Abstract
With the recent spread of the SARS-CoV-2 virus, computer-aided diagnosis (CAD) has received more attention. The most important CAD application is to detect and classify pneumonia diseases using X-ray images, especially, in a critical period as pandemic of covid-19 that is kind of pneumonia. In this work, we aim to evaluate the performance of single and ensemble learning models for the pneumonia disease classification. The ensembles used are mainly based on fined-tuned versions of (InceptionResNet_V2, ResNet50 and MobileNet_V2). We collected a new dataset containing 6087 chest X-ray images in which we conduct comprehensive experiments. As a result, for a single model, we found out that InceptionResNet_V2 gives 93.52% of F1 score. In addition, ensemble of 3 models (ResNet50 with MobileNet_V2 with InceptionResNet_V2) shows more accurate than other ensembles constructed (94.84% of F1 score).
Keywords: Pneumonia disease, Pneumonia multiclass classification, Covid-19, X-ray images, Computer-aided diagnosis, Deep learning, Ensemble deep learning
Introduction
Throughout history, epidemics and chronic diseases have killed numerous individuals and caused significant emergencies that have set aside a long effort to survive [1]. Recently, researchers, specialists, and companies around the world are rolling out CAD systems that can fastly process hundreds of X-ray and computed tomography (CT) images to accelerate the diagnosis of pneumonia such as SARS, MERS, covid-19, and aid in its containment [2]. As the number of patients infected by pneumonia disease increases, it turns out to be increasingly hard for radiologists to finish the diagnostic process in the constrained accessible time [3]. Medical images analysis is one of the most promising research areas [4], and it provides facilities for diagnosis and making decisions of several diseases such as covid-19. Therefore, interpretation of these images requires expertise and necessitates several algorithms in order to enhance, accelerate, and make an accurate diagnosis. Recently, many efforts and more attention are paid to imaging modalities and deep learning (DL) in pneumonia disease [2]. DL is a neural network that consists of five layers which are: input layer, convolutional layers, pooling layers, full-connection layers, and output layer [5] . Following this context, DL models have obtained better performance in the detection and classification of pneumonia disease and demonstrated high accuracy compared with previous state-of-the-art methods [2].
Numerous computer vision applications are intricate that they cannot be solved by the utilization of a single algorithm [6]. This required the need for development of models by combining two or more of the studied algorithms. The selection of models relies on the necessities and characteristics of the issue. Ensemble models combine more than one single model to solve a given task. This methodology was intended to overcome the weaknesses of single models and consolidate their strengths [7]. In the field of medical science, ensemble models are currently served to carry out prediction tasks (e.g., regression and classification) [8]. The single models that comprise the ensemble are trained independently to solve the given task. The last output of the ensemble model is an aggregation of the various outputs given by the single model. Furthermore, ensemble model reduces the variance of predictions and generalization error and significantly improves the computational training and could be utilized with a few training data [8].
Motivated by all advantages cited above, the present work aims to evaluate the performance of the most accurate deep learning models for multiclass classification of X-ray images in order to answer the following research questions (RQ):
(RQ1): What is the diagnostic accuracy that DL can attain based on X-ray images?
(RQ2): Is combining DL to construct ensembles DL will enhance the final accuracy of certain model?
(RQ3): Does the number of DL combined to construct ensembles DL affect the accuracy of the model?
The main contributions of the present paper are summarized as follows:
We implement 3 fined-tuned versions of (InceptionResNet_V2, ResNet50 and MobileNet_V2) according to the technique used by Elasnaoui and Chawki [2], Elasnaoui et al. [3].
We design single and ensemble models based on 3 fined-tuned models.
To avoid over-fitting in different models, we used weight decay and L2-regularizers.
We combine 3 and 2 fined-tuned models to construct single and ensemble models.
We tested single and ensemble on chest X-ray datasets [9, 10].
We evaluate the performance of the single and ensemble models used in this study.
The remainder of this paper is organized as follows. Section 2 deals with some related work. We describe our proposed contribution in Sect. 3. Section 4 presents the experiment material and parameterization. The results obtained and their interpretations are illustrated in Sect. 5. Threats of validity are presented in Sect. 6. Finally, conclusion and future work are given in the last section.
Background
A study of the state of the art reveals that significant works have been published for pneumonia detection and classification from X-ray images in recent years, where most works used several successful deep learning approaches for automatically classifying chest X-ray images into different disease categories [11]. The application of DL in the field of pneumonia leads to the reduction of false-positive and negative errors in the detection and diagnosis of this disease and provides an optimal opportunity to provide fast, cheap, and safe diagnostic services to patients [12]. This section summarizes and discusses the state-of-the-art methods of pneumonia detection and classification using DL.
As listed in Table 1, we observed that:
Table 1.
