SUMMARY
One key aspect of epigenetic inheritance is that chromatin structures can be stably inherited through generations after the removal of the signals that establish such structures. In fission yeast, the RNA interference (RNAi) pathway is critical for the targeting of histone methyltransferase Clr4 to pericentric repeats to establish heterochromatin. However, pericentric heterochromatin cannot be properly inherited in the absence of RNAi, suggesting the existence of mechanisms that counteract chromatin structure inheritance. Here, we show that mutations of components of the INO80 chromatin-remodeling complex allow pericentric heterochromatin inheritance in RNAi mutants. The ability of INO80 to counter heterochromatin inheritance is attributed to one subunit, Iec5, which promotes histone turnover at heterochromatin but has little effects on nucleosome positioning at heterochromatin, gene expression, or the DNA damage response. These analyses demonstrate the importance of the INO80 chromatin-remodeling complex in controlling heterochromatin inheritance and maintaining the proper heterochromatin landscape of the genome.
Graphical Abstract

In Brief
Shan et al. report that the INO80 chromatin-remodeling complex regulates heterochromatin inheritance in wild-type cells and RNAi mutants. Further characterization of the INO80 complex demonstrates that the Iec5 subunit counteracts heterochromatin inheritance through regulating histone turnover but not nucleosome positioning at heterochromatin during DNA replication.
INTRODUCTION
Eukaryotic genomic DNA is folded with histone and non-histone proteins into chromatin. Covalent modifications of histones, chromatin remodeling, and DNA modifications have important roles in setting up the gene expression programs that determine cell identity. Once these chromatin states have been established, they form an epigenetic memory of gene activity that is inherited by subsequent generations of cells (Campos et al., 2014; Moazed, 2011).
Histones have major roles in transmitting epigenetic information (Serra-Cardona and Zhang, 2018; Stewart-Morgan et al., 2020). During DNA replication, the passage of the replication fork disrupts parental nucleosomes. Parental (H3–H4)2 tetramers, which contain original histone modification marks, are deposited at the original location and to both daughter strands to direct the formation of nucleosomes. Because the DNA content increases 2-fold, nucleosomes containing parental (H3–H4)2 cover only half of the replicated DNA. The remaining gaps in the DNA are filled by nucleosomes formed with newly synthesized (H3–H4)2, resulting in the intermingling of nucleosomes containing parental and new (H3–H4)2. The existing histone modifications on parental histones often recruit enzymes responsible for such modifications, leading to modifications of nearby nucleosomes, thereby restoring the original histone modification profiles on both replicated DNA strands. However, the mechanistic details of histone-modification inheritance are still not well understood.
One of the best-studied examples of the inheritance of chromatin states is heterochromatin in the fission yeast Schizosaccharomyces pombe. In that organism, large domains of heterochromatin are formed at the pericentric region, subtelomeres, and the silent mating-type region (Grewal and Jia, 2007). The nucleosomes within those regions all contain methylated histone H3 lysine 9 (H3K9me), which is catalyzed by the histone methyltransferase Clr4 (Nakayama et al., 2001; Rea et al., 2000). In addition to the catalytic SET domain, Clr4 contains a chromodomain that binds H3K9me (Zhang et al., 2008). Once Clr4 is recruited to specific genomic loci to initiate H3K9 methylation, repeated cycles of Clr4 binding to H3K9me and the methylation of adjacent nucleosomes on H3K9 leads to the formation of large H3K9me domains. Such a self-propagating mechanism is also proposed to aid the restoration of H3K9me during DNA replication, with parental histones containing H3K9me serving as seeds for the restoration of the entire H3K9me domain (Allshire and Madhani, 2018; Zhang et al., 2008).
The RNA interference (RNAi) pathway is critical for the recruitment of Clr4 to repetitive DNA elements to establish heterochromatin (Grewal and Jia, 2007; Martienssen and Moazed, 2015). The DNA repeats are transcribed, producing double-stranded RNAs (dsRNAs) with the help of an RNA-dependent RNA polymerase complex (RDRC). The ribonuclease Dicer (Dcr1) processes those dsRNAs into small interfering RNAs (siRNAs), which are loaded onto the RNAi-induced transcriptional silencing complex (RITS) with the help of the Argonaute siRNA chaperone complex (ARC). The Argonaute protein (Ago1) within RITS binds siRNAs and targets RITS to nascent RNA transcripts from repeat regions. RITS then recruits the CLRC complex, which contains the H3K9 methyltransferase Clr4 to initiate H3K9me. H3K9me recruits chromodomain proteins Swi6 and Chp2, which in turn recruit the SHREC complex containing histone deacetylase Clr3 and chromatin-remodeling protein Mit1 to achieve transcriptional silencing.
At the silent mating-type region, heterochromatin persists after removal of RNAi components or the cenH repeat sequence that initiates RNAi, consistent with the idea that heterochromatin can be inherited through mitosis without the initiation signal (Grewal and Klar, 1996; Hall et al., 2002). However, such an explanation is complicated by the presence of an additional, albeit less-robust, mechanism of heterochromatin nucleation involving DNA binding proteins Atf1/Pcr1, which cooperates with RNAi to recruit Clr4 to the silent mating-type region (Jia et al., 2004; Wang and Moazed, 2017). To avoid complications from possible unknown mechanisms at endogenous heterochromatin loci, ectopic heterochromatin is established by the recruitment of Clr4 to tetO binding sites through a TetR-Clr4 fusion protein (Audergon et al., 2015; Ragunathan et al., 2014). This ectopic heterochromatin is unable to maintain itself after the addition of tetracycline to remove TetR-Clr4 from tetO. Therefore, it seems that cells have mechanisms that prevent histone-based heterochromatin maintenance. Indeed, removal of a JmjC domain protein Epe1 allows the inheritance of this artificial heterochromatin established by TetR-Clr4 even after TetR-Clr4 release (Audergon et al., 2015; Ragunathan et al., 2014). Epe1 is a putative H3K9 demethylase, and it is suggested that active H3K9me removal by Epe1 prevents epigenetic inheritance. However, the enzymatic activity of Epe1 has not been demonstrated in vitro, and the commonly used mutations that are expected to abolish Epe1 demethylase activity actually influence heterochromatin through non-enzymatic functions (Raiymbek et al., 2020). Therefore, the mechanisms that prevent histone-based heterochromatin maintenance in fission yeast remain unclear.
At pericentric repeats, heterochromatin is not properly maintained in RNAi mutants, even though RNAi is expected to be only required for heterochromatin establishment but not its subsequent maintenance (Kagansky et al., 2009; Ragunathan et al., 2014). We hypothesized that if there are pathways that actively prevent heterochromatin maintenance, inactivating such pathways will allow heterochromatin to be inherited, even in the absence of RNAi. We, therefore, performed a genetic screen for mutations that allow pericentric heterochromatin inheritance in RNAi mutants (Tadeo et al., 2013). Here, we describe the identification and characterization of mutations of components of the INO80 chromatin-remodeling complex in regulating heterochromatin inheritance. All the mutations identified regulate the stability of one INO80 subunit Iec5, and iec5Δ does not affect nucleosome positioning but affects histone turnover at heterochromatin. This separation-of-function mutation allows us to demonstrate that INO80 promotes histone turnover to regulate epigenetic inheritance and the heterochromatin landscape across the genome.
RESULTS
Mutations of the INO80 Chromatin Remodeling-Complex Rescue Pericentric Heterochromatin Defects of RNAi Mutants
In fission yeast, heterochromatin integrity can be conveniently measured by the silencing of reporter genes inserted within heterochromatin. For example, a ura4+ reporter gene inserted at the outer repeat region of centromere I (otr::ura4+) is silenced, leading to robust cell growth on medium containing 5-fluoroorotic acid (FOA), which is toxic to cells expressing ura4+ (Allshire et al., 1995). Mutations that compromise heterochromatin, such as dcr1Δ, which deletes a gene encoding the ribonuclease Dicer responsible for generating siRNAs, leads to otr::ura4+ silencing defects and little growth on FOA-containing medium. We have previously introduced the otr::ura4+ reporter and dcr1Δ into the fission yeast-deletion library to identify mutations that allow for heterochromatin inheritance in the absence of RNAi (Figure 1A) (Tadeo et al., 2013). The screen also identified three additional mutations, iec1Δ, hap2Δ, and nht1Δ, which harbor deletions of genes whose products are annotated as components of the INO80 chromatin-remodeling complex (Figure 1B). The subunits that make up the catalytic core of the INO80 chromatin-remodeling complex, including Ino80, Rvb1, Rvb2, Ies2, Ies4, Ies6, Act1, Arp4, Arp5, Arp8, and Taf14, are conserved across species (Morrison and Shen, 2009). Organisms analyzed so far all contain species-specific accessory subunits. The three mutations that we identified are all accessory subunits of the fission yeast INO80 complex (Hogan et al., 2010), and we did not identify any subunits of the conserved INO80 core subunits in our screen.
