SUMMARY
Pseudomonas aeruginosa is an opportunistic pathogen that senses and responds to its environment via four chemosensory systems. Oxygen activates the Che2 chemosensory system by binding to the PAS-heme domain of the Aer2 receptor. Ostensibly, the output of Che2 occurs via its response regulator CheY2, but controversy persists over CheY2’s exact role. In this study we show that CheY2 does not interact with the flagellar motor and that the Che2 system does not transfer phosphoryl groups to the chemotaxis (Che) system. We show that CheY2 instead provides feedback control of Aer2 adaptation. In the presence of O2, Aer2 signaling increases the autophosphorylation of the histidine kinase CheA2, followed by CheY2-mediated dephosphorylation. CheY2 does not stably retain phosphate and may not signal the output of the Che2 system. Rather, CheY2 activity enhances the direct interaction of CheY2 with the adaptation protein CheD (a role often facilitated by CheC, which P. aeruginosa lacks). In the absence of O2, Aer2 does not signal, and CheY2/CheD interactions attenuate. This frees CheD to augment CheR2-mediated methylation of Aer2, which enhances Aer2 signaling. CheD does not interact with CheR2, but most likely interacts with Aer2 via conserved CheD-binding motifs to make Aer2 a better methylation substrate.
Keywords: Pseudomonas aeruginosa, signal transduction, chemosensing, response regulator, adaptation, bacterial motility
ABBREVIATED SUMMARY
In this study we explored protein interactions and phosphotransfer reactions amongst the Che2 chemosensory proteins of P. aeruginosa. Our results support the following model: In the presence of O2, Aer2 promotes CheA2 autophosphorylation, CheY2-mediated dephosphorylation, and CheY2/CheD interaction. In the absence of O2, Aer2 does not signal and CheD is freed from CheY2 to augment CheR2-mediated methylation of Aer2, which enhances Aer2 signaling.
INTRODUCTION
Bacterial chemosensory systems sense and respond to stimuli via chemoreceptors that activate phosphotransfer cascades and initiate cellular responses. Pseudomonas aeruginosa has four chemosensory systems, three of which regulate biofilm formation (the Wsp system), twitching motility (the Pil-Chp system), and flagellum-mediated chemotaxis (the Che system) (Kato et al., 2008, Sampedro et al., 2014). The role of P. aeruginosa’s fourth chemosensory system, Che2, is not well understood, although it appears to be involved in stress responses and it impacts virulence (Schuster et al., 2004, Garvis et al., 2009). The P. aeruginosa chemotaxis system receives sensory input from 23 chemoreceptors, whereas the three additional chemosensory systems employ one receptor each (Ortega et al., 2017). The receptor for Che2 is Aer2 (McpB) and it is encoded within the che2 operon [(Hong et al., 2004), Fig. 1A]. Che2 proteins (Y2, A2, W2, Aer2, R2, D and B2) are expressed in stationary phase and form an Aer2-mediated complex near the cell pole that does not co-localize with chemotaxis proteins [(Schuster et al., 2004, Guvener et al., 2006, Yang & Briegel, 2020), Fig. 1B]. The stimulus for Che2 is O2, which binds to the PAS-heme domain of Aer2 with the assistance of a Trp residue that rotates to bond with O2 [(Watts et al., 2011, Airola et al., 2013, Garcia et al., 2017), Fig. 1B]. This initiates a conformational signal in Aer2 that is transmitted to the C-terminal HAMP and kinase control domains. Ostensibly, Aer2 signaling modulates the autophosphorylation of the bound histidine kinase CheA2, which in turn transfers phosphoryl groups to the response regulator CheY2 (Fig. 1B). In the chemotaxis system, CheY has been characterized and, once phosphorylated, it binds to the flagellar motor protein, FliM (Kato et al., 2008). This reverses the direction of flagellar rotation and modulates bacterial swimming direction. In contrast to CheY, the output of CheY2 remains unknown.
Fig. 1. The che2 operon and proposed Che2 phosphorelay in P. aeruginosa.
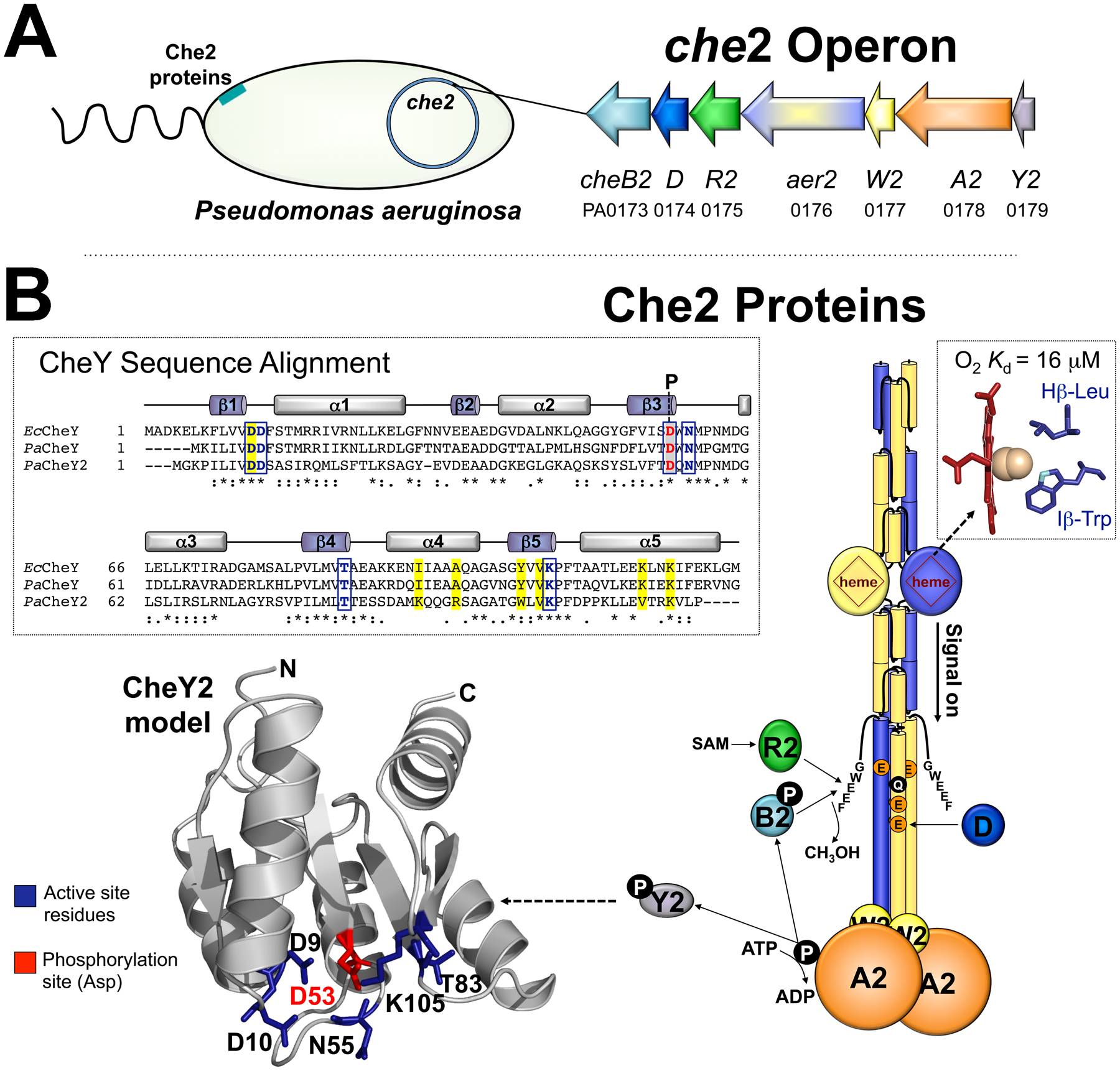
A. The che2 operon encodes a complete chemosensory system including the Aer2 receptor, CheA2 histidine kinase, CheW2 coupling protein, CheY2 response regulator, and three adaptation enzymes CheR2, CheD and CheB2. PAO1 gene numbers are given. The Che2 proteins form an Aer2-mediated complex near the cell pole.
B. Model of an Aer2 dimer and proposed Che2 signaling pathway. When O2 enters the PAS-heme domain of Aer2 (represented by a circle with heme inside), a PAS Hβ-Leu residue moves out of the ligand-binding site and an Iβ-Trp rotates to bond with heme-bound O2 [see inset; the O2-bonding nitrogen is colored light blue in cyanide-bound PAS, PDB entry 3VOL (Sawai et al., 2012)]. This initiates a conformational signal that is transmitted to the C-terminus of Aer2, promoting the autophosphorylation of bound CheA2, with subsequent phosphotransfer to CheY2. Aer2 signaling is regulated by the deamidation, methylation, and demethylation of specific adaptation site residues (QEEE, represented by individual colored circles) after the binding of CheB2-P methylesterase or CheR2 methyltransferase to the C-terminal pentapeptide (GWEEF) of Aer2. A sequence alignment for E. coli CheY, P. aeruginosa CheY and CheY2 is shown (as generated by Clustal Omega; stars indicate conserved residues, colons indicate similar amino acids, and periods indicate amino acids with weakly similar properties). Also shown is a structural model of CheY2 based on E. coli CheY [PDB entry 3CHY (Volz & Matsumura, 1991)] created in SWISS-MODEL. Active site residues in E. coli [for Mg2+ binding and phosphorylation (Volz & Matsumura, 1991, Lee et al., 2001)] are indicated by boxed blue letters and blue side chains, whereas the Asp residue that is phosphorylated in E. coli CheY (Sanders et al., 1989) is indicated by red letters with gray shading and a red side chain (D53 in CheY2). These residues are conserved in P. aeruginosa CheY and in CheY2. CheY residues known to interact with FliM in E. coli [(Lee et al., 2001), highlighted yellow in the sequence alignment] are conserved in P. aeruginosa CheY, but not in CheY2. Abbreviation: SAM, S-adenosylmethionine.
CheY2 is a single domain response regulator that shares 33% sequence identity with chemotaxis CheY. CheY2 contains conserved active site residues, which suggests that it can be phosphorylated (blue and red side chains and boxes in Fig. 1B). However, most of the residues in CheY that are predicted to interact with FliM are not present in CheY2 (highlighted yellow in the Fig. 1B alignment). The CheY2 orthologue from Vibrio cholerae (CheY4) similarly lacks FliM interacting residues and does not bind FliM (Dasgupta & Dattagupta, 2008, Biswas et al., 2013). In P. aeruginosa, deleting cheY2 does not disrupt chemotaxis or aerotaxis (Guvener et al., 2006). Similarly, expressing CheY2 in Escherichia coli does not disrupt E. coli chemotaxis, whereas expressing CheA2, CheW2 or CheB2 does (Ferrandez et al., 2002). Aer2 itself shares significant similarities with E. coli chemoreceptors: the kinase control module of Aer2 is the same length as E. coli Tsr and includes a C-terminal pentapeptide (GWEEF) for adaptation enzyme binding and four putative methylation sites (QEEE; Fig. 1B). The similarities between Che2 proteins (except for CheY2) and the E. coli chemotaxis system helps to explain why Aer2 can hijack E. coli chemotaxis to cause bacterial tumbling in the presence of O2 (Watts et al., 2011). In the absence of O2, Aer2 does not signal and E. coli cells expressing Aer2 as their sole chemoreceptor resume random swimming behavior. The fact that Aer2 can hijack E. coli chemotaxis concurs with recent bioinformatic analyses indicating that Che2 is ancestral to the E. coli chemotaxis system (Ortega et al., 2020).
