Abstract
The severe acute respiratory syndrome coronavirus 2, called a SARS-CoV-2 virus, emerged from China at the end of 2019, has caused a disease named COVID-19, which has now evolved as a pandemic. Amongst the detected Covid-19 cases, several cases are also found asymptomatic. The presently available Reverse Transcription – Polymerase Chain Reaction (RT-PCR) system for detecting COVID-19 lacks due to limited availability of test kits and relatively low positive symptoms in the early stages of the disease, urging the need for alternative solutions.
The tool based on Artificial Intelligence might help the world to develop an additional COVID-19 disease mitigation policy. In this paper, an automated Covid-19 detection system has been proposed, which uses indications from Computer Tomography (CT) images to train the new powered deep learning model- U-Net architecture.
The performance of the proposed system has been evaluated using 1000 Chest CT images. The images were obtained from three different sources – Two different GitHub repository sources and the Italian Society of Medical and Interventional Radiology's excellent collection. Out of 1000 images, 552 images were of normal persons, and 448 images were obtained from COVID-19 affected people. The proposed algorithm has achieved a sensitivity and specificity of 94.86% and 93.47% respectively, with an overall accuracy of 94.10%.
The U-Net architecture used for Chest CT image analysis has been found effective. The proposed method can be used for primary screening of COVID-19 affected persons as an additional tool available to clinicians.
Keywords: SARS-CoV-2, COVID-19, RT-PCR, U-Net architecture, Deep learning
1. Introduction
SARS-CoV-2 has given rise to COVID-19, a widespread disease throughout the world. The virus first emerged in Wuhan, China, in December 2019 and has now become the worldwide health issue [1]. While several cases of COVID-19 are found to be asymptomatic, the majority of cases are reported with typical symptoms of fever, dry cough and tiredness. Many people are found with common onset syndromes like pains and aches, runny nose, sore throat, nasal congestion and diarrhoea [1,2].
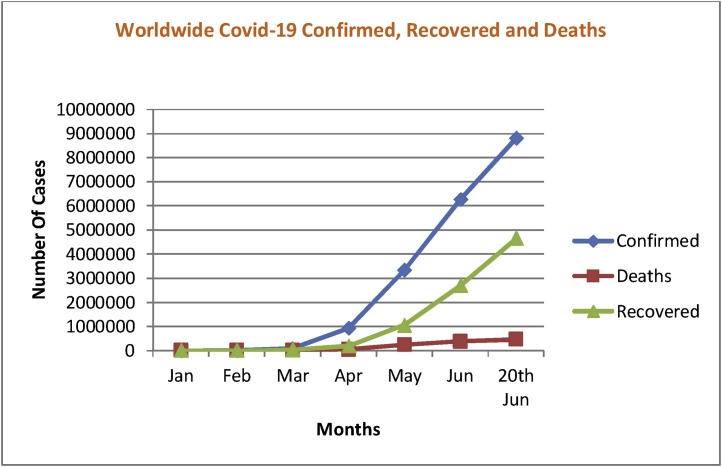
Airborne spread, duration on surfaces and respiratory transmissions from one person to another triggered quick expansion of the pandemic [3]. The COVID-19 epidemic statistics as of 20th June 2020 indicates that there are in total 8,804,268 affected worldwide, 463,510 deaths caused and 4,656,912 are recovered from it.
Many technologically advanced countries fail to manage their medical care systems, as the demand for Intensive Care Unit facilities is increased due to patients with the most severe symptoms of COVID-19 disease.
We have shown the growth statistics of the COVID-19 disease worldwide from 22nd January 2020 to 20th June 2020 in Fig. 1 , including death and recovery rates.
Fig. 1.
Status of the number of confirmed, recovered, and deaths cases of Covid-19 from 22nd January to 20th June 2020 [3].
W.Wang et al. [4] evaluated a total of 1070 samples acquired from 205 people affected by COVID-19; the patients involved in the study were having an average age of 44 years. The patients were suffering from dry cough, fever and weakness; 19% of the cases were suffering from severe syndromes. Bronchoalveolar lavage fluid samples indicated the maximum positive rates (14 out of 15), up next was sputum (72 out of 104), followed by nasal swabs and fibro bronchoscope brush biopsy that is (5 out of 8) and (6 out of 13) respectively, then feces (44 out of 153) after that was blood (3 out of 307). No patient was tested positive through a urine sample. The study concluded that specimen testing from various sites could reduce false-negative rates, thereby improving sensitivity.
RT-PCR detection schemes have shown a low positive rate, i.e. the lower value of sensitivity for the samples tested for COVID-19, in the initial phases of the disease [5]. However, the indicators of CT images of COVID-19 have shown distinct features that are unique when compared with all other viral pneumonia. Considering this fact, doctors have opted for CT images as one of the earlier diagnostic measures for this disease.
Diagnosing COVID-19, with the help of chest CT images, was found more beneficial than initial RT-PCR testing in a case study conducted in China with 1014 affected cases [6]. Chest CT has shown greater sensitivity in the detection of the disease than RT-PCR testing. Out of 1014 cases, 601 cases (59%) were detected as positive with RT-PCR, whereas 888 (88%) were detected using chest CT images.
The clinical symptoms of 41 people affected with COVID-19 were analyzed by Huang et al. [7]. Along with common onset syndromes like cough, fever and fatigue, all 41 cases were found infected with pneumonia, and the abnormalities were shown in their chest CT investigation. Severe respiratory afflictions, chronic heart injury and other secondary contaminations were observed in CT images.