Figure 1. Identification of INO80 Complex Components as Regulators of Heterochromatin Inheritance.

(A) Schematic diagram of the screen with the fission yeast-deletion library for mutations that allow otr::ura4+ silencing to be inherited in a dcr1Δ background.
(B) Images of sections of cells grown on medium containing 5-FOA. Each square represents quadruplicates of one gene deletion. Red boxes highlight the identified mutations.
(C) Serial dilution analyses of the indicated strains to measure the expression of otr::ura4+ and sensitivity to TBZ.
(D) ChIP qPCR analyses of H3K9me3, Swi6, and Rad21-FLAG levels at pericentric dh repeats, normalized to act1+, and qRT-PCR analyses of dh transcript levels, normalized to act1+. Data are presented as means ± SD, n = 3.
To confirm those findings, we generated an nht1Δ dcr1Δ otr::ura4+ strain. Serial dilution analyses show that these cells indeed grow robustly on FOA-containing medium (Figure 1C). In addition, chromatin immunoprecipitation (ChIP) analyses show that levels of H3K9me3 and Swi6 levels at pericentric dh repeats are reduced in dcr1Δ cells but are improved in nht1Δ dcr1Δ cells (Figure 1D). Furthermore, dh transcripts levels are reduced in nht1Δ dcr1Δ compared with dcr1Δ cells (Figure 1D). Therefore, nht1Δ dcr1Δ not only improves silencing of the otr::ura4+ reporter but also restores heterochromatin mark levels at pericentric repeats, even though heterochromatin is not restored to wild-type levels.
Pericentric heterochromatin is responsible for the recruitment of high levels of cohesin to aid the accurate segregation of chromosomes during mitosis (Bernard et al., 2001; Nonaka et al., 2002). When pericentric heterochromatin is compromised, such as in a dcr1Δ strain, the levels of cohesion at pericentric repeats are reduced, and centromeres are defective in attaching to microtubules for biorientation (Hall et al., 2003; Volpe et al., 2003). As a result, these cells are very sensitive to the microtubule poison thiabendazole (TBZ). We found that, in nht1Δ dcr1Δ cells, levels of cohesion subunit Rad21 are higher than in dcr1Δ cells (Figure 1D). Moreover, nht1Δ dcrΔ cells are not as sensitive to TBZ as dcr1Δ cells are (Figure 1C). These results demonstrate that nht1Δ dcr1Δ cells also restore functional pericentric heterochromatin.
INO80 Mutants Bypass RNAi, but Not Histone Modification Pathways, for Pericentric Heterochromatin Inheritance
Heterochromatin formation at pericentric repeats requires a large number of proteins that can be divided into two main categories: RNAi, which is required for the initial targeting of Clr4 to repeats to establish heterochromatin, and histone modifications, which are required for both heterochromatin establishment and maintenance (Figure 2A). Dilution analyses show that nht1Δ rescues pericentric silencing defects and TBZ sensitivity of mutants involved in RNAi, including the deletions of RITS components ago1Δ and chp1Δ, ARC complex component arb1Δ, and RDRC component rdp1Δ (Figure 2B). However, nht1Δ does not rescue silencing defects and TBZ sensitivity of mutants involved in histone modifications, such as CLRC complex components clr4Δ and rik1Δ, HP1 proteins swi6Δ and chp2Δ, and SHREC complex components clr3Δ and mit1Δ (Figure 2C).
Figure 2. Genetic Interactions of INO80 Mutants with Heterochromatin Mutants.
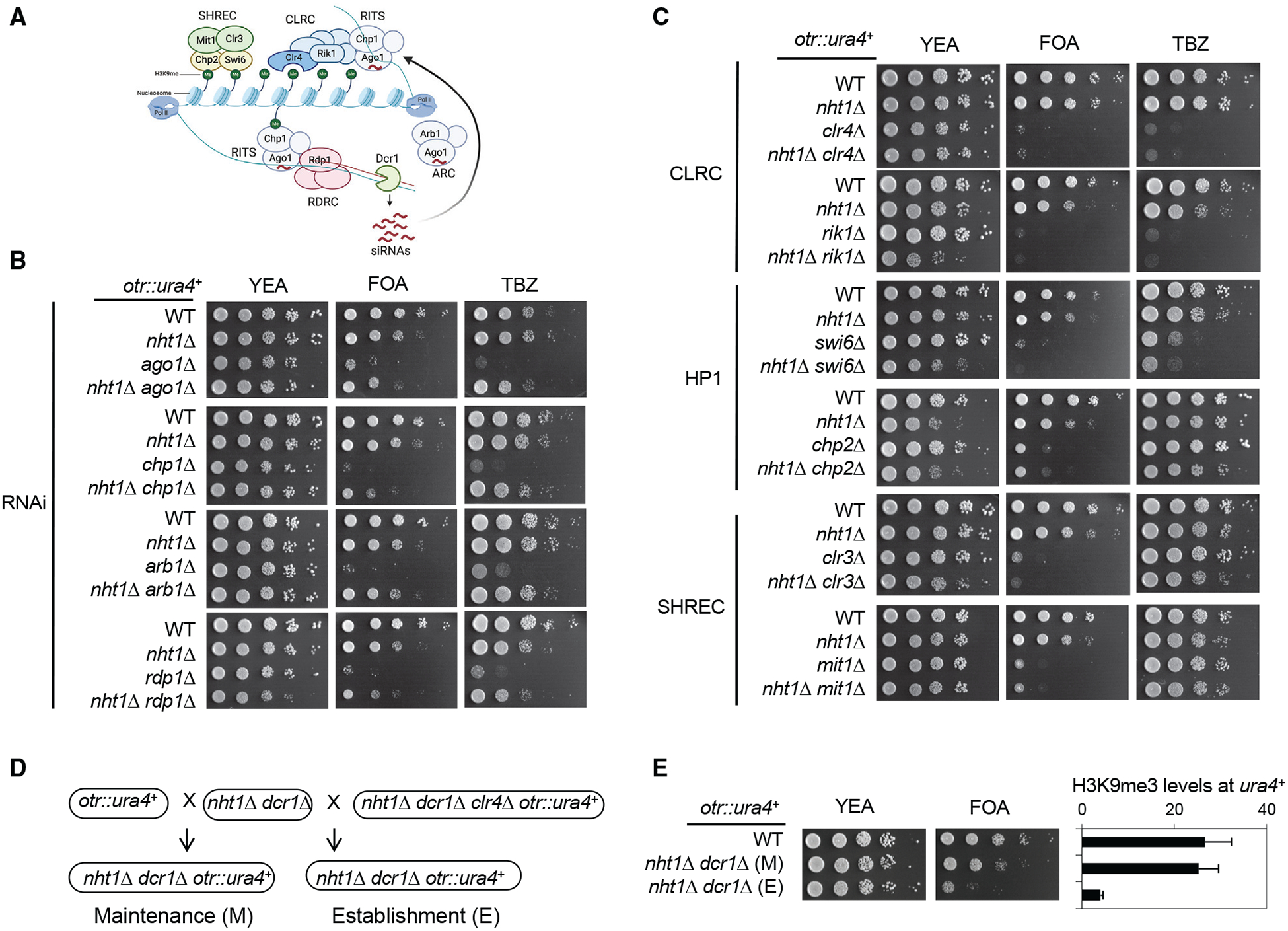
(A) Schematic diagram of RNAi-mediated heterochromatin assembly. RITS, RDRC, Arb1, and Dcr1 are involved in RNAi and the targeting of Clr4 to pericentric repeats. CLRC, Swi6, Chp2, and SHREC are involved in chromatin modifications.
(B and C) Serial dilution analyses of the indicated strains to measure the expression of otr::ura4+ and sensitivity to TBZ. Note that chp2Δ, clr3Δ, and mit1Δ are not sensitive to TBZ.
