Summary
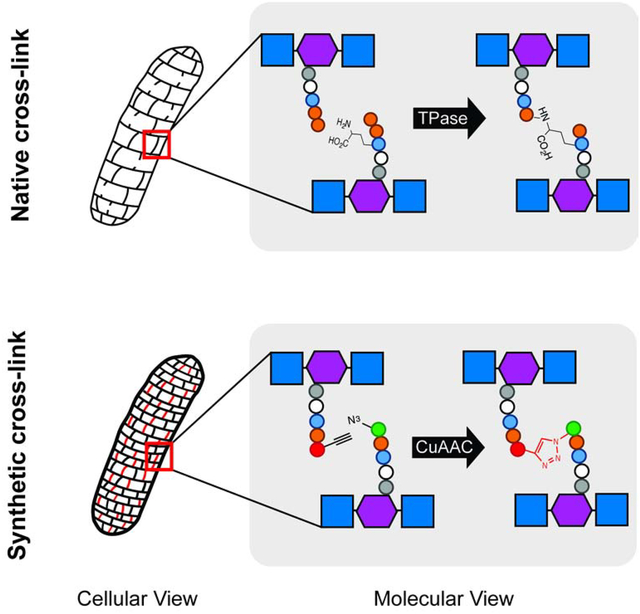
Transpeptidation reinforces the structure of cell wall peptidoglycan, an extracellular heteropolymer that protects bacteria from osmotic lysis. The clinical success of transpeptidase-inhibiting β-lactam antibiotics illustrates the essentiality of these cross-linkages for cell wall integrity, but the presence of multiple, seemingly redundant transpeptidases in many species makes it challenging to determine cross-link function. Here we present a technique to link peptide strands by chemical rather than enzymatic reaction. We employ bio-compatible click chemistry to induce triazole formation between azido- and alkynyl-d-alanine residues that are metabolically installed in the peptidoglycan of Gram-positive or Gram-negative bacteria. Synthetic triazole cross-links can be visualized using azidocoumarin-d-alanine, an amino acid derivative that undergoes fluorescent enhancement upon reaction with terminal alkynes. Cell wall stapling protects Escherichia coli from treatment with the broad-spectrum β-lactams ampicillin and carbenicillin. Chemical control of cell wall structure in live bacteria can provide functional insights that are orthogonal to those obtained by genetics.
Graphical Abstract
eTOC blurb
We developed a chemical method for introducing synthetic cross-links into the cell walls of live Gram-positive and Gram-negative bacteria. The cross-links protect Escherichia coli from broad-spectrum β-lactam antibiotics. Chemically-induced cross-linking complements genetics as an independent way to investigate the physiological roles of cell wall connectivity.
Introduction
Cell wall peptidoglycan is a mesh-like biopolymer that surrounds nearly all bacteria and is required to resist turgor pressure. The macromolecule consists of a glycan backbone and peptides, containing both l- and d-amino acids, that are cross-linked by d,d- and l,d-transpeptidases (Egan et al., 2015). The degree of transpeptidation can vary with species, growth phase and environmental conditions (Vollmer and Seligman, 2010). For example, the peptidoglycan of slow- or non-growing E. coli is more highly cross-linked and less susceptible to in vitro enzymatic turnover than that of actively-replicating E. coli (Glauner et al., 1988; Goodell and Tomasz, 1980; Lee et al., 2013; Pisabarro et al., 1985; Tuomanen and Cozens, 1987; Tuomanen et al., 1988). Cross-linking abundance is also predicted to impact the overall strength and stiffness of the cell envelope (Auer and Weibel, 2017; Huang et al., 2008; Loskill et al., 2014; Vollmer and Bertsche, 2008), cell shape (Huang et al., 2008; Sycuro et al., 2010; Yang et al., 2019), and assembly of macromolecular structures (Scheurwater and Burrows, 2011). The clinical success of transpeptidase-inhibiting β-lactam antibiotics highlights the importance of peptidoglycan cross-linking in bacterial physiology.
Despite the biological and medical significance of peptidoglycan transpeptidation, unraveling the roles of these linkages is challenging. Currently, the standard ways to manipulate cross-linking are to mutate or deplete the expression of the transpeptidase genes or to inhibit these enzymes with small molecules like β-lactams. However, the functional redundancy of transpeptidases and promiscuity of β-lactams (Spratt, 1975) pose challenges to rational control of peptidoglycan connectivity.
d-amino acids bearing reactive groups such as cysteines, alkynes, azides and tetrazines have been used to metabolically label the peptidoglycan stem peptide (de Pedro et al., 1997; Kuru et al., 2012; Pidgeon et al., 2015; Radkov et al., 2018; Siegrist et al., 2015; Siegrist et al., 2013). Once embedded, the presence of these probes can be revealed by chemical reaction with an exogenous label that bears a complementary reactive group (Siegrist et al., 2015). We hypothesized that we might also use functionalized peptide strands to manipulate cell wall cross-linking. More specifically, we reasoned that co-incubation of bacteria with azido- and alkynyl-d-amino acids would result in a subpopulation of labeled muropeptide strands in close enough proximity to undergo copper-catalyzed azide-alkyne cycloaddition (CuAAC) upon introduction of the appropriate reagents. Such structures would serve as synthetic, triazole cross-links.
Results and Discussion
We first tested our hypothesis using a loss-of-fluorescence assay. In this approach, bacteria are co-incubated in the presence of azido- and alkynyl-d-amino acids, washed and subjected to CuAAC (Figure 1A, left). We reasoned that the peptidoglycan-embedded functional groups should either react with each other or with the alkynyl- or azido-fluorophores in CuAAC solution. Bacteria incubated with a single d-amino acid probe, by contrast, should have muropeptides decorated with just one functional group, which in turn should react only with the complementary reactive fluorophore. In this assay, we interpret decreased labeling of co-incubated relative to singly-incubated bacteria to indicate that there are fewer peptidoglycan-embedded functional groups available to react with the fluorophores. This may occur because the reaction between azido- and alkynyl-muropeptides is favored or because there is competition between the d-amino acids for initial incorporation into the muropeptide. To control for the latter possibility, we also subjected metabolically-labeled bacteria to strain-promoted azide-alkyne cycloaddition (SPAAC; Figure 1A, right) with a cyclooctyne-appended fluorophore. In the absence of copper and other reagents, peptidoglycan-embedded azides and alkynes should not react with each other at an appreciable rate and only the azide-cyclooctyne reaction should occur. In the SPAAC reactions, therefore, we interpret changes in labeling to mean that the azido-d-amino acid outcompetes or is outcompeted by other d-amino acids for initial incorporation into the cell wall.
Figure 1. Loss (A-E) and gain (F and G) of fluorescence strategies for detection of synthetic cross-links.
