Abstract
Serine/Arginine splicing factor 1 (SRSF1) is an RNA binding protein abundantly expressed in most tissues. The pleiotropic functions of SRSF1 exert multiple roles in gene expression by regulating major steps in transcription, processing, export through the nuclear pores and translation of nascent RNA transcripts. The aim of this review is to highlight recent findings in the functions of this protein and to describe its role in immune system development, functions and regulation.
Keywords: SR protein, mRNA, transcription, splicing, translation, miRNA, SLE
1. Introduction.
Serine/Arginine splicing factor 1 (SRSF1) is the archetypal member of the serine/arginine rich (SR) family of RNA binding proteins (RBPs), which in homo sapiens is constituted by 12 proteins that share a modular structure, with at least one RNA-recognition motif (RRM) and a C-terminal domain (RS) rich in Ser-Arg dipeptide repeats [1]. SRSF1 was first characterized in the early 90’s as a splicing factor required for constitutive and alternative pre-mRNA splicing [2]. Work carried out in biochemical, cellular and animal systems in the past three decades has characterized SRSF1 functions in mRNA transcription, splicing, nuclear export, decay and translation. SRSF1 is an essential gene, as knockout in mice is lethal in embryos and its overexpression leads to oncogenic transformation in both rodent and human cells [3].
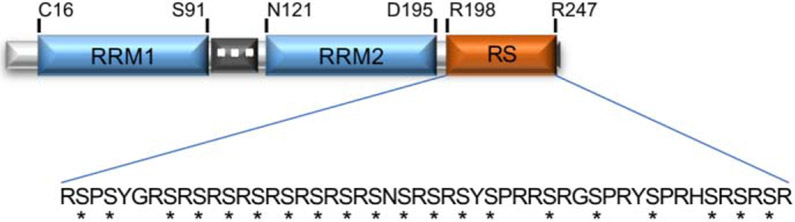
SRSF1 contains two N-terminal RRMs and a short C-terminal RS domain containing 15 Serine/Arginine repeats (Fig. 1). The two RRMs have evolved as part of a double RRM and synergize with each other to determine SRSF1 RNA binding specificity [4]. Cross-linking immunoprecipitation and high-throughput sequencing (CLIP-seq) analysis revealed that SRSF1 preferentially binds to the consensus sequence GGAGA within exonic regions [5, 6]. RRM1 and RRM2 are both required for RNA binding, although RRM2 binding is the main determinant of substrate specificity [7]. In addition to their RNA binding function, the RRMs mediate early spliceosome assembly by serving as a bridge between pre-mRNA and the RRM of the U1-70K component of the U1 small nuclear ribonucleoprotein (snRNP) [8] and playing a role in the subcellular localization of SRSF1 [9]. The RS domain is not required, nor modulates, SRSF1 RNA binding function but has been shown to have a regulatory function in early spliceosome assembly in addition to its role in the nuclear-cytoplasmic shuttling of SRSF1 through interaction with other cellular factors [9].
Figure 1.
SRSF1 is constituted by two RNA Recognition Motifs (RRM1 and RRM2) and a serine arginine rich (SR) domain. The SR domain is heavily phosphorylated (*). Methylated arginine residues in the inter-RRM linker region are indicated with a white dot.
Post-translational modifications of several residues within the RS and RRMs domains modulate SRSF1 functions. Global proteomics analysis revealed that SR proteins may represent an important class of non-histone substrates that are regulated by histone acetyl transferases (HATs) [10]. In addition, three arginine residues in the inter-RRM linker region are methylated [11] and mutations in these three residues lead to the cytoplasmic accumulation of SRSF1, resulting in a decrease in the proteins nuclear functions and an enhancement of its cytoplasmic function in translation. The RS domain is also extensively phosphorylated and its phosphorylation state alters SRSF1’s functions and sub-cellular localization. SR protein kinase 1 and 2 (SRPK1 and SRPK2) phosphorylate SRSF1 in the cytoplasm generating a hypo-phosphorylated form of SRSF1. Transportin-SR (TNPO3) directs the transport of the hypo-phosphorylated SRSF1 into the nucleus [12], where it accumulates in nuclear speckles and is hyper-phosphorylated by the Clk/Sty kinase (CLK1). The hyper-phosphorylated SRSF1 is recruited to the active sites of transcription, where it promotes spliceosome assembly [13] and is subsequently partially dephosphorylated by Protein Phosphatase 1 and 2 Catalytic Subunit Alpha (PPP1CA and PPP2CA); this is a critical step in the progression of the splicing reaction [14, 15].
Given that multiple molecular mechanisms are regulated by SRSF1, it is not surprising that its expression is tightly controlled by several feedback loops. SRSF1 can regulate the expression of its transcript by promoting splicing isoforms that contain premature termination codons (PTCs), which are targets for nonsense mediated decay (NMD) [16]. Additionally, SRSF1 can reduce translation efficiency of its own transcript by shifting the SRSF1 coding mRNAs from polysomes to monosomes [16] and modulating the expression of regulatory miRNAs [17]. Changes in SRSF1 expression levels lead to oncogenesis and have been associated with immune disease [18-20]. Studies aimed at determining the mechanisms of SRSF1-mediated oncogenesis have focused on its functions in alternative splicing [18, 19, 21], although multiple mechanisms are likely to contribute to the physiological effects observed. SRSF1 expression modulates the splicing of several genes that regulate cell proliferation, cell-cycle progression and multiple apoptotic and cellular signaling pathways, [18, 19, 22, 23].
Immune cells can undergo major changes to their transcriptome and proteome in response to a rapidly changing environment. RNA binding proteins can regulate quantitative and qualitative changes in multiple transcripts to rapidly modulate the cell’s response to stimuli [24]. The transcriptome and proteome of cells from the lymphoid or myeloid lineage exhibit quantitative and qualitative changes dependent on the cell’s lineage, activation and disease state [25]. High-throughput genomic approaches have revealed a pervasive role for alternative splicing in T cell, B cell and monocyte differentiation, activation and response to infection [26]. Alternative splicing changes in several genes are also among the causes of autoimmune diseases such as type 1 diabetes [27], multiple sclerosis [28] and systemic lupus erythematosus (SLE) [29]. It is thus not surprising that SRSF1 plays a role in the splicing of key immune response regulators. However, its functions in multiple aspects of RNA biogenesis, decay and translation can also affect immune response, regulation and disease (summarized in Table 1).
Table 1.