Summary of the literature review using DL techniques
| Reference and year | Deep learning technique and application | Imaging modality | Classification task | Findings and results |
|---|---|---|---|---|
| Barrientos et al. [15] | Analysis of patterns present in rectangular segments using neural networks | Ultrasound imaging | Binary classification |
Sensitivity = 91.5% Specificity = 100% |
| Ahmad et al. [16] | Classification of infection and fluid regions into specific abnormalities (infection or fluid or normal) using a block-based approach with Naïve Bayes classifier | X-ray image | Binary classification | – |
| Khobragade et al. [17] | Feedforward and backpropagation neural network are used to detect lung diseases | X-ray image | Multiclass classification | Accuracy = 92% |
| Cicero et al. [18] | GoogLeNet is used to classify images between normal and abnormal | X-ray image | Binary classification |
For class normal: sensitivity = 91%, specificity = 91% AUC = 0.964 For class abnormal: sensitivity, specificity, and AUC, respectively, were between 78%, 78%, 0.861 and 91%, 91%, 0.962 |
| Dong et al. [19] | VGG-16 and ResNet101 are used for binary (normal vs. abnormal) and multiple disease classification | X-ray image | Binary and multiclass classification | VGG-16: accuracy = 82.2%, AUC of 0.88 Resnet-101: accuracy = 90% |
| Rajpurkar et al. [20] | An algorithm named CheXNet with 121-layer convolutional neural network is used to detect pneumonia | X-ray image | Multiclass classification | F1 score = 95% |
| Islam et al. [21] | Ensemble DCCN | X-ray image | Binary classification | Accuracy: 93.0% for cardiomegaly detection and 90% for tuberculosis detection |
| Madani et al. [22] | Generative adversarial networks (GANs) are used to classify images between normal and abnormal | X-ray image | Binary classification | Accuracy = 84.19% |
| Rajaraman et al. [23] | A customized VGG16 is used for detection and classification of the pneumonia disease between bacterial and viral | X-ray image | Binary classification | Accuracy between 93.6% and 96.2% |
| Ausawalaithong et al. [24] | DenseNet-121 is used for lung cancer prediction | X-ray image | Binary classification |
Accuracy = 74.43% Specificity = 74.96% Sensitivity = 74.68% |
| Correa et al. [25] | Detection of pneumonia based on feedforward neural network | Ultrasound image | Binary classification |
Specificity = 100% Sensitivity = 90.9% |
| Gu et al. [26] | Deep convolutional neural network features and manual features are fused together and are put into support vector machines classifier | X-ray image | Binary classification |
Accuracy = 80.48% Sensitivity = 77.55% |
| Ke et al. [27] | Neuro-heuristic approach is used to detect lung diseases | X-ray image | Multiclass classification |
Accuracy = 79.06% Sensitivity = 84.22% Specificity = 66.7% |
| Saraiva et al. [28] | Neural network was used to train the model, and cross-validation was used for the validation of the model | X-ray image | Binary classification | Accuracy = 95.30% |
| Varshni et al. [29] | Xception, VGG16, VGG-19, ResNet-50, DenseNet-121, and DenseNet-169, were used followed by different classifiers including random forest, K-nearest neighbors, Naive Bayes, and support vector machine | X-ray image | Binary classification | Resnet-50 + SVM are the best one with an AUC = 0.7749 |
| Siddiqi et al. [30] | The model performs the ‘normal’ versus ‘pneumonia’ classification using customized sequential convolutional neural network | X-ray image | Binary classification |
Accuracy = 94.39 Sensitivity = 99% Specificity = 86% |
| Ayan and Ünver [31] | Xception and VGG16 are used for diagnosing of pneumonia (normal vs. pneumonia) | X-ray image | Binary classification | Accuracy = 87% For VGG16 and 82% For Xception |
| Sirazitdinov et al. [32] | Ensemble of mask RCNN and RatinaNet | X-ray image | Binary classification |
Precision = 0.75 Recall = 0.79 F-1 score = 0.77 |
| Liang and Zheng [33] | A customized CNN is used with 49 convolutional layers and 2 dense layers for classification of children’s lung patterns | X-ray image | Binary classification | F1 score = 92.7% |
| Bozickovic et al. [34] | ResNet50, InceptionV3, Xception, and InceptionResNet_V2 pretrained models are evaluated: for 4-class problem (normal, viral, bacterial, and COVID-19) | X-ray image | Multiclass classification |
Accuracy of: Resnet50 = 89.97%, InceptionResNet_V2 = 87.96% Xception = 89.48% Inception_V3 = 87.96% |
| Abbas et al. [35] | Classification of covid-19 chest X-ray images using CNN features of pre-trained models and ResNet + decompose, transfer, and compose (DeTraC) | X-ray image | Binary classification |
Accuracy = 95.12% Sensitivity = 97.91% Specificity = 91.87% Precision = 93.36% |
| Wang and Wong [36] | Covid-Net: lightweight residual projection expansion-projection-extension (PEPX) design pattern | X-ray image | Binary classification | Accuracy = 92.4% |
| [37] | Deep features from ResNet50 + SVM classification | X-ray image | Binary classification | Resnet50 + SVM accuracy = 95.38%, F1 = 95.52% |
| Apostolopoulos et al. [38] | Automatic detection from x-ray images using different fine-tuned models: VGG19, MobileNet, inception, InceptionResNet_V2, and Xception. | X-ray image | Binary classification |
Accuracy VGG19 is the highest: Accuracy = 98.75% Sensitivity = 92.85% Specificity = 98.75% |
| Butt et al. [39] | Comparison of multiple convolutional neural network (CNN) models in order to classify computed tomography samples with covid-19, influenza viral pneumonia, or no-infection. | X-ray image | Multiclass classification | Accuracy = 86.7% |
| Narin et al. [40] | Classification of chest X-ray images into coronavirus and normal using ResNet50, InceptionV3, and InceptionResNet_V2 | X-ray image | Binary classification |
Resnet50 was the best with Accuracy = 98% Specificity = 100% Precision = 100% |
| Hemdan et al. [41] | Classification of covid-19 in X-ray images based on seven different of deep learning architectures, namely VGG19, DenseNet201, InceptionV3, InceptionResNet_V2, ResNetV2, MobileNet_V2, and Xception | X-ray image | Binary classification | Densenet201 and VGG19 have achieved: accuracy = 90%, F1 score = 0.89 and 0.91 for normal and Covid-19, respectively |
| Zhang et al. [42] | Classification of covid-19 and non-covid-19 based on X-ray dataset that contains 100 images from 70 covid-19 subjects and 1431 images from 1008 non-covid-19 pneumonia subjects | X-ray image | Binary classification |
Sensitivity = 90.00% Specificity = 87.84% |
| Farooq and Hafeez [43] | Differentiating covid-19 cases from other pneumonia cases using chest X-rays based on fine-tune a pre-trained ResNet-50 | X-ray image | Binary classification | Accuracy = 96.23% |
| Maghdid et al. [44] | Simple convolution neural network (CNN) and modified pre-trained AlexNet model are applied on the prepared X-rays and CT scan images dataset | X-ray image | Binary classification | Accuracy > to 98% via pre-trained network and 94.1% accuracy by using the modified CNN |
| Apostolopoulos et al. [45] | MobileNet_V2 is used and trained from scratch to classify pulmonary diseases based on a large-scale dataset of 3905 X-ray images | X-ray image | Binary classification |
Accuracy = 99.18% Sensitivity = 97.36% Specificity = 99.42% |
| Elasnaoui et al. [2] | A comparison of recent deep learning architectures for classification of pneumonia images based on fined-tuned versions of (VGG16, VGG19, DenseNet201, InceptionResNet_V2, Inception_V3, ResNet50, MobileNet_V2, and Xception), and a retraining of a baseline CNN is proposed | X-ray image | Binary classification | Resnet50 shows highly satisfactory performance > to 96% of accuracy) |
| Elasnaoui et al. [2] | This work conducts a comparative study of the recent deep learning models (VGG16, VGG19, DenseNet201, InceptionResNet_V2, Inception_V3, ResNet50, and MobileNet_V2) to deal with detection and classification of coronavirus pneumonia (bacterial pneumonia, coronavirus, and normal) | X-ray image | Multiclass classification | (92.18% accuracy for InceptionResNet_V2 and 88.09% Accuracy for Densnet201) |
| Habib et al. [46] | CheXNet with 121-layer convolutional neural network andVGG-19 are used for the ensemble | X-ray image | Binary classification | Accuracy = 98.93% |
| Chouhan et al. [47] | Ensemble of different deep learning algorithms (Alexnet, Inception_V3, Resnet, GoogleNet, and Densenet-121) | X-ray image | Binary classification |
Accuracy = 96.4% Sensitivity 99.0% |
(1). Chest X-ray is one of the most common medical imaging modalities used to detect respiratory system diseases. This observation is extended in [12, 13]. (2). Most studies published are focused on the binary classification based on different DL techniques. (3). Elasnaoui et al. [2], Elasnaoui and Chawki [3], Hemdan et al. [41] are deeply compared seven DL for the classification of pneumonia. (4). Some studies have also developed their own customized architecture and methods [segmentation for example (Pulagam 2016)], independent of well-known DL architectures. (5). Sensitivity, specificity, and accuracy are the criteria utilized in several studies for measuring the efficiency of methods. However, F1 score and area under curve (AUC) have been utilized in some studies to determine the efficiency of the method. (6). These studies found out that InceptionResNet_V2, ResNet50, and MobileNet_V2 gave better accuracy. This observation is also proved in [12, 14]. (7). However, few studies reported the use of multiclass classification (4 classes). (8). No study presented an ensemble learning model in the pneumonia disease for multiclass classification. (9). No study reported a comparison between single and ensemble models for multiclass classification.
