Abstract
Hypothalamic-pituitary-adrenal (HPA) axis dysregulation has been associated with altered immune function, but the underlying molecular mechanisms are unclear. Epigenetic processes, including DNA methylation, respond to the glucocorticoid end-products of the HPA axis (cortisol in humans) and could be involved in this neuroendocrine-immune crosstalk. Here we examined the extent to which variations in HPA axis regulation are associated with peripheral blood DNA (CpG) methylation changes in 57 chronically stressed caregivers and 67 control women. DNA methylation was determined with the Illumina 450k array for a panel of genes involved in HPA axis and immune function. HPA axis feedback was assessed with the low-dose dexamethasone suppression test (DST), measuring the extent to which cortisol secretion is suppressed by the synthetic glucocorticoid dexamethasone. After multiple testing correction in the entire cohort, higher post-DST cortisol, reflecting blunted HPA axis negative feedback, but not baseline waking cortisol, was associated with lower DNA methylation at eight TNF and two FKBP5 CpG sites. Caregiver group status was associated with lower methylation at two IL6 CpG sites. Since associations were most robust with TNF methylation (32% of the 450k-covered sites), we further examined functionality of this epigenetic signature in cultured peripheral blood mononuclear cells in 33 participants; intriguingly, lower TNF methylation resulted in higher ex vivo TNF mRNA following immune stimulation. Taken together, our findings link chronic stress and HPA axis regulation with epigenetic signatures at immune-related genes, thereby providing novel insights into how aberrant HPA axis function may contribute to heightened inflammation and disease risk.
Keywords: caregiving, dexamethasone suppression test, DNA methylation, FKBP5, HPA axis, interleukin 6, inflammation, TNF
1. Introduction
The hypothalamic-pituitary-adrenal (HPA) axis is a highly conserved neuroendocrine axis in vertebrates that modulates multiple physiological processes, including stress responses and immune function (Chrousos and Gold, 1992; Horowitz and Zunszain, 2015; Rhen and Cidlowski, 2005). Cortisol, the primary glucocorticoid end product of the HPA axis in humans, and several pro-inflammatory cytokines, such as IL-1β, IL-6, and TNF, increase in response to experimental stress (Marsland et al., 2017; Rohleder, 2014; Seddon et al., 2020). Notably, different timing and context of exposure to cortisol and other glucocorticoids have been reported to either promote or inhibit inflammatory signaling by binding to and activating the glucocorticoid receptor (GR) (Horowitz et al., 2020; Newton et al., 2017). Once activated, the GR functions as a ligand-dependent transcription factor that translocates to the nucleus and interacts with the DNA via glucocorticoid response elements (GREs). GR activation in the hypothalamus and pituitary inhibits HPA axis activity as part of a negative feedback loop, which is essential for homeostatic regulation of stress and immune responses (Cohen et al., 2012; Nikkheslat et al., 2015; Zannas and Chrousos, 2017).
To experimentally assess HPA axis feedback and homeostasis, studies in humans commonly employ the dexamethasone suppression test (DST). DST encompasses systemic administration of the synthetic glucocorticoid dexamethasone (DEX), which triggers the HPA axis negative feedback to decrease levels of circulating cortisol (Carroll et al., 1981). Decreased ability to suppress cortisol – that is, higher cortisol levels after oral DEX administration – reflects diminished HPA axis feedback regulation (Perrin et al., 2019). DST was first used in clinical settings to distinguish among etiologies of Cushing syndrome (Kennedy et al., 1984), but it was subsequently applied in fields as diverse as cardiology, immunology, and psychiatry (Di Dalmazi et al., 2019; Jokinen et al., 2008), highlighting its potential as an endophenotype for multiple disease entities. Among variations of the DST, the use of low oral DEX dose (0.25 mg) has been employed to allow exploration of subtler variations in HPA axis regulation (Direk et al., 2016; Gaffey et al., 2019).
Although previous research has examined possible mechanisms linking HPA axis and immune dysregulation (Barnes and Adcock, 2009), a paucity of studies have investigated the role of epigenetic mechanisms in this neuroimmune interplay (Jung et al., 2015; Misale et al., 2018; Zannas et al., 2019). One critical epigenetic mechanism is DNA methylation, which, with the advent of array technology, has become the most widely studied epigenetic mechanism in humans (Murphy and Mill, 2014). To our knowledge, two studies to date have explored the epigenetic correlates of DST (Kaminsky et al., 2015; Tyrka et al., 2016); hypermethylation at NR3C1, the gene encoding the GR, was associated with increased cortisol response to the DEX/CRH test, a modified version of the DST, whereas DNA methylation at SKA2 gene, encoding a protein that binds to the GR, interacted with childhood trauma to predict post-DST cortisol. Exposure to stress has also been linked with differentially methylated regions at genes regulating HPA axis function, such as NR3C1 (Palma-Gudiel et al., 2015) and FKBP5 (Yehuda et al., 2016). Accordingly, stress-induced DNA methylation changes have been observed in close proximity to GREs (Klengel et al., 2013; Zannas et al., 2015) and have been associated with altered HPA axis regulation (Houtepen et al., 2018; Mulder et al., 2020). Furthermore, stress can influence DNA methylation at several genes with known roles in immune function (Janusek et al., 2017; McDade et al., 2019; Uddin et al., 2010; Zannas et al., 2019). While these studies have assessed cumulative lifetime stress, childhood adversity, and low socioeconomic status, no studies have examined the epigenetic correlates of caregiving burden, a chronic form of adult stress that has been associated with decreased quality of life and adverse health outcomes (Adelman et al., 2014).
In this framework, we examined the extent to which HPA axis negative feedback regulation, assessed with the low-dose DST, as well as caregiving burden are associated with DNA methylation changes at genes involved in HPA axis and immune function. We addressed these questions in a cohort of women caregivers of children with autism spectrum disorders (ASD) and age-matched control mothers of neurotypical children. The functional relevance of the identified DNA methylation patterns was then examined using an ex vivo model of immune-stimulated peripheral blood mononuclear cells (PBMC) in a subset of study participants.
2. Materials and methods
2.1. Participants and sample collection
The current study is derived from a cohort of 183 women who were caregivers of either their children diagnosed with ASD (n = 92) or their neurotypical children (n = 91). Mothers of ASD children will be referred to as caregivers while mothers of neurotypical children will be referred to as controls hereinafter. Perceived stress was assessed in all participants with the Perceived Stress Scale (PSS) at all study timepoints (Cohen et al., 1983). Participants in the caregiver group had to report a PSS score ≥ 13 at enrollment to be included in the study. Further details on this cohort have been described elsewhere (Prather et al., 2018, 2015). This study was approved by the Institutional Review Board at the University of California, San Francisco, and written informed consent was obtained for each study participant prior to enrollment.
At the initial study visit to the laboratory, participants completed a battery of sociodemographic and psychological questionnaires and received instructions and kits to collect, store, and send saliva samples for cortisol determinations. Nine months after enrollment, a cohort subset consisting of the first 20 caregivers and the first 20 controls that were enrolled in the larger study provided a morning blood sample after overnight fasting for ex vivo immune stimulation analysis. PBMCs were isolated from fresh blood collected in EDTA tubes using Ficoll gradient, and collected cells were washed twice with Dulbecco’s PBS, cryopreserved in freezing medium (90% FBS, 10% DMSO), and stored in liquid nitrogen. Blood DNA methylation was assessed at 18 and 24 months after enrollment in 57 caregivers and 67 controls. The study design is schematically depicted in Figure 1.