SRSF1 mediated regulation of the immune system. SRSF1 modulates the expression of several genes involved in immune system functions through multiple mechanisms.
| Mechanism | Regulated Genes | Reference |
|---|---|---|
| Transcription | Upregulates CD3ζ transcription | [36, 39] |
| Upregulates IL-2 transcription | [36, 39] | |
| Upregulates IL-6 transcription | [40, 41] | |
| Alternative Splicing | Exon 5 inclusion in CD6 | [45] |
| Exons 2 and 3 inclusion in IRF3 | [47] | |
| Exon 13 exclusion/exon 14 inclusion in CD46 | [50] | |
| Inclusion of 526 nt in 3’ UTR of CD3ζ | [53] | |
| Exon 11 inclusion in RasGRP1 | [55] | |
| Bcl-xL specific 5’ splice site in Bcl-x | [56] | |
| Inclusion of alternative 3’ exon in BIM | [18-20] | |
| Inclusion of extended C-terminus in ICAD | [59] | |
| Exon 5 inclusion in CYBB | [61] | |
| Stability | Decrease stability of CXCL1 | [66] |
| Decrease stability PTEN | [69] | |
| Export | Block nuclear export of CXCL1 | [74] |
| Block nuclear export of CXCL2 | [74] | |
| Block nuclear export of TNF | [74] | |
| Translation | 1500 mRNA translation upregulated | [80] |
| miRNA | Regulates miR-10b, MICB expression | [83] |
| Regulates miR-10b, IL-17A expression | [84] | |
| Regulates miR-29b, IFN-γ expression | [88] |
2. Transcription.
2a. SRSF1 regulates transcription activation.
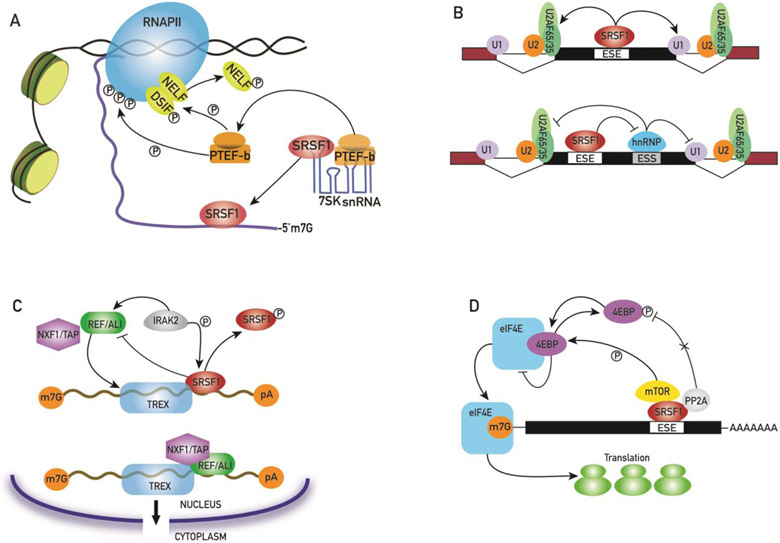
RNA polymerase II (RNAPII) often pauses elongation 20-60 bp downstream of the transcription start site. Additional signals are then required to re-start the paused polymerase and promote productive elongation. This acts as a rate-limiting step found in more than two-thirds of metazoan genes and especially in stimulus-responsive pathways [30]. Pausing is mediated by two factors associated with the polymerase: the negative elongation factor (NELF) and the DRB sensitivity-inducing factor (DSIF). The positive elongation factor b (P-TEFb) is recruited to the stalled polymerase complex to promote its release through phosphorylation of NELF and DSIF; this leads to the dissociation of NELF from the stalled promoter complex and the conversion of DSIF to an elongation-stimulating factor [30, 31]. Over 50% of the P-TEFb within the nucleus exists in a catalytically inactive state sequestered by the 7SK small nuclear ribonucleoprotein (7SK snRNP) [32]. Some SR proteins can be recruited to active promoters as part of the 7SK complex. The transfer of SR proteins onto the nascent transcripts destabilizes the 7SK complex, facilitating the recruitment of P-TEFb from the 7SK particle to the transcription complex and leading to transcription pausing release (Fig 2A). Although a growing number of studies suggest a role for SRSF1 in transcription and its association with the 7SK snRNP at several promoters [33], the mechanism by which SRSF1 modulates cellular transcription is still not well characterized [34].
Figure 2.
A) SRSF1 regulates transcription activation. DSIF and NELF pause transcription shortly after initiation. The kinase activity of P-TEFb phosphorylates DSIF/NELF and the RNAPII to promote elongation and processivity. P-TEFb remains inactive when sequestered within the 7SK snRNP. The binding of SRSF1 to the nascent transcript destabilizes the 7SK snRNP and promotes P-TEFb mobilization leading to the transcription pausing release. B) SRSF1 regulation of alternative splicing. SRSF1 binds to short exonic splicing enhancer (ESE) sequences to either directly promote the recruitment of spliceosomal components (U1 snRNP, U2AF65/35, U2 snRNP) to the nearby 5’ and 3’ splice sites or inhibit the activity of negative regulators of spliceosome assembly (hnRNPs) binding to a nearby sequence. C) SRSF1 regulation of mRNA export. Binding of SRSF1 onto a target mRNAs can lead to the sequestration of these messengers within the nucleus by inhibiting the loading onto the messenger bound export complex TREX of the export factors ALY/REF and NXF1/TAP. Phosphorylation of SRSF1 by IRAK 2 releases SRSF1 from the mRNA and facilitates the loading of ALY/REF and NXF1/TAP onto the TREX complex. D) SRSF1 regulation of mRNA translation. SRSF1 recruits the kinase mTOR and the phosphatase PP2A. This induces the hyperphosphorylation of the translation inhibitor 4E-BP and its release from the translation-initiation factor eIF4E. Binding of the activated eIF4E promotes loading of the mRNAs onto the polyribosomes.
2b. SRSF1-mediated transcription activation in the immune system.
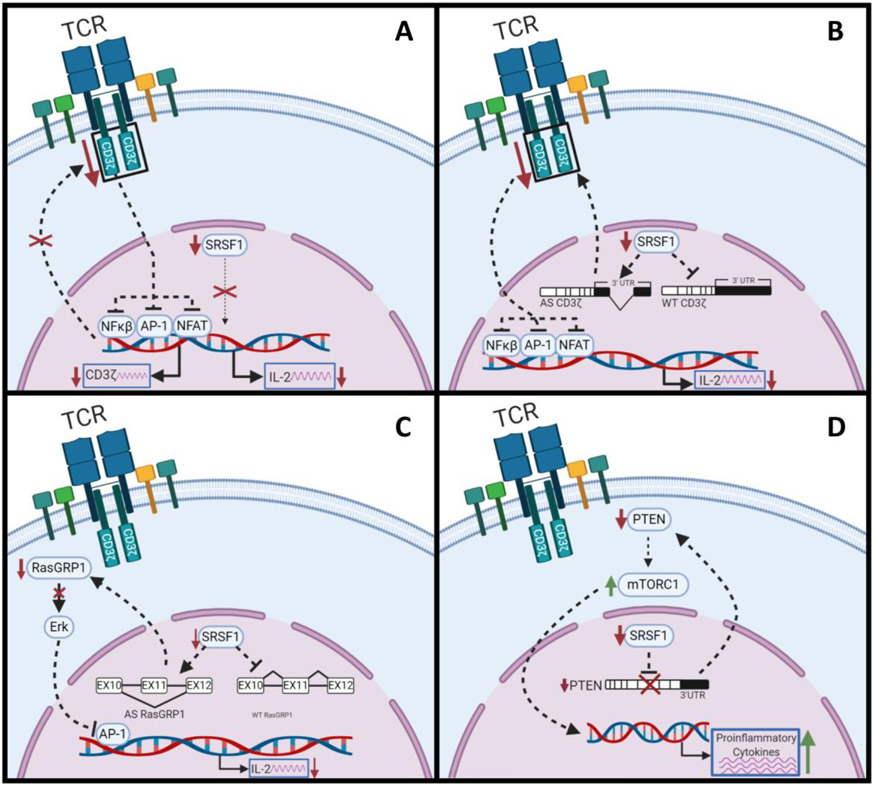
SLE is an autoimmune disease characterized by an abnormal T cell response, the production of autoantibodies and the deposition of immune complexes [35]. T-cells from patients with SLE exhibit lower SRSF1 expression than healthy individuals [20, 36]. This results in the aberrant expression of key immunomodulators, such as interleukin 2 (IL-2) and the T-cell receptor (TCR)-associated CD3ζ [37]. CD3ζ is a component of the T-cell antigen receptor (TCR-CD3) complex and plays an essential role in the initial steps of antigen recognition in TCR signal transduction [38]. The lack of functional CD3ζ leads to impaired immune function and its expression is inversely related to the severity of SLE. IL-2 is an important regulator of T cell activation and its absence correlates with an accumulation of activated T lymphocytes and autoimmune disease. Ex-vivo and biochemical assays show that SRSF1 associates with the promoter of both genes and can drive transcription of CD3ζ and IL-2 [36, 39] (Fig. 3A).