Limited availability of RT-PCR test kits, the time required to process the test, low positive rates in early stages and requirement of considerable human expertise demands an innovative method for detection of COVID-19.
In the current situation of the COVID-19 pandemic, advanced ways of diagnosis for treating and managing the disease are indeed in demand. Exploring approaches based on Artificial Intelligence seems a promising solution for effective analysis of disease. In such an unprecedented situation, the alternative solutions explored should find cheaper approaches for recognizing, controlling and treating this worldwide pandemic. Furthermore, the suggested method should help researchers understand the fundamental reasons and advancement of the disease thoroughly. Engineering techniques such as image processing and innovative machine learning algorithms can facilitate identifying landmark features and occurred lesions, thereby enabling categorization of the input sample as a normal or disease affected case.
One of the methods utilized for the diagnosis of pneumonia is CT images of the chest. We have proposed the use of chest CT images with advanced fully convolutional network architecture developed for biomedical image analysis-U-Net, for detection of abnormalities due to COVID-19. Based on the presence or absence of abnormalities, the input sample is classified as normal or COVID-19 affected case. The rest of the paper is organized as follows. In section 2 Literature survey, limitations identified through it and key contributions of the proposed work are described. In section 3, datasets used, proposed methodology and use of U-NET architecture for the proposed work is presented. Experimental results followed by the comparison of results with four other network architectures performances and with the state-of-the-art methods are presented in section 4. To conclude, the significant contributions of the paper in addition to possible future work approach is discussed.
2. Related work
In the current era of Machine learning and Artificial Intelligence, Convolutional Neural Network (CNN) proves to be the most beneficial and popular algorithm in image processing. We have taken an overview of various types of CNN algorithms and other methods used in recent years for analyzing multiple diseases using medical images such as chest X-ray and chest CT images.
A novel method was proposed by Xiaowei Xu et al. [8] for the screening of the COVID-19 disease. The method has evaluated results with the help of CT images and was implemented using 3D-CNN, including segmentation and feature extraction. A total of 618 samples were from chosen hospitals, of these, 175 samples were of healthy persons, 224 samples were of people with Influenza-A viral pneumonia, and 219 samples were of COVID-19 affected people. Authors reported an overall accuracy of 86.7%.
Baltruschat IM et al. [9] compared different deep learning approaches for identification of various diseases using X-ray images. The comprehensions of powerful architecture ResNet-50 and its extended versions are discussed in this paper. Along with the pathologies visible in X-ray images, such as cardiomegaly, nodule, pneumonia, etc., few non-image parameters such as gender, age and acquisition method are also considered for classification. The performance of algorithms is compared with 5-fold resampling and multi-label loss function using Receiver Operating Characteristics Curve measures and rank correlation. The method reported Area Under Curve (AUC) of 0.795, 0.785 and 0.806 for the architectures ResNet-50, ResNet-101 and ResNet-38 respectively.
The assistance for findings lung cancer is suggested by Nicolas Coudray et al. [10]. The most frequently appearing types of lung cancer – LUAD and LUSC – are classified automatically from normal tissues using Googles CNN - Inception v3. The proposed method was evaluated on whole slide images and reported a sensitivity, specificity and accuracy of 89%, 93% and 97% respectively, in addition to validation of the model for various frozen tissues.
Anthimopoulos et al. [11] defined a CNN for classification of 6 interstitial lung disorders. The proposed CNN includes five convolutional layers with Leaky Rectified Linear Unit (RELU) activation function to avoid overfitting, followed by max-pooling, finally classifying with soft-max function. In total, 14,696 patches of images were derived from 120 CT images obtained from two local hospitals. The cross-entropy was used for minimizing the loss function, and the author reported an overall accuracy of 85.61%.
X-ray equipment is used to scan the vital body parts and identify pathologies such as bone dislocations, fractures, lung infections, tumours and pneumonia. A type of advanced X-ray technique is CT scanning; it inspects the very lenient structure of the active body portion and produces more detailed photographs of the internal soft tissues and organs [12].
The use of chest X-ray images for automatic image analysis and detection of COVID-19 was explored in [13]. The performance of automatic detection is compared using three different convolutional neural networks - InceptionV3, InceptionResNetV2 and ResNet50. The author has used 100 chest X-ray images, 50 each from GitHub and Kaggle repository. A deep transfer knowledge build approach was used for training due to limited availability of image samples. Indian Scientists propose the AI-based tool for discrimination of COVID-19 and other lung-related syndromes at Kyoto, Japan, as per the article in Times of India [14].
To classify the malicious pulmonary nodule, a hybrid CNN based on AlexNet and LeNet was proposed by Zhao et al. [15]. A database of 743 CT images was used for evaluation of the proposed agile CNN. The performance of the proposed CNN was analyzed by the varying learning rate, kernel size, etc. The author reported accuracy of 0.822 and AUC of 0.877. Further improvement was suggested through transferred learning, or feeding derived features to a classifier like Support Vector Machine (SVM).
A 3D weakly superimposed deep CNN was proposed by Zheng et al. [16] for COVID-19 detection using chest CT images. A pre-trained U-Net was used for segmentation of CT images. Lesion annotation is not required in training; out of 630 CT images acquired from samples from hospitals during 13th December to 6th February 2020, 499 were used for training, and 131 were used for testing the proposed CNN architecture. A probability threshold of 0.50 was used to classify samples as COVID-19 affected or normal. The proposed method reported accuracy, Positive Predictive Value (PPV) and Negative Predictive Value (NPV) of 0.901, 0.840 and 0.982, respectively.