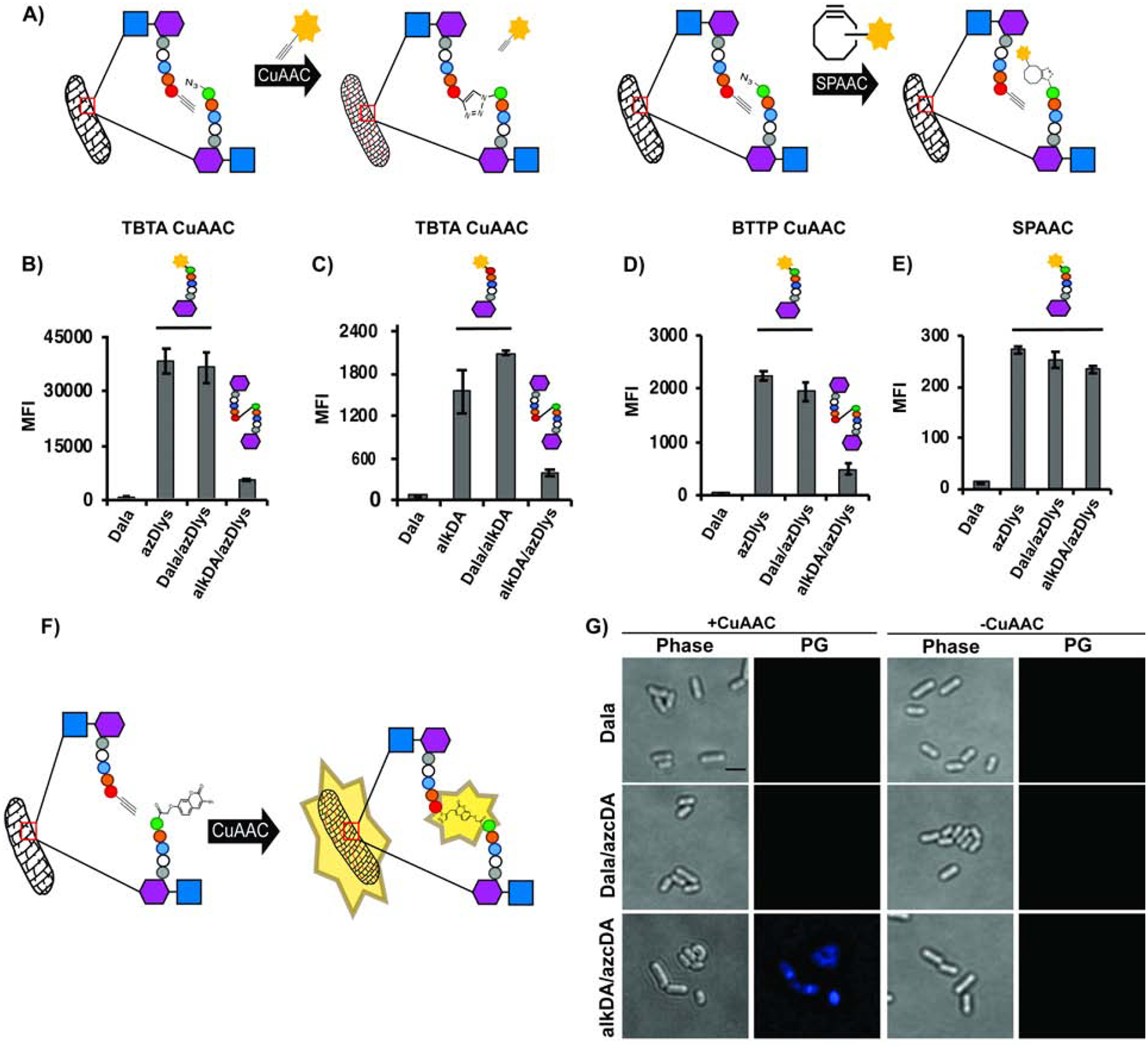
(A) Loss-of-fluorescence logic, including SPAAC control (details in Methods). (B-E) pbp5::tn L. monocytogenes was incubated with the indicated d-amino acids, washed and subjected to CuAAC with TBTA ligand and alkyne-carboxyrhodamine 110 (CR110) (B); TBTA ligand with azido-CR110 (C); BTTP ligand with alkyne-CR110 (D); SPAAC with DBCO-CR110 (E). Fluorescence was quantified by flow cytometry and data are representative of 2–6 biological replicates performed in triplicate. MFI, mean fluorescence intensity. Error bars, +/− standard deviation. (F), Intrapeptidoglycan reaction between alkDA and azidocoumarin-d-alanine (azcDA) results in the fluorescent triazole product. (G) pbp5::tn L. monocytogenes was incubated in the presence of indicated d-amino acids, washed and subjected to BTTP CuAAC with no exogenous fluorescent label. Of the 327 alkDA/azcDA-treated, CuAAC-subjected cells observed in two independent experiments, 310 were fluorescent above Dala/azcDA-treated, CuAAC-subjected background levels. PG, fluorescence derived from peptidoglycan labeling. Scale bar, 1 μM. Images are representative of 4 biological replicates.
We used the loss-of-fluorescence approach to ask whether we could introduce triazole cross-links into the cell wall of Listeria monocytogenes, a Gram-positive, food-borne pathogen. We initially used pbp5::tn L. monocytogenes, a d,d-carboxypeptidase-deficient mutant that we previously showed has high levels of d-amino acid labeling (Siegrist et al., 2013). After incubating the bacteria in the presence of equal amounts of d-alanine (Dala), azido-d-amino acid (azDA or azDlys, the R groups of which respectively have one and four carbons), alkynyl-d-alanine (alkDA) or mixtures thereof, we washed away unincorporated amino acid and subjected the bacteria to CuAAC with either an alkynyl- (Figure 1B) or azido-fluorophore (Figure 1C). We assessed cellular fluorescence by flow cytometry. In both cases, the bacteria that were co-incubated in alkDA/azDlys had lower amounts of fluorescence than those incubated in azDlys (Figure 1B) or alkDA (Figure 1C) alone. We obtained similar results with the more bio-friendly CuAAC reaction (Yang et al., 2014) that employs the 3-[4-({bis[(1-tert-butyl-1H-1,2,3-triazol-4-yl)methyl]amino}methyl)-1H-1,2,3-triazol-1-yl]propanol (BTTP) ligand (Wang et al., 2011) (Figure 1D) or in wild-type L. monocytogenes (Figures S1A and S1B). These data suggested that bacteria co-incubated in azDlys and alkDA had fewer peptidoglycan-embedded functional groups available to react with complementary reactive fluorophores in solution (Figure 1A). Moreover, in bacteria subjected to SPAAC with cyclooctyne-fluorophore, the signal after alkDA/azDlys incubation was similar to that of azDlys alone or Dala/azDlys (Figures 1E and S1C). The SPAAC control reactions suggested that there was no appreciable competition between the d-amino acids for initial incorporation into the peptidoglycan. Taken together these data suggest that cell wall-embedded azides and alkynes can react with each other by CuAAC.
We next sought a more direct read-out for triazole cross-links. Fluorogenic molecules undergo a fluorescence enhancement upon chemical or enzymatic reaction. For example, CuAAC reaction of the non-fluorescent 3-azido-7-hydroxycoumarin (azidocoumarin) with terminal alkynes yields fluorescent triazole products (Sivakumar et al., 2004). As d-amino acids appended to fluorophores, including hydroxycoumarin, incorporate efficiently into peptidoglycan (Kuru et al., 2012), we decided to test whether swapping an azido-coumarin d-amino acid (azcDA) for an azido-d-amino acid would allow us to mark the presence of triazole cross-links (Figure 1F).
We began by synthesizing azcDA, which could be accessed readily by coupling Nα-Boc-d-2,3-diaminopropionic acid to azidocoumarin acid 1 (Weineisen et al., 2017) via pentafluorophenyl ester 2. Trifluoroacetic acid-mediated Boc deprotection afforded azcDA. Next we co-incubated pbp5::tn L. monocytogenes with Dala/azcDA or alkDala/azcDA, washed, and subjected the bacteria or not to a BTTP CuAAC reaction that lacked a complementary alkynyl-fluorophore. By microscopy we found that fluorescence of live, azcDA-labeled bacteria required the inclusion of alkDA in the initial metabolic labeling step as well as the subsequent CuAAC reaction (Figure 1G). These gain-of-fluorescence data were consistent with the loss-of-fluorescence results (Figures 1B–1D, S1A, S1B) and supported the notion that a CuAAC reaction can covalently join azides and alkynes metabolically installed in L. monocytogenes peptidoglycan to form synthetic cross-links.