Figure 3.
Role of SRSF1 in systemic lupus erythematosus (SLE). A) SRSF1 associates with the promoters of CD3ζ and IL-2 to upregulate their expression. CD3ζ and SRSF1 are downregulate in SLE patients (red arrow). B) SRSF1 binds to the 3’UTR of CD3ζ and promotes the inclusion of a 562-nt sequence that is required for stability and translation of the transcript. C) SRSF1 promotes the splicing of a RasGRP1 isoform that is necessary for activation of the Erk/MAPK pathway resulting in a decrease of transcription activation by the transcription factor AP-1 and a decline in IL-2 expression observed in SLE patients. D) SRSF1 promotes stability of the PTEN messenger by binding the PTEN 3’UTR. This results in activation of the mTORC1 pathway, triggering T cell hyperactivity and an increase in proinflammatory cytokines in SLE patients.
SRSF1 can also associate with chromatin in a cell-cycle-specific manner and can be found enriched at active gene loci thanks to its association with the chromatin-associated protein Psip1/Ledgf [40, 41]. Ledgf/Psip1 overexpression or silencing leads to increased or decreased levels of IL-6, respectively [42], suggesting that SRSF1 might also have a function in transcription regulation through its recruitment to active chromatin sites.
3. Alternative splicing.
3a. SRSF1 regulates alternative splicing.
SRSF1 was first characterized as an alternative splicing factor in 1990 [2]. In the past three decades, multiple studies have revealed a pervasive role for SRSF1 in the alternative splicing regulation of vast gene networks and multiple biological pathways. SRSF1 binds to short motifs found in both exonic and intronic sequences to either promote or inhibit the usage of a specific splice site by either i) directly promoting the recognition of the 5’ or 3’ splice site by spliceosomal components such as the U1 small nuclear ribonucleoprotein or the U2AF35/65 heterodimer or ii) inhibiting the functions of other RNA binding proteins, such as heterogenous ribonucleoproteins (hnRNPs), that block spliceosome assembly to the nearby splice sites (Fig. 2B) [1, 3]. Alternative splicing allows a rapid and diversified response to multiple stimuli in cells of the immune compartment. Thus, it is not surprising that SRSF1 plays a prominent role in immune cell activation, functions and autoimmune disease.
3b. SRSF1 regulates alternative splicing in the immune system.
Transmembrane receptors are often coded by alternatively spliced isoforms, whose relative ratios are regulated by SRSF1 and other members of the SR protein family. CD6 is a transmembrane signal-transducing receptor expressed mostly in lymphocytes [43]. CD6 binding to CD166 promotes the formation of a stable immunological synapse between an antigen presenting cell (APC) and a T cell. CD6 binds to the TCR and the associated co-receptor CD3 forming a CD6-TCR/CD3 complex, which promotes T cell activation and proliferation [44]. T cell activation induces the expression of the alternatively spliced isoform CD6Δd3, which lacks exon 5. This exon codes for the CD166 binding domain and without it the isoform can no longer stabilize the immunological synapse. SRSF1 regulates CD6 splicing by binding to intron 4 and promoting the inclusion of exon 5., SRSF1 is downregulated following T cell activation. This leads to the exclusion of exon 5 and the synthesis of CD6Δd3 [45]. Ron, a cell surface receptor for the macrophage-stimulating protein (MSP), regulates cell migration, differentiation and survival in mononuclear phagocytes and other cell types. SRSF1 binds to a short sequence within exon 12 and promotes the skipping of exon 11 generating a shorter transcript coding for the constitutively active isoform ΔRon, which impairs macrophage functions and increases cell motility and metastasis in gastric cancer cell lines [46].
SRSF1 can also modulate the activity of transcription factors such as the interferon regulatory factor-3 (IRF3), a transcriptional activator for type I interferons (IFNα/β) as well as several chemokines [47]. SRSF1 promotes the inclusion of IRF3 exons 2 and 3 by binding to short sequences within intron 1. SRSF1 depletion results in the synthesis of a truncated IRF3 isoform that antagonizes the full-length protein and reduces the expression of downstream IRF3 targets such as IFNα/β and CXCL10.
In the past decade, high throughput genome wide studies combined with animal and cellular models have defined an emerging role for SRSF1-mediated alternative splicing in immune disease. CD46 is a widely expressed type I membrane-bound protein, which protects the host cell from damage by the complement system and modulates the adaptive immune response by controlling CD4+ T cell activation and differentiation from naïve into T helper 1 (Th1) and type 1 Treg (Tr1) cells [48]. Inclusion or skipping of exon 13 in the CD46 mRNA generates two isoforms containing two alternative c-terminal domains coded by either exon 13 or exon 14.
The CYT1 domain is coded by a mRNA containing only exon 13 at its 3’ end while skipping of exon 13 leads to the formation of the CYT2 domain which contains exon 14. CYT1 induces interferon-γ (IFNγ) production in Th1 cells and their differentiation to interleukin 10 (IL-10) secreting Tr1 cells while CYT2 switches off the Tr1 cells [49]. SRSF1 interacts with a short sequence within exon 13 to repress its inclusion and promote exon 14 inclusion instead, thus producing the CYT2 isoform [50]. Balanced expression of the CYT1 and CYT2 isoforms is essential for the modulation of T cell differentiation and homeostasis. Defects in CD46 splicing are associated with autoimmune pathologies like asthma, rheumatoid arthritis, and multiple sclerosis [51, 52].