When Aer2 is expressed in E. coli, it does not adapt to O2 (Watts et al., 2011). This is because the E. coli methyltransferase, CheR, methylates Aer2’s glutamate residues (potentiating the “on” signal), but E. coli’s methylesterase, CheB, does not efficiently deamidate and/or demethylate Aer2’s adaptation site residues (Watts et al., 2011). Ordinarily, the combined activities of CheR and CheB continuously update the methylation record of a receptor and enable cells to detect stimuli over a wide concentration range (Parkinson et al., 2015). In P. aeruginosa, the Che2 system expresses a methyltransferase, CheR2 [which binds to Aer2’s C-terminal pentapeptide and methylates Aer2 (Garcia-Fontana et al., 2014)], a methylesterase CheB2, and an additional enzyme, CheD (Fig. 1). CheD proteins are found in most chemotactic bacteria, although they are absent in enterobacteria like E. coli (Chao et al., 2006). CheD proteins assist adaptation by deamidating or demethylating receptors after binding to the modification site (Kristich & Ordal, 2002, Chao et al., 2006, Glekas et al., 2012). CheD proteins also augment the dephosphorylation of CheY by activating a CheC or CheX phosphatase (Kristich & Ordal, 2002, Chao et al., 2006, Moon et al., 2016). In addition, CheD proteins activate receptors both directly (Walukiewicz et al., 2014) and by increasing receptor methylation, a function that is regulated by CheC in response to the levels of CheY-P (Rosario et al., 1995, Muff & Ordal, 2007). CheY-P increases the affinity of CheD for CheC, thus preventing CheD from interacting with receptors (Yuan et al., 2012). To date, CheD proteins have not been studied in organisms like P. aeruginosa that lack CheC or CheX, and their role remains unknown.
In this study, we analyzed protein interactions and phosphotransfer reactions both within and between the P. aeruginosa Che2 and Che systems. We show that the Che2 system does not transfer phosphoryl groups to the Che system and that CheY2 does not interact with the flagellar motor to modulate chemotaxis. We demonstrate that CheY2 instead functions to provide feedback control of Aer2 signaling though direct interaction with CheD. CheY2/CheD interactions were enhanced by phosphotransfer from CheA2 to CheY2. We also show that free CheD augments both CheR2-mediated methylation of Aer2 and CheR1-mediated methylation of chemotaxis receptors. Lastly, we present evidence that the output of the Che2 system (and hence the cellular response mediated by Che2) may not occur though CheY2.
RESULTS
The Che2 system does not interact or phospho-crosstalk with the Che system
In P. aeruginosa, the Che2 chemosensory proteins form an Aer2-mediated cluster near the cell pole (Guvener et al., 2006). In this cluster, O2 binding to Aer2 is thought to regulate CheA2 autophosphorylation, followed by phosphotransfer to CheY2 (Fig. 1). The role of CheY2-P is unknown. One possibility is that the Che2 system crosstalks with the Che system by shuttling phosphoryl groups to chemotaxis CheY. CheY-P would then bind to the flagellar motor, changing the direction of flagellar rotation and causing cell reversal. To assess Che2-Che crosstalk, we analyzed protein interactions in a Bacterial Adenylate Cyclase Two Hybrid (BACTH) assay. The BACTH system relies on interaction-mediated reconstitution of adenylate cyclase from Bordetella pertussis, the catalytic domain of which is divided into two fragments (T25 and T18) on complementary plasmids (Battesti & Bouveret, 2012). Genes were cloned with the T25 or T18 tag on the 5′ or 3′ end, and plasmid pairs were introduced into E. coli BTH101 (which lacks functional adenylate cyclase; see Table S1 for constructs). Co-transformants were then spotted onto MacConkey agar. Lactose fermentation (red colonies) indicated protein interaction, the strength of which was quantified in a β-galactosidase assay. We used BACTH to assess interactions between the Che2 histidine kinase CheA2 and i) CheY2 (the Che2 response regulator), ii) CheY2-D10K [a potentially signal-on version of CheY2; the E. coli equivalent, EcCheY-D13K, is constitutively active and phosphorylation-independent (Bourret et al., 1993, Jiang et al., 1997)], and iii) P. aeruginosa CheY (the Che system response regulator). In control experiments, we assessed interactions between the CheY proteins and E. coli CheA. As predicted, strong interactions occurred between CheA2 and CheY2, with the strongest association between N-terminally tagged CheA2 and C-terminally tagged CheY2 (Fig. 2A, 34-fold higher β-galactosidase activity than relevant controls). This is congruent with the known interaction face between E. coli CheY and the P2 domain of CheA [(McEvoy et al., 1998), and mapped in Fig. 2B]. Similarly, CheY2-D10K interacted strongly with CheA2, and most efficiently with the same tag order just described (Fig. 2A). In contrast, C-terminally tagged CheY did not interact with CheA2 (Fig. 2A). Likewise, E. coli CheA did not interact with CheY2, CheY2-D10K, or with P. aeruginosa CheY (Fig. 2A), although it is likely that heterologous interactions were masked by T18-EcCheA binding to chromosomally-expressed E. coli CheY in BTH101. A summary of protein interactions is provided in Fig. 2C.
Fig. 2. Protein interactions as demonstrated by BACTH assays.
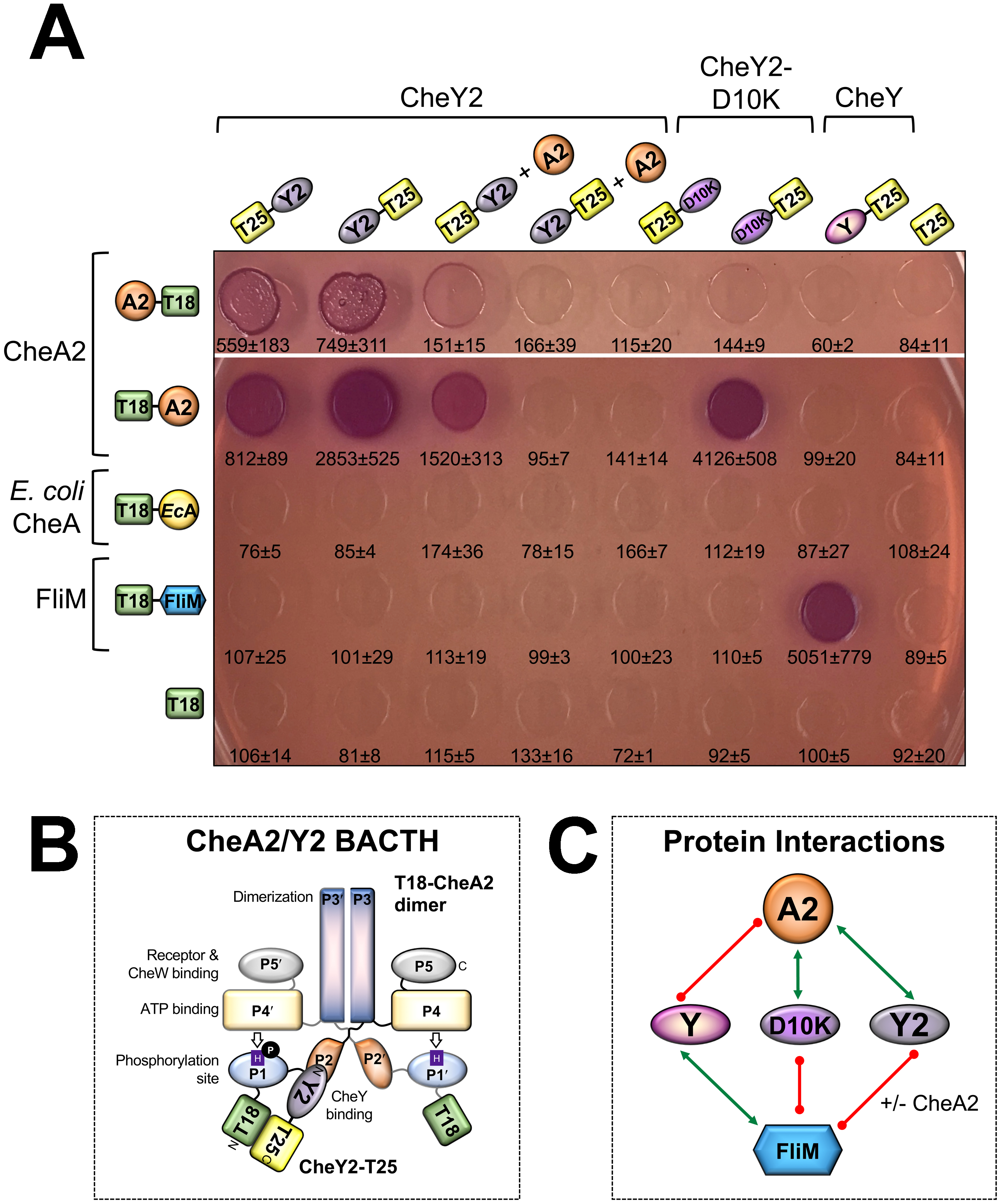
A. E. coli BTH101 cells expressing the indicated fusion proteins were spotted onto MacConkey agar and grown for 48 h at 30 °C (representative photos are shown after 24 h of growth; results were the same at 48 h). Red spots indicate protein interaction, whereas colorless spots indicate weak to no protein interaction. For each co-transformant, the strength of the interaction was quantified in β-galactosidase assays, from which the mean and standard deviation are reported (n>3). T18 and T25 are the two halves of the catalytic domain of B. pertussis adenylate cyclase.
B. Proposed orientation for interaction between a T18-CheA2 dimer and CheY2-T25 monomer (the tag order that produced the strongest adenylate cyclase activity amongst the CheA2/Y2 pairs in Fig. 2A). CheY2 is predicted to interact with the P2 domain of CheA2, analogous to E. coli CheA-CheY.
C. Protein interaction network based on the BACTH results in Fig. 2A. Green arrows link interacting proteins, whereas red lines indicate no interaction. Interactions between E. coli CheA and the various CheY proteins are not shown (see text for details).
BACTH experiments provide a visual indicator of protein interactions. To visualize phosphotransfer between proteins, we purified His-tagged CheA and CheY proteins (see Table S1 for constructs) and performed in vitro γ−32P phosphotransfer assays. In these experiments, CheY2 rapidly dephosphorylated CheA2 (as well as E. coli CheA), but did not stably retain phosphate (Fig. 3A–B). CheY2-P was not usually detected (e.g., Fig. 3A), but could sometimes be observed as a faint band (e.g., in Fig. 3B). The fact that CheY2 dephosphorylated E. coli CheA, even though these proteins did not interact in BACTH assays, further indicates that E. coli CheA preferentially interacts with chromosomally-expressed E. coli CheY in BACTH assays. As expected, E. coli CheY dephosphorylated E. coli CheA and retained phosphate (Fig. 3B). CheY2-D10K interacted with CheA2, but did not dephosphorylate it (Figs. 2 and 3A), similar to reported results for EcCheY-D13K (Bourret et al., 1993, Jiang et al., 1997). P. aeruginosa CheY did not dephosphorylate CheA2 (Fig. 3A; or E. coli CheA, data not shown), indicating that there is no phosphotransfer from the Che2 system into the Che system. A summary of phosphotransfer reactions is provided in Fig. 3C. Overall, the data indicate that in P. aeruginosa, CheY2 specifically interacts with and dephosphorylates CheA2, and that the Che2 system does not transfer phosphoryl groups to the chemotaxis pathway.
Fig. 3. In vitro phosphotransfer between purified CheA and CheY proteins.