Figure 1. Data collection timeline.
Dexamethasone suppression test was performed at study enrollment. Blood samples were additionally collected in subsequent visits 9, 18, and 24 months after enrollment. Peripheral blood mononuclear cells were isolated from the 9-month samples for ex vivo stimulation analyses in the first 20 controls and the first 20 caregivers. DNA was extracted from peripheral whole blood collected at the 18- and 24-month timepoints for DNA methylation assessment in 124 participants. Perceived stress was assessed in all participants with the Perceived Stress Scale at all study timepoints. Abbreviations: PHA, phytohaemagglutinin.
2.2. Cortisol measurements and dexamethasone suppression test (DST)
At study enrollment and for 3 consecutive days, saliva samples were collected at waking with Salivette tubes (Sarstedt, Germany) to obtain baseline and post-DST cortisol levels for each participant. Baseline cortisol was derived by averaging cortisol levels from the first two days. Subsequently, a single oral dose of 0.25 mg dexamethasone was self-administered on the second night, and post-DST cortisol was measured on the third day. We employed the low-dose version of the DST (0.25 mg), because it captures subtler variations in HPA axis suppression in healthy populations (Direk et al., 2016; Labad et al., 2018); in this setting, post-DST cortisol levels offer a continuous measure of the degree of HPA axis suppression (Gaffey et al., 2019). Salivary cortisol concentration was determined by competitive solid phase time-resolved fluorescence immunoassay, as described elsewhere (Dressendörfer et al., 1992). All samples were run in duplicates. Cortisol values were log-transformed to reduce skewness. Baseline and post-DST cortisol levels were available for 90 subjects out of the 124 assessed for DNA methylation. Cortisol values that were three standard deviations above or below the mean cortisol of the entire cohort were considered as outliers; one participant had cortisol values over this threshold and was discarded from further analyses. This resulted in a final total of 89 participants (39 caregivers and 50 controls).
2.3. Ex vivo immune stimulation
Cryopreserved PBMC were thawed at 37°C and washed with RPMI medium supplemented with 10% sterile heat-inactivated FBS, 1% L-glutamine, and 1% sterile penicillin-streptomycin, containing 2 μg/ml DNase I (Sigma Aldrich, St. Louis, MO). Cells used for stimulation were resuspended with 5 μg/ml of phytohaemagglutinin (PHA, Sigma Aldrich, St. Louis, MO) and 2 ng/ml of IL-2 (Thermo Fisher Scientific, Waltham, MA) for 5 days at 37°C with 5% CO2 in a cell culture incubator. Total RNA was extracted from both unstimulated and stimulated PBMC using RNeasy Plus Mini kits (QIAGEN, Hilden, Germany). Gene expression was measured by NanoString technology (Geiss et al., 2008) and analyzed by nCounter SPRINT Profiler according to manufacturer’s instructions (Nanostring, Seattle, WA). Further details about immune stimulation and gene expression analysis are available elsewhere (Lin et al., 2018).
2.4. DNA methylation arrays
DNA methylation was assessed in 57 caregivers and 67 controls at both 18 and 24 months after enrollment. DNA was extracted from peripheral blood with the QIAamp DNA Blood Midi Kit (QIAGEN). Extracted DNA was bisulfite converted using EZ-96 DNA Methylation-Gold™ Kit (Zymo Research). Genome-wide DNA methylation was measured with the Infinium HumanMethylation450 BeadChip (450K), which assays DNA methylation at 482,421 CpG sites (Bibikova et al., 2011). According to standard quality control procedures, the following probes were removed: i) CpG probes in cross-reactive regions and those containing single nucleotide polymorphisms with minor allele frequency above 1% within 10 bp as retrieved from (Chen et al., 2013); ii) CpG probes with less than 3 beads or with a detection p-value over 1% in at least 1% of the samples assayed. Following this standardized procedure, a total of 377,683 CpG were included in subsequent analyses. Raw signal intensities were normalized with subset quantile normalization available in the minfi package (Aryee et al., 2014). Normalized intensity values were then converted into beta values, which were used in all analyses. White blood cell type proportions were estimated from DNA methylation following the Houseman reference method (Houseman et al., 2012).
2.5. Statistical analysis
All statistical analyses were performed with R version 3.6.3 (R Development Core Team, 2011). Sociodemographic differences between caregivers and controls were tested by Student’s t-test for continuous variables and Chi-square test (χ2) for categorical variables. Suppression of cortisol levels by the DST was assessed with paired t-test. Based on prior evidence and to minimize multiple testing (Iwata et al., 2013; Marsland et al., 2017; Ronchetti et al., 2015; Tyrka et al., 2016; Zannas et al., 2019), we selected a panel of candidate genes with putative roles in the interplay between HPA axis and immune function: IL1B, IL6, IL10, TNF, NLRP3, NR3C1, FKBP5, and GILZ. This resulted in a total of 158 CpG sites (annotation in Supplementary Table 1). All associations were tested with linear mixed effects regression models implemented with the lme4 package (Bates et al., 2015). Models examining cortisol and caregiving status included DNA methylation values at both timepoints as the dependent variable, baseline/post-DST cortisol or caregiving status as the independent variable, and subject ID as a random effects term. All models were further adjusted for blood cell type proportions (CD4 T cells, CD8 T cells, B cells, monocytes, NK cells, and granulocytes), array technical variables (slide, stripe, and median intensity of the methylated and unmethylated signals), age, race (White compared with other races), body mass index (BMI), and smoking history (ever-smoked compared with never-smoked), following iterative covariate pruning after PCA as implemented in the RaMWAS pipeline (Guintivano et al., 2020; Shabalin et al., 2018). Additionally, caregiving status and timepoint of DNA methylation assessment (18 or 24 months) were also included as covariates according to study design. Baseline cortisol concentration was included as an additional covariate in models assessing post-DST cortisol effect on DNA methylation as previously described (Direk et al., 2016). Gene-level false discovery rate (FDR) was used to correct for multiple testing, and adjusted q-values under 0.05 were considered statistically significant. Associations of the identified DST-associated DNA methylation patterns with proinflammatory gene mRNA expression were adjusted for unstimulated mRNA expression and DNA methylation timepoint and included subject ID as a random effects term.
2.6. Functional annotation of identified DNA methylation sites
Detailed functional annotation of the identified CpG sites covered by the 450k was performed using the WashU Epigenome Browser (https://epigenomegateway.wustl.edu/) adding the six main histone modification tracks (H3K4me3, H3K4me1, H3K9me1, H3K27me3, H3K27ac, and H3K36me3) as described elsewhere (Zhou et al., 2011). Genomic sequences obtained from NCBI reference sequence (GRCh37.p13 Primary Assembly) were bioinformatically screened for the presence of: (i) canonical glucocorticoid response elements (GREs) according to known chromatin immunoprecipitation (ChIP-exo) signals (Starick et al., 2015); and (ii) previously described “cryptic” potential GR binding sites present in NF-κB response elements (Hudson et al., 2018).