A two-dimensional Deep CNN built on ResNet-50 architecture was proposed by Gozes et al. [17] for detection of COVID-19. The U-Net architecture was used for segmentation of CT images. The CT images of 56 confirmed cases of COVID-19 disease were used. The algorithm has reported sensitivity, specificity and AUC of 0.982, 0.922 and 0.996, respectively.
Barstugan et al. [18] explored COVID-19 detection algorithm by applying features extracted with different feature extraction techniques like Grey Level Co-occurrence Matrix (GLCM), Discrete Wavelet Transform (DWT), Grey-Level Size Zone Matrix (GLSZM), etc. Further, extracted featured were applied as an input to SVM classifier for classification of input sample as normal or COVID-19 affected. The method derived multiple samples of data by extracting 16 × 16, 32 × 32, etc. sized CT images patches. The algorithm was evaluated with different parameters like sensitivity, specificity, F-score and accuracy by implementing 2-fold, 5-fold as well as 10-fold cross-validations and reported overall accuracy of 99.68% on 150 CT abdominal images.
Xie Y et al. [19] proposed a Multi-View Knowledge-Based Collaborative (MV-KBC) deep learning architecture for separation of malignant nodules over benign nodules using CT images. Three pre-trained ResNet-50 architectures were used for analyzing nodules' overall appearance, heterogeneity in shape and voxel. An adaptive weighing scheme updated through backpropagation with 9 KBC sub-modules was used for classification. The reduction in false-negative rate is achieved using a penalty loss function over conventional cross-entropy function. The proposed module is evaluated on 1945 images from the LIDC-IDRI dataset consisting of 1301 benign and 644 malignant nodules; it achieved an overall accuracy of 91.60% and AUC of 95.70%.
It is observed from the literature that for the detection of COVID-19 disease, recognition of abnormalities on chest CT scans plays a significant role and image processing with machine learning algorithms can make it feasible. However, most of the methods in the literature are computationally resource-hungry, where separate mechanisms in terms of a cascade of architectures, are required for segmentation, complex feature extraction and detection of exact lesion locations. These limitations provoked us to imply a new method for detection of COIVID-19 disease. The key contributions of the proposed method are:
-
1.
Proposed method uses an end-to-end system. So, the separate mechanism for segmentation, feature extraction or location detection is not required. The image needs bare pre-processing.
-
2.
The performance evaluation is done on a collection of CT-scan images from three different sources.
-
3.
The performance evaluation is compared with other four architectures with a similar dataset, hardware and software platform.
3. Materials and method
3.1. Dataset
To exercise and evaluate any computational approach for disease diagnosis, collection of a database that is rich in variations is essential. GitHub repository, designed for coordinating the research work amongst all for computational exploration, contains X-ray and Chest CT images of COVID-19 cases. Fifty chest CT images showing the COVID-19 syndromes were obtained from GitHub repository, made available online by Dr Joseph Paul [20]. We have picked up another set of 110 chest CT images, affected with COVID-19, from the Italian Society of Medical and Interventional Radiology's excellent collection. There were about 60 cases, with example Chest X-rays and single slice CT-images, a simple download from these cases resulted in 110 usable, axial CT-images of confirmed COVID-19 cases [21]. The third source of COVID-19 chest affected dataset images is GitHub UCSD-AI4H/COVID-CT, in which 288 chest CT images are available for 169 COVID-19 cases [22]. The 552 normal lung CT scans were acquired from LIDC Dataset, available online [23]. As a result, a dataset of 1000 images were used to train the pre-defined U-Net architecture.
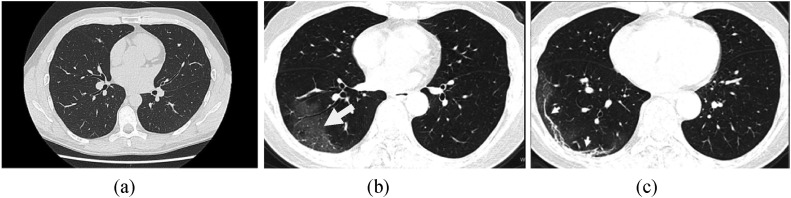
The representative chest CT scans of healthy lungs and lungs affected with SARS-CoV-2 are shown in Fig. 2 (a–c).
Fig. 2.
Lung CT scan images a) CT scan of a healthy Lung b) chest CT scan demonstrating the peripheral right lower lobe ground-glass opacities (arrow). c) Chest CT image progressed with more sub-pleural curvilinear lines, in the extent of Ground-Glass Opacities (GGO), (arrows).
3.2. Proposed methodology
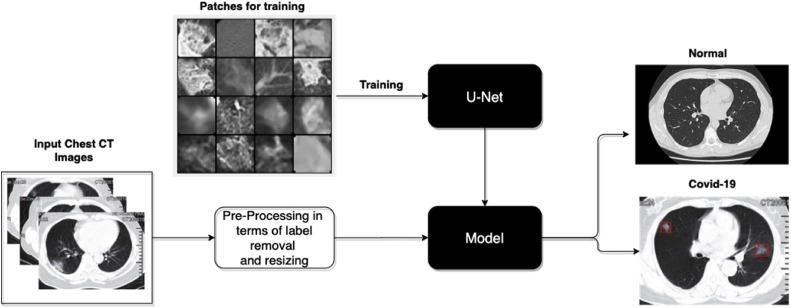
The proposed work, as shown in Fig. 3 , was focused on the use of U-Net architecture for Covid-19 detection using Chest CT scan images. It was done in three phases: 1) Acquiring the Chest CT images and pre-processing for patient's information and other label removals. 2) Training the fully convolutional U-Net Architecture for loss minimization. 3) Testing the applied input image on trained U-Net Architecture to identify COVID-19 affected cases.