We were initially unable to identify peptidoglycan modifications that were specific to alkDA/azDlys-treated, CuAAC-subjected bacteria and that had the exact mass of a triazole cross-link (Figure S1D). We hypothesized that our ability to detect such modifications—which theoretically could include muropeptides with additional, transpeptidase-mediated linkages—was complicated by the pre-existing complexity of L. monocytogenes peptidoglycan, which is both highly-cross-linked and tailored by N-deacetylases, O-acetyltransferases and amidotransferases (Aubry et al., 2011; Boneca et al., 2007; Rae et al., 2011). Therefore, we turned our attention to the model, Gram-negative bacterium Escherichia coli, as its peptidoglycan composition is considerably less complex (Vollmer et al., 2008). To simplify our analysis even more, we employed a strain, CS802–2, in which most of the genes encoding peptide-acting cell wall enzymes were deleted, including all 6 carboxypeptidases (Denome et al., 1999). The lack of tetrapeptides in this background prevents l,d-transpeptidation, so we expected d-amino acid incorporation to occur at the 5th position of the stem peptide (Cava et al., 2011). We first verified that CuAAC-subjected, alkDA/azDA-labeled CS802–2 E. coli, like wild-type E. coli, were less fluorescent than those labeled by azDA or alkDA alone (Figures 2A–2B, S1E–F) The decrease in d-amino acid concentration and labeling time compared to L. monocytogenes (described further below) correlated with a more modest reduction in fluorescence. We next metabolically labeled CS802–2 E. coli with different combinations of d-amino acids, washed away unincorporated amino acid, and performed BTTP CuAAC reactions in the absence of fluorophore. We then separated digested peptidoglycan by ultra-performance liquid chromatography (UPLC) and used MS/MS to identify molecules with the exact masses that corresponded to azDA- and alkDA-terminating pentapeptides in the appropriate samples. We identified peaks that were specific to alkDA/azDA-treated, CuAAC-subjected bacteria (Figure 2C) and had the exact masses of a 5–5 triazole dimer, trimer (+/− anh) or tetramer (Figures 2D–E, S2, S3). The presence of these species increased the total cross-linking by approximately 20% (Table 1). We note that muropeptide incorporation of azDA was ~2-fold more efficient than alkDA and associated with a general decrease in cross-linking (Tables 1 and S2B). Exogenous d-amino acids, including both Dala and non-canonical d-amino acids, have been shown or hypothesized to inhibit d,d-transpeptidation (Caparros et al., 1992; Lam et al., 2009). Importantly, however, CuAAC-dependent cross-linking occurred only in alkDA/azDA-treated samples (Table 1) and not in controls that had been treated with equimolar amounts of Dala/azDA (Figure S2B) or Dala alone (Table 1). These data confirmed our ability to introduce synthetic cross-links into bacterial peptidoglycan in a CuAAC-inducible manner.
Figure 2. Indirect (A-B) and direct (C-E) identification of synthetic triazole cross-links.
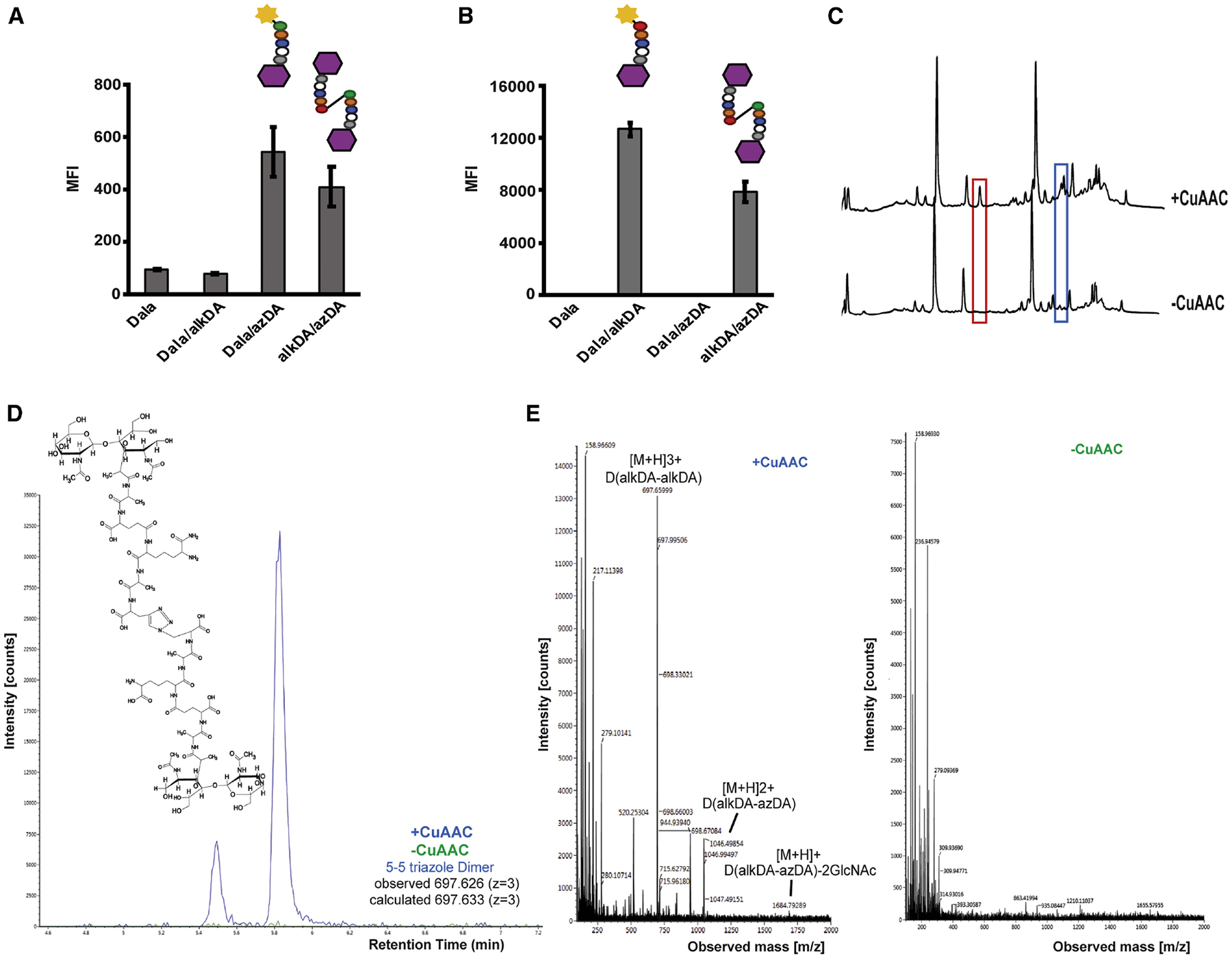
CS802–2 E. coli was incubated +/− d-alanine alone (Dala) or equimolar combinations of d-alanine, azido-d-alanine (azDA), and alkynyl-d-alanine (alkDA) as indicated, washed and subjected to CuAAC with BTTP ligand and complementary fluorophore (A-B) or with the detection reagent omitted (C-E). Peptidoglycan was extracted, digested with mutanolysin and lysozyme, and separated by ultra-performance liquid chromatography (UPLC). We identified several peaks from alkDA/azDA-labeled bacteria that were specific to CuAAC treatment (red and blue boxes, (C)). Chemical structure for 5–5 triazole dimer (D) identified by mass spectrometry (MS) from red boxed peak in (C). (E) Ion detection (left) and MS profile (right) for 5–5 triazole dimer. MS/MS profile and fragmentation shown in Figure S4 and Table S1, respectively. Ion detection and MS profiles for 5–5 triazole trimers and tetramer from blue boxed peaks in (C) shown in Figure S5. Fluorescence in (A-B) was quantified by flow cytometry and data are representative of 2–6 biological replicates performed in triplicate. MFI, mean fluorescence intensity. Error bars, +/− standard deviation. UPLC analysis in (C) was performed on two biological replicates.
Table 1.
Quantification of muropeptides from CS802–2 E. coli treated with Dala or a combination of alkDA/azDA then subjected or not to CuAAC.
| Dala | alkDA/azDA | |||
|---|---|---|---|---|
| -CuAAC | +CuAAC | -CuAAC | +CuAAC | |
| azDA | - | - | 8.21 ± 4.26 | 2.92 ± 0.11 |
| alkDA | - | - | 4.04 ± 2.38 | 1.23 ± 1.32 |
| % PG Modification | - | - | 12.24 ± 1.42 | 11.44 ± 3.39 |
| % Cross-linkage | 53.05 ± 2.04 | 54.31 ± 3.33 | 42.76 ± 1.57 | 51.04 ± 0.23 |
| % Triazole Cross-linkage | - | - | - | 9.98 ± 1.42 |
Data from two biological replicates. One of the alkDA/azDA UPLC traces is shown in Figure 2C. Compare to data for bacteria treated with Dala/alkDA or Dala/azDA, Figure S2B.