3. Results
3.1. Descriptive statistics
Clinicodemographic characteristics of the participants assessed for DNA methylation, stratified by caregiver status, are presented in Supplementary Table 2. Caregivers scored higher in the PSS when compared to controls at all timepoints (all p-values < 0.05). As expected, post-DST cortisol was significantly lower than baseline cortisol (t = 11.4, p < 0.0001). This decrease remained significant when separately analyzing caregivers (t = 7.0, p < 0.0001) and controls (t = 9.2, p < 0.0001). Cortisol measures were not significantly different between caregivers and controls at either baseline or post-DST (all p-values > 0.05). No significant differences in age, BMI, PSS, basal morning cortisol or post-DST cortisol were observed when comparing participants included in the DNA methylation analyses with participants who dropped out of the study.
3.2. Blunted HPA axis negative feedback is associated with lower TNF and FKBP5 methylation
Among the 158 examined CpG sites, and after FDR correction for multiple testing, DST response was associated with DNA methylation at ten individual CpG sites (Supplementary Table 1); eight of these sites were located at the TNF gene and two at FKBP5. Strikingly, all DST-associated CpG sites exhibited negative associations with DST response, i.e. higher post-DST cortisol was associated with a hypomethylated pattern across the identified CpG sites. Given the widespread associations of post-DST cortisol with TNF methylation (involving 32% of the covered TNF CpG sites), we also examined average TNF methylation across all 25 TNF-annotated CpG sites as a simplified summary measure to use in subsequent analyses. This analysis revealed a robust negative association between post-DST cortisol and average TNF methylation (β = −0.0060, SE = 0.0016, t = −3.81, p = 0.0003; Figure 2). To provide a more intuitive effect size for the observed differences in DNA methylation, we further compared the upper and lower tertiles of DST response groups. Average TNF methylation was significantly different between upper and lower post-DST cortisol tertiles (β = −0.0108, SE = 0.0046, t = −2.34, p = 0.0265). Moreover, half of the individual DST-associated CpG sites (cg10717214, cg08553327, cg26729380 and cg17741993) exhibited significant methylation differences when comparing the upper and lower DST tertiles (effect sizes ranging from 1.75% to 2.55%, all p values < 0.05). Within-subject DNA methylation levels robustly correlated across the two timepoints at all identified DST-associated CpG sites (all r > 0.4, all p-values < 0.0001). In striking contrast, no significant associations were found between baseline cortisol and any of the TNF CpG sites (all q-values > 0.18). Moreover, post-DST cortisol did not interact with caregiving status to influence any of the DST-associated CpG sites (all interaction q-values > 0.49).
Figure 2. DST response is negatively associated with average TNF methylation (β = −0.0060, SE = 0.0016, t = −3.81, p = 0.0003).

Response to the dexamethasone suppresion test (DST) was defined as log-transformed cortisol concentration in saliva (measured in nmol/L) at awakening after dexamethasone administration the night before. Higher post-DST cortisol levels reflect blunted HPA axis negative feedback. TNF methylation here corresponds to average DNA methylation of all TNF-annotated CpG sites covered by the 450K array. This analysis was conducted on 89 participants, 39 caregivers and 50 controls.
Further examination of the TNF DNA methylation landscape, as covered by the 450K, revealed two clusters of CpG sites according to both location and DNA methylation levels (Figure 3). Average methylation levels of CpG sites included in Cluster 1 and 2 were thus calculated for further analysis. DST response was significantly associated with average methylation of both Cluster 1 (β = −0.0088, SE = 0.0025, t = −3.54, p = 0.0008) and Cluster 2 (β = −0.0036, SE = 0.0013, t = −2.82, p = 0.0067).
Figure 3. DNA methylation landscape of the TNF gene.

(A) Mean DNA methylation levels (across all subjects) for each of the TNF-annotated CpG sites covered by the 450K array arranged by chromosomal position. The genomic annotation for the CpG sites is detailed in Supplementary Table 1. Each CpG site is depicted as a black dot. Filled dots depict CpG sites significantly associated with response to the dexamethasone suppression test (DST). Blank dots depict CpG sites not associated with DST response. As illustrated, the DST-associated sites fall within one of two of adjacent clusters of CpG sites with similar methylation patterns (enclosed in black circles). The first cluster (Cluster 1) is located at the promoter region/first exon of the TNF gene and encompasses 10 adjacent CpG sites with low-intermediate levels of DNA methylation (range from 15 to 30%). The second cluster (Cluster 2) is located within the TNF gene body and includes 9 CpG sites with intermediate-high DNA methylation levels (range from 68 to 86%). (B) Correlation plot depicting pairwise correlations between mean DNA methylation levels of the TNF-annotated CpG sites. Circle size and color tone correspond to the magnitude of the correlation coefficient. Only significant correlations are depicted. Purple and orange are used to depict positive and negative correlations, respectively.
Examination of chromatin states of peripheral blood mononuclear primary cells using the Epigenome Browser showed that Cluster 1 colocalized with high mono- and tri-methylation at histone 3 lysine 4 (H3K4me1, H3K4me3, respectively) and low trimethylation at histone 3 lysine 36 (H3K36me3), while Cluster 2 colocalized with H3K36me3 peaks (see Figure 4). The whole region encompassing the TNF gene exhibits low levels of repressive marks such as histone 3 trimethylation at lysine 27 (H3K27me3). Based on these patterns, Cluster 1 is consistent with an enhancer/promoter localization and Cluster 2 with a gene body of an actively transcribed gene (Kimura, 2013). Notably, the DST-associated TNF CpG sites colocalized with either H3K4me3 or H3K9me3 peaks. Since unmethylated CpG-dense sequences interact with CpG-binding proteins to establish H3K4me3 domains (Thomson et al., 2010), DNA methylation at DST-associated CpG sites may impact the surrounding chromatin landscape. Furthermore, our search for predicted glucocorticoid binding sites in this region revealed the presence of a canonical GRE (chr6: 31,543,511–31,543,525, forward strand) and two NF-κB response elements with cryptic GR binding motifs (chr6: 31,544,443–31,544,433 and chr6:31,545,418–31,545,408; both in the reverse strand) (see Figure 4).
Figure 4. Functional annotation of the examined CpG sites at the TNF locus.

All chromatin modification tracks correspond to “Peripheral Blood Mononuclear Primary cells” from the publicly available Roadmap Epigenome data. The upper track “Predicted GRBS” displays glucocorticoid receptor binding sites identified by means of logos available in JASPAR (MA0105.3, including a cryptic GRBS as described in Hudson et al., 2018) and Starick et. al. (2015); cryptic sites are shown as black bars while canonical GRE are shown as light grey bars. The track “450k CpG sites” displays the 25 TNF-annotated CpG sites covered by the 450K array according to the genomic coordinates reported in Supplementary Table 1; CpG sites significantly associated with response to the dexamethasone suppression test DST are shown as black bars, whereas non-significant CpG sites are shown as light grey bars. Genomic coordinates (chr6:31,542,401–31,546,250) correspond to human genome assembly 19 (hg19).
Detailed chromatin landscape characterization of the DST-associated FKBP5 CpG sites (cg20813374 and cg00130530) has been described elsewhere (Zannas et al., 2019). Briefly, epigenetic modifications present in this region are consistent with a poised enhancer signature that may regulate FKBP5 expression upon transcription factor binding.