Fig. 3.
Block Diagram of Proposed Methodology.
3.2.1. Pre-processing
The acquired chest images were pre-processed for removal of labels carrying patient's details. Since the images were acquired from different sources, resizing of images was needed. All input images, including training and test CT scans, were resized to 512 × 512.
3.2.2. U-Net architecture
Though object recognition using Convolutional Neural Networks has brought a revolution in the traditional approach of classification [23], yet the practical implementations based on CNN remained unattended due to requirement of large size of training data and availability of high-end hardware. Most of the biomedical image applications, for classification tasks, require at least one class label and the locations of abnormal pixels to be marked.
J. Long and Shelhamer [24] explored the end-to-end training by sharing the image features between down-sample and up-sample routes using the Fully Convolutional Network (FCN) and improved the segmentation output. Concurrently, an architecture named U-Net, specially devised for biomedical image segmentation was published by Olaf Ronneberger et al. [25].
The reasons for choosing U-Net architecture are: i) The U-Net architecture has already been proved superior to other available prior best methods for biomedical image analysis, ii) The end-to-end training requirements can be satisfied with very few samples, which proves undoubtedly beneficial, as a large cohort of training samples may not be available to every researcher, iii) The inclusion of contracting context capture and expansions of symmetric paths enables the U-Net architecture to find precise locations of pixels compared to available Convolutional Neural Networks (CNN) approaches [19,25].
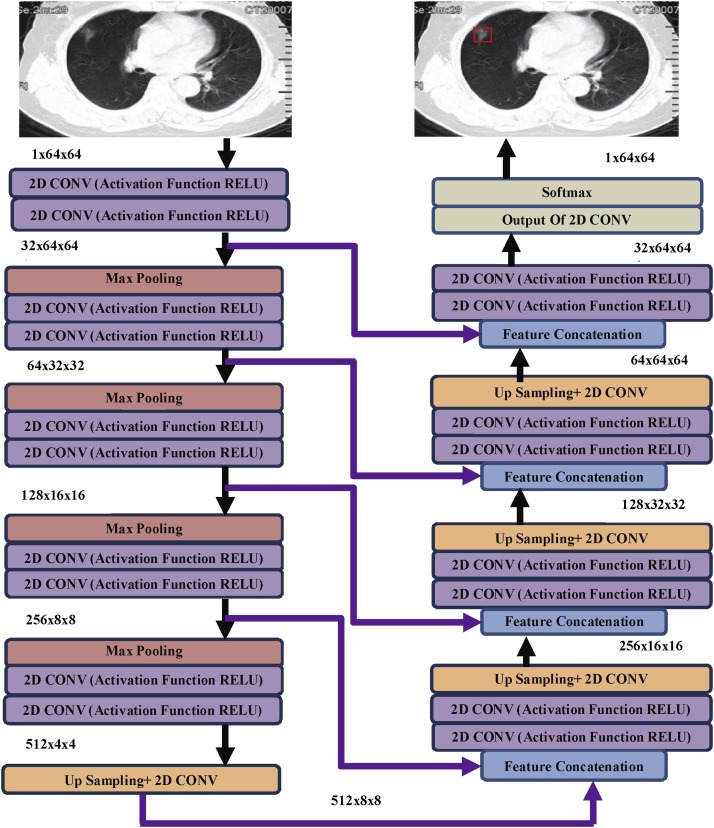
The U-Net architecture was modified in a way that the up-sample section is comprised of many feature blocks, facilitating the transfer of contextual information to upper-resolution layers by the network [8]. Consequently, the expansion path was relatively symmetric to the contraction path, resulting in a U- form of architecture as shown in Fig. 4 .
Fig. 4.
Proposed U-Net Architecture for detection of Normal and COVID-19 affected cases. (Every combination of blocks denotes a multi-channel feature map (mentioned at the upper left corner of contraction path and upper right corner of extraction path).
The implied U-Net model, as shown in Fig. 4, contained multiple layers of convolutions with ReLU activation functions, followed by max-pooling in contraction path and convolutions with up-sampling and feature concatenation followed by softmax layer for final decisions makes the extraction path.
The contraction path was following the conventional architecture of a CNN; there was the repeated use of dual 3 × 3 padded convolutional operations, one layer succeeded by the activation function ReLU. The acquired feature map was down-sampled with consecutive four 2 × 2 max pool operations with a stride of 2. Each block in expansion route consists of an up-sampling the feature maps succeeded by convolutional operation of 2 × 2, a concatenated sequence of equivalent feature vector out of the contraction route and two 3 × 3 convolution operations, each one of it succeeded by activation function ReLU. The mapping of every 32- section feature map to two separate classes was done at the end with a 1 × 1 convolutional operation.
In each convolutional layer at depth , for the input patches applied or from the outputs of earlier levels, locally 3 × 3 convolutions are computed, further added with a bias followed by application of Rectified Linear Unit for producing the results defined in Eq. (1).
| (1) |
Here, the weights of all the 2D convolution kernels and the biases are the trainable parameters. As a max pool layer succeeds each convolutional layer, it computes the max on a 2 × 2 kernel which decreases the size of the feature map by 2.