Cell wall homeostasis is a balance between synthesis and turnover. Peptidoglycan-cleaving enzymes have been implicated directly (Park and Strominger, 1957; Schwarz et al., 1969; Tomasz et al., 1970; Tomasz and Waks, 1975) and indirectly (Cho et al., 2014; Kohlrausch and Höltje, 1991) in β-lactam cidality. Slow- or non-replicating, β-lactam-tolerant E. coli have highly cross-linked cell walls that are more resistant in vitro to lytic enzymes (Glauner et al., 1988; Goodell and Tomasz, 1980; Lee et al., 2013; Pisabarro et al., 1985; Tuomanen and Cozens, 1987; Tuomanen et al., 1988). We wondered whether β-lactam susceptibility might be influenced by pre-existing cell wall cross-linking, either in addition to, or as part of, the well-documented effect of bacterial growth rate (Eng et al., 1991; Lee et al., 2018; Lee et al., 1944; Toumanem et al., 1986). Since exogenous d-amino acids can modify the structure, amount and strength of peptidoglycan and inhibit bacterial growth (Caparros et al., 1992; Cava et al., 2011; Lam et al., 2009), and growth rate in turn correlates with β-lactam lethality (Eng et al., 1991; Lee et al., 2018; Lee et al., 1944; Toumanem et al., 1986), we first optimized d-amino acid concentration and incubation time (Figures S4A and S4B). We then labeled E. coli with different combinations of d-amino acids, washed and performed BTTP CuAAC. After CuAAC reagent washout, bacteria were resuspended in growth medium and challenged with the β-lactam ampicillin. Without CuAAC, ampicillin treatment resulted in similar killing regardless of what d-amino acid(s) the bacteria had been metabolically labeled with (Figure 3A). In E. coli subjected to CuAAC, ampicillin caused ~2–3 logs of killing for bacteria labeled with Dala, Dala/alkDA or Dala/azDA but less than 1 log for those labeled with alkDA/azDA (Figure 3B). These data suggested that triazole cross-links protect E. coli from ampicillin.
Figure 3. Synthetic cross-links protect CS802–2 E. coli from ampicillin (B) but not from kanamycin (C).
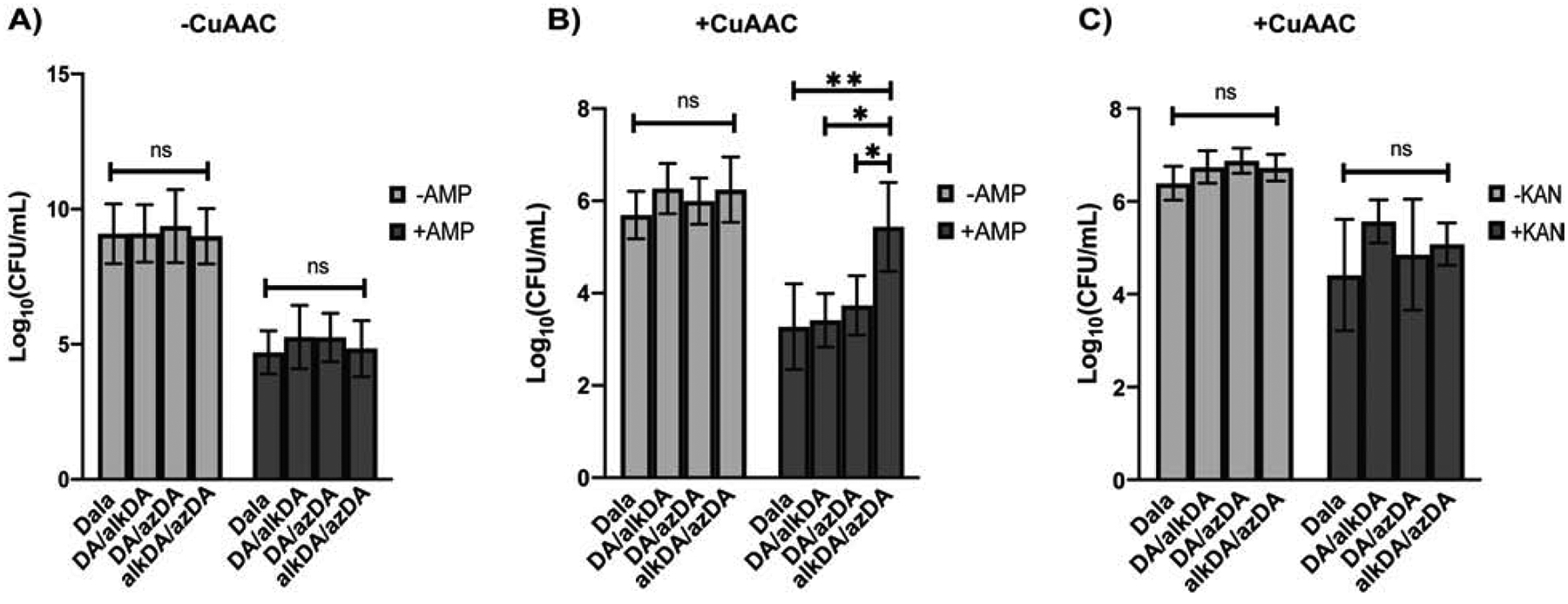
CS802–2 E. coli was incubated +/− the indicated d-amino acids for 6 hrs, washed and subjected (B, C) or not (A) to CuAAC with BTTP ligand (no complementary fluorophore) as in Figure 3. Bacteria were then challenged with antibiotic for 1 hr at 37 °C and plated for colony forming units (CFUs). Data are from three biological replicates performed in triplicate. Error bars, +/− standard deviation. Statistical significance was assessed by two-way ANOVA with Tukey’s multiple comparison test. ns, ≥0.05; *, p<0.05; **, p<0.005.
BTTP-liganded CuAAC (Wang et al., 2011) is more biocompatible than traditional TBTA CuAAC (Yang et al., 2014). While the BTTP CuAAC reaction that we previously optimized for mycobacterial species (Garcia-Heredia et al., 2018) did not change L. monocytogenes cell counts (Figure S4C), they partially inhibited the recovery of E. coli on solid medium (Figure 3). However the effect was consistent across the different d-amino acid combinations, indicating that the synthetic cross-links, not the CuAAC, were responsible for antibiotic rescue. In liquid medium, E. coli subjected to CuAAC had a distinct lag in growth relative to mock-reacted controls (Figures S4D–F). The length of the lag phase was significantly enhanced in CS802–2 E. coli that had been incubated in both azDA and alkDA e.g. bacteria with triazole linkages. Since longer lag phases are associated with antibiotic tolerance (Bertrand, 2019; Fridman et al., 2014) we asked whether the apparent protection afforded by alkDA/azDA labeling followed by CuAAC was transient and whether it was specific to β-lactams. During the post-CuAAC lag phase, synthetic cross-links protected bacteria from ampicillin and the closely-related antibiotic carbenicillin (Figures 3B, S4G, S4H) but not the translation-inhibiting aminoglycoside kanamycin (Figure 3C). This protection was lost following resumption of growth (Figure S4I). Thus an extended, post-CuAAC lag phase correlates with, but is likely not responsible for, the enhanced tolerance of alkDA/azDA-labeled E. coli to β-lactams.