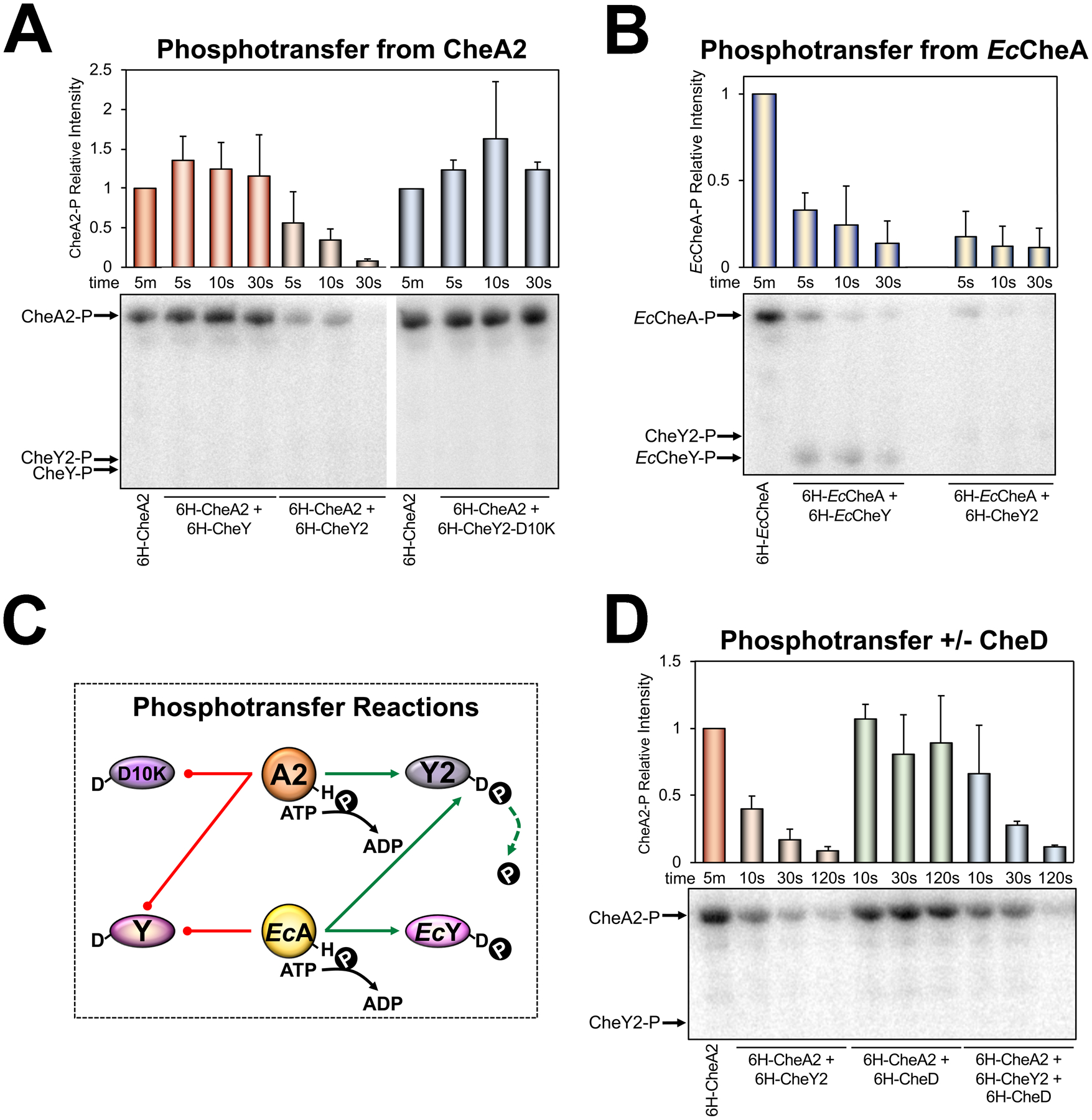
A. Phosphotransfer from P. aeruginosa CheA2 to P. aeruginosa CheY proteins. Bar graphs represent the average intensity of CheA2-P bands normalized to the intensity of CheA2-P after incubating with [γ−32P]-ATP for 5 min. Error bars represent the standard deviation from 4–5 independent experiments. Although CheY2 dephosphorylates CheA2, CheY2 does not stably retain phosphate and a band for CheY2 is not observed on this phospho-image. Bands were observed for all CheY proteins on the Coomassie-stained protein gel (not shown).
B. Phosphotransfer from E. coli CheA to E. coli CheY or P. aeruginosa CheY2. Bar graphs represent the average intensity of EcCheA bands normalized to the intensity of EcCheA after incubating with [γ−32P]-ATP for 5 min. Error bars represent the standard deviation from 3–5 independent experiments. A faint band can be observed for CheY2-P on this phospho-image.
C. Protein phosphotransfer network based on the results of in vitro phosphotransfer experiments in Fig. 3A-B. Green arrows link interacting proteins, whereas red lines indicate no protein interaction.
D. Phosphotransfer from CheA2 to CheY2 in the presence or absence of CheD. Bar graphs represent the average intensity of CheA2 bands normalized to the intensity of CheA2 after incubating with [γ−32P]-ATP for 5 min. Error bars represent the standard deviation from four independent experiments. CheY2-P was not observed on this phospho-image, but was observed on the Coomassie-stained protein gel (not shown).
CheY2 does not interact with the flagellar motor protein FliM
Chemotaxis systems regulate motility via CheY-P binding to the N-terminus of the flagellar switch protein FliM (Welch et al., 1993, Bren & Eisenbach, 1998). Phosphorylation of CheY increases its affinity for FliM 20-fold (Welch et al., 1993). To determine whether CheY2 can interact with FliM, we evaluated protein interactions via BACTH assays. In these assays, N-terminally tagged FliM did not interact with N- or C-terminally tagged CheY2 or with CheY2-D10K (Fig. 2A). However, it could be argued that CheY2 is not sufficiently phosphorylated in E. coli to interact with FliM, even though CheY2 can dephosphorylate E. coli CheA in vitro. To overcome this possible limitation, his-cheA2 was amplified from pProEXHTa-CheA2 along with its Ptrc promoter and cloned into plasmids expressing T25-CheY2 and CheY2-T25 (Table S1). Although His-CheA2 was stably expressed from these plasmids (data not shown), and interacted with tagged CheY2 (His-CheA2 competed with T18-tagged CheA2 in most BACTH assays, Fig. 2A), there was still no evidence of CheY2/FliM interaction (Fig. 2A). This indicates that CheY2 does not interact with FliM, which is consistent with previous data showing that a P. aeruginosa cheY2 mutant had no defect in flagella-mediated chemotaxis or aerotaxis (Guvener et al., 2006). Interestingly, His-CheA2 increased interactions between T18-CheA2 and T25-CheY2 (Fig. 2A). One possible explanation is that His-CheA2/T18-CheA2 heterodimers decrease the steric interference for T25-CheY2 binding compared with T18-CheA2 homodimers. In contrast to the CheY2/FliM data, C-terminally tagged CheY interacted strongly with N-terminally tagged FliM as expected (Fig. 2A).
Identifying the protein partners of CheY2
To identify the protein partners of CheY2 (apart from CheA2), we created two PAO1 genomic DNA (gDNA) libraries: one with an N-terminal T18 tag and one with a C-terminal T18 tag (see Table S1). These libraries were tested in BACTH screens against T25-CheY2 and CheY2-T25 (using the plasmids that also express His-CheA2), and against T25-CheY2-D10K. In total, 1.37 million colonies were screened and 904 red colonies were identified (Table S2). After restreaking all 904 colonies on MacConkey agar, 81 colonies remained strongly red. The plasmids in these 81 clones were then isolated and retested for interaction with the bait CheY2 expression plasmid and against the corresponding empty vector. After these verification steps, just two colonies remained red on MacConkey agar (Table S2) and the gDNA inserts were sequenced. In both cases, the insert corresponded to part of the che2 operon encoding CheR2230–280 (PA0175), CheD1–200 (PA0174), and CheB21–81 (PA0173) (Figs. 1A and 4A). For both clones, only cheD was in frame with the N-terminal T18 tag and all of cheD was present. In the expressed protein, the cheR2 fragment serves as an in-frame 53 residue linker between T18 and CheD (Fig. 4A).
Fig. 4. Interactions between CheD and CheY as demonstrated by BACTH assays.
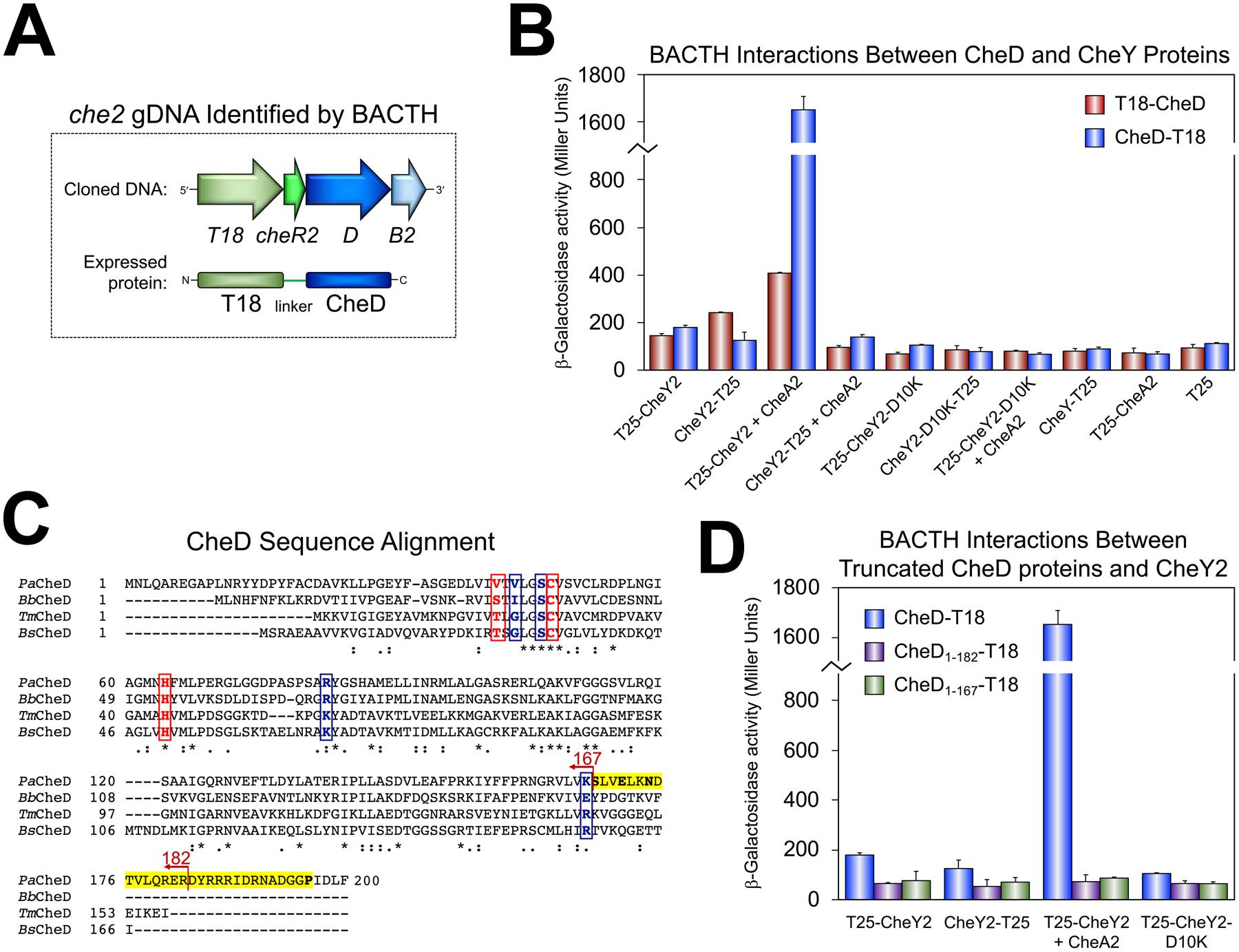
A. The che2 fragment that was identified in the PAO1 gDNA library screen as an interacting partner of T25-CheY2 (in the presence of CheA2). In both clones, only cheD was in frame with T18 and the entire cheD coding sequence was present. In the fusion protein, the cheR2 fragment served as an in-frame 53 residue linker between T18 and CheD.
B. BACTH interactions between CheD and CheY proteins in E. coli BTH101 as quantified in β-galactosidase assays. The mean and standard deviation are shown (n>3).
C. CheD sequence alignment generated in Clustal Omega using CheD sequences from organisms in which CheD has been studied (P. aeruginosa PAO1, Borrelia burgdorferi B31, Thermotoga maritima, and Bacillus subtilis). The C-terminal CheC-like, and possible CheY2-binding, extension in P. aeruginosa CheD (S-X2-E-X2-N-X21-P) is highlighted yellow with conserved residues in bold. Active site residues required for deamidase activity in T. maritima CheD (Chao et al., 2006) are indicated by boxed red letters, whereas residues predicted to interact with receptors (Chao et al., 2006) are indicated by boxed blue letters. The locations of the two CheD truncations created in this study are shown.
D. BACTH interactions between full-length (CheD1–200-T18) and truncated CheD proteins (CheD1–182-T18 and CheD1–167-T18) and CheY2 in E. coli BTH101 as quantified in β-galactosidase assays. The mean and standard deviation are shown (n>3).
