paricio N., Cabut, a C2H2 zinc finger transcription factor, is required during Drosophila dorsal closure downstream of JNK signaling. Dev. Biol. 287, 168–179 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Muñoz-Descalzo S., Belacortu Y., Paricio N., Identification and analysis of Cabut orthologs in invertebrates and vertebrates. Dev. Genes Evol. 217, 289–298 (2007). [DOI] [PubMed] [Google Scholar]
- 48.Suske G., Bruford E., Philipsen S., Mammalian SP/KLF transcription factors: Bring in the family. Genomics 85, 551–556 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Romero M., Blanco E., Paricio N., Serras F., Corominas M., Cabut/dTIEG associates with the transcription factor Yorkie for growth control. EMBO Rep. 16, 362–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartok O., et al. , The transcription factor Cabut coordinates energy metabolism and the circadian clock in response to sugar sensing. EMBO J. 34, 1538–1553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGuire S. E., Roman G., Davis R. L., Gene expression systems in Drosophila: A synthesis of time and space. Trends Genet. 20, 384–391 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Edgar B. A., O’Farrell P. H., Genetic control of cell division patterns in the Drosophila embryo. Cell 57, 177–187 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumagai A., Dunphy W. G., The cdc25 protein controls tyrosine dephosphorylation of the cdc2 protein in a cell-free system. Cell 64, 903–914 (1991). [DOI] [PubMed] [Google Scholar]
- 54.Edgar B. A., O’Farrell P. H., The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string. Cell 62, 469–480 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reis T., Edgar B. A., Negative regulation of dE2F1 by cyclin-dependent kinases controls cell cycle timing. Cell 117, 253–264 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Zielke N., et al. , Fly-FUCCI: A versatile tool for studying cell proliferation in complex tissues. Cell Rep. 7, 588–598 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Jin Y., et al. , EGFR/Ras signaling controls Drosophila intestinal stem cell proliferation via capicua-regulated genes. PLoS Genet. 11, e1005634 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blythe S. A., Wieschaus E. F., Establishment and maintenance of heritable chromatin structure during early Drosophila embryogenesis. eLife 5, e20148 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beckstead R. B., Lam G., Thummel C. S., The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 6, R99 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guertin D. A., Guntur K. V., Bell G. W., Thoreen C. C., Sabatini D. M., Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol. 16, 958–970 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Kadener S., Menet J. S., Schoer R., Rosbash M., Circadian transcription contributes to core period determination in Drosophila. PLoS Biol. 6, e119 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kraut R., Menon K., Zinn K., A gain-of-function screen for genes controlling motor axon guidance and synaptogenesis in Drosophila. Curr. Biol. 11, 417–430 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Neufeld T. P., de la Cruz A. F., Johnston L. A., Edgar B. A., Coordination of growth and cell division in the Drosophila wing. Cell 93, 1183–1193 (1998). [DOI] [PubMed] [Google Scholar]
- 64.Duronio R. J., O’Farrell P. H., Xie J. E., Brook A., Dyson N., The transcription factor E2F is required for S phase during Drosophila embryogenesis. Genes Dev. 9, 1445–1455 (1995). [DOI] [PubMed] [Google Scholar]
- 65.Brook A., Xie J. E., Du W., Dyson N., Requirements for dE2F function in proliferating cells and in post-mitotic differentiating cells. EMBO J. 15, 3676–3683 (1996). [PMC free article] [PubMed] [Google Scholar]
- 66.Joe Song M., Hong C. C., Zhang Y., Buttitta L., Edgar B. A., Comparative generalized logic modeling reveals differential gene interactions during cell cycle exit in Drosophila wing development. GI Ed. Proc. 157, 143–152 (2009). [PMC free article] [PubMed] [Google Scholar]
- 67.Celniker S. E.et al.; modENCODE Consortium , Unlocking the secrets of the genome. Nature 459, 927–930 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duronio R. J., Bonnette P. C., O’Farrell P. H., Mutations of the Drosophila dDP, dE2F, and cyclin E genes reveal distinct roles for the E2F-DP transcription factor and cyclin E during the G1-S transition. Mol. Cell. Biol. 18, 141–151 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Royzman I., Whittaker A. J., Orr-Weaver T. L., Mutations in Drosophila DP and E2F distinguish G1-S progression from an associated transcriptional program. Genes Dev. 11, 1999–2011 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tue N. T., et al. , DREF plays multiple roles during Drosophila development. Biochim. Biophys. Acta. Gene Regul. Mech. 1860, 705–712 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Sadasivam S., DeCaprio J. A., The DREAM complex: Master coordinator of cell cycle-dependent gene expression. Nat. Rev. Cancer 13, 585–595 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korenjak M., Anderssen E., Ramaswamy S., Whetstine J. R., Dyson N. J., RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol. Cell. Biol. 32, 4375–4387 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tapon N., et al. , Salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110, 467–478 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Zhang P., et al. , A balance of Yki/Sd activator and E2F1/Sd repressor complexes controls cell survival and affects organ size. Dev. Cell 43, 603–617.