Taken together, our data suggest that synthetic peptidoglycan cross-links protect against lethality induced by the broad-spectrum β-lactams ampicillin and carbenicillin. The total cross-linking density across CuAAC-treated bacteria is similar (Tables 1 and Figure S2B), suggesting that the unusual linkage (triazole) or position on the stem peptide (5–5) is instead responsible for protection. The classic view of β-lactam activity is that transpeptidase inhibition damages the cell wall by disrupting the balance between peptidoglycan synthases and hydrolases (Park and Strominger, 1957; Schwarz et al., 1969; Tomasz et al., 1970; Tomasz and Waks, 1975). β-lactams also induce a metabolically-taxing, futile cycle of cell wall synthesis and turnover (Cho et al., 2014; Kohlrausch and Höltje, 1991). Both models for β-lactam cidality posit that lethality directly or indirectly depends on the activity of peptidoglycan-degrading enzymes. An artificially-reinforced cell wall may resist β-lactam-induced damage because its structure is partially independent from transpeptidase-mediated synthesis. Additionally, or alternatively, synthetic cross-links may regulate peptidoglycan turnover. Indeed, while this manuscript was under review, Dik and colleagues proposed that non-canonical cell wall cross-links (derived from the reaction of exogenously-incorporated sulfonyl fluoride d-amino acids with endogenous m-DAP) can impede the processivity of lytic transglycosylases (Dik et al., 2020), enzymes that cleave the carbohydrate backbone of peptidoglycan. While we cannot rule out pleiotropic effects on other cell envelope or periplasmic structures, we hypothesize that synthetic triazole cross-links act as molecular speed bumps for lytic transglycosylases, blunting β-lactam cidality by keeping peptidoglycan degradation at bay. Consistent with the diverse roles for these enzymes in peptidoglycan homeostasis (Dik et al., 2017), the prolonged, post-CuAAC recovery in liquid medium of synthetically cross-linked E. coli (Figures S4E–F) may also reflect slowed cell wall turnover. Treatment with unnatural d-amino acids alone modestly enhanced both lag phase and β-lactam tolerance (Figure S4E–I), although the effects were not statistically significant. As noncanonical d-amino acid incorporation is not expected to alter cell wall turnover by lytic transglycosylases (Caparros et al., 1992), we speculate that these more-subtle peptidoglycan modifications impact E. coli physiology by a different mechanism(s) than the synthetic cross-links.
Given the promiscuity with which d-amino acids incorporate into the bacterial peptidoglycan (Radkov et al., 2018; Siegrist et al., 2015) stapling can be readily adapted for a wide variety of species. Molecular control of synthetic cross-link positioning may also be possible. The effect(s) of 5–5 cross-links on cell wall structure may be different from native, 4,3 or 3,3 cross-links. For example, 5,5 cross-links likely allow more flexibility and/or more space between glycan strands, which could in turn change the physical properties of the peptidoglycan. Unlike monopeptides, which can incorporate into the 4th or 5th positions (or both) of stem peptides (Kuru et al., 2012; Siegrist et al., 2013), d-amino acid dipeptides with functional groups on their N- or C-terminus are predicted to install these groups specifically at 4th or 5th position, respectively (Liechti et al., 2013). Our loss-of-fluorescence assay suggests that dipeptides functionalized with N-terminal azides and alkynes permit the introduction of synthetic, 4–4 cross-links into CS802–2 E. coli (Figure S1F), in addition to the 5–5 linkages afforded by monopeptide labeling. The development of alkyne- and azide-bearing DAP derivatives may also enable the introduction of triazole linkages at the 3rd position of the muropeptide. Finally, pulse-chase labeling in species with defined modes of growth can offer sub-cellular control of synthetic cross-links. Independent from and complementary to genetics, cell wall stapling is an orthogonal assay for dissecting the roles of peptidoglycan structure in bacterial physiology.
Significance
Bacteria are surrounded by cell wall heteropolymers that are essential for viability under most circumstances. The structure of the cell wall is well-conserved and consists of a glycan backbone cross-linked by d-amino acid-containing peptides. Cross-link-inhibiting β-lactams account for two-thirds of the global antibiotic market, underscoring the general importance of these linkages to bacterial physiology. For a given species, the density of cross-linking can vary with replication rate and environmental conditions. These changes in cell wall connectivity in turn correlate with other phenotypic properties of the bacterium. However most species have multiple, closely-related enzymes that catalyze cross-links, each with varying susceptibility to different β-lactams, making it difficult to control the density of these linkages by genetics or small molecule inhibition alone. In this work, we present a chemical technique to introduce synthetic cross-links to the cell walls of live bacteria. We use bio-compatible click chemistry to induce a reaction between azido- and alkynyl-d-alanine residues that are metabolically incorporated in the cell wall peptides of Gram-positive and Gram-negative species. The resulting triazole linkages can be visualized by substituting azido-d-alanine with azidocoumarin-d-alanine, an amino acid analogue that becomes more fluorescent after reacting with an alkyne. Stapling the cell wall of Escherichia coli enhances its tolerance to two different broad-spectrum β-lactams. Chemical manipulation complements genetic and small molecule perturbations as an independent means of investigating the role of cell wall connectivity in bacterial physiology.
STAR * Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to Lead Contact, M. Sloan Siegrist (siegrist@umass.edu)
Material Availability
All unique/stable reagents generated in this study are available from the Lead Contact.
Data and Code Availability
This publication did not use unpublished custom code, software, or algorithms.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
E. coli was grown in Luria-Bertani Broth (LB) at 37 °C. L. monocytogenes was grown in Brain Heart Infusion Broth (BHI) at 37 °C.
METHODS DETAILS
Metabolic labeling and CuAAC
E. coli were grown overnight at 37 °C. The next day cultures were back-diluted between 1:50 and 1:500 and d-amino acids (1.25 mM total per sample for monopeptides and 2.5 mM per sample for dipeptides) were added directly in the LB medium. Cells were grown until log phase (OD600 0.6–0.8) then centrifuged for 5 min at 5,000 × g at room temperature (RT). They were then washed with sterile-filtered PBS and subjected to BTTP CuAAC (200 μM CuSO4, 800 μM BTTP [Chemical Synthesis Core Facility, Albert Einstein College of Medicine, Bronx, NY], 2.5 mM sodium ascorbate (freshly prepared), with or without 25 μM of azido or alkynyl fluorescent dye as appropriate) or TBTA CuAAC (1 mM CuSO4, 128 μM TBTA [Click Chemistry Tools, Scottsdale, AZ], 1.2 mM sodium ascorbate (freshly prepared), with or without 25 μM of azido or alkynyl fluorescent dye [Click Chemistry Tools]) for 1 hr at room temperature, shaking. Samples were then centrifuged, washed thrice with PBS, and either fixed with 2% (v/v) formaldehyde or used in assays described below.
L. monocytogenes were grown overnight at 37 °C with the d-amino acids (2.5 mM total per sample) then centrifuged for 5 min at 5,000×g at RT. They were washed with PBS and subjected to CuAAC as described for E. coli.
General fluorescence analysis
Mean fluorescence intensities (MFI) of bacterial cell populations were obtained by flow cytometry from a BD DUAL LSRFortessa instrument.
Samples were imaged on an inverted Nikon Eclipse Ti microscope equipped with a Hamamatsu Orca Flash 4.0 camera and reconstructed with NIS Elements.
Peptidoglycan composition analysis
200 mL cultures of log-phase CS802–2 E. coli were treated with d-amino acids and subjected to BTTP CuAAC as describe above. Bacteria were centrifuged for 5 minutes at 5,000 × g at RT, wash twice with MilliQ water, resuspended in 1 mL MilliQ water then added drop-wise into 80 mL of boiling 4% SDS. Samples were vigorously stirred for 1.5 hr then cooled to RT. The insoluble fraction (PG) was pelleted at 400,000 × g, 15 min, 30 °C (TLA-100.3 rotor; OptimaTM Max ultracentrifuge, Beckman). SDS was washed out and the PG was resuspended in 200 μl of 50 mM sodium phosphate buffer pH 4.9 and digested overnight with 30 μg/mL muramidase (Cellosyl). Samples were incubated at 37 °C. PG digestion was stopped by 5 min incubation in a boiling water bath. Coagulated protein was removed by centrifugation. The supernatants were mixed with 150 μL 0.5 M sodium borate pH 9.5, and subjected to reduction of muramic acid residues into muramitol by sodium borohydride treatment (10 mg/mL final concentration, 30 min at RT). Samples was adjusted to pH 3.5 with phosphoric acid. Chromatographic analyses of muropeptides were performed on AQUITY Ultra Performance Liquid Chromatography (UPLC) BEH C18 column (130 Å, 1.7 μm, 2.1 mm by 150 mm; Waters), and peptides were detected at Abs. 204 nm using ACQUITY UPLC UV-Visible Detector. Muropeptides were separated using a linear gradient from buffer A (0.1% of Formic acid in water) to buffer B (0.1% of Formic acid in acetonitrile) in 218 min, and flow 0.25 mL/min. Muropeptide identity was confirmed by MS/MS analysis, using a Xevo G2-XS QTof system (Waters Corporation, USA). Quantification of muropeptides was based on their relative abundances (relative area of the corresponding peak). Cross-linking was determined by the following formula; crosslinking=dimmer+(trimmer/2).