(D) Schematic diagram of establishment and maintenance crosses.
(E) Left, serial dilution analyses of indicated strains to measure the expression of otr::ura4+. Right, ChIP analysis of H3K9me3 levels at otr::ura4+, normalized to act1+. Data are presented as means ± SD, n = 3.
To further examine whether nht1Δ dcr1Δ rescues heterochromatin establishment or maintenance, we used genetic crosses to introduce otr::ura4+ into nht1Δ dcr1Δ cells from either a silenced state (from wild-type cells) to measure heterochromatin maintenance, or an activated state (from clr4Δ cells) to measure heterochromatin establishment (Figure 2D). It has been shown that dcr1Δ cells failed to establish heterochromatin in both establishment and maintenance crosses (Bayne et al., 2010; Hall et al., 2002). Interestingly, nht1Δ dcr1Δ cells only efficiently silence otr::ura4+ if it is inherited from a wild-type cell (maintenance), but fail to silence otr::ura4+ if it is inherited from a clr4Δ cell (establishment) (Figure 2E). ChIP analyses of H3K9me3 levels at otr::ura4+ also confirm that conclusion (Figure 2E). These results suggest that nht1Δ facilitates inheritance of preexisting heterochromatin in RNAi mutants but cannot establish heterochromatin de novo in the absence of RNAi.
Characterization of the INO80 Complex
To determine whether the accessory subunits function solely as part of the INO80 complex, we generated FLAG-tagged versions of Ino80, Nht1, Iec1, Hap2, and two additional factors, Iec3 and Iec5, which are also annotated as accessory INO80 subunits. Affinity purification of these proteins, followed by mass spectrometry analyses, shows that they have the same associated proteins with similar peptide counts and sequence coverage, suggesting that all five proteins are exclusively in the INO80 complex (Figure 3A).
Figure 3. Characterization of the Role of the Individual Subunit of the INO80 Complex in Heterochromatin Inheritance.

(A) Mass spectrometry analyses of affinity-purified INO80 complex components. The number of peptides identified and the percentage of each protein those peptides cover are indicated.
(B and C) Serial dilution analyses of indicated strains to measure the expression of otr::ura4+ and sensitivity to TBZ.
See also Figure S1.
We then constructed double mutants of dcr1Δ with individual INO80 component mutations and examined their effects on the silencing of otr::ura4+. Interestingly, we found that mutations of every accessory subunit of INO80, such as iec1Δ, hap2Δ, iec3Δ, and iec5Δ, alleviate otr::ura4+ silencing defects in dcr1Δ cells (Figure 3B). We did not identify iec3Δ or iec5Δ in our screen because the iec3Δ in the fission yeast-deletion library is incorrect, and iec5Δ is not present in the library. None of the tested mutations of the conserved subunits, such as ies2Δ, ies4Δ, or arp8Δ, alleviates silencing defects of dcr1Δ, consistent with the fact that these mutations were not identified in our screen (Figure 3C). Other conserved mutations of the INO80 complex are either lethal or extremely sick for further characterization.
Among all chromatin-remodeling proteins, Ino80 shares sequence similarity with Swr1, which catalyzes the incorporation of histone H2A variant H2A.Z into chromatin (Mizuguchi et al., 2004). We found that swr1Δ dcr1Δ cells are still defective in the silencing of otr::ura4+ and TBZ sensitive, similar to dcr1Δ cells (Figure 3C). In addition, ChIP analyses show that Pht1, the fission yeast H2A.Z, is not enriched at pericentric repeats (Figure S1A), consistent with previous analyses (Zofall et al., 2009). Moreover, nht1Δ has no effects on the localization of Pht1 at pericentric repeats or the vid21+ gene promoter. In contrast, swr1Δ abolished Pht1 at the vid21+ gene promoter but has no effects on Pht1 levels at the pericentric repeats (Figure S1A) (Hou et al., 2010). Moreover, the silencing of otr::ura4+ is maintained in swr1Δ nht1Δ dcr1Δ cells (Figure S1B). These results suggest that the ability of INO80 mutants to rescue RNAi is unlikely through regulating H2A.Z localization.
Characterization of the Accessory Subunits of the INO80 Complex
To further analyze the role of the accessory subunits in regulating INO80 complex function, we performed affinity purification of Ino80-FLAG in iec1Δ, hap2Δ, nht1Δ, iec3Δ, and iec5Δ cells. Mass spectrometry analyses show that iec1Δ or hap2Δ results in the loss of association of all accessory subunits from the INO80 complex (Figure 4A). In addition, nht1Δ or iec3Δ results in the loss of Nht1, Iec3, and Iec5 from the INO80 complex but have no effect on the association of Iec1 or Hap2 with Ino80. Finally, iec5Δ results in the loss of only Iec5 from the INO80 complex, but it has no effect on the association of any other accessory subunits with Ino80, suggesting that the effect of mutating other accessory subunits of the INO80 complex on heterochromatin silencing is likely due to a loss of Iec5 subunit of INO80 complex.
Figure 4. Characterization of the Accessory Subunits of INO80.

(A) Mass spectrometry analyses of affinity-purified Ino80-FLAG from different backgrounds. The number of peptides identified and the percentage of each protein those peptides cover are indicated.
(B) Volcano plots of differentially expressed coding (top) or non-coding genes (bottom) in INO80 mutants compared with wild type. Thresholds of 2-fold change and q < 0.1 are used. The q value is the false-discovery-rate-adjusted p value. Red dots indicate RNA with more than a 2-fold increase, and blue dots indicate RNA with more than a 2-fold decrease.
(C) Venn diagram showing the overlaps of RNA that are downregulated in different INO80 complex mutants.
(D) Serial dilution analyses of indicated strains to measure sensitivity to bleomycin.
To examine the functions of the accessory subunits, we performed RNA sequencing (RNA-seq) analyses of RNA purified from iec1Δ, nht1Δ, and iec5Δ cells. We found that all three mutations mildly affect gene expression, with iec1Δ having a stronger effect than nht1Δ, which has a stronger effect than iec5Δ has (Figures 4B and 4C; Tables S1–S6). In addition, iec1Δ, hap2Δ, nht1Δ, and iec3Δ cells are highly sensitive to DNA-damage re-agent bleomycin, but iec5Δ cells are less sensitive (Figure 4D). The severity of effects on gene expression or bleomycin sensitivity is consistent with the mass spectrometry results. Importantly, iec5Δ has almost no effect on transcription programs, and none of the affected genes are involved in heterochromatin assembly. Therefore, the effects of iec5Δ on heterochromatin are unlikely to be due to indirect effects on the transcription of genes involved in heterochromatin inheritance.
Iec5 Is the Critical Component of the INO80 Complex in Regulating Heterochromatin Inheritance
Our results suggest that Iec1 and Hap2 are in close contact with the INO80 catalytic core, Nht1 and Iec3 are linked to INO80 catalytic core through Iec1 and Hap2, and Iec5 is linked to INO80 catalytic core through Nht1 and Iec3 (Figure 5A). Interestingly, western blot analyses show that Iec5 is less stable in iec1Δ, hap2Δ, nht1Δ, and iec3Δ cells (Figure 5B), suggesting that the association of Iec5 with the INO80 catalytic core is required for Iec5 stability and that the effect of iec1Δ, hap2Δ, nht1Δ, and iec3Δ on heterochromatin is through regulating Iec5 stability. To test whether that was the case, we generated a strain expressing Iec1 with a GBP (GFP binding protein) tag (Rothbauer et al., 2008) and Iec5 with a GFP tag. The interaction between GBP and GFP is expected to tether Iec5 back to the INO80 complex, even in the absence of Nht1 or Iec3 (Figure 5C). Iec5-GFP shows a nuclear signal, which is lost in nht1Δ cells (Figure 5D), consistent with the western blot analysis. Interestingly, Iec5-GFP signal is restored in nht1Δ Iec1-GBP cells (Figure 5D). Moreover, nht1Δ dcr1Δ iec5-GFP iec1-GBP cells cannot silence otr::ura4+ and are sensitive to TBZ. Furthermore, ChIP analyses show that H3K9me3 is abolished from pericentric repeats in these cells (Figure 5E). These results suggest that the dissociation of Iec5 from INO80 is critical for the restoration of heterochromatin in RNAi mutant cells. Cells containing nht1Δ iec5-GFP iec1-GBP are still sensitive to bleomycin, similar to nht1Δ cells (Figure 5F), suggesting that the function of Nht1/Iec3 in regulating the DNA damage response is independent of the association of Iec5 with Ino80, and such functions are not involved in heterochromatin regulation.