Activated CheY2 interacts with CheD
To confirm that CheY2 interacts with CheD, cheD was fused to the 5′ or 3′ end of T18 and tested in BACTH assays against CheY2 and CheY constructs. The strongest interaction occurred between N-terminally tagged CheY2 in the presence of His-CheA2 and C-terminally tagged CheD (Fig. 4B, 15-fold higher β-galactosidase activity than the relevant control). In the absence of CheA2, CheD interacted weakly with N-terminally tagged CheY2, suggesting that either the phosphotransfer activity of CheY2 promotes CheY2/CheD interaction, or that CheA2 itself promotes the interaction. Using BACTH, we tested whether N-terminally tagged CheA2 directly interacts with CheD, and found that it does not (Fig. 4B). We next assayed the influence of purified CheD on the rate of CheY2-mediated dephosphorylation of CheA2, and found no consistent differences (Fig. 3D). Moreover, CheD did not stabilize CheY2-P in vitro (Fig. 3D). Curiously, CheY2-D10K, which lacks phosphotransfer activity (Fig. 3A), did not interact with CheD, either in the presence or absence of CheA2 (Fig. 4B). This further supports the hypothesis that phosphotransfer from CheA2 to CheY2 enhances CheY2/CheD interaction.
CheY2 binds to the C-terminus of CheD
In other bacterial genera, CheD has several distinct functions, i) it acts as a chemoreceptor glutamine deamidase or glutamate demethylase, ii) it augments the dephosphorylation of CheY through interactions with a CheC or CheX phosphatase, and iii) once CheD is free from CheC, it activates CheA by binding to chemoreceptors (Kristich & Ordal, 2002, Chao et al., 2006, Yuan et al., 2012, Moon et al., 2016). P. aeruginosa lacks CheC and CheX, and to the best of our knowledge, CheD has not been studied in a system lacking these phosphatases. Compared with well-studied CheD proteins, the C-terminus of P. aeruginosa CheD contains a sequence extension with a CheC-like motif “S-X2-E-X2-N-X21-P” (Fig. 4C, highlighted yellow). This motif is found in CheC, CheX, and FliY proteins that dephosphorylate CheY (Szurmant et al., 2004), and is necessary for CheC binding to CheY-P (Muff & Ordal, 2007). To determine whether the C-terminus of CheD is responsible for CheY2 binding, we created two CheD truncation mutants: CheD1–182-T18, which contains a partially truncated CheC-like motif, and CheD1–167-T18, which lacks the entire CheC-like motif (Fig. 4C). Both constructs had higher steady-state expression levels than WT CheD (data not shown). However, in BACTH assays, neither of the C-terminally truncated CheD proteins interacted with CheY2, in the presence or absence of CheA2 (Fig. 4D). This is consistent with the notion that CheY2 interacts with the C-terminus of CheD, and concurs with the original experiment in which CheD/CheY2 interactions were most efficient with C-terminally tagged CheD (Fig. 4B).
CheD does not deamidate Aer2
In P. aeruginosa, CheD is expressed from che2 with Aer2 (Fig. 1), so Aer2 is likely a primary target of CheD. Chao and colleagues identified a CheD-binding motif in receptors [A/S-X2-Q/E-Q/E-X2-A/S, (Chao et al., 2006)], and this motif surrounds each of the four predicted adaptation sites in Aer2 [Q414, E421, E428, and E610, Fig. 5A–B, (Watts et al., 2011)]. Additionally, CheD residues that are required for deamidation and receptor interaction are reasonably well conserved in P. aeruginosa CheD (Fig. 4C, red and blue letters, respectively). To test for CheD-mediated deamidation of Aer2, His-tagged WT Aer2 (Aer2QEEE) was purified from E. coli UU2610 (tar, tsr, trg, tap, aer, cheRB) and incubated in vitro with His-CheD. Samples were then run by SDS-PAGE to separate deamidated receptors from unmodified receptors. Deamidated receptors migrate more slowly than unmodified receptors because they are negatively charged and bind less SDS. In these tests, we did not detect any mobility differences for Aer2 incubated either with or without CheD (Fig. 5C). This implies that CheD does not directly modify Aer2 in vitro. It was not possible to perform a CheB2 deamidation control in vitro since CheB2 must be activated by phosphotransfer from CheA2, and CheA2 obscures Aer2 on gels. To overcome this, we performed in vivo deamidation assays in E. coli UU2610 in the presence of CheR2, CheD, or CheB2 and visualized Aer2 on Western blots (Fig. 5D). In these assays, CheR2 methylated both Aer2QEEE and Aer2EEEE, increasing their mobility. In contrast, CheB2 deamidated Aer2QQQQ, decreasing its mobility (Fig. 5D). Owing to the large size of Aer2 (76.5 kDa), these size shifts were small, but reproducible. In contrast, no shift was observed for any Aer2 receptor in the presence of CheD. This agrees with the in vitro results and likewise suggests that CheD does not deamidate Aer2. In Thermotoga maritima CheD, three active site residues are essential for deamidase activity: a threonine that stabilizes the positions of the histidine and cysteine residues that mediate the deamidation reaction [(Chao et al., 2006); residues in red in Fig. 4C]. P. aeruginosa CheD lacks the threonine, so we constructed CheD-V41T and tested it in vivo, but it did not deamidate Aer2QQQQ (data not shown).
Fig. 5. Predicted CheD substrate sites and deamidation of Aer2 in vitro and in vivo.
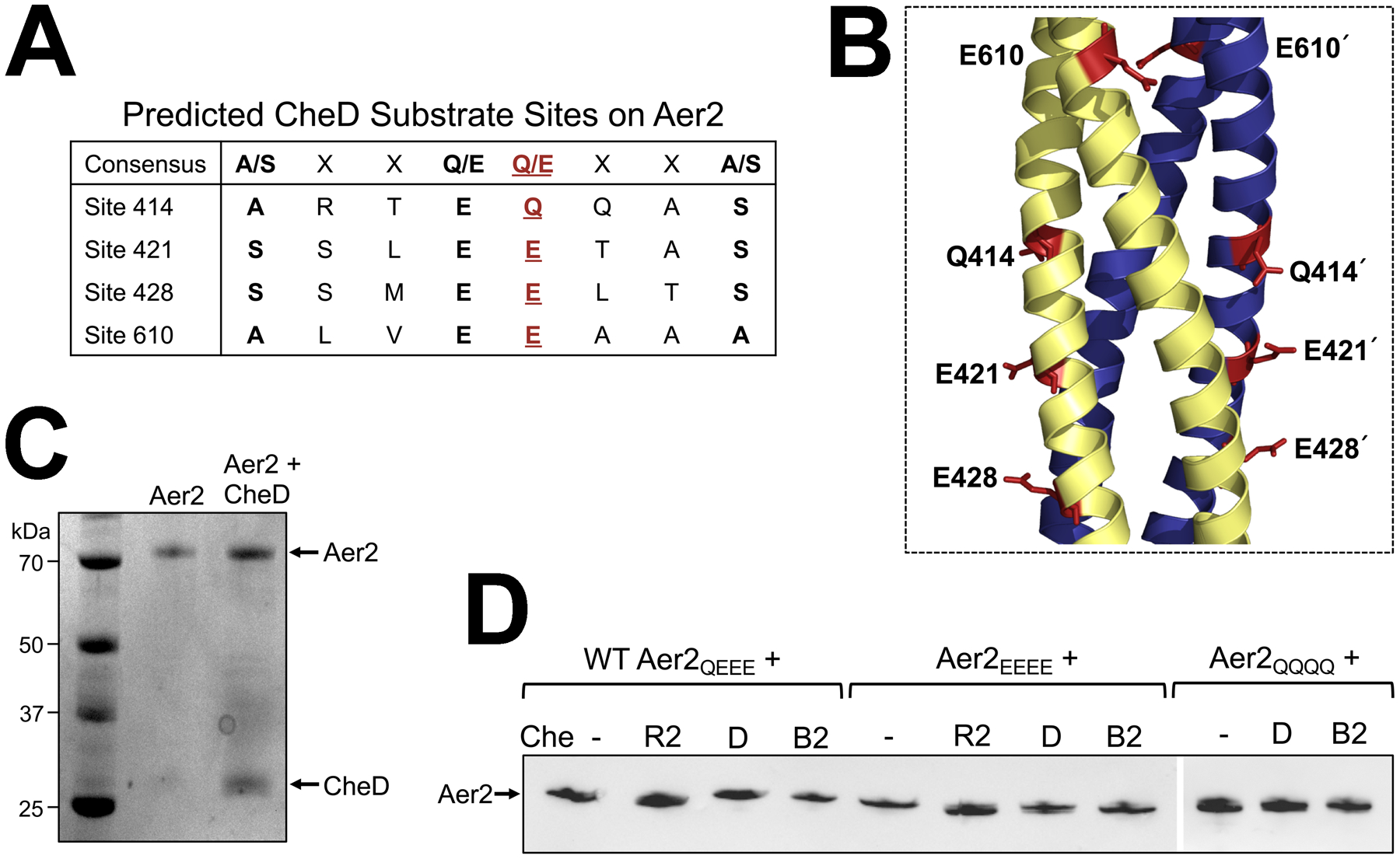
A. Predicted CheD binding sites on Aer2 based on the consensus sequence determined by Chao et al. [A/S-X2-Q/E-Q/E-X2-A/S, (Chao et al., 2006)]. Each of the four predicted methylation sites in Aer2 [Q414, E421, E428, and E610, red font and underlined] are surrounded by the consensus motif for CheD binding.
B. The four predicted adaptation sites in the Aer2 kinase control domain shown as red sticks on an Aer2 dimer model [created in SWISS-MODEL based on the structure of the Tsr kinase control module, PDB entry 3ZX6 (Ferris et al., 2014)].
C. Migration profile of full length purified Aer2 in vitro in the presence or absence of CheD. There was no obvious difference in the migration rate of Aer2 with or without CheD (Coomassie stained gel).
D. Migration profile of full length Aer2 (WT Aer2QEEE, Aer2EEEE, or Aer2QQQQ) after expression in E. coli UU2610 in the presence or absence of CheR2, CheD, or CheB2. Small mobility differences were observed for Aer2 in the presence of CheR2 (Aer2QEEE and Aer2EEEE migrated faster) and CheB2 (Aer2QQQQ migrated more slowly) (Western blots using anti-Tsr antibody). There was no difference in the migration rate of any Aer2 receptor in the presence of CheD.
CheD augments CheR2-mediated methylation and Aer2 signaling
When Aer2 is expressed in chemoreceptorless E. coli, Aer2 hijacks E. coli’s chemotaxis system and directs cell tumbling in the presence of O2, but smooth swimming when O2 is removed (Watts et al., 2011). To determine the effect of the Che2 adaptation proteins on Aer2-mediated behavior in vivo, WT Aer2 (Aer2QEEE) was expressed in E. coli UU2610 in the presence or absence of CheR2, CheD and CheB2 (Fig. 6). Bacteria were then analyzed for tumbling responses in the presence and absence of O2. For UU2610/Aer2, the proportion of bacteria tumbling in air, with or without CheD, was not substantially different (Fig. 6A). This suggests that CheD does not modify Aer2 on its own, congruent with the lack of deamidation observed in deamidation experiments (Fig. 5). UU2610/Aer2 cells expressing CheB2 were signal-off biased compared with cells expressing Aer2 alone, further supporting a role for CheB2-mediated deamidation of Aer2. In contrast, UU2610/Aer2 cells expressing CheR2 were signal-on biased, consistent with methylated Aer2. We next assessed cells expressing various combinations of adaptation proteins. UU2610/Aer2 cells expressing CheDB2 were signal-off biased and tumbled to the same extent as cells expressing only CheB2 (Fig. 6A). However, UU2610/Aer2 cells expressing CheR2D tumbled constantly in both air and in N2 (Fig. 6A). This contrasts with UU2610/Aer2 cells expressing all three adaptation proteins CheR2DB2, where cells tumbled in air and swam smoothly in N2 (a signal-on biased response compared with cells expressing Aer2 alone, Fig. 6A). The Aer2/CheR2D effect (cells tumbling constantly in both air and N2) was also observed in E. coli BT3388, which also expresses E. coli CheR and CheB. Overall, these results suggest that CheD potentiates CheR2-mediated activation of Aer2, possibly through receptor methylation.
Fig. 6. Behavior and methylation of WT Aer2 (Aer2QEEE) in vivo in the presence or absence of the CheB2, CheD and CheR2 adaptation enzymes.