Antibiotic challenge
CS802–2 E. coli that had been pre-labeled with d-amino acids for 6 hrs (OD600 0.6) (Figure 3) or overnight (Figure S4D–F) and subjected or not to BTTP CuAAC were washed with PBS and resuspended in LB medium to a normalized OD600 of 0.3 with or without 125 μg/mL ampicillin, 125 μg/mL carbenicillin, or 6.25 μg/mL kanamycin. After 1–5 hrs incubation at 37 °C, bacteria were washed twice with PBS and plated as 10-fold serial dilutions on LB agar.
Chemical Synthesis and Characterization
Synthesis of azidocoumarin-d-alanine (azcDA)
General Procedures.
Reactions were performed in round bottom flasks fitted with rubber septa under a positive pressure of nitrogen. Gas-tight syringes with stainless steel needles or cannulae were used to transfer air- and moisture-sensitive liquids. Flash column chromatography was performed as described by Still et al. using granular silica gel (60-Å pore size, 40–63 μm, 4–6% H2O content, Silicycle)(Still et al., 1978). Analytical thin layer chromatography (TLC) was performed using glass plates pre-coated with 0.25 mm 230–400 mesh silica gel impregnated with a fluorescent indicator (254 nm). TLC plates were visualized by short wave ultraviolet light (254 nm). Concentration of solutions under reduced pressure were carried out on rotary evaporators capable of achieving a minimum pressure of ~2 torr at 29–30 °C unless noted otherwise.
Dichloromethane, tetrahydrofuran, and N,N-dimethylformamide were were purified by the method of Grubbs et al. under a positive pressure of nitrogen(Pangborn et al., 1996).
Instrumentation.
Proton nuclear magnetic resonance (1H NMR) spectra were recorded with a Bruker Avance III 500 MHz spectrometer, are reported in parts per million, and are referenced to the residual protium in the NMR solvent (CDCl3: δ 7.24 (CHCl3), CD3OD: δ 3.31 (CHD2OD), DMSO-d6: δ 2.50 (DMSO-d5)). Data are reported as follows: chemical shift [multiplicity (s = singlet, d = doublet, t = triplet, sp = septet, m = multiplet), coupling constant(s) in Hertz, integration]. Carbon-13 nuclear magnetic resonance (13C NMR) spectra were recorded with a Bruker Avance III 500 MHz spectrometer, are reported in parts per million, and are referenced from the carbon resonances of the solvent (CDCl3: δ 77.23, CD3OD: δ 49.15, DMSO-d6: δ 39.51). Data are reported as follows: chemical shift. Infrared data (IR) were obtained with a Cary 630 Fourier transform infrared spectrometer equipped with a diamond ATR objective and are reported as follows: frequency of absorption (cm−1), intensity of absorption (s = strong, m = medium, w = weak, br = broad). Optical rotations were measured on a P-2000 JASCO polarimeter and compound concentrations are expressed in units of g/100 mL. High resolution mass spectra (HRMS) were recorded by the Harvard University Small Molecule Mass Spectrometry facility on an Agilent 6210 time-of-flight LCMS using an electrospray ionization (ESI) source.
Overall synthetic scheme.
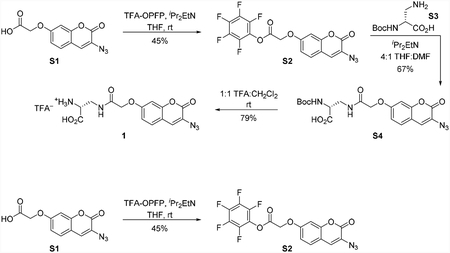
Azidocoumarin pentafluorophenyl ester S3
To a 25 mL round bottom flask charged with azidocoumarin acid S1(Weineisen et al., 2017) (56.0 mg, 214 μmol, 1 equiv) under a nitrogen atmosphere was added tetrahydrofuran (2 mL) at room temperature. N,N-diisopropylethylamine (74.5 μL, 428 μmol, 2.00 equiv) was added to the dark brown solution via syringe. This was followed immediately by addition of pentafluorophenyl trifluoroacetate (73.5 μL, 428 μmol, 2.00 equiv) via syringe and stirred at room temperature. After 30 min, the reaction mixture was concentrated under reduced pressure and the brown residue was purified by flash column chromatography on silica gel (eluent: 10% ethyl acetate in hexanes) to provide the azidocoumarin pentafluorophenyl ester S2 (41.0 mg, 45%) as a white solid. 1H NMR (500 MHz, CDCl3, 25 °C): δ 7.37 (d, J = 8.6 Hz, 1H), 7.15 (s, 1H), 6.93 (dd, J = 8.7, 2.5 Hz, 1H), 6.87 (d, J = 2.5 Hz, 1H), 5.04 (s, 2H). 13C NMR (126 MHz, CDCl3, 25 °C): δ 164.5, 159.1, 157.6, 152.9, 142.2, 141.2, 140.2, 139.2, 137.2, 128.7, 125.9, 124.7, 114.3, 113.3, 102.2, 64.8. 19F NMR (471 MHz, CDCl3, 25 °C): δ −152.2 (m), − 156.4 (m), −161.3 (m). FTIR (thin film, cm−1): 2128 (s), 1804 (m), 1722 (m), 1618 (m), 1521 (s), 1334 (m), 1118 (m), 1070 (m), 995 (m). HRMS (ESI, m/z): 428.0294 (calculated for C17H7F5N3O5 [M+H]+: 428.0300). TLC (15% ethyl acetate in hexanes, Rf): 0.32 (UV).
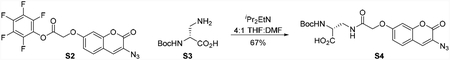
Boc-d-Ala-azidocoumarin S5
To a 25 mL round bottom flask charged with azidocoumarin pentafluorophenyl ester S2 (30.0 mg, 70.2 μmol, 1 equiv) and Nα-Boc-D-2,3-diaminopropionic acid (S3) (28.6 mg, 140 μmol, 2.00 equiv) under a nitrogen atmosphere was added tetrahydrofuran (2 mL) at room temperature. N,N-diisopropylethylamine (14.7 μL, 140 μmol, 2.00 equiv) was then added to the solution via syringe followed by N,N-dimethylformamide (500 μL). After 20 min, the reaction mixture was concentrated under reduced pressure and the residue was purified by flash column chromatography on silica gel (eluent: 20% hexanes, 75% ethyl acetate, 5% acetic acid) to provide Boc-D-Ala-azidocoumarin S4 (21.0 mg, 67%) as a white solid. 1H NMR (500 MHz, d6-DMSO, 25 °C): δ 8.20 (t, J = 6.0 Hz, 1H), 7.63 (s, 1H), 7.59 (d, J = 8.5 Hz, 1H), 7.07–6.98 (m, 3H), 4.60 (s, 2H), 4.13–4.05 (m, 1H), 3.57–3.48 (m, 1H), 3.44–3.36 (m, 1H), 1.38 (s, 9H). 13C NMR (126 MHz, d6-DMSO, 25 °C): δ 172.2, 167.5, 159.7, 157.2, 155.4, 152.4, 128.9, 127.1, 122.6, 113.3, 113.1, 101.7, 78.4, 67.2, 53.4, 39.6, 28.2. FTIR (thin film, cm−1): 3366 (br-s), 2989 (br-m), 2128 (s), 1737 (m), 1670 (m), 1618 (m), 1521 (m), 1148 (m), 1055 (m). HRMS (ESI, m/z): 446.1324 (calculated for C19H20N5O8 [M−H]−: 446.1317). TLC (20:75:5 hexanes:ethyl acetate:acetic acid, Rf): 0.18 (UV). [α]D23 = +78 (c 0.25, DMSO).