Figure 5. Iec5 Is the Critical INO80 Subunit Regulating Heterochromatin Formation.

(A and C) Diagram of the organization of the accessory module of the INO80 complex.
(B and H) Western blot analysis of Iec5-myc levels. Tubulin was used as a loading control.
(D) Live cell imaging of cells expressing Iec5-GFP. Scale bar, 2 μm.
(E) Left, serial dilution analyses of indicated strains to measure the expression of otr::ura4+. Right, ChIP analysis of H3K9me3 levels at pericentric dh repeats, normalized to act1+. Data are presented as means ± SD, n = 3.
(F) Serial dilution analyses of indicated strains to measure sensitivity to bleomycin.
(G) Sequence alignment of fission yeast Iec5 with Drosophila Pho and human YY1. Red box indicates a conserved tryptophan residue.
(I) Serial dilution analyses of indicated strains to measure the expression of otr::ura4+.
Although no clear homologs of Iec5 have been identified in higher organisms, Iec5 shows sequence homology with components of the INO80 complex from other organisms, such as human YY1 and fruit fly Pho (Figure 5G). Mutation of a conserved tryptophan residue in Iec5 (Iec5-W98A), which has no effects on Iec5 protein levels, restores silencing of otr::ura4+ in dcr1Δ cells, similar to iec5Δ (Figures 5H and 5I). Therefore, higher organisms might have evolved functional counterparts of Iec5 into other proteins of the INO80 complex.
Iec5 Regulates Histone Turnover at Heterochromatin
Mass spectrometry analyses of proteins associated with heterochromatin components repeatedly identify the INO80 complex, and ChIP analyses show that INO80 complex components can be detected at pericentric repeats, suggesting that the INO80 complex functions directly at heterochromatin (Fischer et al., 2009; Iglesias et al., 2020; Motamedi et al., 2008; Singh et al., 2020). Indeed, our co-immunoprecipitation analysis of proteins associated with FLAG-Swi6 shows the presence of Nht1-myc (Figure 6A). Moreover, ChIP analysis shows that low levels of Ino80-FLAG are enriched at pericentric dh repeats, and its localization is not affected in clr4Δ (Figure 6B). However, iec5Δ has no effects on Swi6-Nht1 association and does not affect the localization of Ino80-FLAG to dh repeats (Figures 6A and 6C). Therefore, Iec5 is not required for the recruitment of the INO80 complex to heterochromatin.
Figure 6. Iec5 Regulates Histone Turnover but Not Nucleosome Positioning at Pericentric Heterochromatin.

(A) Co-immunoprecipitation analysis of FLAG-Swi6 and Nht1-myc. Immunoprecipitation was performed with a FLAG antibody, and western blots were performed with Myc and FLAG antibodies.
(B and C) ChIP analysis of Ino80-FLAG levels at pericentric dh repeats. Data are presented as means ± SD, n = 3.
(D) Nucleosome positioning within the pericentric repeat region as measured by MNase-seq. The x axis shows the position relative to the center of the pericentric repeat annotated feature. The y axis shows the normalized read counts (counts per million [CPM]). Data are the averages of two independent experiments.
(E) Phasogram of nucleosomes within the pericentric repeat region. The x axis shows the range of recorded phases. The y axis shows the frequencies of the corresponding phases. Data are the averages of two independent experiments.
(F) ChIP analysis of H3-FLAG levels at pericentric dh repeats. Data are presented as means ± SD, n = 3.
(G) A model for the role of the INO80 complex in heterochromatin inheritance during DNA replication. INO80 increases the rate of histone turnover, reducing the amounts of parental histones containing H3K9me. Therefore, RNAi-mediated recruitment of Clr4 is required to counter such dilution effects to maintain heterochromatin.
See also Figures S2 and S3.
One of the key activities of the INO80 complex is to regulate nucleosome positioning (Krietenstein et al., 2016; Udugama et al., 2011; Yen et al., 2012). To test whether Iec5 is required for nucleosome positioning at heterochromatin, we performed micrococcal nuclease digestion with deep sequencing (MNase-seq) analysis to map nucleosome positions. However, iec5Δ or iec5Δ dcr1Δ has little effect on the positioning of nucleosomes at pericentric repeats compared with wild-type or dcr1Δ cells, respectively (Figures 6D, 6E, and S2).
INO80 also regulates histone turnover (Singh et al., 2020; Yen et al., 2013). To further examine the role of Iec5 in regulating histone turnover at heterochromatin, we used a strain in which H3-FLAG is under the control of the urg1 promoter at the endogenous urg1+ locus, which can be quickly induced by the addition of uracil in the growth medium (Wang et al., 2015). We used hydroxyurea (HU) to block cells from entering the cell cycle and thereby preventing replication-coupled histone incorporation before the induction of H3-FLAG. We then used ChIP to measure the incorporation of FLAG-H3 into chromatin as an indication of histone turnover. We found that the enrichment of H3-FLAG at pericentric repeats is low in wild-type cells but increases in dcr1Δ cells, suggesting that histone turnover increases in RNAi mutants, consistent with previous findings (Figure 6F) (Aygün et al., 2013; Wang et al., 2015). In iec5Δ dcr1Δ and nht1Δ dcr1Δ cells, H3-FLAG turnover is reduced compared with dcr1Δ cells (Figures 6F and S3A). Moreover, using Iec1-GFP and Iec5-GFP to reconnect Iec5 to the INO80 complex leads to greater histone turnover in nht1Δ dcr1Δ cells (Figure S3A). Finally, INO80 seems to specifically regulate histone turnover at heterochromatin because, in a clr4Δ background, which completely abolished heterochromatin, histone turnover in clr4Δ iec5Δ is similar to that in clr4Δ cells (Figure S3B).
RNAi is required for the initial recruitment of H3K9 methyltransferase Clr4 to pericentric repeats to establish heterochromatin. Once initiated, heterochromatin can be stably propagated through mitosis because of the deposition of parental histones, which serve as seeds to recruit Clr4 to restore H3K9 methylation. We propose that the presence of the INO80 complex at heterochromatin increases histone turnover rates, thereby reducing the amounts of parental histones on replicated DNA to prevent proper epigenetic inheritance (Figure 6G). As a result, RNAi is consistently needed to reestablish heterochromatin to compensate for the loss of parental histones during DNA replication.
Iec5 Regulates Heterochromatin Inheritance and the Heterochromatin Landscape
We then examined the effects of Iec5 on heterochromatin in otherwise wild-type cells. We used a strain in which the SET domain of Clr4 is targeted to 10 copies of tetO binding sites through a TetR-Clr4-SET fusion protein (Figure 7A), resulting in the formation of a large heterochromatin domain that silences a neighboring GFP reporter gene (Ragunathan et al., 2014). The addition of tetracycline to the medium leads to the quick release of TetR-Clr4-SET, and heterochromatin can be inherited through endogenous Clr4, which binds H3K9me through its chromodomain. In wild-type cells, heterochromatin decays quickly, and within 24 h, H3K9me3 is completely lost around tetO, accompanied by the strong expression of GFP, which can be measured by fluorescence-activated cell sorting (FACS) (Figures 7B, 7C, and S4A). Removal of the JmjC domain protein Epe1 greatly reduces heterochromatin decay, and after 24 h, H3K9me3 levels are only reduced slightly, and most cells still silence GFP expression. In contrast, iec5Δ cells activate GFP expression at a slower rate than wild-type cells do, and H3K9me3 is still detectable at tetO binding sites at 24 h (Figures 7B, 7C, and S4A). Moreover, iec5Δ has little effects on nucleosome positioning around tetO binding sites (Figure S4B) but shows reduced turnover of histone H3 (Figure S4C). These results indicate that Iec5 regulates heterochromatin inheritance through regulating histone turnover in wild-type cells.
Figure 7. INO80 Is Required for Heterochromatin Inheritance and Maintenance of the Heterochromatin Landscape across the Genome.

(A) Diagram of the TetR-Clr4-SET system. The targeting of the SET domain of Clr4 to tetO sites results in the formation of heterochromatin and the silencing of the adjacent GFP+ reporter gene. The addition of tetracycline (TET) results in the release of Clr4 from tetO sites, and heterochromatin is maintained by endogenous Clr4, which contains a chromodomain that recognizes H3K9me.