Taken together, these data suggest that blunted HPA axis suppression in response to DEX is associated with DNA hypomethylation patterns at selected genes involved in immune function and HPA axis response, most notably TNF. Furthermore, our data highlight reduced HPA axis feedback, measured with the DST, as a more robust predictor of site-specific DNA methylation compared to baseline cortisol.
3.3. Caregiving status is associated with minor DNA methylation changes at inflammation-related genes
Among the 158 examined CpG sites and after FDR correction for multiple testing, caregivers exhibited lower methylation at only two CpG sites located within the IL6 first exon (cg00087425; β = −0.012, SE = 0.004, t = −3.14, p = 0.0023, q = 0.02) and first intron (cg15703690; β = −0.017, SE = 0.006, t = −2.92, p = 0.0044, q = 0.02). Overall these findings suggest that chronic stress (caregiving status) may predict a smaller number of distinct epigenetic changes as compared to metrics of HPA axis regulation.
3.4. DST-associated methylation influences TNF expression after ex vivo immune stimulation
To examine the functional relevance of the DST-associated DNA methylation changes, we measured TNF mRNA levels in PBMC derived from 33 study participants with DNA methylation data available (15 caregivers and 18 controls). To determine TNF mRNA responsivity in this context, PBMC were stimulated ex vivo with PHA, a generic and widely used immune stimulus that acutely promotes release of pro-inflammatory cytokines, including TNF (Janefjord et al., 2001; Kartika et al., 2020). Lower average TNF methylation was significantly associated with higher TNF mRNA expression after immune (PHA) stimulation (β = −3.54 × 10−5, SE = 1.22 × 10−5, t = −3.16, p = 0.0036; Figure 5), but not with TNF mRNA expression in unstimulated PBMCs (p = 0.0941). These effects were not moderated by caregiving status (interaction p = 0.17). Analysis by clusters of CpG sites revealed that stimulated TNF mRNA expression was associated with methylation status of Cluster 2 (β = −4.66 × 10−5, SE = 1.43 × 10−5, t = −3.26, p = 0.0028) but not Cluster 1 (p = 0.175). These data suggest that TNF hypomethylation associated with blunted HPA axis feedback may de-repress TNF transcriptional responses, predisposing to heightened peripheral inflammation following immune stimulation.
Figure 5. Average TNF methylation is negatively associated with TNF expression after PHA stimulation.
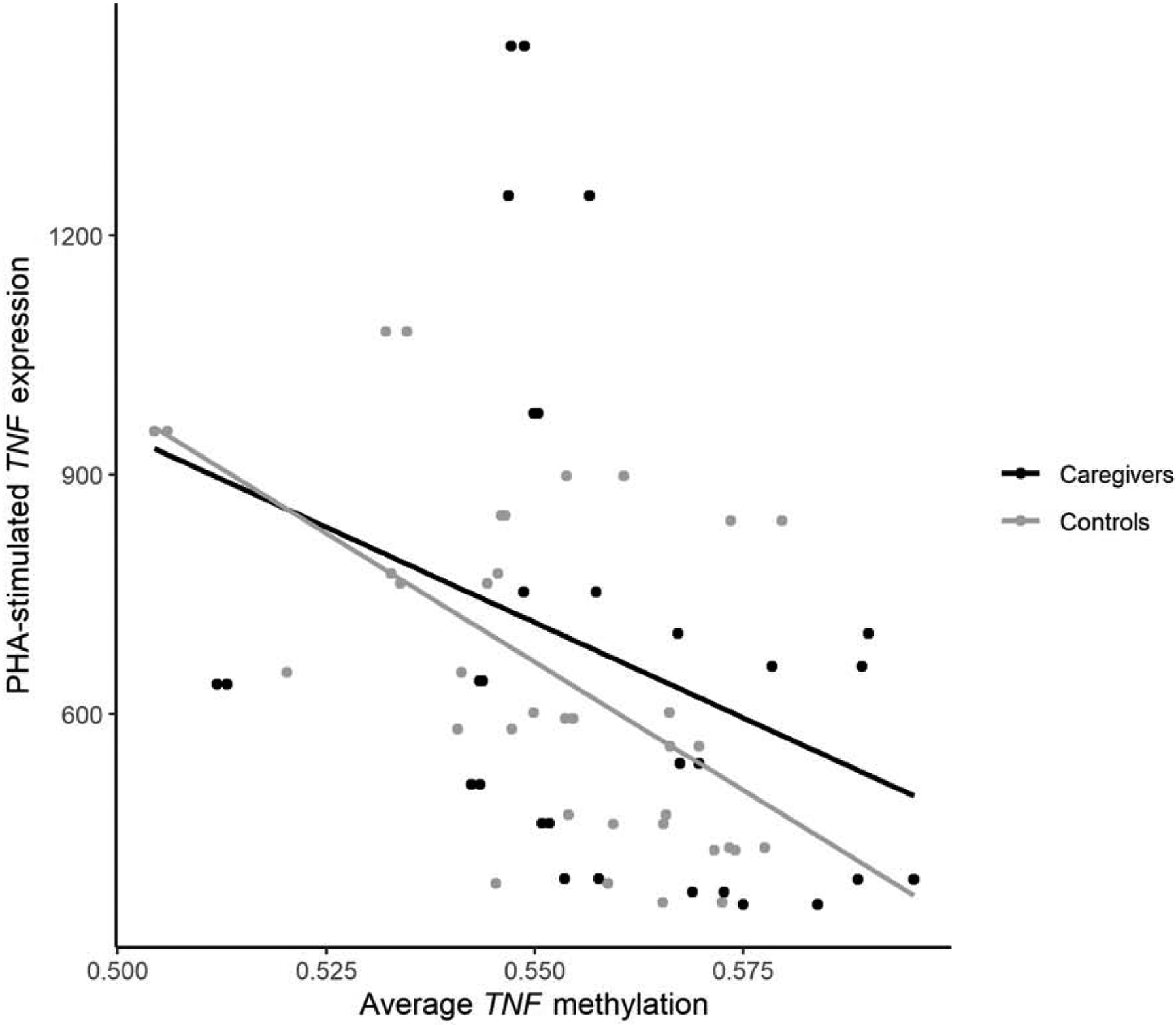
Peripheral blood mononuclear cells from a subset of 33 participants (15 caregivers and 18 controls) were cultured ex vivo to assess TNF expression in response to immune stimulation with phytohaemagglutinin. TNF methylation here corresponds to average DNA methylation of all 25 TNF-annotated CpG sites covered by the 450K array. No interaction was found between caregiving status and TNF expression (p = 0.17).
4. Discussion
The present findings suggest DNA methylation as a mechanistic link between varying degrees of HPA axis negative feedback regulation and immune function. Specifically, blunted HPA axis negative feedback, measured with the DST, was associated with lower methylation levels at eight TNF CpG sites and two FKBP5 CpG sites. Furthermore, decreased methylation at the DST-associated TNF sites was associated with higher TNF expression after ex vivo immune stimulation, indicating functional relevance of the identified epigenetic signatures. In contrast, baseline cortisol was not linked with any DNA methylation changes and caregiving stress was only associated with lower methylation at two IL6 CpG sites.
