Abstract
Background
Natural language processing (NLP) methods have the capability to process clinical free text in electronic health records, decreasing the need for costly manual chart review and improving data quality. We developed rule-base NLP algorithms to automatically extract surgery specific data elements from knee arthroplasty operative notes.
Methods
Within a cohort of 20,000 knee arthroplasty operative notes from 2000 to 2017 at a large tertiary institution, we randomly selected independent pairs of training and test sets to develop and evaluate NLP algorithms to detect five major data elements. The size of the training and test datasets were similar and ranged between 420 to 1592 surgeries. Expert rules using keywords in operative notes were used to implement NLP algorithms capturing: (1) category of surgery (total knee arthroplasty, unicompartmental knee arthroplasty, patellofemoral arthroplasty), (2) laterality of surgery, (3) constraint type, (4) presence of patellar resurfacing, and (5) implant model (catalog numbers). We used institutional registry data as our gold standard to evaluate the NLP algorithms.
Results
NLP algorithms to detect category of surgery, laterality, constraint and patellar resurfacing achieved 98.3%, 99.5%, 99.2% and 99.4% accuracy on test datasets, respectively. The implant model algorithm achieved an F1-score (harmonic mean of precision and recall) of 99.9%.
Conclusions
NLP algorithms are a promising alternative to costly manual chart review to automate extraction of embedded information within knee arthroplasty operative notes. Further validation in other hospital settings will enhance widespread implementation and efficiency in data capture for research and clinical purposes.
Level of Evidence
Level III
Keywords: total knee arthroplasty, natural language processing, artificial intelligence
Introduction
Total knee arthroplasty (TKA) is one of the most common surgical procedures1. Over 700,000 TKA procedures are performed each year in the United States, and almost 5 million Americans are currently living with TKA implants2. Growing demand for improved mobility and quality of life is expected to result in further increases in annual procedure volumes, making TKA the most common elective surgery in the coming decades1,3.
The need for high quality, real-world data is one of the obstacles to improve the quality of TKA research and real-time surveillance efforts. Furthermore, without detailed information on TKA-specific data elements, quality improvement efforts also face a critical obstacle and are limited to imperfect data for TKA classification and risk-stratification4,5. A large amount of clinically relevant information is embedded in unstructured text of electronic health records (EHRs). Manual chart review is labor intensive and requires specialized knowledge of highly trained medical professionals. The cost and infrastructure challenges required to implement this is currently prohibitive for most hospitals. Natural language processing (NLP) is a field in artificial intelligence that offers the ability for the computers to understand, analyze, and retrieve structured data from the unstructured free text of EHRs. NLP methods have been successfully applied to extract surgical data elements in orthopedics including total hip arthroplasty surgical characteristics, periprosthetic fractures and surgical site infections6–8. Yet, operative notes for TKA contain different concepts and data elements.
In collaboration with orthopedic surgeons and data scientists, we developed a series of NLP-based algorithms for ascertainment of five common TKA-specific data elements from operative notes and assessed the accuracy of the NLP algorithms against the gold standard of manual chart review by trained registry specialists.
Materials and Methods
After approval from the Institutional Review Board, we identified all 19,954 knee arthroplasty procedures performed at our institution between 2000 and 2017. These surgeries spanned over two decades and were performed by 48 different surgeons. Surgical details and follow-up data for all procedures were available through the institutional joint registry where trained registry personnel manually review and extract data from the EHR and record them in a structured format according to registry specifications.