(B) FACS analyses of GFP expression before and 24 h after the addition of tetracycline.
(C) ChIP analysis of H3K9me3 levels at tetO before and 24 h after the addition of tetracycline, normalized to act1+. Data are presented as means ± SD, n = 3.
(D) Genome viewer representations of normalized ChIP-seq reads of H3K9me2 across the three chromosomes. Reads are normalized as CPMs.
(E) Examples of H3K9me2 ChIP-seq reads mapped to pericentric repeat regions of chromosome 1 (cen1), subtelomere 1, and heterochromatin islands. Reads are normalized as CPMs.
See also Figures S4–S6.
To further examine the effects of INO80 on heterochromatin across the genome, we performed ChIP-seq analysis of H3K9me2. In wild-type cells, in addition to large constitutive heterochromatin domains at centromeres, telomeres, and the silent mating-type region, H3K9me2 is also present at a number of loci within euchromatin, termed heterochromatin islands (Figure 7D) (Zofall et al., 2012). Interestingly, iec5Δ results in the increase of H3K9me2 levels at most heterochromatin islands that we can detect (Figures 7D and 7E). ChIP-qPCR analysis confirms that H3K9me2 increases at typical heterochromatin islands, such as mei4+, ssm4+, and mcp5+ (Figure S5A).
We then examined the epistasis relationship between epe1Δ and iec5Δ by generating an epe1Δ iec5Δ double mutant. The decay rate of artificial heterochromatin after Clr4 release in epe1Δ iec5Δ cells is similar to that of epe1Δ cells (Figure S4A). Moreover, ChIP analysis shows that H3K9me2 levels at mei4+ are higher in epe1Δ cells than in iec5Δ cells, and epe1Δ iec5Δ cells show no further increase (Figure S5B). These results suggest that Epe1 and INO80 function in the same pathway.
We noticed that the iec1+ locus was annotated as a heterochromatin island in epe1Δ cells (Figure S6A) (Zofall et al., 2012), raising the possibility that the expression of Iec1 might be affected by epe1Δ. However, we found that epe1Δ has no effects on iec1+ mRNA levels or protein levels (Figures S6B and S6C). We also found that epe1Δ has no effects on Iec5 protein levels, and vice versa, iec5Δ has no effects on Epe1 protein levels (Figure S6C). Therefore, Epe1 and INO80 function together through mechanisms other than regulating their protein levels.
DISCUSSION
The dogma for epigenetic inheritance is that, during DNA replication, parental histones are equally distributed into two daughter DNA strands, and existing modifications on parental histones then recruit enzymes that catalyze the same modifications to modify neighboring newly synthesized histones (Figure 6G). Such a self-propagating mechanism allows histone modification patterns to be inherited through generations, even long after the initial signals that establish such patterns disappear. In fission yeast, the RNAi pathway is required for the recruitment of the H3K9 methyltransferase Clr4 to pericentric repeats to establish heterochromatin. However, RNAi mutants cannot maintain pericentric heterochromatin, suggesting the existence of mechanisms that counteract histone-based epigenetic inheritance (Reddy et al., 2011). As a result, RNAi is needed to consistently re-establish heterochromatin. Here, we show that mutations in the INO80 chromatin-remodeling complex allow heterochromatin inheritance even in the absence of RNAi-mediated heterochromatin establishment. INO80 mutations reduce histone turnover at heterochromatin, consistent with the fact that low histone turnover is critical for the preservation of heterochromatin structure (Aygün et al., 2013; Holla et al., 2020; Reddy et al., 2011; Sadeghi et al., 2015).
What then necessitates INO80 to localize at heterochromatin regions and to regulate histone turnover? The repeat regions are transcribed during the S phase of the cell cycle (Chen et al., 2008; Kloc et al., 2008), which inevitably leads to the collision of RNA polymerase II (Pol II) with the advancing replication fork (Zaratiegui et al., 2011). INO80 is critical for resolving transcription replication conflicts by removing Pol II (Lafon et al., 2015; Poli et al., 2016). Therefore, the presence of INO80 at heterochromatin might be essential to ensure the proper replication of heterochromatin. However, an undesired effect is an increase in the histone turnover rate at heterochromatin regions, which reduces the amounts of modified histones on parental and daughter DNA strands, thereby affecting the prorogation of H3K9me required for heterochromatin inheritance (Figure 6F). The mechanism of how INO80 regulates histone turnover is not clear. It is highly likely that such a function requires the enzymatic activity of INO80. However, we could not directly test that idea because components critical for enzymatic activity are essential in fission yeast. The three core subunit mutants, ies2Δ, ies4Δ, and arp8Δ, are all viable and relatively healthy, and therefore, might only mildly affect INO80 activity in vivo.
INO80 not only regulates heterochromatin inheritance in RNAi mutants but also controls the stability of heterochromatin islands in wild-type cells. These heterochromatin islands frequently change under different environmental conditions or genetic perturbations, and in certain cases, the formation of new heterochromatin islands allows cells to adapt to changes (Gallagher et al., 2018; Iglesias et al., 2018; Parsa et al., 2018; Sorida et al., 2019; Torres-Garcia et al., 2020; Wang et al., 2015; Zofall et al., 2012). Interestingly, INO80 is critical for regulating gene expression in response to diverse nutrient environments (Morrison, 2020). Therefore, the ability of INO80 to regulate heterochromatin islands might be part of the response for cells to adapt to the changing environmental conditions.
INO80 regulates diverse cellular processes, such as transcription, DNA replication, and DNA damage repair (Poli et al., 2017). However, the functions of INO80 in different biological processes are not well understood because of the lack of mutations that specifically abolish each function. We found that iec5Δ affects histone turnover, but not nucleosome positioning, at heterochromatin. Moreover, iec5Δ has little effect on transcription or DNA damage response. Therefore, iec5Δ provides a unique separation-of-function mutation that allows us to demonstrate the role of histone turnover in epigenetic inheritance. The molecular function of Iec5 is currently unknown. It has no obvious domain structure that can imply a function. We also generated recombinant Iec5 protein and found it does not interact with histones (data not shown). Therefore, it is unlikely to be a histone chaperone. Given that Iec5 preferentially regulates histone turnover at heterochromatin regions, it is likely that Iec5 interacts with some heterochromatin components to affect INO80 activity. It remains to be tested whether Iec5 directly affects INO80 enzymatic activity or regulates histone turnover through indirect mechanisms.
Subunits of the INO80 complex catalytic core are highly conserved (Morrison and Shen, 2009). Each species also contains several accessory subunits, which are more divergent. One possible explanation is that different organisms have evolved species-specific functions to adapt to their unique chromatin environment. However, we favor the possibility that the accessory subunits share conserved functions but are divergent at the sequence level. Indeed, we found sequence similarities between Iec5 and accessory INO80 subunits from other species and identified conserved residue that is required for Iec5 function. Interestingly, budding yeast INO80 also regulates the stability of heterochromatin (Xue et al., 2015). Therefore, it would be interesting to test whether the INO80 complex regulates the inheritance of epigenetic states in higher organisms.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Songtao Jia (songtao.jia@columbia.edu).
Materials Availability
The materials generated in this study is available upon request and will be shared without restriction.
Data and Code Availability
The accession number for the RNA-seq, MNase-seq, and ChIP-seq data reported in this paper are GEO: GSE150545.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
A list of fission yeast strains used in this study is provided in Table S7. Yeast strains containing ino80+-Flag, iec1+-Flag, hap2+-Flag, nht1+-Flag, iec3+-Flag, iec5+-Flag, iec5+-myc, iec5+-GFP, iec1+-GBP, nht1+-myc, iec3Δ, or iec5Δ were generated by a PCR-based module method. Epitope tags are integrated at the endogenous genetic loci and tagged genes are under the control of their native promoters. Deletion strains iec1Δ, hap2Δ, nht1Δ, ies2Δ, ies4Δ, arp8Δ were derived from the fission yeast deletion library, and the gene deletions were verified by PCR analyses and DNA sequencing. All other strains were constructed through genetic crosses.