Previous studies support a complex interplay between HPA axis activity and the proinflammatory cytokine TNF. The DST-associated CpG sites identified herein are juxtaposed to a canonical GRE and two cryptic GR binding sites located within NF-κB response elements, indicating their potential involvement in epigenetic regulation of the molecular crosstalk between glucocorticoid and inflammatory signaling. Timing is a crucial factor in the bidirectional neuroimmune-neuroendocrine interplay (Horowitz and Zunszain, 2015), and since DNA methylation is a potentially dynamic epigenetic modification, there are two possible scenarios to explain our findings. Firstly, blunted HPA axis negative feedback could contribute to chronically increased circulating levels of cortisol that, in turn, lead to decreased TNF methylation and enhanced TNF signaling. Alternatively, higher cortisol levels after DST could be a consequence of preexisting TNF hyperreactivity. Although study design does not allow us to disentangle these different possibilities, our novel findings point to the potential role of TNF methylation as a mechanistic link between HPA axis and immune function.
Among genes involved in HPA axis function, reduced DST response was only associated with decreased DNA methylation at two closely juxtaposed CpG sites at FKBP5, a glucocorticoid-responsive gene that encodes a cochaperone of the GR complex that inhibits glucocorticoid signaling (Zannas et al., 2016). Notably, decreased DNA methylation at the same two FKBP5 CpG sites was recently shown to contribute to NF-κB-driven peripheral inflammation (Zannas et al., 2019). Taken together, the prior and present findings indicate epigenetic regulation of selected stress- and immune-related genes, such as FKBP5 and TNF, and its effects on NF-κB signaling as potential mediators of the crosstalk between HPA axis and immune function. In contrast, we did not find associations between DST response and other stress-related genes, including NR3C1, though such an association has been reported by a prior study employing the DEX/CRH test (Tyrka et al., 2016). Beyond the use of distinct tests of HPA axis regulation, disparate findings across studies may also be explained by the low overlap between assayed CpG sites due to the sparse CpG coverage by the arrays. Interestingly, however, one CpG site (cg15645634), was positively associated with DST response in the present study at a nominal p-value (Supplementary Table 1), a finding that is consistent with the prior study (Tyrka et al., 2016).
While caregivers did not differ in HPA axis feedback regulation as compared to controls, they exhibited lower IL6 methylation. Notably, our previous work identified upregulated IL6 mRNA in the caregiver group of the present cohort (Lin et al., 2018), and lower IL6 promoter methylation has been associated with greater increases in salivary IL-6 levels following acute laboratory stress (Janusek et al., 2017), overall suggesting the involvement of this epigenetic signature in primed immune responses to stress.
TNF methylation was specifically associated with post-DST cortisol levels, whereas no associations were observed with baseline cortisol, highlighting the importance of measuring HPA axis regulation after experimental activation. Experimental tests of HPA axis regulation, such as the DST, may thus hold promise as a tool to mechanistically dissect the interplay between HPA axis and immune function in humans. While some studies have previously assessed the relationship between candidate gene methylation and HPA axis reactivity under exposure to psychosocial acute stress (Janusek et al., 2017; Ziegler et al., 2015), fewer studies have explored the epigenetic correlates of HPA axis suppression as a measure independent of stress exposure. The subjective measure of perceived stress (PSS) was not associated with morning salivary cortisol in the present sample. Salivary cortisol reflects momentary daily stressors rather than chronic exposure to stress; thus, cortisol measured in saliva has been used in the context of acute challenges such as the Trier Social Stress Test or the DST (Schlotz et al., 2019; Weckesser et al., 2013), and discordance between subjective measures of stress perception and biological metrics of stress response has been previously reported (Kerr et al., 2020; Metz et al., 2020). Notably, differences in TNF and FKBP5 methylation were observed only in association with DST response rather than caregiver status, highlighting the specificity of the reported associations. In the same vein, TNF methylation was associated with TNF expression after immune stimulation but not at baseline, suggesting that this epigenetic pattern de-represses TNF transcription after exposure to a challenge only, rather than increasing baseline TNF expression. These results are in line with previous work in human hippocampal progenitor cells exposed to glucocorticoids, which exhibited widespread long-lasting DNA methylation changes associated with enhanced transcriptional responses but only following subsequent glucocorticoid challenge (Provençal et al., 2019). While site-specific variation in chromatin structures may determine how DNA methylation patterns influence gene expression (Qi et al., 2019), further molecular studies are needed to disentangle the specificity of these associations across distinct genomic regions.
The present study has significant limitations but also notable strengths. Our approach is limited by the sparse coverage of the 450K array, which does not allow a fine-grained assessment of DNA methylation landscapes at the genomic regions of interest. Furthermore, the current study design cannot establish causality, which would require longitudinal studies assessing HPA axis regulation, DNA methylation, and systemic inflammation at repeated timepoints. The relationship between HPA axis suppression and DNA methylation is further obscured by the time lapse between both measures. Due to statistical power constraints, a genome-wide approach was not feasible in the current sample; however, our hypothesis-driven approach is less likely to yield false positives. Finally, there is a standing question of whether the observed DNA methylation differences are biologically meaningful, though the reported effect sizes are comparable to those of similar publications in the field (Chaix et al., 2020; Tyrka et al., 2016). Nevertheless, strengths include: (i) the association of DST response with 8 of the 25 examined TNF CpG sites and with selected disease-relevant FKBP5 CpG sites; (ii) the consistent direction of these findings, showing that lower degrees of HPA axis feedback are associated with decreased DNA methylation at all identified sites; (iii) the stability of the observed DST-associated DNA methylation changes over time; and (iv) the functional relevance of the DST-associated epigenetic pattern of TNF, supported by our ex vivo study examining TNF mRNA expression after immune stimulation.
In conclusion, our findings identify DNA methylation as a novel link between HPA axis regulation and immune function. Intriguingly, lower TNF methylation has been previously observed with increasing age, an epigenetic pattern accompanied with increased TNF expression (Shinozaki et al., 2018), whereas higher TNF methylation has been associated with exposure to supportive parenting and with reported better health in young adults (Beach et al., 2017, 2015). Experimental manipulation of TNF methylation in vitro will help elucidate the functional relevance of this signature. As TNF and IL6 mRNA levels have been recently suggested to discriminate between responders and non-responders to antidepressant treatment (Cattaneo et al., 2020), it would be intriguing to examine HPA axis suppression and DNA methylation in this population. Cell-type specific analysis will be crucial to identify which cell subpopulations drive the observed DNA methylation differences. Prior glucocorticoid exposure has been recently described to prime subsequent immune responses in hippocampal cells (Horowitz et al., 2020); thus, combined stimulation with glucocorticoid and immune stimuli may better reveal the temporal dynamics of the association between HPA axis regulation and TNF methylation. Future longitudinal and experimental studies will be necessary to dissect how HPA axis function contributes to epigenetic programming of immune function and the potential relevance of such programing for health and disease.
Supplementary Material
Highlights.
Blunted HPA axis negative feedback is associated with peripheral blood TNF and FKBP5 hypomethylation
Caregiving stress burden is associated with peripheral blood IL6 hypomethylation
TNF hypomethylation results in higher TNF expression upon ex vivo stimulation of immune cells
Acknowledgements
This work was supported by NIH grants AG030424 and HL142051.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of Interest
None.