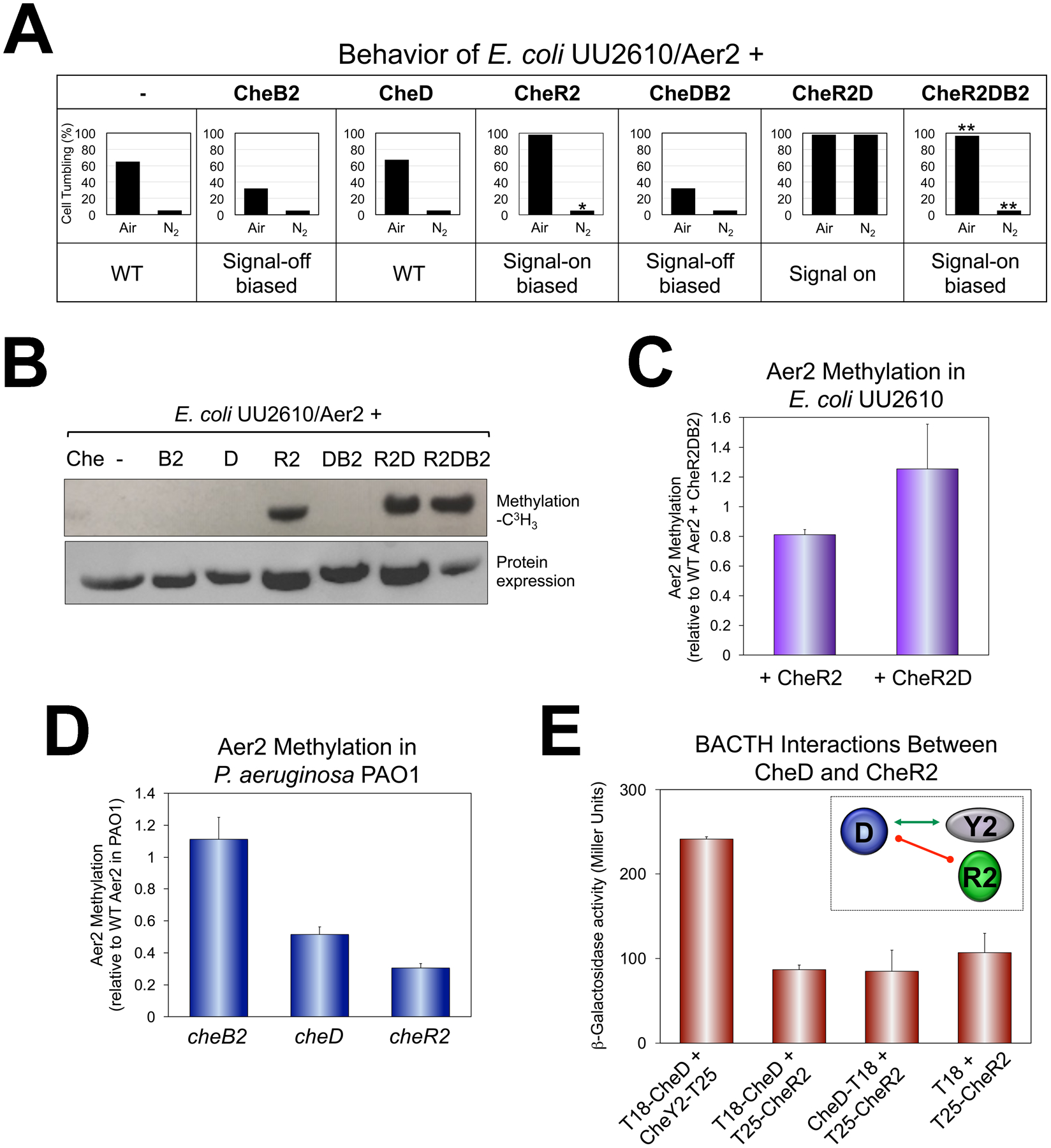
A. Average percent of E. coli UU2610 cells tumbling in the presence of Aer2 and different combinations of CheB2, CheD and CheR2. Percentages represent steady-state tumbling in air and in N2 30 sec after switching to air or N2. CheR2 had a 15 sec delayed smooth-swimming response in N2, which is indicated by an asterisk. CheR2DB2 had a faster smooth-swimming response in N2 (5 sec to smooth-swimming, versus 12 sec for WT), and a slower tumbling response in air (18 sec to tumble versus 7 sec for WT). These are indicated by double asterisks.
B. Methylation of Aer2 in E. coli UU2610 in the presence of different combinations of CheB2, CheD and CheR2 [upper panel, L-(methyl-3H) methionine], and protein expression (lower panel, HisProbe Western blot).
C. Average Aer2 methylation extent in E. coli UU2610 in the presence of CheR2 or CheR2D compared with Aer2 methylation in the presence of CheR2DB2. Error bars represent the standard deviation from three independent experiments. Although Aer2 was more methylated in the presence of CheR2D compared with CheR2 alone, the difference was not highly significant (P = 0.065).
D. Average Aer2 methylation extent in P. aeruginosa PAO1 lacking either CheB2, CheD or CheR2 [all results were significantly different from each other (P < 0.05)]. Error bars represent the standard deviation from three independent experiments.
E. BACTH interactions between CheD and CheR2 in E. coli BTH101 as quantified in β-galactosidase assays (shown as the mean and standard deviation from three independent experiments) and summarized in the inset. The green arrow links interacting proteins, whereas the red line indicates no protein interaction.
To determine whether CheD impacts CheR2-mediated methylation of Aer2, we evaluated methylation levels for Aer2 in both E. coli and in P. aeruginosa. In E. coli, Aer2 methylation was analyzed in UU2610 in the presence of Che2 proteins. As anticipated, Aer2 was methylated by CheR2, but not by CheD, CheB2, or by CheDB2 (Fig. 6B). Aer2 was more highly methylated in the presence of CheR2D compared with CheR2 alone (although the increase was not quite statistically significant, Fig. 6C). However, the trend towards increased methylation in the presence of CheR2D is consistent with the locked tumbling phenotype of these cells. In P. aeruginosa PAO1, we tested the effect of removing these enzymes. His-Aer2 was overexpressed in cheB2, cheD and cheR2 backgrounds and compared with the level of His-Aer2 methylation in WT PAO1. In PAO1, loss of CheD or CheR2 significantly reduced the level of Aer2 methylation, whereas loss of CheB2 moderately increased Aer2 methylation (Fig. 6D). Since Aer2 remained 30% methylated in the cheR2 mutant compared with WT PAO1, it is likely that Aer2 is also methylated in vivo by an alternative methyltransferase.
It is possible that CheD potentiates CheR2 function by direct CheD/CheR2 interaction, or that CheD functions independently, e.g., by binding independently to Aer2 to make Aer2 a better CheR2 substrate. To determine whether CheD directly interacts with CheR2, we performed BACTH assays and found that neither N- or C-terminally tagged CheD interacted with N-terminally tagged CheR2 (Fig. 6E). This implies that the CheD effect is due to direct receptor binding [as modelled by (Chao et al., 2006)].
CheD augments CheR1-mediated methylation of chemotaxis receptors
Lastly, we asked whether CheD could affect the methylation of other P. aeruginosa chemoreceptors. If so, CheD might tune chemotaxis responses according to the level of O2 sensed by Aer2. To explore this, we ascertained which P. aeruginosa chemoreceptors are known to be both methylated by chemotaxis CheR1 (Sheng et al., 2019) and have predicted CheD binding sequences. Two receptors, the aerotaxis receptor Aer and the malate receptor CtpM, fit these criteria. To focus on the methylation regions of these membrane-bound receptors, we cloned the kinase control domains of Aer and CtpM and then performed in vivo methylation assays in the presence of CheR1 or CheR1D. As shown in Fig. 7A, CheD increased the CheR1-mediated methylation levels of both Aer and CtpM (1.7-fold and 1.6-fold, respectively, Fig. 7A). Interestingly, Aer2 was also methylated by CheR1 (although Aer2 was methylated ~3-fold better by CheR2, data not shown). This is congruent with the studies in P. aeruginosa showing that Aer2 remained partially methylated in a cheR2 mutant (Fig. 6D). Notably, CheD did not affect the CheR1-mediated methylation level of Aer2 (Fig. 7A). This contrasts with CheR2, where CheD increased CheR2-mediated methylation by 1.5-fold (Fig. 6C). UU2610/Aer2 cells expressing either CheR1 or CheR1D showed a statistically-significant increase in signal-on bias compared with UU2610/Aer2 cells, consistent with more methylated Aer2 receptors (Fig. 7B). Overall these data indicate that there is crosstalk between the Che and Che2 systems at the level of receptor adaptation that involves both CheR1 and CheD.
Fig. 7. CheR1-mediated methylation and behavior of chemoreceptors in the presence and absence of CheD.
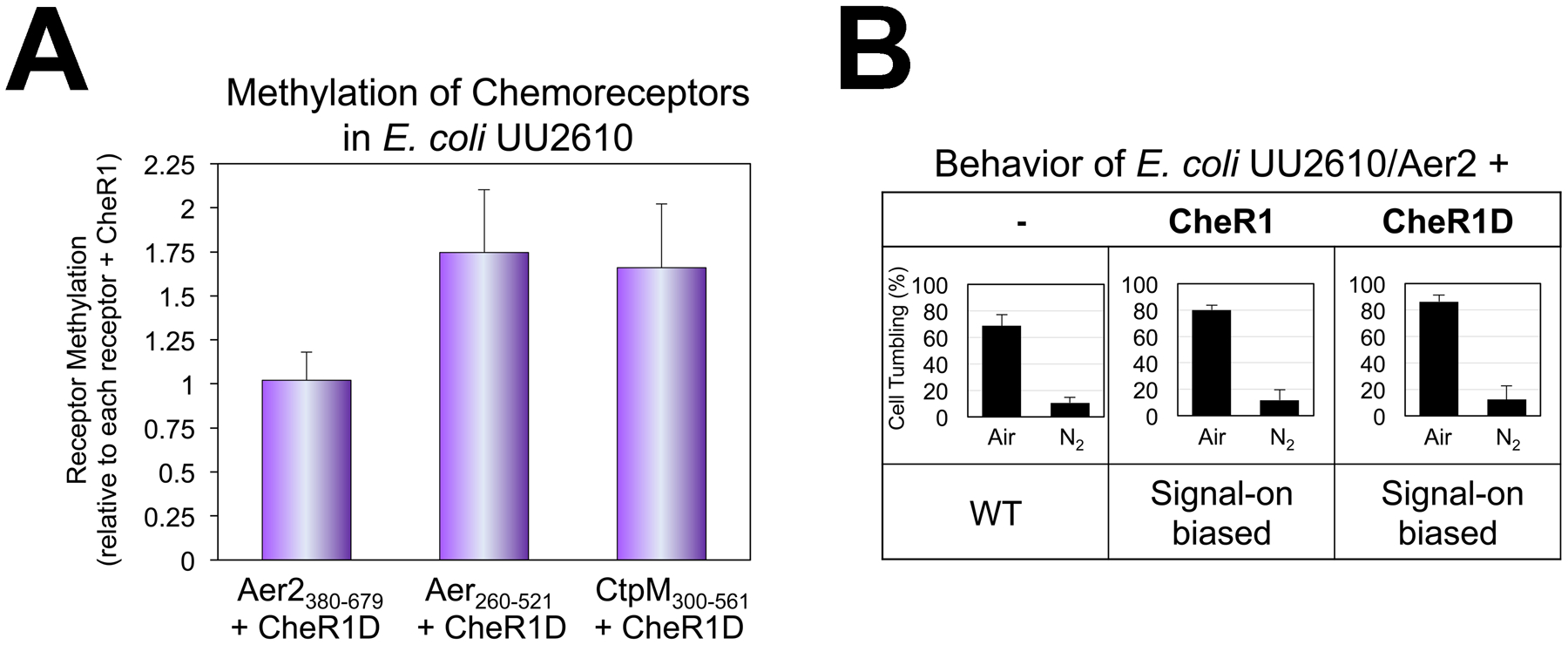
A. Average methylation extents for the kinase control domains of Aer2380–679, Aer260–521 and CtpM300–561 in the presence of CheR1D compared with methylation of the same receptor in the presence of CheR1. Error bars represent the standard deviation from three independent experiments.