METHOD DETAILS
Genetic screen with the fission yeast deletion library
The genetic screen for mutations that alleviate silencing defects of dcr1Δ was performed as previously described (Roguev et al., 2007). The fission yeast deletion library (Bioneer) was constructed by replacing individual genes with a KanMX4 cassette that confers resistance to geneticin. The otr::ura4+ query strain was constructed by inserting a NatMX6 cassette, which confers resistance to nourseothricin, ~400 bp to the right side boundary of centromere I heterochromatin. The otr::ura4+ reporter was introduced into the deletion library first, followed by the introduction of dcr1Δ, using the RoToR HDA pinning robot (Singer). After mating, haploid progeny of desired genotypes were selected by a combination of antibiotics. The colonies were then pinned onto FOA plates to measure colony growth.
Serial dilution analyses
For serial dilution plating assays, ten-fold dilutions of a mid log-phase culture were plated on the indicated medium and grown for 3 days at 30°C.
Protein purification, co-immunoprecipitation, and mass spectrometry analysis
Log phase yeast cells were harvested and washed with 2×HC buffer (300 mM HEPES-KOH at pH 7.6, 2 mM EDTA, 100 mM KCl, 20% glycerol, 2 mM DTT, 1mM PMSF) and frozen in liquid nitrogen. Crude cell extracts were prepared by vigorously blending frozen yeast cells with dry ice using a household blender, followed by incubation with 1×HC buffer containing 250 mM KCl for 30 minutes. The lysate was cleared by centrifugation at 20,000 g for 1 hour. The supernatants were incubated with Flag-agarose (Sigma) overnight, and washed eight times with 1×HC containing 250 mM KCl. For mass spectrometry analysis, bound proteins were eluted with 3×Flag peptides followed by TCA precipitation.
For co-immunoprecipitation analysis, bound proteins were resolved by SDS-PAGE followed by western blot analyses with Myc (Santa Cruz Biotechnology), or Flag (Sigma) antibodies.
For mass spectrometry analyses, protein pellets were dissolved in buffer (8 M urea, 100 mM Tris pH 8.5), reduced with TCEP (Tris[2-Carboxyethyl]-Phosphine Hydrochloride), and alklyated with chloroacetamide. After dilution of urea to 2 M, proteins were digested with trypsin. Digested peptides were analyzed by LC/LC/MS/MS using an LTQ mass spectrometer. Multidimensional chromatography was performed online with 6 salt steps. Tandem mass spectra were collected in a data-dependent manner with up to 5 ms2 scans performed for each initial scan (m/z range 300–2000). The search program Prolucid was used to match data to a fission yeast protein database. Peptide identifications were filtered using the DTASelect program.
Chromatin immunoprecipitation (ChIP) analyses
Log-phase yeast cells grown at 30°C were incubated at 18°C for 2 hours and then fixed for 30 minutes in 3% freshly made formaldehyde. The cells were pelleted and washed with PBS (phosphate-buffered saline) before resuspended in ChIP lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% Deoxycholate). Ice cold glass beads were added, and the mixtures were vigorously disrupted in a beadbeater. The lysates were collected and subjected to sonication to reduce chromatin size to 500–1000 base pairs (bp). The cleared cell lysates were incubated with Flag-agarose beads (Sigma) overnight at 4°C. The beads were then washed with ChIP lysis buffer twice, ChIP lysis buffer containing 0.5 M NaCl, Wash buffer (10 mM Tris, pH 8.0, 250 mM LiCl, 0.5% NP-40, 0.5% Deoxycholate, 1 mM EDTA), and TE (50 mM Tris pH 8.0, 1 mM EDTA). The bound chromatin fragments were eluted with TES (50 mM Tris pH 8.0, 1 mM EDTA, 1% SDS) and the crosslinking was reversed by incubating at 65°C overnight. The protein DNA mixture was then subjected to proteinase K treatment and phenol:chloroform extraction before the DNA was precipitated by ethanol.
For histone turnover assay, cells were cultured in EMM-uracil medium, and then arrested for four hours by 11 mM HU, followed by the addition of 0.25 mg/mL uracil to induce the expression of H3-Flag for 1 hour, before ChIP analysis was performed.
Quantitative real-time PCR (qPCR) was performed with Maxima SYBR Green qPCR Master Mix (Fermentas) in a StepOne Plus Real-Time PCR System (Applied Biosystems). DNA serial dilutions were used as templates to generate a standard curve of amplification for each pair of primers, and the relative concentration of the target sequence was calculated accordingly. An act1 fragment was used to calculate the enrichment of ChIP over whole cell extract (WCE) for each target sequence when indicated.
ChIP-seq
Log-phase yeast cells were crosslinked with 1% formaldehyde for 20 minutes with shaking at room temperature, followed by 5 minutes quenching with 125mM glycine. Cells were harvested, washed with PBS (phosphate-buffered saline), and resuspended in ChIP lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1% Triton X-100, 0.1% Deoxycholate, 1mM PMSF). Ice-cold glass beads were added and the mixtures were vigorously disrupted in a bead-beater with four 30 s rounds. The lysates were collected and NP buffer was added (10 mM Tris, pH 7.4, 1 M sorbitol, 50 mM NaCl, 5 mM MgCl2, 1 mM CaCl2). MNase was added to the reaction and the reactions were incubated at 37°C for 20 minutes. MNase amount was titrated empirically so that the chromatin was digested to yield mainly mono- and di-nucleosomes. The reaction was stopped by the addition of 0.5 M EDTA, and the tubes were placed on ice. 5X ChIP lysis buffer was added to the reaction, mixed by short vortexing, and the tubes were incubated on ice for 30 minutes. The reactions were then cleared by centrifugation at 16,000 × g for 10 minutes. A small fraction of the cleared supernatant was reserved as input and the rest was used for immunoprecipitation. The protocols for immunoprecipitation, reverse-crosslinking, and DNA precipitation are the same as in the previous ChIP section. The precipitated DNA was treated with RNAase A (EN0531, Thermo Fisher Scientific) for 1 hour at 37°C. DNA concentration was determined with the Qubit dsDNA HS Assay Kit (Q33230, Thermo Fisher Scientific). 5–10 ng of ChIP and input DNA were used for library construction using the NEBNext Ultra II DNA Library Prep Kit for Illumina (E7645, NEB). Libraries were pooled and sequenced on a NextSeq500/550 with the Mid-output kit (150 cycles, single-end) at the JP Sulzberger Genome Center at Columbia University.
Sequencing reads were de-multiplexed and aligned to the S. pombe reference genome, obtained from Pombase (Lock et al., 2019) with Bowtie using default parameters (Langmead et al., 2009). Genome-wide coverage was calculated with deepTools2 (Ramírez et al., 2016) and normalized to counts per million (CPM). The coverage plot was visualized with IGV (Robinson et al., 2011). ChIP-seq experiments were performed in duplicates for each genotype.
MNase-seq
MNase-seq sample preparation was the same as the ChIP-seq except that MNase was titrated to yield mainly mononucleosomes (~80% mono-nucleosomes) and the immunoprecipitation step was omitted. 100 ng of extracted DNA was used for library construction as above. Libraries were pooled and sequenced on a NextSeq500/550 with the Mid-output kit (150 cycles, pair-end) at the JP Sulzberger Genome Center at Columbia University. MNase-seq experiments were performed in duplicates for each genotype.
MNase-seq reads of each sample were mapped to the genome assembly of Schizosaccharomyces pombe using HISAT2 (v2.1.0) (Kim et al., 2015). Potential PCR duplicates were removed using the function “MarkDuplicates” of Picard (http://broadinstitute.github.io/picard/v2.22.0, REMOVE_DUPLICATES = true). The fragment size distribution was measured by the function “CollectInsertSizeMetrics” of Picard (v2.22.0). Only mono-nucleosome sized fragments (< 200 bp) were used for the downstream analysis. Nucleosome periodicity in the MNase-seq data was calculated by phasogram (Valouev et al., 2011) using either all fragment centers across the genome or fragment centers on pericentromeric regions. The nucleosome distribution in the promoter and pericentromeric regions are measured and visualized by the functions “computeMatrix” and “plotProfile” of deepTools (v 3.3.2) (Ramírez et al., 2016).
RNA analyses
Total cellular RNA was isolated from log-phase cells using MasterPure yeast RNA purification kit (Epicenter) according to the manufacturer’s protocol. Quantification with real-time RT-PCR was performed with Power SYBR Green RNA-to-CT one-step Kit (Applied Biosystems). RNA serial dilutions were used as templates to generate a standard curve of amplification for each pair of primers, and the relative concentration of the target sequence was calculated accordingly. An act1 fragment served as a reference to normalize the concentration of samples. The concentration of each target gene in wild-type was arbitrarily set to 1 and served as a reference for other samples.