The normalized exponential function, Softmax, for pixel-wise decision defined in Eq.2 was used to find the energy at the output.
| (2) |
The equation takes as input a vector of real numbers and normalizes it into a probability distribution consisting of probabilities. After applying softmax, each component will be in the interval (0,1), and the components will add up to 1 so that they can be interpreted as probabilities. is the estimate of the maximum of activation function, i.e. for that has maximum activation function and for other values of . The softmax classifier resolves the vector as bright (infected) or dark (normal) pixels.
We have used the cross-entropy function for deriving the costs of the decisions and for summing up the total error. Cross-entropy ( is always a positive valued function. Cross entropy is used to modify the weights while training and minimizing the error. As the network gets better trained for acquiring the desired output for applied input samples, the cost function leaned toward zero. Cross-entropy ( is calculated by finding the difference between predicted values (p) and targets (c), as defined in Eq.3.
| (3) |
3.2.3. Image classification
Here, the U-Net architecture is used for object detection, the abnormalities are treated as objects of interests. The U-Net architecture detects the abnormalities as objects, further if the count of detected objects is greater than zero, the image is declared as Covid-19 affected case.
3.3. Evaluation platform and parameters
The implementation of proposed algorithms was done in both Python and MATLAB. A Laptop with Intel-i7 2.4 G CPU, 16 G DDR3 RAM, Windows10, MATLAB R2019a was used for simulation of proposed algorithms. Once the network was trained, testing of images was performed on a single kit using Raspberry Pi.
The performance of U-net model is measured using parameters like sensitivity, specificity, accuracy and precision. The sensitivity indicates the ability to detect true positives cases; specificity shows the ability to detect true normal samples. Accuracy shows the correctness of predictions, whereas precision shows the consistency of correctness. These parameters are defined in the Eqs. (4), (5), (6), (7). Here TP represents COVID-19 cases detected as positives, TN indicates normal sample detected as negatives, FN represents the number of COVID-19 affected cases detected as negative by the FCN and FP indicates the normal chest CT images detected as COVID-19 affected cases by the proposed FCN.
| (4) |
| (5) |
| (6) |
| (7) |
3.4. Training
In the proposed work, a transfer learning method is used to train the defined network architectures. Here, the stored weight of the pre-trained CNN architectures on LIDC dataset was loaded and then further trained with other two datasets used in this study. The benefit of employing the transfer learning method, for training the network is that by this time the primary layers of CNN are trained, and the network is learned with necessary features such as identifying borders, edges and shapes in the image. So, to save time, reduce computational burden and heavy resource demands, transfer learning proves beneficial. A train-test split method is used to randomly divide the available dataset of 1000 images into two parts training set images, and validation set images.
It was learned from the literature of Artificial intelligence [26] that the model parameters should be evaluated on the input samples that were unused to set hyperparameters of the model. Hence, initially, a 90/10 approach was used to divide the input samples into 900 images in the training set and 100 images in the validation set. In the total dataset of 1000 images, the partition of 900 /100 has saved more samples for training. Further, the test set of 100 images was precisely small, facing a lot of variation while evaluating the performance. So, 10-fold validation of a total of 1000 samples was used to reduce the variance and making performance evaluation less sensitive to the partitioning of the samples. The network was trained using 900 images from training samples, and a trained model was used to estimate the test error rate by applying the images from validation samples.
The 900 training images of the total CT scan samples, along with the lesion masks with defined locations, were applied to train the U-Net model in a batch size of 100. Lesions masks were derived by cropping the abnormality patches from training images of the Covid-19 cases. The input images, along with the location references of lesion masks within it, were applied as an input for training. To do robust training, patches with Ground-Glass Opacities (GGO) of varying parenchymal density, patches involving halo signs, patches with the component of consolidation admixed with ground-glass opacities and crazy-paving pattern were used. Patches of multifocal modular with new foci of GGO & consolidation found along with peripheral ground-glass opacities were also used for learning of the model. Additionally, subpleural curvilinear lines were also used for training.
Here, we have used gradient descent to update the parameters (weight and bias) of our model to best estimate the target function. To reduce the time taken, we used a variant of gradient descent called stochastic gradient descent as an optimization function. A randomized training set was used to shuffle the order in which updates are made to the coefficients, to avoid distraction or becoming stuck.
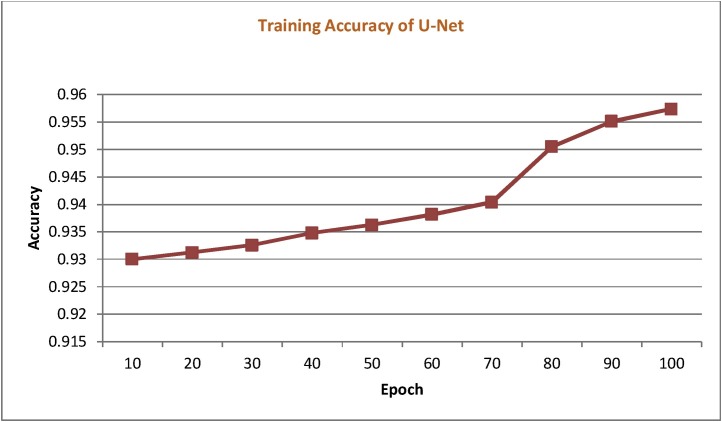
The training accuracy acquired for the first 100 epochs for U-Net architecture was elaborated with the help of graphical representation. Out of the five employed networks, U-Net architecture specifically designed for biomedical image processing applications has produced better accuracy, as shown in Fig. 5 .