Methodology
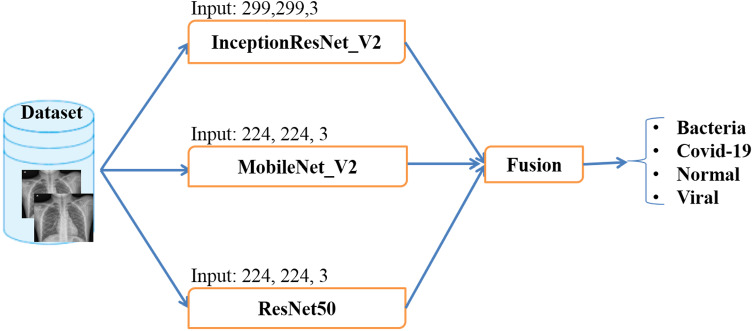
In this section, the methodology used in this study is discussed. According to Table 1 and [12, 14], InceptionResNet_V2, ResNet50, and MobileNet_V2 models give more than 90% of accuracy. Motivated by this conclusion, we compared these pre-trained models for multiclass classification (4 classes) once they have been fine-tuned on a joined image dataset. The steps to do this research are divided into several sections which are shown in Fig. 1. The following sections give out in detail the steps of the proposed methodology.
Fig. 1.
Flow diagram of proposed methodology
Preprocessing dataset of the study
We instantiated the present work with two publicly available image datasets, chest X-ray, and CT dataset [9] and Covid Chest X-ray Dataset [10]. The first one is composed of 5856 images with three classes (1493 viral pneumonia, 2780 bacterial pneumonia, and 1583 normal), while the second one is containing 231 Covid-19 Chest X-ray images. We joined the second dataset to the first one to form a joint dataset which composed of four classes given as follows: bacteria, covid-19, normal, and viral. Finally, as depicted in Table 2, the joined dataset is composed of 6087 images (jpeg format).
Table 2.
Dataset structure
When analyzing several data, a natural question arises on how to efficiently use it. As known, condition of data collected from any area may be contaminated by numerous factors such as sensor/human errors. Using directly such data by the algorithm may conduct to unreliable results. Thus, the next stage is to preprocess input data. The motivation behind data preprocessing is to eliminate or decrease noise present in the original input data, to improve data quality, etc. In the present work, intensity normalization and Contrast Limited Adaptive Histogram Equalization (CLAHE) [2, 3] are used to provide clean data for a successful classification.
Data augmentation
Medical imaging datasets are limited in size due to privacy laws, high cost of obtaining annotations, and considerations [22]. Data augmentation is used for the training process after dataset pre-processing and splitting and has the goal to enrich the data in data-limited scenarios and avoid the risk of overfitting [2, 3, 22]. Moreover, the strategies we used include geometric transforms such as rescaling, rotations, shifts, shears, zooms, and flips [2, 3]. Practically, the images are randomly rotated, shifted vertically or horizontally by a maximum of 90 and 0.2, respectively. Shear and zoom range are set to 0.2 and horizontal flip set to true. Finally, a scale image from integers 0–255 to floats 0–1 is employed. By this way, the models used in this study avoid the risk of over-fitting and learn to be robust to position and orientation variance.
Transfer learning
Research studies in computer vision before 2010 were focused on feature extraction using different techniques such as color [48], texture [49], shape [50]. However, these techniques gradually disappeared between 2010 and 2012 due to the appearance of DL techniques such as convolutional neural networks (CNNs). DL models are highly used for the diagnosis of pneumonia since 2016 [9, 51]. Although the DL models have shown huge achievement in terms of success in medical imaging, they require a large amount of data, which is not yet available in the medical imaging domain due to privacy laws, high cost of obtaining annotations, and considerations [2, 3, 22]. Following the context of non-availability of medical imaging datasets, we use transfer learning (TF). TF is a machine learning technique where we reused a pre-trained model from ImageNet and transfer the learned model into a new model to be trained. In this study, and according to Table 1 and [12, 14], the pre-trained models InceptionResNet_V2, ResNet50, and MobileNet_V2 give more than 90% of accuracy. Following this conclusion, we used these pre-trained models instead of training them from scratch on a small dataset. The following subsections present a brief description of these pre-trained models.
InceptionResNet_V2
InceptionResNet_V2 is a convolutional neural network that is trained on more than a million images from the ImageNet database [52]. It is a hybrid technique combining the inception structure and the residual connection. The model accepts images of 299 × 299 image, and its output is a list of estimated class probabilities. The advantages of InceptionResNet_V2 are converting inception modules to Residual Inception blocks, adding more Inception modules and adding a new type of Inception module (Inception-A) after the Stem module.
ResNet50
ResNet50 is a deep residual network developed by He et al. [53] and is a subclass of convolutional neural networks used for image classification. It is the winner of ILSVRC 2015. The principal innovation is the introduction of the new architecture network-in-network using residual layers. The ResNet50 consists of five steps each with a convolution and identity block; each convolution block and each identity block have 3 convolution layers. ResNet50 has 50 residual networks and accepts images size of 224 × 224 pixels.