References
- Adelman RD, Tmanova LL, Delgado D, Dion S, Lachs MS, 2014. Caregiver Burden: A Clinical Review. JAMA 311, 1052–1060. 10.1001/jama.2014.304 [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, 2014. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30, 1363–1369. 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PJ, Adcock IM, 2009. Glucocorticoid resistance in inflammatory diseases. Lancet 373, 1905–1917. 10.1016/S0140-6736(09)60326-3 [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker BM, Walker SC, 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw 67 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Beach SRH, Lei MK, Brody GH, Dogan MV, Philibert RA, 2015. Higher levels of protective parenting are associated with better young adult health: exploration of mediation through epigenetic influences on pro-inflammatory processes. Front. Psychol 6, 676 10.3389/fpsyg.2015.00676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach SRH, Lei MK, Brody GH, Miller GE, Chen E, Mandara J, Philibert RA, 2017. Smoking in young adulthood among African Americans: Interconnected effects of supportive parenting in early adolescence, proinflammatory epitype, and young adult stress. Dev. Psychopathol 29, 957–969. 10.1017/S0954579416000961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R, 2011. High density DNA methylation array with single CpG site resolution. Genomics. 10.1016/j.ygeno.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Carroll BJ, Feinberg M, Greden JF, Tarika J, Albala AA, Haskett RF, James NM, Kronfol Z, Lohr N, Steiner M, de Vigne JP, Young E, 1981. A Specific Laboratory Test for the Diagnosis of Melancholia: Standardization, Validation, and Clinical Utility. Arch. Gen. Psychiatry 38, 15–22. 10.1001/archpsyc.1981.01780260017001 [DOI] [PubMed] [Google Scholar]
- Cattaneo A, Ferrari C, Turner L, Mariani N, Enache D, Hastings C, Kose M, Lombardo G, McLaughlin AP, Nettis MA, Nikkheslat N, Sforzini L, Worrell C, Zajkowska Z, Cattane N, Lopizzo N, Mazzelli M, Pointon L, Cowen PJ, Cavanagh J, Harrison NA, de Boer P, Jones D, Drevets WC, Mondelli V, Bullmore ET, Pariante CM, 2020. Whole-blood expression of inflammasome- and glucocorticoid-related mRNAs correctly separates treatment-resistant depressed patients from drug-free and responsive patients in the BIODEP study. Transl. Psychiatry 10, 1–14. 10.1038/s41398-020-00874-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaix R, Fagny M, Cosin-Tomás M, Alvarez-López M, Lemee L, Regnault B, Davidson RJ, Lutz A, Kaliman P, 2020. Differential DNA methylation in experienced meditators after an intensive day of mindfulness-based practice: Implications for immune-related pathways. Brain. Behav. Immun 84, 36–44. 10.1016/j.bbi.2019.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R, 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrousos GP, Gold PW, 1992. The Concepts of Stress and Stress System Disorders: Overview of Physical and Behavioral Homeostasis. JAMA 267, 1244–1252. 10.1001/jama.1992.03480090092034 [DOI] [PubMed] [Google Scholar]
- Cohen S, Janicki-Deverts D, Doyle WJ, Miller GE, Frank E, Rabin BS, Turner RB, 2012. Chronic stress, glucocorticoid receptor resistance, inflammation, and disease risk. Proc. Natl. Acad. Sci. U. S. A 109, 5995–5999. 10.1073/pnas.1118355109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, Mermelstein R, 1983. A global measure of perceived stress. J. Health Soc. Behav 24, 385–396. [PubMed] [Google Scholar]
- Di Dalmazi G, Fanelli F, Zavatta G, Ricci Bitti S, Mezzullo M, Repaci A, Pelusi C, Gambineri A, Altieri P, Mosconi C, Balacchi C, Golfieri R, Cosentino ER, Borghi C, Vicennati V, Pasquali R, Pagotto U, 2019. The Steroid Profile of Adrenal Incidentalomas: Subtyping Subjects With High Cardiovascular Risk. J. Clin. Endocrinol. Metab 104, 5519–5528. 10.1210/jc.2019-00365 [DOI] [PubMed] [Google Scholar]
- Direk N, Dekker MJHJ, Luik AI, Kirschbaum C, de Rijke YB, Hofman A, Hoogendijk WJG, Tiemeier H, 2016. The Very Low-Dose Dexamethasone Suppression Test in the General Population: A Cross-Sectional Study. PLoS One 11, e0164348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressendörfer RA, Kirschbaum C, Rohde W, Stahl F, Strasburger CJ, 1992. Synthesis of a cortisol-biotin conjugate and evaluation as a tracer in an immunoassay for salivary cortisol measurement. J. Steroid Biochem. Mol. Biol 43, 683–692. 10.1016/0960-0760(92)90294-s [DOI] [PubMed] [Google Scholar]
- Gaffey AE, Walsh EC, Ladd CO, Hoks RM, Abercrombie HC, 2019. Alterations in Systemic and Cognitive Glucocorticoid Sensitivity in Depression. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 4, 310–320. 10.1016/j.bpsc.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, James JJ, Maysuria M, Mitton JD, Oliveri P, Osborn JL, Peng T, Ratcliffe AL, Webster PJ, Davidson EH, Hood L, Dimitrov K, 2008. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol 26, 317–325. 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- Guintivano J, Shabalin AA, Chan RF, Rubinow DR, Sullivan PF, Meltzer-Brody S, Aberg KA, van den Oord EJCG, 2020. Test-statistic inflation in methylome-wide association studies. Epigenetics 1–4. 10.1080/15592294.2020.1758382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz MA, Cattaneo A, Cattane N, Lopizzo N, Tojo L, Bakunina N, Musaelyan K, Borsini A, Zunszain PA, Pariante CM, 2020. Glucocorticoids prime the inflammatory response of human hippocampal cells through up-regulation of inflammatory pathways. Brain. Behav. Immun 10.1016/j.bbi.2020.03.012 [DOI] [PubMed] [Google Scholar]
- Horowitz MA, Zunszain PA, 2015. Neuroimmune and neuroendocrine abnormalities in depression: two sides of the same coin. Ann. N. Y. Acad. Sci 1351, 68–79. 10.1111/nyas.12781 [DOI] [PubMed] [Google Scholar]
- Houseman E, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT, 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13, 86 10.1186/1471-2105-13-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtepen LC, Hardy R, Maddock J, Kuh D, Anderson EL, Relton CL, Suderman MJ, Howe LD, 2018. Childhood adversity and DNA methylation in two population-based cohorts. Transl. Psychiatry 8, 266 10.1038/s41398-018-0307-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson WH, Vera I.M.S. de, Nwachukwu JC, Weikum ER, Herbst AG, Yang Q, Bain DL, Nettles KW, Kojetin DJ, Ortlund EA, 2018. Cryptic glucocorticoid receptor-binding sites pervade genomic NF-κB response elements. Nat. Commun 9, 1337 10.1038/s41467-018-03780-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M, Ota KT, Duman RS, 2013. The inflammasome: pathways linking psychological stress, depression, and systemic illnesses. Brain. Behav. Immun 31, 105–14. 10.1016/j.bbi.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janusek LW, Tell D, Gaylord-Harden N, Mathews HL, 2017. Relationship of childhood adversity and neighborhood violence to a proinflammatory phenotype in emerging adult African American men: An epigenetic link. Brain. Behav. Immun 60, 126–135. 10.1016/j.bbi.2016.10.006 [DOI] [PubMed] [Google Scholar]