In SLE patients, SRSF1 deficiency leads to a decrease in the transcription of key immunomodulator genes such as IL-2 and CD3ζ [20, 36]. In addition, SRSF1 modulates CD3ζ splicing by binding to the 3’ untranslated region (UTR) of CD3ζ and promoting the inclusion of a 562 nt sequence in the 3’ UTR, which is required for the transcript’s stability and translation. In the absence of SRSF1 the stability and translation of the CD3ζ messenger is reduced [53], ultimately contributing to SLE pathogenesis (Fig. 3B). Additionally, SRSF1 can regulate the splicing of RasGRP1, a guanine nucleotide exchange factor that activates the Ras pathway by engaging T-cell receptors [54]. SRSF1 modulates RasGRP1 splicing by binding to exon 11 and promoting its inclusion. In the absence of SRSF1, exon 11 is skipped producing messenger coding for a RasGRP1 isoform that is unable to activate the Erk/MAP kinase pathway, resulting in a decrease in transcriptional activation by the transcription factor AP-1 and a decline in IL-2 expression [55] (Fig. 3C).
SRSF1 splicing activity can also modulate apoptosis in SLE patients. Studies carried out in both mice and human cell lines showed that SRSF1 modulates the splicing of two members of the Bcl-2 family of apoptotic factors: Bcl-x and Bim. The Bcl-x pre-mRNA is alternatively spliced in two isoforms that encode for the short Bcl-xS, which acts as an apoptotic activator, and the long Bcl-xL, which inhibits apoptosis [23]. SRSF1 binds to a sequence located between the two 5’ splice sites and promotes selection of the Bcl-xL specific splice site [56]. The BIM transcript is alternatively spliced to generate multiple isoforms with different pro-apoptotic potential. SRSF1 promotes the inclusion of a 3’ alternative exon, which limits BIM’s apoptotic functions. Herein, low SRSF1 levels in SLE patients contribute to increased T cell apoptosis and lymphopenia [20]. Changes in SRSF1 expression levels also have the potential of causing thymic defects by affecting the splicing regulation of the inhibitor of caspase-activated DNase (ICAD). ICAD inhibits the caspase-activated DNAse (CAD), which promotes cell differentiation and breaks up the DNA during apoptosis. ICAD codes for two isoforms, ICAD-S and ICAD-L [57]. ICAD-L facilitates the correct folding of CAD and binds to it inhibiting its functions, while ICAD-S cannot promote proper CAD folding [58]. SRSF1 promotes the inclusion of an extended C-terminus in the mRNA coding for the isoform ICAD-L. In the absence of SRSF1, ICAD-S becomes the dominant isoform, leading to a misfolded non-functional CAD, which results in defects in apoptotic DNA fragmentation and thymic developmental defects [59]. This process has been linked to a several autoimmune disorders [60].
SRSF1 contributes to the pathogenesis of granulomatous disease (CGD), an inherited immunodeficiency disease, by modulating the splicing of B-245 Beta Chain (CYBB), a component of cytochrome B that regulates the expression of the NADPH heme binding subunit gp91phox. In patients with chronic CGD, neutrophils have decreased NADPH oxidase activity due to a lack of gp91phox, resulting in an increased susceptibility to bacterial and fungal infections and the formulation of granulomas at sites of inflammation and infection. SRSF1 interacts with a short RNA sequence within exon 5 to promote its inclusion in the mature mRNA. Mutations in the short SRSF1 binding sequence in CGD patients result in exon 5 skipping and the synthesis of a non-functional CYBB protein, leading to a decline in gp91phox expression [61].
SRSF1 spicing activity is essential in immune functions and its de-regulation may lead to several autoimmune diseases by altering the splicing of key immunomodulators and apoptotic factors. In addition, SRSF1 splicing activity modulates multiple signaling pathways and alterations in this factor’s expression levels and activity may lead to oncogenesis as reviewed elsewhere [22].
4. SRSF1 regulates mRNA stability.
Regulation of mRNA stability allows for the fine tuning, rapid mobilization or halting of a gene’s expression. This is of especially true for the messengers that are required to mount a prompt and precise immune response. The rate of mRNA decay is controlled by the polyA and sequences located mostly within the 5’ and 3’ UTR [62]. Regulation of the stability of RNAs coding for cytokines, transcription and signaling factors is required for rapid modification of the protein pool in response to specific stimuli.
Adenine and uridine-rich elements (AREs) within the 3’-UTR are among the most common stability regulatory sequences found in mammals. AREs function by recruiting different sets of RBPs to modulate the stability and translation of the transcript. SRSF1 modulates the activity of the T-cell intracellular antigen-1 (TIA-1) and TIA-1 related protein (TIAR), two RNA binding proteins broadly expressed in eukaryotic cells, which act as translational repressors and affect the stability of ARE-containing mRNAs by assembling into cytoplasmic stress granules [63]. SRSF1 migrates from the nucleus to the cytoplasm and interacts with TIA-1/TIAR containing stress granules in response to environmental stress. Overexpressing or tethering SRSF1 to a reporter transcript bearing an ARE in its 3’ UTR strongly reduces mRNA expression through inhibition of both the transcript’s stability and translation [64]. Interleukin 17A (IL-17A) is a proinflammatory cytokine, which coordinates tissue inflammation by increasing the expression of several proinflammatory cytokines by activating their transcription via the transcription factor NF-κB and stabilizing their mRNA [65]. IL-17 has been shown to stabilize the CXCL1 transcript by engaging the IL-17 receptor (IL-17R) and promoting the formation of a complex between the adaptors TRAF5, TRAF2, Act1 and SRSF1. In the absence of IL-17, SRSF1 binds freely to and destabilizes the CXCL1 messenger [66].
The phosphoinositide- 3 kinase (PI3K)/AKT/mammalian target of rapamycin (mTOR) pathway is a key pathway involved in the activation of T cells. The mTOR pathway determines T cell differentiation, activation and proliferation [67]. The phosphatase and tensin homolog (PTEN) is a negative regulator of the mTOR pathway [68]. SRSF1 regulates stability of the PTEN messenger by interacting with its 3’ UTR [69]. SRSF1 knockout mice exhibit reduced expression of PTEN, which results in increased activation of the mTOR pathway, triggering T cell hyperactivity and an increase in proinflammatory cytokines resulting in autoimmunity [69] (Fig. 3D).
5. mRNA export.
5a. SRSF1 regulates mRNA export.
mRNA transcription, processing and translation are coupled with the export of mRNA from the nucleus to the cytoplasm. This ensures efficient loading of export factors onto the processed mRNA and its loading onto polyribosomes upon entry into the cytoplasm [70]. The factor REF/ALY is initially loaded onto the spliced mRNAs in the nucleus, NXF1/TAP is then recruited into a larger complex named TREX triggering the transfer of the RNA from REF/ALY to TAP/NXF1 [71]. SR proteins such as SRSF1, that shuttle between the nucleus and the cytoplasm have been shown to replace REF/ALY and function as mRNA export adaptors [72]. SRSF1 interacts with NXF1/TAP via a TAP-binding domain in its arginine-rich inter-RRM linker domain and a series of arginine residues in its RS domain [73]. However, the functional significance of such interaction is still unclear since it destabilizes SRSF1 binding onto the target mRNA and in some systems the depletion of SRSF1 increases the export of specific mRNAs [74].
5b. SRSF1 regulates mRNA export in the immune system.