B. Average percent of E. coli UU2610 cells tumbling in the presence of WT full-length Aer2, with or without CheR1 or CheR1D. Percentages represent steady-state tumbling in air and in N2 30 sec after switching to air or N2. Aer2-mediated tumbling in air was statistically higher in the presence of CheR1 or CheR1D (P < 0.05).
Che2 signaling pathway
The data from this study indicate that in P. aeruginosa, CheY2 is an important regulator of Aer2 adaptation. Collectively our results support the following model of Che2 signaling: Aer2 signals in response to O2-binding, increasing the autophosphorylation of bound CheA2. CheY2 then removes phosphoryl groups from CheA2, but does not stably retain phosphate (Fig. 8, left panel). However, phosphotransfer from CheA2 to CheY2 enhances CheY2/CheD interaction so that CheD can’t augment CheR2 function. In the absence of O2, Aer2 does not signal, which attenuates CheY2/CheD interactions. This frees CheD to bind to Aer2 and augment CheR2-mediated methylation, enhancing the likelihood of Aer2 signaling (Fig. 8, right panel). In addition, CheD can augment CheR1-mediated methylation of several different chemoreceptors, suggesting that Che2 might also function to tune chemotaxis responses according to the level of O2 sensed by Aer2. In organisms that contain both CheC and CheD, CheC interacts with CheD to prevent CheD-receptor binding. In P. aeruginosa, which lacks CheC, the role of CheC has been replaced by CheY2. Given that CheY2’s interacting partners are currently limited to CheA2 and CheD, and the fact that CheY2 does not stably retain phosphate, it is possible that the output of the Che2 system occurs via phosphotransfer from CheA2 to an alternative response regulator that has not been identified.
Fig. 8.
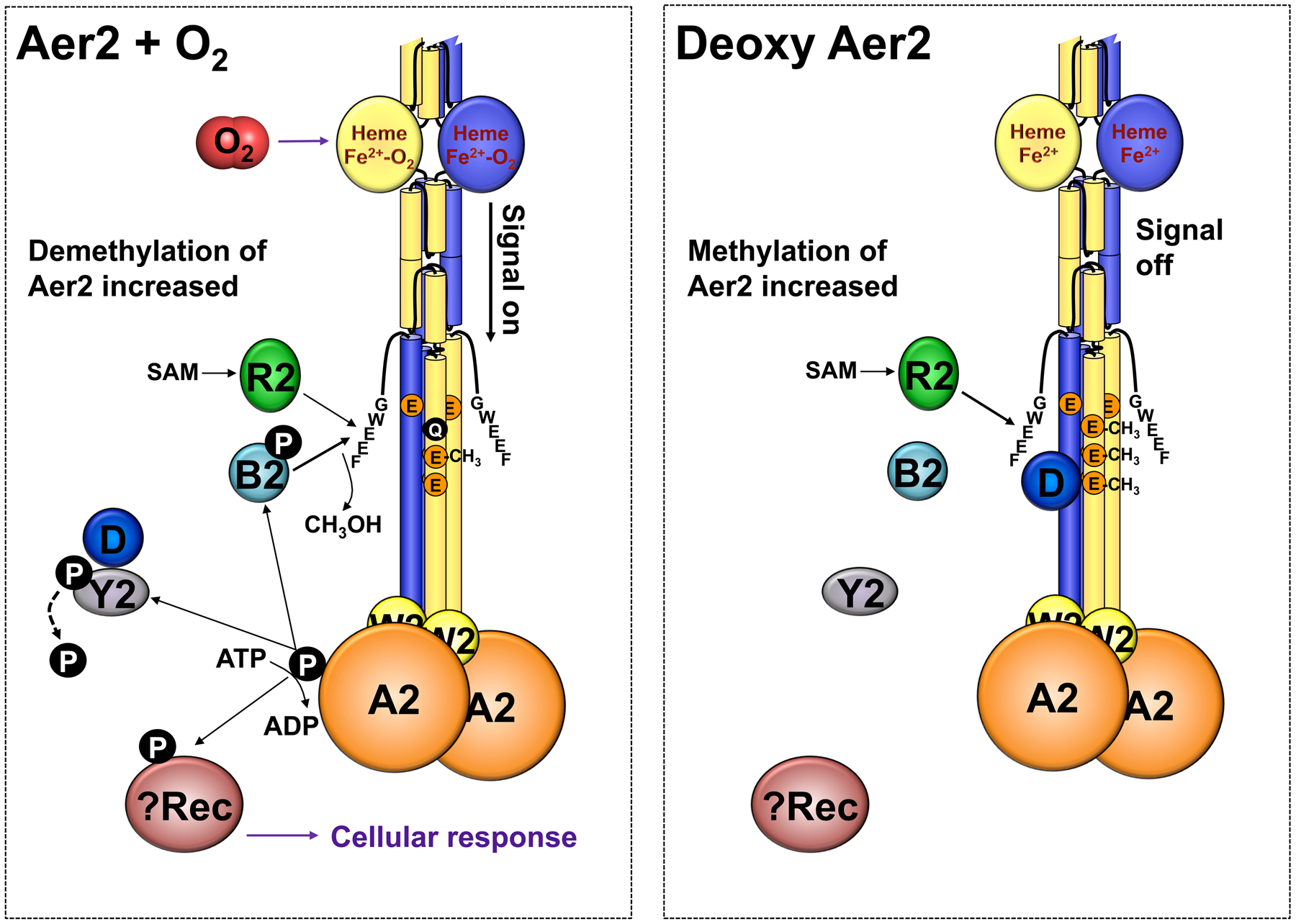
Model summarizing Che2 protein interactions and phosphotransfer reactions consistent with the results of this study. In the presence of O2 (left), Aer2 signaling increases the autophosphorylation of bound CheA2. CheY2 then dephosphorylates CheA2, but does not stably retain phosphate. CheY2 activity enhances CheY2/CheD interactions. Phosphotransfer from CheA2 to CheB2 enhances CheB2 phosphatase activity so that Aer2 is demethylated to terminate signaling. In the absence of O2 (right), Aer2 does not signal, and CheY2/CheD interactions attenuate. This frees CheD to augment CheR2-mediated methylation of Aer2, enhancing the probability of Aer2 signaling. Since CheY2 does not stably retain phosphate, CheA2 possibly transfers a phosphoryl group to an alternative response regulator (Rec) to modulate the cellular response. Abbreviation: SAM, S-adenosylmethionine.
DISCUSSION
In this study we analyzed protein interactions, phosphotransfer reactions and signal adaptation in the Che2 chemosensory system of P. aeruginosa. The Che2 system did not transfer phosphoryl groups to the chemotaxis (Che) system, and the Che2 response regulator, CheY2, did not interact with the flagellar motor protein FliM. This fits with the lack of conserved FliM interacting residues in CheY2 (Fig. 1B), and the absence of Aer2-mediated taxis in P. aeruginosa (Ferrandez et al., 2002, Guvener et al., 2006, Watts et al., 2011). Overall our data fits with a model in which CheY2 is a barometer of Che2 activity and provides feedback control of Aer2 signaling though direct interaction with the adaptation protein CheD (Fig. 8). CheY2/CheD interactions were enhanced by CheY2 activity (which is preceded by Aer2 signaling in response to O2, Fig. 8). In the absence of O2 when Aer2 is signal-off, CheD is free to interact with Aer2 and augment CheR2-mediated methylation, thus enhancing the likelihood of Aer2 signaling (Fig. 8). CheD also enhanced CheR1-mediated methylation of the Aer and CtpM receptors (Fig. 7A). In the case of the aerotaxis receptor Aer, CheD could conceivably increase the likelihood of Aer-mediated cell reversals in a low O2 environment, thus enhancing the aerotaxis response.
In BACTH assays, the strongest CheA2/CheY2 interaction occurred when CheA2 was N-terminally tagged and CheY2 was C-terminally tagged (Fig. 2A). This should orient CheY2 with the P2 (CheY-binding) domain of CheA2, while allowing for reconstitution of adenylate cyclase as modelled in Fig. 2B. Although CheA2 and CheY2 interacted strongly, CheA2 was not identified as an interacting partner of CheY2 in the BACTH gDNA library screen. This might be explained by the fact that cheA2 is a large gene (1920 bp) with 16 Sau3AI recognition sites, and appropriate gene fusions must ligate in frame with T18. In phosphotransfer assays, CheY2 rapidly dephosphorylated CheA2-P, as well as E. coli CheA-P, but CheY2 did not stably retain phosphate (Fig. 3A–B). CheY2-P was occasionally detected as a faint band on phospho-images, but most often was not detected at all (Figs. 3A–B). In contrast, E. coli CheY removed a phosphoryl group from E. coli CheA-P and retained it as expected (Fig. 3B). The half-life of E. coli CheY-P is 15–20 seconds, and can be reduced several orders of magnitude by the CheZ phosphatase (Hess et al., 1988, Lukat et al., 1991). The inability of CheY2 to stably retain phosphate may explain the lack of a dedicated CheY2-phosphatase (CheZ, CheC, or CheX) in the Che2 system. The combination of five active site residues in CheY2 (equivalent to residues 14, 58, 59, 88 and 89 in E. coli CheY) have been associated with fast autodephosphorylation kinetics [(Thomas et al., 2008); residues S11, Q54, N55, T84, and E85 in CheY2, Fig. 1B]. However, the exact same residue combination also exists in Rhodobacter sphaeroides CheY4 and CheY5, both of which have autodephosphorylation rates similar to E. coli CheY (Porter & Armitage, 2002). Thus, the reason why CheY2 does not stably retain phosphate remains to be determined.
Currently, the assigned roles of CheY2 include i) dephosphorylation of CheA2-P (Fig. 3), and ii) feedback control of Aer2 signaling via direct interaction with CheD (Fig. 4). It is possible that these are the major roles of CheY2. CheY2/CheD interactions were enhanced by the presence of CheA2, most likely via phosphotransfer to CheY2. Notably, CheY2-D10K, which did not dephosphorylate CheA2, did not interact with CheD (Fig 4B). Similarly, in B. subtilis and T. maritima, CheY-P specifically increases the affinity of CheC for CheD (Chao et al., 2006, Muff & Ordal, 2007, Yuan et al., 2012). However, unlike the B. subtilis and T. maritima systems, our report provides the first description of a CheY that directly sequesters CheD without the need for CheC, CheX, or any other binding partner. The phosphatase function of CheC or CheX is simply not required by CheY2. CheD contains a C-terminal sequence extension with a “CheC-like motif” (Fig. 4C) that was required for CheD to bind to CheY2 (Fig. 4D). This motif is necessary for CheC to bind to CheY-P in B. subtilis (Muff & Ordal, 2007). However, the CheD proteins from some other Aer2-containing systems, e.g., from V. cholerae (Greer-Phillips et al., 2018) or Vibrio vulnificus, do not have the CheC-like motif, so their CheY proteins may not directly interact with CheD. Rather, their genomes encode CheX, which contains the CheC-like motif. It will be prudent to study the CheY proteins from these systems and compare their CheY autodephosphorylation rates, their ability to interact with CheD, and to determine the role of CheX.
The CheD proteins of B. subtilis and T. maritima deamidate specific glutamine residues in chemoreceptors, thus creating methylatable glutamic acids (Kristich & Ordal, 2002, Chao et al., 2006). Q414 appears to be the only deamidation site in WT Aer2 (Fig. 5A), and this site is not shared by all other Aer2 receptors. For example, V. cholerae Aer2 lacks this adaptation site and the surrounding CheD consensus binding sequence (Greer-Phillips et al., 2018). Moreover, deamidation assays and behavioral assays did not support a role for CheD-mediated deamidation of Aer2 (Figs. 5 and 6). Instead, CheB2 performs that function. We did not directly test whether CheD can demethylate receptors like T. maritima CheD can (Chao et al., 2006), but the results of our methylation assays suggest that CheD does not demethylate Aer2 (Fig. 6C).