We focused on five major data elements documented in operative notes and developed a separate NLP algorithm for each data element: (1) category of knee arthroplasty (total knee arthroplasty, unicompartmental knee arthroplasty, patellofemoral arthroplasty), (2) laterality of surgery (right, left, both), (3) constraint type in three categories (posterior-stabilized [PS], cruciate-retaining [CR], other types (ultra-congruent [UC], medial congruent [MC], constrained condylar knee [CCK]), (4) presence of patellar resurfacing, and (5) implant model numbers. A small number of patients in our cohort received a specific knee design (Persona system, Zimmer-Biomet) that allows use of a CR femoral component with either a CR or MC bearing. In this same system if a PS femoral design is used either a PS, MC or UC bearing insert can be used. Due to small numbers, we decided to keep all knees with a MC and those with a UC insert together (regardless of femoral design used) under other category. We randomly selected training and test datasets for each data element to develop the algorithm and evaluated the performance respectively using the registry data as the gold standard. Table 1 contains the distribution of training and test sets for each data element and demographic information. The size of the training and test datasets were similar and ranged between 420 to 1592 surgeries for most data elements.
Table 1:
Number of operative notes in training and test datasets used for implementing NLP algorithms
| Category of Surgery | Side of Surgery | Patella Resurfacing | Type of Constraint | Implant model number | |
|---|---|---|---|---|---|
| Training dataset | 420 | 574 | 789 | 426 | 1586 |
| Mean age (year) | 65.56 | 66.53 | 68.04 | 68.50 | 68.65 |
| Female (%) | 55.5 | 51.7 | 58.2 | 62.44 | 54.54 |
| Test dataset | 422 | 572 | 796 | 412 | 1592 |
| Mean age (year) | 66.30 | 66.65 | 68.38 | 68.21 | 68.8 |
| Female (%) | 52.4 | 53.2 | 59.6 | 57.28 | 56.16 |
NLP Algorithm Development
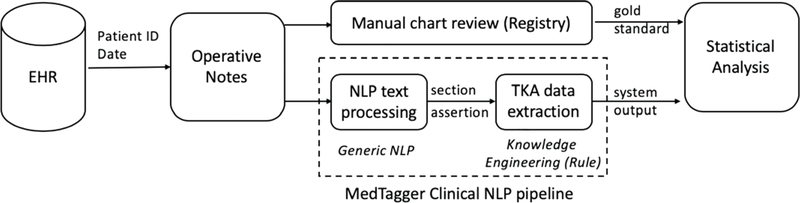
A high-level diagram of the workflow is illustrated in Figure 1. Briefly, we performed the following steps: 1) retrieval of the operative notes from the EHR database, 2) text processing for generic NLP (i.e., sentence segmentation, sectionization, assertion [e.g., positive or negated]) as a preprocessing step to apply expert-based rules, 3) rule development to detect specific data element, and 4) statistical analysis to evaluate the performance of the NLP algorithms. To implement the NLP algorithms, we used MedTaggerIE9, which is an open source information extraction NLP tool specialized for the clinical domain with an ability to detect assertion (e.g., positive, negated, possible) and other attributes (e.g., patient/others, history/present) of the extracted data element. The MedTagger rule engine was used to find keywords and their synonymous variations relevant to each TKA data element based on their description patterns in operative notes. For each data element, our NLP pipeline went through three steps. (1) implementing a prototype system based on the expert knowledge (orthopedic surgeons), (2) implementing an NLP program based on the training dataset, and (3) evaluation of the NLP program within the testing dataset. The final NLP algorithms were evaluated on the independent test datasets. Our NLP programs were based on the rules which were defined by orthopedic surgeons. We applied their proposed rules and terms and then optimized our algorithms based on the performance in training dataset and iterative discussion with experts. The full list of keywords and rules to extract each of the data elements are included in tables Table 2 to 5.
Figure 1:
A Workflow of NLP algorithms to detect TKA data elements
Table 2.
Performance of category of surgery algorithm
| NLP labels⟍Gold standard | Uni | PTKA | PFEM | Keywords |
|---|---|---|---|---|
| Unicompartmental (Uni) | 188 | 1 | 0 | Medial, unicom |
| Primary TKA (PTKA) | 5 | 199 | 0 | TKA, Total knee, Total |
| Patellafemoral (PFEM) | 1 | 0 | 28 | Patellofemoral |
| Accuracy = 98.3% | ||||
|
Rules: Review “PROCEDURE” section If finds any PFEM keywords, assign “PFEM” If finds any PTKA keywords, assign “PTKA” If finds any UNI keywords, assign “UNI” If finds keywords of different types, prioritize to “PTKA,” “UNI,” and “PFEM” in order. | ||||
Table 5.