- Janefjord CK, Jenmalm MC, 2001. PHA-induced IL-12Rβ2 mRNA expression in atopic and non- atopic children. Clin. Exp. Allergy 31, 1493–1500. [DOI] [PubMed] [Google Scholar]
- Jokinen J, Nordström A-L, Nordström P, 2008. ROC analysis of dexamethasone suppression test threshold in suicide prediction after attempted suicide. J. Affect. Disord 106, 145–152. 10.1016/j.jad.2007.06.008 [DOI] [PubMed] [Google Scholar]
- Jung SH, Wang Y, Kim T, Tarr A, Reader B, Powell N, Sheridan JF, 2015. Molecular mechanisms of repeated social defeat-induced glucocorticoid resistance: Role of microRNA. Brain. Behav. Immun 44, 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminsky Z, Wilcox HC, Eaton WW, Van Eck K, Kilaru V, Jovanovic T, Klengel T, Bradley B, Binder EB, Ressler KJ, Smith AK, 2015. Epigenetic and genetic variation at SKA2 predict suicidal behavior and post-traumatic stress disorder. Transl. Psychiatry 5, e627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kartika R, Purnamasari D, Pradipta S, Larasati RA, Wibowo H, 2020. Impact of low interferon-γ and IL-10 levels on TNF-α and IL-6 production by PHA-induced PBMCs in Type 2 Diabetes Mellitus. J. Inflamm. Res 13, 187–193. 10.2147/JIR.S245064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy L, Atkinson AB, Johnston H, Sheridan B, Hadden DR, 1984. Serum cortisol concentrations during low dose dexamethasone suppression test to screen for Cushing’s syndrome. Br. Med. J. (Clin. Res. Ed) 289, 1188–1191. 10.1136/bmj.289.6453.1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr JI, Naegelin M, Weibel RP, Ferrario A, La Marca R, von Wangenheim F, Hoelscher C, Schinazi VR, 2020. The effects of acute work stress and appraisal on psychobiological stress responses in a group office environment. Psychoneuroendocrinology 121, 104837 10.1016/j.psyneuen.2020.104837 [DOI] [PubMed] [Google Scholar]
- Kimura H, 2013. Histone modifications for human epigenome analysis. J. Hum. Genet 58, 439–445. 10.1038/jhg.2013.66 [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TWW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB, 2013. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat. Neurosci 16, 33–41. 10.1038/nn.3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labad J, Armario A, Nadal R, Solé M, Gutiérrez-Zotes A, Montalvo I, Moreno-Samaniego L, Martorell L, Sánchez-Gistau V, Vilella E, 2018. Clinical correlates of hypothalamic-pituitary-adrenal axis measures in individuals at risk for psychosis and with first-episode psychosis. Psychiatry Res. 265, 284–291. 10.1016/j.psychres.2018.05.018 [DOI] [PubMed] [Google Scholar]
- Lin J, Sun J, Wang S, Milush JM, Baker CAR, Coccia M, Effros RB, Puterman E, Blackburn E, Prather AA, Epel E, 2018. In vitro proinflammatory gene expression predicts in vivo telomere shortening: A preliminary study. Psychoneuroendocrinology 96, 179–187. 10.1016/j.psyneuen.2018.06.020 [DOI] [PubMed] [Google Scholar]
- Marsland AL, Walsh C, Lockwood K, John-Henderson NA, 2017. The effects of acute psychological stress on circulating and stimulated inflammatory markers: A systematic review and meta-analysis. Brain. Behav. Immun 64, 208–219. 10.1016/j.bbi.2017.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade TW, Ryan CP, Jones MJ, Hoke MK, Borja J, Miller GE, Kuzawa CW, Kobor MS, 2019. Genome-wide analysis of DNA methylation in relation to socioeconomic status during development and early adulthood. Am. J. Phys. Anthropol 169, 3–11. 10.1002/ajpa.23800 [DOI] [PubMed] [Google Scholar]
- Metz S, Duesenberg M, Hellmann-Regen J, Wolf OT, Roepke S, Otte C, Wingenfeld K, 2020. Blunted salivary cortisol response to psychosocial stress in women with posttraumatic stress disorder. J. Psychiatr. Res 130, 112–119. 10.1016/j.jpsychires.2020.07.014 [DOI] [PubMed] [Google Scholar]
- Misale MS, Witek Janusek L, Tell D, Mathews HL, 2018. Chromatin organization as an indicator of glucocorticoid induced natural killer cell dysfunction. Brain. Behav. Immun 67, 279–289. 10.1016/j.bbi.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder RH, Walton E, Neumann A, Houtepen LC, Felix JF, Bakermans-Kranenburg MJ, Suderman M, Tiemeier H, van IJzendoorn MH, Relton CL, Cecil CAM, 2020. Epigenomics of being bullied: changes in DNA methylation following bullying exposure. Epigenetics 00, 1–15. 10.1080/15592294.2020.1719303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TM, Mill J, 2014. Epigenetics in health and disease: heralding the EWAS era. Lancet 383, 1952–1954. 10.1016/S0140-6736(14)60269-5 [DOI] [PubMed] [Google Scholar]
- Newton R, Shah S, Altonsy MO, Gerber AN, 2017. Glucocorticoid and cytokine crosstalk: Feedback, feedforward, and co-regulatory interactions determine repression or resistance. J. Biol. Chem 292, 7163–7172. 10.1074/jbc.R117.777318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkheslat N, Zunszain PA, Horowitz MA, Barbosa IG, Parker JA, Myint A-M, Schwarz MJ, Tylee AT, Carvalho LA, Pariante CM, 2015. Insufficient glucocorticoid signaling and elevated inflammation in coronary heart disease patients with comorbid depression. Brain. Behav. Immun 48, 8–18. 10.1016/j.bbi.2015.02.002 [DOI] [PubMed] [Google Scholar]
- Palma-Gudiel H, Córdova-Palomera A, Leza JC, Fañanás L, 2015. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neurosci. Biobehav. Rev 55, 520–535. 10.1016/j.neubiorev.2015.05.016 [DOI] [PubMed] [Google Scholar]
- Perrin AJ, Horowitz MA, Roelofs J, Zunszain PA, Pariante CM, 2019. Glucocorticoid resistance: Is it a requisite for increased cytokine production in depression? A systematic review and meta-analysis. Front. Psychiatry 10 10.3389/fpsyt.2019.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather AA, Epel ES, Arenander J, Broestl L, Garay BI, Wang D, Dubal DB, 2015. Longevity factor klotho and chronic psychological stress. Transl. Psychiatry 5, e585–6. 10.1038/tp.2015.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather AA, Epel ES, Portela Parra E, Coccia M, Puterman E, Aiello AE, Dhabhar FS, 2018. Associations between chronic caregiving stress and T cell markers implicated in immunosenescence. Brain. Behav. Immun 73, 546–549. 10.1016/j.bbi.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provençal N, Arloth J, Cattaneo A, Anacker C, Cattane N, Wiechmann T, Röh S, Ködel M, Klengel T, Czamara D, Müller NS, Lahti J, Räikkönen K, Pariante CM, Binder EB, 2019. Glucocorticoid exposure during hippocampal neurogenesis primes future stress response by inducing changes in DNA methylation. Proc. Natl. Acad. Sci 201820842 10.1073/pnas.1820842116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi S, Li Y, Dai Z, Xiang M, Wang G, Wang L, Wang Z, 2019. Uhrf1-Mediated Tnf-α Gene Methylation Controls Proinflammatory Macrophages in Experimental Colitis Resembling Inflammatory Bowel Disease. J. Immunol 203, 3045–3053. 10.4049/jimmunol.1900467 [DOI] [PubMed] [Google Scholar]