MobileNet_V2
MobileNet_V2 [54] is a convolutional neural network being an improved version of MobileNet_V1. It is made of only 54 layers and has an input image size of 224 × 224. Its main characteristic is instead of performing a 2D convolution with a single kernel, instead of performing a 2D convolution with a single kernel. It uses depthwise separable convolutions that consist in applying two 1D convolutions with two kernels. That means, less memory and parameters are required for training leading to a small and efficient model. We can distinguish two types of blocks: first one is residual block with stride of 1; second one is block with stride of 2 for downsizing. For each block, there are three layers: the first layer is 1 × 1 convolution with ReLU6, the second layer is the depthwise convolution, and the third layer is another 1 × 1 convolution but without any nonlinearity.
Training and classification
After data pre-processing, splitting, and data augmentation techniques used, our training dataset size is increased and ready to be passed to the feature extraction step with the proposed models in order to extract the appropriate and pertinent features. The extracted features from each proposed model are flattened together to create the last layer of fully connected and then to classify each image into corresponding classes. Moreover, the single models that comprise the ensemble are trained independently to solve the given task. The last output of the ensemble model is an average/fusion of the various outputs given by the single model. Furthermore, ensemble models reduce the variance of predictions and generalization error and significantly improve the computational training and could be utilized with a few training data [8]. Finally, the performance of single and ensemble models is evaluated on test images using the trained model [2, 3].
Experiment material and parameterization
This section presents experiment settings and performance measure employed in this study in order to predict pneumonia disease using single and ensemble model. We note that single model refers to one model (for example InceptionResNet_V2) trained independently and predicted the output result, while ensemble model stands to combine more than one single model.
Experiment setup
The experimentations were implemented using Python programming language and were carried out based on the following experimental parameters: We employed for data splitting (hold-out) 80% and 20% of the images for training and testing, respectively. We ensure that the images chosen for testing are not used during training. Moreover, we pre-process input images using two different pre-processing techniques (intensity normalization and Contrast Limited Adaptive Histogram Equalization (CLAHE)) [2, 3]. To train the deep transfer learning models, all the images of the dataset were resized to 224 × 224 pixels except those of InceptionResNet_V2 model that were resized to 299 × 299. Furthermore, we set the batch size to 32 with the number of epochs set to 250. β1 = 0.9, β2 = 0.999 are used for Adam optimization, and the learning rate initiated to 0.00001. Furthermore, we employed weight decay and L2-regularizers to reduce over-fitting for the different models. We note that these models are independently trained, and the results of ensemble models are assembled via average/fusion technique. Furthermore, a last dense layer is updated in single and ensemble learning models to output four classes representing bacteria, covid-19, normal, and viral instead of 1000 classes as was utilized for ImageNet. Keras and TensorFlow are used as a deep learning backend. The training and testing steps run using NVIDIA Tesla P40 with 24 Go RAM. Table 3 depicts the parameters used during this study.
Table 3.
Parameterization of the experience
| Parameter name | Value | |
|---|---|---|
| Data splitting | 80% for training (4883 images) and 20% for testing (1181 images) | |
| Input size | 299 × 299 for InceptionResNet_V2 and 224 × 224 for MobileNet_V2 and ResNet50 | |
| Batch size | 32 | |
| Learning rate | 0.00001 | |
| Number of epochs | 250 | |
| Adam optimization | β1 = 0.9, β2 = 0.999 | |
| Number of train samples | 152 | |
| Number of test samples | 36 | |
| Last dense layer | 4 classes | |
| Number of weights | InceptionResNet_V2 |
Total params: 55,125,732 Trainable params: 55,065,188 Non-trainable params: 60,544 |
| ResNet50 |
Total params: 24,769,156 Trainable params: 24,716,036 Non-trainable params: 53,120 |
|
| MobileNet_V2 |
Total params: 5,146,180 Trainable params: 5,112,068 Non-trainable params: 34,112 |
|
Quality assessment
To evaluate the single and ensemble learning models, the present study uses some performance parameters such as: accuracy (ACC), sensitivity (SEN), specificity (SPE), precision (PRE), and F1 score (F1) [2, 3, 12, 14] which are given as follows:
| 1 |
where TN stands to true-negative cases in detection results, while TP denotes the true positive. FP equals the false positive, and FN stands for false negative.
Results and discussion
This section presents and discusses the results obtained based on the experimental setup discussed in the previous section for single and ensemble models.
Results of single model
We firstly present the confusion matrix, accuracy, and loss curves (Figs. 2, 3, 4) given by different deep transfer learning models (InceptionResNet_V2, ResNet50, and MobileNet_V2) and using imbalanced dataset [i.e., most of the datasets containing pneumonia images are class-imbalanced [14] ], then we compared the results of all architectures based on the metrics defined in Eq. (1) in order to determine the best method (Table 2) to use to classify X-ray images between bacteria, covid-19, normal, and viral. The next section shows accuracy and loss curve and confusion matrix of different models used in this study and interpretation of the results obtained. (All figures with high resolution could be find upon request by email to the authors of this study).
InceptionResNet_V2
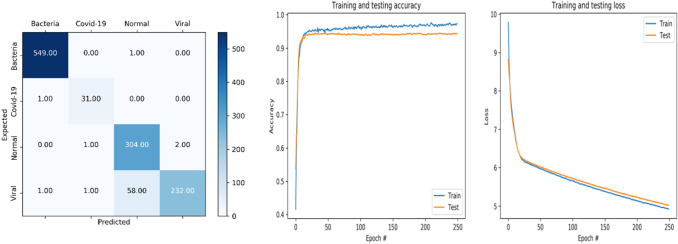
Fig. 2.
Accuracy and loss curve and confusion matrix of InceptionResNet_V2
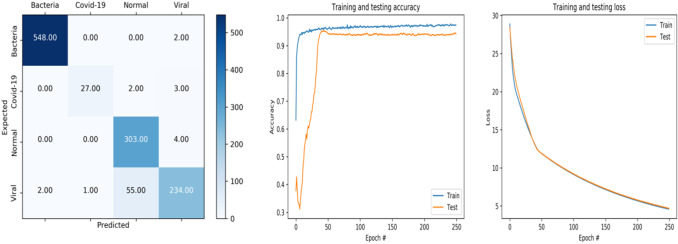
Fig. 3.
Accuracy and loss curve and confusion matrix of MobileNet_V2
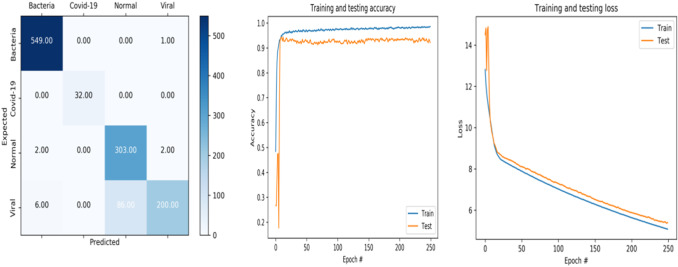
Fig. 4.