Performance of patella resurfacing algorithm
| NLP labels⟍Gold standard | Resurfacing | No Resurfacing | Keywords |
|---|---|---|---|
| Resurfacing | 732 | 0 | (round/oval)... patellar/patella, Patellar... (round/oval/dome), Patellar (was/were) prepared, patellar (were/was) cemented, patellar component, surface of patellar removed, patellar surface was\were osteotomized, patellar was resurfaced, patella was cut, patellar button, patella ream, -patella, -patellar, Postresurfacing |
| No Resurfacing | 5 | 59 | |
| Accuracy = 99.4% | |||
|
Rules: Review “Implant Name” or “PREOP DIAGNOSIS” sections If finds any of the with patella resurfacing keywords, assign “With Patella resurfacing” after excluding all the mentions like “without patella resurfacing”, “unresurfaced patella”, “patella unresurfaced”, “patella was/were not” and “not to resurface”. And, also excludes all “Negative” mentions but keep those within sentences with “no.”, | |||
Depending on the nature of the data element, we focused on different sections of operative notes to extract the information. The operative notes in our institution had the following sections: GRAFT/IMPLANT INFORMATION section which listed the implants used along with catalog/model#, implant name, manufacturer and implant placement, PREOP DIAGNOSIS section which contained surgeon’s description of the primary diagnosis, PROCEDURE section which contained description of the operation, and POSTOP DIAGNOSIS section which contained the diagnosis after the operation. For example, for implant model numbers, we only relied on the GRAFT/IMPLANT INFORMATION section. We extracted keywords related to each data element based on expert rules examining the description patterns of data elements in operative notes. The keywords were initially provided by orthopedic surgeons and updated iteratively as we developed the NLP algorithm on the training dataset. For implant models, we extracted implant catalog numbers within the operative notes by NLP. Then, the catalog numbers were mapped to the Global Unique Device Identification Database (GUDID), allowing access to additional data elements (e.g., manufacturer, brand, etc.). The performance of NLP algorithms was assessed using the data recorded in the registry as the gold standard. Performance was assessed through sensitivity (recall), specificity, positive predictive value (PPV or precision), negative predictive value (NPV) and accuracy (the proportion of correct predictions [both true positives and true negatives] among the total number of cases examined) on the test datasets. For implant catalog numbers, we calculated the f1-score (weighted harmonic mean of precision and recall and calculated as 2* [ (precision*recall)/(precision+recall)].
Results
Tables 2 to 6 show the performance of the NLP algorithms to detect the category of knee arthroplasty, laterality, constraint type, whether patella resurfacing was performed or not, and implant model numbers, respectively. All algorithms achieved an accuracy of greater than 98% on the test datasets.
Table 6.
Performance of the algorithm to detect implant model catalog numbers in 1592 operative notes
| NLP labels⟍Gold standard | Catalog number in gold standard | Catalog number not in gold standard |
|---|---|---|
| Catalog number detected by NLP | 6809 | 12 |
| Catalog number not detected by NLP | 4 | 4† |
| Sensitivity = 99.9% Precision = 99.8% F1-score = 99.9% ( 2*(precision*sensitivity) / (precision + sensitivity) ) | ||
|
Rules:
Review “Implant Name” section Detects implant model number Normalizes model numbers and map to Unique Device Identifier (UDI) | ||
No. of operative notes without catalog number
Category of Knee Arthroplasty
The TKA category algorithm classified TKA into three categories (i.e., unicompartmental, primary TKA, patellafemoral) and produced accuracy of 98.3% (Table 2). Seven misclassified operative notes were due to unusual descriptions in the procedure section (i.e., limited invasive surgery with insertion of hemireplacement of diseased compartment), and/or bilateral surgeries with unicompartmental knee arthroplasty on one side and patellofemoral arthroplasty on the other side.