- R Development Core Team, 2011. R: A Language and Environment for Statistical Computing R Found. Stat. Comput, R Foundation for Statistical Computing; 10.1007/978-3-540-74686-7 [DOI] [Google Scholar]
- Rhen T, Cidlowski JA, 2005. Antiinflammatory action of glucocorticoids - New mechanisms for old drugs. N. Engl. J. Med 353, 1711–1723. 10.1056/NEJMra050541 [DOI] [PubMed] [Google Scholar]
- Rohleder N, 2014. Stimulation of Systemic Low-Grade Inflammation by Psychosocial Stress. Psychosom. Med 76. [DOI] [PubMed] [Google Scholar]
- Ronchetti S, Migliorati G, Riccardi C, 2015. GILZ as a Mediator of the Anti-Inflammatory Effects of Glucocorticoids. Front. Endocrinol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlotz W, 2019. Investigating associations between momentary stress and cortisol in daily life: What have we learned so far? Psychoneuroendocrinology 105, 105–116. 10.1016/j.psyneuen.2018.11.038 [DOI] [PubMed] [Google Scholar]
- Seddon JA, Rodriguez VJ, Provencher Y, Raftery-Helmer J, Hersh J, Labelle PR, Thomassin K, 2020. Meta-analysis of the effectiveness of the Trier Social Stress Test in eliciting physiological stress responses in children and adolescents. Psychoneuroendocrinology 116, 104582 10.1016/j.psyneuen.2020.104582 [DOI] [PubMed] [Google Scholar]
- Shabalin AA, Hattab MW, Clark SL, Chan RF, Kumar G, Aberg KA, van den Oord EJCG, 2018. RaMWAS: fast methylome-wide association study pipeline for enrichment platforms. Bioinformatics 34, 2283–2285. 10.1093/bioinformatics/bty069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki G, Braun PR, Hing BWQ, Ratanatharathorn A, Klisares MJ, Duncan GN, Jellison SS, Heinzman JT, Nagahama Y, Close L, Sabbagh S, Dlouhy BJ, Howard MA, Kawasaki H, Cho HR, 2018. Epigenetics of Delirium and Aging: Potential Role of DNA Methylation Change on Cytokine Genes in Glia and Blood Along With Aging. Front. Aging Neurosci 10, 311 10.3389/fnagi.2018.00311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starick SR, Ibn-Salem J, Jurk M, Hernandez C, Love MI, Chung H-R, Vingron M, Thomas-Chollier M, Meijsing SH, 2015. ChIP-exo signal associated with DNA-binding motifs provides insight into the genomic binding of the glucocorticoid receptor and cooperating transcription factors. Genome Res. 25, 825–835. 10.1101/gr.185157.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr ARW, Deaton A, Andrews R, James KD, Turner DJ, Illingworth R, Bird A, 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464, 1082–1086. 10.1038/nature08924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrka AR, Parade SH, Welch ES, Ridout KK, Price LH, Marsit C, Philip NS, Carpenter LL, 2016. Methylation of the leukocyte glucocorticoid receptor gene promoter in adults: Associations with early adversity and depressive, anxiety and substance-use disorders. Transl. Psychiatry 6 10.1038/tp.2016.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, Aiello AE, Wildman DE, Koenen KC, Pawelec G, de los Santos R, Goldmann E, Galea S, 2010. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc. Natl. Acad. Sci 107, 9470 LP–9475. 10.1073/pnas.0910794107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckesser LJ, Dietz F, Schmidt K, Grass J, Kirschbaum C, Miller R, 2019. The psychometric properties and temporal dynamics of subjective stress, retrospectively assessed by different informants and questionnaires, and hair cortisol concentrations. Sci. Rep 9, 1098 10.1038/s41598-018-37526-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehuda R, Daskalakis NP, Bierer LM, Bader HN, Klengel T, Holsboer F, Binder EB, 2016. Holocaust Exposure Induced Intergenerational Effects on FKBP5 Methylation. Biol. Psychiatry 80, 372–380. 10.1016/j.biopsych.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, Nemeroff CB, Smith AK, Bradley B, Heim C, Menke A, Lange JF, Brückl T, Ising M, Wray NR, Erhardt A, Binder EB, Mehta D, 2015. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: Relevance of glucocorticoid signaling. Genome Biol. 16, 1–12. 10.1186/s13059-015-0828-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas AS, Chrousos GP, 2017. Epigenetic programming by stress and glucocorticoids along the human lifespan. Mol. Psychiatry 22, 640–646. 10.1038/mp.2017.35 [DOI] [PubMed] [Google Scholar]
- Zannas AS, Jia M, Hafner K, Baumert J, Wiechmann T, Pape JC, Arloth J, Ködel M, Martinelli S, Roitman M, Röh S, Haehle A, Emeny RT, Iurato S, Carrillo-Roa T, Lahti J, Räikkönen K, Eriksson JG, Drake AJ, Waldenberger M, Wahl S, Kunze S, Lucae S, Bradley B, Gieger C, Hausch F, Smith AK, Ressler KJ, Müller-Myhsok B, Ladwig K-H, Rein T, Gassen NC, Binder EB, 2019. Epigenetic upregulation of FKBP5 by aging and stress contributes to NF-κB–driven inflammation and cardiovascular risk. Proc. Natl. Acad. Sci 116, 11370 LP–11379. 10.1073/pnas.1816847116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas AS, Wiechmann T, Gassen NC, Binder EB, 2016. Gene-Stress-Epigenetic Regulation of FKBP5: Clinical and Translational Implications. Neuropsychopharmacol. Off. Publ. Am. Coll. Neuropsychopharmacol 41, 261–274. 10.1038/npp.2015.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C, Dannlowski U, Bräuer D, Stevens S, Laeger I, Wittmann H, Kugel H, Dobel C, Hurlemann R, Reif A, Lesch K-P, Heindel W, Kirschbaum C, Arolt V, Gerlach AL, Hoyer J, Deckert J, Zwanzger P, Domschke K, 2015. Oxytocin receptor gene methylation: converging multilevel evidence for a role in social anxiety. Neuropsychopharmacology 40, 1528–38. 10.1038/npp.2015.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Maricque B, Xie M, Li D, Sundaram V, Martin EA, Koebbe BC, Nielsen C, Hirst M, Farnham P, Kuhn RM, Zhu J, Smirnov I, Kent WJ, Haussler D, Madden PAF, Costello JF, Wang T, 2011. The Human Epigenome Browser at Washington University. Nat. Methods 8, 989–990. 10.1038/nmeth.1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