Accuracy and loss curve and confusion matrix of ResNet50
We can observe (Fig. 2) that from epoch 0 to 29, the training and testing accuracy is increasing until the value where the accuracies are equal to 95.18% and 94.01% for training and testing, respectively. After this epoch, the accuracies curves become stable and they are equal to 97.46% and 94.27% for training and testing data, respectively.
For the loss curve of training and testing data, an excellent fit is noticed until the epoch 29. Then the values of these curves are converged toward 5.
Regarding the confusion matrix, the InceptionResNet_V2 model was able to correctly identify 549 images as bacteria class, 31 were classified as covid-19, 304 were correctly labeled as normal, and 232 were identified as viral.
MobileNet_V2
The obtained accuracy curve of training data is speedily increasing until the value of 94.68% (Fig. 3). After epoch 11, the training accuracy enters in the stability stage where it is equivalent to 94.16%. For testing accuracy, the curve is increasing until the epoch 40 where the accuracy value is equal to 94.53; then, it becomes stable.
A good fit can be noticed for the training and testing loss curves. Indeed, the curves are quickly decreasing until the end of training.
The confusion matrix depicts that for the bacteria class, MobileNet_V2 model can correctly recognize 548 images, yet 27 were named as covid-19. The model also was able to correctly identify 303 images as normal, and 234 images were marked as viral class.
ResNet50
It is noted that the accuracy of training data is fastly increasing from epoch 0 to 10 where the accuracy is equal to 93.93% (see Fig. 4). Then it gets stable until the end of training where the accuracy is equal to 98.59%. For the testing data, a quick increasing can be seen from epoch 0 to 9 where the value is 92.97%, after that, it begins to be stable.
For the loss curve of training and testing data, the values are decreasing from epoch 0 to end of training.
When we see this confusion matrix, we can say that for the bacteria class, ResNet50 model has the option to correctly recognize 549 images. Moreover, 32 were selected as covid-19. ResNet50 also can correctly recognize 303 images as normal class; thus, 200 images were marked as viral class.
Results for our experiment classification are depicted in Table 4 based on fine-tuned versions of ResNet50, InceptionResNet_V2, and MobileNet_V2. The table details the classification performances across each experiment using confusion matrices for each model. From the results, it is noted that the accuracy and F1 score when we use InceptionResNet_V2 are higher compared with ResNet50 and MobileNet_V2. Moreover, it can be observed that accuracies of ResNet50 and MobileNet_V2 both are equivalents to 93.73%. However, F1 of ResNet50 (93.47%) is higher than MobileNet_V2 (91.62%).
Table 4.
Evaluations metrics for single model
| Model | Class | TP | TN | FN | FP | ACC | SEN | SPE | PRE | F1 |
|---|---|---|---|---|---|---|---|---|---|---|
| InceptionResNet_V2 | Bacteria | 549 | 629 | 1 | 2 | 94.50 | 93.79 | 98.13 | 94.12 | 93.52 |
| Covid-19 | 31 | 1147 | 1 | 2 | ||||||
| Normal | 304 | 815 | 3 | 59 | ||||||
| Viral | 232 | 887 | 60 | 2 | ||||||
| MobileNet_V2 | Bacteria | 548 | 625 | 2 | 6 | 93.73 | 90.29 | 97.83 | 93.91 | 91.62 |
| Covid-19 | 27 | 1148 | 5 | 1 | ||||||
| Normal | 302 | 815 | 5 | 59 | ||||||
| Viral | 230 | 881 | 62 | 8 | ||||||
| ResNet50 | Bacteria | 548 | 628 | 2 | 3 | 93.73 | 93.07 | 97.85 | 94.89 | 93.47 |
| Covid-19 | 31 | 1149 | 1 | 0 | ||||||
| Normal | 302 | 807 | 5 | 67 | ||||||
| Viral | 226 | 885 | 66 | 4 |
Results of ensemble models
In the rest of this study, we focus on ensembles learning to see whether there is an improvement of performance measures [Eqs. (1)]. Toward this end, we constructed 5 ensembles of different deep transfer learning following the architecture given in Fig. 1 and using 2, and 3 DL models, respectively, those that were fully fine-tuned previously. Then, we evaluate them using quality assessment [Eqs. (1)]. The goal behind using ensemble learning is to show if ensemble learning is more accurate than a single model.
Practically, we based on Table 4 and we construct 3 ensembles using each time 2 models (MobileNet_V2 with InceptionResNet_V2), (ResNet50 with InceptionResNet_V2) and (ResNet50 with MobileNet_V2) followed by 1 ensemble of all models (ResNet50 with MobileNet_V2 with InceptionResNet_V2) (Table 5). The next sections present in detail the results obtained by the different ensembles constructed.
Table 5.