Laterality of Surgery
The laterality algorithm classified surgeries into three categories (right, left, bilateral) with an accuracy of 99.5% (Table 3). Two single-sided surgeries were misclassified as both for laterality because these operative notes described an injection on the contralateral knee in addition to arthroplasty on the one knee.
Table 3.
Performance of the laterality algorithm
| NLP labels⟍Gold standard | Right | Left | Both | Keywords |
|---|---|---|---|---|
| Right | 187 | 0 | 0 | right knee, right gonarthrosis, right arthroplasty, r leg, right leg, right posterior, posterior right |
| Left | 0 | 190 | 0 | left knee, left gonarthrosis, left arthroplasty, l leg, left leg, left posterior, posterior left |
| Both | 2 | 1 | 192 | both knees, bilateral knees, left and right knees, right and left knees, bilateral gonarthrosis, bilateral arthroplasty, bilateral knees, bilateral posterior-stabilized, bilateral posterior, posterior bilateral |
| Accuracy = 99.5% | ||||
|
Rules: Review “PROCEDURE” section If finds any left keywords, assign “left” If find any right keywords, assign “right” If find any both keywords OR meet both left and right conditions, assign “both” | ||||
Constraint Type
The constraint type algorithm identified three categories with an accuracy of 99.2% (Table 4). This was the most challenging algorithm. First, the algorithm relied on the implant names and implant placement to identify constraint type and prioritized the labels in the order of CCK, MC, UC, CR, PS. Three misclassified surgeries were mainly due to no mention of constraint type in implant or procedure descriptions.
Table 4.
Performance of constraint type algorithm
| NLP labels⟍Gold standard | PS | CR | Other | Keywords |
|---|---|---|---|---|
| Posterior Stabilized (PS) | 223 | 0 | 0 | PS,P/S, stabilized posterior, posterior stabilized, tib ins stab, tib insert stab, posterior stabilized, posterior cruciate was excised, cruciate holes made, posterior cruciate substituted, poly stab, post stab, PCL sacrificed, cruciate substituted, box cuts, post cruciate substituting, PCL excised, cruciate ligaments excised, cruciate ligaments removed, posterior stabilized insert, rotating platform |
| Cruciate-retaining (CR) | 0 | 138 | 2 | CR, C/R, cruciate retaining, fixed bearing insert, PCL retained, posterior cruciate was intact and retained, cruciate retaining femoral component, cruc ret |
| Other | 0 | 1 | 48 | CCK (CCK, LCCK, LC/CK, constrained condylar femoral component), MC (Mc, medial congruent liner), UC (UC), and the notes that annotators and NLP didn’t find any clue to determine their categories |
| Accuracy = 99.2% | ||||
|
Rules: Review all sections of a note: If finds any of the relative keyword to each PS/CR/Other categories, assigns the relative label. Prioritize the labels which are found in the “Implant Name” and “Implant Placement” Prioritize the labels by the order of: CCK, MC, UC, CR, PS | ||||
Abbreviations: Ultra-congruent (UC), medial congruent (MC), constrained condylar knee (CCK)
Patella Resurfacing
The patella algorithm classified resurfacing with an accuracy of 99.4% using several commonly used descriptive terms (Table 5). Five operative notes were misclassified because the patella resection and resurfacing was not described in procedure section.
Implant Model
The NLP algorithm to extract catalog numbers and map them to GUDID database produced the F-score of 99.9% achieving almost perfect performance (Table 6). Performance metrics in Table 6 are based on 6825 implants listed in 1592 operative notes. Four operative notes did not contain the catalog numbers. Also, only 4 catalog numbers were not detected by the NLP algorithm because they were entered in unusual format within the operative note. The main challenge with this algorithm was to eliminate invalid numbers detected by NLP.