RNA-seq
For RNA-seq, the total RNA was treated with RiboMinus Transcriptome Isolation Kit (ThermoFisher Scientific, K155003) to deplete rRNA. The rRNA-depleted samples were used to construct strand-specific sequencing libraries with the NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina® (NEB, E7760). Libraries were pooled and sequenced on a NextSeq500/550 with the Mid-output kit (150 cycles, pair-end) at the JP Sulzberger Genome Center at Columbia University. RNA-seq experiments were performed in duplicates for each genotype.
RNA-seq reads of each sample were mapped to the genome assembly of Schizosaccharomyces pombe using HISAT2 (v2.1.0) (Kim et al., 2015). The mapped reads count of each gene was measured by featureCounts (v1.6.1) (Liao et al., 2014). The differential gene expression was calculated and visualized by the R packages DESeq2 (v1.28.0) (Love et al., 2014) and ggplot2 (v3.2.1) (Wickham et al., 2016).
FACS analysis
Cells containing TetR-Clr4-SET and tetO-ura4-GFP reporter were cultured and kept in logarithm phase, and were harvested at various time points after the addition of tetracycline (2.5mg/ml). Cells were collected and fixed by the addition of 70% ethanol for 20 minutes. The cells were then washed twice with PBS (10 mM Na2HPO4, 1.8mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7mM KCl), and resuspended in a FACS tube (BD Falcon). GFP fluorescence was measured using FACSCelesta (Becton Dickinson), and excitation was achieved by using an argon laser emission of 488 nm. Data collection was performed using Cellquest (Becton Dickinson), and a primary gate based on physical parameters (forward and side light scatter) was set to exclude dead cells or debris. Typically, 50,000 cells were analyzed for each sample and time point. Raw data were processed and histograms were drawn using FlowJo (10.6.2, Becton Dickinson).
QUANTIFICATION AND STATISTICAL ANALYSIS
For RNA-seq, FDR adjusted p values (q-values) were generated using the DEseq2 package (v1.28.0) in R. Significantly differentiated genes were defined as having over 2-fold change and a q-value < 0.1. For ChIP-qPCR and RT-qPCR, data are presented as mean ± SD and n represents number of replicates.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG | Sigma | Cat#F13165; RRID:AB_259529 |
| Rabbit Polyclonal anti-c-Myc (A-14) | Santa Cruz Biotechnology | Cat#sc-789; RRID:AB_631274 |
| Rabbit Polyclonal anti-Swi6 | This paper | N/A |
| Rabbit Polyclonal anti-H3K9me3 | Active Motif | Cat# 39161, RRID:AB_2532132 |
| Rabbit Polyclonal anti-H3K9me2 | Abcam | Cat#ab115159, RRID: AB_1090318 |
| Anti-Tubulin | Keith Gull lab | N/A |
| Anti-flag M2 affinity gel | Sigma | Cat# A2220, RRID:AB_10063035 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde, 37% solution | Sigma | Cat#F8775 |
| Tris(2-Carboxyethyl)-Phosphine Hydrochloride | Sigma | Cat#C4706 |
| 5-Fluoroorotic Acid Monohydrate (FOA, 5-FOA) | US Biological | Cat#F5050 |
| RNase A | Thermo Fisher Scientific | Cat#EN0531 |
| MNase | Thermo Fisher Scientific | Cat#88216 |
| Proteinase K | Invitrogen | Cat#10005393 |
| Phenol-chloroform-isoamyl alcohol mixture | Sigma | Cat#77617 |
| Thiabendazole (TBZ) | Sigma | Cat#T5535 |
| Tetracycline | Sigma | Cat#87128 |
| Geneticin (G418) | GIBCO | Cat#11811–098 |
| Nourseothricin Sulfate | Gold Biotechnology | Cat#N-500–1 |
| Hygromycin B | Gold Biotechnology | Cat#H-270–10 |
| EMM Powder | Sunrise Science Products | Cat#2005–1KG |
| Hydroxyurea | Sigma | Cat#H8627 |
| Cycloheximide | Sigma | Cat#01810 |
| Bleomycin Sulfate | Sigma | Cat#B5507 |
| Critical Commercial Assays | ||
| RiboMinus Transcriptome Isolation Kit | ThermoFisher Scientific | Cat#K155003 |
| NEBNext® Ultra II Directional RNA Library Prep Kit for Illumina | New England Biolabs | Cat#E7760 |
| Power SYBR Green RNA-to-CT one-step Kit | Applied Biosystems | Cat#4389986 |
| MasterPure yeast RNA purification kit | Epicenter | Cat# MPY03100 |
| Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat#Q33230 |
| Maxima SYBR Green qPCR Master Mix | ThermoFisher Scientific | Cat#K0223 |
| Deposited Data | ||
| Sequencing Data | This paper | GEO: GSE150545 |
| Experimental Models: Organisms/Strains | ||
| S. pombe strains, see Table S7 | This paper | N/A |
| Fission yeast deletion library | Bioneer | Cat#2030H |
| Oligonucleotides | ||
| Primers, see Table S8 | This paper | N/A |
| Software and Algorithms | ||
| HISAT2 | UT Southwestern (Kim Lab) | v2.1.0 |
| FACSDiva | Becton, Dickinson and Company | v2.1.0 |
| FlowJo | Becton, Dickinson and Company | v10.6.2 |
| Picard | Broad Institute MIT | v2.22.0 |
| DeepTools | Max Planck Institute | v3.3.2 |
| DTASelect | The Scripps Research Institute (Yates Lab) | v2.0 |
| ProLuCID | The Scripps Research Institute (Yates Lab) | v1.3 |
Highlights.
Mutations of INO80 subunits bypass RNAi for pericentric heterochromatin formation
INO80 counteracts heterochromatin inheritance through itsIec5 subunit
Iec5 regulates histone turnover, but not nucleosome positioning, at heterochromatin
Low histone turnover during DNA replication ensures heterochromatin inheritance
ACKNOWLEDGMENTS
We thank Xavier Tadeo and Allison Cohen for early work on the deletion-library-based screen, Danesh Moazed and Kaushik Ragunathan for yeast strains and plasmids, and Zhiguo Zhang and members of the Jia laboratory for discussions and feedback. This work was supported by National Institutes of Health grants R35-GM126910 to S.J. and P41-GM103533 to J.R.Y. C.L. acknowledges support from the Pew-Stewart Scholar for Cancer Research program.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.108561.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Allshire RC, and Madhani HD (2018). Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol 19, 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Nimmo ER, Ekwall K, Javerzat JP, and Cranston G (1995). Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9, 218–233. [DOI] [PubMed] [Google Scholar]
- Audergon PN, Catania S, Kagansky A, Tong P, Shukla M, Pidoux AL, and Allshire RC (2015). Epigenetics: restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aygün O, Mehta S, and Grewal SI (2013). HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol 20, 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayne EH, White SA, Kagansky A, Bijos DA, Sanchez-Pulido L, Hoe KL, Kim DU, Park HO, Ponting CP, Rappsilber J, and Allshire RC (2010). Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell 140, 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, and Allshire RC (2001). Requirement of heterochromatin for cohesion at centromeres. Science 294, 2539–2542. [DOI] [PubMed] [Google Scholar]
- Campos EI, Stafford JM, and Reinberg D (2014). Epigenetic inheritance: histone bookmarks across generations. Trends Cell Biol. 24, 664–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, and Grewal SI (2008). Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature 451, 734–737. [DOI] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, and Grewal SI (2009). Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. USA 106, 8998–9003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PS, Larkin M, Thillainadesan G, Dhakshnamoorthy J, Balachandran V, Xiao H, Wellman C, Chatterjee R, Wheeler D, and Grewal SIS (2018). Iron homeostasis regulates facultative heterochromatin assembly in adaptive genome control. Nat. Struct. Mol. Biol 25, 372–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, and Jia S (2007). Heterochromatin revisited. Nat. Rev. Genet 8, 35–46. [DOI] [PubMed] [Google Scholar]
- Grewal SI, and Klar AJ (1996). Chromosomal inheritance of epigenetic states in fission yeast during mitosis and meiosis. Cell 86, 95–101. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, and Grewal SI (2002). Establishment and maintenance of a heterochromatin domain. Science 297, 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hall IM, Noma K, and Grewal SI (2003). RNA interference machinery regulates chromosome dynamics during mitosis and meiosis in fission yeast. Proc. Natl. Acad. Sci. USA 100, 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan CJ, Aligianni S, Durand-Dubief M, Persson J, Will WR, Webster J, Wheeler L, Mathews CK, Elderkin S, Oxley D, et al. (2010). Fission yeast Iec1-ino80-mediated nucleosome eviction regulates nucleotide and phosphate metabolism. Mol. Cell. Biol 30, 657–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holla S, Dhakshnamoorthy J, Folco HD, Balachandran V, Xiao H, Sun LL, Wheeler D, Zofall M, and Grewal SIS (2020). Positioning heterochromatin at the nuclear periphery suppresses histone turnover to promote epigenetic inheritance. Cell 180, 150–164.e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Wang Y, Kallgren SP, Thompson J, Yates JR 3rd, and Jia S (2010). Histone variant H2A.Z regulates centromere silencing and chromosome segregation in fission yeast. J. Biol. Chem 285, 1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Currie MA, Jih G, Paulo JA, Siuti N, Kalocsay M, Gygi SP, and Moazed D (2018). Automethylation-induced conformational switch in Clr4 (Suv39h) maintains epigenetic stability. Nature 560, 504–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias N, Paulo JA, Tatarakis A, Wang X, Edwards AL, Bhanu NV, Garcia BA, Haas W, Gygi SP, and Moazed D (2020). Native chromatin proteomics reveals a role for specific nucleoporins in heterochromatin organization and maintenance. Mol. Cell 77, 51–66.e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Noma K, and Grewal SI (2004). RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304, 1971–1976. [DOI] [PubMed] [Google Scholar]