The binding of SRSF1 onto mRNAs coding for several pro-inflammatory cytokines (Cxcl1, Tnf and Cxcl2) can lead to the sequestration of these messengers within the nucleus, preventing them from being exported into the cytoplasm and creating a reservoir of transcripts that can be rapidly mobilized in response to extracellular stimuli and the activation of specific signal cascades [74]. Toll-like receptor (TLR) signaling regulates the transcriptional and post-transcriptional expression of chemokines and cytokines. TLRs transduce signals through the adaptor molecule MyD88 and the interleukin 1 receptor associated kinase 2 IRAK2 [75]. Following stimulation, TLRs induces the nuclear localization of IRAK2 to facilitate the nuclear export of mRNAs coding for several pro-inflammatory cytokines. Once in the nucleus IRAK2 promotes the assembly of the nuclear export factors ALY/REF and NXF1/TAP onto the transcripts. SRSF1 has been shown to mediate the nuclear sequestration of these mRNAs, possibly by blocking the binding of ALY/REF and NXF1/TAP to the mRNAs. It is plausible that IRAK2 might mediate the removal of SRSF1 by promoting its phosphorylation, thus facilitating the assembly of the transcription-export (TREX)-NXF1/TAP complex and the export of the bound mRNAs (Fig. 2C) [74].
6. SRSF1 regulates mRNA translation.
Regulation of protein translation allows the induction of immediate changes in the proteome without the need for de novo transcription. Translation can be controlled globally, in response to major physiological changes, or can be selectively applied to single genes or groups of genes. The selective control in the translation of specific genes is emerging as a key mechanism of gene expression to determine a variety of functions in both the adaptive and innate immune response [76]. Transcript-specific translation regulation is often mediated by the interaction of RBPs with short sequences in the UTR of the target messenger. The translation-initiation factor eIF4E enables ribosome assembly and plays a key role in the regulation of both innate and adaptive immune response. The eukaryotic translation initiation factor (eIF)4E-BP kinase mTOR controls the activity of the 4E-binding proteins (4E-BPs), which inhibit the translation-initiation factor eIF4E. The mTOR-dependent regulation of the eIF4E/4E-BP system is one of the principal mechanisms regulating lymphocyte activation, proliferation and migration, and the synthesis of several cytokines during the inflammation process [77].
SRSF1 associates with mRNAs isolated from polyribosomes to enhance the translation of genes containing its cognate binding sites [78]. SRSF1 modulates translation of the bound mRNAs by recruiting both the protein kinase mTOR and the phosphatase PP2A. This induces the hyperphosphorylation of 4E-BP and its release from the inhibitory complex eIF4E/4E-BP, thus activating the translation-initiation factor eIF4E (Fig. 2D) [79]. Over 1500 mRNAs have been identified as translational targets of SRSF1 by high-throughput deep sequencing analysis of polysomal fractions. Several cell cycle regulators and transcription factors are coded by the mRNAs isolated, suggesting that SRSF1-mediated translation control is a key factor in T cell activation, differentiation and proliferation. [80]
7. SRSF1 regulates miRNA processing.
MicroRNAs (miRNAs) are small non-coding RNAs that regulate the degradation and translation of target mRNAs. miRNAs are first transcribed by RNAPII into a longer pri-miRNA, which is cleaved by the RNase Drosha to generate a pre-miRNA. Once exported to the cytoplasm the pre-miRNA is cleaved by the RNase Dicer and is loaded as a mature miRNA into the RNA-induced silencing complex (RISC) where it interacts with its mRNA target [81]. miRNAs negatively regulate the expression of several key immune development genes, fine tuning and conferring robustness to the immune cell response and their aberrant expression can lead to autoimmunity or other pathologies [82].
SRSF1 modulates the expression of miR-10b, which is connected to cancer metastasis and autoimmune disease. miR-10b overexpression in cancer cells downregulates MICB, a stress-induced ligand for the natural killer (NK) cell receptor NKG2D, which is upregulated on the surface of tumor cells. MICB downregulation in breast cancer cells promotes immune escape and initiates robust invasion and metastasis [83]. miR-10b is also upregulated in T helper 17 (Th17) cells of patients with ankylosing spondylitis, an inflammatory arthritic disease that results in fused vertebrae and severe joint pain. miR-10b binds to the 3’ UTR of the MAP3K7 kinase inhibiting its expression, thus suggesting a feedback regulatory mechanism where proinflammatory cytokines upregulate miR-10b expression, which in turn downregulates IL-17A expression to suppress Th17 cell function [84]. SRSF1 binds to the pri-miR-10b sequence inhibiting its processing while miR-10b binds to a sequence in the 3’ UTR of the SRSF1 transcript to suppress its expression in a regulatory feedback loop [85]. A SRSF1-dependent feedback loop also regulates miR-7. The pri-miR-7 transcript is bound by SRSF1 to promote Drosha cleavage, whereby miR-7 represses SRSF1 translation by binding its 3’ UTR [17]. miR-7 inhibition is associated with the impairment of cytotoxic T-lymphocyte (CTL) mediated lysis of breast cancer cells [86], while in patients with type 2 diabetes its expression correlates with failure of insulin-producing cells (pancreatic β cells) [87]. SRSF1 can also promote Drosha cleavage of the miR-29b pri-miRNA, which regulates helper T cell differentiation by repressing multiple target genes including IFN-γ [88]. miR-29b is also downregulated in tumor associated dendritic cells resulting in failure to control proinflammatory pathways and promoting a tumor-permissive phenotype, thus indicating a key role for SRSF1 in both immune response and tumorigenesis [89].
8. Conclusions
SRSF1 exerts a pervasive control on immune cell differentiation, proliferation and adaptive response to various stimuli. Furthermore, the expression of SRSF1 itself is regulated by specific immunomodulators, such as TNFα, in response to inflammation [90]. Although work carried out in the past three decades has identified multiple SRSF1 functions, and its pleiotropic effects on the immune system, the full regulatory potential of this protein is yet to be defined. For example, SRSF1-3, a splice isoform of SRSF1, is necessary for the somatic hypermutation (SHM) of Ig genes, which is initiated by the activation-induced cytidine deaminase (AID) [91, 92]. Nevertheless, the exact molecular mechanism of action of SRSF1 in SHM remains unknown.
Highlights.
SRSF1 regulates the transcription of CD3ζ and IL-2
SRSF1 is downregulation in SLE patients
SRSF1 affects the splicing of cytokines and transmembrane signal-transducing receptor
SRSF1 regulates the stability and nuclear export of mRNAs coding for cytokines
SRSF1 modulates miRNA processing
Acknowledgments
Funding sources:
This work was supported by NIH/NIAID grant R15AI120882 to M.C.
Biography
Sean Paz is currently a Doctoral student at Florida Atlantic University working at the transcriptional functions of SR proteins and on their role in HIV replication.
Anastasia Ritchie received her MS at Florida Atlantic University and is currently pursuing her doctoral degree at the Heinrich-Heine-Universität Düsseldorf. Anastasia work is focused on the understanding of the mechanisms utilized by splicing factors in the regulation of gene expression in cells of the immune compartment.

Christopher Mauer is currently pursuing his MS at Florida Atlantic University where he is investigating the changes induced by HIV infection in the transcriptome and proteonome of T cells.

Massimo Caputi received his PhD at the International Center for Genetic Engineering and Biotechnology in Trieste, Italy. He carried out his postdoctoral training at the University of California, Santa Cruz. He is currently a Professor at the Charles E Schmidt College of medicine at Florida Atlantic University. He has worked extensively on the mechanisms that regulate the alternative splicing of cellular and viral genes.