Discussion
NLP algorithms have the capability to process unstructured EHR data, determine the meaning of sentences, and capture the concepts of interest. In addition, they allow development of user-specific rules for various conditions (e.g., assertion [positive, negated, hypothetical], temporal status [present, history], experiencer [patients or others]), enabling the implementation of expert knowledge concepts. In this study, we successfully developed NLP algorithms to automatically extract five data elements from TKA operative notes and demonstrated high performance in accurately identifying them.
The most common reason for discrepancies in our study was inaccurate or erroneous operative notes many of which were created based on templates. Surgeons who used templates in a very few cases did not edit the templates correctly, and therefore, the NLP algorithms detected signs of multiple conflicting data elements in the same operative note. In some cases, such as patella resurfacing, the “error” produced by the NLP algorithm was actually due to an accurate extraction of information as it is from the operative notes of what was inaccurate or erroneous documentation. Furthermore, some data elements, such as the detection of left and right knee surgery were dependent on frequent terms (“left”, “right”) which misled the algorithms. To tackle these discrepancies, we determined the most relevant and specific sections of operative notes, as described under rules in our tables. Although clinical notes are a standard way of communication and documentation by clinicians, they are composed of a huge amount of unstructured text which can vary between surgeons and even between cases performed by the same surgeon, and this can pose challenges for automated data abstraction. Despite these challenges, NLP tools are distinctive in their ability to extract critical information from unstructured text in EHRs and potentially obviate the barriers of costly manual chart review. Recently, Wyles et al8 evaluated the ability of NLP in identifying common elements of total hip arthroplasty (THA) described by surgeons in operative notes. They showed NLP-enabled algorithms are a promising alternative to the current gold standard of manual chart review for identifying common data elements from orthopedic operative notes. In another study, Murff et al. evaluated the ability of NLP to identify postoperative complications in the EHRs of 2,974 patients10. They noted that NLP had higher sensitivity, but lower specificity, compared with patient-safety indicators based on discharge coding. These studies as well as many others not only shows capability of NLP to serve as a screening tool for queries of large data sets, but also demonstrate a promising alternative to manual chart review for identifying arthroplasty outcomes11.
The NLP systems developed in this study were based on the open-source clinical NLP pipeline (MedTager; https://github.com/OHNLP/MedTagger), which separates generic NLP processes from knowledge engineering (expert rules). The MedTagger is also built under the Apache Unstructured Information Management Architecture (UIMA; https://uima.apache.org), allowing large-volumes unstructured information analyses to discover knowledge relevant to an end user. The use of an open-source tool and modularized architecture increase system portability to other institutions. The tools and algorithms described in this study were deployed as open-source through a GitHub platform to facilitate further development and applications in other institutions (website: https://github.com/OHNLP/TJA/tree/master/module/TKA_NLP). If the NLP tools are coupled with a mobile technology platform as noted in open mHealth architecture12 it will further enhance workflow efficiencies and shared decision-making in orthopedic surgery.
This study has several potential limitations. The NLP algorithms were developed using operative notes in a single institution tailored to a specific EHR system. Although we achieved high performance, we anticipate that the algorithms may not perform similarly in other institutions due to both surgeon usage patterns and EHR structure and documentation variabilities. However, the algorithms should improve as we apply them to operative reports from other institutions because algorithm refinement is an ongoing iterative process. Another limitation is that the success of NLP algorithms depends on the quality and accuracy of the medical records. A major challenge in this study was identifying data elements in operation notes which were created based on “standardized” templates. In such notes, surgeons may forget to delete the irrelevant parts or fail to add unique features specific o that case and this causes NLP algorithms to detect multiple types of a data element in the same note. The potential degradation of medical record accuracy by automated template driven notes and “clip and paste” functions in EHR that create or propagate erroneous information can be a threat to the accuracy of individual medical records and is a larger issue that deserves specific attention. Additionally, some general terms like “right” or “left” also can make NLP algorithm functionality hard. We tackled these challenges by using specific subsections of the operative notes, but this operative report structure may not be present in other institutions. Finally, we extracted all implant model numbers (457 unique models) from implant name section of operative notes, achieving high performance. Those models can be linked to implant databases such as the Global Unique Device Identification Database (GUDID), allowing easy access other data elements and device attributes and enabling post-market TKA device surveillance.