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Subramaniam M., et al. , Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 23, 4907–4912 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cook T., Urrutia R., TIEG proteins join the Smads as TGF-beta-regulated transcription factors that control pancreatic cell growth. Am. J. Physiol. Gastrointest. Liver Physiol. 278, G513–G521 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Cook T., Gebelein B., Mesa K., Mladek A., Urrutia R., Molecular cloning and characterization of TIEG2 reveals a new subfamily of transforming growth factor-beta-inducible Sp1-like zinc finger-encoding genes involved in the regulation of cell growth. J. Biol. Chem. 273, 25929–25936 (1998). [DOI] [PubMed] [Google Scholar]
- 78.Tachibana I., et al. , Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J. Clin. Invest. 99, 2365–2374 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramaniam M., Hawse J. R., Johnsen S. A., Spelsberg T. C., Role of TIEG1 in biological processes and disease states. J. Cell. Biochem. 102, 539–548 (2007). [DOI] [PubMed] [Google Scholar]
- 80.Johnsen S. A., Subramaniam M., Katagiri T., Janknecht R., Spelsberg T. C., Transcriptional regulation of Smad2 is required for enhancement of TGFbeta/Smad signaling by TGFbeta inducible early gene. J. Cell. Biochem. 87, 233–241 (2002). [DOI] [PubMed] [Google Scholar]
- 81.Johnsen S. A., Subramaniam M., Janknecht R., Spelsberg T. C., TGFbeta inducible early gene enhances TGFbeta/Smad-dependent transcriptional responses. Oncogene 21, 5783–5790 (2002). [DOI] [PubMed] [Google Scholar]
- 82.Ellenrieder V., TGFbeta regulated gene expression by Smads and Sp1/KLF-like transcription factors in cancer. Anticancer Res. 28, 1531–1539 (2008). [PubMed] [Google Scholar]
- 83.Lee J. M., et al. , KLF10 is a modulatory factor of chondrocyte hypertrophy in developing skeleton. J. Orthop. Res. 38, 1987–1995 (2020). [DOI] [PubMed] [Google Scholar]
- 84.Kaczynski J., Cook T., Urrutia R., Sp1- and Krüppel-like transcription factors. Genome Biol. 4, 206 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bureau C., et al. , Expression and function of Kruppel like-factors (KLF) in carcinogenesis. Curr. Genomics 10, 353–360 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Black A. R., Black J. D., Azizkhan-Clifford J., Sp1 and Krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188, 143–160 (2001). [DOI] [PubMed] [Google Scholar]
- 87.Safe S., Abdelrahim M., Sp transcription factor family and its role in cancer. Eur. J. Cancer 41, 2438–2448 (2005). [DOI] [PubMed] [Google Scholar]
- 88.Memon A., Lee W. K., KLF10 as a tumor suppressor gene and its TGF-β signaling. Cancers (Basel) 10, 161 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ivanov S. V., et al. , Two novel VHL targets, TGFBI (BIGH3) and its transactivator KLF10, are up-regulated in renal clear cell carcinoma and other tumors. Biochem. Biophys. Res. Commun. 370, 536–540 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Park S. W., et al. , Beta ig-h3 promotes renal proximal tubular epithelial cell adhesion, migration and proliferation through the interaction with alpha3beta1 integrin. Exp. Mol. Med. 36, 211–219 (2004). [DOI] [PubMed] [Google Scholar]
- 91.Hefferan T. E., et al. , Overexpression of a nuclear protein, TIEG, mimics transforming growth factor-beta action in human osteoblast cells. J. Biol. Chem. 275, 20255–20259 (2000). [DOI] [PubMed] [Google Scholar]
- 92.Ji Q., et al. , KLF11 promotes gastric cancer invasion and migration by increasing Twist1 expression. Neoplasma 66, 92–100 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Cheng L., Shi L., Dai H., Bioinformatics analysis of potential prognostic biomarkers among Krüppel-like transcription factors (KLFs) in breast cancer. Cancer Biomark. 26, 411–420 (2019). [DOI] [PubMed] [Google Scholar]
- 94.Xu T., Rubin G. M., Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117, 1223–1237 (1993). [DOI] [PubMed] [Google Scholar]
- 95.Lee T., Luo L., Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24, 251–254 (2001). [DOI] [PubMed] [Google Scholar]
- 96.de la Cruz A. F., Edgar B. A., Flow cytometric analysis of Drosophila cells. Methods Mol. Biol. 420, 373–389 (2008). [DOI] [PubMed] [Google Scholar]
- 97.Livak K. J., Schmittgen T. D., Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and SI Appendix.