Footnotes
Conflicts of interest:
There authors declare that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Jeong S, SR Proteins: Binders, Regulators, and Connectors of RNA, Mol Cells 40(1) (2017) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Krainer AR, Conway GC, Kozak D, The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites, Cell 62(1) (1990) 35–42. [DOI] [PubMed] [Google Scholar]
- [3].Das S, Krainer AR, Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer, Mol Cancer Res 12(9) (2014) 1195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mayeda A, Screaton GR, Chandler SD, Fu XD, Krainer AR, Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements, Mol. Cell. Biol 19 (1999) 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang J, Smith PJ, Krainer AR, Zhang MQ, Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes, Nucleic acids research 33(16) (2005) 5053–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sanford JR, Wang X, Mort M, Vanduyn N, Cooper DN, Mooney SD, Edenberg HJ, Liu Y, Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts, Genome research 19(3) (2009) 381–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Clery A, Sinha R, Anczukow O, Corrionero A, Moursy A, Daubner GM, Valcarcel J, Krainer AR, Allain FH, Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition, Proceedings of the National Academy of Sciences of the United States of America 110(30) (2013) E2802–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Cho S, Hoang A, Sinha R, Zhong X-Y, Fu X-D, Krainer AR, Ghosh G, Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly, PNAS 108(20) (2011) 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Caceres JF, Screaton GR, Krainer AR, A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm, Genes & development 12(1) (1998) 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M, Lysine acetylation targets protein complexes and co-regulates major cellular functions, 2009, pp. 834–840. [DOI] [PubMed] [Google Scholar]
- [11].Sinha R, Allemand E, Zhang Z, Karni R, Myers MP, Krainer AR, Arginine methylation controls the subcellular localization and functions of the oncoprotein splicing factor SF2/ASF, Molecular and cellular biology 30(11) (2010) 2762–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY, A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins, The Journal of biological chemistry 275(11) (2000) 7950–7. [DOI] [PubMed] [Google Scholar]
- [13].Colwill K, Pawson T, Andrews B, Prasad J, Manley JL, Bell JC, Duncan PI, The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution, The EMBO journal 15(2) (1996) 265–75. [PMC free article] [PubMed] [Google Scholar]
- [14].Misteli T, Spector DL, Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors, Molecular biology of the cell 7(10) (1996) 1559–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Novoyatleva T, Heinrich B, Tang Y, Benderska N, Butchbach ME, Lorson CL, Lorson MA, Ben-Dov C, Fehlbaum P, Bracco L, Burghes AH, Bollen M, Stamm S, Protein phosphatase 1 binds to the RNA recognition motif of several splicing factors and regulates alternative pre-mRNA processing, Human molecular genetics 17(1) (2008) 52–70. [DOI] [PubMed] [Google Scholar]
- [16].Sun S, Zhang Z, Sinha R, Karni R, Krainer AR, SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control, Nature structural & molecular biology 17(3) (2010) 306–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wu H, Sun S, Tu K, Gao Y, Xie B, Krainer AR, Zhu J, A splicing-independent function of SF2/ASF in microRNA processing, Molecular cell 38(1) (2010) 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR, The gene encoding the splicing factor SF2/ASF is a proto-oncogene, Nature structural & molecular biology 14(3) (2007) 185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, Muthuswamy SK, Krainer AR, The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation, Nature structural & molecular biology 19(2) (2012) 220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Katsuyama T, Martin-Delgado IJ, Krishfield SM, Kyttaris VC, Moulton VR, Splicing factor SRSF1 controls T cell homeostasis and its decreased levels are linked to lymphopenia in systemic lupus erythematosus, Rheumatology (2020) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gautrey HL, Tyson-Capper AJ, Regulation of Mcl-1 by SRSF1 and SRSF5 in cancer cells, PloS one 7(12) (2012) e51497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gonçalves V, Jordan P, Posttranscriptional regulation of splicing factor SRSF1 and its role in cancer cell biology, BioMed Research International 2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Warren CFA, Wong-Brown MW, Bowden NA, BCL-2 family isoforms in apoptosis and cancer, Cell Death Dis 10(3) (2019) 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Turner M, DÍaz-Muñoz MD, RNA-binding proteins control gene expression and cell fate in the immune system review-article, Nature immunology 19(2) (2018) 120–129. [DOI] [PubMed] [Google Scholar]
- [25].Szabo PA, Levitin HM, Miron M, Snyder ME, Senda T, Yuan J, Cheng YL, Bush EC, Dogra P, Thapa P, Farber DL, Sims PA, Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease, Nature communications 10(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rotival M, Quach H, Quintana-Murci L, Defining the genetic and evolutionary architecture of alternative splicing in response to infection, Nature communications 10(1) (2019) 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dlamini Z, Mokoena F, Hull R, Abnormalities in alternative splicing in diabetes: Therapeutic targets, Journal of Molecular Endocrinology 59(2) (2017) R93–R107. [DOI] [PubMed] [Google Scholar]
- [28].Evsyukova I, Somarelli JA, Gregory SG, Garcia-Blanco MA, Alternative splicing in multiple sclerosis and other autoimmune diseases, RNA biology 7(4) (2010) 462–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lahita R, Textbook of the autoimmune diseases, Lippincott Williams & Wilkins; 2000. [Google Scholar]
- [30].Liu X, Kraus WL, Bai X, Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways, Trends in biochemical sciences 40(9) (2015) 516–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Li Y, Liu M, Chen LF, Chen R, P-TEFb: Finding its ways to release promoter-proximally paused RNA polymerase II, Transcription 9(2) (2018) 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].D'Orso I, 7SKiing on chromatin: Move globally, act locally, RNA biology 13(6) (2016) 545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD, SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase, Cell 153(4) (2013) 855–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mo S, Ji X, Fu XD, Unique role of SRSF2 in transcription activation and diverse functions of the SR and hnRNP proteins in gene expression regulation, Transcription 4(5) (2013) 251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tsokos GC, Mechanisms of disease: Systemic lupus erythematosus, New England Journal of Medicine 365(22) (2011) 2110–2121. [DOI] [PubMed] [Google Scholar]