In conclusion, the NLP algorithms demonstrated excellent performance in identifying five major data elements from TKA operative notes. These algorithms represent a promising alternative to the current gold standard of manual chart review in EHR-based TKA clinical research facilitating large-scale studies. The use of NLP to extract of data elements from EHRs paves a way to advanced analytics coupled with machine learning such as automated surveillance of TKA complications and clinical applications
Supplementary Material
Acknowledgments
Funding: Supported by the National Institutes of Health (NIH) grants R01AR73147 and P30AR76312.
Abbreviations
- TKA
total knee arthroplasty
- NLP
natural language processing
- EHR
electronic health records
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kurtz SM, Ong KL, Lau E, Bozic KJ. Impact of the economic downturn on total joint replacement demand in the United States: updated projections to 2021. J Bone Joint Surg Am. 2014;96(8):624–630. [DOI] [PubMed] [Google Scholar]
- 2.Maradit Kremers H, Larson DR, Crowson CS, et al. Prevalence of Total Hip and Knee Replacement in the United States. J Bone Joint Surg Am. 2015;97(17):1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cram P, Lu X, Kates SL, Singh JA, Li Y, Wolf BR. Total knee arthroplasty volume, utilization, and outcomes among Medicare beneficiaries, 1991–2010. JAMA. 2012;308(12):1227–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrios-Torres SI, Umscheid CA, Bratzler DW, et al. Centers for Disease Control and Prevention Guideline for the Prevention of Surgical Site Infection, 2017. JAMA Surg. 2017;152(8):784–791. [DOI] [PubMed] [Google Scholar]
- 5.Bozic KJ, Grosso LM, Lin Z, et al. Variation in hospital-level risk-standardized complication rates following elective primary total hip and knee arthroplasty. J Bone Joint Surg Am. 2014;96(8):640–647. [DOI] [PubMed] [Google Scholar]
- 6.Thirukumaran CP, Zaman A, Rubery PT, et al. Natural Language Processing for the Identification of Surgical Site Infections in Orthopaedics. J Bone Joint Surg Am. 2019;101(24):2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tibbo ME, Wyles CC, Fu S, et al. Use of Natural Language Processing Tools to Identify and Classify Periprosthetic Femur Fractures. J Arthroplasty. 2019;34(10):2216–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wyles CC, Tibbo ME, Fu S, et al. Use of Natural Language Processing Algorithms to Identify Common Data Elements in Operative Notes for Total Hip Arthroplasty. J Bone Joint Surg Am. 2019;101(21):1931–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, Bielinski SJ, Sohn S, et al. An information extraction framework for cohort identification using electronic health records. AMIA Jt Summits Transl Sci Proc. 2013;2013:149–153. [PMC free article] [PubMed] [Google Scholar]
- 10.Murff HJ, FitzHenry F, Matheny ME, et al. Automated identification of postoperative complications within an electronic medical record using natural language processing. JAMA. 2011;306(8):848–855. [DOI] [PubMed] [Google Scholar]
- 11.Lee GC. More Data Please! The Evolution of Orthopaedic Research: Commentary on an article by Cody C. Wyles MD, et al. : “Use of Natural Language Processing Algorithms to Identify Common Data Elements in Operative Notes for Total Hip Arthroplasty”. J Bone Joint Surg Am. 2019;101(21):e118. [DOI] [PubMed] [Google Scholar]
- 12.Ramkumar PN, Muschler GF, Spindler KP, Harris JD, McCulloch PC, Mont MA. Open mHealth Architecture: A Primer for Tomorrow’s Orthopedic Surgeon and Introduction to Its Use in Lower Extremity Arthroplasty. Journal of Arthroplasty. 2017;32(4):1058–1062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.