Fig. 5.
Training accuracy of U-Net found over the first 100 epochs.
4. Results and discussion
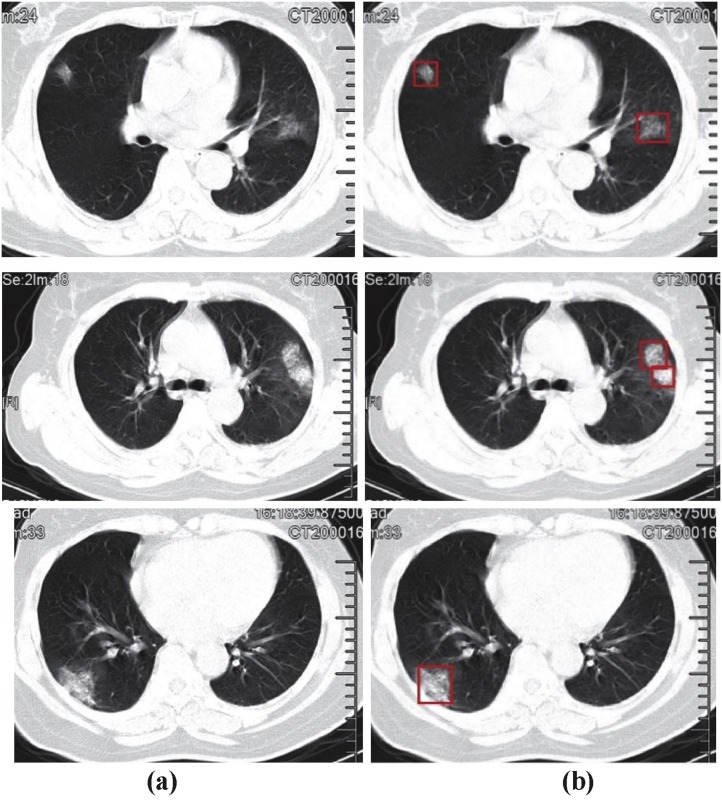
The results of the input chest CT images and abnormalities detected by FCN are as shown in Fig. 6(a & b). In most of the cases, Lung CT showed patchy bilateral ground-glass opacities with per-bronchial and peripheral/subpleural distribution in Lung.
Fig. 6.
Chest CT images a) Input to U-Net architecture b) Infections due to occurrence of COVID-19 disease detected by U-Net.
In some cases, reticular opacities were also found within areas of ground glass (crazy-paving pattern). Fig. 6(a) shows the input chest CT images to U-Net Architecture with different types of infections and Fig. 6(b) shows the images at the output of U-Net Architecture, detected with different abnormalities due to COVID-19 disease.
The validation of the network was performed using 10- fold cross-validation, a total of 1000 images were used for testing. The network was validated with each fold comprising 100 images, and the evaluation parameters achieved during validation of the U-Net architecture are given in Table 1 .
Table 1.
Evaluation parameters achieved during validation of the U-Net architecture using 10-fold cross validation.
| Sensitivity | Specificity | Precision | Accuracy | |
|---|---|---|---|---|
| Fold-1 | 0.9219 | 0.9347 | 0.9486 | 0.941 |
| Fold-2 | 0.9240 | 0.9365 | 0.9508 | 0.943 |
| Fold-3 | 0.9123 | 0.9257 | 0.9531 | 0.938 |
| Fold-4 | 0.9264 | 0.9384 | 0.9553 | 0.946 |
| Fold-5 | 0.9304 | 0.9420 | 0.9553 | 0.948 |
| Fold-6 | 0.9321 | 0.9438 | 0.9508 | 0.947 |
| Fold-7 | 0.9157 | 0.9293 | 0.9464 | 0.937 |
| Fold-8 | 0.9179 | 0.9311 | 0.9486 | 0.939 |
| Fold-9 | 0.9245 | 0.9365 | 0.9575 | 0.946 |
| Fold-10 | 0.9219 | 0.9347 | 0.9486 | 0.941 |
| Average Values | 0.9219 | 0.9347 | 0.9486 | 0.9426 |
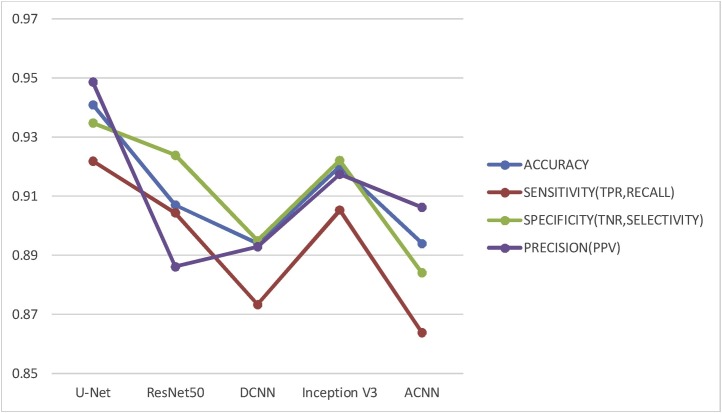
Along with employed U-Net model, standard pre-trained models like ResNet50, DCNN, InceptionV3 and ACNN were trained and tested using similar chest CT-scan images for detection of Covid-19 disease. The images were resized according to the input requirements of the model. Note that all the models mentioned are trained and tested using the similar dataset, hardware and software platform. The performance of the employed U-Net architecture was compared with the implemented ResNet50, DCNN, InceptionV3 and ACNN architectures using the same dataset and same hardware using evaluation parameters like sensitivity, specificity, precision and accuracy results are as shown in Fig. 7 . Comparison of execution time required to process a single test image is given in Table 2 .