In vivo, Aer2 was primarily methylated by CheR2, but it could also be methylated by CheR1 from the Che system (Figs. 6 and 7). This contrasts with previous in vitro studies suggesting that CheR2 alone methylates Aer2 (Garcia-Fontana et al., 2014). The Che and Che2 systems can thus crosstalk at the level of receptor adaptation involving both CheR1 and CheD. The primary role of CheD though is enhancing CheR2-mediated methylation of Aer2 in order to increase the likelihood of Aer2 signaling (Figs. 6 and 8). In the presence of both CheR2 and CheD (but absence of CheB2), Aer2 was constantly signal on, even in the absence of O2, presumably because Aer2 remained highly methylated (Fig. 6). It is probable that CheD binds Aer2 directly via one or more of the four CheD consensus sites [as modelled by (Chao et al., 2006)], thus making Aer2 a better substrate for CheR2-mediated methylation. In contrast, CheR2 binds to the C-terminal GWEEF pentapeptide of Aer2 [Fig. 1B, (Garcia-Fontana et al., 2014)] and moves between methylation sites on a flexible C-terminal tether (Muppirala et al., 2009). We attempted to demonstrate Aer2/CheD interactions in BACTH assays, but they did not interact (data not shown), most likely due to the distance and orientation of the BACTH tags. In any case, the model proposed here for P. aeruginosa is consistent with the model proposed for B. subtilis in which CheD enhances receptor methylation and CheY-P induces CheD sequestration (but via CheC) so that CheD can no longer potentiate methylation (Rosario et al., 1995, Muff & Ordal, 2007). It is also possible that CheD directly modulates Aer2 signaling, as has been shown for McpB in B. subtilis (Walukiewicz et al., 2014). For B. subtilis, the strength of this effect was related to the methylation state of McpB. This could explain why CheD alone did not affect the behavior of WT Aer2QEEE (Fig. 6A).
In this study we sought to identify the interacting partners of CheY2-P. Given that i) no CheY2 partner was identified outside of Che2 after screening 1.37 million colonies, ii) CheY2 did not stably retain phosphate, and iii) CheY2 was involved in feedback control of Aer2 signaling though interaction with CheD, it is probable that the cellular output of Che2 is not mediated by CheY2. Instead, it is possible that CheA2 transfers a phosphoryl group to one or more alternative response regulators (of which P. aeruginosa has 68). This scenario occurs with the CheA protein of Comamonas testosteroni, where CheA not only transfers phosphoryl groups to two CheY proteins (CheY1 and CheY2), but also transfers a phosphoryl group to FlmD, a response regulator involved in biofilm formation (Huang et al., 2019). FlmD is orthologous to the PilH response regulator of the P. aeruginosa Pil-Chp system, and phosphotransfer from CheA to FlmD is slower than phosphotransfer to either CheY protein. In C. testosteroni, CheY2 is the primary chemotaxis response regulator and belongs to the F6 class of chemosensory genes. Interestingly, CheY1 in C. testosteroni has a fast autodephosphorylation rate, similar to CheY2 in P. aeruginosa, and both belong to the same F7 class of chemosensory genes (Huang et al., 2019, Ortega et al., 2020). We propose that C. testosteroni CheY1 might provide feedback control of receptor function by interacting with CheD in a similar way to what we have shown for P. aeruginosa here. To further probe the cellular function of the Che2 system in P. aeruginosa, our future studies will include testing our BACTH gDNA libraries against CheA2 and investigating phosphotransfer from CheA2 to alternative response regulators. It is clear that the major signal input to Aer2-containing chemosensory systems is O2, and that these systems are involved in a stress response during stationary phase, but unmasking how these systems modulate this function remains to be resolved.
EXPERIMENTAL PROCEDURES
Bacterial strains and growth conditions.
The P. aeruginosa and E. coli strains used in this study are described in Table S1. P. aeruginosa strains were grown in lysogeny broth (LB, Lennox) at 30 °C. E. coli strains were grown in LB broth (Lennox), tryptone broth (TB), or on LB agar at 30 °C or 37 °C, supplemented with 0.5 μg ml−1 thiamine, 100 μg ml−1 ampicillin, 50 μg ml−1 kanamycin, or 25 μg ml−1 chloramphenicol, as appropriate. To visualize bacterial two-hybrid interactions, Difco MacConkey agar (Beckton Dickinson and Company, Sparks, MD), supplemented with 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin, was used.
Cloning and mutagenesis.
All plasmids used in this study, and their construction where relevant, are described in Table S1. To generate plasmids expressing CheY2-D10K, CheD-V41T, Aer2EEEE and Aer2QQQQ, site-directed mutagenesis was performed on CheY2, CheD or Aer2 expression constructs using site-specific primers and PfuUltra II Fusion DNA polymerase (Agilent Technologies, Santa Clara, CA). Site-directed mutagenesis products were treated with DpnI (New England Biolabs, Ipswich, MA) to remove template DNA. To generate pUT18 constructs expressing C-terminally truncated CheD (res. 1–167 and res. 1–182), pUT18-CheD (with full-length cheD) was amplified by inverse PCR, gel-purified and ligated with EcoRI. Plasmids were introduced into E. coli or P. aeruginosa by electroporation or heat shock. For all new constructs, protein expression was assessed after inducing with either 600 μM IPTG (pProEXHTa, pUT18, pUT18C, pKT25, and pKNT25 constructs), 2 μM Na salicylate (pKG116 constructs), or 0.25% (w/v) arabinose (pJN105 constructs). Products of the correct size were confirmed by Western blotting with either 1:10,000 HisProbe™-HRP (Thermo Scientific, Rockford, IL) or 1:10,000 anti-CyaA-HRP (Santa Cruz Biotechnology, Dallas, TX). When no antibody was available to confirm protein expression (pKT25 and pKNT25 constructs, and tag-free proteins), proteins of the correct size were visualized by staining SDS-PAGE gels with Coomassie Brilliant Blue. All cloned genes and mutagenesis products were confirmed by sequencing the entire coding sequence of each gene (Eton Bioscience, San Diego, CA).
Construction of gDNA PAO1 libraries.
Genomic libraries were prepared as previously described (Houot et al., 2012). Briefly, P. aeruginosa PAO1 genomic DNA was purified using a DNeasy blood and cell culture kit (Qiagen, Valencia, CA) and then partially digested with 10 to 10−8 units of Sau3AI (New England Biolabs). Digested DNA was separated on a 1% agarose gel to verify the sizes of the digested products and samples containing DNA fragments from 250 to 5000 bp were purified using a QIAquick PCR purification kit (Qiagen). The pUT18 and pUT18C plasmids were digested with BamHI (New England Biolabs), dephosphorylated with calf intestinal phosphatase (New England Biolabs), and ligated to the digested PAO1 gDNA. The resulting ligation mixtures were introduced into E. coli BTH101 by heat-shock. Transformants were collected by pooling colonies, and plasmids were isolated using a QIAprep spin miniprep kit (Qiagen). The average size of the cloned library DNA was 700 bp (as determined by PCR analysis of 80 randomly-selected clones).
Bacterial two-hybrid assays for specific protein interactions.
The Bacterial Adenylate Cyclase Two-Hybrid system (BACTH, Euromedex, France) was used to test protein interactions. For each assay, plasmid pairs encoding T18 and T25 fusion proteins were introduced into E. coli BTH101 and plated onto LB agar supplemented with 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin. Three to five transformants were inoculated into 50 μl of LB containing 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin and incubated for at least 4 h at 37 °C. 2 μl of each culture was then spotted onto pre-warmed MacConkey agar containing both antibiotics. Positive interactions were identified as red colonies on MacConkey agar after incubating plates for 48 h at 30 °C. For each plasmid pair, triplicate experiments were performed.
Bacterial two-hybrid library screens.
The two PAO1 gDNA libraries (in pUT18 and pUT18C) were independently tested against bait proteins (in pKT25 or pKNT25). 50–100 ng of library DNA and 50 ng of plasmid bait were introduced together into BTH101 and plated onto MacConkey agar with 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin. Red colonies were re-streaked onto MacConkey agar. Colonies that remained red after re-plating were then grown overnight in LB broth with both antibiotics and plasmids were isolated using a Zyppy plasmid miniprep kit (Zymo Research, Irvine, CA). The plasmid mix was used to retransform BTH101 and plated onto LB Amp. Colonies were then streaked onto LB with 100 μg ml−1 ampicillin and onto LB with 50 μg ml−1 kanamycin to identify transformants that were AmpR but KanS (i.e., contained only the pUT18 or pUT18C plasmid). The pUT18 or pUT18C plasmid was then isolated and retested for interaction with the bait plasmid and against empty vector. Positive interactions were identified as red colonies on MacConkey agar after 48 h incubation at 30 °C. Plasmids were then sequenced to identify the cloned gene/s. Identified genes were then cloned in their entirety into the pUT18 and pUT18C plasmids and retested for protein interaction against the bait.
β-galactosidase assays.
Co-transformants of interest were grown overnight in LB broth with 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin, and for each culture the OD600nm was recorded. 200 μl of culture was mixed with 800 μl of buffer Z, pH 7 (60 mM Na2HPO4.2H2O, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4.7H2O, and 50 mM β-mercaptoethanol) before adding 35 μl of 0.01% (w/v) SDS and 70 μl of chloroform for permeabilization. 50 μl of permeabilized cells was transferred to a 96 well flat bottom microplate containing 150 μl of buffer Z pre-equilibrated to 30 °C before adding 40 μl of O-nitrophenyl-β-D-galactoside (ONPG, final concentration 0.66 mg ml−1). The kinetic reaction was carried out at 30 °C for 2.5 h in an xMark™ microplate spectrophotometer (Biorad, Hercules, CA). The OD420nm was measured every 2 min for 30 min, and then every 30 min until the end of the experiment. The relative β-galactosidase activity of each sample was expressed as Miller units using the formula: ((OD420nm at t2 – OD420nm at t1)/t2–t1 (min))/OD600nm (Battesti & Bouveret, 2012). The chosen t1 and t2 time points (in minutes) were located in the linear part of the kinetic reaction. All assays were performed in triplicate and results were expressed as the mean ± standard deviation.
His-fusion protein purification.
His-tagged proteins were purified from BL21(DE3) after induction with 600 μM IPTG. Cultures containing pLH1 (Aer2) also included 25 μg ml−1 5-aminolevulinic acid to augment heme synthesis. Proteins were purified on Ni-NTA agarose columns (Qiagen) as previously described (Garcia et al., 2017). Proteins were eluted in 1 ml of elution buffer (50 mM Tris pH 7.5, 500 mM NaCl and 250 mM imidazole), and four elution fractions were collected. Protein concentrations were determined in a BCA™ Protein Assay (Thermo Scientific) and protein quality was assessed by staining SDS-PAGE gels with Coomassie Brilliant Blue. Purified proteins were then dialyzed overnight against storage buffer (50 mM Tris pH 7.5, 500 mM NaCl) and stored at −80°C.
Phosphorylation assays.
For phosphorylation assays, 2–5 μM of purified EcCheA or CheA2 was incubated in phosphorylation buffer (50 mM Tris pH 7.5, 100 mM KCl and 5 mM MgCl2) in the presence of 0.1 mM [γ−32P]-ATP (10 μCi, PerkinElmer, MA) for 5 min at room temperature. Then, 20 μM EcCheY, CheY, CheY2, CheY2-D10K or 4 μM CheD was added and the reaction was carried out for 5s, 10s, 30s, or 120s before adding stop solution (2x sample buffer containing 286 mM β-mercaptoethanol and 10 mM K+EDTA). Reaction products were separated on a 12% polyacrylamide gel and visualized by staining in Coomassie Brilliant Blue for 20 min and then briefly destaining. The gel was dried and placed onto a Storage Phosphor Screen (GE Healthcare, Pittsburgh, PA) for 48 h. The exposed phosphor screen was scanned on a Storm 860 Molecular Dynamics Phosphorimager (GE Healthcare) and bands were quantified using ImageQuant software (GE Healthcare).
Deamidation assays.