Evaluations metrics for ensemble model
| Model | Class | TP | TN | FN | FP | ACC | SEN | SPE | PRE | F1 |
|---|---|---|---|---|---|---|---|---|---|---|
| MobileNet_V2 with InceptionResNet_V2 | Bacteria | 548 | 628 | 2 | 3 | 93.82 | 92.48 | 97.90 | 93.20 | 92.52 |
| Covid-19 | 30 | 1147 | 2 | 2 | ||||||
| Normal | 298 | 816 | 9 | 58 | ||||||
| Viral | 232 | 879 | 60 | 10 | ||||||
| ResNet50 with InceptionResNet_V2 | Bacteria | 548 | 623 | 2 | 8 | 93.65 | 92.31 | 97.79 | 93.13 | 92.43 |
| Covid-19 | 30 | 1147 | 2 | 2 | ||||||
| Normal | 296 | 819 | 11 | 55 | ||||||
| Viral | 232 | 879 | 60 | 10 | ||||||
| ResNet50 with MobileNet_V2 | Bacteria | 548 | 630 | 2 | 1 | 95.17 | 92.47 | 98.37 | 95.46 | 93.79 |
| Covid-19 | 28 | 1149 | 4 | 0 | ||||||
| Normal | 294 | 835 | 13 | 39 | ||||||
| Viral | 254 | 872 | 38 | 17 | ||||||
| ResNet50 with MobileNet_V2 with InceptionResNet_V2 | Bacteria | 549 | 628 | 1 | 3 | 95.09 | 94.43 | 98.31 | 95.53 | 94.84 |
| Covid-19 | 31 | 1149 | 1 | 0 | ||||||
| Normal | 295 | 831 | 12 | 43 | ||||||
| Viral | 248 | 877 | 44 | 12 |
Discussion
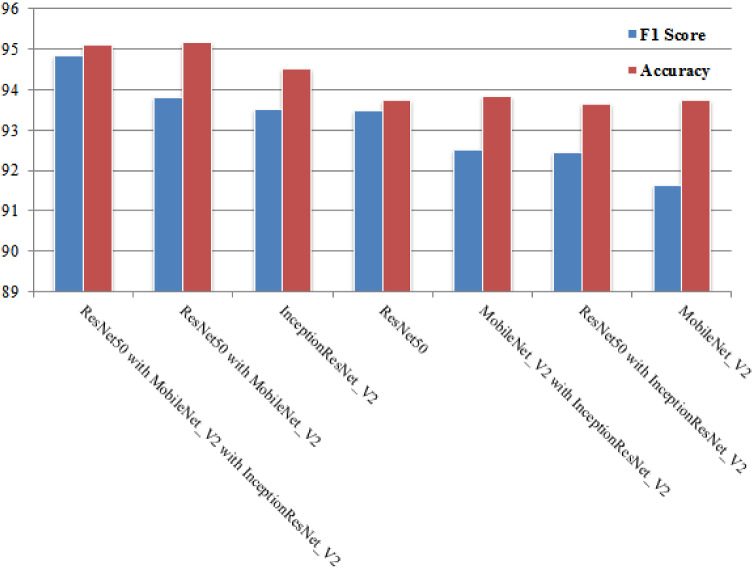
In this study, we investigated the multiclass classification of X-ray images using single and ensemble learning models, in order to identify the best performing architecture based on the several parameters defined in Eq. 1. We note that accuracy is utilized when the TP and TN are more important, while F1 score is utilized when the FN and FP are crucial. Accuracy can be utilized when the class distribution is similar, whereas F1 score is a better metric when there are imbalanced classes. Following this context, this comparison between single and ensemble learning models is based on F1 score since our dataset is highly imbalanced. Figure 5 summarizes F1 score and accuracy obtained during this study for single and ensemble models.
Fig. 5.
Performance measure of single and ensemble models
The findings of this study from Fig. 5 are:
(RQ1): What is the diagnostic accuracy that DL can attain based on X-ray images?
From this study, we can conclude that the results are highly satisfactory. Nevertheless, we observed that InceptionResNet_V2 gives best results (93.52% of F1 score) regardless of the dataset used. In addition, InceptionResNet_V2 has been proven to obtain remarkable results in related tasks [2].
(RQ2): Is combining DL to construct ensembles DL will enhance the final accuracy of certain model?
The analysis of the results depicted in Fig. 5, tells us that there is a slight improvement of accuracy for different ensembles. Since our dataset is highly imbalanced, we focus on F1 score. Indeed, ensemble of (ResNet50 with MobileNet_V2 with InceptionResNet_V2) performs better than single and ensemble models.
(RQ3): Does the number of DL combined to construct ensembles DL affect the accuracy of the model?
There is no strong evidence to prove that the number of DL combined to construct ensembles DL affects the accuracy of the model. In fact, the results are influenced by the type of the ensemble used. As it can be seen in Fig. 5, the ensemble of three models (ResNet50 with MobileNet_V2 with InceptionResNet_V2) is more accurate followed by ensemble (ResNet50 with MobileNet_V2). Moreover, InceptionResNet_V2 performs better than ensembles (MobileNet_V2 with InceptionResNet_V2) and (ResNet50 with InceptionResNet_V2).
Furthermore, Table 6 illustrates the execution time in second for different models tested along this study. For InceptionResnet_V2, the elapsed time for training was 52,586.62 s. ResNet50 has required 31,381.87 s for training, while MobileNet_V2 necessitates 32,976.83 for training.
Table 6.
Computation time
| Model | Training time (s) |
|---|---|
| InceptionResNet_V2 | 52,586.62 |
| MobileNet_V2 | 32,976.83 |
| ResNet50 | 31,381.87 |
| ResNet50 with MobileNet_V2 with InceptionResNet_V2 | 78.86 |
| ResNet50 with InceptionResNet_V2 | 69.63 |
| MobileNet_V2 with InceptionResNet_V2 | 67.89 |
| ResNet50 with MobileNet_V2 | 49.55 |
From Table 6, we observe that IncpetionResNet_V2 even it gives a good result it is not fast because it takes 52,586.62 in training followed by MobileNet_V2. In addition, we notice that ensemble ResNet50 with MobileNet_V2 is fast and provides good results (93.79% of F1 score). In the medical field, the scientist has the choice between the F1 score and the computation time to finally select the DL technique to use. According to [2], the F1 score of the DL techniques stays major selection criteria. But combining both F1 score and computation time remain a good benefit. Consequently, this study shows that ensemble (ResNet50 with MobileNet_V2) can be a good solution to classify X-ray images into 4 classes which are: bacteria, covid-19, normal, and viral.
Threats to validity
The goal of any scientific study is to produce generalizable knowledge about the reality. Without internal and external validity, we cannot apply results obtained from the experiments to the real world. The validity of any research study refers to how well the results obtained represent true findings among similar results outside the study. The validity of the present research study includes internal and external validity.
Internal validity
Threats to internal validity are defined as the extent to which the found results represent the truth in the field we are studying and, thus, are not due to methodological errors. In this study, the goal is to see if ensemble learning is more accurate than a single model by investigating several parameters. Threats to internal validity of this study may concern the criteria utilized to evaluate the model performance. The findings of the present study are mainly based on F1 score since the dataset is highly imbalanced. The choice of deep learning models may be another threat.
External validity
Threats to external validity are the extent to which we generalize the findings, the results and experimental design of a study to other situations. In this study, we used a joined dataset and varying number of models that constitute an ensemble. Some published works in other medical science evaluated their proposed ensembles with only one dataset [55, 56]. However, it will be a good benefit to replicate this study using more datasets, more sophisticated feature extraction techniques.
Conclusion
We reported in this work a classification of chest X-ray images into 4 classes which are: bacteria, covid-19, normal, and viral using single and ensemble learning models based on fine-tuned models (InceptionResNet_V2, ResNet50, and MobileNet_V2). The main goal is to answer the research questions (RQ) defined as follows:
(RQ1): What is the diagnostic accuracy that DL can attain based on X-ray images?
(RQ2): Is combining DL to construct ensembles DL will enhance the final accuracy of certain model?
(RQ3): Does the number of DL combined to construct ensembles DL affect the accuracy of the model?