- [36].Moulton VR, Grammatikos AP, Fitzgerald LM, Tsokos GC, Splicing factor SF2/ASF rescues IL-2 production in T cells from systemic lupus erythematosus patients by activating IL-2 transcription, Proceedings of the National Academy of Sciences of the United States of America 110(5) (2013) 1845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Nambiar MP, Mitchell JP, Ceruti RP, Malloy MA, Tsokos GC, Prevalence of T cell receptor ζchain deficiency in systemic lupus erythematosus, Lupus 12(1) (2003) 46–51. [DOI] [PubMed] [Google Scholar]
- [38].Xu X, Li H, Xu C, Structural understanding of T cell receptor triggering, Cell Mol Immunol 17(3) (2020) 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moulton VR, Gillooly AR, Perl MA, Markopoulou A, Tsokos GC, Serine Arginine-Rich Splicing Factor 1 (SRSF1) Contributes to the Transcriptional Activation of CD3zeta in Human T Cells, PloS one 10(7) (2015) e0131073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Loomis RJ, Naoe Y, Parker JB, Savic V, Bozovsky MR, Macfarlan T, Manley JL, Chakravarti D, Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation, Molecular cell 33(4) (2009) 450–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA, Psip1/Ledgf p52 Binds Methylated Histone H3K36 and Splicing Factors and Contributes to the Regulation of Alternative Splicing, PLoS genetics 8(5) (2012) e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Takeichi T, Sugiura K, Muro Y, Matsumoto K, Ogawa Y, Futamura K, Kaminuma O, Hashimoto N, Shimoyama Y, Saito H, Tomita Y, Overexpression of LEDGF/DFS70 induces IL-6 via p38 activation in HaCaT cells, similar to that seen in the psoriatic condition, Journal of Investigative Dermatology 130(12) (2010) 2760–2767. [DOI] [PubMed] [Google Scholar]
- [43].Consuegra-Fernández M, Lin F, Fox DA, Lozano F, Clinical and experimental evidence for targeting CD6 in immune-based disorders, Autoimmun Rev 17(5) (2018) 493–503. [DOI] [PubMed] [Google Scholar]
- [44].Meddens MBM, Mennens SFB, Celikkol FB, Te Riet J, Kanger JS, Joosten B, Witsenburg JJ, Brock R, Figdor CG, Cambi A, Biophysical Characterization of CD6-TCR/CD3 Interplay in T Cells, Frontiers in immunology 9 (2018) 2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].da Glória VG, Martins de Araújo M, Mafalda Santos A, Leal R, de Almeida SF, Carmo AM, Moreira A, T Cell Activation Regulates CD6 Alternative Splicing by Transcription Dynamics and SRSF1, The Journal of Immunology 193(1) (2014) 391–399. [DOI] [PubMed] [Google Scholar]
- [46].Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G, Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene, Molecular cell 20(6) (2005) 881–90. [DOI] [PubMed] [Google Scholar]
- [47].Solis M, Goubau D, Romieu-Mourez R, Genin P, Civas A, Hiscott J, Distinct functions of IRF-3 and IRF-7 in IFN-alpha gene regulation and control of anti-tumor activity in primary macrophages, Biochem Pharmacol 72(11) (2006) 1469–76. [DOI] [PubMed] [Google Scholar]
- [48].Cardone J, Le Friec G, Vantourout P, Roberts A, Fuchs A, Jackson I, Suddason T, Lord G, Atkinson JP, Cope A, Hayday A, Kemper C, Complement regulator CD46 temporally regulates cytokine production by conventional and unconventional T cells, Nature immunology 11(9) (2010) 862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ni Choileain S, Weyand NJ, Neumann C, Thomas J, So M, Astier AL, The dynamic processing of CD46 intracellular domains provides a molecular rheostat for T cell activation, PloS one 6(1) (2011) e16287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Tang SJ, Luo S, Ho JXJ, Ly PT, Goh E, Roca X, Characterization of the regulation of CD46 RNA alternative splicing, Journal of Biological Chemistry 291(27) (2016) 14311–14323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Xu YQ, Gao YD, Yang J, Guo W, A defect of CD4+CD25+ regulatory T cells in inducing interleukin-10 production from CD4+ T cells under CD46 costimulation in asthma patients, J Asthma 47(4) (2010) 367–73. [DOI] [PubMed] [Google Scholar]
- [52].Martinez-Forero I, Garcia-Munoz R, Martinez-Pasamar S, Inoges S, Lopez-Diaz de Cerio A, Palacios R, Sepulcre J, Moreno B, Gonzalez Z, Fernandez-Diez B, Melero I, Bendandi M, Villoslada P, IL-10 suppressor activity and ex vivo Tr1 cell function are impaired in multiple sclerosis, European journal of immunology 38(2) (2008) 576–86. [DOI] [PubMed] [Google Scholar]
- [53].Moulton VR, Tsokos GC, Alternative splicing factor/splicing factor 2 regulates the expression of the ζ subunit of the human T cell receptor-associated CD3 complex, Journal of Biological Chemistry 285(17) (2010) 12490–12496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Priatel JJ, Teh SJ, Dower NA, Stone JC, Teh HS, RasGRP1 transduces low-grade TCR signals which are critical for T cell development, homeostasis, and differentiation, Immunity 17(5) (2002) 617–27. [DOI] [PubMed] [Google Scholar]
- [55].Kono M, Kurita T, Yasuda S, Kono M, Fujieda Y, Bohgaki T, Katsuyama T, Tsokos GC, Moulton VR, Atsumi T, Decreased Expression of Serine/Arginine-Rich Splicing Factor 1 in T Cells From Patients With Active Systemic Lupus Erythematosus Accounts for Reduced Expression of RasGRP1 and DNA Methyltransferase 1, Arthritis and Rheumatology 70(12) (2018) 2046–2056. [DOI] [PubMed] [Google Scholar]
- [56].Bielli P, Bordi M, Di Biasio V, Sette C, Regulation of BCL-X splicing reveals a role for the polypyrimidine tract binding protein (PTBP1/hnRNP I) in alternative 5' splice site selection, Nucleic acids research 42(19) (2014) 12070–12081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Larsen BD, Sørensen CS, The caspase-activated DNase: apoptosis and beyond, The FEBS journal 284(8) (2017) 1160–1170. [DOI] [PubMed] [Google Scholar]
- [58].Sakahira H, Enari M, Nagata S, Functional differences of two forms of the inhibitor of caspase- activated DNase, ICAD-L, and ICAD-S, Journal of Biological Chemistry 274(22) (1999) 15740–15744. [DOI] [PubMed] [Google Scholar]
- [59].Li X, Wang J, Manley JL, Loss of splicing factor ASF/SF2 induces G2 cell cycle arrest and apoptosis, but inhibits internucleosomal DNA fragmentation, Genes and Development 19(22) (2005) 2705–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ralston SP, Ian; Mark Strachan; Richard Hobson, Davidson’s Principles and Practice of Medicine, Elsevier; 2018. [Google Scholar]
- [61].de Boer M, van Leeuwen K, Geissler J, Belohradsky BH, Kuijpers TW, Roos D, Mutation in an exonic splicing enhancer site causing chronic granulomatous disease, Blood Cells, Molecules, and Diseases 66 (2017) 50–57. [DOI] [PubMed] [Google Scholar]
- [62].Guhaniyogi J, Brewer G, Regulation of mRNA stability in mammalian cells, Gene 265(1-2) (2001) 11–23. [DOI] [PubMed] [Google Scholar]