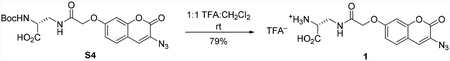
d-Ala-azidocoumarin (1)
To a 25 mL round bottom flask charged with Boc-D-Ala-azidocoumarin S4 (18.0 mg, 40.2 μmol, 1 equiv) was added dichloromethane (2 mL) followed by trifluoroacetic acid (2 mL) at room temperature. After stirring for 15 min, the reaction mixture was concentrated under reduced pressure and the residue was purified by automated C18 reverse phase column chromatography (30 g C18 silica gel, 25 μm spherical particles, eluent: H2O+0.1% TFA (5 CV), gradient 0→100% MeCN/H2O+0.1% TFA (15 CV), tR=10.1 CV) to provide the trifluoroacetic acid salt of D-Ala-azidocoumarin (1) (11.0 mg, 79%) as a white solid. 1H NMR (500 MHz, d6-DMSO, 25 °C): δ 8.46 (t, J = 6.0 Hz, 1H), 8.37 (br-s, 3H), 7.64 (s, 1H), 7.59 (d, J = 8.7 Hz, 1H), 7.07 (d, J = 2.2 Hz, 1H), 7.02 (dd, J = 8.6, 2.4 Hz, 1H), 4.63 (s, 2H), 4.06–4.02 (m, 1H), 3.71–3.65 (m, 1H), 3.62–3.56 (m, 1H). 13C NMR (126 MHz, d6-DMSO, 25 °C): δ 169.2, 168.3, 159.7, 157.2, 152.4, 128.9, 127.2, 122.6, 113.4, 113.1, 101.6, 67.1, 52.2, 38.5. 19F NMR (471 MHz, D2O, 25 °C) δ −75.6. FTIR (thin film, cm−1): 2117 (s), 1707 (m), 1670 (m), 1618 (s), 1536 (m), 1431 (m), 1170 (m), 1141 (s). HRMS (ESI, m/z): 346.0796 (calculated for C14H12N5O6 [M−H]−: 346.0793). [α]D23= +68 (c 0.23, DMSO).
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical significance was evaluated using one- or two-way analysis of variance (ANOVA), followed by Tukey’s multiple comparisons test using GraphPad Prism 8.4.0 software. Details in figure legends.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| L. monocytogenes EGDe | Siegrist et al., 2013 | N/A |
| L. monocytogenes EGDe pbp5::tn (lmo2754:tn) | Siegrist et al., 2013 | N/A |
| CS802–2 E. coli | Demone et al.,, 1999 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| d-alanine (Dala) | Sigma-Aldrich | Cat# A7377 |
| (R)-α-Propargylglycic (alkDA) | Acros Organics | Cat# 441221000 |
| 3-Azido-d-alanine HCl (azDA) | Jena Bioscience | Cat# CLK-AA004 |
| azDlys | Jena Bioscience | Cat# CLK-AA010 |
| Azido-Coumarin-d-alanine (azcDA) | This paper | N/A |
| azDADA (ADADA) | Liechti et al.,, 2014 | https://www.einstein.yu.edu/research/shared-facilities/chemical-biology/ |
| alkDADA (EDADA) | Liechti et al., 2014 | https://www.einstein.yu.edu/research/shared-facilities/chemical-biology/ |
| BTTP | Chemical Synthesis Core Facility, Albert Einstein College of Medicine, Bronx, NY | https://www.einstein.yu.edu/research/shared-facilities/chemical-biology/ |
| TBTA | Click Chemistry Tools, Scottsdale, AZ | Cat# 1061 |
| Copper (II) Sulfate, Anhydrous | Alfa Aesar | Cat# 33308 |
| L-Ascorbic Acid Sodium Salt | Alfa Aesar | Cat# A17759 |
| Carboxyrhodamine 110 Azide (Azide-CR110) | Click Chemistry Tools, Scottsdale, AZ | Cat# AZ105 |
| Carboxyrhodamine 110 Alkyne (Alkyne-CR110) | Click Chemistry Tools, Scottsdale, AZ | Cat# TA106 |
| Carboxyrhodamine 110 DBCO (DBCO-CR110) | Click Chemistry Tools, Scottsdale, AZ | Cat# A127 |
| AFDye 488 Picolyl Azide | Click Chemistry Tools, Scottsdale, AZ | Cat# 1276 |
| Ampicillin Sodium Salt | Fisher Scientific | Cat# BP1760 |
| Kanamycin | Sigma-Aldrich | Cat# K1377 |
| Carbenicillin Disodium Salt | Sigma-Aldrich | Cat# C3416 |
| dichloromethane | Fisher Scientific | N/A |
| tetrahydrofuran | Fisher Scientific | N/A |
| N,N-dimethylformamide | Fisher Scientific | N/A |
| Nα-Boc-D-2,3-diaminopropionic acid | Chem-Impex International, Inc | N/A |
| potassium carbonate | Fisher Scientific | N/A |
| N,N-diisopropylethylamine | Acros Organics | N/A |
| trifluoroacetic acid | Sigma-Aldrich | N/A |
| Software and Algorithms | ||
| ImageJ | Schneider et al., 2012 | https://imagej.nih.gov/ij/ |
| GraphPad Prism 8.4.0 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ChemDraw 18.1 | PerkinElmer Informatics | https://www.perkinelmer.com/es/category/chemdraw |
Highlights.
The bacterial cell wall is cross-linked by d-amino acid-containing peptides.
β-lactam antibiotics interfere with cell wall cross-linking.
We developed a d-amino acid-based method for introducing synthetic cross-links.
The cross-links protect Escherichia coli from broad-spectrum β-lactam antibiotics.
Acknowledgments
No competing financial interests have been declared. This work was supported by NSF RAISE Convergence 1848065 (M.S.S. under “Palmore”), NSF GRFP (A.K.A.), Stanford ChEM-H Chemistry/Biology Interface Predoctoral Training Program (A.K.A.), American Cancer Society Postdoctoral fellowship PF-18-011-01-CDD (P.S.), NIH R25 GM086264 (C.M.-M.) as well as the Swedish Research Council (VR), Knut and Alice Wallenberg Foundation, Laboratory of Molecular Infection Medicine Sweden (MIMS) and Kempe Foundation (F.C.). We gratefully acknowledge K. Young for E. coli strain CS802-2, C. Hill for pbp5::tn (lmo2754::tn) EGD-e L. monocytogenes, and A. Burnside for technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no competing interests.
The authors have a patent related to this work.