- Kagansky A, Folco HD, Almeida R, Pidoux AL, Boukaba A, Simmer F, Urano T, Hamilton GL, and Allshire RC (2009). Synthetic heterochromatin bypasses RNAi and centromeric repeats to establish functional centromeres. Science 324, 1716–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, and Martienssen R (2008). RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol 18, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, and Korber P (2016). Genomic nucleosome organization reconstituted with pure proteins. Cell 167, 709–721.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafon A, Taranum S, Pietrocola F, Dingli F, Loew D, Brahma S, Bartholomew B, and Papamichos-Chronakis M (2015). INO80 chromatin remodeler facilitates release of RNA polymerase II from chromatin for ubiquitin-mediated proteasomal degradation. Mol. Cell 60, 784–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lock A, Rutherford K, Harris MA, Hayles J, Oliver SG, Bähler J, and Wood V (2019). PomBase 2018: user-driven reimplementation of the fission yeast database provides rapid and intuitive access to diverse, interconnected information. Nucleic Acids Res. 47 (D1), D821–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martienssen R, and Moazed D (2015). RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol 7, a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, and Wu C (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348. [DOI] [PubMed] [Google Scholar]
- Moazed D (2011). Mechanisms for the inheritance of chromatin states. Cell 146, 510–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ (2020). Chromatin-remodeling links metabolic signaling to gene expression. Mol. Metab 38, 100973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison AJ, and Shen X (2009). Chromatin remodelling beyond transcription: the INO80 and SWR1 complexes. Nat. Rev. Mol. Cell Biol 10, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, and Moazed D (2008). HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell 32, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, and Grewal SI (2001). Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113. [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, and Watanabe Y (2002). Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol 4, 89–93. [DOI] [PubMed] [Google Scholar]
- Parsa JY, Boudoukha S, Burke J, Homer C, and Madhani HD (2018). Polymerase pausing induced by sequence-specific RNA-binding protein drives heterochromatin assembly. Genes Dev. 32, 953–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Gerhold CB, Tosi A, Hustedt N, Seeber A, Sack R, Herzog F, Pasero P, Shimada K, Hopfner KP, and Gasser SM (2016). Mec1, INO80, and the PAF1 complex cooperate to limit transcription replication conflicts through RNAPII removal during replication stress. Genes Dev. 30, 337–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli J, Gasser SM, and Papamichos-Chronakis M (2017). The INO80 remodeller in transcription, replication and repair. Philos. Trans. R. Soc. Lond. B Biol. Sci 372, 201160290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragunathan K, Jih G, and Moazed D (2014). Epigenetic inheritance un-coupled from sequence-specific recruitment. Science 348, 1258699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiymbek G, An S, Khurana N, Gopinath S, Larkin A, Biswas S, Trievel RC, Cho US, and Ragunathan K (2020). An H3K9 methylation-dependent protein interaction regulates the non-enzymatic functions of a putative histone demethylase. eLife 9, e53155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, and Jenuwein T (2000). Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599. [DOI] [PubMed] [Google Scholar]
- Reddy BD, Wang Y, Niu L, Higuchi EC, Marguerat SB, Bähler J, Smith GR, and Jia S (2011). Elimination of a specific histone H3K14 acetyltransferase complex bypasses the RNAi pathway to regulate pericentric heterochromatin functions. Genes Dev. 25, 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, and Krogan NJ (2007). High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat. Methods 4, 861–866. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, and Leonhardt H (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol. Cell. Proteomics 7, 282–289. [DOI] [PubMed] [Google Scholar]
- Sadeghi L, Prasad P, Ekwall K, Cohen A, and Svensson JP (2015). The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO Rep. 16, 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Cardona A, and Zhang Z (2018). Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem. Sci 43, 136–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PP, Shukla M, White SA, Lafos M, Tong P, Auchynnikava T, Spanos C, Rappsilber J, Pidoux AL, and Allshire RC (2020). Hap2-Ino80-facilitated transcription promotes de novo establishment of CENP-A chromatin. Genes Dev. 34, 226–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorida M, Hirauchi T, Ishizaki H, Kaito W, Shimada A, Mori C, Chikashige Y, Hiraoka Y, Suzuki Y, Ohkawa Y, et al. (2019). Regulation of ectopic heterochromatin-mediated epigenetic diversification by the JmjC family protein Epe1. PLoS Genet. 15, e1008129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Morgan KR, Petryk N, and Groth A (2020). Chromatin replication and epigenetic cell memory. Nat. Cell Biol 22, 361–371. [DOI] [PubMed] [Google Scholar]
- Tadeo X, Wang J, Kallgren SP, Liu J, Reddy BD, Qiao F, and Jia S (2013). Elimination of shelterin components bypasses RNAi for pericentric heterochromatin assembly. Genes Dev. 27, 2489–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Garcia S, Yaseen I, Shukla M, Audergon PNCB, White SA, Pidoux AL, and Allshire RC (2020). Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature 585, 453–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M, Sabri A, and Bartholomew B (2011). The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol. Cell. Biol 31, 662–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, and Sidow A (2011). Determinants of nucleosome organization in primary human cells. Nature 474, 516–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe T, Schramke V, Hamilton GL, White SA, Teng G, Martienssen RA, and Allshire RC (2003). RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11, 137–146. [DOI] [PubMed] [Google Scholar]
- Wang X, and Moazed D (2017). DNA sequence-dependent epigenetic inheritance of gene silencing and histone H3K9 methylation. Science 356, 88–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Reddy BD, and Jia S (2015). Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. eLife 4, e06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Navarro D, and Pedersen TL (2016). ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag; ). [Google Scholar]
- Xue Y, Van C, Pradhan SK, Su T, Gehrke J, Kuryan BG, Kitada T, Vashisht A, Tran N, Wohlschlegel J, et al. (2015). The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev. 29, 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, and Pugh BF (2012). Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149, 1461–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, and Pugh BF (2013). SWR-C and INO80 chromatin remodelers recognize nucleosome-free regions near +1 nucleosomes. Cell 154, 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marín L, Chang AY, Goto D, et al. (2011). RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479, 135–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, and Grewal SI (2008). Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol 15, 381–388. [DOI] [PubMed] [Google Scholar]
- Zofall M, Fischer T, Zhang K, Zhou M, Cui B, Veenstra TD, and Grewal SI (2009). Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature 461, 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zofall M, Yamanaka S, Reyes-Turcu FE, Zhang K, Rubin C, and Grewal SI (2012). RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession number for the RNA-seq, MNase-seq, and ChIP-seq data reported in this paper are GEO: GSE150545.