For in vitro deamidation assays, 20 μM Aer2 (purified from UU2610) and 20 μM CheD were incubated together in 25 mM Tris pH 7.5, 250 mM NaCl at room temperature, or 37 °C for 30 min or 1 h. For assays with CheB2, 20 μM CheB2 was added along with 4 μM CheA2 and 1 mM ATP. Reactions were stopped by adding 2x sample buffer containing 286 mM β-mercaptoethanol. Proteins were separated by SDS-PAGE on 20 cm gels to resolve migration differences between unmodified and modified full length Aer2. Proteins were visualized by straining with Coomassie Brilliant Blue. For in vivo adaptation assays, E. coli UU2610 cultures were grown using the same conditions as behavioral assays. CheR2, D or B2 expression was induced with 2 μM Na salicylate at the time of subculture, whereas Aer2 expression was induced for 45 min with 200 μM IPTG once cells had reached an OD600nm of 0.2–0.25. Cultures were assessed in behavioral assays before sacrificing the bacteria for Western blots using 1:100,000 anti-Tsr antibody (antibody against the E. coli Tsr chemoreceptor, a gift from J.S. Parkinson) for Aer2 detection. As with the in vitro assays, proteins were separated on 20 cm gels to resolve migration differences between unmodified and modified full length Aer2.
Methylation assays.
Methylation assays were performed as previously described (Watts et al., 2011). Briefly, cultures were induced with 200 μM IPTG, centrifuged, washed and resuspended in chemotaxis buffer [0.1 mM K+EDTA, 10 mM KPO4 pH 7.4, 10 mM Na-lactate, 1 mM MgSO4, and 1 mM (NH4)2SO4]. Then, 200 μg ml−1 chloramphenicol was added to inhibit protein synthesis, and methylation was initiated by adding 9.7 μCi ml−1 L-(methyl-3H) methionine (PerkinElmer, Waltham, MA). Reactions were stopped with 2 μl formaldehyde (per 1.02 ml reaction). After SDS-PAGE, gels were soaked for 30 min in Fluorohance™ (Research Products International, Mount Prospect, IL), then dried and exposed to autoradiography film at −80 °C for 2–4 days. Bands were quantified in the linear range using VisionWorks®LS Analysis Software (Analytik Jena, Upland, CA). Band densities were normalized by dividing by the concentration of protein in the formaldehyde-treated samples as determined in a BCA™ Protein Assay (Thermo Scientific). Statistical analyses were carried out using a two-tailed Student’s t-test. A value of P <0.05 was considered statistically significant.
Behavioral assays.
Behavioral assays were performed using E. coli BT3388 or UU2610 cells grown in TB at 30 °C, as previously described (Watts et al., 2011). Adaptation enzymes were induced with 2 μM Na salicylate at the time of subculture, whereas Aer2 expression was induced for 45 min with 200 μM IPTG once cells had reached an OD600nm of 0.2–0.25. Assays were carried out in a gas perfusion chamber and toggled between air (20.9% O2) and N2. Assays were repeated two or more times on at least two separate days. When necessary, mutants were further quantified by calculating the percentage of cells tumbling over a 1 sec period 30 sec after switching to air or N2 (ImageJ; https://imagej.nih.gov/ij/). Significance was determined using a two-tailed Student’s t-test.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Erwin Stuffle and Mark Johnson for constructing several of the mutants in this study and Abigail Rodriguez for assistance with β-galactosidase assays. We also thank Sandy Parkinson, Davi Ortega, and Simon Ringgaard for helpful discussions, and Mark Johnson for constructive comments on the manuscript. This research was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health award number R01GM108655 to K. Watts. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Data sharing statement: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Footnotes
Conflict of interest statement: Both authors declare no known conflict of interest
REFERENCES
- Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ & Crane BR, (2013) Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 425: 886–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battesti A & Bouveret E, (2012) The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58: 325–334. [DOI] [PubMed] [Google Scholar]
- Biswas M, Dey S, Khamrui S, Sen U & Dasgupta J, (2013) Conformational barrier of CheY3 and inability of CheY4 to bind FliM control the flagellar motor action in Vibrio cholerae. PLoS One 8: e73923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourret RB, Drake SK, Chervitz SA, Simon MI & Falke JJ, (1993) Activation of the phosphosignaling protein CheY. II. Analysis of activated mutants by 19F NMR and protein engineering. J Biol Chem 268: 13089–13096. [PMC free article] [PubMed] [Google Scholar]
- Bren A & Eisenbach M, (1998) The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol 278: 507–514. [DOI] [PubMed] [Google Scholar]
- Chao X, Muff TJ, Park SY, Zhang S, Pollard AM, Ordal GW, Bilwes AM & Crane BR, (2006) A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 124: 561–571. [DOI] [PubMed] [Google Scholar]
- Dasgupta J & Dattagupta JK, (2008) Structural determinants of V. cholerae CheYs that discriminate them in FliM binding: comparative modeling and MD simulation studies. J Biomol Struct Dyn 25: 495–503. [DOI] [PubMed] [Google Scholar]
- Ferrandez A, Hawkins AC, Summerfield DT & Harwood CS, (2002) Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol 184: 4374–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris HU, Zeth K, Hulko M, Dunin-Horkawicz S & Lupas AN, (2014) Axial helix rotation as a mechanism for signal regulation inferred from the crystallographic analysis of the E. coli serine chemoreceptor. J Struct Biol 186: 349–356. [DOI] [PubMed] [Google Scholar]
- Garcia D, Orillard E, Johnson MS & Watts KJ, (2017) Gas sensing and signaling in the PAS-heme domain of the Pseudomonas aeruginosa Aer2 receptor. J Bacteriol 199: 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Fontana C, Corral Lugo A & Krell T, (2014) Specificity of the CheR2 methyltransferase in Pseudomonas aeruginosa is directed by a C-terminal pentapeptide in the McpB chemoreceptor. Sci Signal 7: ra34. [DOI] [PubMed] [Google Scholar]
- Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B & Filloux A, (2009) Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5: e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glekas GD, Plutz MJ, Walukiewicz HE, Allen GM, Rao CV & Ordal GW, (2012) Elucidation of the multiple roles of CheD in Bacillus subtilis chemotaxis. Mol Microbiol 86: 743–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer-Phillips SE, Sukomon N, Chua TK, Johnson MS, Crane BR & Watts KJ, (2018) The Aer2 receptor from Vibrio cholerae is a dual PAS-heme oxygen sensor. Mol Microbiol 109: 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guvener ZT, Tifrea DF & Harwood CS, (2006) Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol 61: 106–118. [DOI] [PubMed] [Google Scholar]
- Hess JF, Oosawa K, Kaplan N & Simon MI, (1988) Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell 53: 79–87. [DOI] [PubMed] [Google Scholar]
- Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H & Kato J, (2004) Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol Lett 231: 247–252. [DOI] [PubMed] [Google Scholar]
- Houot L, Fanni A, de Bentzmann S & Bordi C, (2012) A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiology 158: 1964–1971. [DOI] [PubMed] [Google Scholar]
- Huang Z, Wang YH, Zhu HZ, Andrianova EP, Jiang CY, Li D, Ma L, Feng J, Liu ZP, Xiang H, Zhulin IB & Liu SJ, (2019) Cross talk between chemosensory pathways that modulate chemotaxis and biofilm formation. MBio 10, e02876–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Bourret RB, Simon MI & Volz K, (1997) Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY. J Biol Chem 272: 11850–11855. [DOI] [PubMed] [Google Scholar]
- Kato J, Kim HE, Takiguchi N, Kuroda A & Ohtake H, (2008) Pseudomonas aeruginosa as a model microorganism for investigation of chemotactic behaviors in ecosystem. J Biosci Bioeng 106: 1–7. [DOI] [PubMed] [Google Scholar]
- Kristich CJ & Ordal GW, (2002) Bacillus subtilis CheD is a chemoreceptor modification enzyme required for chemotaxis. J Biol Chem 277: 25356–25362. [DOI] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, Huang L, Kustu S, Berry EA & Wemmer DE, (2001) Crystal structure of an activated response regulator bound to its target. Nat Struct Biol 8: 52–56. [DOI] [PubMed] [Google Scholar]
- Lukat GS, Lee BH, Mottonen JM, Stock AM & Stock JB, (1991) Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis. J Biol Chem 266: 8348–8354. [PubMed] [Google Scholar]
- McEvoy MM, Hausrath AC, Randolph GB, Remington SJ & Dahlquist FW, (1998) Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway. Proc Natl Acad Sci U S A 95: 7333–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KH, Hobbs G & Motaleb MA, (2016) Borrelia burgdorferi CheD promotes various functions in chemotaxis and the pathogenic life cycle of the spirochete. Infect Immun 84: 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muff TJ & Ordal GW, (2007) The CheC phosphatase regulates chemotactic adaptation through CheD. J Biol Chem 282: 34120–34128. [DOI] [PubMed] [Google Scholar]
- Muppirala UK, Desensi S, Lybrand TP, Hazelbauer GL & Li Z, (2009) Molecular modeling of flexible arm-mediated interactions between bacterial chemoreceptors and their modification enzyme. Protein Sci 18: 1702–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega DR, Fleetwood AD, Krell T, Harwood CS, Jensen GJ & Zhulin IB, (2017) Assigning chemoreceptors to chemosensory pathways in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 114: 12809–12814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega DR, Subramanian P, Mann P, Kjær A, Chen S, Watts KJ, Pirbadian S, Collins DA, Kooger R, Kalyuzhnaya MG, Ringgaard S, Briegel A & Jensen GJ, (2020) Repurposing a macromolecular machine: Architecture and evolution of the F7 chemosensory system. Nat Commun 11: 2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JS, Hazelbauer GL & Falke JJ, (2015) Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol 23: 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL & Armitage JP, (2002) Phosphotransfer in Rhodobacter sphaeroides chemotaxis. J Mol Biol 324: 35–45. [DOI] [PubMed] [Google Scholar]
- Rosario MM, Kirby JR, Bochar DA & Ordal GW, (1995) Chemotactic methylation and behavior in Bacillus subtilis: role of two unique proteins, CheC and CheD. Biochemistry 34: 3823–3831. [DOI] [PubMed] [Google Scholar]
- Sampedro I, Parales RE, Krell T & Hill JE, (2014) Pseudomonas chemotaxis. FEMS Microbiol Rev 39: 17–46. [DOI] [PubMed] [Google Scholar]
- Sanders DA, Gillece-Castro BL, Stock AM, Burlingame AL & Koshland DE Jr., (1989) Identification of the site of phosphorylation of the chemotaxis response regulator protein, CheY. J Biol Chem 264: 21770–21778. [PubMed] [Google Scholar]
- Sawai H, Sugimoto H, Shiro Y, Ishikawa H, Mizutani Y & Aono S, (2012) Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem Commun (Camb) 48: 6523–6525. [DOI] [PubMed] [Google Scholar]
- Schuster M, Hawkins AC, Harwood CS & Greenberg EP, (2004) The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol 51: 973–985. [DOI] [PubMed] [Google Scholar]
- Sheng S, Xin L, Yam JKH, Salido MM, Khong NZJ, Liu Q, Chea RA, Li HY, Yang L, Liang ZX & Xu L, (2019) The MapZ-mediated methylation of chemoreceptors contributes to pathogenicity of Pseudomonas aeruginosa. Front Microbiol 10: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szurmant H, Muff TJ & Ordal GW, (2004) Bacillus subtilis CheC and FliY are members of a novel class of CheY-P-hydrolyzing proteins in the chemotactic signal transduction cascade. J Biol Chem 279: 21787–21792. [DOI] [PubMed] [Google Scholar]
- Thomas SA, Brewster JA & Bourret RB, (2008) Two variable active site residues modulate response regulator phosphoryl group stability. Mol Microbiol 69: 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz K & Matsumura P, (1991) Crystal structure of Escherichia coli CheY refined at 1.7-Å resolution. J Biol Chem 266: 15511–15519. [DOI] [PubMed] [Google Scholar]
- Walukiewicz HE, Tohidifar P, Ordal GW & Rao CV, (2014) Interactions among the three adaptation systems of Bacillus subtilis chemotaxis as revealed by an in vitro receptor-kinase assay. Mol Microbiol 93: 1104–1118. [DOI] [PubMed] [Google Scholar]
- Watts KJ, Taylor BL & Johnson MS, (2011) PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Mol Microbiol 79: 686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S & Eisenbach M, (1993) Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci U S A 90: 8787–8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W & Briegel A, (2020) Diversity of bacterial chemosensory arrays. Trends Microbiol 28: 68–80. [DOI] [PubMed] [Google Scholar]
- Yuan W, Glekas GD, Allen GM, Walukiewicz HE, Rao CV & Ordal GW, (2012) The importance of the interaction of CheD with CheC and the chemoreceptors compared to its enzymatic activity during chemotaxis in Bacillus subtilis. PLoS One 7: e50689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