Siegrist MS, Jewett JC, Shieh P, Gordon CG, Bertozzi CR. 2016. “D-amino acid derivative-modified peptidoglycan and methods of use thereof.” U.S. patent 9,303,068
References
- Aubry C, Goulard C, Nahori M-A, Cayet N, Decalf J, Sachse M, Boneca IG, Cossart P, and Dussurget O (2011). OatA, a Peptidoglycan O-Acetyltransferase Involved in Listeria monocytogenes Immune Escape, Is Critical for Virulence. The Journal of Infectious Diseases 204, 731–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer GK, and Weibel DB (2017). Bacterial Cell Mechanics. Biochemistry 56, 3710–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand RL (2019). Lag Phase Is a Dynamic, Organized, Adaptive, and Evolvable Period That Prepares Bacteria for Cell Division. Journal of Bacteriology 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boneca IG, Dussurget O, Cabanes D, Nahori M-A, Sousa S, Lecuit M, Psylinakis E, Bouriotis V, Hugot J-P, Giovannini M, et al. (2007). A critical role for peptidoglycan N-deacetylation in Listeria evasion from the host innate immune system. Proceedings of the National Academy of Sciences 104, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparros M, Pisabarro AG, and de Pedro MA (1992). Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. Journal of Bacteriology 174, 5549–5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, de Pedro MA, Lam H, Davis BM, and Waldor MK (2011). Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. The EMBO Journal 30, 3442–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Uehara T, and Bernhardt TG (2014). Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell 159, 1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Pedro MA, Quintela JC, Höltje JV, and Schwarz H (1997). Murein segregation in Escherichia coli. Journal of Bacteriology 179, 2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denome SA, Elf PK, Henderson TA, Nelson DE, and Young KD (1999). Escherichia coli Mutants Lacking All Possible Combinations of Eight Penicillin Binding Proteins: Viability, Characteristics, and Implications for Peptidoglycan Synthesis. Journal of Bacteriology 181, 3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik DA, Marous DR, Fisher JF, and Mobashery S (2017). Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Critical Reviews in Biochemistry and Molecular Biology 52, 503–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik DA, Zhang N, Chen JS, Webb B, and Schultz PG (2020). Semisynthesis of a Bacterium with Non-canonical Cell-Wall Cross-Links. Journal of the American Chemical Society 142, 10910–10913. [DOI] [PubMed] [Google Scholar]
- Egan AJ, Biboy J, van’t Veer I, Breukink E, and Vollmer W (2015). Activities and regulation of peptidoglycan synthases. Philos Trans R Soc Lond B Biol Sci 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng RH, Padberg FT, Smith SM, Tan EN, and Cherubin CE (1991). Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother 35, 1824–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman O, Goldberg A, Ronin I, Shoresh N, and Balaban NQ (2014). Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513, 418–421. [DOI] [PubMed] [Google Scholar]
- Garcia-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Lim HC, Wagner JC, Morita YS, Swarts BM, et al. (2018). Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner B, Holtje JV, and Schwarz U (1988). The composition of the murein of Escherichia coli. J Biol Chem 263, 10088–10095. [PubMed] [Google Scholar]
- Goodell W, and Tomasz A (1980). Alteration of Escherichia coli murein during amino acid starvation. Journal of Bacteriology 144, 1009–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, and Wingreen NS (2008). Cell shape and cell-wall organization in Gram-negative bacteria. Proceedings of the National Academy of Sciences, pnas.0805309105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlrausch U, and Höltje JV (1991). Analysis of murein and murein precursors during antibiotic-induced lysis of Escherichia coli. Journal of Bacteriology 173, 3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, and VanNieuwenhze MS (2012). In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew Chem Int Ed Engl 51, 12519–12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, and Waldor MK (2009). D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AJ, Wang S, Meredith HR, Zhuang B, Dai Z, and You L (2018). Robust, linear correlations between growth rates and β-lactam–mediated lysis rates. Proceedings of the National Academy of Sciences 115, 4069–4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hesek D, Llarrull LI, Lastochkin E, Pi H, Boggess B, and Mobashery S (2013). Reactions of All Escherichia coli Lytic Transglycosylases with Bacterial Cell Wall. Journal of the American Chemical Society 135, 3311–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Foley EJ, and Epstein JA (1944). Mode of Action of Penicillin. Journal of Bacteriology 48, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, and Maurelli AT (2013). A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loskill P, Pereira PM, Jung P, Bischoff M, Herrmann M, Pinho MG, and Jacobs K (2014). Reduction of the Peptidoglycan Crosslinking Causes a Decrease in Stiffness of the Staphylococcus aureus Cell Envelope. Biophys J 107, 1082–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangborn AB, Giardello MA, Grubbs RH, Rosen RK, and Timmers FJ (1996). Safe and Convenient Procedure for Solvent Purification. Organometallics 15, 1518–1520. [Google Scholar]
- Park JT, and Strominger JL (1957). Mode of Action of Penicillin. Science 125, 99. [DOI] [PubMed] [Google Scholar]
- Pidgeon SE, Fura JM, Leon W, Birabaharan M, Vezenov D, and Pires MM (2015). Metabolic Profiling of Bacteria by Unnatural C-terminated D-Amino Acids. Angew Chem Int Ed Engl 54, 6158–6162. [DOI] [PubMed] [Google Scholar]
- Pisabarro AG, de Pedro MA, and Vázquez D (1985). Structural modifications in the peptidoglycan of Escherichia coli associated with changes in the state of growth of the culture. Journal of Bacteriology 161, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkov AD, Hsu Y-P, Booher G, and VanNieuwenhze MS (2018). Imaging Bacterial Cell Wall Biosynthesis. Annual Review of Biochemistry 87, 991–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae CS, Geissler A, Adamson PC, and Portnoy DA (2011). Mutations of the Listeria monocytogenes Peptidoglycan N-Deacetylase and O-Acetylase Result in Enhanced Lysozyme Sensitivity, Bacteriolysis, and Hyperinduction of Innate Immune Pathways. Infection and Immunity 79, 3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater EM, and Burrows LL (2011). Maintaining network security: how macromolecular structures cross the peptidoglycan layer. FEMS Microbiology Letters 318, 1–9. [DOI] [PubMed] [Google Scholar]
- Schwarz U, Asmus A, and Frank H (1969). Autolytic enzymes and cell division of Escherichia coli. J Mol Biol 41, 419–429. [DOI] [PubMed] [Google Scholar]
- Siegrist MS, Swarts BM, Fox DM, Lim SA, and Bertozzi CR (2015). Illumination of growth, division and secretion by metabolic labeling of the bacterial cell surface. FEMS Microbiology Reviews 39, 184–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist MS, Whiteside S, Jewett JC, Aditham A, Cava F, and Bertozzi CR (2013). (D)-amino acid chemical reporters reveal peptidoglycan dynamics of an intracellular pathogen. ACS Chemical Biology 8, 500–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, and Wang Q (2004). A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org Lett 6, 4603–4606. [DOI] [PubMed] [Google Scholar]
- Spratt BG (1975). Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proceedings of the National Academy of Sciences 72, 2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Still WC, Kahn M, and Mitra A (1978). Rapid chromatographic technique for preparative separations with moderate resolution. The Journal of Organic Chemistry 43, 2923–2925. [Google Scholar]
- Sycuro LK, Pincus Z, Gutierrez KD, Biboy J, Stern CA, Vollmer W, and Salama NR (2010). Peptidoglycan Crosslinking Relaxation Promotes Helicobacter pylori’s Helical Shape and Stomach Colonization. Cell 141, 822–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A, Albino A, and Zanati EVE (1970). Multiple Antibiotic Resistance in a Bacterium with Suppressed Autolytic System. Nature 227, 138–140. [DOI] [PubMed] [Google Scholar]
- Tomasz A, and Waks S (1975). Mechanism of action of penicillin: triggering of the pneumococcal autolytic enzyme by inhibitors of cell wall synthesis. Proc Natl Acad Sci U S A 72, 4162–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toumanem E, Cozens R, Tosch W, Zak O, and Tomasz A (1986). The Rate of Killing of Escherichia coli by B-Lactam Antibiotics Is Strictly Proportional to the Rate of Bacterial Growth. Journal of General Microbiology 132, 1297–1304. [DOI] [PubMed] [Google Scholar]
- Tuomanen E, and Cozens R (1987). Changes in peptidoglycan composition and penicillin-binding proteins in slowly growing Escherichia coli. J Bacteriol 169, 5308–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuomanen E, Markiewicz Z, and Tomasz A (1988). Autolysis-resistant peptidoglycan of anomalous composition in amino-acid-starved Escherichia coli. Journal of Bacteriology 170, 1373–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, and Bertsche U (2008). Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli. Biochim Biophys Acta 1778, 1714–1734. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Blanot D, and de Pedro MA (2008). Peptidoglycan structure and architecture. FEMS Microbiol Rev 32, 149–167. [DOI] [PubMed] [Google Scholar]
- Vollmer W, and Seligman SJ (2010). Architecture of peptidoglycan: more data and more models. Trends Microbiol 18, 59–66. [DOI] [PubMed] [Google Scholar]
- Wang W, Hong S, Tran A, Jiang H, Triano R, Liu Y, Chen X, and Wu P (2011). Sulfated ligands for the copper(I)-catalyzed azide-alkyne cycloaddition. Chem Asian J 6, 2796–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weineisen NL, Hommersom CA, Voskuhl J, Sankaran S, Depauw AMA, Katsonis N, Jonkheijm P, and Cornelissen JJLM (2017). Photoresponsive, reversible immobilization of virus particles on supramolecular platforms. Chemical Communications 53, 1896–1899. [DOI] [PubMed] [Google Scholar]
- Yang DC, Blair KM, Taylor JA, Petersen TW, Sessler T, Tull CM, Leverich CK, Collar AL, Wyckoff TJ, Biboy J, et al. (2019). A Genome-Wide Helicobacter pylori Morphology Screen Uncovers a Membrane-Spanning Helical Cell Shape Complex. J Bacteriol 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Jalloh AS, Wei W, Zhao J, Wu P, and Chen PR (2014). Biocompatible click chemistry enabled compartment-specific pH measurement inside E. coli. Nat Commun 5, 4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This publication did not use unpublished custom code, software, or algorithms.