Fig. 7.
Comparison of evaluation parameters for Covid-19 disease detection using different CNN's and FCN.
Table 2.
Comparison of Execution time required for five Network architectures.
| Network Architecture | Execution Time |
|---|---|
| U-Net | 1.06 s |
| ResNet50 | 2.34 s |
| DCNN | 2.41 s |
| Inception V3 | 2.11 s |
| ACNN | 1.46 s |
From the details of Fig. 7 and Table 2, it is clear that U-Net performance is better in multiple aspects as compared to other conventional neural networks. Further comparison of the proposed method with state-of-the-art methodologies is given in Table 3 .
Table 3.
Comparison of the proposed method with State-of-the-Art Methods.
| Author and Year | Deep Learning Module Used | Type of Diseases Involved | Dataset and type of image | Results |
|
|---|---|---|---|---|---|
| Metric Name | Metric Value | ||||
| Zheng C et al. (Jan 2020) [16] | Deep CNN | COVID-19 | Local Hospitals 630 CT Images | Accuracy | 0.9 |
| Xiaowei Xu et al. (Feb 2020) [8] | 3D-CNN | COVID-19 | CT images (Hospitals in China) 618 samples |
Accuracy | 0.87 |
| Influenza-A viral pneumonia | |||||
| Healthy People | |||||
| Ali Naren et al. (March 2020) [13] | ResNet50 | COVID-19 | GitHub and Kaggle repository 100 chest X-ray Images |
Accuracy | 0.98 |
| Inception -ResNetV2 | 0.87 | ||||
| InceptionV3 | 0.97 | ||||
| Gozes et al. (March 2020) [17] | ResNet-50 based 2-D CNN | Covid-19 | 56 CT Images from Local Hospitals | Sensitivity | 0.98 |
| Specificity | 0.92 | ||||
| AUC | 0.99 | ||||
| Barstuga et al. (March 2020) [18] | Feature Extraction –GLCM, LDP, GLRLM, GLSZM, DWT | COVID-19 | Local Hospitals 150 CT abdominal Images | Accuracy | 0.99 |
| Classifier- SVM | |||||
| Proposed Method | U-Net | COVID-19 |
1000 Chest CT Images GitHub Repositories & SIRM |
Sensitivity | 0.92 |
| Specificity | 0.93 | ||||
| Accuracy | 0.94 | ||||
| Precision | 0.95 | ||||
We have demonstrated a novel method with the use of advanced U-Net architecture for segmentation and classification of chest CT images for normal or COVID-19 affected cases. To illustrate the performance of our method, we have collected chest CT samples from three different sources. The pixel-based evaluation of the proposed method has achieved a sensitivity of 92.19%, specificity of 93.47%, precision of 94.86% and an accuracy of 94.26% as shown in Table 3. The values of performance parameters obtained using dataset acquired from three different sources prove the robustness of our algorithm.
Our method showed better performance when compared to the architectures proposed by (Xiaowei Xu et al.[10] and Zheng C et al. [18]. Though the performance of ResNet-50 was better for X-ray images in terms of overall accuracy - the experimentation was performed on the very small size of a dataset as compared to our method. Even in the case of chest CT images, the dataset mentioned is small in size. Though the value of sensitivity is more, the value of specificity is smaller compared to our method.
The segmentation results of available CNN approaches get hindered due to the use of coarse mapping for up-sampling the down-sampled pixels. In U-Net architecture, the receptive parts next to convolution operations are connected by the receptive parts in the up-convolving method. This aspect permits the U-Net model, to utilize original descriptions along with features from up-convolution. So, the overall performance observed with U-Net is better than other Convolutional architectures. Barstuga et al. [11] used different feature extraction methods with SVM classifier and achieved the highest accuracy of 99%. While the comparison is performed to get a rough idea about the performance of various methods, a detailed introspection is needed on number and types of images used, sources of samples, pre and post-processing required, number of samples used for training and testing, computational complexity involved and time taken by each method.
5. Conclusions
In an effort to fulfil the urgent necessity which has emerged to fight against the COVID-19 pandemic, we have devised an AI-based tool for automatic detection of the COVID-19 disease. The higher performance parameters achieved in terms of sensitivity, specificity and accuracy, for the input samples from different sources, proves the robustness of the proposed algorithm. In the scenarios of availability of chest CT samples, the recommended U-Net architecture proves a better choice being able to work with little data. The flexibility of U-Net to work with various image sizes makes the algorithm scale-invariant. A comparison of the employed U-Net architecture against state-of-the-art CNN's shows that the proposed network outperforms in terms of (small) number of training samples, precise pixel locations- as it detects anomalies with their locations and overall accuracy. Additionally, to demonstrate the robustness of the algorithm - the database was prepared with CT images from three different sources. Hence, the suggested tool can be promising as an assistive tool to clinical teams for diagnosis of the COVID-19 disease, thereby reducing the global burden on them and will help to reduce the chances of cross contaminations at hospitals. Further, it can be trained for various types of lung infections and more possible abnormal patches which will allow recognizing the exact reason for the infection.