As a result, for a single model, we found out that InceptionResNet_V2 gives 93.52% of F1 score. Besides, ensemble of 3 models (ResNet50 with MobileNet_V2 with InceptionResNet_V2) performs better than other ensembles constructed (94.84% of F1 score). Moreover, there is no strong evidence to prove that the number of DL combined to construct ensembles DL affects the accuracy of the model.
Future work intends to develop a full system for pneumonia by combining deep learning and feature extraction using different techniques such as color [48], texture [49], shape [50], Ouhda [57]). In addition, the performance may be improved using more datasets, more sophisticated feature extraction techniques; also other fusion approaches would be interesting.
Acknowledgements
We thank the reviewer for his/her thorough review and highly appreciate the comments, corrections, and suggestions that ensued, which significantly contributed to improving the quality of the publication.
Compliance with ethical standards
Conflicts of interest
We declare that we have no conflicts of interest to disclose. Author has no received research grants from any company.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Orbann C, Sattenspiel L, Miller E, Dimka J. Defining epidemics in computer simulation models: how do definitions influence conclusions? Epidemics. 2017;19:24–32. doi: 10.1016/j.epidem.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Elasnaoui K, Chawki Y, Radeva P, Idri A (2020) Automated methods for detection and classification pneumonia based on X-ray images using deep learning. arXiv preprint arXiv:2003.14363
- 3.Elasnaoui K, Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J Biomol Struct Dyn. 2020 doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zerouaoui H, Idri A, El Asnaoui K (2020) Machine learning and image processing for breast cancer: a systematic map. In: World conference on information systems and technologies. Springer, Cham, pp 44–53
- 5.Ouhda M, El Asnaoui K, Ouanan M, Aksasse B (2017) Content-based image retrieval using convolutional neural networks. In: First international conference on real time intelligent systems. Springer, Cham, pp 463–476
- 6.Janghel RR, Shukla A, Sharma S, Gnaneswar AV (2014) Evolutionary ensemble model for breast cancer classification. In: International conference in swarm intelligence. Springer, Cham, pp 8–16
- 7.Kwon H, Park J, Lee Y. Stacking ensemble technique for classifying breast cancer. Healthc Inf Res. 2019;25(4):283–288. doi: 10.4258/hir.2019.25.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zilly J, Buhmann JM, Mahapatra D. Glaucoma detection using entropy sampling and ensemble learning for automatic optic cup and disc segmentation. Comput Med Imaging Graph. 2017;55:28–41. doi: 10.1016/j.compmedimag.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Kermany DS, Zhang K, Goldbaum M (2018) Labeled optical coherence tomography (oct) and chest X-ray images for classification. 10.17632/rscbjbr9sj.2
- 10.Cohen JP, Morrison P, Dao L (2020) COVID-19 image data collection. arXiv:2003.11597. https://github.com/ieee8023/covid-chestxray-dataset
- 11.Taghanaki SA, Das A, Hamarneh G (2018) Vulnerability analysis of chest X-ray image classification against adversarial attacks. In: Understanding and interpreting machine learning in medical image computing applications. Springer, Cham, pp 87–94
- 12.Ghaderzadeh M, Asadi F (2020) Deep learning in detection and diagnosis of covid-19 using radiology modalities: a systematic review. arXiv preprint arXiv:2012.11577 [DOI] [PMC free article] [PubMed]
- 13.Guendel S, Grbic S, Georgescu B, Liu S, Maier A, Comaniciu D (2018) Learning to recognize abnormalities in chest x-rays with location-aware dense networks. In: Iberoamerican congress on pattern recognition. Springer, Cham, pp 757–765
- 14.Khan W, Zaki N, Ali L (2020) Intelligent pneumonia identification from chest X-rays: a systematic literature review. medRxiv
- 15.Barrientos R, Roman-Gonzalez A, Barrientos F, Solis L, Correa M et al (2016) Automatic detection of pneumonia analyzing ultrasound digital images. In: 2016 IEEE 36th Central American and Panama Convention (CONCAPAN XXXVI). IEEE, pp 1–4
- 16.Ahmad WSHMW, Zaki WMDW, Fauzi MFA, Tan WH (2016) Classification of infection and fluid regions in chest X-ray images. In: 2016 international conference on digital image computing: techniques and applications (DICTA). IEEE, pp 1–5
- 17.Khobragade S, Tiwari A, Patil CY, Narke V (2016) Automatic detection of major lung diseases using chest radiographs and classification by feed-forward artificial neural network. In: 2016 IEEE 1st international conference on power electronics, intelligent control and energy systems (ICPEICES). IEEE, pp 1–5
- 18.Cicero M, Bilbily A, Colak E, Dowdell T, Gray B, Perampaladas K, Barfett J. Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol. 2017;52(5):281–287. doi: 10.1097/RLI.0000000000000341. [DOI] [PubMed] [Google Scholar]
- 19.Dong Y, Pan Y, Zhang J, Xu W (2017) Learning to read chest X-ray images from 16000 + examples using CNN. In: 2017 IEEE/ACM international conference on connected health: applications, systems and engineering technologies (CHASE). IEEE, pp 51–57
- 20.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T et al (2017) Chexnet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225
- 21.Islam MT, Aowal MA, Minhaz AT, Ashraf K (2017) Abnormality detection and localization in chest X-rays using deep convolutional neural networks. arXiv preprint arXiv:1705.09850
- 22.Madani A, Moradi M, Karargyris A, Syeda-Mahmood T (2018) Chest X-ray generation and data augmentation for cardiovascular abnormality classification. In: Medical imaging 2018: image processing, vol 10574. International Society for Optics and Photonics, p 105741 M
- 23.Rajaraman S, Candemir S, Kim I, Thoma G, Antani S. Visualization and interpretation of convolutional neural network predictions in detecting pneumonia in pediatric chest radiographs. Appl Sci. 2018;8(10):1715. doi: 10.3390/app8101715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ausawalaithong W, Thirach A, Marukatat S, Wilaiprasitporn T (2018) Automatic lung cancer prediction from chest X-ray images using the deep learning approach. In: 2018 11th biomedical engineering international conference (BMEiCON). IEEE, pp 1–5