- [63].Pullmann R Jr., Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M, Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs, Molecular and cellular biology 27(18) (2007) 6265–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Delestienne N, Wauquier C, Soin R, Dierick JF, Gueydan C, Kruys V, The splicing factor ASF/SF2 is associated with TIA-1-related/TIA-1-containing ribonucleoproteic complexes and contributes to post-transcriptional repression of gene expression, The FEBS journal 277(11) (2010) 2496–514. [DOI] [PubMed] [Google Scholar]
- [65].Hartupee J, Liu C, Novotny M, Li X, Hamilton T, IL-17 enhances chemokine gene expression through mRNA stabilization, J Immunol 179(6) (2007) 4135–41. [DOI] [PubMed] [Google Scholar]
- [66].Sun D, Novotny M, Bulek K, Liu C, Li X, Hamilton T, Treatment with IL-17 prolongs the half-life of chemokine CXCL1 mRNA via the adaptor TRAF5 and the splicing-regulatory factor SF2 (ASF), Nature immunology 12(9) (2011) 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chapman NM, Chi H, mTOR Links Environmental Signals to T Cell Fate Decisions, Frontiers in immunology 5 (2014) 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Chi H, Regulation and function of mTOR signalling in T cell fate decisions, Nat Rev Immunol 12(5) (2012) 325–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Katsuyama T, Li H, Comte D, Tsokos GC, Moulton VR, Splicing factor SRSF1 controls T cell hyperactivity and systemic autoimmunity, Journal of Clinical Investigation (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Carmody SR, Wente SR, mRNA nuclear export at a glance, Journal of cell science 122(Pt 12) (2009) 1933–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Viphakone N, Hautbergue GM, Walsh M, Chang CT, Holland A, Folco EG, Reed R, Wilson SA, TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export, Nature communications 3 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Huang Y, Gattoni R, Stévenin J, Steitz JA, SR Splicing Factors Serve as Adapter Proteins for TAP-Dependent mRNA Export, Molecular cell 11(3) (2003) 837–843. [DOI] [PubMed] [Google Scholar]
- [73].Tintaru AM, Hautbergue GM, Hounslow AM, Hung ML, Lian LY, Craven CJ, Wilson SA, Structural and functional analysis of RNA and TAP binding to SF2/ASF, EMBO Rep 8(8) (2007) 756–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Zhou H, Bulek K, Li X, Herjan T, Yu M, Qian W, Wang H, Zhou G, Chen X, Yang H, Hong L, Zhao J, Qin L, Fukuda K, Flotho A, Gao J, Dongre A, Carman JA, Kang Z, Su B, Kern TS, Smith JD, Hamilton TA, Melchior F, Fox PL, Li X, IRAK2 directs stimulus-dependent nuclear export of inflammatory mRNAs, eLife 6 (2017) 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Kawasaki T, Kawai T, Toll-like receptor signaling pathways, Frontiers in immunology 5 (2014) 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Mazumder B, Li X, Barik S, Translation control: a multifaceted regulator of inflammatory response, J Immunol 184(7) (2010) 3311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O, Translational control of immune responses: from transcripts to translatomes, Nature immunology 15(6) (2014) 503–11. [DOI] [PubMed] [Google Scholar]
- [78].Sanford JR, Gray NK, Beckmann K, Caceres JF, A novel role for shuttling SR proteins in mRNA translation, Genes & development 18(7) (2004) 755–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Michlewski G, Sanford JR, Cáceres JF, The Splicing Factor SF2/ASF Regulates Translation Initiation by Enhancing Phosphorylation of 4E-BP1, Molecular cell 30(2) (2008) 179–189. [DOI] [PubMed] [Google Scholar]
- [80].Maslon MM, Heras SR, Bellora N, Eyras E, Caceres JF, The translational landscape of the splicing factor SRSF1 and its role in mitosis, eLife (2014) e02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].O'Brien J, Hayder H, Zayed Y, Peng C, Overview of microRNA biogenesis, mechanisms of actions, and circulation, Frontiers in Endocrinology 9(AUG) (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Mehta A, Baltimore D, MicroRNAs as regulatory elements in immune system logic, Nature Reviews Immunology 16(5) (2016) 279–294. [DOI] [PubMed] [Google Scholar]
- [83].Ma L, Teruya-Feldstein J, Weinberg RA, Tumour invasion and metastasis initiated by microRNA-10b in breast cancer, Nature 449(7163) (2007) 682–8. [DOI] [PubMed] [Google Scholar]
- [84].Chen L, Al-Mossawi MH, Ridley A, Sekine T, Hammitzsch A, De Wit J, Simone D, Shi H, Penkava F, Kurowska-Stolarska M, Pulyakhina I, Knight JC, Kim TJ, Bowness P, MiR-10b-5p is a novel Th17 regulator present in Th17 cells from ankylosing spondylitis, Annals of the Rheumatic Diseases 76(3) (2017) 620–624. [DOI] [PubMed] [Google Scholar]
- [85].Sokół E, Kędzierska H, Czubaty A, Rybicka B, Rodzik K, Tański Z, Bogusławska J, Piekiełko-Witkowska A, microRNA-mediated regulation of splicing factors SRSF1, SRSF2 and hnRNP A1 in context of their alternatively spliced 3′UTRs, Experimental cell research 363(2) (2018) 208–217. [DOI] [PubMed] [Google Scholar]
- [86].Akalay I, Tan TZ, Kumar P, Janji B, Mami-Chouaib F, Charpy C, Vielh P, Larsen AK, Thiery JP, Sabbah M, Chouaib S, Targeting WNT1-inducible signaling pathway protein 2 alters human breast cancer cell susceptibility to specific lysis through regulation of KLF-4 and miR-7 expression, Oncogene 34(17) (2015) 2261–71. [DOI] [PubMed] [Google Scholar]
- [87].Horsham JL, Ganda C, Kalinowski FC, Brown RAM, Epis MR, Leedman PJ, MicroRNA-7: A miRNA with expanding roles in development and disease, International Journal of Biochemistry and Cell Biology 69 (2015) 215–224. [DOI] [PubMed] [Google Scholar]
- [88].Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM, MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells, Immunity 35(2) (2011) 169–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Botta C, Cuce M, Pitari MR, Caracciolo D, Gulla A, Morelli E, Riillo C, Biamonte L, Gallo Cantafio ME, Prabhala R, Mignogna C, Di Vito A, Altomare E, Amodio N, Di Martino MT, Correale P, Rossi M, Giordano A, Munshi NC, Tagliaferri P, Tassone P, MiR-29b antagonizes the pro-inflammatory tumor-promoting activity of multiple myeloma-educated dendritic cells, Leukemia 32(4) (2018) 1003–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Xiong Z, Shaibani A, Li YP, Yan Y, Zhang S, Yang Y, Yang F, Wang H, Yang XF, Alternative splicing factor ASF/SF2 is down regulated in inflamed muscle, J Clin Pathol 59(8) (2006) 855–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Kanehiro Y, Todo K, Negishi M, Fukuoka J, Gan W, Hikasa T, Kaga Y, Takemoto M, Magari M, Li X, Manley JL, Ohmori H, Kanayama N, Activation-induced cytidine deaminase (AID)-dependent somatic hypermutation requires a splice isoform of the serine/arginine-rich (SR) protein SRSF1, Proceedings of the National Academy of Sciences of the United States of America 109(4) (2012) 1216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Kawaguchi Y, Nariki H, Kawamoto N, Kanehiro Y, Miyazaki S, Suzuki M, Magari M, Tokumitsu H, Kanayama N, SRSF1-3 contributes to diversification of the immunoglobulin variable region gene by promoting accumulation of AID in the nucleus, Biochemical and biophysical research communications 485(2) (2017) 261–266. [DOI] [PubMed] [Google Scholar]