Authors contribution
All the authors have contributed equally.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.Yan L., Zhang H.T., Xiao Y., Wang M., Guo Y., Sun C., Tang X., Jing L., Li S., Zhang M., Xiao Y., Cao H., Chen Y., Ren T., Jin J., Wang F., Xiao Y., Huang S., Tan X., Huang N., Jiao B., Zhang Y., Luo A., Cao Z., Xu H., Yuan Y. Prediction of criticality in patients with severe Covid-19 infection using three clinical features: a machine learning-based prognostic model with clinical data in Wuhan. medRxiv preprint. 2020:1–18. doi: 10.1101/2020.02.27.20028027. [DOI] [Google Scholar]
- 2.Espy M.J., Uhl J.R., Sloan L.M., Buckwalter S.P., Jones M.F., Vetter E.A., Yao J.D.C., Wengenack N.L., Rosenblatt J.E., Cockerill F.R., III, Smith T.F. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 2006;19(January 1):165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.worldometers.info/coronavirus/.
- 4.Wang W., Xu Y., Gao R., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. Published online 11th March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao Wei, Zhong Zheng, Xie Xingzhi, Yu Qizhi, Liu Jun. Relation between chest CT findings and clinical conditions of coronavirus disease (COVID-19) pneumonia: a multicenter study. Am. J. Roentgenol. 2021:1–6. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 6.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020 doi: 10.1016/S0140-6736(20)30183-5. 24th January. pii: S0140-6736(20)30183-30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X., Jiang X., Ma C., Du P., Li X., Lv S., Yú L., Chen Y., Su J., Lang G., Li Y., Zhao H., Xu K., Ruan L., Wu W. Deep learning system to screen coronavirus disease 2019 pneumonia. ArXiv. 2020 doi: 10.1016/j.eng.2020.04.010. abs/2002.09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baltruschat I.M., Nickisch H., Grass M., Knopp T., Saalbach A. Comparison of deep learning approaches for multi-label chest X-Ray classification. Sci. Rep. 2019;9(April 1):6381. doi: 10.1038/s41598-019-42294-8. PMID: 31011155; PMCID: PMC6476887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coudray Nicolas, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24(10) doi: 10.1038/s41591-018-0177-5. p. 1559. Gale Academic OneFile, Accessed 27th April. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anthimopoulos M., Christodoulidis S., Ebner L., Christe A., Mougiakakou S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging. 2016;35(May 5) doi: 10.1109/TMI.2016.2535865. [DOI] [PubMed] [Google Scholar]
- 12.https://biodifferences.com/difference-between-x-ray-and-ct-scan.html.
- 13.Narin Ali, Kaya Ceren, Pamuk Ziynet. 2020. Automatic Detection of Coronavirus Disease (COVID-19) Using X-ray Images and Deep Convolutional Neural Networks.https://arxiv.org/abs/2003.10849v1 24 March. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.http://toi.in/4TAgLY/a31gj.
- 15.Zhao X., Liu L., Qi S., et al. Agile convolutional neural network for pulmonary nodule classification using CT images. Int. J. CARS. 2018;13:585–595. doi: 10.1007/s11548-017-1696-0. [DOI] [PubMed] [Google Scholar]
- 16.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for COVID-19 from chest CT using weak label. medRxiv. 2020 1st January. [Google Scholar]
- 17.Gozes O., Frid-Adar M., Greenspan H., Browning Pd, Zhang H., Ji W., Bernheim A., Siegel E. Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv preprint arXiv. 2003 050372020 10th March. [Google Scholar]
- 18.Barstugan M., Ozkaya U., Ozturk S. Coronavirus (COVID-19) classification using CT images by machine learning methods. arXiv preprint arXiv. 2003:09424. 2020 20th March. [Google Scholar]
- 19.Xie Y., Xia Y., Zhang J., et al. Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans. Med. Imaging. 2019;38(April 4):991–1004. doi: 10.1109/tmi.2018.2876510. [DOI] [PubMed] [Google Scholar]
- 20.Cohen Joseph Paul, et al. COVID-19 image data collection. ArXiv. 2020;11597 abs/2003 n. pag. [Google Scholar]
- 21.https://www.sirm.org/category/senza-categoria/covid-19/.
- 22.Zhao Jinyu, et al. COVID-CT-Dataset: a CT scan dataset about COVID-19. ArXiv. 2020;13865 abs/2003. n. pag. [Google Scholar]
- 23.(a) Armato, Samuel G., 3rd, et al. The Lung Image Database Consortium (LIDC) and Image Database Resource Initiative (IDRI): a completed reference database of lung nodules on CT scans. Med. Phys. 2011;38(2):915–931. doi: 10.1118/1.3528204Krizhevsky. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sutskever A., Hinton I., G.E . NIPS; 2012. Imagenet Classification With Deep Convolutional Neural Networks; pp. 1106–1114. [Google Scholar]
- 24.Long J., Shelhamer E., Darrell T. Fully convolutional networks for semantic segmentation. 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Boston, MA; 2015. pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 25.Ronneberger O., Fischer P., Brox T. U-Net: convolutional networks for biomedical image segmentation. Proc. MICCAI. 2015 pp. 234-24. [Google Scholar]
- 26.James Gareth, Witten Daniela, Hastie Trevor, Tibshirani Robert. 1st ed. 2013. An Introduction to Statistical Learning: With Applications in R (Springer Texts in Statistics) Corr. 7th printing 2017 Edition. [Google Scholar]