- 25.Correa M, Zimic M, Barrientos F, Barrientos R, Román-Gonzalez A, Pajuelo MJ, et al. Automatic classification of pediatric pneumonia based on lung ultrasound pattern recognition. PLoS ONE. 2018;13(12):e0206410. doi: 10.1371/journal.pone.0206410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gu X, Pan L, Liang H, Yang R (2018) Classification of bacterial and viral childhood pneumonia using deep learning in chest radiography. In: Proceedings of the 3rd international conference on multimedia and image processing, pp 88–93
- 27.Ke Q, Zhang J, Wei W, Połap D, Woźniak M, Kośmider L, Damaševĭcius R. A neuro-heuristic approach for recognition of lung diseases from X-ray images. Expert Syst Appl. 2019;126:218–232. doi: 10.1016/j.eswa.2019.01.060. [DOI] [Google Scholar]
- 28.Saraiva AA, Ferreira NMF, de Sousa LL, Costa NJC, Sousa JVM, Santos DBS et al (2019) Classification of images of childhood pneumonia using convolutional neural networks. In: BIOIMAGING, pp 112–119
- 29.Varshni D, Thakral K, Agarwal L, Nijhawan R, Mittal A (2019) Pneumonia detection using CNN based feature extraction. In: 2019 IEEE international conference on electrical, computer and communication technologies (ICECCT). IEEE, pp 1–7
- 30.Siddiqi R (2019) Automated pneumonia diagnosis using a customized sequential convolutional neural network. In: Proceedings of the 2019 3rd international conference on deep learning technologies, pp 64–70
- 31.Ayan E, Ünver HM (2019) Diagnosis of pneumonia from chest X-ray images using deep learning. In: 2019 scientific meeting on electrical-electronics & biomedical engineering and computer science (EBBT). IEEE, pp 1–5
- 32.Sirazitdinov I, Kholiavchenko M, Mustafaev T, Yixuan Y, Kuleev R, Ibragimov B. Deep neural network ensemble for pneumonia localization from a large-scale chest X-ray database. Comput Electr Eng. 2019;78:388–399. doi: 10.1016/j.compeleceng.2019.08.004. [DOI] [Google Scholar]
- 33.Liang G, Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed. 2020;187:104964. doi: 10.1016/j.cmpb.2019.06.023. [DOI] [PubMed] [Google Scholar]
- 34.Bozickovic J, Lazic I, Turukalo TL (2020) Pneumonia detection and classification from X-ray images—a deep learning approach
- 35.Abbas A, Abdelsamea MM, Gaber MM (2020) Classification of COVID-19 in chest X-ray images using DeTraC deep convolutional neural network. arXiv preprint arXiv:2003.13815 [DOI] [PMC free article] [PubMed]
- 36.Wang L, Wong A (2020) COVID-Net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images. arXiv preprint arXiv:2003.09871 [DOI] [PMC free article] [PubMed]
- 37.Sethy PK, Behera SK (2020) Detection of coronavirus disease (covid-19) based on deep features
- 38.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43:635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butt C, Gill J, Chun D, et al. Deep learning system to screen coronavirus disease 2019 pneumonia. Appl Intell. 2020 doi: 10.1007/s10489-020-01714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narin A, Kaya C, Pamuk Z (2020) Automatic detection of coronavirus disease (covid-19) using X-ray images and deep convolutional neural networks. arXiv preprint arXiv:2003.10849 [DOI] [PMC free article] [PubMed]
- 41.Hemdan EED, Shouman MA, Karar ME (2020) Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv preprint arXiv:2003.11055
- 42.Zhang J, Xie Y, Li Y, Shen C, Xia Y (2020) Covid-19 screening on chest X-ray images using deep learning based anomaly detection. arXiv preprint arXiv:2003.12338
- 43.Farooq M, Hafeez A (2020) Covid-resnet: a deep learning framework for screening of covid19 from radiographs. arXiv preprint arXiv:2003.14395
- 44.Maghdid HS, Asaad AT, Ghafoor KZ, Sadiq AS, Khan MK (2020) Diagnosing COVID-19 pneumonia from X-ray and CT images using deep learning and transfer learning algorithms. arXiv preprint arXiv:2004.00038
- 45.Apostolopoulos I, Aznaouridis S, Tzani M (2020) Extracting possibly representative COVID-19 biomarkers from X-ray images with deep learning approach and image data related to pulmonary diseases. arXiv preprint arXiv:2004.00338 [DOI] [PMC free article] [PubMed]
- 46.Habib N, Hasan MM, Reza MM, Rahman MM. Ensemble of CheXNet and VGG-19 feature extractor with random forest classifier for pediatric pneumonia detection. SN Comput Sci. 2020;1(6):1–9. doi: 10.1007/s42979-020-00373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chouhan V, Singh SK, Khamparia A, Gupta D, Tiwari P, Moreira C, et al. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl Sci. 2020;10(2):559. doi: 10.3390/app10020559. [DOI] [Google Scholar]
- 48.El Asnaoui K, Chawki Y, Aksasse B, Ouanan M. A new color descriptor for content-based image retrieval: application to coil-100. J Digit Inf Manag. 2015;13(6):473. [Google Scholar]
- 49.El Asnaoui K, Chawki Y, Aksasse B, Ouanan M. Efficient use of texture and color features in content-based image retrieval (CBIR) Int J Appl Math Stat. 2016;54(2):54–65. [Google Scholar]
- 50.Chawki Y, El Asnaoui K, Ouanan M, Aksasse B. Content frequency and shape features based on CBIR: application to color images. Int J Dyn Syst Differ Eqn. 2018;8(1–2):123–135. [Google Scholar]
- 51.Bhandary A, Prabhu GA, Rajinikanth V, Thanaraj KP, Satapathy SC, Robbins DE, Shasky C, Zhang YD, Tavares JMRS, Raja NSM. Deep-learning framework to detect lung abnormality—a study with chest X-ray and lung CT scan images. Pattern Recogn Lett. 2020;129:271–278. doi: 10.1016/j.patrec.2019.11.013. [DOI] [Google Scholar]
- 52.Szegedy C, Ioffe S, Vanhoucke V, Alemi A (2016) Inception-v4, inception-ResNet and the impact of residual connections on learning, rXiv:1602.07261
- 53.He K, Zhang X, Ren S, Sunet J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition CVPR’2016, pp 770–778
- 54.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C (2018) MobileNetV2: inverted residuals and linear bottlenecks. In: Proceeding of the IEEE conference on computer vision and pattern recognition (CVPR), Salt Lake City, UT, pp 4510–4520
- 55.Braga PL, Oliveira AL, Ribeiro GH, Meira SR (2007) Bagging predictors for estimation of software project effort. In: 2007 international joint conference on neural networks. IEEE, pp 1595–1600
- 56.Azzeh M, Nassif AB, Minku LL. An empirical evaluation of ensemble adjustment methods for analogy-based effort estimation. J Syst Softw. 2015;103:36–52. doi: 10.1016/j.jss.2015.01.028. [DOI] [Google Scholar]
- 57.Ouhda M, El Asnaoui K, Ouanan M, Aksasse B (2017) Using image segmentation in content-based image retrieval method. In: International conference on advanced information technology, services and systems. Springer, Cham, pp 179–195